Abstract
Recent advances in the burgeoning field of genome engineering are accelerating the realization of personalized therapeutics for cardiovascular disease. In the post-genomic era, sequence-specific gene-editing tools enable the functional analysis of genetic alterations implicated in disease. In partnership with high-throughput model systems, efficient gene manipulation provides an increasingly powerful toolkit to study phenotypes associated with patient-specific genetic defects. Herein, this review emphasizes the latest developments in genome engineering and how applications within the field are transforming our understanding of personalized medicine with an emphasis on cardiovascular diseases.
Keywords: TALENs, zebrafish, iPS cells, genome engineering, cardiovascular disease modeling
Introduction
Cardiovascular disease affects approximately 83.6 million American adults and is the leading cause of death in the United States.1 Recent advances in the ease and affordability of genome sequencing have illuminated a spectrum of genetic underpinnings linked to various cardiovascular diseases.2–4 From the initial discovery of genetic mutations, further study of the molecular pathways underlying cardiac dysfunction utilizes both in vitro and in vivo model systems. However, unprecedented numbers of candidate genes from whole-genome screens commonly overwhelm the capacity of downstream functional analyses. The prioritization of candidate mutations and identification of causative mutations will require the continued development of high-throughput tools and platforms to efficiently and effectively assess candidate genes. Genomic engineering is a rapidly evolving field that can now facilitate gene manipulation in a sequence-specific manner in most genomic contexts. The disruptive nature of this technology, when coupled with high-throughput model systems, is accelerating the study of cardiovascular disease as well as the development of personalized therapeutics by revealing innate genotype-phenotype mechanisms.
With site-specific gene editing, researchers manipulate experimental model systems by modulating the genomic mutation of interest. Novel genetic toolkits can be employed to generate targeted gene lesions or ‘knock-outs’, genomic-based reporter systems, and single nucleotide changes at gene loci across the genome. One prominent gene-editing strategy harnesses the sequence-specificity of transcription activator-like effector nucleases (TALENs) to introduce site-specific mutations in gene loci-of-interest.5 TALENs are attracting well-deserved attention for their simplicity and flexibility of design as well as their gene-targeting efficiency.6 TALEN technology can be maximized in the setting of personalized cardiovascular research when paired with high-sensitivity and high-throughput cell- and tissue-based model systems.
Zebrafish (Daniorerio) offer an established vertebrate model that is well positioned to capitalize on TALEN-based precision engineering. One pair of adult zebrafish can produce 100+ embryos weekly, and the relatively inexpensive maintenance of fish colonies facilitates a large number of specimens to be mutagenized, characterized, and raised. Once the appropriate mutations are introduced, the cardiac phenotype can be studied in fish larvae, as zebrafish undergo full cardiac development within 48 hours, or in the adult fish for cardiovascular diseases that develop over an extended time. Importantly, zebrafish bring the power of forward genetic screens into a vertebrate system.
TALEN technology also enables genetic manipulation of induced pluripotent stem (iPS) cells. Characterized by pluripotent capacity and unlimited self-renewal, iPS cells can provide a long-term cell culture platform with the ability to generate functional cardiomyocytes from primitive to mature developmental stages. Importantly, iPS cells can be derived from patients with a variety of cardiovascular diseases and therefore offer a novel way to model disease in vitro. Genetic manipulation of iPS cells via TALENs promises to further advance the utility of this platform to interrogate cell-autonomous genotype-phenotype relationships in the absence of physiological compensation. In addition, gene correction of patient-derived iPS cells using TALENs could generate “disease-free” autologous cells with potential therapeutic promise.
In this review, we highlight the technical aspects of TALEN design and assembly as well as current applications of TALEN technology in the study of cardiovascular diseases. With a focus on zebrafish and pluripotent stem cell model systems, we will explore the advantages of both “disease-in-a-fish” and “disease-in-a-dish” applications. Our objective is to highlight how TALEN-based genome engineering in partnership with biological model systems accelerates the advancement of personalized medicine for cardiovascular diseases.
Origin of TALENs
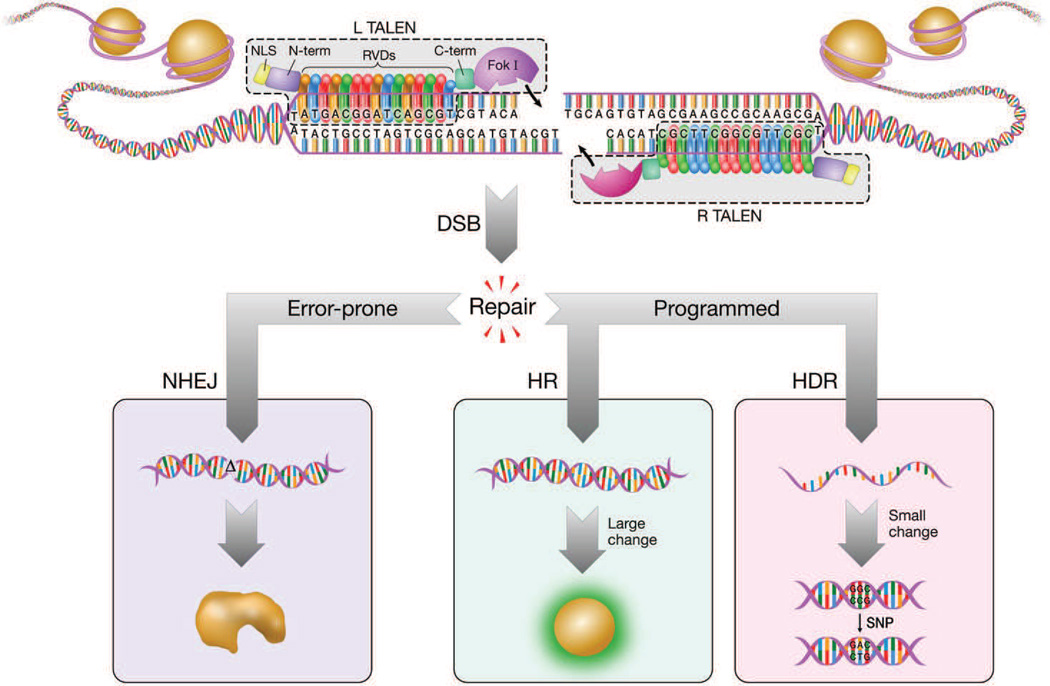
Transcription activator-like (TAL) proteins were originally discovered by studying secreted proteins of the bacterial plant pathogen Xanthomonas and named for their innate ability to activate transcription of endogenous plant genes essential for pathogenicity.7 The DNA binding domain of TAL proteins was found to comprise several 33–35 amino acid repeats with differences at residues 12 and 13, a position that was named the repeat variable di-residue (RVD).8 A one-to-one correspondence between the RVDs and nucleotide binding was discovered, a structural feature used as the basis for the assembly of predictable DNA binding domains (NI = A, NN/NK = G, NG = T, HD = C).9, 10 The last RVD in the string of repeats is only similar to the others in its first 20 amino acids, and is therefore often referred to as a half-repeat. For simplification in this review, the half-repeat will be included as a whole RVD in the final TAL protein targeting length (i.e. 14.5 repeats= 15 RVDs). Like zinc finger nucleases before11, TAL proteins were fused to a type IIS endonuclease, FokI, assembling the transcription activator-like effector nucleases, TALENs, a new class of artificial targeted DNA nucleases (Figure 1).5, 12
Figure 1.
Overview of TALEN genome engineering. Left and right TALEN arms are directed to a specific sequence of genomic DNA by the RVD repeats comprising the DNA binding domain. Specificity is achieved by the one-to-one interactions between the RVDs and nucleotides, as well as a 5’T that is recognized by the N-terminus of the TAL protein. The length of the TALEN arms and spacer region contribute to TALEN activity; here is shown a 15-15-15 (left arm-spacer-right arm) design that has been shown to be effective.25 The C-terminus domain brings the FokI obligate heterodimer (or homodimer) endonuclease in proximity to dimerize. The resulting double-strand break (DSB) is repaired by either error-prone non-homologous end joining (NHEJ) or programmed gene repair by homologous recombination (HR) or homology-directed repair (HDR) facilitated by an exogenous DNA donor. Repair by NHEJ causes unpredictable insertions/deletions (indels) at the cut site, which can lead to protein loss-of-function. Repair by HR requires long, double-stranded DNA donors that can be used to direct large changes in the genome, such as the fluorescent labeling of an endogenous protein by GFP. Conversely, HDR using short, single-stranded oligonucleotides can be used to make small, subtle changes like a single nucleotide polymorphism (SNP). NLS = nuclear localization sequence; RVD = repeat variable di-residue.
Harnessing DNA repair mechanisms for genome engineering
TALENs enable locus-specific mutagenesis by inducing targeted double-strand breaks (DSBs) after two TALEN arms each bring half of the FokI endonuclease within proximity to dimerizein the spacer, or the stretch of DNA between the TALEN arms (Figure 1).5 Eukaryotic cells repair the resulting DSB by either error-prone non-homologous end joining (NHEJ), resulting in insertions or deletions (indels) at the cut site, or by homologous recombination (HR) or homology-directed repair (HDR) in the presence of anexogenous DNA donor (Figure 1).13 If targeted to a conserved exon, the indels from NHEJ may induce a frameshift mutation that can result in a protein knock-out, a useful tool that has broad applications to any gene-of-interest.14
In contrast to NHEJ, both HR and HDR require exogenous DNA in the form of double-stranded DNA (dsDNA; plasmid, adenoviral vector, PCR amplicon, etc.) or a synthesized single-stranded oligodeoxyribonucleotide (ssODN)to serve as a template for programming of site-specific knock-ins(Figure 1). The choice of this ‘donor’ DNA depends on the experiment and model system. In general, to introduce larger-stretches of DNA changes such as coding exons or artificial expression cassettes, dsDNA with long homologous arms that will induce HR is effective.15 For example, researchers are able to make substantial changes to the genome that may include fluorescent labeling of a protein to better understand its function. In the context of cardiovascular disease modeling, increased understanding of protein function may provide insight into the mechanism of disease and inform the development of novel therapeutics. Efficacy can vary greatly between model systems or even between loci within the same system; furthermore, HR is a unanimously rarer event than NHEJ-based mutagenesis due to the much slower efficiency and speed of this mechanism of repair.16
While smaller DNA changes may be accomplished using traditional HR, there is a growing appreciation for using ssODNs for this application (Figure 1).17 The efficiency of editing using ssODNs can be high enough to not necessarily require enrichment even when used in vivo.18 To date, ssODNs have been used to introduce short DNA fragments such as single nucleotide polymorphisms (SNPs), small insertions, or epitope tags.18–21 Unlike dsDNA, which is used as a template in the canonical HR pathway, ssODNs appear to use a related but distinct homology-directed repair mechanism that is not well understood. One hallmark of this HDR pathway is the insertion imprecision that can arise by using ssODNs in zebrafish,18 mammalian cell culture,21 and mouse embryos.22 This asymmetric repair process, however, has not been reported in all studies using ssODNs as HDR donors.20, 23 Gaining a better understanding of this HDR mechanism, especially in in vivo models such as zebrafish, is an important next step in the field. In particular, given the ease of ssODN synthesis and delivery compared to dsDNA, ssODNs may be more amenable to the high-throughput genome editing required to screen the numerous candidate genes identified in whole-genome sequencing projects.
TALEN scaffold
TALEN proteins can be conceptually broken down into the DNA binding domain (composed of RVDs), the N- and C-termini of the protein, and the FokI endonuclease domain (Figure 1). The repeats that make up the RVD domain can be assembled in multiple ways, which will be discussed further below. For clarity, the N-, C-termini, and FokI domain together will be referred to as the “TAL scaffold” in this review.
Interestingly, there has been a divergence of TAL proteins chosen to make TALENs. The difference between the proteins is species-specific; one is originally from Xanthomonas oryzae, a rice pathogen, and another is from Xanthomonas axonopodis, a soybean pathogen.24 While both are effective mediators of their respective pathogens, there are minor amino acid substitutions between the proteins that may or may not impact artificial TALEN efficiencies.18, 25 The natural TAL scaffold that originated from X. axonopodis was originally nicknamed TALE13 and was the first to undergo an architecture change of the N- and C-termini, which was found to improve activity.26 The original scaffold using the X. oryzae TAL protein, pTAL27, has also undergone similar architectural changes. Table 1 describes commonly used TALEN scaffolds in each of these classes to provide insight into the critical characteristics of TALEN scaffolds.
Table 1.
TALEN Scaffolds
| Species | Architecture† | FokI | Epitope Tags |
Promoters | Addgene Name |
Assembly compatibility |
Reference |
|---|---|---|---|---|---|---|---|
| Xanthomona saxonopodis (TALE1326) | NΔ152/C63 | WT | 3X Flag | CMV and T7 | JDS 70, 71, 74, 78 | FLASH, REAL, REAL-Fast | 26 |
| NΔ152/C63 | EL/KK or ELD/KKR35 | 3X Flag | CMV and T7 | N/A | FLASH, REAL, REAL-Fast | 106 | |
| NΔ152/C63 | RR/DD33 or AS/RR34 | 3X Flag and HA | CMV and SP6 | N/A | Unit Assembly | 44 | |
| NΔ152/C63 | WT | 3X Flag | CMV and T7 | TALEN-ID12-A (C, T, and G) | Ligation- independent cloning | 42 | |
| Xanthomona soryzae (pTAL27) | NΔ152/C63 | WT | None | miniCAGGs or T3 | pC-GoldyTALEN, RCIscript-GoldyTALEN | Golden Gate 2.0 | 18,67 |
| NΔ152/C63 | RR/DD | HA or Flag | CMV and SP6 | pCS2TAL3RR, pCS2TAL3DD | Golden Gate 2.0 | 29 | |
| NΔ134/C63 | WT or ELD/KKR and AS/RR | None | CAG and T7 | pCAG-T7-TALEN(Sangamo)- Destination, -FokI-KKR-Destination, -FokI-ELD-Destination | Golden Gate 2.0 | N/A | |
| NΔ134/C47 | WT | Flag | CMV and T7 or CAG | pcDNA-TAL-NC2, pCAGGS-TAL-NC2 | Golden Gate 2.0 | 43 | |
| C63* | WT | 3X Flag | CMV | pTALEN_v2 (NI), (NN), (HD), and (NI) | Hierarchical PCR/ligation | 68 |
= N-terminal truncation not reported
= All TALEN scaffolds have a nuclear localization sequence at the N-terminus
The primary overt differences between scaffolds are based on a combination of four factors: the expression system, the epitope tag(s), the N- and C-termini sequences, and the FokI domain. It should also be noted that all TAL scaffolds have a nuclear localization sequence (NLS) that traffics these artificial proteins to the nucleus. The choice of promoter will determine whether constitutive expression of a DNA plasmid post-gene transfer (CMV or CAG) or invitro transcription of RNA (T7, T3, or SP6) and subsequent delivery will be used in the model-of-interest. There is an emphasis placed on the promoter choice when deciding on an expression construct, but it is important to remember there may also be different untranslated regions or polyadenylation signals that may impact TALEN expression. Traditionally, RNA injection is most common in zebrafish, whereas DNA transfection/transduction is most often used in mammalian cell lines. However, due to the high mutagenesis caused by nuclease activity, a transient burst of expression may be more desirable for TALEN applications, and therefore RNA delivery to cells is an on-going area of investigation.
For functional studies to characterize protein activity, epitope tags are commonly added to the TAL scaffold. While the primary reason for this addition is convention, as the TAL scaffolds are often cloned into pre-existing plasmids that already include the epitope tag, it is unclear whether or not this feature impacts TALEN activity. The most commonly included epitopes are the hemagglutinin (HA) and flag (or 3x flag) tag (Table 1). The architecture of the TAL scaffold includes truncations of the N- and C-termini, most commonly a 152 amino acid truncation of the N-terminus with a 63 amino acid C-terminus (NΔ152/C63) that was initially established by Miller et al.18 There is a balance in how an architecture change impacts TALEN activity versus the specificity of binding. For example, shortening the C-terminus appears to restrict the TALEN spacer length required for activity (described below), which may increase the specificity of the TALEN. However, there have also been reports of a shorter C-terminus having more14, 28 and less26 activity than the C63 design. Keeping this balance in mind, the preferred scaffold architecture for different models and applications is a continuing area of research.
An important consideration of any homing nuclease is off-target mutagenesis. TALENs appear to have minimal off-target effects,15, 23, 29, 30 particularly when compared to ZFNs.31 Mutations that have been detected thus far are at loci with high homology to the target, and there is much work to be done to appreciate the rate of genome-wide off-targeting of TALENs. One approach to further reduce off-targeting has been to alter the catalytic properties of the FokI endonuclease. Wild type FokI requires homodimerization for activity, and in an attempt to improve specificity obligate Fok1 heterodimers were developed by changing their amino acid composition.32 Today, there are several heterodimer FokI variants32–35 and TALEN constructs with eitherhomodimer or heterodimer FokI domains.
While epitope tags facilitate protein analysis and the FokI heterodimer may reduce off-target effects, the impact of scaffold modifications on TALEN activity continues to be elucidated. The efficiency differences between architecture changes, notably the truncation of the C-terminus, have been well documented. The GoldyTALEN scaffold, for example, has been demonstrated to improve activity over the original, full-length pTAL scaffold.18 It should be noted, however, that it is difficult to compare activities of different scaffolds with the same architecture both within and between model systems, as different groups often use different methods of quantification and few direct activity comparisons have been made. Nonetheless, Table 3 compiles the TALEN scaffolds used and the reported efficacy in zebrafish and stem cells to date. In zebrafish, it should be noted that both somatic and germline efficiencies using published TALEN scaffolds are sufficient to generate mutants. Furthermore, the efficiency of any homology-directed repair is markedly less efficient than NHEJ mutagenesis in this in vivo system. In stem cells, mutation rates tend to be lower; however co-selection using puromycin or GFP can drastically improve activity (Table 3).Perhaps the most important conclusion that can be drawn is that there is great variability in TALEN activity and, in general, each scaffold has the potential to produce high activity. However, it is becoming clear is that the design of the TALEN in combination with the scaffold choice can be optimized to improve the average activity.25 While Table 3 provides a snapshot of current progress in this field, it also suggests there is room for further optimization.
Table 3.
Efficacy of TALEN scaffolds
| Model System |
TALEN Scaffold | Assembly Protocol |
TALEN Efficiency Notes |
Reference |
|---|---|---|---|---|
| Zebrafish | JDS series | REAL Assembly | 0–55% somatic mutation rate, 10–100% germline efficiency (11 loci) | 39 |
| GoldyTALEN | Golden Gate | 7–100% somatic mutation rate, 17–100% germline efficiency (5 loci) – HDR | 18 | |
| GoldyTALEN | Golden Gate | 24–86% somatic mutation rate, 18–100% germline efficiency (10 loci) | 25 | |
| TALE13 NΔ152/C63-FokI AS/RR | Unit Assembly | 9–33% germline efficiency (3 loci) | 44 | |
| JDS series | “Iterative assembly” | 11–33% somatic mutation rate (4 loci) | 158 | |
| TALE13 NΔ152/C63 - FokI AS/RR | Unit Assembly | 70–98% somatic mutation rate (1 locus) – HR | 107 | |
| pCS2TAL3-DD, pCS2TAL3-RR | Golden Gate | 98–100% somatic positive embryos, 77–100 germline efficiency (4 loci) | 29 | |
| JDS series | FLASH | 20–77% somatic mutation rate (6 loci) | 159 | |
| JDS-FokIEL/KK or ELD/KKR* | FLASH | 0–76% somatic mutation rate, 8–63% (10 loci), germline efficiency (8 loci) | 106 | |
| Human Stem cells | JDS series | REAL Assembly | HR = 100% with co- selection† (1 loci) | 152 |
| pTAL-Δ152/C63- FokI ELD/KKR and AS/RR | Modified Golden Gate | NHEJ = 2% to 34% HDR = 1.6% without co- selection (16 loci) | 23 | |
| JDS series | DNAWorks160 | HR = 1–100% with co-selection† (5 loci) | 15 |
= Heterodimer FokI with JDS scaffold
= HR-positive cells were enriched for by co-selecting with puromycin or GFP
Considerations for TALEN design
While TALEN sequence targeting has few known absolute constraints, some best-practice rules governing the efficiency of mutagenesis have been identified. Early in silico data provided the first insights into the importance of some key components. For example, endogenous TAL protein sequence targets were preceded by a 5’ thymine (5’T) due to an interaction between the N-terminus of the TAL protein and the 5’T.36 There have been reports emphasizing25, 37 and negating26, 38, 39 the importance of the 5’T when designing the targeted nuclease, however. Most current TALEN design tools recommend including the 5’T.
In addition, the length of both the TALEN arms and spacer region impacts efficiency. The length of the TALEN binding domain has a minimal requirement of 11 RVDs for activity.9 Depending on the scaffold, different groups have generated TALENs with binding arms ranging from 13–40 RVDs. To date, only three groups have systematically tested the effect of arm length on TALEN activity.9, 38, 40 Collectively, 14–20 RVDs are sufficient to maintain high activity with minimal toxicity. More specifically, one study suggests that 15-RVD TALEN pairs maintain high activity with a spacer length between 13 and 20 basepairs.25 There is currently no evidence that longer sequences reduce off-target effects; and structural evidence suggests that RVDs closer to the N-terminus may play a larger role in binding than more C-terminal RVDs.41
In contrast to the 5’T recommendation for the target sequence and the nucleotide binding length that may to be applicable to most TALEN systems, the optimal spacer length can be TALEN scaffold-specific. Different backbones have different C-termini lengths that modulate the proximity between the Fok1 nuclease domains and impact dimerization. Conservatively, a spacer region from 14–20 basepairs will lead to efficient TALEN activity for scaffolds with a C-terminus length of 63 amino acids or above.5, 25, 26, 28, 31, 42, 43 Other scaffold architectures with even more truncated C-termini, from 17–28 amino acids, function more efficiently with a shorter spacer of 12–14 basepairs.14, 26, 28, 31
RVD designs are based on several naturally occurring RVDs, each with differing specificities and binding strengths. Three of the four nucleotides (A, C, and T) have established RVDs that are commonly used (NI, HD, and NG, respectively).6 There is less consensus on how best to target guanine. NN, one of the first RVDs identified to target G, will also recognize and bind A.9, 10, 26 The other early-identified RVD targeting G, NK, appears to be more specific, but incorporation of NK instead of NN leads to a lower activity TALEN.28, 44 Subsequently, NH was identified, which appears to be more specific to G than either NK or NN.45 The impact of using NH in place of NN or NK has not been thoroughly documented; however, one study suggests it has higher specificity but lower activity than NN, much like NK.46 An additional consideration is that TALEN activity can be more influenced by CpG methylation compared to zinc finger nucleases (ZFNs).39, 47 To overcome this issue, NG, HG, and N* RVDs (* = deletion of residue 13) are able to bind 5-methyl cytosine efficiently and should be considered when designing TALENs to regions of high CpG methylation.48, 49 To make identifying target TALEN sites simple, software that can probe DNA for potential target sequences using several of the rules governing TALEN design has been developed.50–52 After identifying the target region within the locus-of-interest, the RVD repeats can be systematically assembled using established protocols.
TALEN DNA binding domain assembly
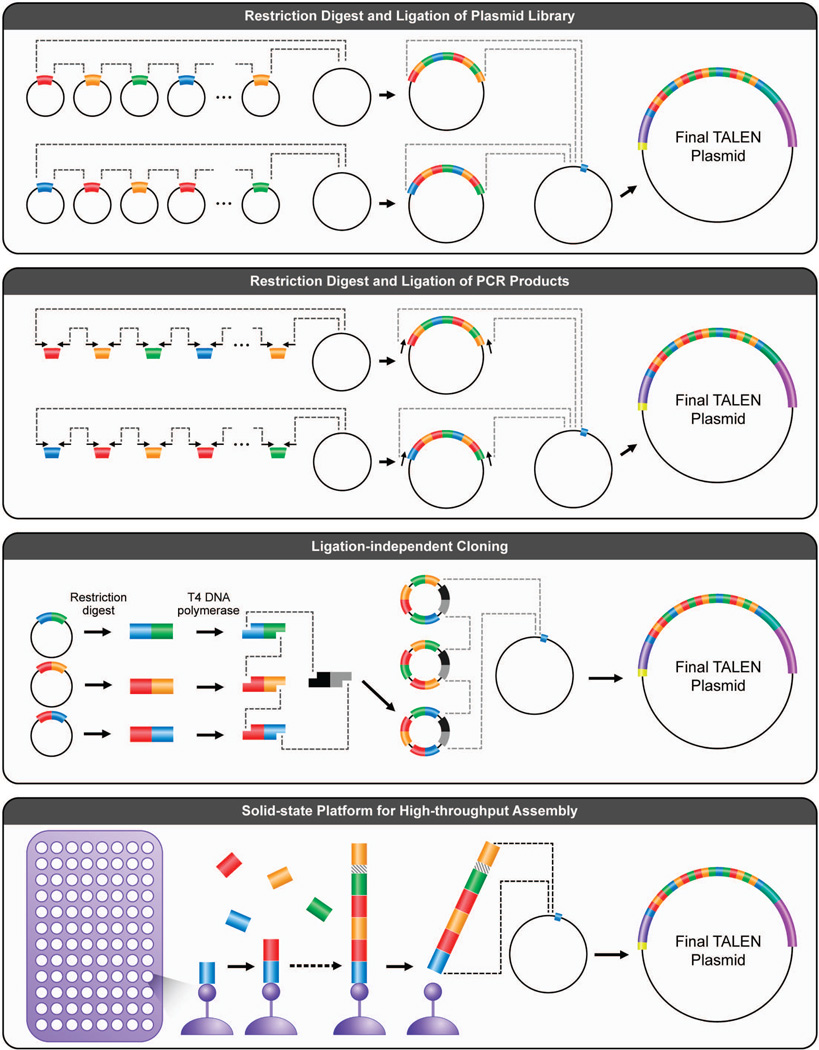
Several methods have been developed to generate the RVD modules of the TAL protein DNA binding domain prior to insertion into the TAL scaffold (for references, see Table 2). Broadly, the methodologies can be broken down into four unique classes: plasmid-based restriction digest and ligation, PCR-based restriction digest and ligation, ligation-independent assembly, and solid-state assembly (Figure 2). Each of the diverse assembly methods may encompass one or more of these classes, and the TAL scaffolds that complement each method are not always interchangeable; the assembly will dictate the selection of TAL scaffolds and vice-versa (Table 1). Table 2 is a list of common assembly options, including the method used to build repeats, the minimum length of time to build, and the targeting length limitations. In addition, the availability of the method on the non-profit plasmid repository, Addgene, and the corresponding name has been included for ease of identification.
Table 2.
TALEN Assembly Methods
| Class/Name | Description | Minimum Length of Protocol† |
Size RVD assembly (bps) |
Addgene Name | Reference |
|---|---|---|---|---|---|
| Golden Gate (GG) | Original Golden Gate | 5 days | 12–31 | Golden Gate 2.0 | 27 |
| Modified Golden Gate | Original GG with FusB4 library | 5 days | 15 | TBA | 25 |
| Original GG with 6-RVD FusA | 5 days | 12–31 | TALEN construction and evaluation accessory pack | 43 | |
| Original GG with tetramer/trimer library | 3 days | 15 | N/A | 23 | |
| GG/PCR assembly of multimers | 3 days | 14–19 | N/A | 156 | |
| GG tetramer/dimer/monomer library | 3 days | 15–19 | N/A | 14 | |
| Hierarchical PCR/ligation | GG followed by PCR/ligation (human codon optimized) | 3 days | 14–25 | Zhang lab TALE Toolbox | 68 |
| Modular Assembly | GG-like/8-mer assembly | 5 days | 16 or 24 | N/A | 157 |
| Unit Assembly | Units assembled via icosaudamer restriction digest/ligation | 7 days | 10–17 | N/A | 44 |
| FLASH | Libraries of 2-, 3-, and 4-mer repeats/solid-phase magnetic beads | 3 days | Unlimited | N/A | 40 |
| REAL* | Restriction digest/ligation (non-GG) | 9 days | 13–17 | Joung Lab REAL assembly TALEN kit | 54 |
| REAL-Fast* | FLASH libraries/cloning hybrid | 7 days | 13–17 | N/A | 54 |
| Ligation-independent cloning | T4 DNA polymerase endonuclease activity to assemble multimers | 3 days | 16 or 19 | LIC TAL effector assembly kit | 42 |
| Iterative Cap Assembly | Sequential construction using monomers on solid- state beads | 3 days | Unlimited | N/A | 38 |
= For TALENs longer than 17 RVDs, two additional days are needed
= Differences in lengths a result of the number of overnight bacterial cultures
Figure 2.
Classes of TAL repeat assembly methodologies. The top panel shows a plasmid-based restriction digest and ligation method that moves TAL repeat monomers (in blue, red, yellow and green) in a pre-determined order from one plasmid to a receiver plasmid by one round of restriction digest and ligation, and then into the destination TAL backbone by another round. The second panel from the top shows a PCR-based restriction digest and ligation method that is conceptually similar to a plasmid-based assembly, however a PCR amplicon replaces whole plasmids as the starting material, and PCR amplification can replace bacterial plasmid propagation. The restriction sites necessary for the digest and ligation reactions are built into the primer design. The next panel outlines a ligation-free assembly method, in which T4 DNA polymerase activity in the presence of dNTPs‘chewsback’ free double-stranded DNA ends to make long single-stranded overhangs. These overhangs anneal efficiently, and bacteria ligate together the DNA backbone if a bacteria origin of replication (grey) and antibiotic resistance gene (black) are included in the reaction. This method eliminates the need for in vitro ligation. The bottom panel demonstrates how solid-state technology can facilitate high-throughput TAL repeat assembly. A biotinylated repeat monomer is first tethered to a plate coated with streptavidin beads, and then monomers are sequentially added until the desired repeat length is achieved. The final assembly is cloned into the final TAL scaffold.
The Cermak et al.27 Golden Gate RVD assembly method is among the most popular approaches used to date and can be classified as a plasmid-based restriction digest and ligation reaction. Golden Gate reactions utilize a type II restriction endonuclease that cuts outside of its recognition sequence and leaves a predictable four basepair overhang, to which the next plasmid can be added sequentially by designing ends which complement and eliminate the original restriction site.53 This allows a single enzyme to cut all the necessary plasmids in a one-pot reaction. The original Golden Gate method takes a minimum of five days to complete. Several groups have attempted to streamline the TALEN-building process for more rapid assembly using variations of an RVD multimer platform and/or a PCR-based restriction digest and ligation reaction to overcome longer individual steps like overnight bacterial cultures (Table 2, Figure 2). Importantly, not all Golden Gate-like methods are compatible with the same TAL scaffolds (Table 1).
Another iteration of the plasmid-based restriction digest and ligation assembly uses hierarchical digestion/ligation, but does not use the multi-fragment ligation reaction characteristic of Golden Gate. This assembly class includes the “unit assembly” method44 and the restriction enzyme and ligation (REAL) method.54 Unit assembly relies on simple isocaudamer restriction sites that leave complementary overhangs for each subsequent ligation. The REAL method uses a combination of type I and II restriction enzymes to make complementary overhangs without requiring a multistep reaction to assemble the final product.
As opposed to each of the steps thus far requiring ligation to assemble RVD repeats, a ligation independent cloning reaction has been described.42 This method utilizes DNA T4 polymerase, a 3’ → 5’ exonuclease, which will ‘chewback’ free dsDNA ends and leave 5’ overhangs. To control the length of the overhangs, dNTPs can be added to compete with DNA T4 polymerase and stop its activity. For instance, DNA T4 polymerase in the dTTP will ‘chewback’ the free 3’ends until the first thymine is reached. This method facilitates the synthesis of predictable complementary overhangs, however it does not require specific site recognition characteristic of restriction digest. In addition, the overhangs can be much longer for direct transformation of the fragments along with both an antibiotic resistance gene and an origin of replication into bacteria without in vitro ligation.
TALEN assembly on solid-state platforms has also been demonstrated. The ligation-based automatable solid-phase high-throughput (FLASH) assembly method uses a library of TAL effector repeat plasmids encoding one to four RVD repeats, which can be assembled in a directed fashion on solid-phase magnetic beads.40 The process was scaled up to a 96-well plate, from which 96 different TALENs could in principle be made in less than one day. The REAL-Fast assembly54 complements both the REAL and the FLASH methods by enabling the FLASH plasmids to be assembled by basic restriction digest/ligation strategies. Like the FLASH method, iterative cap assembly (ICA) was developed on a solid-state platform.38 ICA enables rapid construction of TALENs using RVD repeat monomers that are sequentially assembled on solid-state beads. Thus, this method is unique from the others in that it will be able to utilize advances in microarray printing technology to build TALENs.
Which TALEN assembly method to choose depends on the needs of each project. For example, the original Golden Gate and REAL assembly methods have translated well into practice for individual laboratories interested in making TALENs on an as-needed basis. These methods only require routine molecular cloning reagents and expertise, making it relatively straightforward to initiate without purchasing expensive equipment. However, unless modified these methods are not able to scale-up like the FLASH or ICA methods. Generally, there are limited kinds of TAL scaffolds (and often only one) compatible with each assembly method, decreasing the flexibility of many methods for the user. One notable exception is the Golden Gate method, which has been popular with individual labs and subsequently additional compatible scaffolds and accessory systems have been established.
While TALENs remain under active research and development, the promise of this technology to accelerate the field of cardiovascular biology hinges on the coupling of TALENs with in vivo and in vitro platforms. The complementary nature of whole animal models and cell culture systems allows researchers to interrogate questions of both cardiovascular disease physiology and cellular phenotypes. This review focuses primarily on TALEN applications within zebrafish and human iPSCs; however, these platforms are among many others that have been genetically engineered using TALENs. Recently, TALENS have been harnessed to engineer a variety of whole animal models via direct embryo injection or transfection of fibroblast cells followed by reproductive cloning. The utility of this gene-editing tool has been demonstrated in diverse organisms including roundworms,55 silkworms,56 fruit flies,57 mosquitoes58, crickets,59 frogs,60, 61 mice,22, 62–64 rats,65 pigs66, 67 and cows.67 In addition, TALENs have also shown efficacy in a variety of cellular model systems including both primary and transformed human cells.26, 31, 37, 40, 68–70 When combined with these cell culture platforms, TALENs can facilitate a variety of disease modeling applications. Recent reports have demonstrated the efficacy of TALENs to induce mutations within human miRNAs70 and generate oncogenic translocations in human mesenchymal progenitors.71 With growing numbers of publications using TALEN technology, this genome editing tool is gaining well-deserved popularity within diverse fields of research. The following sections will highlight the utility of zebrafish and induced pluripotent stem cells to model cardiac diseases and facilitate the development of novel therapeutics by capitalizing on the power of TALENs.
Zebrafish: Physiological models of cardiovascular disease
Zebrafish occupy a unique niche for investigating cardiac diseases as a high-throughput in vivo vertebrate model system. With expressional annotation via whole mount in situ data for 1/3 of the genome,72–74 zebrafish are an increasingly well-characterized model system that can be further enriched through the power of TALEN technology. High fecundity, rapid cardiogenesis, and the ability to conduct forward genetic screens have already established zebrafish as the premier non-mammalian model for developmental genetic studies. Importantly, this system is quite economic with the maintenance cost of adult fish at one tenth or less than the maintenance cost of the same number of mice. As an inexpensive vertebrate model system, zebrafish are a quality platform to study late-onset or progressive cardiovascular diseases that require extended observation time. In addition, with impressive capacity to regenerate injured myocardium, zebrafish offer an ideal model system to study cardiac regeneration.75–78
Although comprised of a single atrium and a single ventricle, the two-chamber zebrafish heart is an established in vivo platform for modeling several forms of human cardiac diseases.79–85 By means of positional cloning, pickwick and tnnt2, truncation mutations of cardiac genes titin 82 and Tnnt2 86, were considered the first two embryonic zebrafish models for cardiomyopathy. Later, a series of loss-of-function studies of known cardiomyopathy genes were conducted, including actn2, mlc, rlc, cypher, and mlp, underscoring the value of embryonic zebrafish for annotating cardiac gene expression and function.83, 87–89 Phenotypic analysis of ilk and nexilin mutants in zebrafish embryos supported the corresponding human genes as novel dilated cardiomyopathy-causative genes.84, 90 Zebrafish embryos have also been used to model arrhythmia defects such as Long QT syndrome,91, 92 as well as pathogenic events during atherosclerosis.93, 94 These innovative, proof-of-principle studies have established zebrafish embryos as a unique platform capable of revealing novel insights into human cardiovascular diseases.95, 96 A major incentive for generating disease models in zebrafish embryos is the feasibility to directly identify therapeutic compounds via high-throughput in vivo pharmaceutical screens because of their small size and ability to absorb small molecules from their environment. Several chemical screens have already been employed using zebrafish as a platform to identify compounds of potential therapeutic value for treating Long QT syndrome and hypertrophic cardiomyopathy.97, 98
Although recognized as a useful model for certain aspects of cardiac disease phenotypes, inherent limitations in embryonic zebrafish models will continue to restrict their extensive use to predict genotype-phenotype relationships in humans. For example, many of the genes associated with human cardiomyopathies are completely depleted in embryonic zebrafish models. This makes it difficult to study cardiovascular diseases that develop later in life, especially those with autosomal dominant inheritance. In addition, the short developmental time restricts fish embryos from precisely recapitulating cardiac pathogenesis, which typically exhibits age-dependent penetrance and gradual progression to overt heart failure in adulthood. Given the intrinsic limitations of zebrafish embryos, several groups have explored adult fish as a model for cardiac diseases. For example, anemia imposes a high-output stress on the heart and can subsequently induce classical cardiomyopathy-like phenotypes in adult zebrafish including myofibril disarray and reactivation of fetal gene transcription.80, 81 Injection of doxorubicin (DOX), a widely-used chemotherapeutic with cardiotoxic side effects99, 100, can also induce these cardiomyopathy-like phenotypes in adult fish.79 Cryoinjury of the adult zebrafish heart has recently been demonstrated as a useful model to study pathogenesis during myocardial infarction.76, 101 As a complement to physically-induced disease models, continued development in gene editing technologies will further extend the capacity of this model system to recapitulate genetically-linked cardiovascular diseases.
Zebrafish: Platform for genomic engineering
Classic genetic tools employed in zebrafish include morpholino technology, targeted induced local lesions in the genome (TILLING)102, 103, and zinc-finger nucleases (ZNFs).104 However, important caveats to each of these techniques limit their utility as genomic tools. When morpholinos are used to knock-down expression of a certain transcript, the downstream phenotype is largely restricted to the first few days of zebrafish development.105 TILLING has been an important reverse genetics tool for identifying zebrafish mutants; however, it is a relatively expensive technique that can only feasibly be conducted in large research centers and the generated mutations carry a wide range of other, linked sequence changes as well.105 ZFNs have also notably contributed to genomic engineering in zebrafish, but the complexity of ZFN design and divergent range of activity have limited the utility and adoption of this technology by most individual laboratories.105 In contrast, TALENs offer a more rapid and less expensive way to genetically engineer stable mutant zebrafish lines.29, 106 TALEN technology coupled with zebrafish as a high-throughput, inexpensive in vivo discovery platform enables the ready generation of mutant lines for individual labs.
The experimental strategies that can be employed using TALENs differ based on which mechanism of genomic repair is induced downstream of the double-strand DNA break (Figure 3). When the genomic DNA is repaired by NHEJ, a frameshift mutation could result in a truncated gene product or abolish gene expression. The resulting mutant zebrafish lines can be easily screened for physiological genotype-phenotype relationships. In addition, TALENs can facilitate HR repair mechanisms using large dsDNA donors to insert selection cassettes or reporter systems downstream of candidate genes.107 A third application for TALEN technology in zebrafish is the use of small ssODNs to insert epitope tags, loxP sites, or alter single base pairs via homology-directed repair.18, 19 Each of these three applications will be highlighted below in a discussion of current TALEN-based genome engineering in zebrafish.
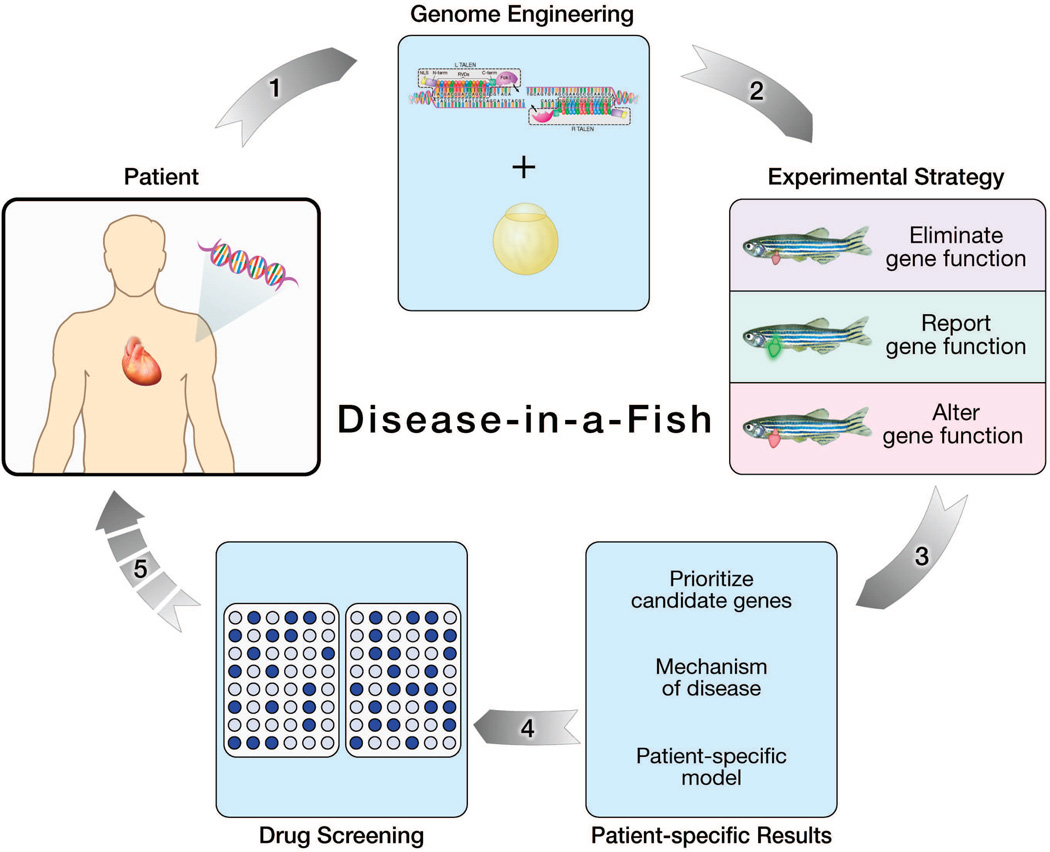
Figure 3.
Genome engineering in zebrafish: Modeling the physiology of gene-linked cardiovascular disease. 1) Whole-genome sequencing approaches reveal genetic mutations underlying cardiac disease in an individual patient. Identification of zebrafish orthologs enables TALEN-based gene modification in zebrafish to study the patient-specific defect. 2) Error-prone or programmed gene repair mechanisms facilitate the generation of adult zebrafish with different experimental phenotypes. 3) Engineered zebrafish models provide patient-specific insights into the physiological mechanism of disease. 4) Drug screens offer the potential to identify novel therapeutic agents. 5) Pharmaceuticals are then tested and eventually delivered back to the patient to complete the individualized medicine loop.
1) Eliminate gene function: Truncational mutations to prioritize candidate genes from human genetic studies
One application of TALENs in this model system is to generate numerous truncational, loss-of-function zebrafish lines that could prioritize candidate genes generated from whole-genome screens (Figure 3). As indicated in the Online Mendelian Inheritance in Man (OMIM) database,108 many human cardiac diseases are caused by nonsense mutations that result in truncated proteins. For such dominantly inherited alleles, the underpinning pathogenesis mechanism could be due to either haploinsufficiency or the generation of a dominant loss-of-function peptide, it is desirable to generate truncational mutations at the corresponding amino acid that is found in human patients. The flexibility of the TALEN-based system can effectively meet this challenge. A large number of disease-relevant truncational mutants will be generated via TALENs in the near future, which shall significantly expand the scope of cardiac diseases that adult zebrafish can model. This knowledge will lay the foundation for zebrafish as an in vivo functional assay to prioritize candidate genes identified from human cohort-based genetic studies, genome-wide association studies (GWAS), and/or quantitative trait locus (QTL) analysis in rodents. As a complement to current mutagenesis techniques in zebrafish, TALENs can be especially useful to probe the genes that were missed by previous non-specific approaches including mutagenesis screening and TILLING.103 In this way, TALEN-based gene editing will contribute to the zebrafish phenome project as an efficient tool to generate novel mutant lines.
2) Report gene function: Insertion of reporter systems to selectively study candidate genes
The feasibility of TALEN-based gene editing in zebrafish will enable the insertion of reporter systems to any genomic loci, which can be used to tag proteins-of-interest (Figure 3). Both tissue-specific and subcellular localization of the tagged protein can be tracked during cardiac development or disease progression. The strength of this gene editing strategy can be invaluable for interrogating the molecular underpinnings of cardiovascular diseases caused by the affected protein. The potential of inserting a targeted GFP reporter system in zebrafish via TALEN-mediated homologous recombination has been recently demonstrated.107 This method utilizes dsDNA with long homologous arms as a template to repair a double strand break induced by TALENs and opens the door to the possibility of directing large fragments of DNA into the zebrafish genome.
3) Alter gene function: Genetic manipulation of zebrafish via homology-directed repair
Smaller targeted modifications like loxP sites, epitope tags or specific sequence changes can also be helpful when studying gene function (Figure 3). Recently, a loxP site was inserted into an intron using an ssODN as a donor for homology-directed repair (HDR) in zebrafish.18 Although dsDNA may also facilitate this modification, ssODNs can be more simply synthesized in a high-throughput manner without requiring a time-consuming molecular cloning process. Approximately the same size as loxP sites, epitope tags may also be inserted using this same technique. Both of these genetic modifications could provide valuable tools for studying the function of cardiac genes. In addition, TALENs and ssODNs may also enable the insertion of SNPs into the zebrafish genome. By developing ways to modify single basepairs in zebrafish, researchers can study numerous SNPs identified through whole genome sequencing in an in vivo context.
There are several clinically relevant benefits to engineering SNPs in a high-throughput model like zebrafish. Unlike bigger vertebrates, zebrafish are amenable to large-scale drug screening efforts. Conceptually, zebrafish engineered with a patient-specific SNP can be subjected to FDA-approved drugs to search for those that ameliorate the physiological disease phenotype. This exciting prospect would allow for identification of drugs that are appropriate for that individual patient based on his/her personal genetic variability; true individualized medicine (Figure 3). While this idea highlights the massive potential of using TALENs and zebrafish in cardiovascular medicine, there is much more work to be done to evolve precision genome engineering to meet such high demand.
Induced pluripotent stem cells: Cell-autonomous models of cardiovascular disease
Where high-throughput animal models are essential for interrogating physiological phenotypes of cardiac gene defects, questions regarding the cell-autonomous nature of cardiovascular diseases can now be probed within the native genomic context using patient-specific cell culture models. To address such clinical and patient-specific questions, induced pluripotent stem (iPS) cells stand out as an ideal model system to harness TALEN technology and interrogate the cell-autonomous features of cardiac disease. Originally described by Takahashi and Yamanaka in 2006, iPS cells can be derived via nuclear reprogramming of mature cells back to the pluripotent stem cell state.109–111 Following the advent of nuclear reprogramming, it is now possible to derive patient-specific pluripotent stem cells that have the capacity to differentiate to any mature cell phenotype in the adult body, including functional cardiomyocytes. Thus through in vitro cardiac differentiation, researchers can probe the molecular underpinnings of cardiac dysfunction as it relates to cell-autonomous genotype-phenotype perturbations. Perhaps the most exciting advantage of this platform is the capacity of iPS-derived cardiomyocytes to potentially serve as an autologous cellular therapy for cardiovascular diseases.
Several unique qualities of iPS cells make them an unprecedented platform to harness TALEN technology for cardiovascular research (Figure 4). The pluripotent capacity of iPS cell lines enables unlimited in vitro generation of cardiac lineages that would otherwise be unavailable or inconvenient for cell culture. Primary cardiomyocytes are difficult to obtain from patients, have limited proliferative capacity, and can only be cultured for several days to weeks while maintaining their contractile properties.112, 113 In contrast, iPS cells can be derived from easily accessible cell sources, most often a skin biopsy, and can be expanded indefinitely as a renewable, high-throughput source of patient-specific cardiomyocytes. Also, by modeling normal developmental stages in the absence of physiological compensatory mechanisms, iPS cell platforms are well positioned to interrogate the cell-autonomous nature of genotype-phenotype relationships. Upon genetic manipulation of iPS cells, researchers can probe the impact of specific gene defects on cardiomyocytes, rather than on the heart as a whole organ. For cardiomyopathies characterized by variable penetrance, the generation of patient-specific iPS cells allows researchers to investigate genotype-phenotype relationships in the context of other unidentified modifier mutations, which may unmask the complexity of inheritance for these diseases. Importantly, cardiomyocytes derived from patient-specific iPS cells offer a novel way to model disease in vitro and test the efficacy and safety profile of pharmacological therapeutics on the patient’s own cells outside his/her body (Figure 4).
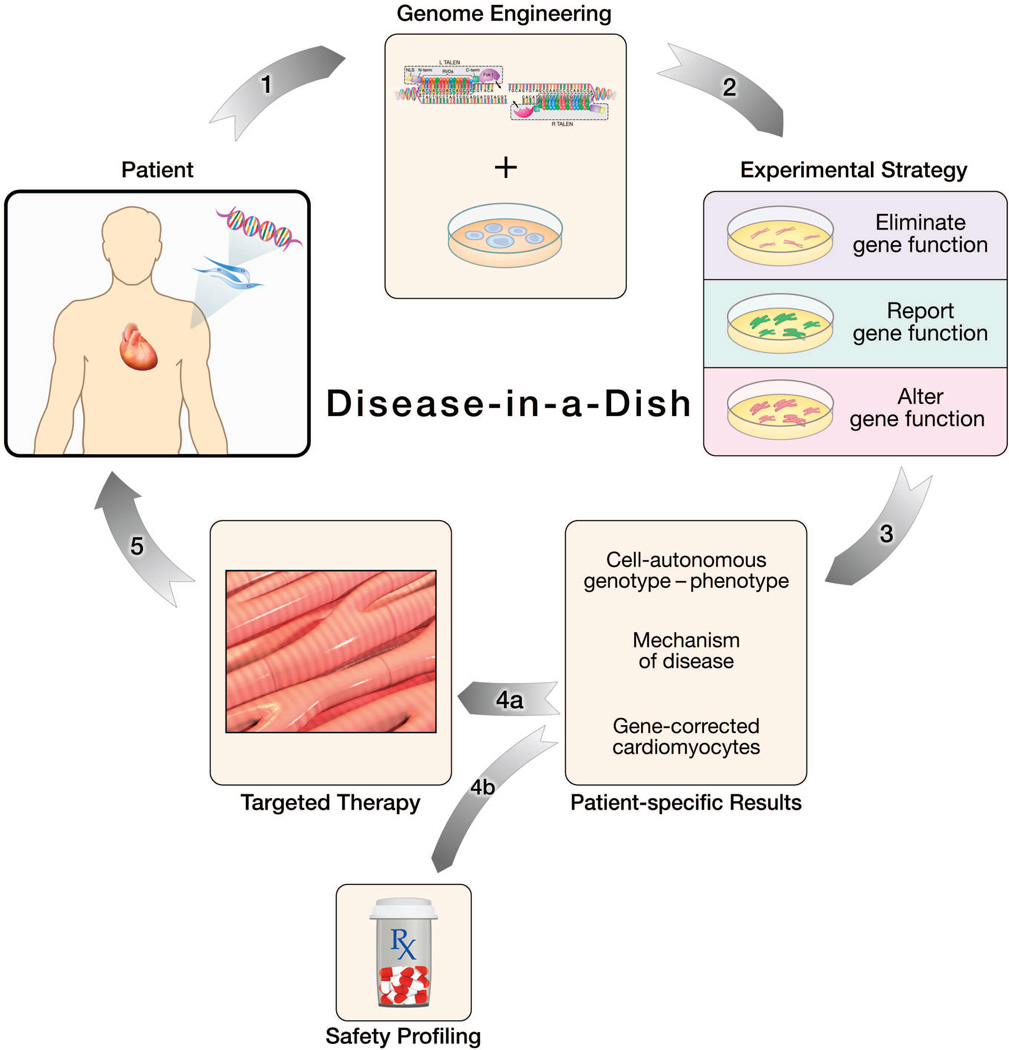
Figure 4.
Genome engineering in induced pluripotent (iPS) cells: Modeling the cell-autonomous nature of gene-linked cardiovascular disease. 1) Fibroblasts are derived from skin biopsy of patient with an inherited cardiovascular disease. Nuclear reprogramming generates induced pluripotent stem cells that are genetically engineered via TALENs. 2) Error-prone or programmed gene repair mechanisms followed by in vitro cardiac differentiation yield patient-specific cardiomyocytes with different experimental phenotypes. 3) Engineered cardiomyocytes provide patient-specific insights into the cell-autonomous mechanism of disease. 4a) Gene-corrected cardiomyocytes offer an autologous cellular therapy and 4b) safety profiling screens enable the identification of potential arrhythmogenic pharmaceutical agents5) Cellular therapies are delivered back to the patient to complete the individualized medicine loop.
The value of patient-specific iPS cells as an autologous surrogate to model cardiac disease has been established by numerous studies to date. Although iPS-derived cardiac cells in vitro do not recapitulate all the structural organization of adult cardiomyocytes isolated directly from heart tissue, the functional characteristics of these bioengineered cells have demonstrated great potential as an in vitro disease-modeling platform. Several detailed reviews capture the current state of this field with an emphasis on arrhythmogenic cardiac diseases.114–118 Notably, iPS-based disease modeling has revolutionized the field of cardiovascular medicine by providing an ex vivo drug screening platform. Applying high-throughput model systems such as zebrafish and now patient-specific iPS-derived cardiomyocytes, whole libraries of small molecules can be screened for functional improvement of cellular disease phenotypes. The patient-specific nature of this model system promises to advance the field of personalized medicine and drug development.
Previous studies using iPS cells derived from patients with genetically linked arrhythmogenic cardiac diseases have demonstrated the utility of this platform to recapitulate cellular disease phenotypes and pharmaceutical responses. iPS cell lines derived from patients with Long QT syndrome (Types 1, 2, and 8) have been differentiated in vitro to cardiomyocytes that show prolonged action potentials due to mutations in KCNQ1, KCNH2, or CACNA1C, respectively.119–124 Mechanistically, these studies have shed light onto the cell-autonomous disease phenotype displayed by iPS-derived cardiomyocytes. In addition to Long QT syndrome, other arrhythmogenic cardiac diseases have been modeled in vitro using patient-specific iPS cells, including catecholaminergic polymorphic ventricular tachycardia (CPVT) and an overlap syndrome caused by a defect in SCN5A.125–128 The utility of iPS cells to model non-arrhythmogenic cardiac diseases has also been demonstrated in patient-derived iPS cells to model the pathognomonic features of LEOPARD syndrome and dilated cardiomyopathy.129, 130
Despite the documented capacity of iPS cells to model cardiac disease in vitro, an important limitation of current disease-modeling applications is the use of allogeneic healthy controls for comparison with patient-derived iPS cells. In addition to the clonal heterogeneity between iPS cell lines from a single individual, there is increased genetic and epigenetic variability when using allogeneic iPS cells.131–136 This genetic diversity may be unrelated to the disease phenotype and thereby confound comparative analysis of cardiac phenotypes derived from different iPS cells. To address this confounding variability, genome engineering strategies have been employed to generate isogenic controls by inducing site-specific repair of disease-causing mutations in iPS cell lines.137 These gene editing technologies ideally have minimal off-target effects to limit the mutational risk of unintentional genome modifications. The standard in vitro processing of iPS cells already poses a risk of altering the genotype of these cells, which must not be further increased by non-specific mutations as a result of genomic engineering.
Induced pluripotent stem cells: Platform for genomic engineering
Genome editing in iPS cells was pioneered by custom zinc finger nucleases (ZFNs).The utility of this gene editing tool in iPS cells was recently reviewed in Cheng et al.138 Briefly, ZFNs have been used to induce site-specific knock-out139–142, knock-in141, 142, or correction of disease-related genes in human iPS cells.141, 143–147 The power of genome engineering in iPS cells may be extended even further through the use of TALEN technology. With greater flexibility of design and ease of assembly, TALENs offer an increasingly popular alternative to ZFNs with high targeting efficiency in iPS cells.15, 148 The capacity of TALENs to facilitate site-specific genome modifications in iPS cells through NHEJ, HR, and HDR repair mechanisms will be highlighted in the following sections.
1) Eliminate gene function: Null mutations to probe cell-autonomous genotype-phenotype relationships
A recent study by Ding et al.23 reveals the power of TALENs to rapidly and efficiently induce genetic knock-outs as a strategy to study cell-autonomous genotype-phenotype relationships upon differentiation of human pluripotent stem cells. Although this group demonstrates similar efficiency of TALEN-targeted genetic mutations between human embryonic stem (hES) cells and iPS cells, the phenotypic characterization was restricted to genetically engineered hES cells. The power and versatility of this platform is highlighted by the successful knock-out of SORT1, a gene recently implicated in several disease states. Upon differentiation to hepatocyte-like cells, adipocytes, or motor neurons, the engineered cell lines displayed disease-related phenotypes when compared to isogenic controls.23 Through genetic manipulation at the pluripotent state, this group modeled a variety of disease phenotypes by guiding the differentiation of hES cells to different somatic lineages. Although this study focuses on non-cardiac cell phenotypes, TALEN-based knock-down of critical cardiac genes would also provide mechanistic insights for cardiac disease modeling using iPS cells (Figure 4).
2) Report gene function: Insertion of reporter systems to identify and study cell lineages
An additional application for TALEN-based engineering in iPS cells was recently demonstrated by Hockemeyer et al.15 This study demonstrates the utility of TALENs to facilitate transgene insertion, including a GFP reporter downstream of pluripotency marker Oct4 in hES and hiPS cells. The fidelity of this reporter system was demonstrated by the disappearance of GFP expression following in vitro differentiation. TALEN technology provides an effective genomic editing tool to selectively label pluripotency genes, which could be used for the isolation of residual iPS cells from a heterogeneous population of differentiating iPS progeny.149 By selectively purging the remaining pluripotent cells from a differentiating population, this strategy may complement other approaches to increase the safety profile of iPS-derived progeny for future therapeutic applications.150, 151 In addition, TALENs could also be utilized to insert reporter constructs downstream of critical cardiac genes to follow their expression and localization during in vitro differentiation. Such a reporter system would also enable selection of transcriptionally defined cellular lineages from a heterogeneous population of differentiating iPS cells (Figure 4). A population of stage-matched cells would significantly improve the analytical power of this platform to study cell-autonomous mechanisms of cardiovascular disease.
3) Alter gene function: Genetic correction of disease-causing mutations in iPS cells
For monogenic diseases, TALEN-based gene correction can generate disease-free autologous iPS cells for potential delivery back to the patient (Figure 4). The efficiency of TALENs to facilitate gene correction in patient-derived iPS cells was recently demonstrated by Choi et al.152 In this study, traceless genomic repair of a disease-causing gene was reported in iPS cell lines derived from two patients with α1-antitrypsin (AAT)-deficiency related liver disease. TALENs targeting the mutated AAT efficiently induced biallelic HDR correction in 25–33% of puromycin-selected iPS cells. Upon differentiation to hepatocyte-like cells, the gene-corrected iPS cells displayed restoration of cellular function to the level of healthy controls. In addition, this group also conducted a high-throughput pharmaceutical screen and identified five clinically relevant compounds that restored cellular function to the same level of rescue as the gene-corrected iPS cells. This study highlights the utility of iPS cells for pharmaceutical screening and the efficiency of TALENs for genomic repair. In therapeutic applications, the gene-corrected cells may have increased viability that could translate to improved repair upon re-introduction into the patient.
Overall, the unique advantages of TALEN-based genomic engineering paired with the unlimited potential of bioengineered patient-specific tissues with isogenic controls, reporter systems, and disease mapping mutagenesis have the potential to transform personalized cardiovascular research towards cell-based theranostics.
Future Directions
As TALEN technology continues to develop, there are questions being raised regarding the optimal methods of synthesis and distribution. One common option allows individual laboratories to assemble TALENs on anas-needed basis. Methods like Golden Gate assembly are ideal for this approach as they are straightforward for those comfortable with molecular cloning and readily accessible to the community. While several labs have taken the approach, others aim to assemble more TALENs than feasible using standard cloning techniques and are primarily interested in downstream applications rather than the TALEN building process. In response, scalable assembly platforms such as FLASH or ICA may function well in core facilities at institutions in which TALENs are consistently being made in an efficient manner and distributed on-site. Several companies have also been established to assemble and distribute TALENs, and this is yet another avenue through which labs can obtain TALEN technology. With the simplicity of making TALENs, especially compared to ZFNs, the cost of TALENs is relatively low and continues to decrease as more companies enter the market.
Genetic engineering continues to be a fast-growing field, with new technologies regularly in development. The clustered regularly interspaced short palindromic repeats(CRISPR)/CRISPR-associated (Cas9) system is one such technology.153 Unlike ZFNs and TALENs, which are protein-guided, the specificity of CRISPR/Cas9 targeting is RNA-guided.148 While CRISPR/Cas9 applications are currently being actively investigated, its full range of activity – targeted and otherwise – is also being defined. Early studies indicate that on-site CRISPR/Cas9 targeting is roughly comparable to TALEN-based targeting; however, off-target activity is comparable or even greater than on-target activity in some cases.154 Just like ZFNs and TALENs before, there is likely room for optimization of the CRISPR/Cas9 system that could improve both specificity and efficacy.
As disease modeling applications are rapidly emerging in both zebrafish and iPS cell culture, genome engineering tools will also need to be optimized for both in vivo and in vitro model systems. For example, efficient donor templates will need to be developed to generate single basepair changes in zebrafish. Achieving this milestone will allow researchers to develop patient-specific zebrafish with subtle changes in the genome to model the physiological phenotype of specific SNPs. To further advance disease-modeling in iPS cells, whose epigenetic profiles drastically change during in vitro differentiation, further studies will be needed to decipher the role that epigenetic environment of the target gene has on TALEN and CRISPR efficacy. Increased TALEN cutting efficiency at several gene loci has been demonstrated upon treatment with a DNA methyltransferase inhibitor.14 This observation may indicate a decreased TALEN efficiency at somatic gene loci that are in a heterochromatic state in iPS cells. It has been postulated that the helicase activity of Cas9 may facilitate more efficient gene editing at epigenetically silenced genes.155 Future studies that compare both gene editing tools will help address some of these remaining questions and further advance cardiac disease modeling in both zebrafish and iPS cells.
Conclusions
Here we have highlighted the current state of TALEN technology as a novel genome editing strategy to advance cardiovascular research applications. The relative simplicity and flexibility of TALEN design and assembly enable efficient gene manipulation in a sequence-specific manner. When coupled with high-throughput model systems such as zebrafish and patient-specific iPS cells, TALENs can facilitate disease-modeling applications to study the cardiac phenotype and molecular underpinnings of individual genetic defects. The disruptive nature of this technology will accelerate the development of patient-specific cellular and pharmaceutical therapeutics for the personalized treatment of cardiovascular disease.
Acknowledgements
The authors thank Mayo Clinic Media Support Services for assistance in figure production and Alvin Ma, Han Lee, Hind Fadel, Patrick Blackburn and Karl Clark for critical review of the manuscript.
Sources of Funding
This project is supported by the State of Minnesota (University of Minnesota Mayo Clinic Partnership grant to SCE), NIH grant GM63904 to SCE, National Institute of Diabetes and Digestive and Kidney Diseases grant P30DK084567 to SCE, National Heart, Lung, and Blood Institute grants HL107304 and HL81753 to XX, the Todd and Karen Wanek Family Program for Hypoplastic Left Heart Syndrome (TJN), and the Mayo Foundation.
Nonstandard Abbreviations and Acronyms
- iPS cells
induced pluripotent stem cells
- TALEN
transcription activator-like effector nuclease
- RVD
repeat variable di-residue
- DSB
double-strand break
- HR
homologous recombination
- NHEJ
non-homologous end joining
- HDR
homology-directed repair
- ssODN
single-stranded oligodeoxyribonucleotide
- SNP
single nucleotide polymorphism
- TILLING
targeted induced local lesions in the genome
- ZFN
zinc finger nuclease
- CRISPR
clustered regularly interspaced short palindromic repeats
- Cas
CRISPR-associated
- GWAS
genome wide association study
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
None for JMC, KAH, XX, and SCE. TJN and Mayo Clinic have a financial interest in the stem cell technology referenced in this review.
References
- 1.Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Borden WB, Bravata DM, Dai S, Ford ES, Fox CS, Franco S, Fullerton HJ, Gillespie C, Hailpern SM, Heit JA, Howard VJ, Huffman MD, Kissela BM, Kittner SJ, Lackland DT, Lichtman JH, Lisabeth LD, Magid D, Marcus GM, Marelli A, Matchar DB, McGuire DK, Mohler ER, Moy CS, Mussolino ME, Nichol G, Paynter NP, Schreiner PJ, Sorlie PD, Stein J, Turan TN, Virani SS, Wong ND, Woo D, Turner MB. Heart Disease and Stroke Statistics—2013 Update: A Report From the American Heart Association. Circulation. 2013;127:e6–e245. doi: 10.1161/CIR.0b013e31828124ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Faita F, Vecoli C, Foffa I, Andreassi MG. Next generation sequencing in cardiovascular diseases. World J Cardiol. 2012;4:288–295. doi: 10.4330/wjc.v4.i10.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maron BJ, Maron MS, Semsarian C. Genetics of Hypertrophic Cardiomyopathy After 20 Years: Clinical Perspectives. J Am Coll Cardiol. 2012;60:705–715. doi: 10.1016/j.jacc.2012.02.068. [DOI] [PubMed] [Google Scholar]
- 4.Kimura A. Molecular Etiology and Pathogenesis of Hereditary Cardiomyopathy. Circ J. 2008;72:A38–A48. doi: 10.1253/circj.cj-08-0050. [DOI] [PubMed] [Google Scholar]
- 5.Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. Targeting DNA Double-Strand Breaks with TAL Effector Nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bogdanove AJ, Voytas DF. TAL Effectors: Customizable Proteins for DNA Targeting. Science. 2011;333:1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- 7.Bogdanove AJ, Schornack S, Lahaye T. TAL effectors: finding plant genes for disease and defense. Curr Opin Plant Biol. 2010;13:394–401. doi: 10.1016/j.pbi.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 8.Schornack S, Meyer A, Römer P, Jordan T, Lahaye T. Gene-for-gene-mediated recognition of nuclear-targeted AvrBs3-like bacterial effector proteins. J Plant Physiol. 2006;163:256–272. doi: 10.1016/j.jplph.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 9.Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the Code of DNA Binding Specificity of TAL-Type III Effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 10.Moscou MJ, Bogdanove AJ. A Simple Cipher Governs DNA Recognition by TAL Effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 11.Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci USA. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li T, Huang S, Jiang WZ, Wright D, Spalding MH, Weeks DP, Yang B. TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res. 2011;39:359–372. doi: 10.1093/nar/gkq704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol. 2013;14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim Y, Kweon J, Kim A, Chon JK, Yoo JY, Kim HJ, Kim S, Lee C, Jeong E, Chung E, Kim D, Lee MS, Go EM, Song HJ, Kim H, Cho N, Bang D, Kim S, Kim J-S. A library of TAL effector nucleases spanning the human genome. Nat Biotech. 2013;31:251–258. doi: 10.1038/nbt.2517. [DOI] [PubMed] [Google Scholar]
- 15.Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, Zeitler B, Cherone JM, Meng X, Hinkley SJ, Rebar EJ, Gregory PD, Urnov FD, Jaenisch R. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mao Z, Bozzella M, Seluanov A, Gorbunova V. Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair. 2008;7:1765–1771. doi: 10.1016/j.dnarep.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aarts M, te Riele H. Progress and prospects: oligonucleotide-directed gene modification in mouse embryonic stem cells: a route to therapeutic application. Gene Ther. 2011;18:213–219. doi: 10.1038/gt.2010.161. [DOI] [PubMed] [Google Scholar]
- 18.Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug Ii RG, Tan W, Penheiter SG, Ma AC, Leung AYH, Fahrenkrug SC, Carlson DF, Voytas DF, Clark KJ, Essner JJ, Ekker SC. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang N, Sun C, Gao L, Zhu D, Xu X, Zhu X, Xiong J-W, Xi JJ. Genome editing with RNA-guided Cas9 nuclease in Zebrafish embryos. Cell Res. 2013;23:465–472. doi: 10.1038/cr.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen F, Pruett-Miller SM, Huang Y, Gjoka M, Duda K, Taunton J, Collingwood TN, Frodin M, Davis GD. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat Methods. 2011;8:753–755. doi: 10.1038/nmeth.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Radecke S, Radecke F, Cathomen T, Schwarz K. Zinc-finger nuclease-induced gene repair with oligodeoxynucleotides: wanted and unwanted target locus modifications. Mol Ther. 2010;18:743–753. doi: 10.1038/mt.2009.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wefers B, Meyer M, Ortiz O, Hrabé de Angelis M, Hansen J, Wurst W, Kühn R. Direct production of mouse disease models by embryo microinjection of TALENs and oligodeoxynucleotides. Proc Natl Acad Sci USA. 2013;110:3782–3787. doi: 10.1073/pnas.1218721110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding Q, Lee Y-K, Schaefer Esperance AK, Peters Derek T, Veres A, Kim K, Kuperwasser N, Motola Daniel L, Meissner Torsten B, Hendriks William T, Trevisan M, Gupta Rajat M, Moisan A, Banks E, Friesen M, Schinzel Robert T, Xia F, Tang A, Xia Y, Figueroa E, Wann A, Ahfeldt T, Daheron L, Zhang F, Rubin Lee L, Peng Lee F, Chung Raymond T, Musunuru K, Cowan Chad A. A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell. 2012 doi: 10.1016/j.stem.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scholze H, Boch J. TAL effectors are remote controls for gene activation. Curr Opin Microbiol. 2011;14:47–53. doi: 10.1016/j.mib.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 25.Ma AC, Lee HB, Clark KJ, Ekker SC. High efficiency in vivo genome engineering with a simplified 15-RVD GoldyTALEN design. PLoS One. 2013;8:e65259. doi: 10.1371/journal.pone.0065259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, Dulay GP, Hua KL, Ankoudinova I, Cost GJ, Urnov FD, Zhang HS, Holmes MC, Zhang L, Gregory PD, Rebar EJ. A TALE nuclease architecture for efficient genome editing. Nat Biotech. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 27.Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christian ML, Demorest ZL, Starker CG, Osborn MJ, Nyquist MD, Zhang Y, Carlson DF, Bradley P, Bogdanove AJ, Voytas DF. Targeting G with TAL Effectors: A Comparison of Activities of TALENs Constructed with NN and NK Repeat Variable Di-Residues. PLoS ONE. 2012;7:e45383. doi: 10.1371/journal.pone.0045383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dahlem TJ, Hoshijima K, Jurynec MJ, Gunther D, Starker CG, Locke AS, Weis AM, Voytas DF, Grunwald DJ. Simple Methods for Generating and Detecting Locus-Specific Mutations Induced with TALENs in the Zebrafish Genome. PLoS Genet. 2012;8:e1002861. doi: 10.1371/journal.pgen.1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hisano Y, Ota S, Arakawa K, Muraki M, Kono N, Oshita K, Sakuma T, Tomita M, Yamamoto T, Okada Y, Kawahara A. Quantitative assay for TALEN activity at endogenous genomic loci. Biology Open. 2013 doi: 10.1242/bio.20133871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mussolino C, Morbitzer R, Lütge F, Dannemann N, Lahaye T, Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39:9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller JC, Holmes MC, Wang J, Guschin DY, Lee Y-L, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, Gregory PD, Pabo CO, Rebar EJ. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 33.Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo J, Gaj T, Barbas CF. Directed evolution of an enhanced and highly efficient FokI cleavage domain for zinc finger nucleases. J Mol Biol. 2010;400:96–107. doi: 10.1016/j.jmb.2010.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doyon Y, Vo TD, Mendel MC, Greenberg SG, Wang J, Xia DF, Miller JC, Urnov FD, Gregory PD, Holmes MC. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat Methods. 2011;8:74–79. doi: 10.1038/nmeth.1539. [DOI] [PubMed] [Google Scholar]
- 36.Mak AN-S, Bradley P, Cernadas RA, Bogdanove AJ, Stoddard BL. The Crystal Structure of TAL Effector PthXo1 Bound to Its DNA Target. Science. 2012;335:716–719. doi: 10.1126/science.1216211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun N, Liang J, Abil Z, Zhao H. Optimized TAL effector nucleases (TALENs) for use in treatment of sickle cell disease. Mol Biosyst. 2012;8:1255–1263. doi: 10.1039/c2mb05461b. [DOI] [PubMed] [Google Scholar]
- 38.Briggs AW, Rios X, Chari R, Yang L, Zhang F, Mali P, Church GM. Iterative capped assembly: rapid and scalable synthesis of repeat-module DNA such as TAL effectors from individual monomers. Nucleic Acids Res. 2012;40:e117. doi: 10.1093/nar/gks624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen S, Oikonomou G, Chiu CN, Niles BJ, Liu J, Lee DA, Antoshechkin I, Prober DA. A large-scale in vivo analysis reveals that TALENs are significantly more mutagenic than ZFNs generated using context-dependent assembly. Nucleic Acids Res. 2013;41:2769–2778. doi: 10.1093/nar/gks1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotech. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mak AN-S, Bradley P, Bogdanove AJ, Stoddard BL. TAL effectors: function, structure, engineering and applications. Curr Opin Struct Biol. 2013;23:93–99. doi: 10.1016/j.sbi.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmid-Burgk JL, Schmidt T, Kaiser V, Honing K, Hornung V. A ligation-independent cloning technique for high-throughput assembly of transcription activator-like effector genes. Nat Biotech. 2013;31:76–81. doi: 10.1038/nbt.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sakuma T, Hosoi S, Woltjen K, Suzuki K-i, Kashiwagi K, Wada H, Ochiai H, Miyamoto T, Kawai N, Sasakura Y, Matsuura S, Okada Y, Kawahara A, Hayashi S, Yamamoto T. Efficient TALEN construction and evaluation methods for human cell and animal applications. Genes Cells. 2013;18:315–326. doi: 10.1111/gtc.12037. [DOI] [PubMed] [Google Scholar]
- 44.Huang P, Xiao A, Zhou M, Zhu Z, Lin S, Zhang B. Heritable gene targeting in zebrafish using customized TALENs. Nat Biotech. 2011;29:699–700. doi: 10.1038/nbt.1939. [DOI] [PubMed] [Google Scholar]
- 45.Streubel J, Blucher C, Landgraf A, Boch J. TAL effector RVD specificities and efficiencies. Nat Biotech. 2012;30:593–595. doi: 10.1038/nbt.2304. [DOI] [PubMed] [Google Scholar]
- 46.Cong L, Zhou R, Kuo Y-c, Cunniff M, Zhang F. Comprehensive interrogation of natural TALE DNA-binding modules and transcriptional repressor domains. Nat Commun. 2012;3:968. doi: 10.1038/ncomms1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bultmann S, Morbitzer R, Schmidt CS, Thanisch K, Spada F, Elsaesser J, Lahaye T, Leonhardt H. Targeted transcriptional activation of silent oct4 pluripotency gene by combining designer TALEs and inhibition of epigenetic modifiers. Nucleic Acids Res. 2012;40:5368–5377. doi: 10.1093/nar/gks199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valton J, Dupuy A, Daboussi F, Thomas S, Maréchal A, Macmaster R, Melliand K, Juillerat A, Duchateau P. Overcoming Transcription Activator-like Effector (TALE) DNA Binding Domain Sensitivity to Cytosine Methylation. J Biol Chem. 2012;287:38427–38432. doi: 10.1074/jbc.C112.408864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deng D, Yin P, Yan C, Pan X, Gong X, Qi S, Xie T, Mahfouz M, Zhu J-K, Yan N, Shi Y. Recognition of methylated DNA by TAL effectors. Cell Res. 2012;22:1502–1504. doi: 10.1038/cr.2012.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neff K, Argue D, Ma A, Lee H, Clark K, Ekker S. Mojo hand, a TALEN design tool for genome editing applications. BMC Bioinformatics. 2013;14 doi: 10.1186/1471-2105-14-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sander JD, Maeder ML, Reyon D, Voytas DF, Joung JK, Dobbs D. ZiFiT (Zinc Finger Targeter): an updated zinc finger engineering tool. Nucleic Acids Res. 2010;38:W462–W468. doi: 10.1093/nar/gkq319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Doyle EL, Booher NJ, Standage DS, Voytas DF, Brendel VP, VanDyk JK, Bogdanove AJ. TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 2012;40:W117–W122. doi: 10.1093/nar/gks608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Engler C, Kandzia R, Marillonnet S. A One Pot, One Step, Precision Cloning Method with High Throughput Capability. PLoS ONE. 2008;3:e3647. doi: 10.1371/journal.pone.0003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reyon D, Khayter C, Regan MR, Joung JK, Sander JD. Curr Protoc Mol Biol. John Wiley & Sons, Inc; 2012. Engineering Designer Transcription Activator--Like Effector Nucleases (TALENs) by REAL or REAL-Fast Assembly. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wood AJ, Lo T-W, Zeitler B, Pickle CS, Ralston EJ, Lee AH, Amora R, Miller JC, Leung E, Meng X, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Meyer BJ. Targeted Genome Editing Across Species Using ZFNs and TALENs. Science. 2011;333:307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ma S, Zhang S, Wang F, Liu Y, Liu Y, Xu H, Liu C, Lin Y, Zhao P, Xia Q. Highly Efficient and Specific Genome Editing in Silkworm Using Custom TALENs. PLoS ONE. 2012;7:e45035. doi: 10.1371/journal.pone.0045035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu J, Li C, Yu Z, Huang P, Wu H, Wei C, Zhu N, Shen Y, Chen Y, Zhang B, Deng W-M, Jiao R. Efficient and Specific Modifications of the Drosophila Genome by Means of an Easy TALEN Strategy. J Genet Genomics. 2012;39:209–215. doi: 10.1016/j.jgg.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 58.Aryan A, Anderson MAE, Myles KM, Adelman ZN. TALEN-Based Gene Disruption in the Dengue Vector Aedes aegypti. PLoS ONE. 2013;8:e60082. doi: 10.1371/journal.pone.0060082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watanabe T, Ochiai H, Sakuma T, Horch HW, Hamaguchi N, Nakamura T, Bando T, Ohuchi H, Yamamoto T, Noji S, Mito T. Non-transgenic genome modifications in a hemimetabolous insect using zinc-finger and TAL effector nucleases. Nat Commun. 2012;3:1017. doi: 10.1038/ncomms2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lei Y, Guo X, Liu Y, Cao Y, Deng Y, Chen X, Cheng CHK, Dawid IB, Chen Y, Zhao H. Efficient targeted gene disruption in Xenopus embryos using engineered transcription activator-like effector nucleases (TALENs) Proc Natl Acad Sci USA. 2012;109:17484–17489. doi: 10.1073/pnas.1215421109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lei Y, Guo X, Deng Y, Chen Y, Zhao H. Generation of gene disruptions by transcription activator-like effector nucleases (TALENs) in Xenopus tropicalis embryos. Cell Biosci. 2013;3:21. doi: 10.1186/2045-3701-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sung YH, Baek I-J, Kim DH, Jeon J, Lee J, Lee K, Jeong D, Kim J-S, Lee H-W. Knockout mice created by TALEN-mediated gene targeting. Nat Biotech. 2013;31:23–24. doi: 10.1038/nbt.2477. [DOI] [PubMed] [Google Scholar]
- 63.Qiu Z, Liu M, Chen Z, Shao Y, Pan H, Wei G, Yu C, Zhang L, Li X, Wang P, Fan H-Y, Du B, Liu B, Liu M, Li D. High-efficiency and heritable gene targeting in mouse by transcription activator-like effector nucleases. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Davies B, Davies G, Preece C, Puliyadi R, Szumska D, Bhattacharya S. Site Specific Mutation of the Zic2 Locus by Microinjection of TALEN mRNA in Mouse CD1, C3H and C57BL/6J Oocytes. PLoS ONE. 2013;8:e60216. doi: 10.1371/journal.pone.0060216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tesson L, Usal C, Menoret S, Leung E, Niles BJ, Remy S, Santiago Y, Vincent AI, Meng X, Zhang L, Gregory PD, Anegon I, Cost GJ. Knockout rats generated by embryo microinjection of TALENs. Nat Biotech. 2011;29:695–696. doi: 10.1038/nbt.1940. [DOI] [PubMed] [Google Scholar]
- 66.Carlson DF, Tan W, Fahrenkrug SC. 328 versatile transcription activator-like effector nuclease (TALEN)-mediated engineering of ossabaw miniature swine. Reprod Fertil Dev. 2012;25:312. [Google Scholar]
- 67.Carlson DF, Tan W, Lillico SG, Stverakova D, Proudfoot C, Christian M, Voytas DF, Long CR, Whitelaw CBA, Fahrenkrug SC. Efficient TALEN-mediated gene knockout in livestock. Proc Natl Acad Sci USA. 2012;109:17382–17387. doi: 10.1073/pnas.1211446109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sanjana NE, Cong L, Zhou Y, Cunniff MM, Feng G, Zhang F. A transcription activator-like effector toolbox for genome engineering. Nat. Protocols. 2012;7:171–192. doi: 10.1038/nprot.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Holkers M, Maggio I, Liu J, Janssen JM, Miselli F, Mussolino C, Recchia A, Cathomen T. Gonçalves MAFV. Differential integrity of TALE nuclease genes following adenoviral and lentiviral vector gene transfer into human cells. Nucleic Acids Res. 2013;41:e63. doi: 10.1093/nar/gks1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hu R, Wallace J, Dahlem TJ, Grunwald DJ, O'Connell RM. Targeting Human MicroRNA Genes Using Engineered Tal-Effector Nucleases (TALENs) PLoS ONE. 2013;8:e63074. doi: 10.1371/journal.pone.0063074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Piganeau M, Ghezraoui H, De Cian A, Guittat L, Tomishima M, Perrouault L, René O, Katibah GE, Zhang L, Holmes MC, Doyon Y, Concordet J-P, Giovannangeli C, Jasin M, Brunet E. Cancer translocations in human cells induced by zinc finger and TALE nucleases. Genome Res. 2013 doi: 10.1101/gr.147314.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protocols. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- 73.Ouyang M, Garnett AT, Han TM, Hama K, Lee A, Deng Y, Lee N, Liu H-Y, Amacher SL, Farber SA, Ho S-Y. A web based resource characterizing the zebrafish developmental profile of over 16,000 transcripts. Gene Expr Patterns. 2008;8:171–180. doi: 10.1016/j.gep.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bradford Y, Conlin T, Dunn N, Fashena D, Frazer K, Howe DG, Knight J, Mani P, Martin R, Moxon SAT, Paddock H, Pich C, Ramachandran S, Ruef BJ, Ruzicka L, Bauer Schaper H, Schaper K, Shao X, Singer A, Sprague J, Sprunger B, Van Slyke C, Westerfield M. ZFIN: enhancements and updates to the zebrafish model organism database. Nucleic Acids Res. 2011;39:D822–D829. doi: 10.1093/nar/gkq1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.González-Rosa JM, Martín V, Peralta M, Torres M, Mercader N. Extensive scar formation and regression during heart regeneration after cryoinjury in zebrafish. Development. 2011;138:1663–1674. doi: 10.1242/dev.060897. [DOI] [PubMed] [Google Scholar]
- 76.Gonzalez-Rosa JM, Mercader N. Cryoinjury as a myocardial infarction model for the study of cardiac regeneration in the zebrafish. Nat. Protocols. 2012;7:782–788. doi: 10.1038/nprot.2012.025. [DOI] [PubMed] [Google Scholar]
- 77.Chablais F, Veit J, Rainer G, Jazwinska A. The zebrafish heart regenerates after cryoinjury-induced myocardial infarction. BMC Dev Biol. 2011;11:21. doi: 10.1186/1471-213X-11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schnabel K, Wu C-C, Kurth T, Weidinger G. Regeneration of Cryoinjury Induced Necrotic Heart Lesions in Zebrafish Is Associated with Epicardial Activation and Cardiomyocyte Proliferation. PLoS ONE. 2011;6:e18503. doi: 10.1371/journal.pone.0018503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ding Y, Sun X, Huang W, Hoage T, Redfield M, Kushwaha S, Sivasubbu S, Lin X, Ekker S, Xu X. Haploinsufficiency of target of rapamycin attenuates cardiomyopathies in adult zebrafish. Circ Res. 2011;109:658–669. doi: 10.1161/CIRCRESAHA.111.248260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sun X, Hoage T, Bai P, Ding Y, Chen Z, Zhang R, Huang W, Jahangir A, Paw B, Li Y-G, Xu X. Cardiac Hypertrophy Involves Both Myocyte Hypertrophy and Hyperplasia in Anemic Zebrafish. PLoS ONE. 2009;4:e6596. doi: 10.1371/journal.pone.0006596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sun X, Xu X. Anemic Zebrafish Models of Cardiomyopathy. In: Szallasi A, Bíró T, editors. TRP Channels in Drug Discovery. Humana Press; 2012. pp. 41–54. [Google Scholar]
- 82.Xu X, Meiler SE, Zhong TP, Mohideen M, Crossley DA, Burggren WW, Fishman MC. Cardiomyopathy in zebrafish due to mutation in an alternatively spliced exon of titin. Nat Genet. 2002;30:205–209. doi: 10.1038/ng816. [DOI] [PubMed] [Google Scholar]
- 83.Vogel B, Meder B, Just S, Laufer C, Berger I, Weber S, Katus HA, Rottbauer W. In-vivo characterization of human dilated cardiomyopathy genes in zebrafish. Biochem Biophys Res Commun. 2009;390:516–522. doi: 10.1016/j.bbrc.2009.09.129. [DOI] [PubMed] [Google Scholar]
- 84.Knoll R, Postel R, Wang J, Kratzner R, Hennecke G, Vacaru AM, Vakeel P, Schubert C, Murthy K, Rana BK, Kube D, Knoll G, Schafer K, Hayashi T, Holm T, Kimura A, Schork N, Toliat MR, Nurnberg P, Schultheiss HP, Schaper W, Schaper J, Bos E, Den Hertog J, van Eeden FJ, Peters PJ, Hasenfuss G, Chien KR, Bakkers J. Laminin-alpha4 and integrin-linked kinase mutations cause human cardiomyopathy via simultaneous defects in cardiomyocytes and endothelial cells. Circulation. 2007;116:515–525. doi: 10.1161/CIRCULATIONAHA.107.689984. [DOI] [PubMed] [Google Scholar]
- 85.Milan DJ, MacRae CA. Zebrafish genetic models for arrhythmia. Prog Biophys Mol Biol. 2008;98:301–308. doi: 10.1016/j.pbiomolbio.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sehnert AJ, Huq A, Weinstein BM, Walker C, Fishman M, Stainier DY. Cardiac troponin T is essential in sarcomere assembly and cardiac contractility. Nat Genet. 2002;31:106–110. doi: 10.1038/ng875. [DOI] [PubMed] [Google Scholar]
- 87.Chen Z, Huang W, Dahme T, Rottbauer W, Ackerman MJ, Xu X. Depletion of zebrafish essential and regulatory myosin light chains reduces cardiac function through distinct mechanisms. Cardiovasc Res. 2008;79:97–108. doi: 10.1093/cvr/cvn073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.van der Meer DL, Marques IJ, Leito JT, Besser J, Bakkers J, Schoonheere E, Bagowski CP. Zebrafish cypher is important for somite formation and heart development. Dev Biol. 2006;299:356–372. doi: 10.1016/j.ydbio.2006.07.032. [DOI] [PubMed] [Google Scholar]
- 89.Yang J, Xu X. α-Actinin2 is required for the lateral alignment of Z discs and ventricular chamber enlargement during zebrafish cardiogenesis. FASEB J. 2012;26:4230–4242. doi: 10.1096/fj.12-207969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hassel D, Dahme T, Erdmann J, Meder B, Huge A, Stoll M, Just S, Hess A, Ehlermann P, Weichenhan D, Grimmler M, Liptau H, Hetzer R, Regitz-Zagrosek V, Fischer C, Nurnberg P, Schunkert H, Katus HA, Rottbauer W. Nexilin mutations destabilize cardiac Z-disks and lead to dilated cardiomyopathy. Nat Med. 2009;15:1281–1288. doi: 10.1038/nm.2037. [DOI] [PubMed] [Google Scholar]
- 91.Arnaout R, Ferrer T, Huisken J, Spitzer K, Stainier DYR, Tristani-Firouzi M, Chi NC. Zebrafish model for human long QT syndrome. Proc Natl Acad Sci USA. 2007;104:11316–11321. doi: 10.1073/pnas.0702724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jou CJ, Barnett SM, Bian J-T, Weng HC, Sheng X, Tristani-Firouzi M. An In Vivo Cardiac Assay to Determine the Functional Consequences of Putative Long QT Syndrome Mutations. Circ Res. 2013;112:826–830. doi: 10.1161/CIRCRESAHA.112.300664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stoletov K, Fang L, Choi S-H, Hartvigsen K, Hansen LF, Hall C, Pattison J, Juliano J, Miller ER, Almazan F, Crosier P, Witztum JL, Klemke RL, Miller YI. Vascular Lipid Accumulation, Lipoprotein Oxidation, and Macrophage Lipid Uptake in Hypercholesterolemic Zebrafish. Circ Res. 2009;104:952–960. doi: 10.1161/CIRCRESAHA.108.189803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fang L, Green SR, Baek JS, Lee S-H, Ellett F, Deer E, Lieschke GJ, Witztum JL, Tsimikas S, Miller YI. In vivo visualization and attenuation of oxidized lipid accumulation in hypercholesterolemic zebrafish. J Clin Invest. 2011;121:4861–4869. doi: 10.1172/JCI57755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dahme T, Katus HA, Rottbauer W. Fishing for the genetic basis of cardiovascular disease. Dis Model Mech. 2009;2:18–22. doi: 10.1242/dmm.000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bakkers J. Zebrafish as a model to study cardiac development and human cardiac disease. Cardiovasc Res. 2011 doi: 10.1093/cvr/cvr098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Becker JR, Robinson TY, Sachidanandan C, Kelly AE, Coy S, Peterson RT, MacRae CA. In vivo natriuretic peptide reporter assay identifies chemical modifiers of hypertrophic cardiomyopathy signalling. Cardiovasc Res. 2012;93:463–470. doi: 10.1093/cvr/cvr350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Peal DS, Mills RW, Lynch SN, Mosley JM, Lim E, Ellinor PT, January CT, Peterson RT, Milan DJ. Novel Chemical Suppressors of Long QT Syndrome Identified by an In Vivo Functional Screen. Circulation. 2011;123:23–30. doi: 10.1161/CIRCULATIONAHA.110.003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hayward R, Hydock DS. Doxorubicin Cardiotoxicity in the Rat: An In Vivo Characterization. J Am Assoc Lab Anim Sci. 2007;46:20–32. [PubMed] [Google Scholar]
- 100.Christiansen S, Autschbach R. Doxorubicin in experimental and clinical heart failure. Eur J Cardiothorac Surg. 2006;30:611–616. doi: 10.1016/j.ejcts.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 101.Chablais F, Ja wi, ska A. Induction of myocardial infarction in adult zebrafish using cryoinjury. J Vis Exp. 2012:e3666. doi: 10.3791/3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Stainier DY, Fouquet B, Chen JN, Warren KS, Weinstein BM, Meiler SE, Mohideen MA, Neuhauss SC, Solnica-Krezel L, Schier AF, Zwartkruis F, Stemple DL, Malicki J, Driever W, Fishman MC. Mutations affecting the formation and function of the cardiovascular system in the zebrafish embryo. Development. 1996;123:285–292. doi: 10.1242/dev.123.1.285. [DOI] [PubMed] [Google Scholar]
- 103.Kettleborough RNW, Busch-Nentwich EM, Harvey SA, Dooley CM, de Bruijn E, van Eeden F, Sealy I, White RJ, Herd C, Nijman IJ, Fenyes F, Mehroke S, Scahill C, Gibbons R, Wali N, Carruthers S, Hall A, Yen J, Cuppen E, Stemple DL. A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature. 2013;496:494–497. doi: 10.1038/nature11992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lawson Nathan D, Wolfe Scot A. Forward and Reverse Genetic Approaches for the Analysis of Vertebrate Development in the Zebrafish. Developmental Cell. 2011;21:48–64. doi: 10.1016/j.devcel.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 105.Huang P, Zhu Z, Lin S, Zhang B. Reverse Genetic Approaches in Zebrafish. J Genet Genomics. 2012;39:421–433. doi: 10.1016/j.jgg.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 106.Cade L, Reyon D, Hwang WY, Tsai SQ, Patel S, Khayter C, Joung JK, Sander JD, Peterson RT, Yeh JR. Highly efficient generation of heritable zebrafish gene mutations using homo- and heterodimeric TALENs. Nucleic Acids Res. 2012;40:8001–8010. doi: 10.1093/nar/gks518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zu Y, Tong X, Wang Z, Liu D, Pan R, Li Z, Hu Y, Luo Z, Huang P, Wu Q, Zhu Z, Zhang B, Lin S. TALEN-mediated precise genome modification by homologous recombination in zebrafish. Nat Meth. 2013;10:329–331. doi: 10.1038/nmeth.2374. [DOI] [PubMed] [Google Scholar]
- 108.McKusick-Nathans Institute of Genetic Medicine John Hopkins University B, MD. Online Mendelian Inheritance in Man, OMIM. 2013 [Google Scholar]
- 109.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 110.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced Pluripotent Stem Cell Lines Derived from Human Somatic Cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 111.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 112.Nguyen PD, Hsiao ST, Sivakumaran P, Lim SY, Dilley RJ. Enrichment of neonatal rat cardiomyocytes in primary culture facilitates long-term maintenance of contractility in vitro. Am J Physiol Cell Physiol. 2012;303:C1220–C1228. doi: 10.1152/ajpcell.00449.2011. [DOI] [PubMed] [Google Scholar]
- 113.Bird SD, Doevendans PA, van Rooijen MA, Brutel de la Riviere A, Hassink RJ, Passier R, Mummery CL. The human adult cardiomyocyte phenotype. Cardiovasc Res. 2003;58:423–434. doi: 10.1016/s0008-6363(03)00253-0. [DOI] [PubMed] [Google Scholar]
- 114.Sinnecker D, Goedel A, Laugwitz K-L, Moretti A. Induced Pluripotent Stem Cell-Derived Cardiomyocytes: A Versatile Tool for Arrhythmia Research. Circ Res. 2013;112:961–968. doi: 10.1161/CIRCRESAHA.112.268623. [DOI] [PubMed] [Google Scholar]
- 115.Priori SG, Napolitano C, Di Pasquale E, Condorelli G. Induced pluripotent stem cell–derived cardiomyocytes in studies of inherited arrhythmias. J Clin Invest. 2013;123:84–91. doi: 10.1172/JCI62838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Narsinh K, Narsinh KH, Wu JC. Derivation of Human Induced Pluripotent Stem Cells for Cardiovascular Disease Modeling. Circ Res. 2011;108:1146–1156. doi: 10.1161/CIRCRESAHA.111.240374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kawaguchi N, Hayama E, Furutani Y, Nakanishi T. Prospective In Vitro Models of Channelopathies and Cardiomyopathies. Stem Cells Int. 2012;10 doi: 10.1155/2012/439219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yoshida Y, Yamanaka S. Recent stem cell advances: induced pluripotent stem cells for disease modeling and stem cell-based regeneration. Circulation. 2010;122:80–87. doi: 10.1161/CIRCULATIONAHA.109.881433. [DOI] [PubMed] [Google Scholar]
- 119.Moretti A, Bellin M, Welling A, Jung CB, Lam JT, Bott-Flügel L, Dorn T, Goedel A, Höhnke C, Hofmann F, Seyfarth M, Sinnecker D, Schömig A, Laugwitz K-L. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N Engl J Med. 2010;363:1397–1409. doi: 10.1056/NEJMoa0908679. [DOI] [PubMed] [Google Scholar]
- 120.Egashira T, Yuasa S, Suzuki T, Aizawa Y, Yamakawa H, Matsuhashi T, Ohno Y, Tohyama S, Okata S, Seki T, Kuroda Y, Yae K, Hashimoto H, Tanaka T, Hattori F, Sato T, Miyoshi S, Takatsuki S, Murata M, Kurokawa J, Furukawa T, Makita N, Aiba T, Shimizu W, Horie M, Kamiya K, Kodama I, Ogawa S, Fukuda K. Disease characterization using LQTS-specific induced pluripotent stem cells. Cardiovasc Res. 2012;95:419–429. doi: 10.1093/cvr/cvs206. [DOI] [PubMed] [Google Scholar]
- 121.Itzhaki I, Maizels L, Huber I, Zwi-Dantsis L, Caspi O, Winterstern A, Feldman O, Gepstein A, Arbel G, Hammerman H, Boulos M, Gepstein L. Modelling the long QT syndrome with induced pluripotent stem cells. Nature. 2011;471:225–229. doi: 10.1038/nature09747. [DOI] [PubMed] [Google Scholar]
- 122.Matsa E, Rajamohan D, Dick E, Young L, Mellor I, Staniforth A, Denning C. Drug evaluation in cardiomyocytes derived from human induced pluripotent stem cells carrying a long QT syndrome type 2 mutation. Eur Heart J. 2011;32:952–962. doi: 10.1093/eurheartj/ehr073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lahti AL, Kujala VJ, Chapman H, Koivisto A-P, Pekkanen-Mattila M, Kerkelä E, Hyttinen J, Kontula K, Swan H, Conklin BR, Yamanaka S, Silvennoinen O, Aalto-Setälä K. Model for long QT syndrome type 2 using human iPS cells demonstrates arrhythmogenic characteristics in cell culture. Dis Model Mech. 2012;5:220–230. doi: 10.1242/dmm.008409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yazawa M, Hsueh B, Jia X, Pasca AM, Bernstein JA, Hallmayer J, Dolmetsch RE. Using induced pluripotent stem cells to investigate cardiac phenotypes in Timothy syndrome. Nature. 2011;471:230–234. doi: 10.1038/nature09855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Davis RP, Casini S, van den Berg CW, Hoekstra M, Remme CA, Dambrot C, Salvatori D, Oostwaard DW-v, Wilde AAM, Bezzina CR, Verkerk AO, Freund C, Mummery CL. Cardiomyocytes Derived From Pluripotent Stem Cells Recapitulate Electrophysiological Characteristics of an Overlap Syndrome of Cardiac Sodium Channel Disease / Clinical Perspective. Circulation. 2012;125:3079–3091. doi: 10.1161/CIRCULATIONAHA.111.066092. [DOI] [PubMed] [Google Scholar]
- 126.Fatima A, Xu G, Shao K, Papadopoulos S, Lehmann M, Arnáiz-Cot JJ, Rosa AO, Nguemo F, Matzkies M, Dittmann S, Stone SL, Linke M, Zechner U, Beyer V, Hennies HC, Rosenkranz S, Klauke B, Parwani AS, Haverkamp W, Pfitzer G, Farr M, Cleemann L, Morad M, Milting H, Hescheler J, Šaric T. In vitro Modeling of Ryanodine Receptor 2 Dysfunction Using Human Induced Pluripotent Stem Cells. Cell Physiol Biochem. 2011;28:579–592. doi: 10.1159/000335753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jung CB, Moretti A, Mederos y Schnitzler M, Iop L, Storch U, Bellin M, Dorn T, Ruppenthal S, Pfeiffer S, Goedel A, Dirschinger RJ, Seyfarth M, Lam JT, Sinnecker D, Gudermann T, Lipp P, Laugwitz K-L. Dantrolene rescues arrhythmogenic RYR2 defect in a patient-specific stem cell model of catecholaminergic polymorphic ventricular tachycardia. EMBO Mol Med. 2012;4:180–191. doi: 10.1002/emmm.201100194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Novak A, Barad L, Zeevi-Levin N, Shick R, Shtrichman R, Lorber A, Itskovitz-Eldor J, Binah O. Cardiomyocytes generated from CPVTD307H patients are arrhythmogenic in response to β-adrenergic stimulation. J Cell Mol Med. 2012;16:468–482. doi: 10.1111/j.1582-4934.2011.01476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Carvajal-Vergara X. Patient-specific induced pluripotent stem-cell-derived models of LEOPARD syndrome. Nature. 2010;465:808–812. doi: 10.1038/nature09005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sun N, Yazawa M, Liu J, Han L, Sanchez-Freire V, Abilez OJ, Navarrete EG, Hu S, Wang L, Lee A, Pavlovic A, Lin S, Chen R, Hajjar RJ, Snyder MP, Dolmetsch RE, Butte MJ, Ashley EA, Longaker MT, Robbins RC, Wu JC. Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci Transl Med. 2012;4:130–147. doi: 10.1126/scitranslmed.3003552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Burridge PW, Anderson D, Priddle H, Barbadillo Muñoz MD, Chamberlain S, Allegrucci C, Young LE, Denning C. Improved human embryonic stem cell embryoid body homogeneity and cardiomyocyte differentiation from a novel V-96 plate aggregation system highlights interline variability. Stem Cells. 2007;25:929–938. doi: 10.1634/stemcells.2006-0598. [DOI] [PubMed] [Google Scholar]
- 132.Martinez-Fernandez A, Nelson T, Ikeda Y, Terzic A. c-MYC-Independent Nuclear Reprogramming Favors Cardiogenic Potential of Induced Pluripotent Stem Cells. J Cardiovasc Transl Res. 2010;3:13–23. doi: 10.1007/s12265-009-9150-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Martinez-Fernandez A, Nelson T, Terzic A. Nuclear Reprogramming Strategy Modulates Differentiation Potential of Induced Pluripotent Stem Cells. J Cardiovasc Transl Res. 2011;4:131–137. doi: 10.1007/s12265-010-9250-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LIR, Yabuuchi A, Takeuchi A, Cunniff KC, Hongguang H, McKinney-Freeman S, Naveiras O, Yoon TJ, Irizarry RA, Jung N, Seita J, Hanna J, Murakami P, Jaenisch R, Weissleder R, Orkin SH, Weissman IL, Feinberg AP, Daley GQ. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kim K, Zhao R, Doi A, Ng K, Unternaehrer J, Cahan P, Hongguang H, Loh Y-H, Aryee MJ, Lensch MW, Li H, Collins JJ, Feinberg AP, Daley GQ. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nat Biotech. 2011;29:1117–1119. doi: 10.1038/nbt.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kaichi S, Hasegawa K, Takaya T, Yokoo N, Mima T, Kawamura T, Morimoto T, Ono K, Baba S, Doi H, Yamanaka S, Nakahata T, Heike T. Cell line-dependent differentiation of induced pluripotent stem cells into cardiomyocytes in mice. Cardiovasc Res. 2010;88:314–323. doi: 10.1093/cvr/cvq189. [DOI] [PubMed] [Google Scholar]
- 137.Soldner F, Jaenisch R. iPSC Disease Modeling. Science. 2012;338:1155–1156. doi: 10.1126/science.1227682. [DOI] [PubMed] [Google Scholar]
- 138.Cheng L-T, Sun L-T, Tada T. Genome editing in induced pluripotent stem cells. Genes Cells. 2012;17:431–438. doi: 10.1111/j.1365-2443.2012.01599.x. [DOI] [PubMed] [Google Scholar]
- 139.Yao Y, Nashun B, Zhou T, Qin L, L Q, Zhao S, Xu J, Esteban MA, Chen X. Generation of CD34+ cells from CCR5-disrupted human embryonic and induced pluripotent stem cells. Hum Gene Ther. 2012;23:238–242. doi: 10.1089/hum.2011.126. [DOI] [PubMed] [Google Scholar]
- 140.Ramalingam S, London V, Kandavelou K, Cebotaru L, Guggino W, Civin C, Chandrasegaran S. Generation and genetic engineering of human induced pluripotent stem cells using designed zinc finger nucleases. Stem Cells Dev. 2013;22:595–610. doi: 10.1089/scd.2012.0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zou J, Mali P, Huang X, Dowey SN, Cheng L. Site-specific gene correction of a point mutation in human iPS cells derived from an adult patient with sickle cell disease. Blood. 2011;118:4599–4608. doi: 10.1182/blood-2011-02-335554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B, Meng X, Miller JC, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Jaenisch R. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotech. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sebastiano V, Maeder ML, Angstman JF, Haddad B, Khayter C, Yeo DT, Goodwin MJ, Hawkins JS, Ramirez CL, Batista LFZ, Artandi SE, Wernig M, Joung JK. In Situ Genetic Correction of the Sickle Cell Anemia Mutation in Human Induced Pluripotent Stem Cells Using Engineered Zinc Finger Nucleases. Stem Cells. 2011;29:1717–1726. doi: 10.1002/stem.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wang Y, Zheng C-G, Jiang Y, Zhang J, Chen J, Yao C, Zhao Q, Liu S, Chen K, Du J, Yang Z, Gao S. Genetic correction of [beta]-thalassemia patient-specific iPS cells and its use in improving hemoglobin production in irradiated SCID mice. Cell Res. 2012;22:637–648. doi: 10.1038/cr.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chang C-J, Bouhassira EE. Zinc-finger nuclease-mediated correction of α-thalassemia in iPS cells. Blood. 2012;120:3906–3914. doi: 10.1182/blood-2012-03-420703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Soldner F, Laganière J, Cheng Albert W, Hockemeyer D, Gao Q, Alagappan R, Khurana V, Golbe Lawrence I, Myers Richard H, Lindquist S, Zhang L, Guschin D, Fong Lauren K, Vu BJ, Meng X, Urnov Fyodor D, Rebar Edward J, Gregory Philip D, Zhang HS, Jaenisch R. Generation of Isogenic Pluripotent Stem Cells Differing Exclusively at Two Early Onset Parkinson Point Mutations. Cell. 2011;146:318–331. doi: 10.1016/j.cell.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yusa K, Rashid ST, Strick-Marchand H, Varela I, Liu P-Q, Paschon DE, Miranda E, Ordonez A, Hannan NRF, Rouhani FJ, Darche S, Alexander G, Marciniak SJ, Fusaki N, Hasegawa M, Holmes MC, Di Santo JP, Lomas DA, Bradley A, Vallier L. Targeted gene correction of [agr]1-antitrypsin deficiency in induced pluripotent stem cells. Nature. 2011;478:391–394. doi: 10.1038/nature10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Blackburn PR, Campbell JM, Clark KJ, Ekker SC. The CRISPR system-keeping zebrafish gene targeting fresh. Zebrafish. 2013;10:116–118. doi: 10.1089/zeb.2013.9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Tang C, Lee AS, Volkmer J-P, Sahoo D, Nag D, Mosley AR, Inlay MA, Ardehali R, Chavez SL, Pera RR, Behr B, Wu JC, Weissman IL, Drukker M. An antibody against SSEA-5 glycan on human pluripotent stem cells enables removal of teratoma-forming cells. Nat Biotech. 2011;29:829–834. doi: 10.1038/nbt.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Smith AJ, Nelson NG, Oommen S, Hartjes KA, Folmes CD, Terzic A, Nelson TJ. Apoptotic Susceptibility to DNA Damage of Pluripotent Stem Cells Facilitates Pharmacologic Purging of Teratoma Risk. Stem Cells Transl Med. 2012;1:709–718. doi: 10.5966/sctm.2012-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ben-David U, Gan Q-F, Golan-Lev T, Arora P, Yanuka O, Oren Yifat S, Leikin-Frenkel A, Graf M, Garippa R, Boehringer M, Gromo G, Benvenisty N. Selective Elimination of Human Pluripotent Stem Cells by an Oleate Synthesis Inhibitor Discovered in a High-Throughput Screen. Cell Stem Cell. 2013;12:167–179. doi: 10.1016/j.stem.2012.11.015. [DOI] [PubMed] [Google Scholar]
- 152.Choi SM, Kim Y, Shim JS, Park JT, Wang R-H, Leach SD, Liu JO, Deng C-X, Ye Z, Jang Y-Y. Efficient drug screening and gene correction for treating liver disease using patient-specific stem cells. Hepatology. 2013 doi: 10.1002/hep.26237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotech. 2013 doi: 10.1038/nbt.2623. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-Guided Human Genome Engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Uhde-Stone C, Gor N, Chin T, Huang J, Lu B. A do-it-yourself protocol for simple transcription activator-like effector assembly. Biol Proced Online. 2013;15:3. doi: 10.1186/1480-9222-15-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Li T, Huang S, Zhao X, Wright DA, Carpenter S, Spalding MH, Weeks DP, Yang B. Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Res. 2011;39:6315–6325. doi: 10.1093/nar/gkr188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sander JD, Cade L, Khayter C, Reyon D, Peterson RT, Joung JK, Yeh J-RJ. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat Biotech. 2011;29:697–698. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Moore FE, Reyon D, Sander JD, Martinez SA, Blackburn JS, Khayter C, Ramirez CL, Joung JK, Langenau DM. Improved Somatic Mutagenesis in Zebrafish Using Transcription Activator-Like Effector Nucleases (TALENs) PLoS ONE. 2012;7:e37877. doi: 10.1371/journal.pone.0037877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hoover DM, Lubkowski J. DNA Works: an automated method for designing oligonucleotides for PCR-based gene synthesis. Nucleic Acids Res. 2002;30:e43. doi: 10.1093/nar/30.10.e43. [DOI] [PMC free article] [PubMed] [Google Scholar]