Abstract
Equol, one of the intestinal microflora metabolites of daidzein, has gained much attention for having greater bioactivity than its precursor (daidzein and daidzin) and seeming to be promoted by hydrogen gas. The effects of lactulose on the equol-producing capacity and liver antioxidant status of barrows treated with daidzein were investigated in this study. Male castrated piglets (barrows) of Landrace×Duroc, aged 40 days, were randomly divided into the following three groups: control group (C, n = 12, fed an isoflavones-free basic diet), daidzein group (D, n = 12, fed an isoflavones-free basic diet with 50 mg/kg of daidzein supplementation) and daidzein+lactulose group (D+L, n = 12, fed an isoflavones-free basic diet with 1% of lactulose and 50 mg/kg of daidzein supplementation). After 20 days, the profile of short-chain fatty acids in the colon digesta showed that lactulose significantly increased the fermented capacity in the gastrointestinal tract of the barrows. First-void urinary equol concentrations were significantly higher in the D+L group than in the D group (3.13±0.93 compared to 2.11±0.82 μg/ml, respectively). Furthermore, fecal equol levels were also significantly higher in the D+L group than in the D group (12.00±2.68 compared to 10.00±2.26 μg/g, respectively). The population of bacteroidetes and the percentage of bacteroidetes to bacteria in feces were higher in the D+L group than in the D group. The DGGE profiles results indicate that lactulose might shift the pathways of hydrogen utilization, and changing the profiles of SRB in feces. Moreover, the D+L group had weak enhancement of T-SOD and CuZn-SOD activities in the livers of barrows treated with daidzein.
Introduction
Daidzein, the main isoflavones found in soybeans and most soy foods, can be transformed to equol [1]. Equol is exclusively produced by intestinal microflora [2] and may be superior to all other isoflavones in terms of its antioxidant [3] and anti-inflammatory properties [4]. Among humans, 30–50% has the intestinal bacteria capable of producing equol [5]. Equol producers are advantageous in terms of enhancing health benefits, and gained much attention since the postulation of the equol hypothesis [6]. Although there is conflicting evidence available regarding the potential health benefits associated with the ability to produce equol, there is a growing interest in dietary applications that might enhance equol production in humans and animals.
Hydrogen gas was reported to stimulate equol production in an equol-producing mixed culture (EPC4), likely by acting as an electron donor in the formation of equol [7]. In the presence of daidzein, EPC4 significantly decreased the methanogenesis and sulphidogenesis in the cultures of faecal samples with methanogenic or sulphate-reducing abilities [8]. A stronger decrease was observed with increasing amounts of EPC4 and constant equol production, suggesting daidzein to act partly as a hydrogen sink.
Lactulose, a disaccharide formed from one molecule each of the simple sugars fructose and galactose, cannot be digested and absorbed by the body, but can be digested by bacteria colonizing the gastrointestinal tract, especially the colon. Oral administration of lactulose can significantly increase breath H2 production [9]. We hypothesized that equol production could be regulated by adding a hydrogen-producing prebiotic in vivo/in vitro. So far, no relationship between a hydrogen-producing prebiotic and equol production has been studied in vivo. The current study was designed to examine the effects of lactulose on the equol production and liver antioxidant status of castrated male piglets treated with daidzein.
Materials and Methods
Statement of Ethics
Animal care and use were conducted in accordance with the Animal Research Institute Committee guidelines of Nanjing Agricultural University, China. The study’s castrated male piglets (barrows), aged 40 days and initially weighing 9.35±0.17 kg, were crossed from Landrace and Duroc (weaned at 28±1d of age). All pigs were housed in an environment-controlled room and maintained under the care of the Laboratory Animal Unit from the Institute of Experimental Animal Center, Jiangsu Academy of Agricultural Sciences, China. Within the house, each of the six pigs were housed in a pen (4×4 m) with one feeder and two nipple drinkers to allow them ab libitum access to feed and water. This study was approved by the Committee of Animal Research Institute, Nanjing Agricultural University, China.
Animal Husbandry, Diets, Experimental Design, and Sampling
Barrows were given free access to an isoflavones-free diet for five days before the feeding trial and then randomly divided into the following three groups: control (C, n = 12, fed an isoflavones-free basic diet), daidzein (D, n = 12, fed an isoflavones-free basic diet with 50 mg/kg of daidzein supplementation), and daidzein+lactulose (D+L, n = 12, fed an isoflavones-free basic diet with 1% of lactulose- and 50 mg/kg of daidzein supplementation). Table S1 shows the composition and nutrient levels of the experimental diets, which were prepared according to the nutrient requirements [10] for weaned pigs.
After 20 days of the feeding trial, first-void urine and fecal samples were collected and stored at −70°C until assay. On the 22nd day, six pigs from each treatment were randomly selected and killed humanely by an intramuscular injection of sodium pentobarbital (40 mg/kg body weight). The liver, colon mucosa, and digesta were sampled and stored at −70°C until assay.
Extraction and Quantification of Daidzein and Equol
Urine samples were prepared as previously described [11]. In brief, each 2 ml of centrifuged urine sample (centrifuged at 850 g for 15 minutes) was mixed with 7 ml of sodium acetate buffer (pH 5.0) and 30 μl β-glucuronidase (GUS) from Escherichia coli(100 U/ml)/30 μl Sulfatate from Helix pomatia (500 U/ml) and incubated for three hours at 37°C, followed by extraction with 7 ml of ethyl acetate. The solvent was removed under a gentle stream of nitrogen, and the sample was re-dissolved in 500 μl 80% (V/V) methanol prior to analysis.
Faecal daidzein and equol were analyzed according to a method previously mentioned [12]. Faecal samples (1 g) were extracted with 7 ml of ethyl acetate. The solvent was removed under a gentle stream of nitrogen, and the sample was re-dissolved in 3 ml 80% (V/V) methanol prior to analysis.
Concentrations of daidzein and equol were determined using a HPLC system consisting of a Surveyor MS Pump Plus, Surveyor Autosampler, Surveyor PDA Plus (Thermo Fisher Scientific, Finnigan™, USA), and the Thermo Fisher Xcalibur™ software package (version 1.4 SR1). A 20 μl sample was injected and separated over a YMC-Pack Pro C18 column with a particle size of 5 μm (YMC Inc., Japan) kept at 30°C. Elution was isocratic with a mobile phase consisting of 20% acetonitrile, 30% methanol, and 45% water and the flow rate was maintained at 300 μl/min. The eluate was analyzed with Surveyor photodiode array (PDA) Plus detection system (Thermo Fisher Scientific, Finnigan™, USA), UV spectra of the peaks for equol was detected at 205 nm, and daidzein was detected at 260 nm. UV-absorption spectra allowed identification of the peaks after comparison with pure standards. Calibration curves for quantification of daidzein and equol were constructed using pure standards obtained from Sigma (New York). The concentration of equol was calculated following the equation Y = 85653.6+1.23123e+006*X (y: HPLC peak area; x: concentration of equol) [13]. The concentration of daidzein was calculated following the equation Y = 36529.5+844385*X (y: HPLC peak area; x: concentration of daidzein) [13].
DNA Isolation, PCR Amplification, and DGGE Analysis
A QIAamp® DNA Stool Mini Kit was purchased from QIAGEN Germany and used to extract DNA from the fecal, colon digesta, and mucosa samples according to the manufacturer’s instructions. DNA extracts were stored at −20°C until ready for use.
Primers U968-GC and L1401 [14] were used to amplify the V6–V8 regions of the bacterial 16 S rRNA gene. Primers 519f and 915rGC [15] were used to amplify the 16S rDNA of methanogenic Archaea. Primers APS-FW and APS-RV-GC [16] were used to amplify a 436-bp fragment of the adenosine-5′-phosphosulfate (APS) reductase subunit A gene. The primers used in this study are listed in Table S2. Oligonucleotide primers were ordered from Invitrogen (Shanghai, China). PCR amplification was performed in a T1 Thermal Cycler (Biometra, Germany). PCR products (5 μl) were analyzed by electrophoresis on 1.5% agarose gel (w/v) to check the sizes and amounts of the amplicons.
PCR products of the total bacteria, methanogenic Archaea, and sulfate-reducing bacteria were separated by DGGE [17] using a Dcode™ System (Bio-Rad, Hercules, CA) in 8%, 6%, and 8% polyacrylamide gels with a denaturing gradient of 38–55%, 30–60%, and 32–65% (a 100% denaturant is defined as 7 M urea and 40% (v/v) deionized-formamide)respectively. The gels were run in a 0.5×TAE buffer. The electrophoresis was performed at 60°C for 10 minutes at a voltage of 200 V, and subsequently run at a fixed voltage of 85 V for 12 h at 60°C (with methanogenic Archaea for 16 h). After completion of electrophoresis, the gels were stained with silver nitrate (AgNO3) [18] and scanned by a GS-800 Calibrated Densitometer (Bio-Rad) for analysis with GelCompare II 5.0 (Applied Maths, Kortrijk, Belgium).
Real-time PCR Assay on the Number of Selected Bacteria
Firmicutes [19], bacteroidetes [20], [21], methanogen (mcrA) [22], and subunit A of the adenosine-5′-phosphosulfate (APS) reductase gene [16] from our previous experimental DNA samples (unpublished data) were amplified with the primers shown in Table S2. Purified PCR products were cloned in Escherichia coli JM109 using the pGEM-T vector system (Invitrogen, Shanghai, China). Plasmid DNA with correct size inserts was extracted and used for constructing standard curves. The standard curves were generated using triplicate ten-fold dilutions of plasmid DNA. For all real-time PCR assays, there was a linear relationship between the log of the plasmid DNA copy number and the calculated CT value across the specified concentration range (R2 = 0.99 in all cases) (data not shown). Real time PCR was carried out using an ABI 7300 real-time PCR machine (Applied Biosystems, Foster City, CA, USA) in a total volume of 20 μl containing 10 μl 2 × SYBR Premix Ex Taq (Takara Dalian China), 2 μl DNA (1∶9 diluted with PCR-grade H2O), 0.4 μl each of 10 μmol/L forward and reverse primers, 0.4 μl ROX, and 4.2 μl PCR-grade water. The program was set at 95°C for 10 seconds, followed by 40 cycles of 95°C for 5 seconds, and 60°C for 31 seconds. The melting curve analysis of amplification products was performed at the end of each PCR reaction to confirm that only one PCR product was amplified and detected.
Biochemical Analysis of Oxidative and Anti-oxidative Biomarkers in Liver, and the Biochemical Analysis of SCFA in Colonic Digesta
The antioxidant assays were conducted using assay kits purchased from the Nanjing Jiancheng Institute of Bioengineering (Nanjing, Jiangsu, China). One hundred milligrams of frozen liver tissue in 1 ml of homogenization buffer (0.9% cool physiological saline) was homogenized on ice with a Polytron-aggregate homogenizer (POLYTRON PT-1200E, Lucerne, Switzerland) for 30 seconds at 12500 rpm. The homogenate was centrifuged at 2500 rpm for 10 minutes at 4°C, and the resultant supernatant was a 10% concentration. In following, 0.9% of physiological saline was used to dilute the supernatant to different concentrations, which were then stored at −20°C until analysis. All following assays were conducted according to the manufacture’s procedures.
The VFAs in the colon digesta were measured by using a capillary column gas chromatograph (GC-14A; Shimadzu, Japan) according to Mao et.al [23].
Calculations and Statistical Analysis
DGGE analysis of all samples was repeated twice. All gels were scanned at 400 dpi. The number of DGGE bands and similarity indices were calculated from the densitometric curves of the scanned DGGE profiles using the GelCompare II 5.0 software (Applied Maths, Kortrijk, Belgium) with the Pearson product-moment correlation coefficient [24]. Similarity indices were calculated for pairs of DGGE profiles for the colon mucosa, digesta, and feces samples, respectively. As a parameter for the structural diversity of the microbial community, the Shannon index of general diversity, H′ [25], was calculated [26].
All statistical analyses were performed using a one-way ANOVA with SPSS statistical software (ver. 18.0 for Windows, SPSS Inc., Chicago, IL) The differences were considered significant at P<0.05. Differences among treatments for parameters were tested for significance using Duncan's test. Data are presented as means±SD.
Results
SCFA Profile in the Colon Digesta
Table 1 shows the effect of lactulose and daidzein on the SCFA profile in colon digesta. Although little difference was found between the D and C groups, large effects of lactulose were found. The acetic acid, propionic acid, butyric acid, and valeric acid concentrations in the colon digesta between the D and D+L groups showed no significant difference (Table 1). However, isobutyric acid, isovaleric acid, and branched chain fatty acid (BCFA), as well as the total VFA levels in the colon digesta, were significantly higher in the D+L group than in the D and C groups (Table 1).
Table 1. Effects of lactulose on SCFA levels in the colonic digesta of barrows fed with daidzein.
| Items | Control(n = 6) | Daidzein(n = 5) | D+L(n = 6) | P |
| Acetic acid (mM) | 31.39±5.11 | 31.69±3.51 | 36.44±2.56 | 0.069 |
| Propionic acid (mM) | 10.39±0.75 | 11.35±1.41 | 11.86±1.70 | 0.191 |
| Isobutyric acid (mM) | 1.17±0.35b | 1.25±0.22b | 1.80±0.40a | 0.010 |
| Butyric acid (mM) | 4.07±0.63b | 4.98±0.63ab | 5.44±1.03a | 0.025 |
| Isovaleric acid (mM) | 1.76±0.18b | 1.87±0.58b | 2.66±0.30a | 0.002 |
| Valeric acid (mM) | 1.18±0.14b | 1.53±0.34a | 1.56±0.19a | 0.025 |
| BCFA (mM) | 2.93±0.40b | 3.12±0.77b | 4.46±0.67a | 0.001 |
| TVFA (mM) | 49.96±6.16b | 52.66±4.66b | 59.77±4.38a | 0.013 |
Note: values with different small letter superscripts in the same row mean that there was a significant difference (P<0.05), and the same small letter superscripts mean that there was no difference.
Levels of Daidzein and Equol in the Urine and Feces
Thirty-two individuals completed urine and feces sample collection (from 10 pigs in C group, 11 in D group, and 11 in the D+L group, respectively). The concentrations of daidzein and equol in the urine and feces in the barrows treated with daidzein were significantly higher than those in the control pigs (Table 2). The urinary equol concentrations and the ratio of equol to daidzein were significantly higher in the D+L group than in the D group. However, no significant difference was observed in daidzein concentration in the urine and feces between the D and D+L groups. Fecal equol levels were significantly higher in the D+L group compared to the D group (Table 2). No difference was found in the ratio of fecal equol to daidzein concentration between the D and D+L groups (Table 2).
Table 2. Effects of lactulose on the concentrations of urinary and fecal daidzein and equol, and the ratio of urinary and fecal equol to daidzein concentrations in barrows fed with daidzein.
| Items | Control | Daidzein | D+L | P |
| (n = 10) | (n = 11) | (n = 11) | ||
| Urine | ||||
| Daidzein (μg/ml) | NDb | 9.70±2.69a | 8.49±2.18a | <0.0001 |
| Equol (μg/ml) | NDc | 2.11±0.82b | 3.13±0.93a | <0.0001 |
| Equol to daidzein ratio | NDb | 0.21±0.06b | 0.38±0.10a | <0.0001 |
| Faecal | ||||
| Faecal daidzein (μg/g) | NDb | 3.92±1.48a | 3.84±1.93a | <0.0001 |
| Faecal equol (μg/g) | NDc | 10.00±2.26b | 12.00±2.68a | <0.0001 |
| Equol to daidzein ratio | NDb | 2.90±1.40a | 3.77±1.57a | <0.0001 |
Note: values with different small letter superscripts in the same row mean that there was a significant difference (P<0.05), and the same small letter superscripts mean that there was no difference. ND = not detected.
Microflora Profiles of Total Bacteria, Methanogenic Archaea, and Sulfate-reducing Bacteria (SRB) in Feces, Colon Digesta, and Mucosa
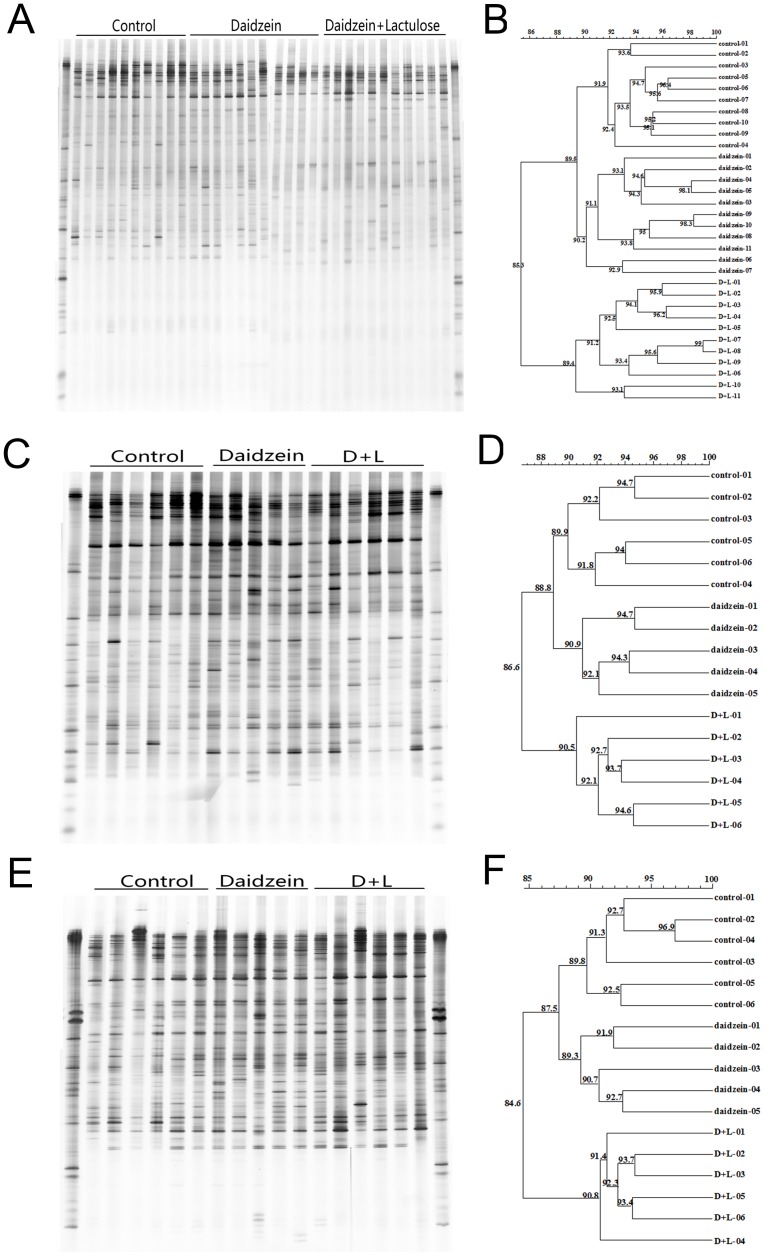
The representative DGGE analysis of the PCR fragment generated with primers U968GC and L1401, 519f and 915rGC, and APS-FW and APS-RV-GC are shown in Figure 1 (A, C, and E), Figure 2 (A, C, and E), and Figure 3 (A, C, and E), respectively. For the total bacteria DGGE profile, the predominant bands in the fecal DNA samples were on the upper gels (Figure 1 A), whereas in the colon digesta and mucosa, the predominant bands were evenly separated in the gels (Figure 1 C and E). Although some bands changed for different groups, no particular bands were commonly changed by dietary treatment. Cluster analysis revealed that the overall similarity of bacteria DGGE profiles in feces, colon digesta, and mucosa were 85.3%, 86.6%, and 84.6%, respectively (Figure 1 B, D, and F). Interestingly, in three different sites the C and D groups were all in the same cluster with a similarity of 89.5%, 88.8%, and 87.5%, respectively (Figure 1 B, D, and F).
Figure 1. DGGE profiles and similarities of bacterial communities in the feces (A)(B), colonic digesta (C)(D), and colonic mucosa (E)(F).
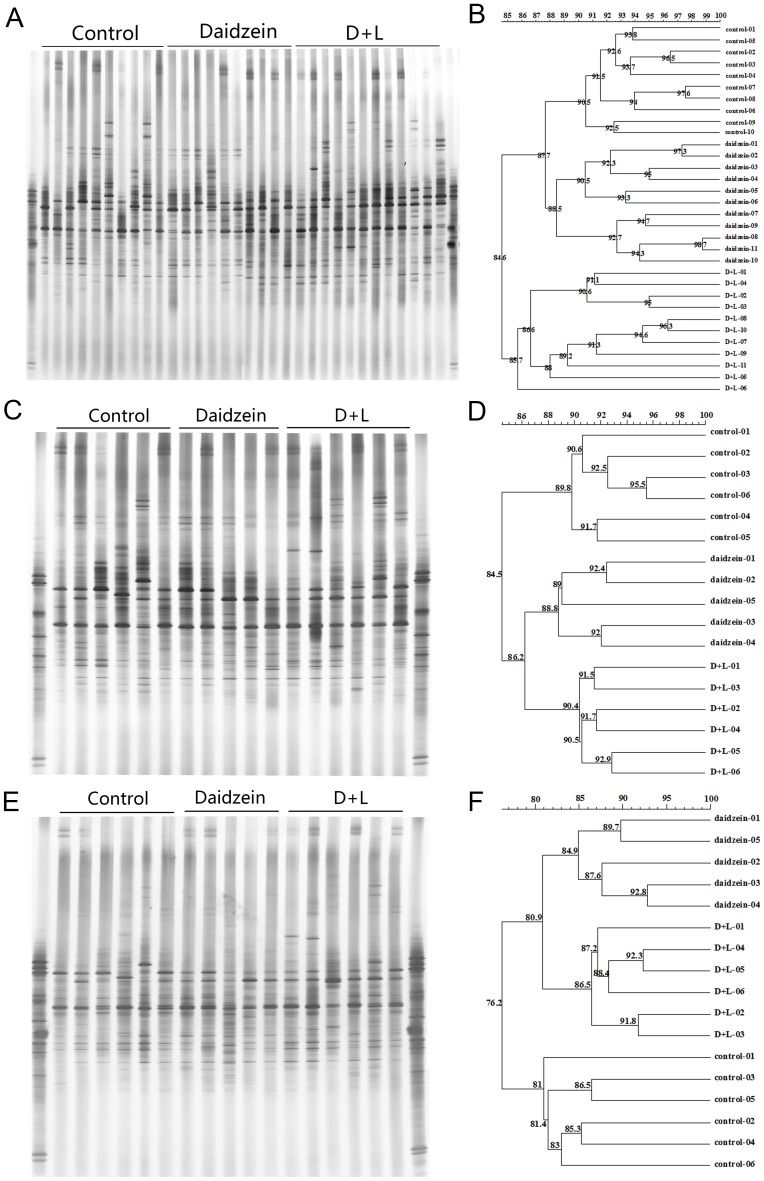
Figure 2. DGGE profiles and similarities of methanogenic Archaea in the feces (A)(B), colonic digesta (C)(D), and colonic mucosa (E)(F).
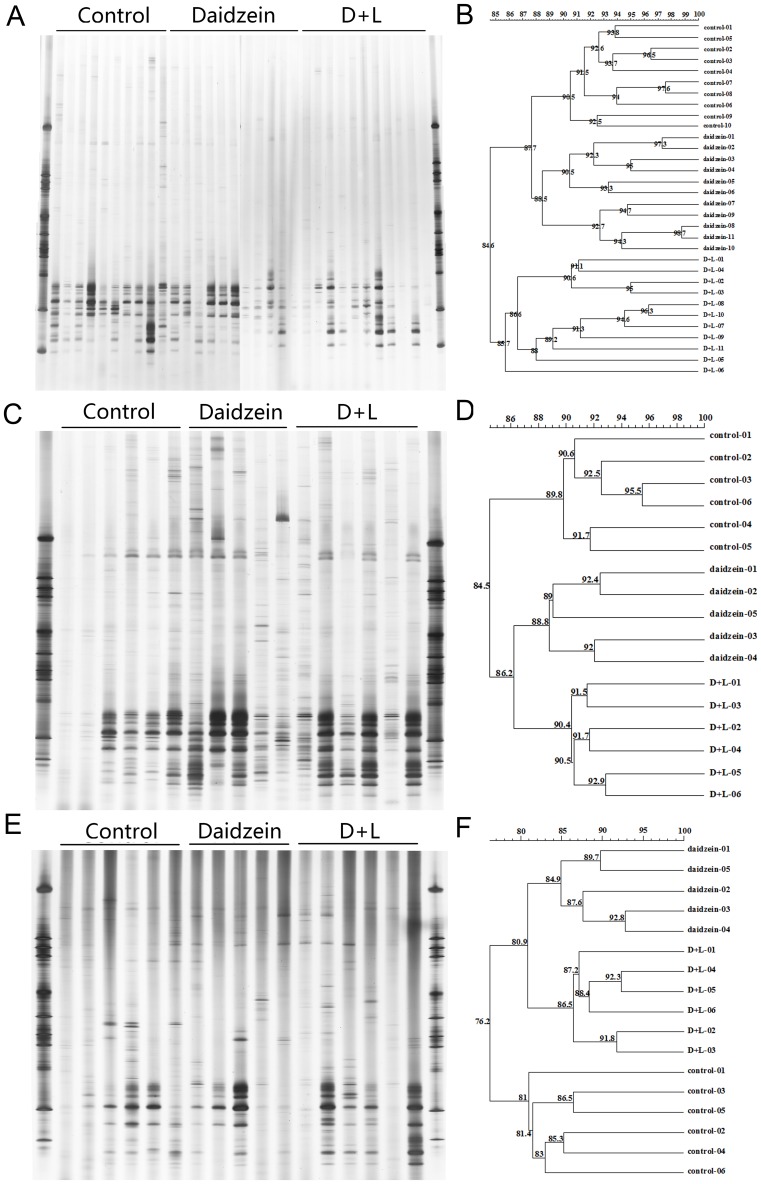
Figure 3. DGGE profiles and similarities of APS reductase subunit A gene in the feces (A)(B), colonic digesta (C)(D), and colonic mucosa (E)(F).
For the methanogenic Archaea composition in the fecal, colon digesta, and mucosa of the barrows, the predominant bands were all in the middle of the gels (Figure 2 B, D and F). There were no special bands influenced by daidzein or the D+L treatment. Cluster analysis revealed that the overall similarity of the MPB DGGE profiles in feces, colon digesta, and mucosa were 84.6%, 84.5%, and 76.2%,respectively (Figure 2 B, D, and F). The C and D samples were found in the coherent cluster with a similarity of 87.7% in the feces of the barrows. However, the D and D+L samples were found in one cluster with a similarity of 86.2% and 80.9% respectively in the colon digesta and mucosa samples of the barrows (Figure 2 B, D, and F).
For the DGGE analysis of the APS reductase DNA amplicons in three different locations, the bands number seemed less than the number of MPB and bacteria. The predominant bands were all at the bottom of the DGGE gels. Still, no particular bands were found to be influenced by daidzein or the D+L treatment. Cluster analysis showed that the overall similarity of the APS reductase DGGE profiles in feces, colon digesta, and mucosa were 50.4%, 69.6%, and 67.7%, respectively. The C and D groups were in the same cluster with a similarity of 55.8% in the feces samples. With the DNA samples from the colon digesta, the D and D+L groups were in one cluster with a similarity of 77.1%. Lastly, the cluster analysis for the colon mucosa in the C and D+L groups were in the same cluster with a similarity of 71.9% (Figure 3 B, D, and F).
To further examine the effects of lactulose and daidzein, the number of predominant bands in the DGGE profiles and their Shannon index of general diversity were analyzed (Table 3). For the fecal samples, major effects were found in the DGGE bands for bacteria, MPB, and SRB, while no differences were found in the Shannon diversity of selected bacteria. The band number of the DGGE gels for bacteria was significantly lower in the D+L group than the C and D groups, and the band number of the DGGE gels for MPB in the C and D+L groups was significantly higher than in the D group. The DGGE band number for SRB in the D+L group was significantly lower than in the C and D groups, and the C group had a higher DGGE band number for SRB than in the D group (Table 3).
Table 3. Effects of lactulose on the number of DGGE bands and Shannon diversity of the feces and colonic samples of pigs fed with daidzein.
| Target group | DNA sample | Items | Control | Daidzein | D+L | P |
| Total bacteria | FecesA | DGGE bands number | 57.80±3.52a | 57.81±3.87a | 51.00±2.32b | <0.0001 |
| Shannon Diversity | 3.53±0.15 | 3.55±0.16 | 3.42±0.11 | 0.095 | ||
| Colon digestaB | DGGE bands number | 76.00±2.61a | 77.40±2.88a | 69.50±4.32b | 0.003 | |
| Shannon Diversity | 3.71±0.28 | 3.94±0.08 | 3.81±0.16 | 0.200 | ||
| Colon mucosaC | DGGE bands number | 67.17±5.85ab | 71.20±3.35a | 63.17±2.22b | 0.021 | |
| Shannon Diversity | 3.81±0.16 | 3.91±0.04 | 3.82±0.11 | 0.350 | ||
| MPB | FecesA | DGGE bands number | 41.40±1.58a | 38.55±1.64b | 41.18±1.66a | <0.0001 |
| Shannon Diversity | 3.07±0.24 | 3.09±0.16 | 3.21±0.15 | 0.164 | ||
| Colon digestaB | DGGE bands number | 55.33±1.75 | 57.20±3.42 | 58.33±2.58 | 0.170 | |
| Shannon Diversity | 3.32±0.26 | 3.32±0.11 | 3.26±0.19 | 0.827 | ||
| Colon mucosaC | DGGE bands number | 35.67±4.84b | 43.80±4.82a | 44.83±2.48a | 0.004 | |
| Shannon Diversity | 3.16±0.22 | 3.25±0.27 | 3.34±0.14 | 0.333 | ||
| SRB | FecesA | DGGE bands number | 42.40±7.44a | 30.00±7.95b | 23.45±5.65c | <0.0001 |
| Shannon Diversity | 2.58±0.26a | 2.34±0.28ab | 2.20±0.35b | 0.031 | ||
| Colon digestaB | DGGE bands number | 55.83±12.19b | 78.20±3.96a | 67.67±6.31a | 0.002 | |
| Shannon Diversity | 3.26±0.25 | 3.52±0.16 | 3.28±0.33 | 0.237 | ||
| Colon mucosaC | DGGE bands number | 50.50±6.67a | 39.20±3.90b | 49.83±4.54a | 0.006 | |
| Shannon Diversity | 3.02±0.28 | 2.80±0.23 | 3.01±0.29 | 0.353 |
Note: values with different small letter superscripts in the same row mean that there was a significant difference (P<0.05), and the same small letter superscripts mean that there was no difference.
The fecal sample replications in C group n = 10, D group n = 11, D+L group n = 11.
The colon content sample replications in C group n = 6, D group n = 5, D+L group n = 6.
The mucosa of colon sample replications in C group n = 6, D group n = 5, D+L group n = 6.
For the colon digesta samples, the results showed that there was no influence on the Shannon diversity of the DGGE profile for bacteria, MPB, and SRB, and that the DGGE band numbers for bacteria and SRB were of significant difference (Table 3). The band numbers of the DGGE profile for bacteria in the D+L groups were significantly lower than in the C and D groups, and the DGGE band numbers for SRB in the D and D+L groups were significantly lower than in the C group (Table 3).
There was no difference in the Shannon diversity of the DGGE profile for bacteria, MPB, and SRB in the samples of colon mucosa. However, the DGGE band numbers for bacteria were found to be significantly lower in the D+L group than the in the D group (Table 3). The DGGE band numbers for the MPB in the C group were significantly higher lower than the D and D+L groups, whereas no difference was found between the D and D+L groups. The DGGE band numbers for SRB in the D group were significantly lower than in the C and D+L groups (Table 3).
Enumeration of Selected Bacteria in Feces, Colon Digesta, and Mucosa
To explore the further influence of lactulose and daidzein on the microorganisms, real-time RCR was used to quantify the abundance of total bacteria, firmicutes, bacteroidetes, methanogen-producing bacteria, and sulfate-reducing bacteria in the fecal, colon digesta, and mucosa. Table 4 shows the population and abundance of selected bacteria in the three different locations of the C, D, and D+L groups. For the fecal DNA samples, the number of total bacteria, firmicutes, and methanogen-producing bacteria in the three groups there was no difference. However, the population of bacteroidetes and sulfate-reducing bacteria in the D+L group was significantly higher than in the C and D groups. No difference was found between the C and D groups (Table 4). Still, the percentage of firmicutes and bacteroidetes to total bacteria was found to be significantly higher in the D+L group than in the C and D groups. Unexpectedly, excluding the sulfate-reducing bacteria in the mucosa of the colon, a significant difference was not found in the population and abundance of selected bacteria in the mucosa of the colon, or in the digesta of the colon in the barrows (Table 4). The population of sulfate-reducing bacteria in the mucosa of the colon was found to be significantly higher in C group than in the D and D+L groups (Table 4).
Table 4. The abundance of selected bacteria in the feces, colonic digesta, and mucosa of barrows.
| DNA sample | Items | Control | Daidzein | D+L | P |
| FaecalA | Bacteria | (5.38±2.86)×1011 | (5.10±1.50)×1011 | (6.01±1.44)×1011 | 0.56 |
| Firmicutes | (3.24±1.68)×1011 | (3.17±0.92)×1011 | (4.18±1.16)×1011 | 0.133 | |
| Bacteroidetes | (3.86±1.68)×1010b | (4.02±1.83)×1010b | (7.09±1.77)×1010a | <0.0001 | |
| Firmicutes% | 61.10±5.97b | 62.15±4.46b | 69.30±5.39a | 0.002 | |
| Bacteroidetes% | 7.74±1.54b | 7.89±3.21b | 11.86±1.43a | <0.0001 | |
| Methanogen | (4.06±0.91)×106 | (4.43±2.32)×106 | (3.57±2.67)×106 | 0.641 | |
| SRB | (4.89±1.49)×104a | (3.68±0.64)×104b | (2.71±1.12)×103c | <0.0001 | |
| Colon digestaB | Bacteria | (3.88±2.75)×1010 | (3.90±1.17)×1010 | (4.07±2.32)×1010 | 0.9879 |
| Firmicutes | (2.45±1.68)×1010 | (2.47±0.65)×1010 | (2.67±1.60)×1010 | 0.9589 | |
| Bacteroidetes | (5.75±2.86)×109 | (6.74±0.50)×109 | (7.54±3.18)×109 | 0.4974 | |
| Firmicutes% | 63.62±3.93 | 63.85±2.75 | 65.19±3.25 | 0.6996 | |
| Bacteroidetes% | 16.13±3.41 | 18.20±3.90 | 19.71±4.01 | 0.2878 | |
| MPB | (5.24±2.48)×106 | (3.57±1.99)×106 | (5.35±3.23)×106 | 0.493 | |
| SRB | (2.80±2.36)×104 | (3.98±1.30)×104 | (8.87±7.04)×104 | 0.06 | |
| Colon mucosaC | Bacteria | (5.24±2.36)×108 | (4.91±1.78)×108 | (5.05±1.75)×108 | 0.9636 |
| Firmicutes | (3.35±1.71)×108 | (3.09±1.12)×108 | (3.18±1.13)×108 | 0.9498 | |
| Bacteroidetes | (9.42±5.63)×107 | (8.81±2.27)×107 | (9.70±3.09)×107 | 0.9353 | |
| Firmicutes% | 62.72±5.72 | 62.82±5.32 | 62.67±1.78 | 0.9983 | |
| Bacteroidetes% | 17.64±4.04 | 18.45±2.60 | 19.45±2.07 | 0.5906 | |
| MPB | (4.50±3.00)×105 | (2.19±1.20×105 | (3.10±2.06)×105 | 0.2663 | |
| SRB | (3.66±1.48)×102a | (1.79±0.65)×102b | (1.39±0.33)×102b | 0.0028 |
Note: values with different small letter superscripts in the same row mean that there was a significant difference (P<0.05), and the same small letter superscripts mean that there was no difference.
The fecal sample replications in C group n = 10, D group n = 11, D+L group n = 11.
The colon digesta sample replications in C group n = 6, D group n = 5, D+L group n = 6.
The colon mucosa sample replications in C group n = 6, D group n = 5, D+L group n = 6.
Oxidative Biomarkers and Antioxidant Enzymes in the Liver of Barrows
The data for liver oxidative biomarkers and antioxidant enzymes are summarized in Table 5. Hydrogen peroxide (H2O2) levels, inhibiting hydroxyl radical ability, NO concentrations, protein carbonyl, and MDA status were of no significant difference among the C, D, and D+L groups. However, significant effects were found on the total carbonyl and 8-OH-dG levels in the liver of barrows. Total carbonyl and 8-OH-dG levels in the D and D+L groups were significantly lower than in the C group, and no difference was found between D and D+L groups (Table 5). For the oxidant and anti-oxidant relative enzymes, only total-SOD, CuZn-SOD, and Mn-SOD were influenced by dietary treatment. The T-SOD levels in the D+L group were significantly higher than in the C and D groups. The CuZn-SOD enzyme activity in the D+L group was significantly higher than in the D group, and to have no difference compared to the C group. Finally, Mn-SOD activity in the D and D+L groups was significantly higher than in the C group. The CAT, GSH-px, GR, GSH, GSSH, and the ratio of GSH to GSSG were not influenced by dietary treatment (Table 5).
Table 5. Oxidative biomarkers and antioxidant enzymes status in the liver of barrows.
| Items | Control (n = 6) | Daidzein (n = 5) | D+L (n = 6) | P |
| Total carbonyl (mg/g wt liver) | 7.07±0.78a | 4.69±1.10b | 5.09±1.04b | 0.002 |
| Hydrogen Peroxide(H2O2) (mmol/g protein) | 5.77±0.65 | 5.50±0.81 | 5.40±0.88 | 0.699 |
| Inhibiting Hydroxyl Radical (U/mg protein) | 25.15±6.34 | 26.19±3.47 | 30.42±6.52 | 0.280 |
| NO (μmol/g protein) | 0.41±0.04 | 0.32±0.05 | 0.36±0.07 | 0.079 |
| Protein Carbonyl (nmol/mg protein) | 5.16±0.73 | 4.24±0.80 | 4.40±0.44 | 0.076 |
| 8-OH-dG (ng/g wt liver) | 337.00±47.43a | 258.53±75.22b | 244.54±26.67b | 0.017 |
| CAT (U/g protein) | 78.34±10.54 | 78.34±9.68 | 84.23±8.93 | 0.509 |
| Total-SOD (U/mg protein) | 239.08±12.06b | 251.43±15.11b | 299.38±35.46a | 0.002 |
| CuZn-SOD (U/mg protein) | 156.04±13.49ab | 134.32±5.90b | 169.73±29.03a | 0.030 |
| Mn-SOD (U/mg protein) | 83.04±5.36b | 117.11±15.13a | 129.65±14.96a | <0.0001 |
| MDA (nmol/mg protein) | 2.62±0.51 | 2.27±0.59 | 2.11±0.30 | 0.174 |
| GSH-px (activity unit) | 20.08±1.56 | 21.70±2.13 | 20.49±20.70 | 0.417 |
| GR (U/g protein) | 7.34±2.07 | 5.81±1.33 | 6.17±1.50 | 0.310 |
| GSH (μmol/g wt liver) | 0.84±.09 | 0.81±0.10 | 0.77±0.07 | 0.383 |
| GSSG (μmol/g wt liver) | 0.17±0.03 | 0.17±0.02 | 0.19±0.02 | 0.300 |
| GSH/GSSG | 5.1±0.98 | 4.74±0.92 | 4.05±0.34 | 0.090 |
Note: values with different small letter superscripts in the same row mean that there was a significant difference (P<0.05), and the same small letter superscripts mean that there was no difference.
Discussion
Several studies have attempted to manipulate equol production by supplementing prebiotics or probiotics with daidzein or soy isoflavones in humans and animals [27], [28]. Our previous studies showed that 1% (10g/L) of lactulose was able to improve equol production in vitro [13]. In the current study, lactulose showed clear promotion of equol production in the feces and urine of the barrows treated with daidzein.
Hydrogen, in particular, was found to stimulate equol production in an equol-producing mixed microbial culture from a human fecal sample [7]. Previous study investigated the interactions between methanogens, sulphate-reducing, and equol-producing bacteria under in vitro simulated intestinal conditions [8]. Results showed that supplementation equol-producing bacteria EPC4 significantly decreased the methanogenesis and sulphidogenesis in the faecal samples with methanogenic or sulphate-reducing abilities. Daidzein was also found to act as an electron acceptor in equol production that likely diverts hydrogen away from CH4 or H2S formation [8]. Our data suggests that lactulose may be a novel way of promoting equol production. Although the status of hydrogen gas in the colon was not measured, the isobutyric acid, isovaleric acid, branched chain fatty acid (BCFA), and total VFA levels in the colon digesta were improved by lactulose supplement, which has been reported the VFA levels in the colon digesta manipulated by lactulose corresponded with an increase of hydrogen gas production [29].
It is thought that certain bacteria in the intestinal microflora are greatly involved in equol bioconversion. Recently, bacteria that convert daidzein to equol were isolated from human, pig [7], [30]–[33] and rat [34], [35] feces or cecal contents. Although these equol-producing bacteria are different in individual and species, they play the comparable role in improving the greater efficacy of a soy food diet. So our dietary lactulose application to improve intestinal equol production in swine might be applied to human and other animals. To explore the intestinal microflora composition manipulation by lactulose in barrows treated with daidzein as well as the interactions between equol production and hydrogen-utilizing bacteria (methanogenic Archaea and sulfate-reducing bacteria), and due to the fact that equol formation largely occurs in the colon and distal intestine [36], the fecal, colon digesta, and mucosa samples were tested. Cluster results of DGGE profiles obtained from the colon digesta and mucosa showed that daidzein but not lactulose had selective pressure on the community of SRB and MPB, which is aggreed with previous study [8]. Moreover, our data also indicates that the hydrogen-producing prebiotic lactulose might shift the pathways of hydrogen utilization, thus changing the profiles of SRB in feces.
The real-time PCR results showed that the abundance of bacteroidetes and firmicutes in the feces were significantly higher in the D+L group than in the D and C groups. These findings are partly in agreement with our previous study [37], which reported that the fecal population of bacteroidetes in rates treated with daidzein and equol-producing bacterium ZX-7 had a strong correlation with fecal equol concentration. Lactulose was also found to have a reduction effect on the SRB population in the feces and colon mucosa. This result runs contrary to previous results [38] that reported that a strong equol producer phenotype correlated positively with the abundance of sulfate-reducing bacteria. The results from this study indicate a strong relationship between equol production and hydrogen-utilization bacteria. However, the relationship between equol-producing, hydrogen-utilizing, and the selected bacteria seems to be more complex than previously suggested [7], [8]. Further research is needed to explain the exact mechanism that lactulose serve as a hydrogen-producing prebiotic to enhance equol production in vivo.
It is well established that isoflavones have estrogenic and antioxidant capacities [3]. The conversion of daidzein to equol by intestinal microflora may be physiologically important, as equol has significantly greater antioxidant activity and estrogenic activity compared to daidzein [39]. In our experiment, dietary supplements with 1% lactulose significantly increased the T-SOD and CuZn-SOD activity in the liver of barrows treated with daidzein (Table 5). The antioxidant ability of pure equol in humans, cells, and rats is widely established [40], [41]. However, to our knowledge, no information is available on the effects of a hydrogen-producing prebiotic that increases equol production and improves antioxidants status in vivo. The effects of equol on oxidative stress and the antioxidant defense system in the livers of mice suggests that equol may act as an antioxidant through an inhibition of oxidative stress and the stimulation of catalase and SOD [40]. Considering our experimental term was only 22 days and the forming of equol in barrows’ intestinal was sustained and in a slow rate, and that the homeostasis of oxidative and antioxidant status is relatively stable in healthy pigs, it is not surprising that only T-SOD and CuZn-SOD activity in the liver were improved by lactulose manipulation. The results also indicated that using a hydrogen-producing prebiotic as an equol-promoting activator may be a novel way to improve the bioavailability of daidzein, which may be of benefit to broader populations of humans and animals.
In conclusion, we have shown that dietary supplemental lactulose can significantly improve equol-producing ability by changing microbial profile, the population and abundance of selected bacteria, especially the SRB population in the digesta and mucosa of colon. In addition, lactulose results in SCFAs profile shifting in the colon digesta. Furthermore, the addition of lactulose in the barrows’ diet improved the antioxidant status of the livers of barrows treated with daidzein, showing the potential of the hydrogen-producing prebiotic lactulose for enhancing equol production in animals and humans.
Supporting Information
Formula composition and nutrient levels of animal diet.
(DOCX)
Primer sequences used in this study.
(DOCX)
Acknowledgments
We thank Prof. Xiaojing Yang for her general advice, we also thank the technical assistance from Yingjie Xu.
Funding Statement
This study was funded by Ph.D. Programs Foundation of Ministry of Education of China (B0201100681), Scientific Research Foundation for Returned Overseas Chinese Scholars (G0201100315) and the Special Fund for Agro-scientific Research in the Public Interest (201003011). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Setchell KD, Clerici C (2010) Equol: history, chemistry, and formation. J Nutr 140: 1355S–1362S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bowey E, Adlercreutz H, Rowland I (2003) Metabolism of isoflavones and lignans by the gut microflora: a study in germ-free and human flora associated rats. Food Chem Toxicol 41: 631–636. [DOI] [PubMed] [Google Scholar]
- 3. Rimbach G, De Pascual-Teresa S, Ewins BA, Matsugo S, Uchida Y, et al. (2003) Antioxidant and free radical scavenging activity of isoflavone metabolites. Xenobiotica 33: 913–925. [DOI] [PubMed] [Google Scholar]
- 4. Blay M, Espinel AE, Delgado MA, Baiges I, Blade C, et al. (2010) Isoflavone effect on gene expression profile and biomarkers of inflammation. J Pharm Biomed Anal 51: 382–390. [DOI] [PubMed] [Google Scholar]
- 5. Possemiers S, Bolca S, Eeckhaut E, Depypere H, Verstraete W (2007) Metabolism of isoflavones, lignans and prenylflavonoids by intestinal bacteria: producer phenotyping and relation with intestinal community. FEMS Microbiol Ecol 61: 372–383. [DOI] [PubMed] [Google Scholar]
- 6. Setchell KD, Brown NM, Lydeking-Olsen E (2002) The clinical importance of the metabolite equol-a clue to the effectiveness of soy and its isoflavones. J Nutr 132: 3577–3584. [DOI] [PubMed] [Google Scholar]
- 7. Decroos K, Vanhemmens S, Cattoir S, Boon N, Verstraete W (2005) Isolation and characterisation of an equol-producing mixed microbial culture from a human faecal sample and its activity under gastrointestinal conditions. Arch Microbiol 183: 45–55. [DOI] [PubMed] [Google Scholar]
- 8. Bolca S, Verstraete W (2010) Microbial equol production attenuates colonic methanogenesis and sulphidogenesis in vitro. Anaerobe 16: 247–252. [DOI] [PubMed] [Google Scholar]
- 9. Florent C, Flourie B, Leblond A, Rautureau M, Bernier JJ, et al. (1985) Influence of chronic lactulose ingestion on the colonic metabolism of lactulose in man (an in vivo study). J Clin Invest 75: 608–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.NRC (1998) Nutrient requirements of swine, tenth ed. National Academines Press, Washington, DC.
- 11. Maubach J, Bracke ME, Heyerick A, Depypere HT, Serreyn RF, et al. (2003) Quantitation of soy-derived phytoestrogens in human breast tissue and biological fluids by high-performance liquid chromatography. J Chromatogr B Analyt Technol Biomed Life Sci 784: 137–144. [DOI] [PubMed] [Google Scholar]
- 12. Xu X, Wang HJ, Murphy PA, Cook L, Hendrich S (1994) Daidzein is a more bioavailable soymilk isoflavone than is genistein in adult women. J Nutr 124: 825–832. [DOI] [PubMed] [Google Scholar]
- 13. Zheng W, Hou Y, Su Y, Yao W (2014) Lactulose promotes equol production and changes the microbial community during in vitro fermentation of daidzein by fecal inocula of sows. Anaerobe 25: 47–52. [DOI] [PubMed] [Google Scholar]
- 14. Nubel U, Engelen B, Felske A, Snaidr J, Wieshuber A, et al. (1996) Sequence heterogeneities of genes encoding 16 S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J Bacteriol 178: 5636–5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cheng YF, Mao SY, Liu JX, Zhu WY (2009) Molecular diversity analysis of rumen methanogenic Archaea from goat in eastern China by DGGE methods using different primer pairs. Lett Appl Microbiol 48: 585–592. [DOI] [PubMed] [Google Scholar]
- 16. Deplancke B, Hristova KR, Oakley HA, McCracken VJ, Aminov R, et al. (2000) Molecular ecological analysis of the succession and diversity of sulfate-reducing bacteria in the mouse gastrointestinal tract. Appl Environ Microbiol 66: 2166–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhu W-Y, Williams BA, Konstantinov SR, Tamminga S, De Vos WM, et al. (2003) Analysis of 16 S rDNA reveals bacterial shift during in vitro fermentation of fermentable carbohydrate using piglet faeces as inoculum. Anaerobe 9: 175–180. [DOI] [PubMed] [Google Scholar]
- 18. Sanguinetti CJ, Dias Neto E, Simpson AJ (1994) Rapid silver staining and recovery of PCR products separated on polyacrylamide gels. Biotechniques 17: 914–921. [PubMed] [Google Scholar]
- 19.Lane D (1991) 16 S/23 S rRNA sequencing. Nucleic acid techniques in bacterial systematics.
- 20. Suzuki MT, Taylor LT, DeLong EF (2000) Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5'-nuclease assays. Appl Environ Microbiol 66: 4605–4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Layton A, McKay L, Williams D, Garrett V, Gentry R, et al. (2006) Development of Bacteroides 16 S rRNA gene TaqMan-based real-time PCR assays for estimation of total, human, and bovine fecal pollution in water. Appl Environ Microbiol 72: 4214–4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Denman SE, Tomkins NW, McSweeney CS (2007) Quantitation and diversity analysis of ruminal methanogenic populations in response to the antimethanogenic compound bromochloromethane. FEMS Microbiol Ecol 62: 313–322. [DOI] [PubMed] [Google Scholar]
- 23. Mao S, Zhu W, Wang Q, Yao W (2007) Effect of daidzein on in vitro fermentation by microorganisms from the goat rumen. Animal feed science and technology 136: 154–163. [Google Scholar]
- 24. Hane BG, Jager K, Drexler HG (1993) The Pearson product-moment correlation coefficient is better suited for identification of DNA fingerprint profiles than band matching algorithms. Electrophoresis 14: 967–972. [DOI] [PubMed] [Google Scholar]
- 25.Shannon-Weaver (1963) Mathematical theory of communication: University Illinois Press.
- 26. Konstantinov SR, Zhu WY, Williams BA, Tamminga S, Vos WM, et al. (2003) Effect of fermentable carbohydrates on piglet faecal bacterial communities as revealed by denaturing gradient gel electrophoresis analysis of 16 S ribosomal DNA. FEMS Microbiol Ecol 43: 225–235. [DOI] [PubMed] [Google Scholar]
- 27. Tousen Y, Abe F, Ishida T, Uehara M, Ishimi Y (2011) Resistant starch promotes equol production and inhibits tibial bone loss in ovariectomized mice treated with daidzein. Metabolism 60: 1425–1432. [DOI] [PubMed] [Google Scholar]
- 28. Tousen Y, Uehara M, Kruger MC, Ishimi Y (2012) Effects of dietary fibre and tea catechin, ingredients of the Japanese diet, on equol production and bone mineral density in isoflavone-treated ovariectomised mice. J Nutr Sci 1(e13): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gibson GR, Cummings JH, Macfarlane GT, Allison C, Segal I, et al. (1990) Alternative pathways for hydrogen disposal during fermentation in the human colon. Gut 31: 679–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ueno T, Uchiyama S (2001) Identification of the specific intestinal bacteria capable of metabolising soy isoflavone to equol. Ann Nutr Metab 45: 114. [Google Scholar]
- 31. Wang XL, Kim HJ, Kang SI, Kim SI, Hur HG (2007) Production of phytoestrogen S-equol from daidzein in mixed culture of two anaerobic bacteria. Arch Microbiol 187: 155–160. [DOI] [PubMed] [Google Scholar]
- 32. Yu ZT, Yao W, Zhu WY (2008) Isolation and identification of equol-producing bacterial strains from cultures of pig faeces. FEMS Microbiol Lett 282: 73–80. [DOI] [PubMed] [Google Scholar]
- 33. Jin JS, Kitahara M, Sakamoto M, Hattori M, Benno Y (2010) Slackia equolifaciens sp. nov., a human intestinal bacterium capable of producing equol. Int J Syst Evol Microbiol 60: 1721–1724. [DOI] [PubMed] [Google Scholar]
- 34. Minamida K, Tanaka M, Abe A, Sone T, Tomita F, et al. (2006) Production of equol from daidzein by gram-positive rod-shaped bacterium isolated from rat intestine. J Biosci Bioeng 102: 247–250. [DOI] [PubMed] [Google Scholar]
- 35. Minamida K, Ota K, Nishimukai M, Tanaka M, Abe A, et al. (2008) Asaccharobacter celatus gen. nov., sp. nov., isolated from rat caecum. Int J Syst Evol Microbiol 58: 1238–1240. [DOI] [PubMed] [Google Scholar]
- 36. Setchell KD, Borriello SP, Hulme P, Kirk DN, Axelson M (1984) Nonsteroidal estrogens of dietary origin: possible roles in hormone-dependent disease. Am J Clin Nutr 40: 569–578. [DOI] [PubMed] [Google Scholar]
- 37. Zhang X, Zheng W, Huang S, Yao W (2012) Microbial conversion of daidzein affects fecal equol concentration and bacterial composition of rats with or without ovariectomy. Wei sheng wu xue bao = Acta microbiologica Sinica 52: 866–874. [PubMed] [Google Scholar]
- 38. Bolca S, Possemiers S, Herregat A, Huybrechts I, Heyerick A, et al. (2007) Microbial and dietary factors are associated with the equol producer phenotype in healthy postmenopausal women. J Nutr 137: 2242–2246. [DOI] [PubMed] [Google Scholar]
- 39. Setchell KD, Clerici C (2010) Equol: pharmacokinetics and biological actions. J Nutr 140: 1363S–1368S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Choi EJ (2009) Evaluation of equol function on anti- or prooxidant status in vivo. J Food Sci 74: H65–71. [DOI] [PubMed] [Google Scholar]
- 41. Wei XJ, Ni YD, Lu LZ, Grossmann R, Zhao RQ (2011) The effect of equol injection in ovo on posthatch growth, meat quality and antioxidation in broilers. Animal 5: 320–327. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Formula composition and nutrient levels of animal diet.
(DOCX)
Primer sequences used in this study.
(DOCX)