Abstract
High-resolution diffusion MRI (dMRI) is useful for resolving complex microstructures in the mouse brain, but technically challenging for in vivo studies due to the long scan time. In this study, selective excitation and a three-dimensional fast imaging sequence were used to achieve in vivo high-resolution dMRI of the mouse brain at 11.7 Tesla. By reducing the field of view using spatially selective radio frequency pulses, we were able to focus on targeted brain structures and acquire high angular resolution diffusion imaging (HARDI) data at an isotropic resolution of 0.1 mm and 30 diffusion encoding directions in approximately one hour. We investigated the complex tissue microstructures of the mouse hippocampus, cerebellum, and several cortical areas using this localized dMRI approach, and compared the results with histological sections stained with several axonal and dendritic markers. In the mouse visual cortex, the results showed predominately radially arranged structures in an outer layer and tangentially arranged structures in an inner layer, similar to observations from postmortem human brain specimens.
1. Introduction
Diffusion magnetic resonance imaging (dMRI) is a useful tool in the ongoing investigation of the organization, functions, and diseases of the brain. By measuring the properties of water diffusion, which are affected by local tissue barriers inside the brain, e.g., the cell membrane and myelin sheath (Beaulieu, 2002), dMRI provides image contrasts that contain rich microstructural information and allow noninvasive reconstruction of major white matter pathways (Le Bihan, 2003; Mori and Zhang, 2006). For example, diffusion tensor imaging (DTI) has been widely used to examine white matter morphology and structural integrity (Basser et al., 1994; Mori and Zhang, 2006). More advanced dMRI techniques, such as high angular resolution diffusion imaging (HARDI) (Frank, 2001; Tuch et al., 2003) and diffusion spectrum imaging (DSI) (Wedeen et al., 2005), have been designed to investigate the complex structural organization of the brain. The capability of these dMRI techniques in visualizing complex brain structures and connectivity has been demonstrated by several studies of post-mortem brain specimens (Aggarwal et al., 2013; Calamante et al., 2012; Leuze et al., 2012; Takahashi et al., 2013; Wedeen et al., 2012), as well as studies of the live human brain (Granziera et al., 2009; McNab et al., 2013; Setsompop et al., 2013; Verstynen et al., 2011). Recently, multi-parameter models based on customized dMRI acquisition schemes have been reported to extract specific microstructural information from neuronal tissues (Assaf and Basser, 2005; Wang et al., 2011; Zhang et al., 2012a).
Further developments of dMRI hinge on our understanding of the relationships between dMRI signals and the underlying brain microstructure (Barazany et al., 2009; Budde et al., 2009; Hansen et al., 2011; Leergaard et al., 2010; Wang et al., 2011; Zhang et al., 2012a). Such knowledge can be obtained through studying the relationships between dMRI signals and histopathological findings in experimental animal models. The laboratory mouse brain is an ideal subject for examining the microstructural basis of dMRI signals, because there is a wealth of histology-based information on its microstructure. In addition, there are many well-established mouse models that mimic various pathological conditions in patients with neurological diseases. Knowledge gained through studying the mouse brain using dMRI may be readily translated to study the microstructural organization and pathology of the human brain.
In the last decade, dMRI has emerged as a useful tool to examine anatomy and pathology of the mouse brain (Mori and Zhang, 2006; Zhang et al., 2012c). Ex vivo high-resolution dMRI provides superior image quality at high spatial resolution and is well-suited for visualizing complex microstructures in the mouse brain (Aggarwal et al., 2010; Calamante et al., 2012; Flint et al., 2010; Jiang and Johnson, 2010; Moldrich et al., 2010). At spatial resolutions of 100 μm or higher, ex vivo dMRI can reveal exquisite anatomical details in the mouse brain, and with high angular resolution, reconstruct complex tract trajectories, such as crossing or branching fiber tracts (Kurniawan et al., 2013; Moldrich et al., 2010). In vivo dMRI, while cannot match ex vivo dMRI in terms of image quality and spatial resolution, is key to investigate the relationships between dMRI signals and microstructures in live tissues, especially under various pathological conditions presented in mouse models of diseases. This is because death and chemical fixation can alter the microstructural properties of brain tissues, resulting in significant changes in apparent diffusion coefficient measurements (Shepherd et al., 2009; Sun et al., 2005; Wu et al., 2013) and changes in the sensitivity of related dMRI-based markers to detect certain tissue pathology (Sun et al., 2006a; Zhang et al., 2012b). The main technical challenges of in vivo high-resolution mouse brain dMRI are limited imaging resolution and lengthy acquisition time, which limits our ability to examine small structures or perform longitudinal studies. With recent advances in MR instruments (e.g., high field magnet, high performance gradient and coils, and fast imaging sequences), several groups have reported in vivo dMRI studies with increasing spatial and angular resolutions. For example, Harsan et al. reported in vivo mouse brain HARDI with 30 diffusion encoding directions (b = 1000 s/mm2) and a resolution of 0.156 mm x 0.156 mm x 0.5 mm in 99 minutes at 9.4 Tesla (Harsan et al., 2013), and we demonstrated in vivo DTI of the mouse brain with 12 diffusion encoding directions (b = 1000 s/mm2)and an isotropic resolution of 0.125 μm in about 2.5 hours at 11.7 Tesla (Wu et al., 2013). In vivo dMRI with both high spatial and angular resolutions will be ideal if the total imaging time falls within one or two hours.
In this work, we demonstrate that in vivo HARDI of the mouse brain can be performed at approximately 0.1 mm isotropic resolution and 30 diffusion directions in one hour using spatially selective RF pulses and a three-dimensional (3D) gradient and spin echo (GRASE) (Oshio and Feinberg, 1991) imaging sequence, modified from (Aggarwal et al., 2010). By selectively exciting a portion of the brain that contains the structures of interest, a dramatic reduction in imaging volume, and therefore, imaging time, was possible. Further acceleration was achieved using the 3D GRASE sequence, which obtained images twenty times faster than conventional spin echo sequences in this study. High-resolution dMRI data of the cerebellum, hippocampus, and several cortical regions of the mouse brain were compared to histology, focusing on the capability of dMRI to resolve microstructures and connectivity in these regions.
2. Material and methods
2.1 Experimental animals
All experimental procedures were approved by the Animal Use and Care Committee at the Johns Hopkins University School of Medicine. Seven adult mice (C57BL/6, three-month old, female, Jackson Laboratory, Bar Harbor, ME, USA) were used in this study.
2.2 RF pulse design
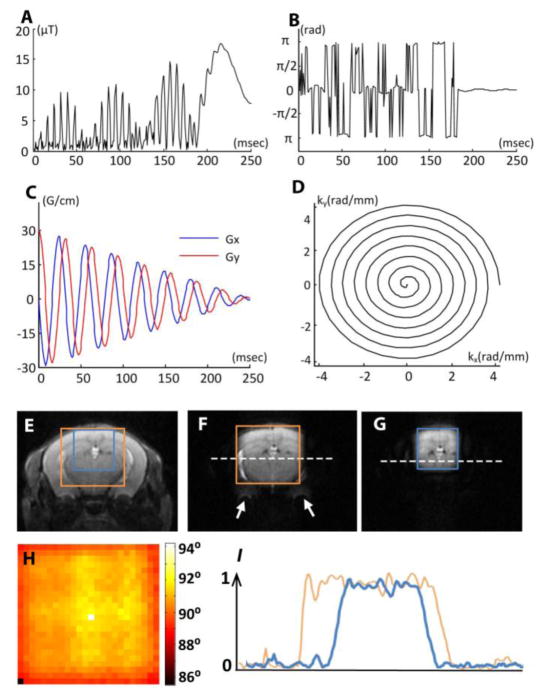
The 90° localized excitation RF pulses used in this study were designed based on a linear class of large tip-angle (LCLTA) pulses (Pauly et al., 1989), with spiral k-space trajectories that start and end at the origin. Under the “incoherently refocused” condition (Pauly et al., 1989), a two-dimensional spatially selective 90° RF pulse can be derived by inverse Fourier transform of the desired excitation profile. Although B1 field weighting can be incorporated in the Fourier kernel, we assumed a homogeneous B1 field across the mouse brain because the mouse brains were always positioned near the center of a large volume coil, and the inner diameter (72 mm) of the volume coil was far larger than the size of the mouse brain (10–12 mm in diameter). The pulse length was set at 2.5 ms, and the pulse amplitudes ranged from 15 to 20 μT, depending on the size, position, and shape of the desired field of excitation (FOE) (Pauly et al., 1989). The typical waveform of an RF pulse to excite a square region in the mouse brain is shown in Fig. 1A–B. We chose an eight-turn spiral-in excitation k-space trajectory (Fig. 1C–D), which resulted in an excitation resolution of 1 x 1 mm in the x-y plane, with a maximum gradient strength of about 210 mT/m.
Figure 1.
Spatially selective excitation RF pulse and its experimental validation. A–B: The pulse amplitude and phase of a typical 90° selective excitation pulse. C–D: an eight-turn spiral-in gradient waveform and excitation k-space trajectory in the x-y plane. The gradient waveform was synchronized with the RF waveform to achieve selective excitation. (E) A T2-weighted image of an adult mouse brain overlaid with a 8 mm x 8 mm field of excitation (FOE) (orange) and a 6 mm x 6 mm FOE (blue). (F–G) The results of selective excitation corresponding to the 8 mm x 8 mm and 6 mm x 6 mm FOEs shown in (E), respectively. H: the measured flip angle map of the excited region (corresponding to the FOE in F). I: the normalized (with respect to the maximal value) intensity profiles along the dashed lines in F and G.
Typical FOEs used in this study included the anterior, middle, and posterior brain FOEs (Table 1). The anterior and middle FOEs were designed to cover only half the brain, assuming relative symmetry in normal brains. The imaging field of view (FOV) was chosen to be slightly larger than the field of excitation (FOE) to accommodate the transition area between the excited region and the suppressed region (approximately 1/10 of the FOE). The performance of the spatially selective RF pulses was tested using the standard, double flip-angle B1 mapping (Cunningham et al., 2006). While the FOE in the x-y plane was controlled by the selective excitation pulses, a slab-selective refocusing pulse (Mao pulse (Mao et al., 1988)) was applied to restrict the imaging slab in the z direction.
Table 1.
Definition of the anterior, middle and posterior brain fields of excitation (FOEs), and the SNR of non-diffusion weighted images from these FOEs.
| Field of Excitation (FOE) | Anterior | Middle | Posterior |
|---|---|---|---|
| FOE width & height (mm) | 8 x 5 | 8 x 5 | 8 x 5 |
| FOV size (readout x phase x slice) (mm) | 9.6 x 5.6 x 4 | 9.6 x 5.6 x 4 | 9.6 x 5.6 x 4 |
| Readout direction | Superior-inferior | Superior-inferior | Left-right |
| Slice position with respect to Bregma (mm) | +2 to −2 | 0 to −4 | −5 to −9 |
| Structures of interest | Motor/Sensory cortex Striatum (right hemisphere) | Sensory/visual cortex Hippocampus (right hemisphere) | Cerebellum |
| SNR of non-diffusion image | 42 ± 0.6 (n=5) | 44 ± 2 (n=5) | 42 ± 2.6 (n=3) |
2.3 Image acquisition
In vivo imaging was performed on a horizontal 11.7 Tesla MR scanner (Bruker Biospin, Billerica, MA, USA) with an integrated shim and active shielded triple-axis gradient (B-GA 9S, Burker Biospin, Billerica, MA, USA, inner diameter = 90 mm, maximum gradient strength = 740 mT/m). During imaging, mice were anesthetized with isoflurane (1%), together with air and oxygen mixed at a 3:1 ratio, via a vaporizer, and were positioned in an animal holder (Bruker Biospin, Billerica, MA, USA). Respiration was monitored via a pressure sensor (SAII, Stony Brook, NY, USA) and maintained at 40–60 breaths per minute. After imaging, animals recovered within five minutes.
The selective excitation pluses were transmitted through a quadrature volume excitation coil (72 mm diameter, Bruker Biospin, Billerica, MA, USA), and signal was acquired using a 10 mm diameter, planar surface receive-only coil (with active decoupling). The planar receive surface coil was placed immediately above the specific FOE to ensure optimal sensitivity. Using the Fieldmap method (Schneider and Glover, 1991; van Zijl et al., 1994; Wen and Jaffer, 1995), B0 homogeneity in a region containing the FOE was optimized using the first and second order shim to reduce image distortion and a T2* signal decay. A water line width of less than 30 Hz could be routinely achieved within the FOE. Before each dMRI experiment, a multi-slice, T2-weighted image (echo time (TE) = 50 ms, repetition time (TR) of 2000 ms) was first acquired as an anatomical reference to define the position of the FOE. The selective excitation RF pulse was then calculated based on the reference image and desired FOE in Matlab (Mathworks, Natick, MA, USA).
HARDI data were acquired with a modified 3D diffusion-weighted GRASE sequence (Aggarwal et al., 2010), which acquired 40 echoes in each repetition. A double-sampled EPI readout (Yang et al., 1996), which samples the same k-space line twice, with both positive and negative gradients, was incorporated into the GRASE sequence to enhance SNR, as well as to reduce artifacts from imperfect read-out gradients. A twin-navigator (Mori and van Zijl, 1998) was implemented to correct motion-induced phase errors, and no respiration trigger was used. The imaging parameters of the HARDI protocol were: TE/TR = 23/500ms; two signal averages; spectral width = 150 kHz; 30 diffusion directions (Jones et al., 1999); b = 2335 s/mm2. The anterior and posterior brain FOEs were imaged at 0.1 x 0.1 x 0.1 mm resolution, while the middle brain FOE was imaged at 0.125 x 0.125 x 0.125 mm, 0.1 x 0.1 x 0.1 mm, and 0.08 x 0.08 x 0.08 mm resolution. The imaging parameters used to acquire data at these resolutions are summarized in Table 2. We measured the point-spread function (PSF) of the imaging sequence to evaluate the effects of the T2 and T2* decays on image resolutions as described in (Wu et al., 2013). The full-width-half-maximum (FWHM) of the PSF is 0.119 ± 0.003 mm along the phase encoding direction (due to T2 decay) and 0.073 ± 0.004 mm (n=5) along the slice selection (due to T2* decay) direction. The signal-to-noise ratio (SNR) was evaluated as the ratio of the mean signal intensity in the brain cortex against the standard deviation of the background noise.
Table 2.
Imaging parameters to acquire 30-direction HARDI data at 0.125 mm, 0.1 mm and 0.08 mm isotropic resolution from the middle brain FOE. Because of the use of blip gradients in the slice selection direction, the maximum slice selection gradients were larger than the maximum phase encoding gradients.
| Resolution (isotropic) | 0.125 mm | 0.1 mm | 0.08 mm |
|---|---|---|---|
| Scan time (minutes) | 42 | 63 | 102 |
| SNR of non-diffusion image | 57 (n=1) | 44 ± 2 (n=5) | 33 ± 2 (n=5) |
| Duty cycle (%) | 13.2 | 18.5 | 24.8 |
| Maximum readout gradient (mT/m) | 286 | 358 | 429 |
| Maximum phase encoding gradient (mT/m) | 144 | 180 | 216 |
| Maximum slice selection gradient (mT/m) | 360 | 444 | 562 |
Ex vivo whole brain HARDI data were acquired from a postmortem C57BL/6 mouse brain specimen, which was perfusion fixed with 4% paraformaldehyde (PFA), and later transferred to phosphate buffered saline with 2 mM gadopentetate dimeglumine (Magnevist, Berlex Imaging, Wayne, NJ) (Chuang et al., 2011) to enhance MR signals. Specimens were scanned using a vertical 11.7 Tesla NMR spectrometer (Bruker Biospin, Billerica, MA, USA) and a birdcage volume coil (15 mm inner diameter). The same imaging sequence as that used in vivo MRI with whole-brain coverage was used, but with slightly different parameters: TE/TR = 27/1000ms; two signal averages; spectral width = 125 kHz; 30 diffusion directions; b = 4000 s/mm2; 0.1 x 0.1 x 0.1 mm resolution. The total scan time was about 20 hours.
2.4 Data processing
Images were reconstructed from k-space data using Matlab (Mathworks) and processed using MRtrix (Tournier et al., 2012), which used the constrained spherical deconvolution (CSD) method to reconstruct fiber orientation distribution (FOD). FOD-based probabilistic streamline tracking was performed to generate super-high-resolution track density images (TDI) (Calamante et al., 2011; Calamante et al., 2010). Fiber-tracking was performed using a step size of 0.01 mm and a maximum angle of 45° between steps, according to (Calamante et al., 2012). Tracking terminated when the FOD amplitude became less than 0.01, or when fibers exited the specific brain regions. Probabilistic streamlines with a length between 0.4 – 1 mm were selected (Calamante et al., 2012), and about one million such streamlines were tracked to generate TDI at a grip size of 10 μm isotropic resolution.
2.5 Immunohistochemistry
Mice were anesthetized and perfused with 0.1M phosphate-buffered saline (PBS), followed by 4% paraformaldehyde in 0.1M PBS, and then were post-fixed with 4% paraformaldehyde for 12–18 hours. Brains were cryo-protected in a 30% sucrose solution in 0.1M PBS. The forebrain was cryostat-sectioned at 40 μm coronally and the cerebellum was sectioned at 20 μm parasagitally. Representative sections were mounted and adjacent slides were used for immunohistochemical stainings with different antibodies. Endogenous peroxidase was quenched using 3% hydrogen peroxide in 0.1M PBS. Antigen-retrieval was performed with 0.01 M citrate buffer containing 0.05% Tween-20 (pH 6). Slides were then incubated in blocking solution, followed by primary antibody incubation at 4°C overnight: Anti-Pan-Axonal Neurofilament Marker (SMI312R, Covance, Princeton, NJ, USA, 1:2000) and Anti-Parvalbumin antibody (ab11427, Abcam, MA, USA, 1:4000) for the detection of large GABAergic neurons and their processes (e.g., Purkinje-Neurons of the cerebellum); Anti-Glial Fibrillary Acidic Protein antibody (Anti-GFAP Z0334, Dako, Richmond, VA, USA, 1:2500) for the detection of astrocytes; and Anti-Microtubule-associated Protein 2 (Anti-MAP2, M1406, Sigma-Aldrich, St.Louis, MO, USA, 1:1000) for the detection of neuronal dendrites. Negative control slides were put in blocking solution. Slides were rinsed and visualized using the ABC ELITE kit (Vector Labs, Burlingame, CA, USA) and 3,3′-Diaminobenzidine (DAB, Sigma-Aldrich, St. Louis, MO, USA). Images were acquired using a Zeiss Observer.Z1 microscope equipped with an AxioCam MRc camera at 20X.
3. Results
The 90° spatial excitation RF pulse designed in this study produced satisfactory 2D excitation profiles using a conventional quadrature volume coil at 11.7 Tesla. Images acquired with an 8 mm x 8 mm FOE and a 6 mm x 6 mm FOE in the center of the brain (Fig. 1E) are shown in Fig. 1F–G. The residual outer-volume signal intensity was less than 6% of the average inner-volume signal intensity (Fig. 1I). For large FOEs that include air-tissue interfaces, e.g., the area close to the ear canal, which have severe B0 inhomogeneity and thereby distort the excitation k-space trajectory, residual signals outside the FOE could be observed near the air-tissue interfaces (indicated by the white arrows in Fig. 1F). The artifact disappeared when the FOE fell completely within the brain (Fig. 1G). Within the selected region, the measured flip angles were relatively uniform (90°±2.2°, Fig. 1H).
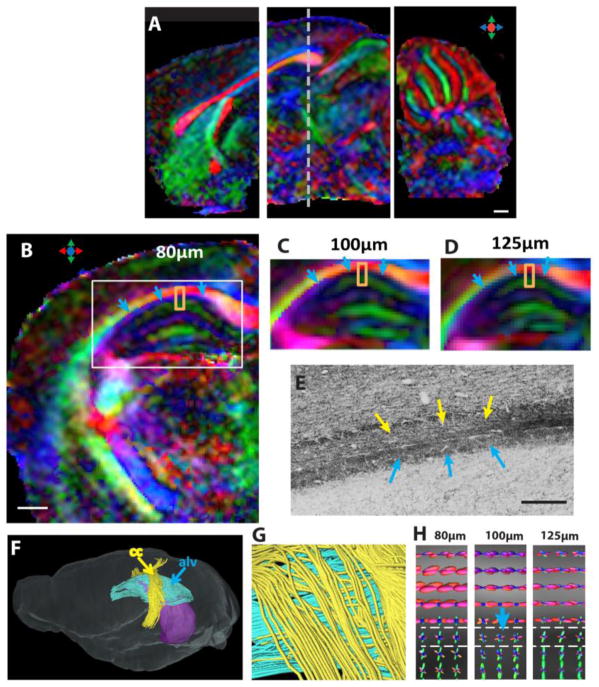
Using selective excitation, high-resolution dMR images of local brain regions could be acquired within a relatively short time period. Figure 2A shows three sagittal direction-encoded colormap (DEC) images acquired separately with the anterior, middle, and posterior FOEs at 100 μm isotropic resolution. Figures 2B–D compare DEC images acquired at 80 μm isotropic resolution with images acquired at 100 μm and 125 μm isotropic resolution. With the standard 30-direction diffusion-encoding scheme (Jones and Leemans, 2011), the imaging time ranged from 40 minutes for the 125 μm resolution data to two hours for the 80 μm resolution data in a local FOE (Table 2). Major white matter tracts, e.g., the corpus callosum, and several small white matter structures could be resolved in the high-resolution data (Fig. 2A–B). For example, the alveus of the hippocampus, a thin sheet of axonal fibers (indicated by the blue arrows in Fig. 2B–E), could be distinguished from the neighboring corpus callosum/external capsule and hippocampus in the DEC image at 80 μm resolution (Fig. 2B). Tensor-based fiber-tracking results based on the 80 μm data could separate the alveus from the corpus callosum (Fig. 2F–G). In comparison, images acquired at 100 and 125 μm resolutions (Fig. 2C–D) showed the alveus with reduced FA and a loss of clearly defined diffusion orientation, due to partial volume effects. In order to reliably resolve small white matter tracts and crossing fibers, it is necessary to use more sophisticated approaches than the conventional diffusion tensor model. Figure 2H demonstrates that fiber orientation distribution (FOD) estimated from the dMRI data, using spherical deconvolution, could resolve the fibers in the alveus (blue FOD surface) and corpus callosum/external capsule (red FOD surface) at 100 resolution.
Figure 2.
High spatial resolution diffusion MRI of the live mouse brain. A: Sagittal views of the DTI colormaps taken from the anterior, middle, and posterior FOEs used in this study. B–D: Axial direction-encoded colormaps of the hippocampus at 80 μm, 100 μm, and 125 μm isotropic resolutions, respectively. The axial location of these images is indicated by the dashed line in A. E: A Pan-Neurofilament-stained section that shows the alveus (alv) of the hippocampus (indicated by the blue arrows) and the corpus callosum (indicated by the yellow arrows). F–G: Fiber-tracking results at 80 μm isotropic resolution, where the corpus callosum fibers (yellow) and the alv fibers (blue) can be separated. The purple shading in F indicates the position of hippocampus. H: The fiber orientation distribution (FOD) maps of the selected regions (orange squares in B–D) at three resolutions. Scale bars in A and B are 500 μm, and the scale bar in E is 200 μm.
Figure 3 compares localized in vivo dMRI data with ex vivo whole-brain dMRI data, both acquired at 100 μm resolution. For white matter structures, the in vivo and ex vivo DEC images (Fig. 3A and B) showed similar tissue contrasts, and several small white matter tracts, e.g., the dorsal hippocampal commissure and optic tracts, could be delineated in both datasets. In the hippocampus, the in vivo and ex vivo data both showed the radiating pattern (indicated by the orange arrows) reported previously (Calamante et al., 2012; Zhang et al., 2002). This is further illustrated using the track-density imaging (TDI) technique (Fig. 3B and E), which renders fiber tracking results at a resolution higher than the native imaging resolution, and provides an intuitive way to visualize tissue microstructures. This pattern was in good agreement with the spatial arrangement of small axons and dendrites in the hippocampus, as shown by the Parvalbumin-stained histological sections (Fig. 3C and F). Small differences in gray matter tissue contrasts, however, could be observed in several regions. For example, the in vivo data showed more prominent radially organized structures near the surface of the visual cortex (Fig. 3D, indicated by the yellow arrows) and higher diffusion anisotropy in a region immediately above the dentate gyrus (Fig. 3D, indicated by the blue arrows) than the ex vivo data.
Figure 3.
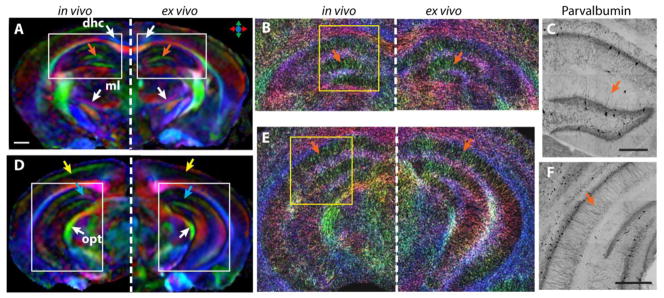
Comparison of the localized in vivo dMRI of the mouse hippocampus with the ex vivo results from a whole-brain scan. A–B: DTI colormaps (scale bar 500 μm) and TDI maps from the anterior hippocampus from the in vivo (left) and ex vivo (right, mirrored) dMRI experiments. C: Parvalbumin-stained section of a selected area in the anterior hippocampus. D–F: DTI colormaps, TDI map, and Parvalbumin-stained section of the posterior hippocampus, following the same order as in A–C. Abbreviations: dhc—dorsal hippocampal commissure; ml—medial lemniscus; opt—optical tract. Scale bars in C and F are 200 μm.
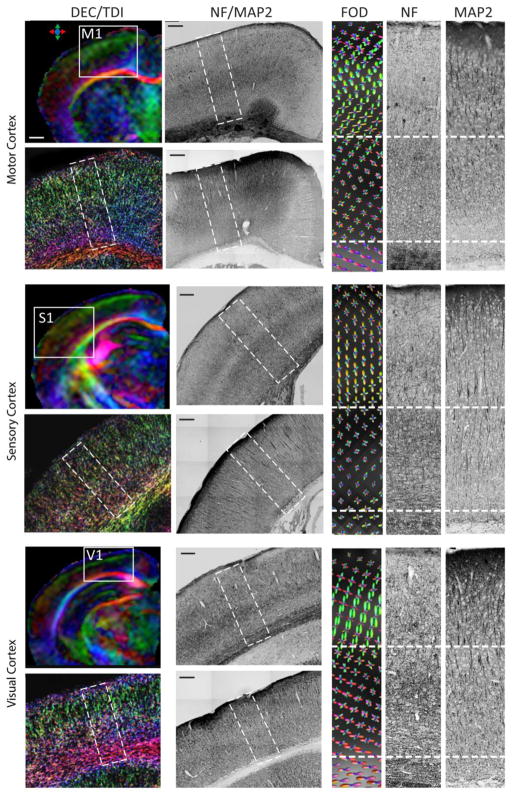
The in vivo high resolution HARDI data could provide detailed tissue microstructural information in the mouse brain. As shown in Figure 4A, individual cerebellar folium and white matter tracts in the mouse cerebellum could be delineated in the DEC image based on the in vivo HARDI data. While the DEC image shows only the dominant structural components in each pixel, e.g., parallel fibers in the cerebellar cortex (indicated by the white arrow in Fig. 4A), spherical deconvolution of the HARDI data revealed additional structural components (represented by the blue FOD surfaces in Fig. 4D) that were arranged perpendicular to the parallel fibers (represented by the red FOD surfaces in Fig. 4D). Neurofilament (NF)-, Glial Fibrillary Acidic Protein (GFAP)-, and Parvalbumin-stained sections (Fig. 4C, E–G) showed that the secondary structural components might include small axons, dendrites, and processes of glial cells in the cerebellar cortex, as they all run orthogonal to the parallel fibers (perpendicular to these sections). The high-resolution TDI of the cerebellum (Fig. 4B) demonstrated the spatial arrangement of axonal fibers as they fanned out near the end of the cerebellar folia, as shown by the neurofilament staining (Fig. 4E). Figure 5 compares MRI and histological images of the mouse motor, sensory, and visual cortices. In all three cortical regions, MAP2-stained sections showed radially oriented long-running apical dendrites in the cortex. In comparison, NF-stained sections showed radially oriented axons in the outer layer of the cortex and densely populated axons with no clear dominant orientation in the inner layer. FOD maps derived from the in vivo HARDI data showed predominantly radially oriented FODs in the outer layer of the three cortices and both radially and tangentially oriented FODs in the inner layer of the cortices. Compared to the motor cortex, TDI-generated images showed that the inner layer of the visual cortices have a large amount of axons running tangentially to the cortical surface, and the estimated FOD maps showed these tangential fibers, especially in the visual cortex. In all three cortices, the outer layers had higher FA values than the inner layers, with most significant difference in the visual cortex (Table 3).
Figure 4.
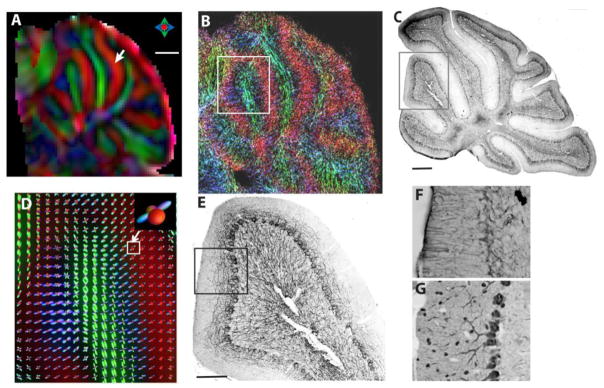
Localized HARDI of the mouse cerebellum. A–B: DTI colormap and TDI of the mouse cerebellum on a mid-sagittal view. C: Neurofilament-stained section corresponding to B. D: Fiber orientation distribution (FOD) map of the selected area indicated in B. The inset plot indicates two groups of crossing fiber coexisting in the cerebellar cortex. E: Neurofilament-stained section corresponding to D. F–G: Glial fibrillary acidic protein (GFAP)- and Parvalbumin-stained sections of the cerebellar cortex. Scale bars in A and C are 500 μm, and the scale bar in E is 200 μm.
Figure 5.
In vivo HARDI of the mouse motor (M1), sensory (S1), and visual (V1) cortices and corresponding neurofilament (NF) and microtubule-associated protein 2 (MAP2)-stained sections. Images in the left-most column are DTI colormaps of localized anterior / middle / posterior mouse brain FOEs and high-resolution TDI maps of the M1 / S1 / V1 cortices in selected areas, as indicated on the colormaps. The right-most three columns show FOD maps estimated from the HARDI data from each cortex and matching NF- and MAP2-stained sections. The regions shown in the FOD maps are indicated by the dashed rectangular boxes in the NF and MAP2 sections. The dashed lines in the rightmost three columns separate the cortex into outer and inner layers. Scale bar in A is 500 μm, and all the other scale bars are 200 μm.
Table 3.
FA values measured from the outer and inner layers of the motor, sensory, and visual cortices.
| Regions of interest | FA | p-value | |
|---|---|---|---|
| Motor cortex | Outer | 0.143 ± 0.016 | 0.07 |
| Inner | 0.136 ± 0.015 | ||
| Sensory cortex | Outer | 0.142 ± 0.018 | 0.02 |
| Inner | 0.122 ± 0.009 | ||
| Visual cortex | Outer | 0.159 ± 0.009 | 9.5 x 10−5 |
| Inner | 0.090 ± 0.005 | ||
4. Discussion
Spatially localized imaging is a tried-and-true approach to obtain high-resolution images from a target region within a reasonable time. Even though imaging with whole-brain coverage is important for examining the overall anatomy or neuropathology, many studies that focus on a particular brain region, e.g., the mouse cortex and hippocampus, may benefit from the increased imaging resolution or speed provided by localized imaging techniques. Studies that use mouse models of diseases, such as stroke (Wang et al., 2008), traumatic brain injury (Mac Donald et al., 2007), and demyelination (Sun et al., 2006b), often have localized pathology, and may benefit from the techniques as well. Several techniques have been utilized to achieve spatial localization. For example, 2D localized imaging can be achieved by using a slice-selective 90° RF pulse and a 180° RF pulse in an orthogonal fashion (Hiepe et al., 2009; Jeong et al., 2005), or by using direct 2D RF pulses as demonstrated by Finsterbusch et al. (Finsterbusch, 2010, 2013). While the former requires no specially designed RF pulses, the latter offers more flexibility in terms of pulse sequence design and shape of the region to be excited. The LCLTA pulses used in this study achieved satisfactory in-volume homogeneity (90° ± 2.2°) and outer-volume suppression within a relatively short duration (2.5 ms), which was ideal for our experiments, as tissue T2s shorten at a high magnetic field. Moreover, the LCLTA method provides an analytical approach to pulse design and is compatible with parallel transmission. For example, Xu, et al. (Xu et al., 2007) extended the LCLTA framework to an eight-channel parallel transmitter and achieved about a four-fold acceleration for 2D selective excitation profiles in a phantom experiment. Ullman et al. (Schneider et al., 2012) demonstrated 3D localized imaging of the live rat brain based on a numerical optimization of small tip-angle pulses (Grissom et al., 2006; Yip et al., 2005). In this study, the single-channel LCLTA pulse was integrated with our DW-GRASE sequence to improve the spatial and angular resolutions of in vivo dMRI experiments. Simple rectangular FOEs were sufficient here, given that the imaging volume is rectangular with conventional Cartesian k-space acquisition. Using the FOEs in our experiments, the imaging volumes were reduced to approximately one-sixth of the whole-brain volume, and the scan time was reduced proportionally. To mitigate the SNR loss associated with high spatial resolution, heavy diffusion attenuation, and short scan time, we placed a sensitivity surface receive coil as close to the FOE as possible. This, however, is only effective when the target regions are close to the head surface, such as the cortex, whereas the deep brain structures, e.g., the thalamus and the hypothalamus, still suffer from low SNR.
Localized high-resolution dMRI, using selective excitation and fast imaging sequences, allowed us to examine the live mouse brain in greater detail than ever before. While ultra-high resolution (<50 μm) ex vivo dMRI of the mouse brain has been reported (Aggarwal et al., 2010; Flint et al., 2010; Jiang and Johnson, 2010), spatial and diffusion angular resolutions of the live mouse brain dMRI have been hindered by SNR and imaging time. For example, given a typical FOV of 16 mm x 16 mm x 18 mm for the adult mouse brain (Wu et al., 2013), whole brain dMRI performed using comparable settings as described here (100 μm isotropic resolution, 30 diffusion directions, b = 2335 s/mm2) would take about 10 hours. In this study, we were able to acquire in vivo HARDI of the mouse hippocampus at 125 μm, 100 μm, and 80 μm isotropic resolution within one to two hours, and the level of microstructural details available in the in vivo 100 μm resolution data were comparable to ex vivo whole brain data acquired at the same resolution (Fig. 3). One limitation of the GRASE sequence used here was image blurring as measured by the FWHMs of the point-spread function of the in vivo dMRI experiments. The broadening of the point-spread function was caused by T2 an d T2* decays experienced by the spin and gradient echoes, imperfect refocusing pulses, and subject motions, and can be reduced by using higher spectral width, which requires strong gradient systems, improved refocusing pulses, and more sophisticated shimming and motion suppression techniques.
We tested the capability of the technique to resolve microstructures in the mouse cerebellum, which has been well documented. In the mouse cerebellar cortex, the in vivo data showed two orthogonally arranged groups of fibers: one parallel to the surface of the cerebellar cortex (represented by the red FOD surfaces in Fig. 4D), and the other perpendicular to the cerebellar cortex (represented by the blue FOD surfaces in Fig. 4D), similar to previous reports based on post-mortem mouse brain specimens (Zhang et al., 2006). The first group may reflect the densely packed parallel fibers in the cerebellar cortex, and the second group may reflect axons/dendrites of the cerebellar Purkinje cells (Fig. 4G) and processes of glia cells (Fig. 4F). We then examined the microstructural organization of the mouse neo-cortex. There have been several reports on imaging cortical microstructures in the human brain. Diffusion anisotropy in the cortex was revealed in in vivo DTI studies of both developing (McKinstry et al., 2002) and mature (Heidemann et al., 2010) human brains. Several recent studies showed that different cortical regions have unique microstructural signatures. For example, McNab et al. showed that the human motor cortex contained mostly radially organized structures, whereas part of the sensory cortex showed tangentially organized structures (McNab et al., 2013). High-resolution HARDI of postmortem brain samples further delineated the layered patterns in the human cortex, e.g., Dyrby et al. separated the cortex into two depth layers based on the different fiber orientations in the inner and outer rims of the cortex (Dyrby et al., 2011); Leuze et al. divided the visual cortex into four layers using the FOD patterns and tractography feature (Leuze et al., 2012). Other studies have attempted to parcellate cortical regions using raw diffusion signals (Deoni and Jones, 2006) or FOD-derived contrasts (Haroon et al., 2010; Nagy et al., 2013). These post-mortem studies demonstrated that high-resolution HARDI signals could be used to characterize the neo-cortical microstructure, especially the layered organization with radial or tangential patterns. It is not clear whether in vivo high-resolution data will show consistent cortical tissue contrasts and how the contrasts correlate with microstructures. As the human and mouse neocortex, despite their differences, share a similar columnar organization (DeFelipe et al., 2002), high-resolution studies on the mouse cortex could bring insight into these questions. In this study, high-resolution dMRI of the mouse cortex showed contrast patterns similar to those seen in postmortem human brain specimens. For example, the observation that radially arranged structures dominate the outer layer of the visual cortex, while tangentially arranged structures dominate in the inner layer (Fig. 5), is similar to the observations reported by previous post-mortem human brain studies (Dyrby et al., 2011; Leuze et al., 2012). A comparison between HARDI results and histological data suggested that multiple structural components could contribute to these unique patterns. MAP2-stained histological sections displayed relatively uniform radiating dendritic fibers in all three cortices, whereas neurofilament-stained sections showed a layered axonal organization, with the inner layer of the cortex more densely stained than the outer layer. While it is still difficult to quantitatively measure the contribution of each structure’s components, the results demonstrate the potential of our technique in assisting future detailed analysis.
5. Conclusions
In summary, we demonstrate that high spatial resolution and angular resolution dMRI of the live mouse brain is now feasible with localized imaging, in combination with fast imaging sequences. The dMRI contrast derived from the in vivo high-resolution data revealed complex microstructures in both the gray matter and white matter. The techniques presented in this study could be potentially used to investigate longitudinal changes in local neuroanatomy and connectivity under normal or diseased states.
Acknowledgments
This work was supported by NIH grants R01NS070909 (J.Z.), R01HD074593 (J.Z.), R01AG20012 (S.M.), R01EB003543 (S.M.), 1S10 RR028955 (P.V.Z.), and P41EB015909 (P.V.Z.). The authors thank Mary McAllister (Department of Radiology, Johns Hopkins University School of Medicine) for her editorial assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aggarwal M, Mori S, Shimogori T, Blackshaw S, Zhang J. Three-dimensional diffusion tensor microimaging for anatomical characterization of the mouse brain. Magn Reson Med. 2010;64:249–261. doi: 10.1002/mrm.22426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal M, Zhang J, Pletnikova O, Crain B, Troncoso J, Mori S. Feasibility of creating a high-resolution 3D diffusion tensor imaging based atlas of the human brainstem: a case study at 11.7 T. Neuroimage. 2013;74:117–127. doi: 10.1016/j.neuroimage.2013.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaf Y, Basser PJ. Composite hindered and restricted model of diffusion (CHARMED) MR imaging of the human brain. Neuroimage. 2005;27:48–58. doi: 10.1016/j.neuroimage.2005.03.042. [DOI] [PubMed] [Google Scholar]
- Barazany D, Basser PJ, Assaf Y. In vivo measurement of axon diameter distribution in the corpus callosum of rat brain. Brain. 2009;132:1210–1220. doi: 10.1093/brain/awp042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basser PJ, Mattiello J, LeBihan D. MR diffusion tensor spectroscopy and imaging. Biophysical journal. 1994;66:259–267. doi: 10.1016/S0006-3495(94)80775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu C. The basis of anisotropic water diffusion in the nervous system - a technical review. NMR Biomed. 2002;15:435–455. doi: 10.1002/nbm.782. [DOI] [PubMed] [Google Scholar]
- Budde MD, Xie M, Cross AH, Song SK. Axial Diffusivity Is the Primary Correlate of Axonal Injury in the Experimental Autoimmune Encephalomyelitis Spinal Cord: A Quantitative Pixelwise Analysis. Journal of Neuroscience. 2009;29:2805–2813. doi: 10.1523/JNEUROSCI.4605-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calamante F, Tournier JD, Heidemann RM, Anwander A, Jackson GD, Connelly A. Track density imaging (TDI): validation of super resolution property. Neuroimage. 2011;56:1259–1266. doi: 10.1016/j.neuroimage.2011.02.059. [DOI] [PubMed] [Google Scholar]
- Calamante F, Tournier JD, Jackson GD, Connelly A. Track-density imaging (TDI): super-resolution white matter imaging using whole-brain track-density mapping. Neuroimage. 2010;53:1233–1243. doi: 10.1016/j.neuroimage.2010.07.024. [DOI] [PubMed] [Google Scholar]
- Calamante F, Tournier JD, Kurniawan ND, Yang Z, Gyengesi E, Galloway GJ, Reutens DC, Connelly A. Super-resolution track-density imaging studies of mouse brain: comparison to histology. Neuroimage. 2012;59:286–296. doi: 10.1016/j.neuroimage.2011.07.014. [DOI] [PubMed] [Google Scholar]
- Chuang N, Mori S, Yamamoto A, Jiang H, Ye X, Xu X, Richards LJ, Nathans J, Miller MI, Toga AW, Sidman RL, Zhang J. An MRI-based atlas and database of the developing mouse brain. Neuroimage. 2011;54:80–89. doi: 10.1016/j.neuroimage.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham CH, Pauly JM, Nayak KS. Saturated double-angle method for rapid B1+ mapping. Magn Reson Med. 2006;55:1326–1333. doi: 10.1002/mrm.20896. [DOI] [PubMed] [Google Scholar]
- DeFelipe J, Alonso-Nanclares L, Arellano JI. Microstructure of the neocortex: comparative aspects. J Neurocytol. 2002;31:299–316. doi: 10.1023/a:1024130211265. [DOI] [PubMed] [Google Scholar]
- Deoni S, Jones D. Time–Series Analysis of the Diffusion Weighted Signal as a Model–Free Approach to Segmenting Tissue. ISMRM 14th Scientific Meeting & Exhibition, Seattle; Washington, USA. 2006. p. 2734. [Google Scholar]
- Dyrby TB, Baare WFC, Alexander DC, Jelsing J, Garde E, Sogaard LV. An Ex Vivo Imaging Pipeline for Producing High-Quality and High-Resolution Diffusion-Weighted Imaging Datasets. Human Brain Mapping. 2011;32:544–563. doi: 10.1002/hbm.21043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finsterbusch J. Fast-spin-echo imaging of inner fields-of-view with 2D-selective RF excitations. J Magn Reson Imaging. 2010;31:1530–1537. doi: 10.1002/jmri.22196. [DOI] [PubMed] [Google Scholar]
- Finsterbusch J. Functional neuroimaging of inner fields-of-view with 2D-selective RF excitations. Magn Reson Imaging. 2013 doi: 10.1016/j.mri.2013.03.005. [DOI] [PubMed] [Google Scholar]
- Flint JJ, Hansen B, Fey M, Schmidig D, King MA, Vestergaard-Poulsen P, Blackband SJ. Cellular-level diffusion tensor microscopy and fiber tracking in mammalian nervous tissue with direct histological correlation. Neuroimage. 2010;52:556–561. doi: 10.1016/j.neuroimage.2010.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank LR. Anisotropy in high angular resolution diffusion-weighted MRI. Magn Reson Med. 2001;45:935–939. doi: 10.1002/mrm.1125. [DOI] [PubMed] [Google Scholar]
- Granziera C, Schmahmann JD, Hadjikhani N, Meyer H, Meuli R, Wedeen V, Krueger G. Diffusion Spectrum Imaging Shows the Structural Basis of Functional Cerebellar Circuits in the Human Cerebellum In Vivo. PLoS One. 2009;4 doi: 10.1371/journal.pone.0005101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissom W, Yip CY, Zhang Z, Stenger VA, Fessler JA, Noll DC. Spatial domain method for the design of RF pulses in multicoil parallel excitation. Magn Reson Med. 2006;56:620–629. doi: 10.1002/mrm.20978. [DOI] [PubMed] [Google Scholar]
- Hansen B, Flint JJ, Heon-Lee C, Fey M, Vincent F, King MA, Vestergaard-Poulsen P, Blackband SJ. Diffusion tensor microscopy in human nervous tissue with quantitative correlation based on direct histological comparison. Neuroimage. 2011;57:1458–1465. doi: 10.1016/j.neuroimage.2011.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haroon HA, Binney RJ, Parker GJ. Probabilistic quantification of regional cortical microstructural complexity. Joint Annual Meeting ISMRM-ESMRMB; 2010; Stockholm, Sweden. 2010. [Google Scholar]
- Harsan LA, David C, Reisert M, Schnell S, Hennig J, von Elverfeldt D, Staiger JF. Mapping remodeling of thalamocortical projections in the living reeler mouse brain by diffusion tractography. Proc Natl Acad Sci U S A. 2013;110:E1797–1806. doi: 10.1073/pnas.1218330110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidemann RM, Porter DA, Anwander A, Feiweier T, Heberlein K, Knosche TR, Turner R. Diffusion Imaging in Humans at 7T Using Readout-Segmented EPI and GRAPPA. Magnetic Resonance in Medicine. 2010;64:9–14. doi: 10.1002/mrm.22480. [DOI] [PubMed] [Google Scholar]
- Hiepe P, Ros C, Reichenbach JR, Herrmann KH. Diffusion Weighted ZOOM Imaging in the Lumbar Spine Based on Single-Shot STEAM. World Congress on Medical Physics and Biomedical Engineering, Vol 25, Pt 2 - Diagnostic Imaging. 2009;25:670–672. [Google Scholar]
- Jeong EK, Kim SE, Guo J, Kholmovski EG, Parker DL. High-resolution DTI with 2D interleaved multislice reduced FOV single-shot diffusion-weighted EPI (2D ss-rFOV-DWEPI) Magn Reson Med. 2005;54:1575–1579. doi: 10.1002/mrm.20711. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Johnson GA. Microscopic diffusion tensor imaging of the mouse brain. Neuroimage. 2010;50:465–471. doi: 10.1016/j.neuroimage.2009.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DK, Horsfield MA, Simmons A. Optimal strategies for measuring diffusion in anisotropic systems by magnetic resonance imaging. Magn Reson Med. 1999;42:515–525. [PubMed] [Google Scholar]
- Jones DK, Leemans A. Diffusion tensor imaging. Methods Mol Biol. 2011;711:127–144. doi: 10.1007/978-1-61737-992-5_6. [DOI] [PubMed] [Google Scholar]
- Kurniawan ND, Richards KL, Yang Z, She D, Ullmann JF, Moldrich RX, Liu S, Yaksic JU, Leanage G, Kharatishvili I, Wimmer V, Calamante F, Galloway GJ, Petrou S, Reutens DC. Visualization of mouse barrel cortex using ex-vivo track density imaging. Neuroimage. 2013 doi: 10.1016/j.neuroimage.2013.09.030. [DOI] [PubMed] [Google Scholar]
- Le Bihan D. Looking into the functional architecture of the brain with diffusion MRI. Nature Reviews Neuroscience. 2003;4:469–480. doi: 10.1038/nrn1119. [DOI] [PubMed] [Google Scholar]
- Leergaard TB, White NS, de Crespigny A, Bolstad I, D’Arceuil H, Bjaalie JG, Dale AM. Quantitative histological validation of diffusion MRI fiber orientation distributions in the rat brain. PLoS One. 2010;5:e8595. doi: 10.1371/journal.pone.0008595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuze CW, Anwander A, Bazin PL, Dhital B, Stuber C, Reimann K, Geyer S, Turner R. Layer-Specific Intracortical Connectivity Revealed with Diffusion MRI. Cereb Cortex. 2012 doi: 10.1093/cercor/bhs311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mac Donald CL, Dikranian K, Song SK, Bayly PV, Holtzman DM, Brody DL. Detection of traumatic axonal injury with diffusion tensor imaging in a mouse model of traumatic brain injury. Exp Neurol. 2007;205:116–131. doi: 10.1016/j.expneurol.2007.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J, Mareci TH, Andrew ER. Experimental-Study of Optimal Selective 180-Degrees Radiofrequency Pulses. Journal of Magnetic Resonance. 1988;79:1–10. [Google Scholar]
- McKinstry RC, Mathur A, Miller JH, Ozcan A, Snyder AZ, Schefft GL, Almli CR, Shiran SI, Conturo TE, Neil JJ. Radial organization of developing preterm human cerebral cortex revealed by non-invasive water diffusion anisotropy MRI. Cereb Cortex. 2002;12:1237–1243. doi: 10.1093/cercor/12.12.1237. [DOI] [PubMed] [Google Scholar]
- McNab JA, Polimeni JR, Wang R, Augustinack JC, Fujimoto K, Stevens A, Janssens T, Farivar R, Folkerth RD, Vanduffel W, Wald LL. Surface based analysis of diffusion orientation for identifying architectonic domains in the in vivo human cortex. Neuroimage. 2013;69:87–100. doi: 10.1016/j.neuroimage.2012.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldrich RX, Pannek K, Hoch R, Rubenstein JL, Kurniawan ND, Richards LJ. Comparative mouse brain tractography of diffusion magnetic resonance imaging. Neuroimage. 2010;51:1027–1036. doi: 10.1016/j.neuroimage.2010.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, van Zijl PC. A motion correction scheme by twin-echo navigation for diffusion-weighted magnetic resonance imaging with multiple RF echo acquisition. Magn Reson Med. 1998;40:511–516. doi: 10.1002/mrm.1910400403. [DOI] [PubMed] [Google Scholar]
- Mori S, Zhang J. Principles of diffusion tensor imaging and its applications to basic neuroscience research. Neuron. 2006;51:527–539. doi: 10.1016/j.neuron.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Nagy Z, Alexander DC, Thomas DL, Weiskopf N, Sereno MI. Using High Angular Resolution Diffusion Imaging Data to Discriminate Cortical Regions. PLoS One. 2013;8 doi: 10.1371/journal.pone.0063842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshio K, Feinberg DA. Grase (Gradient-Echo and Spin-Echo) Imaging - a Novel Fast Mri Technique. Magnetic Resonance in Medicine. 1991;20:344–349. doi: 10.1002/mrm.1910200219. [DOI] [PubMed] [Google Scholar]
- Pauly J, Nishimura D, Macovski A. A linear class of large-tip-angle selective excitation pulses. Journal of Magnetic Resonance (1969) 1989;82:571–587. [Google Scholar]
- Schneider E, Glover G. Rapid in vivo proton shimming. Magn Reson Med. 1991;18:335–347. doi: 10.1002/mrm.1910180208. [DOI] [PubMed] [Google Scholar]
- Schneider JT, Kalayciyan R, Haas M, Herrmann SR, Ruhm W, Hennig J, Ullmann P. Inner-volume imaging in vivo using three-dimensional parallel spatially selective excitation. Magn Reson Med. 2012 doi: 10.1002/mrm.24381. [DOI] [PubMed] [Google Scholar]
- Setsompop K, Kimmlingen R, Eberlein E, Witzel T, Cohen-Adad J, McNab JA, Keil B, Tisdall MD, Hoecht P, Dietz P, Cauley SF, Tountcheva V, Matschl V, Lenz VH, Heberlein K, Potthast A, Thein H, Van Horn J, Toga A, Schmitt F, Lehne D, Rosen BR, Wedeen V, Wald LL. Pushing the limits of in vivo diffusion MRI for the Human Connectome Project. Neuroimage. 2013;80:220–233. doi: 10.1016/j.neuroimage.2013.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd TM, Thelwall PE, Stanisz GJ, Blackband SJ. Aldehyde fixative solutions alter the water relaxation and diffusion properties of nervous tissue. Magn Reson Med. 2009;62:26–34. doi: 10.1002/mrm.21977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SW, Liang HF, Le TQ, Armstrong RC, Cross AH, Song SK. Differential sensitivity of in vivo and ex vivo diffusion tensor imaging to evolving optic nerve injury in mice with retinal ischemia. Neuroimage. 2006a;32:1195–1204. doi: 10.1016/j.neuroimage.2006.04.212. [DOI] [PubMed] [Google Scholar]
- Sun SW, Liang HF, Trinkaus K, Cross AH, Armstrong RC, Song SK. Noninvasive detection of cuprizone induced axonal damage and demyelination in the mouse corpus callosum. Magn Reson Med. 2006b;55:302–308. doi: 10.1002/mrm.20774. [DOI] [PubMed] [Google Scholar]
- Sun SW, Neil JJ, Liang HF, He YY, Schmidt RE, Hsu CY, Song SK. Formalin fixation alters water diffusion coefficient magnitude but not anisotropy in infarcted brain. Magn Reson Med. 2005;53:1447–1451. doi: 10.1002/mrm.20488. [DOI] [PubMed] [Google Scholar]
- Takahashi E, Song JW, Folkerth RD, Grant PE, Schmahmann JD. Detection of postmortem human cerebellar cortex and white matter pathways using high angular resolution diffusion tractography: a feasibility study. Neuroimage. 2013;68:105–111. doi: 10.1016/j.neuroimage.2012.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournier JD, Calamante F, Connelly A. MRtrix: Diffusion tractography in crossing fiber regions. International Journal of Imaging Systems and Technology. 2012;22:53–66. [Google Scholar]
- Tuch DS, Reese TG, Wiegell MR, Wedeen VJ. Diffusion MRI of complex neural architecture. Neuron. 2003;40:885–895. doi: 10.1016/s0896-6273(03)00758-x. [DOI] [PubMed] [Google Scholar]
- van Zijl PCM, Sukumar S, Johnson MO, Webb P, Hurd RE. Optimized Shimming for High-Resolution Nmr Using 3-Dimensional Image-Based Field-Mapping. Journal of Magnetic Resonance Series A. 1994;111:203–207. [Google Scholar]
- Verstynen T, Jarbo K, Pathak S, Schneider W. In Vivo Mapping of Microstructural Somatotopies in the Human Corticospinal Pathways. Journal of Neurophysiology. 2011;105:336–346. doi: 10.1152/jn.00698.2010. [DOI] [PubMed] [Google Scholar]
- Wang S, Wu EX, Tam CN, Lau HF, Cheung PT, Khong PL. Characterization of white matter injury in a hypoxic-ischemic neonatal rat model by diffusion tensor MRI. Stroke. 2008;39:2348–2353. doi: 10.1161/STROKEAHA.107.509927. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang Q, Haldar JP, Yeh FC, Xie M, Sun P, Tu TW, Trinkaus K, Klein RS, Cross AH, Song SK. Quantification of increased cellularity during inflammatory demyelination. Brain. 2011;134:3590–3601. doi: 10.1093/brain/awr307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedeen VJ, Hagmann P, Tseng WY, Reese TG, Weisskoff RM. Mapping complex tissue architecture with diffusion spectrum magnetic resonance imaging. Magn Reson Med. 2005;54:1377–1386. doi: 10.1002/mrm.20642. [DOI] [PubMed] [Google Scholar]
- Wedeen VJ, Rosene DL, Wang R, Dai G, Mortazavi F, Hagmann P, Kaas JH, Tseng WY. The geometric structure of the brain fiber pathways. Science. 2012;335:1628–1634. doi: 10.1126/science.1215280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen H, Jaffer FA. An in-Vivo Automated Shimming Method Taking into Account Shim Current Constraints. Magnetic Resonance in Medicine. 1995;34:898–904. doi: 10.1002/mrm.1910340616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Xu J, McMahon MT, van Zijl PC, Mori S, Northington FJ, Zhang J. In vivo high-resolution diffusion tensor imaging of the mouse brain. Neuroimage. 2013 doi: 10.1016/j.neuroimage.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, King KF, Zhu Y, McKinnon GC, Liang ZP. A noniterative method to design large-tip-angle multidimensional spatially-selective radio frequency pulses for parallel transmission. Magn Reson Med. 2007;58:326–334. doi: 10.1002/mrm.21314. [DOI] [PubMed] [Google Scholar]
- Yang QX, Posse S, LeBihan D, Smith MB. Double-sampled echo-planar imaging at 3 tesla. Journal of Magnetic Resonance Series B. 1996;113:145–150. doi: 10.1006/jmrb.1996.0167. [DOI] [PubMed] [Google Scholar]
- Yip CY, Fessler JA, Noll DC. Iterative RF pulse design for multidimensional, small-tip-angle selective excitation. Magn Reson Med. 2005;54:908–917. doi: 10.1002/mrm.20631. [DOI] [PubMed] [Google Scholar]
- Zhang H, Schneider T, Wheeler-Kingshott CA, Alexander DC. NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage. 2012a;61:1000–1016. doi: 10.1016/j.neuroimage.2012.03.072. [DOI] [PubMed] [Google Scholar]
- Zhang J, Jones MV, McMahon MT, Mori S, Calabresi PA. In vivo and ex vivo diffusion tensor imaging of cuprizone-induced demyelination in the mouse corpus callosum. Magn Reson Med. 2012b;67:750–759. doi: 10.1002/mrm.23032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, van Zijl PC, Mori S. Three-dimensional diffusion tensor magnetic resonance microimaging of adult mouse brain and hippocampus. Neuroimage. 2002;15:892–901. doi: 10.1006/nimg.2001.1012. [DOI] [PubMed] [Google Scholar]
- Zhang JY, Aggarwal M, Mori S. Structural insights into the rodent CNS via diffusion tensor imaging. Trends in Neurosciences. 2012c;35:412–421. doi: 10.1016/j.tins.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JY, van Zijl PCM, Mori S. Image contrast using the secondary and tertiary eigenvectors in diffusion teinsor imaging. Magnetic Resonance in Medicine. 2006;55:439–449. doi: 10.1002/mrm.20767. [DOI] [PubMed] [Google Scholar]