Abstract
Nanos is an essential factor of germ line success in all animals tested. This gene encodes a Zn-finger RNA-binding protein that in complex with its partner pumilio, binds to and changes the fate of several known transcripts. We summarize here the documented functions of nanos in several key organisms, and then emphasize echinoderms as a working model for how nanos expression is regulated. Nanos presence outside of the target cells is often detrimental to the animal, and in sea urchins, nanos expression appears to be regulated at every step of transcription, and post-transcriptional activity, making this gene product exciting, every which way.
Keywords: Sea urchin, Starfish, Sea star, Primordial germ cells, Germ line
Introduction
Nanos is an essential element in the formation, development, and/or maintenance of the germ line of all animals tested. It is an RNA binding protein first characterized in Drosophila as a translational repressor of the hunchback gene, a gap gene involved in anterior-posterior polarity. Nanos subsequently functions in the germ line of this organism (Cho et al., 2006; Irish et al., 1989). The Nanos protein is structurally conserved amongst animals, with two Cys-Cys-His-Cys zinc finger motifs that are indispensable for its function (Curtis et al., 1997); its conserved basic surface is directly involved in the RNA binding (Hashimoto et al., 2010). Its sequence, however, is remarkably divergent, only 19% amongst several dipteran species (Curtis et al., 1995). Current models argue that Nanos functions through its interaction with Pumilio, which binds RNAs containing a conserved motif in their 3’UTR, the Nanos Response Element (NRE), or more effectively to document the Pumilio role in this interaction, the Pumilio Response Element (PRE) (Gerber et al., 2006; Sonoda and Wharton, 1999; Wharton et al., 1998; Wharton and Struhl, 1991). Only a few mRNAs, however, have been identified as Nanos/Pumilio targets. These include: hunchback (a transcription factor) (Murata and Wharton, 1995; Wreden et al., 1997), cyclin B (an interactor of a cell cycle kinase) (Asaoka-Taguchi et al., 1999; Dalby and Glover, 1993; Kadyrova et al., 2007; Lai et al., 2011), hid (a pro-apoptotic protein) (Hayashi et al., 2004; Sato et al., 2007), VegT (a T-box transcription factor) (Lai et al., 2012) and fem-3 (a unique protein structure that associates with the ubiquitination complex) (Ahringer and Kimble, 1991; Zhang et al., 1997) (Table 1). A database has recently been generated using Xenopus laevis to predict potential Pumilio targeted RNAs based on prevalence and concordance of PREs in their 3’UTR (Lai and King, 2013).
Table1.
The transcripts documented for targeting by the Nanos/Pumilio complex for translational repression. The name of the transcript is presented on the left column, the name of the species in which the mechanism was discovered is indicated in parenthesis. The corresponding cellular function of these transcripts is presented on the right.
| Nanos/Pumilio targets | Functions |
|---|---|
| hunchback (Drosophila) | Establishment of the anterior-posterior body axis |
| cyclin B (Drosophila) | Cell cycle |
| hid (Drosophila) | Apoptosis |
| vegT (Xenopus) | Endoderm formation, Mesoderm induction |
| fem-3 (C.elegans) | Switch from sperm to oocyte |
The expression of nanos genes is highly regulated, and mis-expression of these genes often induces cell cycle and developmental defects. Studies in several model organisms showed that a loss of Nanos results in diverse abnormal phenotypes including precocious cell divisions, ectopic expression of somatic genes, abnormal germ cell migration, and eventual loss of primordial germ cells (PGCs) through apoptosis (Forbes and Lehmann, 1998; Kobayashi et al., 1996; Koprunner et al., 2001; Sato et al., 2007; Tsuda et al., 2003). Ectopic expression of Nanos often leads to embryonic lethality, giving rise to the concept of nanos toxicity (M.L. King, personal communication) in some embryos.
Nanos/Pumilio functions
In Drosophila, maternal hunchback mRNA is uniformly distributed throughout the early embryo, yet its translation is inhibited in the posterior region, resulting in an anterior-posterior protein concentration gradient. This system is a model for how morphogen gradients can form, and for how Nanos functions by being concentrated in the posterior pole of the embryo (Lehmann and Nusslein-Volhard, 1987; Struhl et al., 1992; Wharton and Struhl, 1991). Inhibition of hunchback mRNA translation at the posterior pole to create this gradient requires an mRNP complex that consists of the NRE in its 3’UTR, and the protein complex Nanos, Pumilio and Brain tumor. d4EHP, a 7-Me-Guanosine RNA cap-binding protein inhibits hunchback mRNA translation by interacting simultaneously with the mRNA 5’ cap structure and Brain tumor within the Nanos/Pumilio complex (Cho et al., 2006). In addition to their role in abdominal patterning, Nanos and Pumilio subsequently function in several roles in the PGCs.
Generally PGCs cease proliferating shortly after their formation at the posterior pole of the embryo, emerging from quiescence only after migrating to, and arriving in, the presumptive gonad in late embryogenesis (Su et al., 1998). In the pole cells, cyclin B translation, as in most cells, is needed for cell cycle progression but in the germ cells its translation is directly inhibited by the binding of Nanos and Pumilio to its 3’UTR in a Brain Tumor independent mechanism (Sonoda and Wharton, 2001). Further, ectopic repression of cyclin B in the presumptive somatic cytoplasm causes nuclear division defects that are lethal (Kadyrova et al., 2007). Nanos probably acts in the germ line, at least in part, by recruiting the CCR4-Pop2-NOT deadenylase complex, interacting directly with the NOT4 subunit (Kadyrova et al., 2007). Nanos and Pumilio are also required for the survival of the germ line by repressing the translation of hid through PREs in its 3’UTR. Hid is a member of the RHG gene family required for caspase activation (Sato et al., 2007). Pole cells lacking maternal Nanos enter the apoptotic pathway by precocious hid activity and do not populate the embryonic gonads (Hayashi et al., 2004). In Xenopus, Nanos1-depleted PGCs inappropriately express somatic genes characteristic of endoderm regulated by maternal VegT, including Xsox17α, Bix4, Mixer, GATA4 and Edd. Pumilio specifically binds VegT RNA in vitro and represses, along with Nanos1, VegT translation within the PGCs (Lai et al., 2012). In C. elegans, normally, hermaphrodites make sperm first and then switch to oogenesis. The translational repression of fem-3 controls this switch (Ahringer and Kimble, 1991). In mutants that disrupt a regulatory element in the fem-3 3′UTR, the switch does not occur, and sperm are made continuously. FBF, a pumilio homolog, binds the 3’UTR of fem-3, and can form a regulatory complex with Nanos-3 to regulate the sperm-oocyte switch (Kraemer et al., 1999; Zhang et al., 1997).
Nanos function in the germline
The essential nature of Nanos function in germ line development is seen widespread. In the planarian flatworms, nanos is expressed in the testes and the ovaries of juvenile and mature hermaphrodites, and in the presumptive testes primordia of asexual individuals that reproduce strictly by fission. RNA interference experiments show that Nanos is required for proper germ cell development, regeneration and maintenance in both sexual and asexual planarians (Wang et al., 2007). In the nematode Caenorhabditis elegans, three nanos-related genes have been identified. Nanos-2 is required maternally for efficient incorporation of the PGCs into the somatic gonad and has overlapping functions with Nanos-1 to regulate survival and proliferation of PGC descendants during larval development (Subramaniam and Seydoux, 1999). Nanos-3 functions in the sperm-to-oocyte switch in hermaphrodites (Kraemer et al., 1999).
In Drosophila, Nanos is required in the male and female germ line (Bhat, 1999). The protein is required for the migration (Forbes and Lehmann, 1998; Kobayashi et al., 1996) and the survival (Sato et al., 2007) of the pole cells which give rise to the primordial germ cells. PGCs are normally transcriptionally repressed at times when somatic cells are initially expressing their gene program. In Drosophila as well as in C. elegans, loss of Nanos results in prematurely active transcription and the failure to establish germ line-specific histone modifications typical of transcriptionally inactive chromatin (Schaner et al., 2003).
In zebrafish, three nanos homologs are present in the genome. Nanos3 function is required for the maintenance of GSCs, and Nanos2 and Nanos3 functions are partially overlapping (Beer and Draper, 2013). A null mutation in the zebrafish homolog of nanos3 (formerly nanos1) has been described (Draper et al., 2007). In contrast to wild-type ovaries, which contain both mitotic and meiotic germ cells, juvenile nanos3- mutant ovaries contain only meiotic germ cells, and no oocytes are present in these ovaries even after 5 months of age. The zebrafish Nanos3 also regulates PGC migration and survival during embryonic development (Koprunner et al., 2001).
Xenopus nanos RNA (nanos1; formerly called Xcat2) is expressed in the germ line (Houston and King, 2000; Mosquera et al., 1993). This nanos mRNA is most abundant in the oocyte and is localized to the vegetal cortex. Misexpression of Nanos in oocytes from un-restricted RNA (expression of nanos in the somatic cells) results in abnormal development and embryonic lethality, indicating that restriction of nanos translation to the germ line is crucial (Luo et al., 2011). nanos RNA and protein persists until PGCs leave the Xenopus endoderm at late tailbud stages. Nanos1-depleted PGCs fail to migrate out of the endoderm, inappropriately express somatic genes characteristic of the endoderm, and undergo apoptosis (Lai et al., 2012).
The mouse genome contains three nanos genes and knock-out of Nanos1 did not reveal a discernible germ cell function for this ortholog (Haraguchi et al., 2003). Nanos2 mutant mice however, have decreased testis size and are infertile due to a loss of germ cells following PGCs incorporation into the gonad, while female mice appear developmentally normal and retain fertility (Saga, 2010; Tsuda et al., 2003). Nanos3 knockout mice present a decrease in gonad size and both males and females are infertile.
Analysis of nanos mRNA expression in human fetal and adult tissues indicated that expression of nanos2 and 3 was enriched in the fetal ovary, testis, and brain, as well as in the adult ovary and testis (Curtis et al., 1995; Julaton and Reijo Pera, 2011). Nanos 3 protein is expressed during multiple stages of human oogenesis, including primordial, primary, secondary, and antral follicles, with the highest expression in the oocytes. A decrease in Nanos3 expression by morpholino nucleofection in hESCs resulted in reduced germ cell numbers and aberrant expression of germ cell markers (Julaton and Reijo Pera, 2011). Recently, a novel mutation in the human nanos3 open reading frame (Arg153Trp) was found on a screen of “primary ovarian insufficiency (POI)” patients (Wu et al., 2013). POI is defined as the cessation of ovarian function before the age of 40. The human Nanos3 protein has a half-life of 3h in HEK293 cells, whereas the mutated protein had an even shorter half-life of 1.5h. The mutant Nanos3 protein tends to be structurally unstable and form aggregates which are cleared by the ubiquitin-proteasome system. The results suggest that the dosage of Nanos3 has an important role in the maintenance and survival of PGCs in mouse model, and establishes a possible link between Nanos3 and POI.
In spite of the many animals studied for germ line determination and for Nanos function, remarkably little conservation exists for each of the regulatory steps in Nanos expression. Clearly a broad perspective in Nanos regulation is important in order to see the deep trends in Nanos functionality and transition in its regulation and function.
Nanos regulation in Echinoderms
Echinodermata is a diverse Phylum, a sister group to Chordates, and contains varied organisms that may be useful to understand the mechanisms of the germ-line specification. The sea star P. miniata contains a nanos mRNA ortholog in the oocyte and then later in development in the posterior enterocoel. This later structure in sea stars is important as it appears to be a source of germ cells. This suggestion is based on these cells accumulating nanos, vasa, and piwi – each important in germ line determination, excluding transcripts important for somatic cell fates, and upon removal of the posterior enterocoel, many fewer germ cells form later in development (Fresques et al., 2013)(Inoue et al., 1992). The function of nanos, if any, in formation of the posterior enterocoel or germ line of this organism has not been tested.
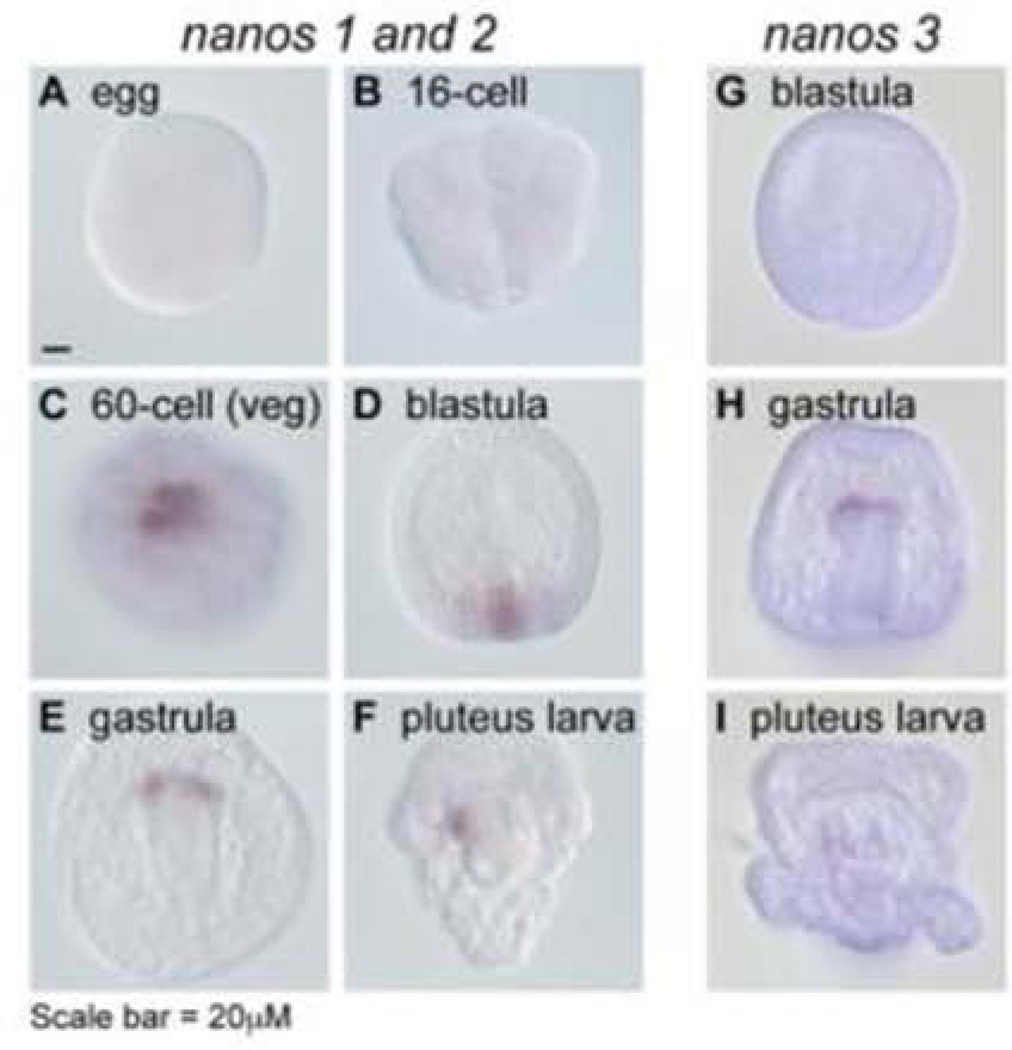
Significant testing of nanos in the sea urchin though has been accomplished. Sea urchin embryos contain three nanos homologs, each expressed in the small micromeres (Figure 1) (Juliano et al., 2010). This is the lineage in sea urchins that contributes to the germ line (Yajima and Wessel, 2011). Four small micromeres arise from an asymmetric division of the micromeres as the embryo develops from the 16-cell to 32-cell stage. Subsequently, the small micromeres divide once when in the vegetal plate of the blastula and then they remain quiescent for the rest of embryogenesis. Thus, eight small micromeres travel at the tip of the invaginating archenteron and then are incorporated into the coelomic pouches of the pluteus. In the sea urchin Strongylocentrotus purpuratus (Sp), Sp-Nanos1 and 2 knockdown gastrulae show a precocious increased number of Sp-vasa cells at the tip of the archenteron suggesting that Sp-nanos1 and 2 may be required to maintain the mitotically quiescent state of the small micromeres during embryogenesis. These embryos also accumulate increased Sp-nanos1 and 2 transcripts, suggesting a feedback mechanism to maintain steady levels of nanos protein. Further, Sp-Nanos1 and 2 are required to maintain the small micromere lineage – nanos knockdown results in apoptosis of the small micromeres. Additionally, nanos is required for the formation of the adult rudiment – nanos knockdown larvae develop guts, skeletal systems and larval shape but the coelomic pouches do not form and the larvae, although swimming and feeding, do not develop beyond a stunted early larva (Juliano et al., 2010). At this point, it is not known if the small micromeres contribute to additional fates in the larvae (as a multipotent cell), induce additional fates in cells of the larvae to develop into coeloms, or whether nanos is present at low levels in the developing larvae that is required for diverse tissue morphogenesis, but which is inhibited by the knockdown strategy used. In the In the sea urchin Hemicentrotus pulcherrimus (Hp), knockdown experiments also showed that Hp-Nanos2 is involved in the ingression of primary mesenchyme cells (PMCs) at the mesenchyme blastula stage, and is required for the survival of small micromere descendants after gastrulation. Hp-Nanos2 knockdown caused a decreased in the number of cells comprising the left coelomic pouch, resulting from a caspase-dependent apoptotic cell death of small micromeres at the tip of the archenteron (Fujii et al., 2009).
Figure 1.
Expression of Sp-nanos1/nanos2 and Sp-nanos3 mRNA in the small micromere lineage by in situ hybridization (Juliano et al., 2010)
In sea urchins, FoxY positively regulates nanos transcription (Song and Wessel, 2012). It is a member of the forkhead transcription factor family that is transiently enriched in the presumptive germ line of sea urchins (Ransick et al., 2002). Two splice forms of FoxY protein are present in the ovary and in early development. Both forms of foxy mRNA accumulate in the small micromeres lineage and in the adjacent non-skeletogenic mesoderm. Embryos injected with a FoxY morpholino appear to develop normally up to pluteus stage, but contain reduced levels of nanos mRNA and protein. Moreover, after two weeks of Foxy kncokdown, the coelomic pouches regressed and the embryos present a phenotype similar to the Nanos-knockdown (Juliano et al., 2010). Interestingly, most of the forkhead transcription factors bind to a seven-nucleotide core consensus sequence (RYMAAYA [R=A or G; Y=C or T; M= A or C]) that is found several times in the nanos promoter. In the future this region of the putative promoter should be dissected and tested for the factors essential for its expression. Further, if FoxY is essential for this transcription, how is it selectively functional in the small micromeres at this time? The timing of FoxY expression is more consistent with it functioning to maintain nanos transcription more than to initiate it.
In vitro differentiation of isolated micromeres resulted in expression of both vasa and nanos on schedule (Yajima and Wessel, 2012). This is particularly noteworthy since although the skeletogeneic genes of the PMC lineage (large micromeres) have been seen to be regulated in vitro similarly to the intact embryo, suggesting an autonomy of PMC differentiation, this is the first example of autonomous development and gene expression in the small micromeres. This result means that at least by the 16 cell stage, the micromere is committed to gene expression by each of the subsequent lineages in the asymmetric division. Although nanos gene regulation is yet to be understood, we can assume that at least sufficient machinery is in place for the small micromeres by this time to enable activation of the nanos gene independent of the rest of the embryo.
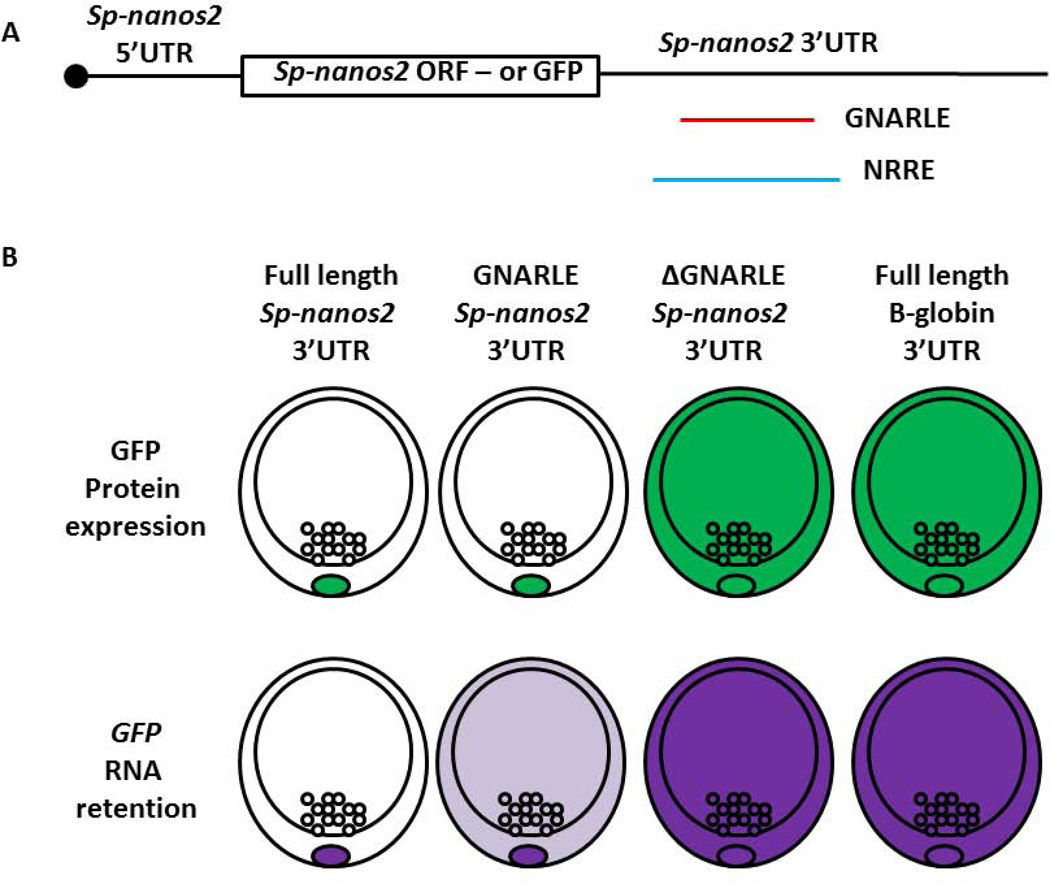
Mere RNA presence is insufficient to conclude translational activity and protein accumulation encoded by the gene. Indeed, nanos mRNAs are translationally repressed in many cases (Drosophila (Gavis et al., 1996), C. elegans (D'Agostino et al., 2006), Xenopus (Luo et al., 2011)). In sea urchins however, Nanos2 protein selectively accumulates in the small micromeres shortly following accumulation of its mRNA. No broad translational delay appears in this embryo as in seen in many other organisms. The initiation of translation, however, is not standard. Nanos translation requires an element in its 3’UTR (Oulhen et al., 2013). This element is composed of 388 nucleotides and is referred to as GNARLE (Global Nanos Associated RNA Lability Element) (Figure 2). It contains two regions that are highly conserved between S. purpuratus (Sp) and H. pulcherrimus (Hp), two sea urchin species separated by 20 million years, but not in L. variegatus, separated by a last common ancestor of ~50 million years (Smith et al., 2006). GNARLE is required for the selective retention of an injected RNA in the small micromeres but is not as effective as the full length 3’UTR. Even though GNARLE does not give a strong selective RNA retention, this element is sufficient for high protein enrichment in the small micromeres, suggesting a role of the GNARLE in inhibiting the translation in the non-small micromeres, and/or stimulating the translation in the small micromeres. Deletion of GNARLE leads to a stabilization of the protein expression throughout the embryo (Sp nanos2 5’UTR – GFP ORF- Sp nanos2 3’UTR ΔGNARLE) in comparison with a control mRNA bearing only GNARLE in its 3’UTR. Moreover, the Sp nanos2 5’UTR is insufficient for selective RNA retention or protein enrichment but does strongly increase the level of protein synthesis compared to a control Xenopus β-globin 5’UTR. This regulation is independent of the 3’UTR used. These data suggest that in sea urchins also, nanos is regulated at the translational level through elements in both its 5’ and 3’ UTRs (Oulhen et al., 2013).
Figure 2.
Nanos regulation in the sea urchin at the mesenchyme blastula stage. (A) Schematic of the Sp-nanos2 transcript: it contains a cap, Sp-nanos2 5’UTR, Sp-nanos2 ORF and Sp-nanos2 3’UTR. The regulatory elements, GNARLE and NRRE, located in Sp-nanos2 3’UTR, are presented with a red and a blue line respectively. To test nanos regulation, Sp-nanos2 ORF was exchanged by the GFP ORF, and the 3’UTR was modified. Constructs were injected in Sp fertilized eggs. (B) Schematic of the GFP protein expression and the retention of the RNA obtained in mesenchyme blastula after injection of the construct presented in (A). Four different 3’UTRs were tested: Sp-nanos2 3’UTR full length, GNARLE, ΔGNARLE, and Xenopus β-globin 3’UTR.
RNA stability is also an important regulator of selective nanos expression in the small micromeres. Any ectopic mRNA injected into the early sea urchin embryo retains the mRNA selectively in the small micromeres (Oulhen et al., 2013). The nanos2 mRNA also selectively accumulates in the small micromeres, but it requires an element in its 3’UTR (Oulhen et al., 2013). Designated as NRRE (Nanos RNA Retention Element), it contains the GNARLE region plus additional flanking sequences and is sufficient to drive mRNA retention dynamics similar to the one obtained with the full length 3’UTR (Figure 3). The mechanism of selective retention in the small micromeres – both the mechanism for such long lived mRNA in small micromeres, and the rapid turnover in non-small micromeres – is yet unknown. It is clear though that the degradation of nanos RNA outside of the small micromeres is independent of the miRNA pathway (Oulhen et al., 2013).
Using sea urchins as a model, nanos has been found to be regulated by transcription, translation and RNA stability. Moreover, the expression of nanos mRNA is inducible in other cell lineages in the Nanos knockdown and micromere-deleted embryos (Fujii et al., 2009; Juliano et al., 2010), suggesting that the small micromere descendants repress nanos mRNA expression in other cell lineage. It is unclear, however, if the effect is direct or indirect. This repressive function of the micromere lineage was also observed for Vasa protein expression in sea urchin (Voronina et al., 2008).
Key elements for examination of nanos function in the future include the mechanism of transcriptional regulation in the sea urchin. Nanos is an important, select, and early transcriptional product of the small micromeres, and understanding how it is activated is important for an understanding of this lineage. The RNA retention mechanism for nanos (and other transcripts) in the small micromeres is important. Clearly the small micromeres, as PGCs, have unique characteristics in the embryo, many of which are shared with other animals. The character of RNA retention is likely a shared strategy by PGCs in many animals and understanding how RNA is differentially treated in the germ line is important for understanding the establishment and maintenance of these cells. Finally, the Nanos protein sequence is remarkably diverse in echinoderms and other animals. Understanding this marked diversification may illuminate differences in the PGC program and perhaps in the promiscuous functionality of its key regulators.
Abbreviations
- NRE
Nanos response element
- PRE
Pumilio response element
- PGCs
primordial germ cells
- GNARLE
Global Nanos Associated RNA Lability Element
- NRRE
Nanos RNA Retention Element
References
- Ahringer J, Kimble J. Control of the sperm-oocyte switch in Caenorhabditis elegans hermaphrodites by the fem-3 3' untranslated region. Nature. 1991;349:346–348. doi: 10.1038/349346a0. [DOI] [PubMed] [Google Scholar]
- Asaoka-Taguchi M, Yamada M, Nakamura A, Hanyu K, Kobayashi S. Maternal Pumilio acts together with Nanos in germline development in Drosophila embryos. Nat Cell Biol. 1999;1:431–437. doi: 10.1038/15666. [DOI] [PubMed] [Google Scholar]
- Beer RL, Draper BW. nanos3 maintains germline stem cells and expression of the conserved germline stem cell gene nanos2 in the zebrafish ovary. Dev Biol. 2013;374:308–318. doi: 10.1016/j.ydbio.2012.12.003. [DOI] [PubMed] [Google Scholar]
- Bhat KM. The posterior determinant gene nanos is required for the maintenance of the adult germline stem cells during Drosophila oogenesis. Genetics. 1999;151:1479–1492. doi: 10.1093/genetics/151.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho PF, Gamberi C, Cho-Park YA, Cho-Park IB, Lasko P, Sonenberg N. Cap-dependent translational inhibition establishes two opposing morphogen gradients in Drosophila embryos. Curr Biol. 2006;16:2035–2041. doi: 10.1016/j.cub.2006.08.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis D, Apfeld J, Lehmann R. nanos is an evolutionarily conserved organizer of anterior-posterior polarity. Development. 1995;121:1899–1910. doi: 10.1242/dev.121.6.1899. [DOI] [PubMed] [Google Scholar]
- Curtis D, Treiber DK, Tao F, Zamore PD, Williamson JR, Lehmann R. A CCHC metal-binding domain in Nanos is essential for translational regulation. EMBO J. 1997;16:834–843. doi: 10.1093/emboj/16.4.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Agostino I, Merritt C, Chen PL, Seydoux G, Subramaniam K. Translational repression restricts expression of the C. elegans Nanos homolog NOS-2 to the embryonic germline. Dev Biol. 2006;292:244–252. doi: 10.1016/j.ydbio.2005.11.046. [DOI] [PubMed] [Google Scholar]
- Dalby B, Glover DM. Discrete sequence elements control posterior pole accumulation and translational repression of maternal cyclin B RNA in Drosophila. EMBO J. 1993;12:1219–1227. doi: 10.1002/j.1460-2075.1993.tb05763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper BW, McCallum CM, Moens CB. nanos1 is required to maintain oocyte production in adult zebrafish. Dev Biol. 2007;305:589–598. doi: 10.1016/j.ydbio.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes A, Lehmann R. Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development. 1998;125:679–690. doi: 10.1242/dev.125.4.679. [DOI] [PubMed] [Google Scholar]
- Fresques T, Zazueta-Novoa V, Reich A, Wessel GM. Selective accumulation of germ-line associated gene products in early development of the sea star and distinct differences from germ-line development in the sea urchin. Dev Dyn. 2013 doi: 10.1002/dvdy.24038. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii T, Sakamoto N, Ochiai H, Fujita K, Okamitsu Y, Sumiyoshi N, Minokawa T, Yamamoto T. Role of the nanos homolog during sea urchin development. Dev Dyn. 2009;238:2511–2521. doi: 10.1002/dvdy.22074. [DOI] [PubMed] [Google Scholar]
- Gavis ER, Lunsford L, Bergsten SE, Lehmann R. A conserved 90 nucleotide element mediates translational repression of nanos RNA. Development. 1996;122:2791–2800. doi: 10.1242/dev.122.9.2791. [DOI] [PubMed] [Google Scholar]
- Gerber AP, Luschnig S, Krasnow MA, Brown PO, Herschlag D. Genome-wide identification of mRNAs associated with the translational regulator PUMILIO in Drosophila melanogaster. Proc Natl Acad Sci USA. 2006;103:4487–4492. doi: 10.1073/pnas.0509260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi S, Tsuda M, Kitajima S, Sasaoka Y, Nomura-Kitabayashid A, Kurokawa K, Saga Y. nanos1: a mouse nanos gene expressed in the central nervous system is dispensable for normal development. Mech Dev. 2003;120:721–731. doi: 10.1016/s0925-4773(03)00043-1. [DOI] [PubMed] [Google Scholar]
- Hashimoto H, Hara K, Hishiki A, Kawaguchi S, Shichijo N, Nakamura K, Unzai S, Tamaru Y, Shimizu T, Sato M. Crystal structure of zinc-finger domain of Nanos and its functional implications. EMBO Rep. 2010;11:848–853. doi: 10.1038/embor.2010.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Hayashi M, Kobayashi S. Nanos suppresses somatic cell fate in Drosophila germ line. Proc Natl Acad Sci USA. 2004;101:10338–10342. doi: 10.1073/pnas.0401647101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston DW, King ML. Germ plasm and molecular determinants of germ cell fate. Curr Top Dev Biol. 2000;50:155–181. doi: 10.1016/s0070-2153(00)50008-8. [DOI] [PubMed] [Google Scholar]
- Irish V, Lehmann R, Akam M. The Drosophila posterior-group gene nanos functions by repressing hunchback activity. Nature. 1989;338:646–648. doi: 10.1038/338646a0. [DOI] [PubMed] [Google Scholar]
- Julaton VT, Reijo Pera RA. NANOS3 function in human germ cell development. Hum Mol Genet. 2011;20:2238–2250. doi: 10.1093/hmg/ddr114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano CE, Yajima M, Wessel GM. Nanos functions to maintain the fate of the small micromere lineage in the sea urchin embryo. Dev Biol. 2010;337:220–232. doi: 10.1016/j.ydbio.2009.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyrova LY, Habara Y, Lee TH, Wharton RP. Translational control of maternal Cyclin B mRNA by Nanos in the Drosophila germline. Development. 2007;134:1519–1527. doi: 10.1242/dev.002212. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Yamada M, Asaoka M, Kitamura T. Essential role of the posterior morphogen nanos for germline development in Drosophila. Nature. 1996;380:708–711. doi: 10.1038/380708a0. [DOI] [PubMed] [Google Scholar]
- Koprunner M, Thisse C, Thisse B, Raz E. A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes Dev. 2001;15:2877–2885. doi: 10.1101/gad.212401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer B, Crittenden S, Gallegos M, Moulder G, Barstead R, Kimble J, Wickens M. NANOS-3 and FBF proteins physically interact to control the sperm-oocyte switch in Caenorhabditis elegans. Curr Biol. 1999;9:1009–1018. doi: 10.1016/s0960-9822(99)80449-7. [DOI] [PubMed] [Google Scholar]
- Lai F, King ML. Repressive translational control in germ cells. Mol Reprod Dev. 2013;80:665–676. doi: 10.1002/mrd.22161. [DOI] [PubMed] [Google Scholar]
- Lai F, Singh A, King ML. Xenopus Nanos1 is required to prevent endoderm gene expression and apoptosis in primordial germ cells. Development. 2012;139:1476–1486. doi: 10.1242/dev.079608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F, Zhou Y, Luo X, Fox J, King ML. Nanos1 functions as a translational repressor in the Xenopus germline. Mech Dev. 2011;128:153–163. doi: 10.1016/j.mod.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann R, Nusslein-Volhard C. hunchback, a gene required for segmentation of an anterior and posterior region of the Drosophila embryo. Dev Biol. 1987;119:402–417. doi: 10.1016/0012-1606(87)90045-5. [DOI] [PubMed] [Google Scholar]
- Luo X, Nerlick S, An W, King ML. Xenopus germline nanos1 is translationally repressed by a novel structure-based mechanism. Development. 2011;138:589–598. doi: 10.1242/dev.056705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosquera L, Forristall C, Zhou Y, King ML. A mRNA localized to the vegetal cortex of Xenopus oocytes encodes a protein with a nanos-like zinc finger domain. Development. 1993;117:377–386. doi: 10.1242/dev.117.1.377. [DOI] [PubMed] [Google Scholar]
- Murata Y, Wharton RP. Binding of pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell. 1995;80:747–756. doi: 10.1016/0092-8674(95)90353-4. [DOI] [PubMed] [Google Scholar]
- Oulhen N, Yoshida T, Yajima M, Song JL, Sakuma T, Sakamoto N, Yamamoto T, Wessel GM. The 3'UTR of nanos2 directs enrichment in the germ cell lineage of the sea urchin. Dev Biol. 2013;377:275–283. doi: 10.1016/j.ydbio.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransick A, Rast JP, Minokawa T, Calestani C, Davidson EH. New early zygotic regulators expressed in endomesoderm of sea urchin embryos discovered by differential array hybridization. Dev Biol. 2002;246:132–147. doi: 10.1006/dbio.2002.0607. [DOI] [PubMed] [Google Scholar]
- Saga Y. Function of Nanos2 in the male germ cell lineage in mice. Cell Mol Life Sci. 2010;67:3815–3822. doi: 10.1007/s00018-010-0456-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Hayashi Y, Ninomiya Y, Shigenobu S, Arita K, Mukai M, Kobayashi S. Maternal Nanos represses hid/skl-dependent apoptosis to maintain the germ line in Drosophila embryos. Proc Natl Acad Sci USA. 2007;104:7455–7460. doi: 10.1073/pnas.0610052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaner CE, Deshpande G, Schedl PD, Kelly WG. A conserved chromatin architecture marks and maintains the restricted germ cell lineage in worms and flies. Dev Cell. 2003;5:747–757. doi: 10.1016/s1534-5807(03)00327-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AB, Pisani D, Mackenzie-Dodds JA, Stockley B, Webster BL, Littlewood DT. Testing the molecular clock: molecular and paleontological estimates of divergence times in the Echinoidea (Echinodermata) Mol Biol Evol. 2006;23:1832–1851. doi: 10.1093/molbev/msl039. [DOI] [PubMed] [Google Scholar]
- Song JL, Wessel GM. The forkhead transcription factor FoxY regulates Nanos. Mol Reprod Dev. 2012;79:680–688. doi: 10.1002/mrd.22073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J, Wharton RP. Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev. 1999;13:2704–2712. doi: 10.1101/gad.13.20.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J, Wharton RP. Drosophila Brain Tumor is a translational repressor. Genes Dev. 2001;15:762–773. doi: 10.1101/gad.870801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G, Johnston P, Lawrence PA. Control of Drosophila body pattern by the hunchback morphogen gradient. Cell. 1992;69:237–249. doi: 10.1016/0092-8674(92)90405-2. [DOI] [PubMed] [Google Scholar]
- Su TT, Sprenger F, DiGregorio PJ, Campbell SD, O'Farrell PH. Exit from mitosis in Drosophila syncytial embryos requires proteolysis and cyclin degradation, and is associated with localized dephosphorylation. Genes Dev. 1998;12:1495–1503. doi: 10.1101/gad.12.10.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam K, Seydoux G. nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development. 1999;126:4861–4871. doi: 10.1242/dev.126.21.4861. [DOI] [PubMed] [Google Scholar]
- Tsuda M, Sasaoka Y, Kiso M, Abe K, Haraguchi S, Kobayashi S, Saga Y. Conserved role of nanos proteins in germ cell development. Science. 2003;301:1239–1241. doi: 10.1126/science.1085222. [DOI] [PubMed] [Google Scholar]
- Voronina E, Lopez M, Juliano CE, Gustafson E, Song JL, Extavour C, George S, Oliveri P, McClay D, Wessel G. Vasa protein expression is restricted to the small micromeres of the sea urchin, but is inducible in other lineages early in development. Dev Biol. 2008;314:276–286. doi: 10.1016/j.ydbio.2007.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zayas RM, Guo T, Newmark PA. nanos function is essential for development and regeneration of planarian germ cells. Proc Natl Acad Sci USA. 2007;104:5901–5906. doi: 10.1073/pnas.0609708104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton RP, Sonoda J, Lee T, Patterson M, Murata Y. The Pumilio RNA-binding domain is also a translational regulator. Mol Cell. 1998;1:863–872. doi: 10.1016/s1097-2765(00)80085-4. [DOI] [PubMed] [Google Scholar]
- Wharton RP, Struhl G. RNA regulatory elements mediate control of Drosophila body pattern by the posterior morphogen nanos. Cell. 1991;67:955–967. doi: 10.1016/0092-8674(91)90368-9. [DOI] [PubMed] [Google Scholar]
- Wreden C, Verrotti AC, Schisa JA, Lieberfarb ME, Strickland S. Nanos and pumilio establish embryonic polarity in Drosophila by promoting posterior deadenylation of hunchback mRNA. Development. 1997;124:3015–3023. doi: 10.1242/dev.124.15.3015. [DOI] [PubMed] [Google Scholar]
- Wu X, Wang B, Dong Z, Zhou S, Liu Z, Shi G, Cao Y, Xu Y. A NANOS3 mutation linked to protein degradation causes premature ovarian insufficiency. Cell Death Dis. 2013;4:e825. doi: 10.1038/cddis.2013.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima M, Wessel GM. Small micromeres contribute to the germline in the sea urchin. Development. 2011;138:237–243. doi: 10.1242/dev.054940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima M, Wessel GM. Autonomy in specification of primordial germ cells and their passive translocation in the sea urchin. Development. 2012;139:3786–3794. doi: 10.1242/dev.082230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Gallegos M, Puoti A, Durkin E, Fields S, Kimble J, Wickens MP. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature. 1997;390:477–484. doi: 10.1038/37297. [DOI] [PubMed] [Google Scholar]