Abstract
Plant type II arabinogalactan (AG) polysaccharides are attached to arabinogalactan proteins (AGPs) at hydroxyproline residues, and they are very diverse and heterogeneous structures. The AG consists of a β-(1→3)-linked galactan backbone with β-(1→6)-galactan side chains that are modified mainly with arabinose, but they may also contain glucuronic acid, rhamnose or other sugars. Here, we studied the positions of fucose substitutions in AGPs, and we investigated the functions of this fucosylation. Monosaccharide analysis of Arabidopsis leaf AGP extracts revealed a significant reduction in L-Fucose content in the fut4 mutant, but not in the fut6 mutant. In addition, Fucose was reduced in the fut4 mutant in root AGP extracts and was absent in the fut4/fut6 mutant. Curiously, in all cases reduction of fucose was accompanied with a reduction in xylose levels. The fucosylated AGP structures in leaves and roots in wild type and fut mutant plants were characterised by sequential digestion with AG specific enzymes, analysis by Polysaccharide Analysis using Carbohydrate gel Electrophoresis, and Matrix Assisted Laser Desorption/Ionisation (MALDI)-Time of Flight Mass spectrometry (MS). We found that FUT4 is solely responsible for the fucosylation of AGPs in leaves. The Arabidopsis thaliana FUT4 and FUT6 genes have been previously proposed to be non-redundant AG-specific fucosyltransferases. Unexpectedly, FUT4 and FUT6 enzymes both fucosylate the same AGP structures in roots, suggesting partial redundancy to each other. Detailed structural characterisation of root AGPs with high energy MALDI-Collision Induced Dissociation MS and NMR revealed an abundant unique AG oligosaccharide structure consisting of terminal xylose attached to fucose. The loss of this structure in fut4/fut6 mutants explains the reduction of both fucose and xylose in AGP extracts. Under salt-stress growth conditions the fut4/fut6 mutant lacking AGP fucosylation exhibited a shorter root phenotype than wild type plants, implicating fucosylation of AGPs in maintaining proper cell expansion under these conditions.
Introduction
Proteins glycosylated with arabinogalactan (AGPs) have been implicated in growth and a wide variety of developmental processes [1], [2], [3]. The arabinogalactan (AG) may influence cell surface protein trafficking and stability, perform functions such as chaperoning polysaccharides or Ca+ chelation, and it may generate signalling molecules [4], [5].
The AGs can be large branched polysaccharides and may consist of as many as 100 to 150 sugar residues [6], [7]. The AGs show vast heterogeneity not only in size but also in composition and structure. The AG consists of a β-(1→3)-galactan backbone with β-(1→6)-linked galactan side chains modified by α-(1→3)-L-arabinosyl (L-Ara) residues (known as type II AG; [8], [9]. Tan et al., have reported an alternative modular structure for AGPs heterologously expressed in tobacco (Nicotiana tabacum; [10], consisting of repeating blocks of 15 sugar residues of decorated β-(1→3)-trigalactosyl subunits connected with β-(1→6)-linkages. In both models, the β-(1→6)-galactan side chains can be further modified with other less abundant sugars such as glucuronic acid (GlcA) and its 4-O-methylated counterpart (4-O-Me-GlcA), L-rhamnose (Rha) and L-fucose (Fuc) [7], [9], [11], [12], [13]. Recently GlcA or 4-O-Me-GlcA modifications of the β-(1→3)-galactan backbone were reported [14]. The AGP APAP-1 from Arabidopsis is reported to have short arabinoxylan and pectin side chains [15]. However, any function of the different sugar moieties on the AG are unclear.
Radish leaf AGPs are modified with Fuc residues [16] but this sugar is missing from radish roots [9]. Fuc is reported in AGPs of Arabidopsis leaves (Arabidopsis thaliana; [14], radish [16], [17], [18], thyme (Thymus vulgaris; [19] and celery (Apium graveolens; [20]. In Arabidopsis and radish leaf AGPs, Fuc has been reported in the structure α-L-Fuc-(1→2)-α-D-Ara-(1→) [14], [17]. The biological roles of fucosylation however, remain unclear although there are some lines of evidence suggesting that extracellular Fuc may be involved in plant cell signalling and development: Fuc has been detected in root exudates from maize (Zea mays; [21]) and wheat (Triticum aestivum L.; [22]). Experiments using Fuc specific lectins or fucosidases have suggested that this sugar may play a role in root-microbe interactions [23]. In addition, the Arabidopsis mur1 (murus 1) mutant which is affected in the biosynthesis of Fuc has been reported to lack Fuc from the cell wall polymers rhamnogalacturonan-II (RG-II) and xyloglucan (XG) [24], as well as from leaf AGPs [14]. Mur1 plants were slightly dwarfed and the mechanical strength of the cell wall was reduced and they had shorter roots compared to wild-type [25]. Root AGP preparations were found to have 40% less Fuc than wild-type plants [25]. Addition of the Fuc-binding eel lectin or the AG β-(1→3)-galactan backbone reactive β-Glc-Yariv reagent [26] to the growth medium almost completely phenocopied mur1, suggesting that fucosylated root AGPs may play a role in root elongation. However, Fuc is present in several plant glycans, including xyloglucan, RG-I, RG-II and some N-glycans, complicating the interpretation of the mur1 phenotype.
Two members of the plant GT37 family (FUT4 and FUT6) have been biochemically characterised to transfer Fuc to AG in vitro [27]. It was proposed that they have different fucosyl transfer activities, but it remains unclear what fucosylated structures were generated by these enzymes. The phenotype of the corresponding fut mutants has not been reported. One of the major bottlenecks in identifying the roles of AGP biosynthetic glycosyltransferases is the difficulty of characterising changes in AG structure in putative mutants. Recently, we reported a powerful approach for the structural characterisation of the carbohydrate components of AGPs which combines the use of AG-specific enzymes, high-energy matrix assisted laser desorption/ionisation (MALDI)-collision induced dissociation (CID) mass spectrometry (MS) and polysaccharide analysis using carbohydrate gel electrophoresis (PACE; [7]). This technique allowed the characterisation of the structure of Arabidopsis leaf AG polysaccharides and the Fuc modifications on those molecules [14].
In this work, we characterised the structure of AGPs to determine the function of AtFUT4 and AtFUT6 in the synthesis of Arabidopsis leaf and root fucosylated AGPs, and show that complete loss of Fuc in root AGPs leads to increased salt sensitivity in root growth.
Results
Identification of T-DNA Insertion Mutants
To investigate the functional roles of AtFUT4 and AtFUT6, T-DNA insertional lines were identified (Figure S1 a and b). Arabidopsis plant lines fut4 (SAIL_284_B05) and fut6 (SALK_078357) homozygous for the insertion were identified by PCR analysis (Figure S1 a and b). The morphology of the fut4 and fut6 plants appeared similar to the wild-type plants (this is discussed later) and hence a fut4/fut6 double mutant was generated (Figure 1). FUT4 transcripts were not detected in the homozygous mutant by RT-PCR. Similarly, absence of FUT6 transcript in the corresponding mutant was confirmed, and no FUT4 and FUT6 transcripts were detected in the double mutant (Figure 1).
Figure 1. RNA transcript levels of FUT4 and FUT6 genes by RT-PCR.
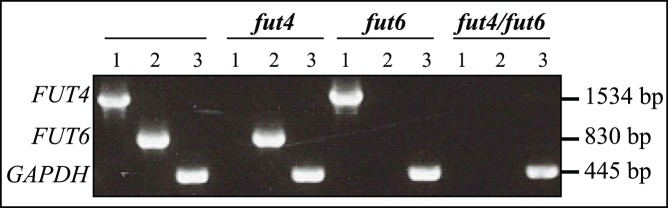
RNA was isolated from roots 14 days after germination from homozygous fut4, fut6, fut4/fut6 and wild-type seedlings and RT-PCR was performed using the primers listed in Table S1. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a control. In lane 1 the fut4 left (LP) and right (RP) primers were used. In lanes 2 and 3, the fut6 and gapdh primers (LP and RP) respectively, were used.
AGPs from Arabidopsis fut4 Leaves Lack Fuc Modifications
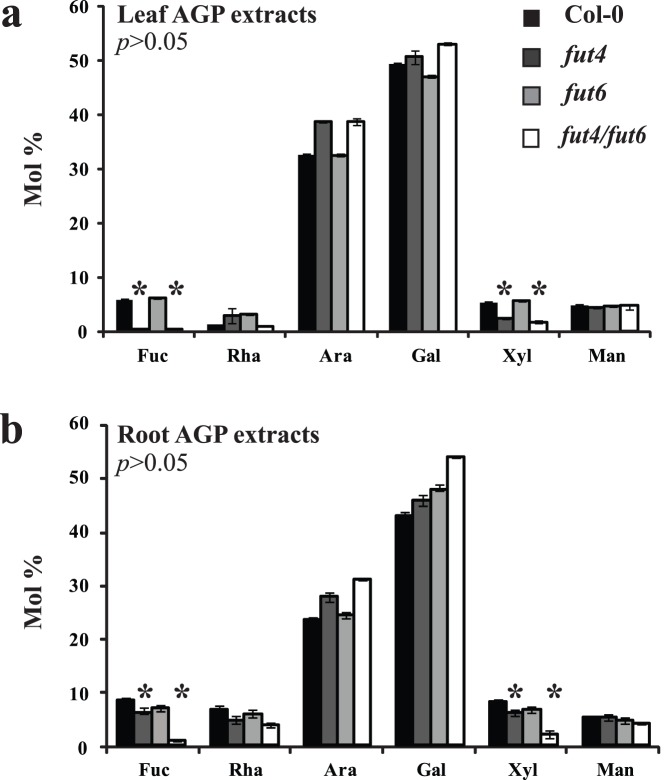
AtFUT4 is expressed in all tissues but has higher levels of transcript accumulation in leaves, although it is also expressed in roots [28]. AtFUT6 on the other hand, is expressed in roots but not leaves [28]. To determine any changes in AGP sugar composition in the fut4, fut6 and fut4/fut6 mutants, AGP-enriched extracts were prepared from both leaves and roots and their monosaccharide content was analysed by High pH Anion Exchange Chromatography (HPAEC) with Pulse Amperometric Detection (PAD; Figure 2). High Ara and Gal amounts were detected in both leaf and root samples, consistent with the high AGP content of the sample. Significant differences in neutral sugar composition were found between the mutants and the wild-type samples from both leaves (Figure 2a) and roots (Figure 2b). Fuc was absent in AGP extracts from fut4 mutant leaves and was also reduced in fut4 root AGP extracts. The double fut4/fut6 mutant contained no detectable Fuc in leaf and very low levels of Fuc in root extracts. These results are consistent with a role for FUT4 in AGP fucosylation in leaves and a role for both FUT4 and FUT6 enzymes in roots. Interestingly, the amount of Xyl was reduced in AGP extracts from fut4 and fut4/fut6 leaves and from fut4/fut6 roots. To confirm that the compositional changes in leaf AGP extracts is due to the fut4 mutation, the FUT4 gene was reintroduced into the corresponding fut4 mutant. The decreases in Fuc and Xyl in AGP extracts were reversed by complementation of fut4 with the wild type FUT4 gene (Table S2).
Figure 2. HPAEC-PAD monosaccharide analysis of neutral sugars of AGP extracts from Arabidopsis wild-type, fut4, fut6 and fut4/fut6 double mutant (a) leaves and (b) roots.
Bars represent mean ± SD (n = 3). A significant difference was identified between wild type and fut4 single- and fut4/fut6 double-mutant plants for Fuc and Xyl sugars both in leaves and roots as indicated by p>0.05 in students t-test.
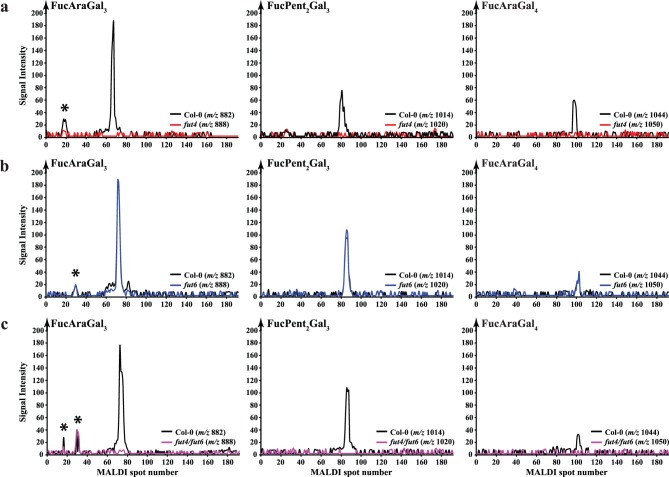
Recently we identified the fucosylated oligosaccharides released by AG specific hydrolases from Arabidopsis leaf AGP as α-L-Fuc-(1→2)-α-L-Araf-(1→3)-β-Galp-(1→6)-β-Galp-(1→6)-Galp (FucAraGal3) and α-L-Fuc-(1→2)-α-L-Araf-(1→3)-β-Galp-(1→6)-β-Galp-(1→6)-β-Galp-(1→6)-Galp (FucAraGal4) [14]. An oligosaccharide composed of two pentoses, Fuc and three Galp residues was also released, and we proposed that the structure of that oligosaccharide might be α-L-Fuc-(1→2)-α-L-Araf-(1→?)-α-L-Araf-(1→3)-β-Galp-(1→6)-β-Galp-(1→6)-Galp (FucPent2Gal3) [14]. We investigated whether these fucosylated leaf AG oligosaccharides were released by digestion of AGP extracts from the fut4, fut6 and fut4/fut6 double mutants with the Oligosaccharide relative Quantitation using stable Isotope Tagging (OliQuIT) method [29]. The OliQuIT method takes advantage of the fact that the chromatographic elution positions of an oligosaccharide labelled with 13C6- or 12C6-aniline are identical. This property can therefore be used for comparison of the abundance of oligosaccharides between samples. Hence, purified AGP extracts from wild type and mutant leaves were hydrolysed with AG-specific α-arabinofuranosidase followed by endo-β-(1→6)-galactanase and the purified oligosaccharide products were reductively aminated with 13C6- or 12C6-aniline. The isotope tagged oligosaccharides were then separated by HILIC coupled off-line to MALDI-ToF-MS and the abundances of the differently labelled oligosaccharides were compared. The extracted ion chromatograms (EICs; Figure 3 a, b and c) indicate that all three of the Fuc-modified oligosaccharides are absent from fut4 and the fut4/fut6 double mutant but were present in fut6 mutant leaves. Together with the monosaccharide composition data, this MS data suggests that FUT4 is solely responsible for the fucosylation of all three oligosaccharides in Arabidopsis leaf AG polysaccharides.
Figure 3. Capillary HILIC-MALDI-ToF-MS using stable isotope tagging of fucosylated oligosaccharides from Arabidopsis leaf AGP extracts of Columbia wild-type (Col-0; black line), fut4 (red line), fut6 (blue line) and fut4/fut6 (magenta line) plants.
Purified leaf AGP extracts were subjected to sequential digestion with α-arabinofuranosidase, exo-β-(1→3)-galactanase, β-glucuronidase and endo-β-(1→6)-galactanase. The oligosaccharide products were purified on a C18 cartridge (elution with 5% acetic acid) and cation exchange resin (Dowex; elution with 5% acetic acid) and were reductively aminated with [12C6]-aniline (wild-type oligosaccharides; black line) and [13C6]-aniline (fut4, fut6 and fut4/fut6; red, blue and magenta lines respectively). The labelled oligosaccharides were purified from the reductive amination reagents on a normal phase cartridge (Glyko clean S) and the purified glycans were separated by HILIC and analysed by MALDI-ToF-MS. Although all three fucosylated oligosaccharides were detected for wild-type and fut6 samples, no corresponding glycans were detected from AGP leaf extracts from fut4 and fut4/fut6 plants. Background peaks are marked with an asterisk (*). On panel b the Col-0 trace (black line) partly coincides with, and therefore obscures, the fut6 (blue line) trace.
Arabidopsis AGP Fucosylation differs between Leaves and Roots
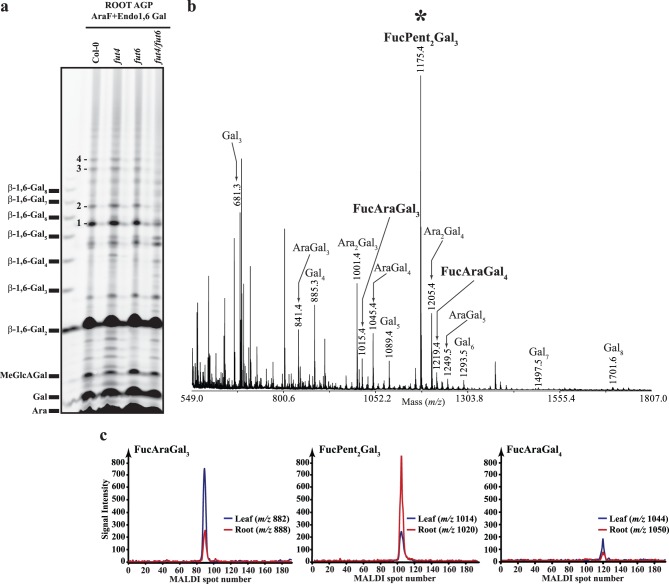
The fucosylated oligosaccharides released by digestion of root AGPs have not previously been determined. Therefore, we structurally characterised the AG oligosaccharides by PACE and MALDI-ToF-MS. Root AGP extracts were hydrolysed with α-arabinofuranosidase followed by endo-β-(1→6)-galactanase to remove terminal Ara modifications and to cleave the β-(1→6)-galactan side chains, releasing the fucosylated residues. The resulting oligosaccharides were analysed by PACE (Figure 4a). From Figure 4a it is clear that root AGP extracts are susceptible to the enzymatic treatment releasing a ladder of oligosaccharides with degree of polymerisation (DP) extending from 1 to 12. Some of the oligosaccharides that do not comigrate with β-(1→6)-galactooligosaccharide standards (marked with numbers 1 to 4) are substantially reduced or missing from the fut4/fut6 double mutant suggesting that they may be fucosylated. To characterise further the released oligosaccharides, enzyme digested samples were analysed by MALDI-ToF-MS (Figure 4b). The α-arabinofuranosidase and endo-β-(1→6)-galactanase sequential digestion released oligosaccharides consisting predominantly of hexose (Hex), pentose (Pent) and deoxyhexose (Deoxyhex). A series of oligosaccharides corresponding to Hex3–8 and Pent1–2Hex3–5 were seen, and three Deoxyhex-containing oligosaccharides corresponding to DeoxyhexPent1–2Hex3–4. The hexosyl and the pentosyl residues in the Hex3–8 and Pent1–2Hex3–5 oligosaccharides, respectively are Gal and Ara since the oligosaccharides arise from AG-specific enzyme digestion.
Figure 4. Characterisation of oligosaccharides released by the sequential digestion with the AG-specific enzymes α-arabinofuranosidase and endo-β-(1→6)-galactanase from Arabidopsis root AGP extracts.
(a) Polysaccharide Analysis using Carbohydrate gel Electrophoresis (PACE). Oligosaccharide products from wild-type (Col-0), fut4, fut6 and fut4/fut6 were reductively aminated with 2-aminonaphthaline trisulfonic acid and separated by electrophoresis on acrylamide gels. An oligosaccharide ladder prepared from β-(1→6)-galactan was used as migration marker. The numbers indicate putatively fucosylated oligosaccharides with altered abundance in the wild-type and fut mutant samples. (b) MALDI-ToF-MS spectrum of per-methylated oligosaccharide products from wild-type plants. Peaks marked with an asterisk (*) were selected for high-energy MALDI-CID structural analysis. (c) Extracted ion chromatograms (EICs) for the fucosylated oligosaccharides originating from Arabidopsis leaf (blue lines) and root (red lines) AGP extracts hydrolysed sequentially by α-arabinofuranosidase, exo-β-(1→3)-galactanase, β-glucuronidase and endo-β-(1→6)-galactanase. Arabidopsis root AGP extracts contain the same three fucosylated oligosaccharides as leaves albeit in different relative abundances.
To determine whether the Deoxyhex containing oligosaccharides were the same fucosylated structures found in leaf AGP, we used the comparative approach using the [13C6]aniline/[12C6]aniline labelling (OliQuIT). The HILIC-MALDI-ToF-MS EICs for FucAraGal3, FucAraGal4 and FucPent2Gal3 oligosaccharides from leaves (m/z 882, 1044 and 1014, respectively labelled with the light isotope [12C6]) and the root (m/z 888, 1050 and 1020, respectively labelled with the heavy isotope [13C6]) are shown in Figure 4c. For each of the three oligosaccharides the same main structural isomer was detected in both leaf and root tissues. However, the relative abundance of these oligosaccharides varies between leaves and roots. In particular, the FucPent2Gal3 oligosaccharide was much more abundant in roots than in leaves.
Arabidopsis fut4/fut6 AGPs from Roots Lack Fuc Modifications
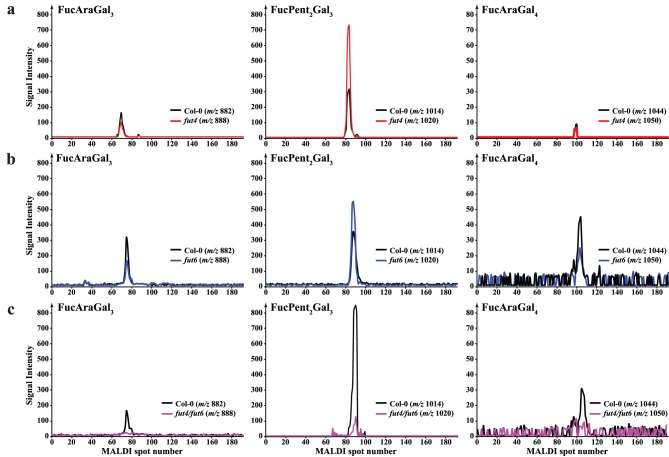
Since both FUT4 and FUT6 are expressed in Arabidopsis roots and the monosaccharide analysis from root AGP extracts indicated a reduction in Fuc levels in both fut4 and fut6 mutants and near absence in the fut4/fut6 double mutant, we hypothesised that both AtFUT4 and AtFUT6 enzymes may be responsible for the fucosylation of AGPs in this tissue. Root AGPs for fut4, fut6 and fut4/fut6 double mutants were extracted and subjected as before to α-arabinofuranosidase and endo-β-(1→6)-galactanase sequential digestion to release the fucosylated oligosaccharides resistant to this treatment. The enzymatic hydrolysis products were compared with those prepared from the wild-type plants using the OliQuIT approach. Figure 5 shows the HILIC-MALDI-ToF-MS EICs for FucAraGal3, FucAraGal4 and FucPent2Gal3 oligosaccharides from wild-type (m/z 882, 1044 and 1014 respectively labelled with the light isotope) and the fut4, fut6 and fut4/fut6 mutants (m/z 888, 1050 and 1020 respectively labelled with the heavy isotope). Comparison of the EICs for all three oligosaccharides from wild-type and fut4 mutants indicates a reduction in the relative abundance of FucAraGal3 and FucAraGal4 and an increase in the relative abundance of FucPent2Gal3 oligosaccharide relative to the wild-type. Similar changes were seen in fut6 mutant samples. FucAraGal3 and FucAraGal4 oligosaccharides were absent from fut4/fut6 samples. A small amount of FucPent2Gal3 oligosaccharide was detectable in root AGP extracts from fut4/fut6 plants but its abundance relative to wild-type is significantly reduced. This data suggests that both FUT4 and FUT6 enzymes contribute to the fucosylation of Arabidopsis root AG polysaccharides, and that they are both able to generate all three fucosylated structures.
Figure 5. EIC for the L-Fuc modified oligosaccharides from Arabidopsis root AGP extracts of wild-type (black line), fut4 (red line), fut6 (blue line) and fut4/fut6 (magenta line) plants.
Although all three fucosylated oligosaccharides were detected for wild-type, fut4 and fut6 samples, no FucAraGal3 and FucAraGal4 were detected from AGP root extracts from fut4/fut6 plants and very small amount of FucPent2Gal3 oligosaccharide was detectable in root AGP extracts from fut4/fut6 plants but its abundance relative to wild-type is significantly reduced.
One of the Fucosylated Oligosaccharides from Arabidopsis AGPs Contains a Xyl Residue
The reduction in Xyl from AGP extracts from the fut4 and fut4/fut6 mutants in leaves and fut4, fut6 and fut4/fut6 in roots, closely followed the changes in abundance of the FucPent2Gal3 oligosaccharide (Figure 2, 4). This suggests that a Xyl residue is present on the AGP in these tissues, and the addition of the Xyl is dependent on the presence of Fuc. For example, fucosylation could provide the context to either add a terminal Xyl to a side chain containing Fuc or to add a Xyl residue to the galactan backbone in close proximity to Fuc. To test these hypotheses we investigated whether Xyl was detectable in the oligosaccharide with unknown structure FucPent2Gal3. Thus root AGP extracts from wild-type plants were hydrolysed with arabinofuranosidase followed by endo-β-(1→6)-galactanase, the oligosaccharide products were purified and labelled with 2-AA and separated with HILIC. The fractions containing the FucPent2Gal3 oligosaccharide were collected from the HILIC column and hydrolysed by TFA. Even though very low amounts of the oligosaccharide were available, the sugar content was identified by HPAEC-PAD monosaccharide analysis. The data in Table 1 is consistent with the FucPent2Gal3 oligosaccharide composition being FucXylAraGal3. The mannose is likely to arise from contamination of the oligosaccharide, as there is no mannose in the oligosaccharide structure detected by NMR (see below).
Table 1. HILIC purified oligosaccharide: HPAEC-PAD neutral monosaccharide analysis (% Mol).
| Sugar | Root oligosaccharide |
| Fuc | 8.02±2.29 |
| Ara | 24.79±0.02 |
| Gal | 42.29±0.57 |
| Xyl | 18.05±0.59 |
| Man | 6.45±2.33 |
Values represent mean ± SD (n = 2).
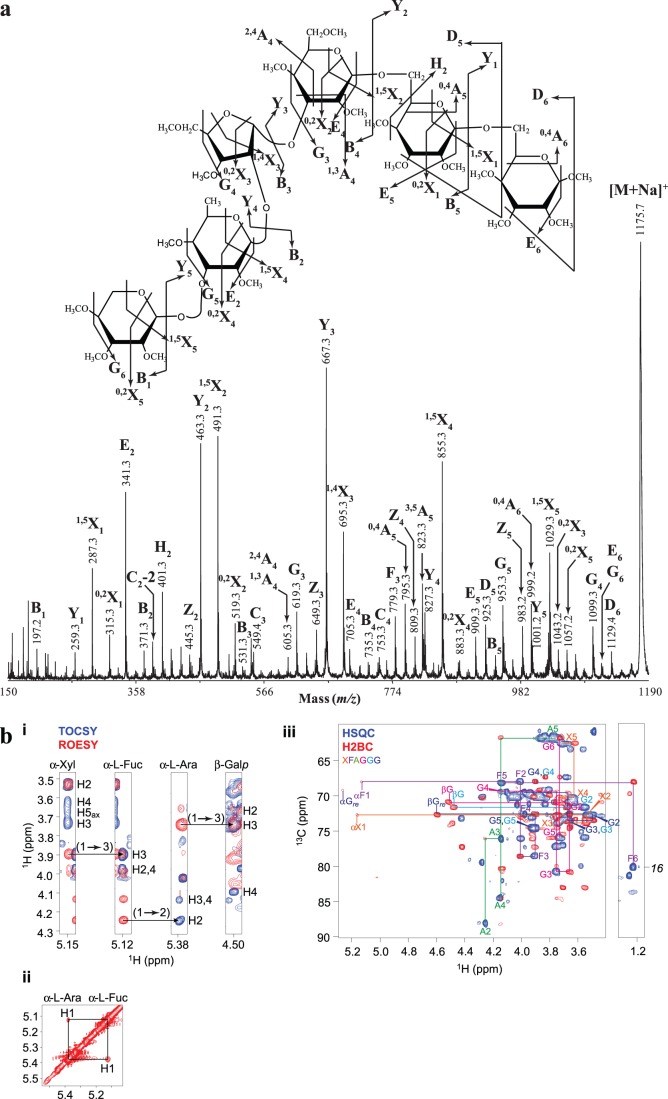
To identify the structure of the FucXylAraGal3 hexasaccharide, we next carried out high energy MALDI-CID. The tandem mass spectrometry (MS/MS) spectrum of the per-methylated FucXylAraGal3 (m/z 1175.7) oligosaccharide is shown in Figure 6a. The series of Y and 1,5X ions [30] reveals the sequence of the sugars on the oligosaccharide chain. Starting from the reducing end the order of sugars was as follows: Gal, Gal, Gal, Ara, Fuc and Xyl. The 0,4A5 and 0,4A6 ions, which are unique to the non-reducing end, indicate that the three Gal residues are connected via a (1→6)-linkage. The presence of the G3 and E4 [31] ‘elimination ions’ together with the 1,3A4 and 2,4A4 non-reducing end ions, indicate a linkage of the Ara at the C-3 of the third Gal residue from the reducing end. Similarly, the G4 ‘elimination ion’ and the 0,2X3 reducing-end ion are indicative of a branching at the C-2 of the Ara. Finally, the presence of a G4 ‘elimination ion’ indicated that an Ara in furanose form is present on the hexasaccharide. To study the structure further by NMR, AGPs were extracted from hydroponically grown roots and were subjected to sequential digestion with the AG-specific enzymes α-arabinofuranosidase and endo-β-(1→6)-galactanase. The hydrolysis products were separated on a Size Exclusion Chromatography (SEC) column, and the fractions containing the FucPent2Gal3 hexasaccharide as judged by PACE, were combined and analysed by NMR (Figure 6b). Chemical-shift assignments were obtained using 2D 1H-1H TOCSY and ROESY alongside 2D 13C HSQC, H2BC, HSQC-TOCSY and HSQC-ROESY experiments. The non-reducing-end Xyl residue was readily identified and the chemical shifts of the H-1 and C-1 were consistent with an α configuration. The (1→3) linkage to Fuc was apparent from the intense NOE from Xyl H-1 to Fuc H-3 taken together with the downfield shift of Fuc C-3 characteristic of a glycosidic link. The Fuc-(1→2)-Ara and Ara-(1→3)-Gal linkages were similarly confirmed by a combination of NOEs and downfield 13C chemical shifts of Ara C-2 and Gal C-3. Although the chemical-shift positions of the remaining Gal C-5/H-5 and C-6/H-6 were severely overlapped, 2D 13C HMBC connections were observed across the glycosidic linkages from the H-1 to the C-6, confirming the (1→6) linkages. Otherwise, the assignment was complete and is shown in Table 2. No mannose was present in this oligosaccharide, although mannose was observed as part of a separate, contaminating oligosaccharide co-purifying by SEC, along with free 4-OMe-β-D-GlcpA.
Figure 6.
(a) Structural characterisation of the per-methylated FucPent2Gal3 oligosaccharide by high-energy MALDI-CID. Arabidopsis root AGP extracts were sequentially hydrolysed with α-arabinofuranosidase and endo-β-(1→6)-galactanase and the hydrolysis products were purified, per-methylated and analysed by MALDI-ToF-MS as shown in Figure 4b. The oligosaccharide with m/z 1175.3 as selected for MALDI-CID analysis and was identified as Xyl-(1→3)-α-L-Fuc-(1→2)-α-L-Ara-(1→3)-β-Galp-(1→6)-β-Galp-(1→6)-Galp. Glycosidic cross-ring fragments were identified according to the Domon and Costello nomenclature (1988). (b) HPAEC-PAD monosaccharide analysis of neutral sugars for the HILIC purified root oligosaccharide. Arabidopsis root AGP extracts were sequentially hydrolysed with α-arabinofuranosidase and endo-β-(1→6)-galactanase and the hydrolysis products were reductively aminated with 2-AA and separated with HILIC. The FucPent2Gal3 oligosaccharide was collected from the HILIC column and hydrolysed by TFA. The sugar content was identified by HPAEC-PAD monosaccharide analysis. (b) NMR analysis of the FucPent2Gal3 hexasaccharide. (i) H-1 strip plots from 2D 1H-1H TOCSY (blue) and ROESY (red) spectra showing the NOE connectivity arising from the Xyl-(1→3)-α-L-Fuc-(1→2)-α-L-Ara-(1→3)-β-Galp glycosidic linkages. (ii) 2D 1H-1H ROESY spectrum of the α-H1 region showing the Fuc H-1/Ara H-1 NOE arising from their close proximity due to the α-L-Fuc-(1→2)-α-L-Ara glycosidic linkage. (iii) 2D 13C HSQC and H2BC spectra showing the assignment of 1H,13C HSQC peaks using H2BC, which exclusively reveals sequential connections over two covalent bonds (Nyberg et al., 2005). The H-1 chemical shift of the non-reducing-end Xyl is consistent with an α configuration; the 13C resonance positions of Fuc C-3, Ara C-2 and Gal C-3 are downfield shifted consistent with their involvement in glycosidic linkages. (NB: Fuc C-6 was aliased in the spectra; the actual resonance frequency c. 16 ppm is shown for clarity.).
Table 2. 1H and 13C NMR assignments of α-Xyl-(1→3)-α-L-Fuc-(1→2)-α-L-Ara-(1→3)-β-Galp-(1→6)-β-Galp-(1→6)-Galp. at 25°C in D2O.
| Residue | Assignment | ||||||
| 1 | 2 | 3 | 4 | 5 | 6 | ||
| α-Xylnr | 1H | 5.150 | 3.526 | 3.724 | 3.617 | 3.666, 3.698 | |
| 13C | 101.47 | 72.61 | 73.68 | 70.15 | 62.49 | ||
| α-L-Fuc | 1H | 5.119 | 3.995 | 3.894 | 3.964 | 4.128 | 1.211 |
| 13C | 98.95 | 67.88 | 78.46 | 72.54 | 68.05 | 16.08 | |
| α-L-Ara | 1H | 5.374 | 4.242 | 4.132 | 4.132 | 3.722, 3.739 | |
| 13C | 108.54 | 88.05 | 76.03 | 84.42 | 61.71 | ||
| β-Galp | 1H | 4.502 | 3.651 | 3.740 | 4.090 | 3.727 | 3.765, 3.837 |
| 13C | 103.90 | 70.81 | 80.68 | 69.36 | 75.81 | 61.68 | |
| β-Galp | 1H | 4.467 | 3.540 | 3.663 | 3.968 | ∼3.901 | ∼3.915, 4.053 |
| 13C | 104.01 | 71.52 | 73.37 | 69.40 | ∼74.50 | 69.86 | |
| β-Galp re | 1H | 4.586 | 3.485 | 3.650 | 3.952 | ∼3.901 | ∼3.901, 4.051 |
| 13C | 97.21 | 72.59 | 73.39 | 69.51 | ∼74.50 | 70.16 | |
The compositional data taken together with the MALDI-CID and NMR data allow the identification of the FucPent2Gal3 hexasaccharide as α-Xyl-(1→3)-α-L-Fuc-(1→2)-α-L-Ara-(1→3)-β-Galp-(1→6)-β-Galp-(1→6)-Galp.
The Fucose-deficient fut4/fut6 Double Mutant is Salt-sensitive
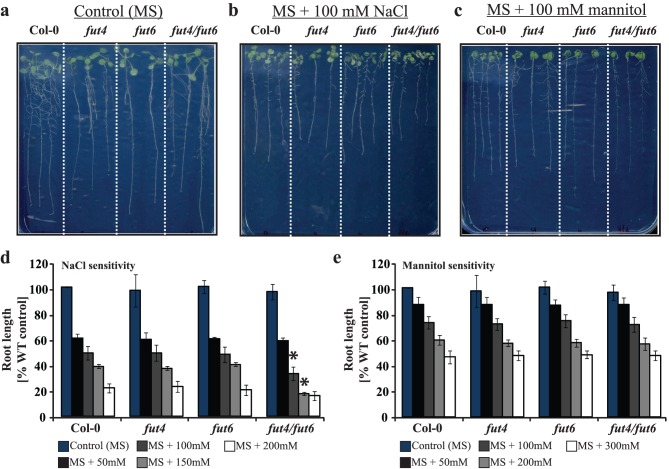
The importance of AGP fucosylation in controlling root cell elongation has been proposed from studies of the Arabidopsis mutant mur1 [32] where alterations in root elongation and morphology were observed and also with the salt overly sensitive 5 (sos5/fla4) mutant which exhibited root tip swelling and arrest in root growth under salinity stress [33]. We therefore investigated whether fut mutants may also exhibit a root phenotype. The roots of both fut4 and fut6 single mutants grew comparably to the wild-type, and they did not exhibit any response to salt growth (Figure 7 a, b and d). The roots of the fut4/fut6 double mutant also grew to the same length as the wild-type under no salt stress conditions. However, the root length of the mutant was significantly reduced compared to wild-type under salt stress conditions (100 mM and 150 mM NaCl) (Figure 7b and d). Under salt stress conditions a difference in the lateral root formation between the fut mutants and wild-type may also be apparent (Figure 7b), but any difference was not quantified. To investigate whether the short root phenotype was due to salt sensitivity or to the osmotic shock caused by the high salt concentration, seedlings from the wild-type, fut4, fut6 and fut4/fut6 double mutant were grown on solid MS medium supplemented with mannitol. Figure 7c and e shows that fut4/fut6 root growth was not hypersensitive to osmotic stress caused by mannitol. Together, these findings suggest that absence of fucosylation on root AGPs causes salt sensitivity of root growth in Arabidopsis.
Figure 7. Phenotypic analysis of Arabidopsis fut mutants compared to wild-type plants grown on solid medium.
Wild-type, fut4, fut6 and fut4/fut6 seedlings were grown on MS medium for 7 days and then transferred to cellophane layered MS-agar medium plates without any supplements (a), with 100 mM NaCl (b) and with 100 mM mannitol (c) and grown for additional 7 days. (d) fut4/fut6 double mutants are salt sensitive. The root growth response of fut4, fut6 and fut4/fut6 mutant plants to various NaCl concentrations was compared with those of wild-type. Data are presented as percentage relative to the growth of wild-type on MS medium. Bars represent mean ± SD (n = 3). A significant difference was identified between wild type and fut4/fut6 mutant plants as indicated by p>0.05 in students t-test. (e) fut4/fut6 double mutants are not sensitive to osmotic stress. The root growth response of fut4, fut6 and fut4/fut6 mutant plants to various mannitol concentrations was compared with those of wild-type. Data are presented as percentage relative to the growth of wild-type on MS medium. Bars represent mean ± SD (n = 3).
Discussion
We have shown that three fucosylated oligosaccharides are released by enzymatic digestion of AG from root AGPs. These are the same structures as those previously shown in leaf AGP [14]. Our data indicate that FUT4 is required for the fucosylation of all these structures in leaf AGPs and that both FUT4 and FUT6 contribute to the fucosylation of the root AGPs.
All three fucosylated oligosaccharides (FucAraGal3, FucAraGal4 and FucPent2Gal3) previously identified in leaves [14] were found in roots, but the relative abundance of these oligosaccharides in leaves relative to roots was different. To our knowledge, this is the first time that the AGP fucosylated structures for Arabidopsis roots are reported. We investigated which fucosylated structures are affected in fut4, fut6 and fut4/fut6 mutants. Leaf and root AGPs for fut4, fut6 and fut4/fut6 double mutants were extracted and subjected as before to α-arabinofuranosidase and endo-β-(1→6)-galactanase sequential digestion to release the fucosylated oligosaccharides resistant to this treatment. The enzymatic hydrolysis products were compared with those prepared from the wild-type plants by PACE and using the OliQuIT approach. Our analysis showed that FucAraGal3 and FucAraGal4 oligosaccharides were clearly absent from leaf fut4, and root fut4/fut6 samples. A small amount of FucPent2Gal3 oligosaccharide was detectable in root AGP extracts from fut4/fut6 plants but its abundance relative to wild-type was significantly reduced. These data suggest therefore, that the fucosylation of AGPs in leaves is entirely dependent on FUT4. In contrast, in roots fucosylation of AGPs is the result of the action of both FUT4 and FUT6 enzymes, which apparently fucosylate the same structures. The view that AtFUT4 and AtFUT6 are functionally non-redundant enzymes that transfer Fuc residues onto different acceptor structures on AGP molecules [27] is apparently not consistent with our findings. A possible explanation for this inconsistency may be the great heterogeneity of the AGP molecules. The glycan structure of AGPs varies greatly between different cell-types, developmental stage or species [34]. Therefore it is conceivable that the FUT6 enzyme is fucosylating a specific group of root AGPs that is not fucosylated by FUT4. However, the view that FUT4 and FUT6 are the only enzymes fucosylating AGPs [27] is consistent with our findings. Similarly to FUT4, the FUT2, FUT5, FUT7 and FUT10 proteins are expressed in leaves [28], however when FUT4 was disrupted by the T-DNA insertion in fut4 mutant, no compensation by the other enzymes was observed suggesting that they may not be involved in the biosynthesis of the AGP molecules.
The HPAEC-PAD monosaccharide compositional analysis of leaf and root AGPs showed not only a reduction in Fuc levels (in fut4 and fut4/fut6 double mutant in leaves and in fut4, fut6 and fut4/fut6 in roots) but also a concomitant reduction in Xyl levels. This coincident reduction in Xyl from AGPs from the fut mutants suggests that fucosylation provides the context to add a terminal Xyl residue to a side chain containing Fuc or to add a Xyl residue in close proximity to Fuc. Since the structure of the FucPent2Gal3 oligosaccharide detected both in leaves and roots, was unknown we investigated the presence of Xyl in this oligosaccharide by HPAEC-PAD. Indeed, the composition of this oligosaccharide was identified as FucXylAraGal3. High energy MALDI-CID combined with NMR analysis for the FucXylAraGal3 hexasaccharide allowed the identification of the above oligosaccharide as α-Xyl-(1→3)-α-L-Fuc-(1→2)-α-L-Araf-(1→3)-β-Galp-(1→6)-β-Galp-(1→6)-Galp. This finding supports the hypothesis that Fuc provides the context to add a terminal Xyl to the side chain containing the Fuc residue. To our knowledge this is the first time that the α-Xyl-(1→3)-α-L-Fuc oligosaccharide is reported for AGPs. α-D-Xylp-(1→3)-α-L-Fucp is found in pectin, RG-II [35] but there the Xyl residue is methylated, and Fuc is linked to a Rha residue via an α-(1→4)-linkage. The presence of Xyl on AGPs has been reported recently for the APAP-1 structure [15]. However, the structure reported is different, and the APAP-1 AGP is described covalently linked to short arabinoxylan.
Even though AGPs account for less than 10% of the wall matrix, they have been implicated in a number of cellular processes [12] such as programmed cell death, somatic embryogenesis [3], cell expansion, root formation and development [36]. Treatment of Arabidopsis seedlings with Yariv, a phenylglycoside that binds to the β-(1→3)-galactan backbone of AGPs [26], causes a disruption of root growth and abnormal morphology [36]. A phenotype similar to the one caused by Yariv binding has been reported for the root epidermal bulger (reb-1) mutant [37]. The REB-1 gene encodes for a UDP D-glucose 4-epimerase that converts D-Glc to D-Gal providing Gal to AGPs [38]. This finding confirms the hypothesis that AGPs are important for the control of cell expansion in elongating roots. Further confirmation of the involvement of AGPs in controlling cell root elongation comes from the Arabidopsis mutant mur1 which is deficient in the biosynthesis of Fuc [32]. The mur1 mutant is lacking fucosylated AGPs in leaves [14] and roots and exhibits an alteration in root morphology and elongation that has been ascribed to AGP fucosylation [25]. However, the fut4/fut6 mutant did not show a root phenotype under normal growth conditions suggesting that the mur1 short root phenotype can not be attributed entirely to the lack of Fuc on root AGPs. In the mur1 mutant, besides AGPs other Fuc-containing glycans were affected such as xyloglucan, RG-I and RG-II and N-linked glycans, therefore the root phenotype may be the effect of under-fucosylation of multiple cell wall components.
Absence of the SOS5/FLA4 protein, a putatively cell surface adhesion protein with AGP-like and fasciclin-like domains in the sos5/fla4 mutant leads to root tip swelling and arrest in root growth [33] under salt stress conditions. This suggested that FUT mutants may also exhibit a root phenotype, and indeed salt-sensitivity assays revealed that the fut4/fut6 double mutant has shorter roots under high-salt concentrations. Experiments on solid media supplemented with mannitol instead of NaCl at equivalent osmotic pressure showed that the short root phenotype is not due to osmotic stress but is caused by the salt. Since the short root phenotype was observed only in seedlings from the double fut4/fut6 mutant and not in either of the single fut mutants, the severe reduction of Fuc on root AGPs rather than absence on a subgroup of AGPs seems to cause the phenotype. It is not possible at this time to assign the salt-sensitivity phenotype to the lack of a specific fucosylated oligosaccharide structure, since the fut4/fut6 mutants were found to lack both the terminal fucose-containing oligosaccharides and the Xylp-(1→3)-α-L-Fucp oligosaccharide. The identification of a xylosyltransferase for AGPs may help to shed a light onto the salt sensitivity phenotype. However, these findings are suggesting that fucosylated AGPs have an important role in maintaining proper cell expansion under salt stress.
As this manuscript was being submitted, a report was published on the fut4 and fut6 mutants [39]. Their AGP sugar composition data is consistent with the data presented here, but that work did not address the structures of the AG oligosaccharides affected by the mutations or show the functional redundancy of FUT4 and FUT6.
Materials and Methods
Plant Material, Preparation of AG Proteins and AG Specific Enzymes
Arabidopsis (Arabidopsis thaliana) seeds were surface sterilised and sown on soil as previously described [14]. Six weeks post stratification healthy green leaves from the rosettes were collected and frozen immediately in liquid nitrogen and then stored in −80°C before further processing. Tissue used to isolate AGPs from roots was collected from six week old plants grown hydroponically [40], frozen in liquid nitrogen and stored in −80°C. Arabidopsis leaf and root AGPs were prepared using a previously established protocol [14]. For complementation analysis the AGPs were prepared as described above and were precipitated with the addition of 2 mg ml−1 (β-D-Glc)3-Yariv reagent made according to [14]. The Yariv-AGP pellet was “salted-out” according to [41], [42] before being dried and analysed for sugar composition. When AGPs were precipitated by Yariv the reagent was not removed from the AGPs and as a result glucose was not included in the analysis. α-L-Arabinofuranosidase (EC 3.2.1.55); exo-β-(1→3)-galactanase (EC 3.2.1.145); β-glucuronidase (EC 3.2.1.31) and endo-β-(1→6)-galactanase (EC 3.2.1.164) were prepared by methods described previously [43], [44], [45], [46].
Identification of T-DNA by PCR and RT-PCR
Seeds of T-DNA insertional lines SAIL_284_B05 (fut4), SALK_078357 (fut6) and the fut4/fut6 double knock out construct were obtained from NASC (Nottingham, UK). Plants homozygous for the insertion were detected by PCR and the corresponding primer sequences and PCR conditions are listed in Table S1. To analyse transcript levels of FUT4 and FUT6, RNA was extracted using the Qiagen PNeasy kit according to the manufactures instructions. The RT-PCR primers are listed in Table S1.
Complementation
A genomic 7.8 Kb Pstl fragment from Bac clone F26H6 containing 2.773 kb upstream the FUT4 coding sequence and 3.574 kb downstream was cloned into the plant transformation vector pCAMBIA 1300 and introduced into Agrobacterium (Agl-O). Constructs were confirmed by PCR and restriction analysis both in E. coli and in Agrobacterium. Homozygous mutant fut4 plants were transformed with either the genomic rescue construct or the empty vector plasmid and transgenic seeds were selected on hygromycin, The genotypes of the T1 plants were confirmed by PCR (rescued plants were positive for the presence of the fut4 T-DNA insertion in the genomic copy whereas the plants containing the empty vector only showed the presence of the T-DNA insertion). T2 seeds from individual lines were screened on hygromycin selective plates to estimate the number of insertions. Resistant seedlings from the empty vector line and seven genomic rescue lines were transplanted to soil and rosette leaves were harvested from plants of each line for AGP isolation and subsequent sugar analysis.
Enzymatic Hydrolysis and PACE Analysis
Arabinogalactan peptide preparations (1 mg) were digested with the AG-specific enzymes as described by Tryfona et al. [14]. The derivatization of carbohydrates was performed according to previously developed protocols [47]. Carbohydrate electrophoresis and PACE gel scanning and quantification was performed as described by Goubet et al. [47]. Control experiments without substrates or enzymes were performed under the same conditions to identify any non-specific compounds in the enzymes, polysaccharides/cell walls or labelling reagents.
AGP Oligosaccharide Sample Desalting and Clean-up
Following the enzyme digestions and prior to per-methylation released peptides and enzymes were removed using reverse-phase Sep-Pak C18 cartridges (Waters) as described previously [7]. Briefly, the AG oligosaccharides were eluted with 3 ml of 5% acetic acid and were lyophilised. Dry samples were dissolved in 0.5 ml of 5% acetic acid and were desalted using 2 ml Dowex beads (50×8, H+ form, 50–100 mesh) as previously described [48]. Purified samples were lyophilised.
Per-methylation of Arabinogalactan Polysaccharides
Per-methylation of glycans was performed according to previously established protocols [49]. Per-methylated samples were resuspended in 100 μl MeOH and were kept at room temperature for MALDI-ToF-MS analysis.
MALDI-ToF/ToF-MS/MS
Per-methylated samples were analysed by MALDI-ToF/ToF-MS/MS (4700 Proteomics Analyser, Applied Biosystems, Foster City, CA, USA) as previously described [50], using 2,5-dihydroxybenzoic acid (2,5-DHB) matrix (10 mg ml−1 dissolved in 50% MeOH). The above tandem mass spectrometer uses a 200 Hz frequency triple Nd-YAG laser operating at 355 nm wavelength. High energy MALDI-CID spectra were acquired with an average 10,000 laser shots/spectrum, using a high collision energy (1 kV). The oligosaccharide ions were allowed to collide in the CID cell with argon at a pressure of 2×10−6 Torr.
Reductive Amination of AG Oligosaccharides and Purification
The AG oligosaccharides were reductively aminated with 2-aminobenzoic acid (2-AA, Sigma) or 12C6- and 13C6-aniline (Sigma) using optimised labelling conditions described previously [29]. The saccharides were then purified from the reductive amination reagents using a Glyko Clean S cartridge (Prozyme, San Leandro, CA) as described previously [48]. When isotopic labelling was performed samples labelled with the two aniline isotopes were mixed equally prior to purification from the reductive amination reagents.
HILIC-MALDI-ToF/ToF-MS/MS
Capillary HILIC was carried out using an LC-Packings Ultimate system (Dionex, CA, USA) equipped with an amide-80 column (300 μm×25 cm; 3 μm particle size; Dionex) as previously described [14]. The column eluent passed through a capillary UV detector (set at 254 nm) to the MALDI sample spotter. For HILIC-MALDI-ToF/ToF tandem mass spectrometry a Probot sample fraction system (Dionex) was employed for automated spotting of the HPLC eluent onto a MALDI target at 20 s intervals. After air drying, the sample spots were overlaid with 2,5-DHB matrix and analysed by MALDI-ToF-MS. The MS spectra were obtained in automatic mode with an average 1500 laser shots/spectrum. The oligosaccharide molecular ions [M+Na]+ were identified in the MALDI data and their HILIC elution positions were determined by carrying out an extracted ion chromatogram (EIC). High energy MALDI-CID spectra were obtained as described above.
Purification of XylFucAraGal3 Oligosaccharide by Size Exclusion Chromatography (SEC)
Root AGPs were extracted from hydroponically grown plants as described above and were subjected to sequential enzyme hydrolysis with α-arabinofuranosidase and endo-β-(1→6)-galactanase. SEC was performed on a gravity driven BioGel P-2 (190×2.5 cm; BioRad) column. 2 ml of concentrated sample were applied onto the column and eluted with distilled water; 2 ml fractions were collected and analysed by PACE as described above. The fractions with the XylFucAraGal3 oligosaccharide were combined, freeze dried and resuspended in 0.6 ml deuterium oxide for NMR analysis.
NMR Analysis
NMR spectra were recorded at 298 K with a Bruker AVANCE III spectrometer operating at 600 MHz equipped with a TCI CryoProbe. Two-dimensional 1H-1H TOCSY, ROESY, 13C HSQC, H2BC, HMBC, HSQC-TOCSY and HSQC-ROESY experiments were performed, using established methods [51], [52]; the mixing times were 70 ms and 200 ms for the TOCSY and ROESY experiments, respectively. Chemical shifts were measured relative to internal acetone (δH = 2.225, δC = 31.07 ppm). Data were processed using the Azara suite of programs (v. 2.8, copyright 1993–2014, Wayne Boucher and Department of Biochemistry, University of Cambridge, unpublished) and chemical-shift assignment was performed using Analysis v2 [53].
TFA Hydrolysis and HPAEC-PAD Monosaccharide Analysis
Samples were hydrolysed in 2 M trifluoroacetic acid (TFA) for 1h at 120°C. Following evaporation under vacuum for the removal of TFA, the samples were resuspended in 200 μL water and the monosaccharide sugars were separated using protocols adapted from Currie and Perry [54] on a Dionex ICS3000 system equipped with a PA20 column, a PA20 guard column and a borate trap (Dionex) [14].
Growth Assay for Salt Stress Tolerance
Salt sensitivity assays were performed as described by Kang et al. [55]. Briefly, surface sterilised seeds were sown onto cellophane membrane (BioRad), placed on solid medium containing 0.5x Murashige and Skoog (MS) [56] salts including vitamins (Sigma) and sucrose (1% w/v), stratified for 48h at 4°C and then incubated at 21°C for 7 days. The membranes with seedlings were transferred onto MS medium supplemented with NaCl (50, 100, 150 and 200 mM) or mannitol (50, 100, 200 and 300 mM) and the plates were incubated vertically. Root growth was scored 7 days later. Plants were scanned using an Epson document scanner and root lengths were determined by using Image J software.
Supporting Information
FUT T-DNA insertional mutants. (A) Modified image from TAIR showing the locations of T-DNA insertions within the FUT genes. The arrows represent the location of the primers and the orange triangles represent the T-DNA insertions. The fut6 line was found to contain two T-DNA insertions in opposite directions. Flanking regions of the left border of the T-DNA insertion were amplified with the corresponding primer (left border primer, LB) and fut4- and fut6-specific right primers (4RP and 6RP, respectively). Similarly, PCRs with the T-DNA left borders and the fut4- and fut6-specific left primers (4LP and 6LP, respectively) were performed. (B) Confirmation by PCR that fut4 and fut6 genes are disrupted by the T-DNA insertions.
(EPS)
Primers and PCR/RT-PCR conditions used in this study.
(PDF)
Complementation of the fut4 mutant: HPAEC-PAD neutral monosaccharide analysis (% Mol) of leaf AGPs. Values represent mean ± SD (n = 2).
(PDF)
Acknowledgments
The authors thank Prof. Yuichi Tsumuraya and Dr. Toshihisa Kotake for the gift of the α-L-Arabinofuranosidase (EC 3.2.1.55); exo-β-(1→3)-galactanase (EC 3.2.1.145); β-glucuronidase (EC 3.2.1.31), endo-β-(1→6)-galactanase (EC 3.2.1.164) and the β-(1→6)-galactooligosaccharide standards. We thank Mrs Zhinong Zhang for technical support. The NMR facility infrastructure is funded by the BBSRC and the Wellcome Trust.
Funding Statement
The authors are grateful for the financial support for this research from the United Kingdom BBSRC Sustainable Bioenergy Centre (BSBEC, http://www.bbsrc.ac.uk/), working within the Cell Wall Sugars Programme (Grant BB/G016240/1), and the BioMASS Programme of the centre (Grant BB/G016216/1). This work was supported in part by grants from the Plant Genome Research Program at the National Science Foundation and the Energy Biosciences Program at the U.S. Department of Energy. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Cheung AY, Wang H, Wu HM (1995) A floral transmitting tissue-specific glycoprotein attracts pollen tubes and stimulates their growth. Cell 82: 383–393. [DOI] [PubMed] [Google Scholar]
- 2. Ellis M, Egelund J, Schultz CJ, Bacic A (2010) Arabinogalactan-Proteins: Key Regulators at the Cell Surface? Plant Physiology 153: 403–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. van Hengel AJ, van Kammen A, De Vries SC (2002) A relationship between seed development, arabinogalactan-proteins (AGPs) and the AGP mediated promotion of somatic embryogenesis. Physiologia Plantarum 114: 637–644. [DOI] [PubMed] [Google Scholar]
- 4. Borner GHH, Lilley KS, Stevens TJ, Dupree P (2003) Identification of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A proteomic and genomic analysis. Plant Physiology 132: 568–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamport DTA, Várnai P (2012) Periplasmic arabinogalactan glycoproteins act as a calcium capacitor that regulates plant growth and development. New Phytologist: n/a-n/a. [DOI] [PubMed]
- 6. Gane AM, Craik D, Munro SLA, Howlett GJ, Clarke AE, et al. (1995) Structural analysis of the carbohydrate moiety of arabinogalactan-proteins from stigmas and styles of Nicotiana alata . Carbohydrate Research 277: 67–85. [DOI] [PubMed] [Google Scholar]
- 7. Tryfona T, Liang HC, Kotake T, Kaneko S, Marsh J, et al. (2010) Carbohydrate structural analysis of wheat flour arabinogalactan protein. Carbohydrate Research 345: 2648–2656. [DOI] [PubMed] [Google Scholar]
- 8. Fincher GB, Sawyer WH, Stone BA (1974) Chemical and physical properties of an arabinogalactan-peptide from wheat endosperm. Biochem J 139: 535–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tsumuraya Y, Ogura K, Hashimoto Y, Mukoyama H, Yamamoto S (1988) Arabinogalactan-proteins from primary and mature roots of Radish (Raphanus sativus L.). Plant Physiology 86: 155–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tan L, Varnai P, Lamport DTA, Yuan C, Xu J, et al. (2010) Plant O-Hydroxyproline Arabinogalactans Are Composed of Repeating Trigalactosyl Subunits with Short Bifurcated Side Chains. Journal of Biological Chemistry 285: 24575–24583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clarke AE, Anderson RL, Stone BA (1979) Form and function of arabinogalactans and arabinogalactan-proteins. Phytochemistry 18: 521–540. [Google Scholar]
- 12. Seifert GJ, Roberts K (2007) The Biology of Arabinogalactan Proteins. Annual Review of Plant Biology 58: 137–161. [DOI] [PubMed] [Google Scholar]
- 13. Tan L, Qiu F, Lamport DTA, Kieliszewski MJ (2004) Structure of a Hydroxyproline (Hyp)-Arabinogalactan Polysaccharide from Repetitive Ala-Hyp Expressed in Transgenic Nicotiana tabacum . Journal of Biological Chemistry 279: 13156–13165. [DOI] [PubMed] [Google Scholar]
- 14. Tryfona T, Liang HC, Kotake T, Tsumuraya Y, Stephens E, et al. (2012) Structural Characterization of Arabidopsis Leaf Arabinogalactan Polysaccharides. Plant Physiology 160: 653–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan L, Eberhard S, Pattathil S, Warder C, Glushka J, et al.. (2013) An Arabidopsis Cell Wall Proteoglycan Consists of Pectin and Arabinoxylan Covalently Linked to an Arabinogalactan Protein. The Plant Cell Online. [DOI] [PMC free article] [PubMed]
- 16. Tsumuraya Y, Hashimoto Y, Yamamoto S, Shibuya N (1984) Structure of l-arabino-d-galactan-containing glycoproteins from radish leaves. Carbohydrate Research 134: 215–228. [Google Scholar]
- 17. Tsumuraya Y, Nakamura K, Hashimoto Y, Yamamoto S (1984) Immunological properties of arabinogalactan proteins from leaves of cruciferous plants. Agricultural and Biological Chemistry 48: 2915–2917. [Google Scholar]
- 18. Nakamura K, Tsumuraya Y, Hashimoto Y, Yamamoto S (1984) Arabinogalactan-proteins reacting with Eel anti-H agglutinin from leaves of cruciferous plants. Agric Biol Chem 48: 753–760. [Google Scholar]
- 19. Chun H, Shin DH, Hong BS, Cho HY, Yang HC (2001) Purification and biological activity of acidic polysaccharide from leaves of Thymus vulgaris L. Biological & Pharmaceutical Bulletin. 24: 941–946. [DOI] [PubMed] [Google Scholar]
- 20. Lin LY, Ker YB, Chang CH, Chen KC, Peng RY (2011) Arabinogalactan present in the mountain celery seed extract potentiated hypolipidemic bioactivity in hamsters. Pharmaceutical Biology 49: 319–326. [DOI] [PubMed] [Google Scholar]
- 21. Bacic A, Moody SF, Clarke AE (1986) Structural Analysis of Secreted Root Slime from Maize (Zea mays L.). Plant Physiology 80: 771–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Roy SS, Mittra B, Sharma S, Das TK, Babu CR (2002) Detection of root mucilage using an anti-fucose antibody. Annals of Botany 89: 293–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Donaldson SP, Deacon JW (1993) Differential encystment of zoospores of Pythium species by saccharides in relation to establishment on roots. Physiological and Molecular Plant Pathology 42: 177–184. [Google Scholar]
- 24. Reiter WD, Chapple CCS, Somerville CR (1993) Altered Growth and Cell Walls in a Fucose-Deficient Mutant of Arabidopsis. Science 261: 1032–1035. [DOI] [PubMed] [Google Scholar]
- 25. van Hengel AJ, Roberts K (2002) Fucosylated arabinogalactan-proteins are required for full root cell elongation in arabidopsis. The Plant Journal 32: 105–113. [DOI] [PubMed] [Google Scholar]
- 26. Kitazawa K, Tryfona T, Yoshimi Y, Hayashi Y, Kawauchi S, et al. (2013) β-Galactosyl Yariv Reagent Binds to the β-1,3-Galactan of Arabinogalactan Proteins. Plant Physiology 161: 1117–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu Y, Williams M, Bernard S, Driouich A, Showalter AM, et al. (2010) Functional Identification of Two Nonredundant Arabidopsis alpha(1,2)Fucosyltransferases Specific to Arabinogalactan Proteins. Journal of Biological Chemistry 285: 13638–13645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sarria R, Wagner TA, O'Neill MA, Faik A, Wilkerson CG, et al. (2001) Characterization of a Family of Arabidopsis Genes Related to Xyloglucan Fucosyltransferase1. Plant Physiology 127: 1595–1606. [PMC free article] [PubMed] [Google Scholar]
- 29. Ridlova G, Mortimer JC, Maslen SL, Dupree P, Stephens E (2008) Oligosaccharide relative quantitation using isotope tagging and normal-phase liquid chromatography/mass spectrometry. Rapid Communications in Mass Spectrometry 22: 2723–2730. [DOI] [PubMed] [Google Scholar]
- 30. Domon B, Costello CE (1988) A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconjugate Journal 5: 397–409. [Google Scholar]
- 31. Spina E, Sturiale L, Romeo D, Impallomeni G, Garozzo D, et al. (2004) New fragmentation mechanisms in matrix-assisted laser desorption/ionization time-of-flight/time-of-flight tandem mass spectrometry of carbohydrates. Rapid Communications in Mass Spectrometry 18: 392–398. [DOI] [PubMed] [Google Scholar]
- 32. Bonin CP, Potter I, Vanzin GF, Reiter WD (1997) The MUR1 gene of Arabidopsis thaliana encodes an isoform of GDP-D-mannose-4,6-dehydratase, catalyzing the first step in the de novo synthesis of GDP-L-fucose. Proceedings of the National Academy of Sciences 94: 2085–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shi H, Kim Y, Guo Y, Stevenson B, Zhu JK (2003) The Arabidopsis SOS5 Locus Encodes a Putative Cell Surface Adhesion Protein and Is Required for Normal Cell Expansion. The Plant Cell Online 15: 19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nguema-Ona E, Coimbra S, Vicre-Gibouin M, Mollet JC, Driouich A (2012) Arabinogalactan proteins in root and pollen-tube cells: distribution and functional aspects. Annals of Botany 110: 383–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stevenson TT, Darvill AG, Albersheim P (1988) Structural features of the plant cell-wall polysaccharide rhamnogalacturonan-II. Carbohydrate Research 182: 207–226. [Google Scholar]
- 36. Willats WGT, Knox JP (1996) A role for arabinogalactan-proteins in plant cell expansion: evidence from studies on the interaction of β-glucosyl Yariv reagent with seedlings of Arabidopsis thaliana . The Plant Journal 9: 919–925. [DOI] [PubMed] [Google Scholar]
- 37. Baskin T, Betzner A, Hoggart R, Cork A, Williamson R (1992) Root Morphology Mutants in Arabidopsis thaliana . Functional Plant Biology 19: 427–437. [Google Scholar]
- 38. Seifert GJ, Barber C, Wells B, Dolan L, Roberts K (2002) Galactose Biosynthesis in Arabidopsis: Genetic Evidence for Substrate Channeling from UDP-D-Galactose into Cell Wall Polymers. Current Biology 12: 1840–1845. [DOI] [PubMed] [Google Scholar]
- 39. Liang Y, Basu D, Pattathil S, Xu Wl, Venetos A, et al. (2013) Biochemical and physiological characterization of fut4 and fut6 mutants defective in arabinogalactan-protein fucosylation in Arabidopsis. Journal of Experimental Botany 64: 5537–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gibeaut DM, Hulett J, Cramer GR, Seemann JR (1997) Maximal Biomass of Arabidopsis thaliana Using a Simple, Low-Maintenance Hydroponic Method and Favorable Environmental Conditions. Plant Physiology 115: 317–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Serpe MD, Nothnagel EA (1995) Fractionation and Structural Characterization of Arabinogalactan-Proteins from the Cell Wall of Rose Cells. Plant Physiology 109: 1007–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Serpe MD, Nothnagel EA (1996) Heterogeneity of Arabinogalactan-Proteins on the Plasma Membrane of Rose Cells. Plant Physiology 112: 1261–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tsumuraya Y, Mochizuki N, Hashimoto Y, Kováč P (1990) Purification of an exo-beta-(1–>3)-D-galactanase of Irpex lacteus (Polyporus tulipiferae) and its action on arabinogalactan-proteins. Journal of Biological Chemistry 265: 7207–7215. [PubMed] [Google Scholar]
- 44. Takata R, Tokita K, Mori S, Shimoda R, Harada N, et al. (2010) Degradation of carbohydrate moieties of arabinogalactan-proteins by glycoside hydrolases from Neurospora crassa . Carbohydrate Research 345: 2516–2522. [DOI] [PubMed] [Google Scholar]
- 45. Kotake T, Kaneko S, Kubomoto A, Haque MA, Kobayashi H, et al. (2004) Molecular cloning and expression in Escherichia coli of a Trichoderma viride endo-β-(1–>6)-galactanase gene. Biochemical Journal 377: 749–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Konishi T, Kotake T, Soraya D, Matsuoka K, Koyama T, et al. (2008) Properties of family 79 β-glucuronidases that hydrolyze β-glucuronosyl and 4-O-methyl-β-glucuronosyl residues of arabinogalactan-protein. Carbohydrate Research 343: 1191–1201. [DOI] [PubMed] [Google Scholar]
- 47. Goubet F, Jackson P, Deery MJ, Dupree P (2002) Polysaccharide Analysis Using Carbohydrate Gel Electrophoresis: A Method to Study Plant Cell Wall Polysaccharides and Polysaccharide Hydrolases. Analytical Biochemistry 300: 53–68. [DOI] [PubMed] [Google Scholar]
- 48. Tryfona T, Stephens E (2010) Analysis of carbohydrates on proteins by offline Normal-Phase Liquid Chromatography MALDI-ToF/ToF-MS/MS. Methods in Molecular Biology 658: 137–151. [DOI] [PubMed] [Google Scholar]
- 49. Ciucanu I, Kerek F (1984) A simple and rapid method for the permethylation of carbohydrates. Carbohydrate Research 131: 209–217. [Google Scholar]
- 50. Maslen SL, Goubet F, Adam A, Dupree P, Stephens E (2007) Structure elucidation of arabinoxylan isomers by normal phase HPLC-MALDI-TOF/TOF-MS/MS. Carbohydrate Research 342: 724–735. [DOI] [PubMed] [Google Scholar]
- 51. Nyberg NT, Duus JÃ, Sörensen OW (2005) Heteronuclear Two-Bond Correlation: Suppressing Heteronuclear Three-Bond or Higher NMR Correlations while Enhancing Two-Bond Correlations Even for Vanishing 2JCH . Journal of the American Chemical Society 127: 6154–6155. [DOI] [PubMed] [Google Scholar]
- 52.Cavanagh J, Fairbrother WJ, Palmer AG, Skelton NJ (1996) Protein NMR Spectroscopy: Principles and Practice. San Diego, CA, USA: Academic Press.
- 53. Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, et al. (2005) The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins: Structure, Function, and Bioinformatics 59: 687–696. [DOI] [PubMed] [Google Scholar]
- 54. Currie HA, Perry CC (2006) Resolution of complex monosaccharide mixtures from plant cell wall isolates by high pH anion exchange chromatography. Journal of Chromatography A 1128: 90–96. [DOI] [PubMed] [Google Scholar]
- 55. Kang JS, Frank J, Kang CH, Kajiura H, Vikram M, et al. (2008) Salt tolerance of Arabidopsis thaliana requires maturation of N-glycosylated proteins in the Golgi apparatus. Proceedings of the National Academy of Sciences 105: 5933–5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Murashige T, Skoog F (1962) A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiologia Plantarum 15: 473–497. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FUT T-DNA insertional mutants. (A) Modified image from TAIR showing the locations of T-DNA insertions within the FUT genes. The arrows represent the location of the primers and the orange triangles represent the T-DNA insertions. The fut6 line was found to contain two T-DNA insertions in opposite directions. Flanking regions of the left border of the T-DNA insertion were amplified with the corresponding primer (left border primer, LB) and fut4- and fut6-specific right primers (4RP and 6RP, respectively). Similarly, PCRs with the T-DNA left borders and the fut4- and fut6-specific left primers (4LP and 6LP, respectively) were performed. (B) Confirmation by PCR that fut4 and fut6 genes are disrupted by the T-DNA insertions.
(EPS)
Primers and PCR/RT-PCR conditions used in this study.
(PDF)
Complementation of the fut4 mutant: HPAEC-PAD neutral monosaccharide analysis (% Mol) of leaf AGPs. Values represent mean ± SD (n = 2).
(PDF)