Abstract
Dynamic contrast enhanced magnetic resonance imaging (DCE-MRI) can quantitatively and qualitatively assess physiological characteristics of tissue. Quantitative DCE-MRI requires an estimate of the time rate of change of the concentration of the contrast agent in the blood plasma, the vascular input function (VIF). Measuring the VIF in small animals is notoriously difficult as it requires high temporal resolution images limiting the achievable number of slices, field-of-view, spatial resolution, and signal-to-noise. Alternatively, a population-averaged VIF could be used to mitigate the acquisition demands in studies aimed to investigate, for example, tumor vascular characteristics. Thus, the overall goal of this manuscript is to determine how the kinetic parameters estimated by a population based VIF differ from those estimated by an individual VIF. Eight rats bearing gliomas were imaged before, during, and after an injection of Gd-DTPA. Ktrans, ve, and vp were extracted from signal-time curves of tumor tissue using both individual and population-averaged VIFs. Extended model voxel estimates of Ktrans and ve in all animals had concordance correlation coefficients (CCC) ranging from 0.69 to 0.98 and Pearson correlation coefficients (PCC) ranging from 0.70 to 0.99. Additionally, standard model estimates resulted in CCCs ranging from 0.81 to 0.99 and PCCs ranging from 0.98 to 1.00, supporting the use of a population based VIF if an individual VIF is not available.
Keywords: MRI, dynamic contrast, DCE, tumor, pharmacokinetic modeling, input function
Introduction
Dynamic contrast enhanced magnetic resonance imaging (DCE-MRI) can be used to investigate vascular properties of tumors including blood vessel perfusion and permeability, and tissue volume fractions. DCE-MRI probes these characteristics by rapidly acquiring heavily T1-weighted images before, during, and after injection of a paramagnetic contrast agent (CA) such as gadopentetate dimeglumine (Gd-DTPA). Pharmacokinetic modeling of the resulting signal-intensity time courses allow for estimating the local physiological properties (1) making it a valuable tool in preclinical and clinical studies of, for example, anti-angiogenic treatments (2).
One challenge with pharmacokinetic modeling of DCE-MRI data is estimating the time rate of change of the concentration of the CA in a nearby feeding vessel, the so-called vascular input function (VIF). Accurately estimating the VIF is particularly challenging in small animals where small vessel sizes impedes direct VIF measurement (3). Imaging based approaches to measuring the VIF typically require a technical balance between high temporal resolution acquisitions to capture the VIF’s rapid uptake and high spatial resolution to observe tumor heterogeneity (4,5). Two alternative imaging derived approaches that alleviate the technical constraints of acquiring both high spatial and temporal resolution images are reference region methods and the use of a population-averaged VIF. In a reference region analysis, characterization of the VIF can be avoided by using the parameters and the signal-intensity time course of a reference tissue, which can be acquired at a lower temporal resolution, to calibrate CA uptake in tumors (6,7). Rather than acquire a VIF for each individual subject, some investigators have explored using a population-averaged VIF built from a cohort of subjects and subsequently used for pharmacokinetic analysis (8-13). Replacing an individual based VIF with a population-averaged VIF can reduce the need for high temporal resolution and alleviate SNR restrictions required for individual VIF measurements, thereby potentially allowing improved characterization of the tumor environment.
Following the work of Loveless et al (11), the goal of this study is to determine, in a cohort of tumor bearing rats, the differences in kinetic parameters resulting from replacing an individually measured VIF with a population based VIF. The resulting parameters were statistically compared at both the voxel and ROI level.
Methods
Animal Model
Female Sprague-Dawley rats (n = 8, 234-270 g) were anesthetized, given analgesic and inoculated with C6 glioma cells (1×105 cells) via stereotaxic injection. CA was injected through a jugular catheter placed within 24 h prior to imaging. During each MRI procedure body temperature was maintained near 37 °C by a flow of warm air directed over the animal and respiration was monitored using a pneumatic pillow. Each rat was anesthetized using 2% isoflurane in oxygen for all surgical and imaging procedures. All experimental procedures were approved by Vanderbilt University’s Institutional Animal Care and Use Committee.
DCE-MRI
Imaging procedures are described in more detail elsewhere (14). Briefly, MRI was performed using a 9.4 T horizontal-bore magnet (Agilent, Santa Clara, CA, USA). A pre-contrast T1 map was generated with data from an inversion-recovery snapshot experiment-repetition time (TR) = 12s, with 10 TIs logarithmically spaced between 0.250 and 11 s, two averaged excitations, 128 × 128 samples, 32 × 32 mm field of view. Two slices were acquired during the DCE-MRI experiment, one through the center of the brain tumor and another through the neck containing the major vessels feeding the brain. The DCE-MRI protocol consisted of a standard spoiled gradient-echo sequence with TR/TE/α = 10 ms/2.1 ms/15°, a 96 × 96 acquisition matrix, and two averaged excitations resulting in a temporal resolution of approximately 2 seconds. Dynamic images were collected before, during, and for 20 minutes after manually delivering a 200 μl bolus of 0.05 mmol kg−1 Gd-DTPA (Magnevist, Wayne, NJ) over 5 s.
Data Analysis
For each animal, the pre-contrast T1 map was used to identify the tumor ROI from which the concentration of the CA in tissue space (Ct) was calculated. A voxel ROI was then manually drawn around the linguofacial artery in the neck from which voxels exhibiting partial volume effects with tissue were eliminated by visual inspection and the remaining voxels (5-10 voxels) were averaged to form the VIF, from which the concentration of the CA in the blood/plasma space (Cp) was calculated. After aligning the arrival times of the individual VIFs (VIFind), the mean of the eight VIFind was used to generate the population-averaged VIF (VIFpop). Following the method of Li et al, each animal’s VIFind and VIFpop were scaled so that the concentration time course of a 50 voxel ROI in the temporalis muscle yielded a ve of 0.11 (15-19). The scaled VIFind and VIFpop for each animal were used with the Ct and fit to both the standard (ST) and the extended (EX) models in MATLAB R2012a (Natick, MA, USA). The ST model is given by:
| [1] |
where Ktrans is the volume transfer constant (in units of min−1) and ve is the is extravascular-extracellular volume fraction. The EX includes a vascular volume fraction, vp, within the tissue space:
| [2] |
After fitting the voxel data to the ST and EX models the coefficient of determination, r2, was calculated to measure the goodness of fit to the voxel data. Voxel fits with a returned r2 < 0.6, or estimated parameters outside the physiological range (0 < Ktrans < 1 min−1, 0 < ve < 1, and 0 < vp < 1) were removed from subsequent analyses.
Statistical Analysis
Agreement between pharmacokinetic parameters extracted from the ST (Ktrans and ve) and EX (Ktrans,ve, and vp) models were examined. The parameters estimated with VIFpop and VIFind were paired. The paired voxel and ROI data for each animal was then compared based on its Pearson correlation coefficient (PCC) and concordance correlation coefficient (CCC) (20).
Results
Population-averaged VIF
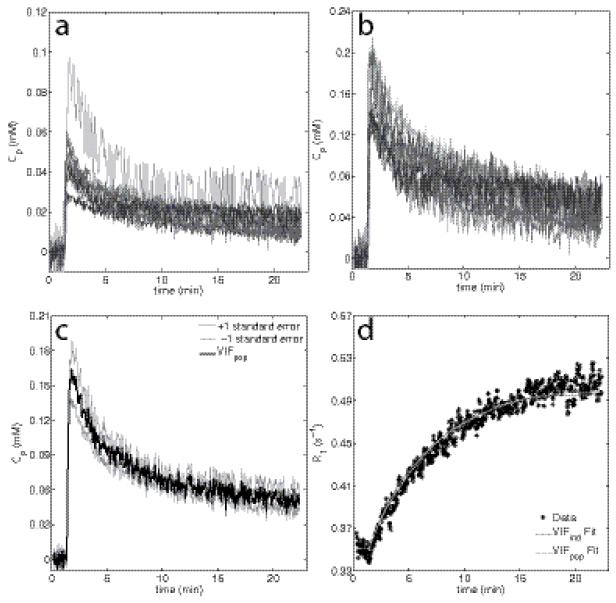
The eight individual VIFs are shown in Figure 1a. Figure 1b shows the same individual VIFs after the application of a multiplicative scaling factor as described by Li et al and others (16,21,22), to minimize flow effects. The population-averaged VIF is presented in Figure 1c (black line) where the gray lines represent one standard error from the population-averaged VIF. Both the peak and the washout share a similar standard error (18.0% and 14.73% of the mean, respectively). Figure 1d shows an example dynamic R1 time course obtained from a single voxel. The VIFind and VIFpop based fits both resulted in r2 values of 0.97.
Figure 1.
Panels (a-b) display the eight individual VIFs un-scaled and scaled, respectively. Panel (c) displays a population averaged VIF and the standard error at each time point; individual VIFs were tightly grouped resulting in a small difference between the standard error at the peak and during the washout period. Panel (d) shows an example voxel R1 time course within a tumor fit with both the individual and population based VIFs, both of which show a good fit to the measured R1 time course (r2 = 0.97, for both).
ROI Analysis
The results of ROI analysis are presented in the bottom rows of Tables 1 and 2. The ROI value for the PCC and CCC were calculated from the individual rat ROIs. Table 1 presents the calculated PCCs for both the ST Model and the EX model, while table 2 displays the calculated CCC’s from the ST and EX model parameter estimation. For the ROI analysis, a strong linear relationship was observed between parameters estimated using VIFpop and VIFind with Ktrans and ve having PCCs greater than 0.94. Likewise, ROI analysis resulted in CCCs greater than 0.74 for ST and EX model estimates of Ktrans and ve. vp had a lower PCC (0.56) and a lower CCC (0.18) compared to Ktrans and ve results. ST estimates of Ktrans and ve had greater PCCs (PCC = 0.98 and 0.98, respectively) compared to EX estimates of Ktrans and ve (PCC = 0.97 and 0.94, respectively). Likewise, ST estimates of Ktrans and ve also had greater CCCs (CCC = 0.76 and 0.86, respectively) compared to EX estimates of Ktrans and ve (CCC = 0.74 and 0.82, respectively).
Table 1.
PCC Results for ST and EX Model Voxel and ROI Estimates
| Rat # | PCC (95% CI) for Standard Model |
PCC (95% CI) for Extended Model |
|||
|---|---|---|---|---|---|
| Ktrans | ve | Ktrans | ve | vp | |
| 1 | 0.99 (0.99 0.99) | 0.99 (0.99 1.00) | 0.98 (0.97 0.99) | 0.98 (0.97 0.98) | 0.52 (0.37 0.64) |
| 2 | 0.99 (0.98 0.99) | 1.00 (0.99 1.00) | 0.98 (0.97 0.99) | 0.99 (0.99 0.99) | 0.27 (0.09 0.42) |
| 3 | 1.00 (0.99 1.00) | 1.00 (0.99 1.00) | 0.97 (0.96 0.98) | 0.99 (0.98 0.99) | 0.54 (0.42 0.63) |
| 4 | 0.99 (0.99 1.00) | 0.99 (0.99 1.00) | 0.98 (0.98 0.99) | 0.95 (0.93 0.96) | 0.55 (0.42 0.66) |
| 5 | 1.00 (0.99 1.00) | 0.98 (0.98 0.99) | 0.99 (0.99 0.99) | 0.97 (0.96 0.98) | 0.77 (0.67 0.84) |
| 6 | 0.99 (0.99 1.00) | 1.00 (0.99 1.00) | 0.97 (0.95 0.98) | 0.98 (0.96 0.99) | 0.61 (0.45 0.74) |
| 7 | 1.00 (0.99 1.00) | 0.99 (0.98 0.99) | 0.70 (0.62 0.77) | 0.98 (0.97 0.99) | 0.40 (0.27 0.52) |
| 8 | 1.00 (0.99 1.00) | 0.99 (0.99 0.99) | 0.92 (0.90 0.94) | 0.99 (0.99 0.99) | 0.72 (0.64 0.77) |
|
| |||||
| ROI | 0.98 (0.88 1.00) | 0.98 (0.91 1.00) | 0.98 (0.88 1.00) | 0.94 (0.71 0.99) | 0.56 (−0.24 0.91) |
Table 2.
CCC Results for ST and EX Model Voxel and ROI Estimates
| Rat # | CCC (95% CI) for Standard Model |
CCC (95%) for Extended Model |
|||
|---|---|---|---|---|---|
| Ktrans | ve | Ktrans | ve | vp | |
| 1 | 0.90 (0.87 0.92) | 0.96 (0.95 0.97) | 0.92 (0.90 0.99) | 0.91 (0.88 0.93) | 0.51 (0.49 0.53) |
| 2 | 0.93 (0.91 0.95) | 0.97 (0.97 0.98) | 0.95 (0.94 0.97) | 0.97 (0.96 0.98) | 0.25 (0.24 0.26) |
| 3 | 0.96 (0.95 0.96) | 0.99 (0.98 0.99) | 0.92 (0.90 0.94) | 0.96 (0.95 0.97) | 0.39 (0.36 0.42) |
| 4 | 0.98 (0.98 0.99) | 0.99 (0.98 0.99) | 0.97 (0.97 0.98) | 0.93 (0.90 0.95) | 0.18 (0.15 0.21) |
| 5 | 0.97 (0.97 0.98) | 0.97 (0.97 0.98) | 0.98 (0.97 0.99) | 0.93 (0.91 0.95) | 0.72 (0.68 0.75) |
| 6 | 0.81 (0.77 0.86) | 0.98 (0.98 0.99) | 0.74 (0.68 0.81) | 0.95 (0.93 0.97) | 0.21 (0.17 0.26) |
| 7 | 0.99 (0.98 0.99) | 0.98 (0.98 0.98) | 0.69 (0.62 0.77) | 0.97 (0.96 0.98) | 0.35 (0.33 0.36) |
| 8 | 0.87 (0.85 0.89) | 0.97 (0.96 0.97) | 0.76 (0.72 0.79) | 0.94 (0.93 0.95) | 0.68 (0.67 0.70) |
|
| |||||
| ROI | 0.76 (0.56 0.96) | 0.86 (0.69 1.03) | 0.74 (0.53 0.94) | 0.82 (0.64 1.01) | 0.18 (0.09 0.27) |
Voxel Analysis
The estimated parameters obtained by fitting the voxel level Ct time courses to the ST and EX models with both VIFind and VIFpop were compared across all animals. The results are presented in Tables 1 and 2. The PCC along with its 95% confidence intervals computed for each animal and parameter are presented in Table 1. ST and EX estimates of Ktrans and ve returned from voxel data analysis resulted in PCCs ranging from 0.70 to 1.00. The PCC between the VIFind and VIFpop estimates of vp was < 0.77.
The high level of agreement between parameters estimated with VIFpop and those estimated with VIFind is presented in Table 2 by the CCC and its 95% confidence interval. For most rats, ST estimates of Ktrans and ve resulted in increased PCCs (16 of 16) and CCCs (12 of 16) over their EX model counterparts. In general, a weak correlation (CCC < 0.72) was observed between estimates of vp estimated using VIFpop and VIFind.
Discussion and Conclusions
A substantial challenge to performing quantitative DCE-MRI in small animals is the measurement of the VIF. While there are imaging and non-imaging approaches that can provide a reasonable estimate of the VIF, each method has its limitations. The necessity for high temporal resolution for imaging based estimates of the VIF significantly restricts the SNR, FOV, and spatial resolution available to characterize the tumor itself. Additionally, to better investigate tumor growth, angiogenesis, or treatment response, multiple slice or 3D data sets are desired which will result in either a decreased spatial resolution and/or FOV in order to maintain a temporal resolution appropriate for VIF estimation. One way to acquire multi-slice, high spatial resolution DCE-MRI data—without forfeiting a high temporal resolution VIF—is to use a population based VIF. This work has reported on the implementation and agreement between a population-averaged VIF and an individually acquired VIF in tumor bearing rats.
The motivation of using a population-averaged over an individual VIF is the potential for high spatial resolution images to characterize tumor heterogeneity. Initially, high temporal resolution images are still needed to generate the VIFpop, but future experiments using the resulting population based VIF benefit by the decrease in required temporal resolution and the increases in spatial resolution, and/or SNR, and/or FOV. Investigating tumor microstructure through ROI analysis, however, is a useful tool and can be used clinically, for example, in grading gliomas using extracted Ktrans values (23). In this study, ROI analysis of ST and EX estimates of Ktrans and ve suggests that using the VIFpop provided parameters statistically similar, as observed by CCCs > 0.74, to those produced with the VIFind. Both the voxel and ROI estimates of vp resulted in lower PCCs and CCCs compared to estimates of Ktrans and ve. The poorer correlation and linear relationships between VIFpop and VIFind estimates of vp may arise from its increased sensitivity to deviations from the “true VIF”. Additionally, when a rapid bolus injection is used (as was done here) errors in vp estimation generally increase as temporal resolution, SNR, and vp decrease (24,25). If VIFind compared to VIFpop was assumed to result in more accurate estimates of vp, the “true VIF” would be more similar to the VIFind resulting in a decreased correlation between the two estimates. When the “true VIF” is less biased towards either the VIFind or the VIFpop, the resulting estimates of vp should have an increased correlation, which could account for the higher CCCs observed in rats 5 and 8 compared to the remaining rats.
The results of this study also suggest that ROI and voxel estimates of ve using the VIFpop produced more statistically similar values and stronger linear relationships to VIFind estimates than estimates of Ktrans. The increased agreement of ve over Ktrans in this study may be traced back to the use of a scaled VIF to avoid inflow effects. Because the VIF is scaled to produce an assumed ve value of a reference tissue, error may be induced if the actual ve of the reference tissue is significantly different due to physiological conditions and/or disease states. Additionally, the higher PCC and CCC observed in ST model estimates over their EX model counterparts at both a voxel and ROI level, suggest an increased sensitivity to variations between VIFpop and VIFind due to the inclusion of the plasma space within the tissue space.
Although using a population-averaged VIF may allow improved characterization of the tumor environment by reducing the temporal resolution and FOV constraints, it may not capture physiological differences between animals due to disease progression. In other studies, experimental or physiological variation between animals was shown to affect VIFpop estimates of model parameters (10,26). However, assuming that experimental procedures remained consistent between animals, physiological variation did not seem to affect the degree to which VIFpop estimated parameters agree with VIFind parameters. Thus, if the injection protocol reported here is followed, the reported VIFpop may be used in lieu of a VIFind.
In summary, the results of this study indicate that both Ktrans and ve estimated using a population based VIF have a high level of agreement to those estimated using an individual VIF. Additionally, the results of this study validate the work done previously in mice (11). Thus, we propose that individual VIFs can be obtained and averaged to form a population VIF providing a suitable replacement for an individual VIF when one is not readily available.
Acknowledgements
We thank the National Institute of Health for funding through NIBIB EB001744, NCI R21CA169387, NCI P30CA068485, NCI R01CA138599, NCI 2R25CA092043, and NCI R25CA136440. The authors thank Zou Yue for performing the animal surgeries. The Authors also thank Dr. Mary Loveless, Ph.D. and Dr. Jennifer Whisenant, Ph.D. for informative discussions.
Grant Sponsor: NIBIB EB001744, NCI R21CA169387, NCI P30CA068485, NCI R01CA138599, NCI 2R25CA092043, and NCI R25CA136440
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yankeelov TE, Gore JC. Dynamic Contrast Enhanced Magnetic Resonance Imaging in Oncology: Theory, Data Acquisition, Analysis, and Examples. Curr. Med. Imaging Rev. 2009;3:91–107. doi: 10.2174/157340507780619179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wedam SB, Low Ja, Yang SX, Chow CK, Choyke P, Danforth D, Hewitt SM, Berman A, Steinberg SM, Liewehr DJ, Plehn J, Doshi A, Thomasson D, McCarthy N, Koeppen H, Sherman M, Zujewski J, Camphausen K, Chen H, Swain SM. Antiangiogenic and antitumor effects of bevacizumab in patients with inflammatory and locally advanced breast cancer. J. Clin. Oncol. 2006;24:769–77. doi: 10.1200/JCO.2005.03.4645. [DOI] [PubMed] [Google Scholar]
- 3.Barnes SL, Whisenant JG, Loveless ME, Ayers GD, Yankeelov TE. Assessing the reproducibility of dynamic contrast enhanced magnetic resonance imaging in a murine model of breast cancer. Magn. Reson. Med. 2012:000. doi: 10.1002/mrm.24422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paulus W, Peiffer J. Intratumoral Histologic Heterogeneity of Gliomas - A Quantitative Study. Cancer. 1989;64:442–447. doi: 10.1002/1097-0142(19890715)64:2<442::aid-cncr2820640217>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 5.Wagner M, Nafe R, Jurcoane A, Pilatus U, Franz K, Rieger J, Steinbach JP, Hattingen E. Heterogeneity in malignant gliomas: a magnetic resonance analysis of spatial distribution of metabolite changes and regional blood volume. J. Neurooncol. 2011;103:663–72. doi: 10.1007/s11060-010-0443-y. [DOI] [PubMed] [Google Scholar]
- 6.Yang C, Karczmar GS, Medved M, Stadler WM. Estimating the Arterial Input Function Using Two Reference Tissues in Dynamic Contrast-Enhanced MRI Studies: Fundamental Concepts and Simulations. Magn. Reson. Med. 2004;1117:1110–1117. doi: 10.1002/mrm.20243. [DOI] [PubMed] [Google Scholar]
- 7.Yankeelov T, Luci J, Lepage M. Quantitative pharmacokinetic analysis of DCE-MRI data without an arterial input function: a reference region model. Magn. Reson. Med. 2005;23:519–529. doi: 10.1016/j.mri.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 8.Parker GJM, Roberts C, Macdonald A, Buonaccorsi Ga, Cheung S, Buckley DL, Jackson A, Watson Y, Davies K, Jayson GC. Experimentally-derived functional form for a population-averaged high-temporal-resolution arterial input function for dynamic contrast-enhanced MRI. Magn. Reson. Med. 2006;56:993–1000. doi: 10.1002/mrm.21066. [DOI] [PubMed] [Google Scholar]
- 9.Just N, Koh D-M, D’Arcy J, Collins DJ, Leach MO. Assessment of the effect of haematocrit-dependent arterial input functions on the accuracy of pharmacokinetic parameters in dynamic contrast-enhanced MRI. NMR Biomed. 2011;24:902–15. doi: 10.1002/nbm.1648. [DOI] [PubMed] [Google Scholar]
- 10.Steingoetter A, Svensson J, Kosanke Y, Botnar RM, Schwaiger M, Rummeny E, Braren R. Reference region-based pharmacokinetic modeling in quantitative dynamic contract-enhanced MRI allows robust treatment monitoring in a rat liver tumor model despite cardiovascular changes. Magn. Reson. Med. 2011;65:229–38. doi: 10.1002/mrm.22589. [DOI] [PubMed] [Google Scholar]
- 11.Loveless ME, Halliday J, Liess C, Xu L, Dortch RD, Whisenant J, Waterton JC, Gore JC, Yankeelov TE. A quantitative comparison of the influence of individual versus population-derived vascular input functions on dynamic contrast enhanced-MRI in small animals. Magn. Reson. Med. 2012;67:226–36. doi: 10.1002/mrm.22988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meng R, Chang SD, Jones EC, Goldenberg SL, Kozlowski P. Comparison between population average and experimentally measured arterial input function in predicting biopsy results in prostate cancer. Acad. Radiol. 2010;17:520–5. doi: 10.1016/j.acra.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGrath DM, Bradley DP, Tessier JL, Lacey T, Taylor CJ, Parker GJM. Comparison of model-based arterial input functions for dynamic contrast-enhanced MRI in tumor bearing rats. Magn. Reson. Med. 2009;61:1173–84. doi: 10.1002/mrm.21959. [DOI] [PubMed] [Google Scholar]
- 14.Skinner JT, Yankeelov TE, Peterson TE, Does MD. Comparison of dynamic contrast-enhanced MRI and quantitative SPECT in a rat glioma model. Contrast Media Mol. Imaging. 2012;7:494–500. doi: 10.1002/cmmi.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donahue KM, Weisskoff RM, Parmelee DJ, Callahan RJ, Wilkinson Ra, Mandeville JB, Rosen BR. Dynamic Gd-DTPA enhanced MRI measurement of tissue cell volume fraction. Magn. Reson. Med. 1995;34:423–32. doi: 10.1002/mrm.1910340320. [DOI] [PubMed] [Google Scholar]
- 16.Li X, Rooney WD, Várallyay CG, Gahramanov S, Muldoon LL, Goodman Ja, Tagge IJ, Selzer AH, Pike MM, Neuwelt Ea, Springer CS. Dynamic-contrast-enhanced-MRI with extravasating contrast reagent: rat cerebral glioma blood volume determination. J. Magn. Reson. 2010;206:190–9. doi: 10.1016/j.jmr.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Braren R, Curcic J, Remmele S, Altomonte J, Ebert O, Rummeny EJ, Steingoetter A. Free-Breathing Quantitative Dynamic Contrast-Enhanced Magnetic Resonance Imaging in a Rat Liver Tumor Model Using Dynamic Radial T-1 Mapping. Invest. Radiol. 2011;46:624–631. doi: 10.1097/RLI.0b013e31821e30e7. [DOI] [PubMed] [Google Scholar]
- 18.Han SH, Ackerstaff E, Stoyanova R, Carlin S, Huang W, Koutcher JA, Kim JK, Cho G, Jang G, Cho H. Gaussian mixture model-based classification of dynamic contrast enhanced MRI data for identifying diverse tumor microenvironments: preliminary results. NMR Biomed. 2013;26:519–532. doi: 10.1002/nbm.2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson EM, Pishko GL, Muldoon LL, Neuwelt Ea. Inhibition of SUR1 Decreases the Vascular Permeability of Cerebral Metastases. Neoplasia. 2013;15:535–543. doi: 10.1593/neo.13164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin LI. A concordance correlation coefficient to evaluate reproducibility. Biometrics. 1989;45:255–68. [PubMed] [Google Scholar]
- 21.Hansen AE, Pedersen H, Rostrup E, Larsson HBW. Partial volume effect (PVE) on the arterial input function (AIF) in T1-weighted perfusion imaging and limitations of the multiplicative rescaling approach. Magn. Reson. Med. 2009;62:1055–9. doi: 10.1002/mrm.22098. [DOI] [PubMed] [Google Scholar]
- 22.Sakaie KE, Shin W, Curtin KR, McCarthy RM, Cashen TA, Carroll TJ. Method for improving the accuracy of quantitative cerebral perfusion imaging. J. Magn. Reson. Imaging. 2005;21:512–9. doi: 10.1002/jmri.20305. [DOI] [PubMed] [Google Scholar]
- 23.Zhang N, Zhang L, Qiu B, Meng L, Wang X, Hou BL. Correlation of volume transfer coefficient Ktrans with histopathologic grades of gliomas. J. Magn. Reson. Imaging. 2012;36:355–363. doi: 10.1002/jmri.23675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henderson E, Rutt BK, Lee T-Y. Temporal sampling requirements for the tracer kinetics modeling of breast disease. Magn. Reson. Imaging. 1998;16:1057–1073. doi: 10.1016/s0730-725x(98)00130-1. [DOI] [PubMed] [Google Scholar]
- 25.Zwick S, Brix G, Tofts P, Strecker R, Kopp-Schneider A, Laue H, Semmler W, Kiessling F. Simulation-based comparison of two approaches frequently used for dynamic contrast-enhanced MRI. Eur. Radiol. 2010;20:432–442. doi: 10.1007/s00330-009-1556-6. [DOI] [PubMed] [Google Scholar]
- 26.Simpson NE, He Z, Evelhoch JL. Deuterium NMR tissue perfusion measurements using the tracer uptake approach: I. Optimization of methods. Magn. Reson. Med. 1999;42:42–52. doi: 10.1002/(sici)1522-2594(199907)42:1<42::aid-mrm8>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]