Abstract
While a number of studies have documented the persistent presence of chikungunya virus (CHIKV) in muscle tissue with primary fibroblast as the preferable cell target, little is known regarding the alterations that take place in muscle tissue in response to CHIKV infection. Hence, in the present study a permissive mouse model of CHIKV infection was established and characterized in order to understand the pathophysiology of the disease. The two dimensional electrophoresis of muscle proteome performed for differential analysis indicated a drastic reprogramming of the proteins from various classes like stress, inflammation, cytoskeletal, energy and lipid metabolism. The roles of the affected proteins were explained in relation to virus induced myopathy which was further supported by the histopathological and behavioural experiments proving the lack of hind limb coordination and other loco-motor abnormalities in the infected mice. Also, the level of various pro-inflammatory mediators like IL-6, MCP-1, Rantes and TNF-α was significantly elevated in muscles of infected mice. Altogether this comprehensive study of characterizing CHIKV induced mouse myopathy provides many potential targets for further evaluation and biomarker study.
Introduction
Chikungunya fever (CHIKF) is an enervating, but non-fatal, arthropod-borne disease transmitted to humans by the bite of infected Aedes aegypti and Aedes albopictus mosquitoes. It is a debilitating viral illness of global concern due to its escalating outbreaks in different parts of the world particularly in Africa, Europe and South East Asia [1]–[3]. There have been a number of epidemics associated with severe morbidity in Philippines, Thailand, Cambodia, Vietnam, India, Myanmar, Sri Lanka and on the islands of the Indian Ocean, including Madagascar, Comoros, Mauritius, and Reunion Island [4]–[6].
Chikungunya virus (CHIKV), an Alphavirus belonging to the family Togaviridae, is an enveloped virus with a positive sense RNA of approximately 11.8 kb. The virus is responsible for an acute infection characterized by high fever, arthralgia, myalgia, head-ache, chills, photophobia and rash [7]–[9]. However, the infection is classically self- limiting and usually resolves within 3-4 days except for the joint symptoms that may persist for a longer period. Despite the pathological significance of chikungunya, the physiological and molecular mechanisms occurring during viral infection are still not well defined. With respect to the alphavirus induced myopathy, Ross River Virus (RRV) pathogenesis has been studied in detail. The arthritic disease is thought to be initiated by viral replication and inflammatory infiltrates that is comprised of monocytes/macrophages in the affected joints [10], [11], [12]. Like other alphaviruses of old world class, CHIKV is also primarily arthritogenic in nature and muscle tissue has been proposed to be their target [13]–[17]. Studies done on animal models have identified muscle fibres and/or infiltrating cells as the cellular target of alphavirus infection. However, a study done on muscle biopsies of CHIKV infected patients with myositic syndrome has identified muscle satellite cells and not muscle fibres as the primary target of viral infection [18]. Recently, evidence of arthritis, tenosynovitis, and myositis in CHIKV infected neonatal and adult mice of different strains has been reported by several groups [19]–[25].
It is important to investigate the changes that occur in response to the infection in laboratory animal models that can simulate clinical infection in humans. Viral infections generally result in alterations in the host proteome that can determine the fate of the infected host cells or tissues, which can impact the disease progression and outcome. Hence, in order to uncover the host protein responses to CHIKV infection, a comprehensive study of interaction between CHIKV and a permissive mouse model was undertaken. In one of our previous studies, protein profiling of CHIKV infected mouse liver and brain tissues was reported explaining the implications of differentially affected proteins in disease manifestation [26]. The neonatal mice (2–3 day old) serve as a useful system for understanding the arthritic manifestations and identify host factors that may contribute to CHIKV induced inflammation and myositis. Using two dimensional electrophoresis (2-DE) method, the differential proteome analysis of muscle tissue was undertaken. The roles of the affected proteins were explained in relation to virus induced myopathy which was further supported by the histopathological and behavioural experiments proving the lack of hind limb coordination and other loco-motor abnormalities in the infected mice. Altogether this comprehensive study of characterizing CHIKV induced mouse myopathy provides many potential targets for further evaluation and biomarker study.
Materials and Methods
Virus propagation and titration
An Indian strain of CHIKV DRDE-06 (Genbank accession no: EF- 210157) of ECSA genotype was used in the present study [27]. The virus was isolated during 2006 epidemic from a serologically confirmed human patient. The clinical virus isolate was cultured in BHK-21 cells and further passaged in C6/36 cell line to increase the viral titre. The virus stock was harvested from the culture supernatant and stored at −80°C. CHIKV was titrated by plaque formation on monolayers of Vero cells (obtained from NCCS, Pune, India).
Ethics statement
All animal experiments were carried out strictly following the approved study protocol of Institutional Animal Ethics Committee (IAEC) of the Defence Research and Development Establishment (DRDE) Gwalior, India (Protocol Number: Viro-07/50/MMP). The institutional committee is registered with committee for the purpose of control and supervision of experiments on animals (CPCSEA), Ministry of Environment and Forestry, Government of India (Regd. No – 37/GO/c/1999/CPCSEA). Balb/c mice of 2–3 days of age along with their mothers, obtained from the institutional animal facility, were housed in polypropylene cages with dust-free rice husk as bedding material at an ambient temperature of 25–26°C with a standard 12-hour light/12-hour dark cycle and the mothers were allowed ad libitum access to food and water while the pups were allowed to suckle freely.
Animal experimentation
Mice (n = 8) were inoculated subcutaneously with CHIKV in the loose skin on their back (106 pfu in 50 ul of infected C6/36 cell culture supernatant). In parallel, mock-infected group (n = 8) was inoculated with uninfected C6/36 cell supernatant. Following virus inoculation, the mice were monitored at 24 hour interval for disease signs like retarded growth, hair loss, skin rashes and hind limb paralysis. The clinical signs of the disease were also determined by behavioural parameters of surface righting reflex and walking traits. For surface righting reflex, mice pups were placed on their back and were allowed some time to turn over to upright position.
Clinical scoring of the mice inoculated with CHIKV was done as follows: 0, no disease signs; 1, ruffled fur; 2, mild hind limb weakness; 3, moderate hind limb weakness; 4, severe hind limb weakness and dragging and 5, moribund. The clinical score of 5 was set as the terminal point. When the mice inoculated with CHIKV reached the terminal point, they were euthanized by cervical dislocation and tissues were processed for further studies.
Determination of viral titre
To determine the viral titre in muscles, mice were perfused with PBS and hind limbs were removed by dissection. Upon removal of bone from hind limbs, tissue was homogenized in 1× minimum essential media (MEM) and viral RNA was extracted using Qiagen kit according to manufacturer's protocol. Viral presence in muscle was assessed by SYBR green I based one-step real-time quantitative RT-PCR using primers specific to a region of 200 base pairs of the viral envelope (E1) gene. Standard curve was constructed with Ct (threshold cycle) values obtained using dilutions of the titrated CHIKV virus, and genome equivalent of CHIKV in muscle was determined from respective Ct values obtained from standard curve as reported [28].
Histopathology and histochemistry
At necropsy, skin, spleen and hind limbs from each mouse were cut and fixed in 10% neutral buffered formalin. After 48 h of fixation, muscles were transferred to 70% ethanol, processed by automated tissue processor (Leica TP1020), dehydrated and embedded in paraffin wax. Multiple sections of 12 μm thickness were prepared using automatic microtome (Microm HM360) and stained with hematoxylin and eosin in Leica Autostain-XL. Muscle sections of 4 μm thickness were used for the localization of viral antigen. Anti-CHIKE2 antibody raised in rabbit immunized with purified recombinant envelope protein (rCHIKE2) was used to recognize the CHIKV antigen.
Protein extraction
During the peak symptomatic phase (i.e 9–10 days post infection), when the animals were in moribund condition with severe paralysis, four of the suckling mice (n = 4) from each group (mock-infected and CHIKV-infected) were dissected and tissues were frozen in liquid nitrogen and stored at −80°C until further processed for proteome analysis.
Tissue samples were homogenized according to a previously described method with some modifications [29]. Briefly, the muscle tissue was thawed on ice, separated from the bones using scalpel and combined with an equivalent volume of solubilization buffer [7M urea, 2% CHAPS, 1× concentration of protease inhibitor cocktail (Sigma, St. Louise, MO, USA)]. Thereafter, the tissue was continuously homogenized for 3 min on ice using Tissue ruptor (Qiagen, Hilden, Germany). The clear supernatant was collected by centrifugation and subjected to five pulses of sonication on ice. Total soluble protein fraction was recovered by centrifugation at 40,000×g for 30 min and the protein concentration was determined using Bradford reagent (Sigma, USA). The solubilized protein in the supernatant was then precipitated with 10% (wt/vol) trichloroacetic acid, kept at 4°C overnight and centrifuged at 10,000×g for 10 min at 4°C. The resulting protein precipitate was washed twice with cold acetone containing 0.07% β-mercapto-ethanol, air-dried, and stored at −80°C until use.
Two dimensional electrophoresis (2-DE) and image analysis
2-DE was performed using 7 cm Readystrip IPG strips (Linear, pI 4–7, Biorad, Hercules, CA) in the PROTEAN IEF Cell and PROTEAN plus Dodeca Cell (Biorad). Before proceeding for isoelectric focusing, the IPG strips were passively rehydrated for 16 hours with 150 μl of rehydration buffer (8 M urea, 2% CHAPS, 15 mM DTT and 0.5% v/v IPG buffer pH 3–10) which contained 500 μg of protein. The isoelectric focusing of the rehydrated strips was automatically processed using the following parameters: 250 V rapid, 15 min; 4000 V rapid 2 h; 8000–10000 Vh at 20°C under mineral oil. After focusing, the strips were incubated for 10 min in equilibration buffer (6 M urea, 30% w/v glycerol, 2% w/v SDS and 0.375 M Tris/HCl buffer, pH 8.8) containing 1% w/v DTT, followed by additional equilibration for 15 min in equilibration buffer containing 4% w/v iodoacetamide. The equilibrated strips were then further resolved with 12% SDS PAGE gels keeping constant current of 10 mA per gel until the dye front reached the bottom of the gel. Gels were then stained with Coomassie Brilliant Blue G-250 and scanned at 300 dpi using GS800 densitometer (Biorad). Comparative analysis of protein spots was performed using PD Quest 2D analysis software (Biorad). The gels were normalized according to the total quantity in the analysis set. The spots were checked manually to eliminate any possible artifacts and spots that were consistently reproducible in all gel images, including both the biological and technical replicates, were chosen for subsequent analysis. The student's t-test was used for statistical analysis of data. Expression intensity ratios of CHIKV-infected/mock-infected values greater than 1.8 (p≤0.05) or less than 0.5 (p≤0.05) were set as the thresholds to indicate significant changes. Two such independent experiments were carried out. In each experiment a total of 8 gels derived from pooled protein samples of four animals from each group were analysed.
Identification of differentially expressed proteins by MALDI-TOF/TOF Mass Spectrometry and database search
The protein spots of interest were manually excised from the coomassie stained gels and subjected to ingel digestion as previously described [30]. Tryptic digests were extracted using 10 μL of Milli-Q water initially, followed by two extractions with a total of 20 μL of 0.1% TFA. The combined extracts were dried in a vacuum concentrator at room temperature, and then dissolved in 1 μL of 5% acetonitrile with 0.5% TFA. The prepared MS samples were mixed with equal volume of the α-cyano-4-hydroxycinnamic acid (CHCA) matrix solution (10 mg/mL) and then spotted onto the MALDI plate. Samples were allowed to air dry and analysed by a 4800 MALDI-TOF-TOF Analyzer (Applied Biosystems, Framingham, MA). A default calibration was applied using a six component peptide standard spotted onto 13 calibration spot spread on the plate and four corners of 384 well MALDI plate. The UV laser was operated at a 200-Hz repetition rate at a wavelength of 355 nm. The accelerated voltage was operated at 20 kV. The MS/MS mass spectra were acquired by the data dependent acquisition method with the 20 strongest precursors selected from one MS scan. All MS and MS/MS spectra were obtained by accumulation of at least 800 and 2100 laser shots, respectively. MS and MS/MS data were analysed and peak list were generated using Protein Pilot 2.0 (Applied Biosystems). Twenty MS peaks were selected between 850 and 4000 Da and filtered with a signal-to-noise ratio greater than 20. A peak intensity filter was used with no more than 50 peaks per 200 Da in the setting parameter of MASCOT search after acquisition. MS/MS peaks were selected based on a signal-to-noise ratio greater than 10 over a mass range of 60 to 20 Da below the precursor mass. MS and MS/MS were analysed using MASCOT 2.0 search engine (Matrix Science, London, UK) to search against the MSDB protein sequence database in the taxonomy group of Mus musculus. Searching parameters were as follows: trypsin digestion with one missed cleavage, variable modifications (oxidation of methionine and carbamidomethylation of cysteine), and the peptide mass tolerance of 50 ppm for precursor ion and mass tolerance of ±0.6 for fragment ion for +1 charged state. For all proteins successfully identified by Peptide Mass Fingerprint and/or MS/MS, Mascot score greater than 62 was accepted as significant (p value <0.05).
Real-Time qRT-PCR
Total RNA was extracted using the Qiagen (GmbH, Hilden, Germany) RNAEasy Mini kit. The quantitative real-time RT-PCR was carried out for the analysis of host gene expression in muscles using gene-specific primers from Quanti Tect primer assay kit and Quanti Fast one-step RT-PCR kit (Qiagen, Germany). The thermal profile consists of 10 min of reverse transcription at 50°C one cycle and 5 min of polymerase activation at 95°C, followed by 40 cycles of PCR at 95°C for 10 s, 60°C for 30 s for combined annealing/extension. Following amplification, a melting curve analysis was performed to verify the authenticity of the amplified product by its specific melting temperature (Tm) with the melting curve analysis software of the Mx3005p. The threshold cycle (Ct) of gene of interest (GOI) and housekeeping gene (HKG) and the difference between their Ct values (Ct) were determined. Relative changes of gene expression were calculated using delta delta Ct method [31] and the data were represented as fold up-regulation/down-regulation compared to the mock-infected group.
Western blot analysis
Total protein was extracted from hind limb muscle tissues of the mock and CHIKV infected animals after 9 days post infection. Muscle tissue was homogenized in lysis buffer (Tris-Cl 50 mM (pH 7.4), NaCl 150 mM, EDTA 1 mM, Triton X-100 1%, SDS 0.1%, sodium deoxycholate 1%, Na3VO4 1 mM) and centrifuged at 12000×g for 20 min. Protein concentration was quantified in supernatants by Bradford reagent. Equivalent amounts of the tissue lysates (40 μg) were subjected to 12% SDS-PAGE and then transferred to PVDF–immobilon membrane (Millipore, Bedford, MA, USA), incubated with respective primary antibodies (1∶1000) at 4°C overnight, thoroughly washed and incubated with the recommended horseradish peroxidise (HRP) conjugated secondary antibodies (1∶30000) at 37°C for 1 hr. The bound antibodies were detected using chemiluminiscence kit (Life technologies, Carlsbad, CA, USA).
Statistical analysis
For the statistical analysis of 2-DE gels, student's unpaired t-test was done. The data were expressed as mean±SE of four gels per group. Expression intensity ratios of CHIKV-infected/mock-infected values greater than 1.8 (p≤0.05) or less than 0.5 (p≤0.05) were set as the thresholds to indicate significant changes. Two such independent experiments were carried out. In each experiment, a total of 8 gels derived from pooled protein samples of four animals from each group were analysed.
For all other experiments, the data were expressed as mean±SE of three animals per group. The level of significance was set at p<0.05. All experiments were repeated at least twice.
Results and Discussion
Animal experimentation
Among the available CHIKV disease animal models, newborn mice represent an ideal model to unravel the host response during CHIKV infection because CHIKV-infected newborn mice exhibit muscle and joint pathologies similar to those observed in humans. Although CHIKV infected suckling mice exhibit 10–20% mortality, majority of the animals eventually recover from the disease with serum virus clearance after 12 days of infection except in the muscles where the viral antigen may persist for a longer period [19], [20]. The neurotropic and neuroinvasive nature of CHIKV has also been explored in different strains of newborn mice [20], [26]. The behavioral traits, loco-motor abnormalities and microscopic observations reported in this study collectively supported the fact that CHIKV infected newborn mice clinically mimic human CHIKV patients in terms of symptoms and disease progression. Based on the clinical similarity between CHIKV infected patients and newborn suckling mice, we aim to explore the host cellular pathophysiological processes that occur during viral infection.
The CHIKV infected new-born mice developed symptoms, such as retarded growth, alopecia, lethargy, dragging of hind limbs on 8 day post infection (dpi). The animals developed stiffness in the hind limbs as they failed to bend their limbs at the joints (Figure 1A). The loco-motor behaviour of CHIKV infected mice was severely affected and exhibited a loss of coordination. When tested for surface righting reflex, the infected animals showed difficulty in turning back to the upright position confirming loco-motor disturbance of the hind limbs. The video clips demonstrating the walking traits and surface righting reflex of CHIKV infected mice are provided as supplementary materials (Movie S1, S2 and S3). In addition, the average walking stride length of CHIKV infected mice was significantly decreased. The details of this parameter are given in Figure S1. These data implicate CHIKV induced severe myopathic symptoms in infected new-born mice.
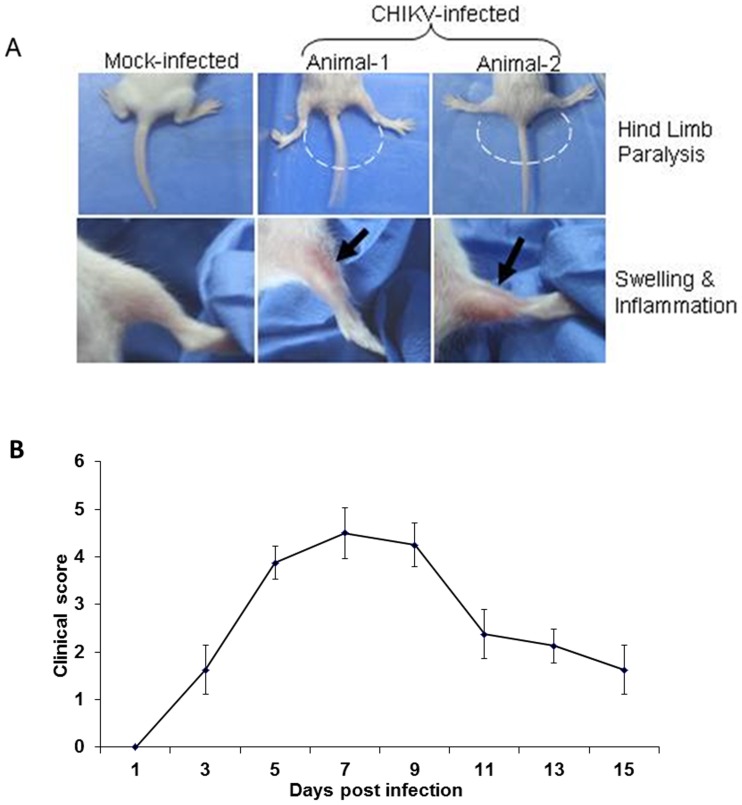
Figure 1. Chikungunya virus (CHIKV) induced disease signs.
A. Pictures of the hind limbs of mock- and CHIKV-infected animals. Chikungunya virus infection induced severe hind limb disease in new born mice. 2–3 day old mice inoculated with 106 PFU (50 ul) of CHIKV by subcutaneous injection in the loose fold of skin on the back of the animal developed peak clinical signs on 8 day post inoculation whereas mock-infected group injected with uninfected tissue culture supernatant remained healthy. B. Mice were scored for the development of hind limb dysfunction and disease based on the following scale: 0, no disease signs; 1, ruffled fur; 2, mild hind limb weakness; 3, moderate hind limb weakness; 4, severe hind limb weakness and dragging and 5, moribund. Each data point represents the arithmetic mean ± SD for eight animals. Data is representative of three independent experiments.
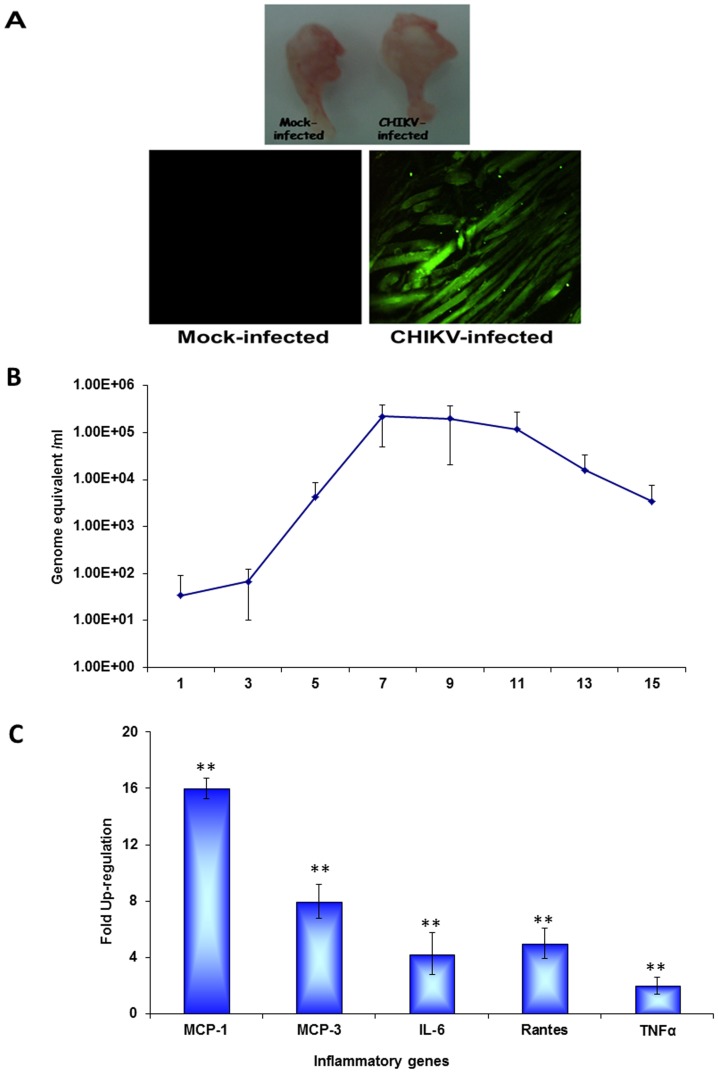
It was also noted that the severity of the symptoms subsided at 12 dpi. With this observation, all subsequent experiments were conducted when animals exhibited peak clinical signs between 8–9 dpi. The disease severity in terms of clinical score is depicted in Figure 1B. The gross pathology of the CHIKV infected muscle showed swelling due to possibly to overt inflammation of the tissue (Figure 1A). Using immunoflourescence analysis, we showed that high levels of CHIKV antigen were localised in sections of muscle tissue at 9 dpi (Figure 2A). The muscle sections showing the localization of CHIKV antigen by immunofluorescence at different days of infection are provided as Figure S2. Viral load in muscles was determined on different days post infection to indicate that the virus titre peaked at day 7 post infection and started to decline at day 11 (Figure 2B). In order to confirm the virus induced inflammation in swollen muscles, we conducted gene expression profiling of pro-inflammatory cytokines at 9 dpi. As shown in Figure 2C, significant elevated levels of pro-inflammatory cytokines, such as IL-6, TNF-α, MCP-1, MCP-3 and RANTES, were observed (Figure 2C). Collectively, our data showed that CHIKV successfully invaded the mouse muscle tissue, which led to enhanced inflammation followed by myopathy.
Figure 2. Gross pathology, viral replication and profile of inflammatory cytokines.
A. Surgically removed hind limb muscles from mock- and CHIKV-infected mice showing gross pathology of the muscles (in terms of swelling) along with the 4 μm sections showing the localization of CHIKV antigen in the hind limb skeletal muscle on day 9 post infection. B. Virus titre in the hind limb muscles. C. Real time analysis showing relative fold change in inflammatory genes (MCP-1, MCP-3, IL-6, TNF-α, Rantes) in muscle tissue on day 9 post infection. ** Genes were considered significantly up-regulated if the change in their relative expression level was ≥2 fold at p<0.05 by student's t test.
Microscopic observation
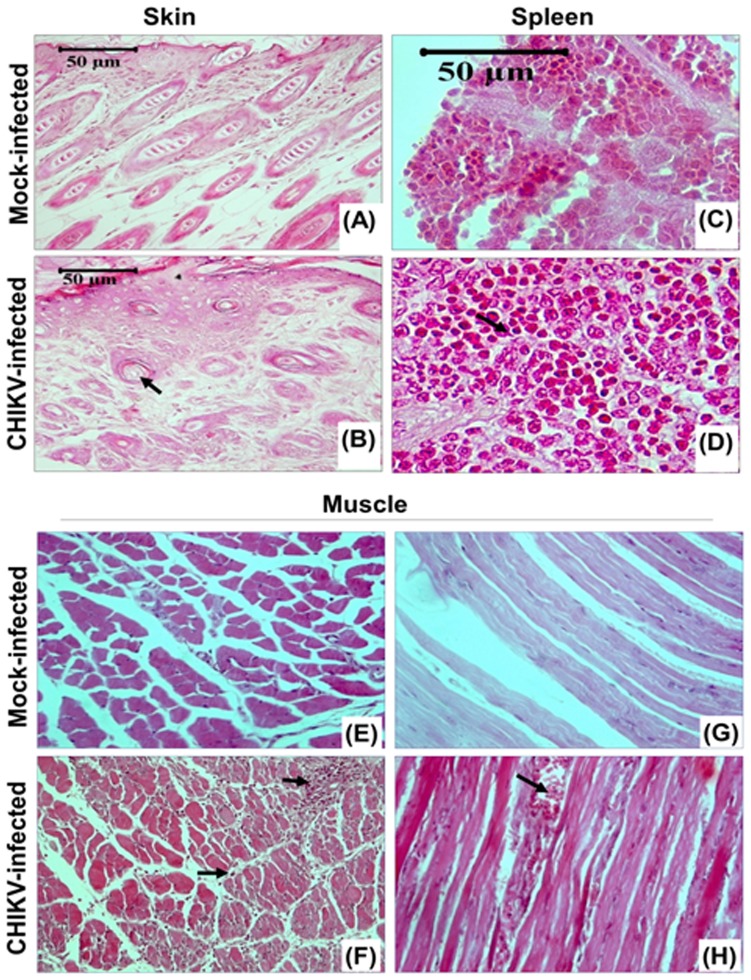
Histological analyses of skin, spleen and muscle tissue at 8 dpi showed differential changes that took place in response to CHIKV infection as compared to tissues collected from uninfected animals. The skin sections the of mock-infected mice showed that the matrix of hair follicle consisted of dividing cells and the growing hair shaft was surrounded with multi-layered root sheath whereas the skin of CHIKV-infected mice showed hyperplasia of the basal keratinocytes along with swelling and hyperkeratinisation. Hair follicles of the infected animals showed atrophy with no dividing cells in the matrix that was eosinophilic in nature. Contrary to the uninfected tissue, the external root sheath in infected animals was single layered (compared panel A to B in Figure 3). The spleen of the CHIKV-infected animals showed considerable lymphoid proliferation and haemorrhage (Figure 3D, arrow). The most significant pathologic changes in the new born mice were in the skeletal muscle and adjacent connective tissues. The CHIKV infected mice showed degenerative changes with dark pink staining indicating atrophied and necrotic muscle fibres. In addition, neutrophils and monocytoid cells heavily infiltrated the muscle fibres (Figure 3F and H, arrows).
Figure 3. Skin, spleen and muscle pathology of CHIKV infected mice.
Representative picture showing the pathology staining of the tissues (skin, spleen and hind limb muscle) from mock-infected and CHIKV-infected mice on 8th day of post infection. Characteristic histological features are indicated by arrows. Skin of CHIKV-infected mice showed hyperplasia of basal keratinocytes and hyperkeratinisation. Hair follicles showed atrophy with no dividing cells in the matrix. Spleen of CHIKV-infected mice showed considerable lymphoid proliferation and haemorrhage. Muscle sections showed degenerative changes with dark pink stained, atrophied and necrotic muscle fibres, infiltration of neutrophiles and monocytoid cells between the muscle fibres.
Analysis of differentially expressed proteins after CHIK infection
Whole cell lysates of muscle tissue from mock and CHIKV-infected mice were subjected to 2-DE analysis and their proteomic profiles were obtained. More than 800-900 protein spot features were matched upon inspection of 2-DE gels between mock and CHIKV infected muscle samples.
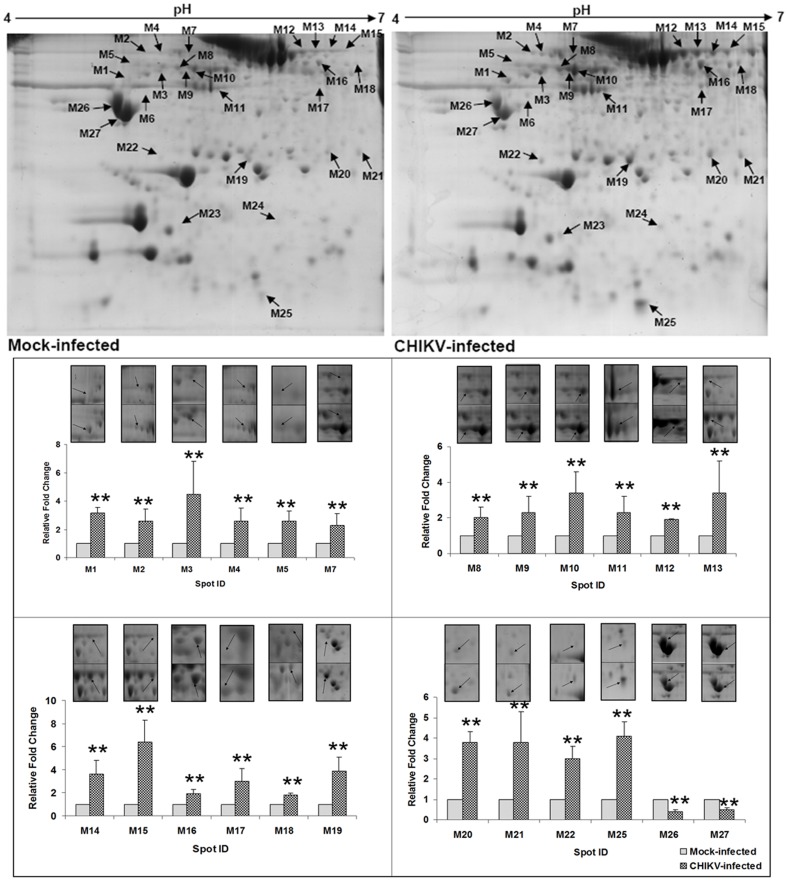
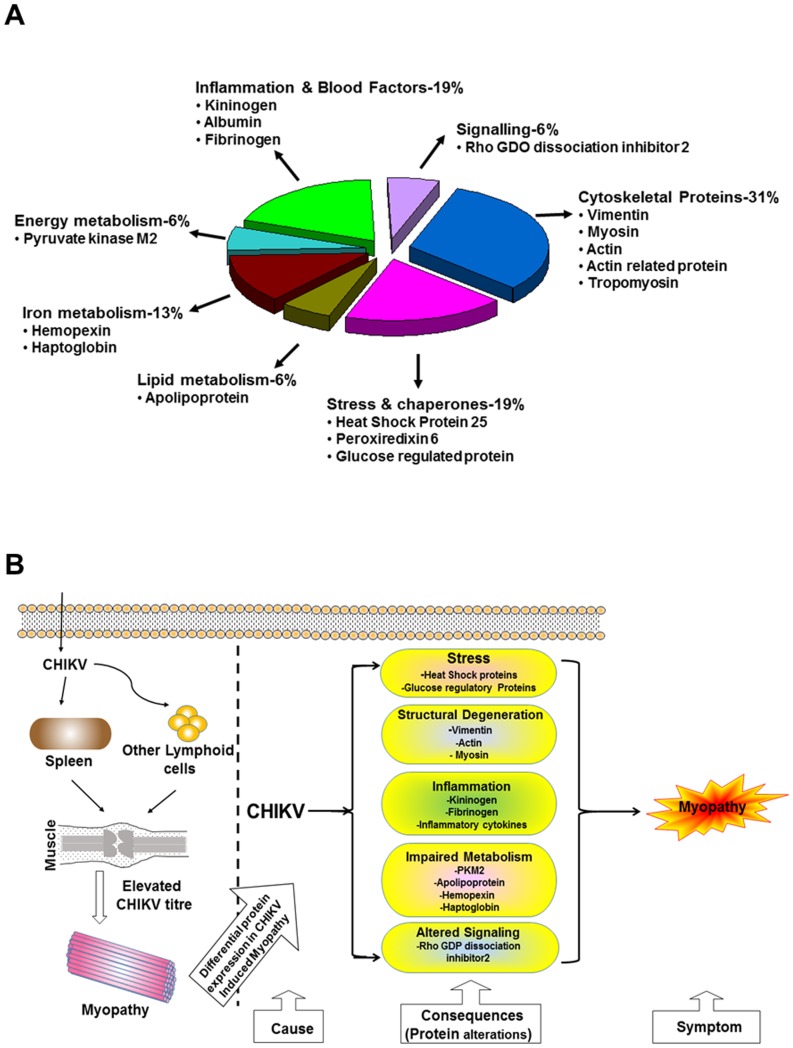
A total of 27 protein spots showing significant changes in mean intensity upon CHIKV infection in muscle tissue were observed (Figure 4). Twenty five spots showed up regulation of gene expressions whereas 2 spots showed down regulation at the ratio of CHIK/mock ≥|1.8|, p<0.05). These protein spots were excised, digested in-gel with trypsin and identified by MALDI TOF MS analysis. The MS and MS/MS spectra are listed in File S1. The resulting peptide profiles and fragment ion spectra were searched against Mus musculus protein sequence database (MSDB). Table 1 provides the identity of each of the protein spots including MOWSE score, sequence coverage, number of peptides matched/searched, theoretical/observed Mr and pI obtained after tandem MS analysis. The identified proteins could be functionally classified into various groups (http://ca.expasy.org/), including those involved in inflammation, iron metabolism, cytoskeletal, energy metabolism, fatty acid metabolism, and stress chaperons. The 27 differentially expressed protein spots correspond to 15 proteins that include cytoskeleton-associated (structural) proteins (31%), stress proteins (19%), iron metabolism (13%), energy metabolism (6%), lipid metabolism (6%), inflammation and blood factors (19%) and signal transduction proteins (6%) (Figure 5A). The importance of the differentially expressed proteins in disease manifestation has been hypothesized in Figure 5B.
Figure 4. Representative 2-D gel image obtained from muscle tissue of mock-infected and CHIKV-infected mice.
Equal amount of protein sample (750 μg) was first separated in a linear gradient of pH 4–7, followed by separation in SDS-PAGE (12%) and coomassie staining. A total of 27 protein spots were found to be significantly altered. Fold changes (mean values± SD) of the identified proteins are illustrated graphically (** signifies p≤0.05). Spots M6, M23 and M24 were found to be affected qualitatively and hence were not presented graphically.
Table 1. Summary of proteins differentially expressed in mouse muscle tissue infected by Chikungunya virus.
| Spot intensity Mean± SD | ||||||||||||
| S.No | Spot ID | Protein Identified | Accessiona | Mowse Scoreb PMF/MS-MS | % Covc | Peptides Matched (searched) | Mr/pI theoretical | Mr/pI observed | Mock Infected | CHIKV Infected | Fold change Mean± SD | P value |
| 1 | M1 | Vimentin | P20152 | 290/530 | 77 | 59 (132) | 53.68/5.06 | 63/4.9 | 5.8±1.7 | 18.5±6.4 | 3.15±0.41 | 8×10−3 |
| 2 | M2 | a.Albumin b.Kininogen | P07724 O08677 | 132/NA 86/77 | 59 51 | 36 (119) 31 (119) | 53.17/4.88 | 95/5.2 | 3.6±1.2 | 8.2±1.7 | 2.6±0.82 | 1×10−2 |
| 3 | M3 | Vimentin | P20152 | 241/241 | 85 | 61 (176) | 53.5/5.06 | 70/5.2 | 5.4±2.3 | 26±11.4 | 4.5±2.3 | 3×10−2 |
| 4 | M4 | Kininogen | O08677 | 105/NS | 56 | 28 (147) | 47.8/5.74 | 95/5.2 | 6.1±1.3 | 14.9±1.7 | 2.6±0.9 | 9×10−3 |
| 5 | M5 | Vimentin | P20152 | 214/73 | 73 | 54 (173) | 51.5/4.9 | 75/5.0 | 6.9±1.6 | 17.6±5.8 | 2.6±0.7 | 1×10−2 |
| 6 | M6 | Haptoglobin | Q3UBS3 | 64/117 | 54 | 21 (129) | 38.8/6.08 | 52/5.0 | 0 | 5.4±1.4 | DIV/0e | 6×10−3 |
| 7 | M7 | Myosin | Q5SX40 | 120/58 | 48 | 95 (167) | 60.9/5.4 | 92/5.4 | 4.9±1.9 | 12.2±3.0 | 2.3±0.8 | 1×10−2 |
| 8 | M8 | Vimentin | P20152 | 262/219 | 89 | 65 (162) | 51.5/4.96 | 72/5.3 | 27.7±4.3 | 52±9.2 | 2±0.6 | 5×10−3 |
| 9 | M9 | Vimentin | Q5FWJ3 | 265/474 | 89 | 77 (181) | 53.6/5.06 | 72/5.3 | 16.2±2.5 | 35.9±8.1 | 2.3±0.9 | 5×10−3 |
| 10 | M10 | Vimentin | P20152 | 217/337 | 83 | 64 (178) | 53.6/5.06 | 72/5.4 | 30±1.6 | 101±37.4 | 3.4±1.2 | 9×10−3 |
| 11 | M11 | Actin | P60710 | 160/215 | 82 | 37 (125) | 41.7/5.3 | 57/5.6 | 17.5±4.3 | 36.4±3.9 | 2.3±0.9 | 1×10−3 |
| 12 | M12 | a.Albumin b.Hemopexin | P07724 Q91X72 | 154/NA 98/417 | 71 56 | 54 (164) 27 (164) | 50.51/7.32 | 95/6.3 | 41.5±12 | 81±27.05 | 1.9±0.04 | 4×10−2 |
| 13 | M13 | a.Hemopexin b.Albumin | Q91X72 P07724 | 152/404 114/NA | 63 69 | 31 (126) 46 (126) | 51.3/7.92 | 95/6.4 | 17.5±6.8 | 53±14.6 | 3.4±1.8 | 4×10−3 |
| 14 | M14 | Hemopexin | Q91X72 | 170/55 | 51 | 25 (108) | 51.3/7.92 | 95/6.5 | 13±7.9 | 40±12.2 | 3.6±1.2 | 9×10−3 |
| 15 | M15 | Hemopexin | Q91X72 | 211/115 | 58 | 29 (111) | 51.3/7.92 | 95/6.7 | 5.67±0.7 | 35.5±6.3 | 6.4±1.9 | 2×10−2 |
| 16 | M16 | Glucose regulated protein | P27773 | 203/421 | 69 | 45 (170) | 56.59/5.78 | 72/6.4 | 21.4±3.6 | 40.1±6.8 | 1.9±0.4 | 2×10−3 |
| 17 | M17 | Pyruvate kinase isozyme M2 | P52480 | 120/84 | 83 | 35 (159) | 43.1/5.88 | 58/6.4 | 3.9±0.43 | 11.5±3.2 | 3.0±1.1 | 3×10−3 |
| 18 | M18 | Fibrinogen | Q3TGR2 | 110/124 | 66 | 42 (182) | 54.7/6.68 | 78/6.7 | 12±1.7 | 22±1.0 | 1.85±0.18 | 3×10−2 |
| 19 | M19 | Apolipoprotein A-1 | Q3V2G1 | 120/268 | 60 | 24 (147) | 30.66/5.64 | 34/5.9 | 18.7±7.1 | 66.3±4.0 | 3.9±1.2 | 2×10−5 |
| 20 | M20 | Peroxiredoxin-6 | O08709 | 176/306 | 88 | 30 (119) | 24.7/5.72 | 36/6.5 | 7.2±0.9 | 23.6±5.9 | 3.8±0.5 | 1×10−4 |
| 21 | M21 | Heat shock protein 25 | P14602 | 117/160 | 71 | 18 (110) | 23/6.12 | 36/6.7 | 11.4±3.8 | 40.6±10 | 3.8±1.6 | 9×10−3 |
| 22 | M22 | Rho GDP dissociation inhibitor 2 | Q61599 | 92/37 | 81 | 17 (97) | 22.7/4.97 | 34/5.1 | 3.4±0.7 | 10±0.96 | 3.0±0.6 | 3×10−5 |
| 23 | M23 | Actin | P60710 | NS/55 | NA | NA | 39/5.8 | 26/5.3 | 0 | 7.6±1.2 | DIV/0e | 1×10−5 |
| 24 | M24 | Actin related protein | Q6ZQK2 | 64/90 | 37 | 65 (180) | 191.2/6.15 | 26/6.3 | 0 | 4.7±0.95 | DIV/0e | 5×10−4 |
| 25 | M25 | NAd | NA | NA | NA | NA | NA | NA | 23.8±5.3 | 94.5±11 | 4.1±0.7 | 2×10−5 |
| 26 | M26 | Tropomyosin alpha chain | P58772 | 167/420 | 70 | 35 (129) | 32.8/4.6 | 50/4.8 | 512.6±22 | 208±15 | 0.4±0.3 | 5×10−3 |
| 27 | M27 | Tropomyosin alpha chain | P58772 | 126/323 | 69 | 31 (142) | 32.6/4.8 | 45/4.9 | 637±128 | 284.5±52 | 0.5±0.1 | 2×10−3 |
a = Swiss Prot accession number. b = By MASCOT. c = % Coverage of the identified sequence. NAd = not available. DIV/0e = Divided by zero. Sopt ID represents the number on 2-DE gels. NS = Non significant.
Figure 5. Functional classification of differentially affected proteins and their possible role in disease pathogenesis.
A. Functional classification of the differentially affected proteins of muscle tissue in CHIKV infection. B. Schematic representation of the possible roles of identified proteins of different classes showing metabolic and rheumatic implications in CHIKV induced myopathy.
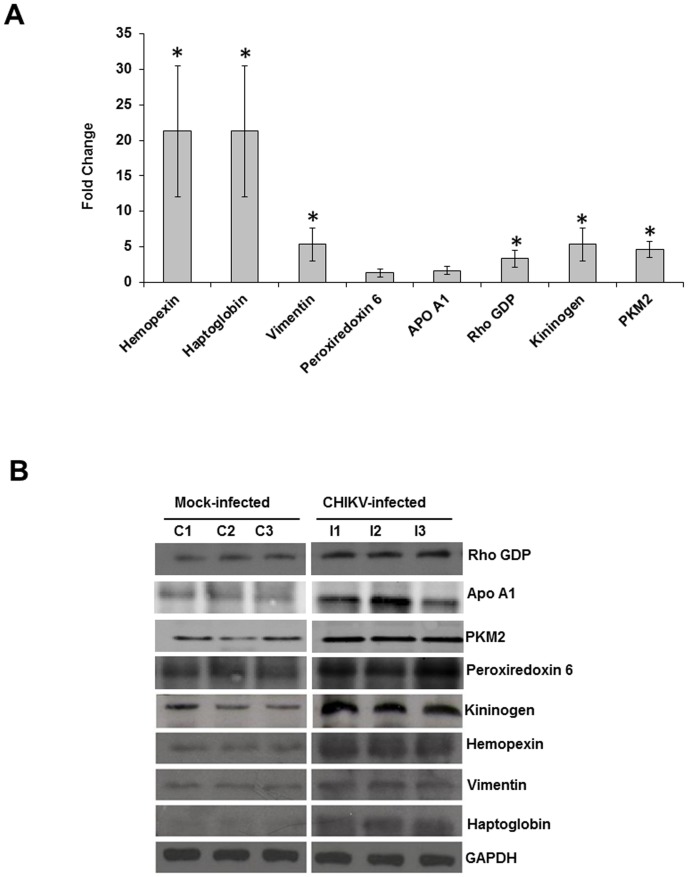
The differentially affected proteins were validated by Q-PCR and western blotting. The transcriptional alteration of eight selected genes was analysed at the mRNA transcript level using the housekeeping GAPDH gene for normalization purpose. The candidate proteins were selected as being representative of structural (vimentin), stress (peroxiredoxin 6), lipid metabolism (ApoA1), signalling (Rho GDP), iron metabolism (hemopexin and haptoglobin), inflammation (kininogen), and energy metabolic (pyruvate kinase M2) pathways. Out of eight selected genes, six genes (hemopexin, haptoglobin, vimentin, kininogen, PKM2 and Rho GDP) exhibited the same trend of up-regulation as observed in 2-DE gels of the corresponding proteins (Figure 6A). On the contrary, the transcript levels of ApoA1 and peroxiredoxin 6 remained unchanged. To further confirm the alterations of protein expression during CHIKV infection, all the selected proteins (i.e., hemopexin, haptoglobin, vimentin, ApoA1, peroxiredoxin 6, Rho GDP, PKM2, and kininogen) were analysed by western blotting. Equal amount of mock and CHIKV infected muscle-tissue protein were examined by western blot analysis with specific antibodies. The data obtained agreed with the expression changes by the 2-DE analysis as all the proteins were up-regulated (Figure 6B). The discrepancy at the transcriptional and protein levels of ApoA1 and peroxiredoxin 6 could be due to the altered regulation of transcription and translation of these genes.
Figure 6. Validation of proteomic results using Q-PCR and immunoblotting.
A. Transcript alteration of the differentially expressed proteins in muscle tissue upon CHIKV infection. Total RNA of muscle tissue (infected/uninfected) was analysed by real time RT-PCR. House-keeping GAPDH gene was used for normalization purpose. RNA expression changes of vimentin, hemopexin, haptoglobin, Rho GDP, PKM2 and kininogen were in concordance with protein expression changes and were determined to be statistically significant (p≤0.05). *Genes were considered to be significantly up-regulated if the change in their relative expression levels was ≥2 fold. No significant difference in the RNA expression of ApoA1 and peroxiredoxin 6 was found. B. Immunoblot of representative proteins showing increased expressions in muscle tissue upon CHIKV infection.
Among the differentially expressed proteins, 31% of proteins were assigned to the structural protein category. Vimentin, actin, actin related protein, myosin and tropomyosin were among the differentially expressed cytoskeleton-associated proteins. Vimentin, an intermediate filament protein normally expressed in cells of mesenchymal origin, plays important roles in many biological reactions [32], including but not necessarily limited to cell growth [33], cellular motility and invasiveness of some cancers [34], and as a signal transducer, relaying information from extracellular matrix to the nucleus [35]. Interestingly, vimentin is also expressed on the cell surface of activated macrophages and apoptotic neutrophils. Hence, one possible reason for the increased expression of vimentin could be due to the increased neutrophil infiltration in CHIKV infected muscles as observed in histological analysis.
Many of other differentially expressed structural proteins may play important roles in the development of arthritis and myositis in CHIKV-infected individuals. For example, there are several reports documenting that the over-expression of cytoskeletal proteins such as actin, myosin and vimentin lead to development of auto-antibodies in rheumatoid arthritis and other inflammatory diseases [36], [37]. Likewise, overexpression of proteins, such as haptoglobin and hemopexin, has been reported as a result of inflammation and injury [38], [39] and in myopathic disorders [40]. We also observed a significant level of increase of kininogen and fibrinogen expression in the muscle of CHIKV infected mice. Kininogen and fibrinogen have been shown to be associated with inflammation and arthritis [41], [42] and kininogen is also one of the known regulators of stress/energy metabolic proteins [43]. Among the differentially affected proteins, 19% of the proteins were stress-related proteins that included heat shock protein 25 (HSP25), peroxiredoxin 6 and glucose regulated protein (Grp). The upregulation of heat shock proteins in skeletal muscles have been shown to provide some protection against a variety of stressors, such as disuse atrophy [44], [45], oxidative stress [45], [46], increased calcium concentrations [47] and muscle damage [48], [49], [50]. Furthermore several observations suggest that the small HSPs, such as HSP25 play crucial roles in the normal function and integrity of skeletal muscles as well in arthritis [51], [52]. Glucose regulated protein (Grp), an ER molecular chaperone that assists in protein folding and assembly has been reported to be differentially expressed in rheumatoid synovitis [53]. Similarly, we observed a significant increase in expression of peroxiredoxin-6, another antioxidant enzyme that functions to catalyze peroxides and protects cells against reactive oxygen species. Apart from the antioxidant properties, peroxiredoxins have also been shown to be important in various cellular processes such as regulation of apoptosis [54] and antiviral activities [55]. The possible involvement of stress-related proteins in CHIKV infection has been reported in our previous studies [26], [56] and by other researchers [57].
The metabolic proteins that were overexpressed in CHIKV-infected muscles were PKM2, an isoenzyme of the glycolytic enzyme pyruvate kinase, and ApoA1, a protein associated with lipid metabolism. PKM2 protein expression has been reported to be increased in rheumatic diseases and has also been shown to support the replication of RNA viruses [58], [59]. Hence, it is possible that the elevated level of PKM2 might be responsible for supporting CHIKV replication in infected muscle. Interestingly, expressions of ApoA1 and Rho GDP dissociation inhibitor 2 have also been shown to be elevated in synovial fluid of patients with rheumatoid arthritis and osteoarthritis [60], [61]. Collectively, elevated expressions of many of the proteins observed in our study might be directly involved in CHIKV infection and disease pathogenesis and directly or indirectly correlated with muscle-related disorders such as arthritis and myositis.
Further studies are needed for in depth characterization of affected proteins and their interactions with CHIKV that will likely open new avenues for the design of antiviral therapies against CHIKV and related disorders caused by alphaviruses. Also, the validation of differentially expressed proteins in serum samples of CHIKV infected patients may reveal relevant clinical biomarkers and thereby allow for early detection of the disease that can improve outcomes by providing better medical treatment and/or disease management strategies.
Supporting Information
Average walking stride length. This parameter was measured manually by allowing the control (mock-infected) and CHIKV-infected mice to walk on a sheet of paper with their foot-steps being marked. The total distance covered was then divided by the number of steps taken. The data was expressed as mean±SE of five animals per group and the experiment was repeated twice (p≤0.05).
(TIF)
Muscle sections showing the localisation of CHIKV antigen by immunofluorescence at different days of infection.
(TIF)
The MS and MS/MS spectra of the differentially affected proteins.
(PDF)
Walking behavior of CHIKV infected mice showing lack of coordination and hind limb paralysis.
(WMV)
Surface righting reflex of control (mock-infected) mice.
(MPG)
Surface righting reflex of CHIKV-infected mice. The infected mice demonstrated difficulty in turning to the upright position confirming weakness of the hind limbs.
(MPG)
Acknowledgments
We gratefully acknowledge Dr. R. Vijayaraghwan, Director, DRDE for providing all the infrastructure necessary for this study, Dr. S.I. Alam, DRDE, Gwalior for his fruitful suggestions and Dr. S.C. Pant, DRDE, Gwalior for providing the facility for histological examination.
Funding Statement
The study was funded by Defence Research and Development Organisation (DRDO), Govt. of India. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Enserink M (2008) Entomology. A mosquito goes global. Science 320: 864–866. [DOI] [PubMed] [Google Scholar]
- 2. Rezza G, Nicoletti L, Angelini R, Romi R, Finarelli AC, et al. (2007) Infection with chikungunya virus in Italy: an outbreak in a temperate region. Lancet 370: 1840–1846. [DOI] [PubMed] [Google Scholar]
- 3. Santhosh SR, Dash PK, Parida M, Khan M, Tiwari M, et al. (2008) Comparative full genome analysis revealed E1:A226V shift in 2007 Indian Chikungunya virus isolates. Virus Research 135: 36–41. [DOI] [PubMed] [Google Scholar]
- 4. Jain M, Rai S, Chakravarti A (2008) Chikungunya: a review. Trop Doct 38: 70–72. [DOI] [PubMed] [Google Scholar]
- 5. Arankalle VA, Shrivastava S, Cherian S, Gunjikar RS, Walimbe AM (2007) Genetic divergence of Chikungunya viruses in India (1963–2006) with special reference to the 2005–2006 explosive epidemic. J Gen Virol 88: 1967–1976. [DOI] [PubMed] [Google Scholar]
- 6. Sam IC, AbuBakar S (2006) Chikungunya virus infection. Med J Malaysia 61: 264–269. [PubMed] [Google Scholar]
- 7. Queyriaux B, Simon F, Grandadam M, Michel R, Tolou H, et al. (2008) Clinical burden of chikungunya virus infection. Lancet Infect Dis 8: 2–3. [DOI] [PubMed] [Google Scholar]
- 8. Tesh RB (1982) Arthritides caused by mosquito-borne viruses. Annu Rev Med 33: 31–40. [DOI] [PubMed] [Google Scholar]
- 9. Robin S, Ramful D, Le Seach F, Jaffar-Bandjee MC, Rigou G, et al. (2008) Neurologic manifestations of pediatric chikungunya infection. J Child Neurol 23: 1028–1035. [DOI] [PubMed] [Google Scholar]
- 10. Hazelton RA, Hughes C, Aaskov JG (1985) The inflammatory response in the synovium of a patient with Ross River arbovirus infection. Aust N Z J Med 15: 336–339. [DOI] [PubMed] [Google Scholar]
- 11. Fraser JRE, Cunningham AL, Clarris BJ, Aaskov JG, Leach R (1981) Cytology of synovial effusions in epidemic polyarthritis. Aust N Z J Med 11: 168–173. [DOI] [PubMed] [Google Scholar]
- 12. Soden M, Vasudevan H, Roberts B, Coelen R, Hamlin G, et al. (2000) Detection of viral ribonucleic acid and histologic analysis of inflamed synovium in Ross River virus infection. Arthiritis Rheum 43: 365–369. [DOI] [PubMed] [Google Scholar]
- 13. Hiruma M, Ide S, Hohdatsu T, Yamagishi H, Tanaka Y, et al. (1990) Polymyositis in mice experimentally inoculated with Getah virus. Nihon Juigaku Zasshi 52: 767–772. [DOI] [PubMed] [Google Scholar]
- 14. Harley D, Sleigh A, Ritchie S (2001) Ross River virus transmission, infection, and disease: a cross-disciplinary review. Clin Microbiol Rev 14: 909–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Morrison TE, Whitmore AC, Shabman RS, Lidbury BA, Mahalingam S, et al. (2006) Characterization of Ross River virus tropism and virus-induced inflammation in a mouse model of viral arthritis and myositis. J Virol 80: 737–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Trgovcich J, Aronson JF, Johnston RE (1996) Fatal Sindbis virus infection of neonatal mice in the absence of encephalitis. Virology 224: 73–83. [DOI] [PubMed] [Google Scholar]
- 17. Villoing S, Bearzotti M, Chilmonczyk S, Castric J, Bremont M (2000) Rainbow trout sleeping disease virus is an atypical alphavirus. J Virol 74: 173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ozden S, Huerre M, Riviere J-P, Coffey LL, Afonso PV, et al. (2007) Human muscle satellite cells as targets of Chikungunya virus infection. Plos One 2: e527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Couderc T, Chretien F, Schilte C, Disson O, Brigitte M, et al. (2008) A mouse model for chikungunya: young age and inefficient type-I interferon signaling are risk factors for severe disease. PLoS Pathog 4: e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ziegler SA, Lu L, Travassos da Rosa APA, Xiao S-Y, Tesh RB (2008) An animal model for studying the pathogenesis of chikungunya virus infection. Am J Trop Med Hyg 79: 133–139. [PubMed] [Google Scholar]
- 21. Morrison TE, Oko L, Montgomery SA, Whitmore AC, Lotstein AR, et al. (2011) A mouse model of chikungunya virus–induced musculoskeletal inflammatory disease. Am J Pathol 178: 32–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gardner J, Anraku I, Le TT, Larcher T, Major L, et al. (2010) Chikungunya virus arthritis in adult wild-type mice. J Virol 84: 8021–8032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nakaya HI, Gardner J, Poo YS, Major L, Pulendran B, et al. (2012) Gene profiling of chikungunya virus arthritis in a mouse model reveals significant overlap with rheumatoid arthritis. Arthritis Rheum 64: 3553–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Patil DR, Hundekar SL, Arankalle VA (2012) Expression profile of immune response genes during acute myopathy induced by chikungunya virus in a mouse model. Microbes Infect (14) 457–469. [DOI] [PubMed] [Google Scholar]
- 25. Rulli NE, Rolph MS, Srikiatkhachorn A, Anantapreecha S, Guglielmotti A, et al. (2011) Protection from arthritis and myositis in a mouse model of acute chikungunya virus disease by Bindarit, an inhibitor of monocyte chemotactic protein-1 synthesis. J Infect Dis 204: 1026–1030. [DOI] [PubMed] [Google Scholar]
- 26. Dhanwani R, Khan M, Alam SI, Rao PVL, Parida MM (2011) Differential proteome analysis of Chikungunya virus infected new born mice tissues reveal implication of stress, inflammatory and apoptotic pathways in disease pathogenesis. Proteomics 11: 1936–1951. [DOI] [PubMed] [Google Scholar]
- 27. Dash PK, Parida MM, Santhosh SR, Verma SK, Tripathi NK, et al. (2007) East Central South African genotype as causative agent in re-emergence of Chikungunya outbreak in India. Vector Borne Zoonotic Dis 7: 719–727. [DOI] [PubMed] [Google Scholar]
- 28. Santhosh SR, Parida MM, Dash PK, Pateriya A, Pattnaik B, et al. (2007) Development and evaluation of SYBR Green I based one step real time RT-PCR assay for detection and quantification of Chikungunya virus. J. Clin. Virol 39: 188–193. [DOI] [PubMed] [Google Scholar]
- 29. Butt RH, Coorssen JR (2005) Postfractionation for enhanced proteomic analyses: routine electrophoretic methods increase the resolution of standard 2D-PAGE. J. Proteome Res 4: 982–991. [DOI] [PubMed] [Google Scholar]
- 30. Fernandez-Arenas E, Cabezon V, Bermejo C, Arroyo J, Nombela C, et al. (2007) Integrated proteomics and genomics strategies bring new insight into Candida albicans response upon macrophage interaction. Mol Cell Proteomics 6: 460–478. [DOI] [PubMed] [Google Scholar]
- 31. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real time quantitative PCR and the 2 delta Ct method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 32. Rittling SR, Baserga R (1987) Functional analysis and growth factor regulation of the human vimentin promoter. Mol Cell Biol 7: 3908–3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Franke WW, Schmid E, Winter S, Osborn M, Weber K (1979) Widespread occurrence of intermediate-sized filaments of the vimentin-type in cultured cells from diverse vertebrates. Exp Cell Res 123: 25–46. [DOI] [PubMed] [Google Scholar]
- 34. Hendrix MJ, Seftor EA, Chu YW, Trevor KT, Seftor RE (1996) Role of intermediate filaments in migration, invasion and metastasis. Cancer Metastasis Rev 15: 507–525. [DOI] [PubMed] [Google Scholar]
- 35. Skalli O, Goldman RD (1991) Recent insights into the assembly, dynamics, and function of intermediate filament networks. Cell Motil Cytoskeleton 19: 67–79. [DOI] [PubMed] [Google Scholar]
- 36. Shrivastav M, Mittal B, Aggarwal A, Misra R (2002) Autoantibodies against Cytoskeletal Proteins in Rheumatoid Arthritis. Clinical Rheumatology 21: 505–510. [DOI] [PubMed] [Google Scholar]
- 37. Moisan E, Girard D (2006) Cell surface expression of intermediate filament proteins vimentin and lamin B1 in human neutrophil spontaneous apoptosis. Journal of Leukocyte Biology 79: 489–498. [DOI] [PubMed] [Google Scholar]
- 38. Miller I, Haynes P, Eberini I, Gemeiner M, Aebersold R, et al. (1999) Proteins of rat serum: III. Gender-related differences in protein concentration under baseline conditions and upon experimental inflammation as evaluated by two dimensional electrophoresis. Electrophoresis 20: 836–845. [DOI] [PubMed] [Google Scholar]
- 39. Marinkovic S, Jahreis GP, Wong GG, Baumann H (1989) IL-6 modulates the synthesis of a specific set of acute phase plasma proteins in vivo. J Immunol 142: 808–812. [PubMed] [Google Scholar]
- 40. Niedermeier W, Cross R, Beetham WP (1965) The concentration of haptoglobin in synovial fluid of patients with rheumatoid arthritis. Arthritis & Rheumatism 8: 355–360. [DOI] [PubMed] [Google Scholar]
- 41. Sharma JN, Zeitlin IJ, Brooks PM, Dick WC (1976) A novel relationship between plasma kininogen and rheumatoid disease. Inflammation Res 6: 147–153. [DOI] [PubMed] [Google Scholar]
- 42. Ho PP, Lee LY, Zhao X, Tomooka BH, Paniagua RT, et al. (2010) Autoimmunity against Fibrinogen Mediates Inflammatory Arthritis in Mice. J Immunol 184: 379–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Seematter G, Binnert C, Martin JL, Tappy L (2004) Relationship between stress, inflammation and metabolism. Curr Opin Clin Nutr Metab Care 7: 169–173. [DOI] [PubMed] [Google Scholar]
- 44. Naito H, Powers SK, Demirel HA, Sugiura T, Dodd SL, et al. (2000) Heat stress attenuates skeletal muscle atrophy in hindlimb-unweighted rats. J Appl Physiol 88: 359–363. [DOI] [PubMed] [Google Scholar]
- 45. Selsby JT, Dodd SL (2005) Heat treatment reduces oxidative stress and protects muscle mass during immobilization. Am J Physiol Regul Integr Comp Physiol 289: R134–R139. [DOI] [PubMed] [Google Scholar]
- 46. Escobedo J, Pucci AM, Koh TJ (2004) HSP25 protects skeletal muscle cells against oxidative stress. Free Radic Biol Med 37: 1455–1462. [DOI] [PubMed] [Google Scholar]
- 47. Welch WJ (1992) Mammalian stress response: cell physiology, structure/function of stress proteins, and implications for medicine and disease. Physiol Rev 72: 1063–1081. [DOI] [PubMed] [Google Scholar]
- 48. Escobedo J, Pucci AM, Koh TJ (2004) HSP25 protects skeletal muscle cells against oxidative stress. Free Radic Biol Med 37: 1455–62. [DOI] [PubMed] [Google Scholar]
- 49. Koh TJ, Escobedo J (2004) Cytoskeletal disruption and small heat shock protein translocation immediately after lengthening contractions. Am J Physiol Cell Physiol 286: C713–C722. [DOI] [PubMed] [Google Scholar]
- 50. Liu Y, Steinacker JM (2001) Changes in skeletal muscle heat shock proteins: pathological significance. Front Biosci 6: D12–D25. [DOI] [PubMed] [Google Scholar]
- 51. Concannon CG, Gorman AM, Samali A (2003) On the role of Hsp27 in regulating apoptosis. Apoptosis 8: 61–70. [DOI] [PubMed] [Google Scholar]
- 52. Al-Shamma MRR, McSharry C, McLeod K, McCruden EAB, Stack BHR (1997) Role of heat shock proteins in the pathogenesis of cystic fibrosis arthritis Thorax. 52: 1056–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dong W, Li X, Feng Y, Fan C, Chen Z, et al. (2009) The differential expressions of 78-kDa glucose-regulated protein of infiltrating plasma cells in peripheral joints with the histopathological variants of rheumatoid synovitis Arthritis Research & Therapy. 11: R4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang P, Liu B, Kang SW, Seo MS, Rhee SG, et al. (1997) Thioredoxin peroxidase is a novel inhibitor of apoptosis with a mechanism distinct from that of Bcl-2. J Biol Chem 272: 30615–30618. [DOI] [PubMed] [Google Scholar]
- 55. Geiben-Lynn R, Kursar M, Brown NV, Addo MM, Shau H, et al. (2003) HIV-1 antiviral activity of recombinant natural killer cell enhancing factors, NKEFA and NKEF-B, members of the peroxiredoxin family. J Biol Chem 278: 1569–1574. [DOI] [PubMed] [Google Scholar]
- 56. Dhanwani R, Khan M, Bhaskar ASB, Singh R, Patro IK, et al. (2012) Characterization of Chikungunya virus infection in human neuroblastoma SH-SY5Y cells: Role of apoptosis in neuronal cell death Virus Research. 163: 563–572. [DOI] [PubMed] [Google Scholar]
- 57. Abere B, Wikan N, Ubol S, Auewarakul P, Paemanee A, et al. (2012) Proteomic Analysis of Chikungunya Virus Infected Microgial Cells. PLoS ONE 7: e34800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wu X, Zhou Y, Zhang K, Liu Q, Guo D (2008) Isoform-specific interaction of pyruvate kinase with hepatitis C virus NS5B. FEBS Lett 582: 2155–60. [DOI] [PubMed] [Google Scholar]
- 59. Oremek GM, Müller R, Sapoutzis N, Wigand R (2003) Pyruvate kinase type tumor M2 plasma levels in patients afflicted with rheumatic diseases. Anticancer Res. 23: 1131–4. [PubMed] [Google Scholar]
- 60. Oliviero F, Sfriso P, Baldo G, Dayer JM, Giunco S, et al. (2009) Apolipoprotein A-I and cholesterol in synovial fluid of patients with rheumatoid arthritis, psoriatic arthritis and osteoarthritis. Clin Exp Rheumatol 27: 79–83. [PubMed] [Google Scholar]
- 61. Baatartsogt O, Choi K, Mandal PK, Lim HK, Oh JD, et al. (2009) Candidate molecules identified by proteomic study of synovial cells in experimental post traumatic arthritis of knee in swine. Biotech 8: 13–23. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Average walking stride length. This parameter was measured manually by allowing the control (mock-infected) and CHIKV-infected mice to walk on a sheet of paper with their foot-steps being marked. The total distance covered was then divided by the number of steps taken. The data was expressed as mean±SE of five animals per group and the experiment was repeated twice (p≤0.05).
(TIF)
Muscle sections showing the localisation of CHIKV antigen by immunofluorescence at different days of infection.
(TIF)
The MS and MS/MS spectra of the differentially affected proteins.
(PDF)
Walking behavior of CHIKV infected mice showing lack of coordination and hind limb paralysis.
(WMV)
Surface righting reflex of control (mock-infected) mice.
(MPG)
Surface righting reflex of CHIKV-infected mice. The infected mice demonstrated difficulty in turning to the upright position confirming weakness of the hind limbs.
(MPG)