Abstract
Non-coding RNAs have been implicated in the regulation of expression of numerous genes, however, the mechanism is not fully understood. We identified bidirectional, long non-coding RNAs upstream of the TNF gene using five different methods. They arose in a region where the repressors LRRFIP1, EZH2, and SUZ12 were demonstrated to bind, suggesting a role in repression. The non-coding RNAs were polyadenylated, capped, and chromatin-associated. Knock-down of the non-coding RNAs was associated with de-repression of TNF mRNA and diminished binding of LRRFIP1 to both RNA targets and chromatin. Over-expression of the non-coding RNAs led to diminished expression of TNF and recruitment of repressor proteins to the locus. One repressor protein, LRRFIP1, bound directly to the non-coding RNAs. These data place the non-coding RNAs upstream of TNF gene as central to the transcriptional regulation. They appear to serve as a platform for the assembly of a repressive complex.
Introduction
TNF is a member of a family of proteins that regulate immunologically competent cells. It is predominantly produced by myeloid cells, activated T cells and natural killer cells. The major roles of TNF include killing of tumor cells, the induction of adhesion molecule expression at sites of inflammation, stimulation of bone resorption, induction of fever, and activation of B cells, neutrophils, and monocytes (1–3). TNF inhibition is used therapeutically for arthritis and inflammatory bowel disease and inhibition is associated with an increased risk of infection (4–8). Conversely, over-expression of TNF in murine models is associated with pathologic inflammation (9–14). Thus, regulation of TNF is of paramount importance.
TNF is regulated at the level of chromatin, transcription, splicing, message turnover, and cleavage from the membrane (15–19). DNA methylation inhibitors and HDAC inhibitors can induce TNF expression, supporting a role for chromatin in the regulation of TNF transcription (20–22). Priming of TNF transcription requires PU.1 and CEBP proteins as is true for most lineage-specific transcripts in monocytes (23–26). After stimulation, NFκB, AP1, and ETS family members bind to specific promoter motifs and drive active elongation (27–29). As is true for many highly inducible genes, message turnover is highly regulated with tristetraprolin and TIA-1 predominantly responsible for destabilizing and repressing translation, respectively (30–33). HuR, TIAR, KSRP, and miRNAs have also been implicated in TNF mRNA turnover (34–36). Collectively, these studies demonstrate that the regulation of TNF is rigorous and redundant, presumably to limit the adverse consequences related to under or over-expression.
In our studies of chromatin at the TNF locus, we identified a region 300bp upstream of the transcription start site where the majority of the transcriptionally relevant histone modifications were found (19). We also identified a transcriptional repressor called LRRFIP1 (previously called GCF2) (37). Further evidence that this region might be important in the regulation of TNF came from a study of patients with systemic lupus erythematosus which found that the histone modifications at this site were different in patients compared to controls (38). This led us to examine the upstream region of the TNF promoter more carefully. We found significant levels of non-coding RNAs that mapped to this region.
Non-coding RNAs (ncRNAs) are common in the genome, with approximately 8000 identified (39). In general, their abundance, conservation and correlation with transcription have argued for functionality but there are relatively few specific examples known (40–43). The best known ncRNA that regulates chromatin conformation is Xist, which coats the X-chromosome destined for inactivation (44, 45). Long non-coding RNAs have been implicated in pluripotency and innate immune responses (46–48). Several studies have found that chromatin-associated RNAs are bound to chromatin-modifying complexes on chromatin, suggesting a role in epigenetic regulation (49–51). In general, the ncRNA is thought to confer locus specificity and alter local histone modifications but the specific mechanisms for each gene appear to be diverse and are largely not understood (52, 53).
Several groups have manipulated ncRNAs in an effort to dissect their exact function. The most common model is one where the ncRNAs regulate H3K9me2 and H3K27me3 marks in cis and mediate transcriptional repression (54–60). Nevertheless, a recent study found that many ncRNAs regulate gene expression in trans, suggesting that there are many more mechanisms yet to be identified (47). In spite of the rapid increase in our understanding of RNA-mediated transcriptional repression, much remains to be learned regarding the mechanisms of repression.
This study was undertaken to examine ncRNAs upstream of the TNF gene. We found a tightly linked choreography of ncRNAs and repressors on intergenic chromatin upstream of TNF. Furthermore, we identified a novel function of the transcriptional repressor LRRFIP1.
Materials and Methods
Cells, Transfections and reagents
All cell types are human. K562 is a hematopoietic stem cell-like line. THP1 is an immature monocytic leukemia line. MonoMac6 cells are a more highly differentiated monocytic cell line (61). Each was maintained in RPMI with 10% cosmic calf serum (Fisher Scientific, Pittsburgh, PA). Primary monocytes were obtained from normal human donors and purified by elutriation at the Penn Center for AIDS Research and then further purified by adherence. They were approximately 90% pure as defined by CD14 expression. Transfection of cells was performed by electroporation with the Amaxa Cell Line Lonza Nucleofector Kit (Amaxa Biosystems, Gaithersburg, MD). HPLC-purified lipopolysaccharide (LPS) and phorbol myristate acetate (PMA) were from Sigma Aldrich (St. Louis, MO). The SMART vector 2.0 Lentiviral LRRFIP1-shRNA or non-targeting negative control viral particles were purchased from Dharmacon (Chicago, IL). The target sequences of three LRRFIP1-shRNA used in the studies were: 5′-GGUUAUCACCCAGAUUAGA-3′, 5′-AAUGGAGAGACUUCCGACA-3′, and 5′-GUAGGGAUCACAACGAAGA-3′. Three shRNAs were evaluated and the most effective was used for the experiments. Transduction of cells and stable cell line development were done following the manufacturer’s protocol. The EZH2 siRNA sequence was GGAUGGUACUUUCAUUGAATT and the SUZ12 sequence was GGAUAGAUGUUUCUAUCAATT (Ambion Intvitrogen, Grand Island, NY). The ncRNA over-expression system utilized the PMK CMV vector with the ncRNA ligated downstream of the CMV promoter.
RNA extraction, Quantitative Real-Time PCR and NanoString nCounter Assay
RNA was prepared, DNase treated, and reverse transcribed using the Advantage RT for PCR kit (Clontech, Mountainview, Calif). Nuclear RNA was extracted with the PARIS™ Kit (Ambion, Grand Island, NY). Primer-probe combinations for all targets are listed in the Supplemental Table. Custom primers and labeled probes were synthesized by IDT (Coralville, IA). Spliced TNF mRNA was detected by proprietary gene-specific primers from Applied Biosystems on a Taqman SDS 7900HT. Primers to amplify 18S rRNA were included in each amplification and served as the internal standard for normalization.
NanoString nCounter analysis was performed using NanoString custom-synthesized probes (NanoString, Seattle, WA). Total RNA, nuclear RNA, or lysate was directly hybridized with gene-specific color-coded probes and data collection was carried out in the nCounter Digital Analyzer as described by the manufacturer. Transcript numbers for each gene were normalized to the mean of housekeeping genes. In addition, six positive-control and eight negative-control probes were added to each reaction. Normalization then used the top 100 geometric mean. All the reaction counts were within the linear dynamic range of the standard curve.
5′ and 3′ RACE were performed with the FirstChoice RLM-RACE Kit (Ambion) according to the manufacturer’s instructions. Total RNA was extracted from K562 and treated with DNase I. The PCR products were purified and cloned into a pGEM-T vector (Promega) for sequencing.
Subcellular fractionation, m7-cap analysis and poly A+ RNA purification
Subcellular fractions were prepared as described with a few modifications (62, 63). 5–10×106 K562 cells were lysed in RSB-100 buffer (100mM Tris-HCl pH 7.4, 100mM NaCl, 2.5mM MgCl2, 50 μg/ml digitonin, 100 U/ml RNasin, 1X phosphatase inhibitor cocktail (Sigma, St. Louis, MO) followed by centrifugation at 2,000g for 8 min. The supernatant was collected as the cytosolic fraction. The nuclear pellet was then resuspended in RSB-100 T (0.5% Triton X-100 in RSB-100). After centrifugation at 2,000g for 8 min, the supernatant was collected as the nuclear fraction. The resulting chromatin pellet was resuspended in RSB-100T and sonicated. The soluble DNA-bound RNA fraction was collected after centrifugation at 4,000g for 15 min. RNA was extracted with Trizol (Invitrogen, Grand Island, NY) and treated with RNase-free DNase I (Qiagen, Valencia, CA). Antibody for the m7G-cap was from Synaptic Systems (Goettingen, Germany) and we performed the immunoprecipitation according to the manufacturer’s instructions with 30μg total K562 RNA and protein G beads (GeneScript, Piscataway, NJ.) (64, 65). The IP-RNA and the non-IP-RNA were reverse transcribed and analyzed by qPCR. PolyA+ RNA was purified by oligo (dT)-cellulose (Sigma) with 150μg K562 RNA according to the manufacturer’s recommendation. The RNA extracted from the supernatant or wash buffer was used as a negative control. Controls using 18S (non-capped, non-polyadenylated) and IL-1β (capped, polyadenylated) were used to confirm the appropriate recovery.
Northern blot, run on assay, and RNA binding assay
For Northern analysis, total RNA from K562 was isolated with Trizol reagent (Invitrogen). A 674bp P32-labeled sense or antisense RNA probe was generated by in vitro transcription with the MAXIscript kit (Invitrogen). Hybridization was performed using QuickHyb (Agilent, La Jolla, CA) following the manufacturer’s instructions.
The modified run-on assay was performed as described (66). Nuclei were isolated and transcription allowed to proceed in the presence of biotin-16-UTP (Roche, Indianapolis, IN). The nascent transcripts were collected on avidin magnetic beads (Dynal, Invitrogen) and AMV reverse transcriptase used to generate cDNA. The proximal cDNA was quantitated by qRT-PCR using custom primers with actin primers (Applied Biosystems) for normalization.
Three different size ncRNAs (674, 377 and 150) were produced by different primers (Supplemental Table) and then ligated into the pGEM-T Easy vector. The sense and antisense ncRNAs were generated by in vitro transcription with T7 or SP6 polymerase using MaxiScript (Invitrogen). The 300bp actin RNA derives from the 3′ end of ACTB and the GFP RNA derives from 735bp of GFP cDNA. The RNA binding assay was modified from that described (67). In brief, different size ncRNAs were incubated with purified protein in gel shift buffer (10mM MOPS, pH7.0, 50mM KCl, 5mM MgCl2, 1mM, DTT, 10% glycerol and heparin) at 4°C for 30 min. The binding reactions were loaded onto 0.5% agarose gel (prepared by TB buffer) and run in TB buffer (45mM Tris, 45mM boric acid).
Chromatin and nuclear RNA immunoprecipitations
Chromatin immunoprecipitation (ChIP) assays were carried out as previously described (68, 69) and utilized the antibodies for SUZ12 (Abcam, Cambridge, MA), LRRFIP1 (Sigma, St. Louis, MO), H3K27me3 and EZH2 (Millipore, Billerica, MA). A negative control antibody (anti-glutathione S-transferase [GST]; Abcam) was always included but is omitted from some of the figures for simplicity. Duplicates or triplicates were analyzed from each experiment. ChIP data was normalized to input according to the formula: .
Nuclear RNA immunoprecipitation (RNA-IP) was prepared as described by Rinn et al., 2007 with a few modifications (70). Briefly, 4x107 cells were harvested and resuspended in 2 ml PBS, 2 ml nuclear isolation buffer (1.28 M sucrose; 40 mM Tris-HCl pH 7.5; 20 mM MgCl2; 4% Triton X-100), and 6 ml water on ice for 20 min. The nuclei were obtained by douncing with pestle B. Nuclei were pelleted by centrifugation at 2,500g for 15 min and resuspended in 1 ml RIP buffer (150 mM KCl, 25 mM Tris pH 7.4, 5 mM EDTA, 0.5 mM DTT, 0.5% IGEPAL, 9 μg/ml leupeptin, 9 μg/ml pepstatin, 10 μg/ml chymostatin, 3 μg/ml aprotinin, 1 mM PMSF, 100 U/ml RNasin). Nuclear membranes and debris were pelleted by centrifugation at 13,000 RPM for 10 min. Antibody was added to supernatant and incubated for 2 hr at 4°C. Protein A beads were added and incubated for 1 hr at 4°C with gentle rotation. The beads were pelleted at 2,500 RPM for 30 seconds and resuspended in 500 μl RIP buffer and repeated for a total of three RIP washes, followed by one wash in PBS. The beads were resuspended in 1 ml of Trizol. Co-precipitated RNAs were isolated and reverse transcribed as above. Real time PCR was performed with TNF1–4 primers and normalized to 18s rRNA.
Protein analysis
The TNF ELISAs utilized the BD Biosciences’ BD OptEIA TNF ELISA kit. A BIA3000 instrument (Biacore, Piscataway, NJ) was used to detect binding to the TNF promoter RNA or DNA sequences. Biotinylated probes centered on −308, consisting of 25-bp double-stranded RNA or DNA or DNA: RNA and a 6-bp spacer, were captured to the CM5 sensor chip via streptavidin. The proteins were suspended in running buffer (10 mM HEPES [pH 7.4], 150 mM NaCl, 0.005% Tween 20). The experiments were performed at 25°C with a flow rate of 30 μL/min. Kinetic constants were obtained by fitting the data to a simple one-to-one binding model using BIA EVALUATION 3.1 software.
The co-immunprecipitations utilized EZH2 and SUZ12 antibodies. Protein A beads were used for collection (Invitrogen). Protein was quantitated using a Bradford assay and were equally loaded on 4–12% NuPAGE gel (Invitrogen Life Technologies, Carlsbad, Calif.), and blotted with LRRFIP1 antibody (BD, Franklin Lakes, NJ).
Results
Identification of TNF upstream ncRNAs
We previously identified a transcriptional repressor of TNF, LRRFIP1, that was a known RNA-binding protein (37, 71). To understand potential mechanisms, UCSC genome browser was used to explore the existence of ncRNAs upstream of the TNF transcriptional start site (TSS). GM12878 and K562 tracks both showed low abundance ncRNAs upstream of the TNF TSS. We also explored our own RNA-seq data for evidence of transcription upstream of the TNF TSS and found low levels of transcripts identified, similar to those seen in the UCSC tracks (Supplemental Figure S1A). We examined the coding potential of these transcripts through in silico translation and found a very limited potential for peptide production.
To directly identify ncRNAs, we initially designed qRT-PCR amplimers across this region: TNF1, 2, 3, 4, 5 (Figure 1A). RNA species were detected in the LTA-TNF intergenic region in K562 cells after reverse transcription of RNA with directional primers. RNase treatment abrogated the signal, demonstrating that the amplification was truly RNA. The 3′ end of the LTA gene lies only 1.24Kb upstream of the TNF TSS and RNA species were found throughout this region (Figure 1B and 1C).
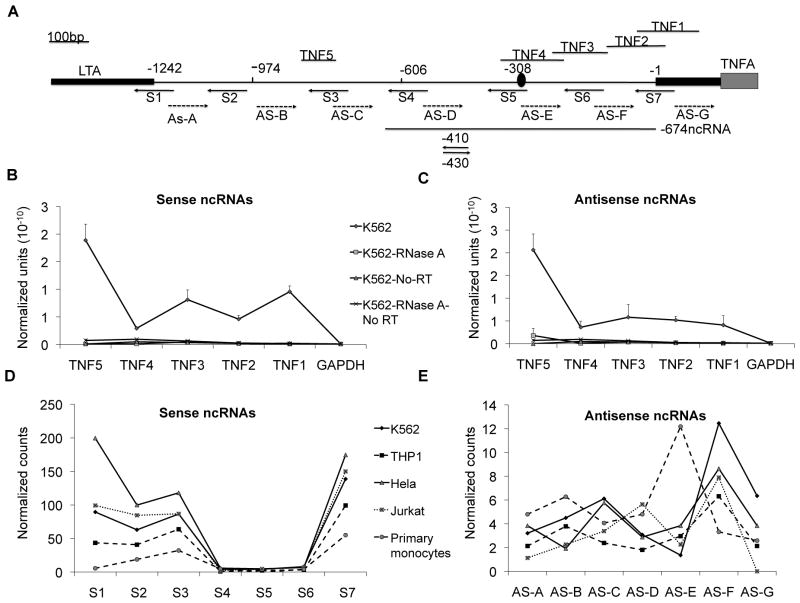
Figure 1. ncRNAs identified upstream of TNF.
RT-PCR and direct detection were used to characterize the ncRNAs upstream of TNF. A) A map demonstrates the location of the primers and probes to detect ncRNAs. Nanostring nCounter probes for digital detection are labeled S1–S7 (for sense targeting) and AS-A–G (for antisense targeting). TNF1–5 represent the qRT-PCR amplimers. The 674bp ncRNA used for over-expression studies is shown near the bottom of the map. The 410 and 430 phosphorothioate oligonucleotides used for knockdown of sense and antisense ncRNAs respectively are shown. The LRRFIP1 binding site at −308 previously identified is indicated with an oval (37). The arrows indicate the direction of the oligonucleotide sequences. B) Sense ncRNAs and C) antisense ncRNAs were identified by qRT-PCR from total RNA with or without RNase A treatment. Directional reverse transcription primers at exon 1 (for sense) and at −991 (for antisense) were used. The chart legend applies to both sense and antisense graphs. All the differences between K562 RNA and RNase A-treated K562 RNA and the No-RT K562 RNA (No RT) were significant (p<0.05, n=3). Error bars denote SE. NanoString nCounter technology was used to quantify the (D) sense or (E) antisense ncRNAs in five different types of cells
We wanted to confirm our detection of the ncRNAs without the potential confounder of skewing due to PCR amplification and we therefore used a digital method called nCounter. Seven sense or antisense probes were designed across the intergenic region: S 1–7 and AS A–G (Figure 1A). We compared the ncRNAs in several cell types of differing competence for TNF expression. The K562 cell line does not make TNF protein although it produces very low levels of TNF message. HeLa cells are fully repressed for TNF expression. Jurkat, THP1, and primary monocytes do not produce TNF protein at baseline, but in each case, the cells can be induced to express significantly more TNF protein and mRNA after stimulation. All probes detected ncRNA signal in five different cell types, confirming the presence of ncRNAs (Figure 1D and 1E).
We further confirmed the presence of the ncRNAs by rapid amplification of cDNA ends (RACE) which showed heterogeneous lengths of sense and antisense ncRNAs. The shortest product was about 200bp and longest product was >900bp (Supplemental Figure S1B). To better define the sizes of the RNAs, we used directional primers and RT-PCR (shown on the map in Figure 1). This showed strong bands of 600bp and smaller in both sense and antisense primed cDNAs (Figure 2A). An 800bp band was seen faintly in the sense-primed sample. Northern blot confirmation was attempted but the signal was dispersed around 700bp and no clear band was visualized (not shown). Therefore, the ncRNAs are bidirectional and heterogeneous in length with a peak length of about 600–700bp.
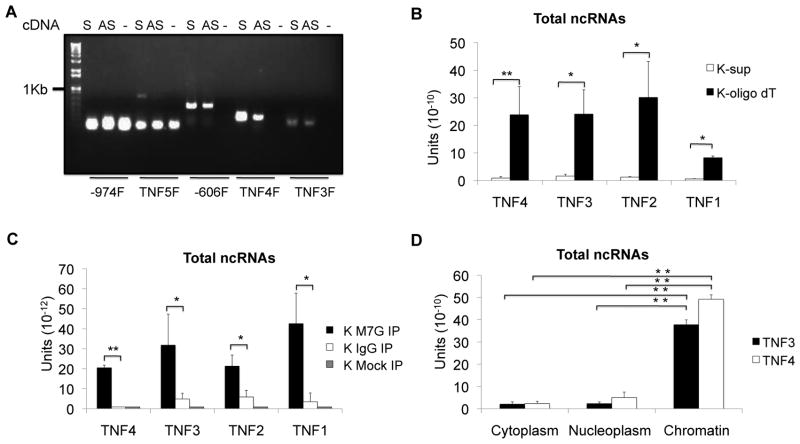
Figure 2. ncRNA structure and localization.
The biochemical structure of the ncRNAs was defined. A) Detection of ncRNA transcripts by RT-PCR. RNA from K562 cells was reverse transcribed using either no primer (-), sense (S), or antisense (AS)-specific primers. For the PCR amplification, the reverse primer was TNF3R, the locations of the forward primers are shown on the bottom of the gel. The primers TNF3, 4, 5, −974 and −606 refer to positions as shown in Figure1A. The strong bands at the bottom of the gel for the −974F and TNF5F amplifications represent primer dimers and not authentic amplification. This gel is representative of three experiments. B) Poly A+ RNA was purified by oligo dT-cellulose and the RNA extracted from supernatant was used as the negative control. Poly A+ selection enriched for ncRNA recovery detected by qRT-PCR. N=4 C) qRT-qPCR was performed for the RNA immunoprecipitated by M7G, IgG, or mock immunoprecipitated. The M7G cap-precipitated material enriched the ncRNA. N=4. D) RNA was extracted from K562 cytoplasm, nucleoplasm and chromatin and qRT-PCR was performed after reverse transcription with random primers. The ncRNAs localized primarily to the chromatin fraction. N=3. Error bars in panels denote SE. One asterisk (p<0.05); two asterisks (p<0.01), three asterisks (p<0.001).
The ncRNA structure
To examine the structure of the ncRNAs, we used an antibody for the 5′ cap m7G structure to determine whether the ncRNAs are capped and oligo dT cellulose to capture polyadenylated RNAs. These two strategies determined that the ncRNAs were largely capped and polyadenylated (Figure 2B and 2C). Controls for the oligo dT and antibody capture demonstrated the appropriate modification patterns for 18S and IL-1β mRNAs (data not shown). Different species of ncRNAs localize to different cellular compartments and are associated with different post-transcriptional modifications. To localize the ncRNAs, we fractionated K562 cells into cytoplasm, nucleoplasm and chromatin. The ncRNAs were detected by qRT-PCR and were found almost exclusively in the chromatin fraction (Figure 2D). Thus, the ncRNAs are chromatin-associated, capped and polyadenylated.
Function of the ncRNAs
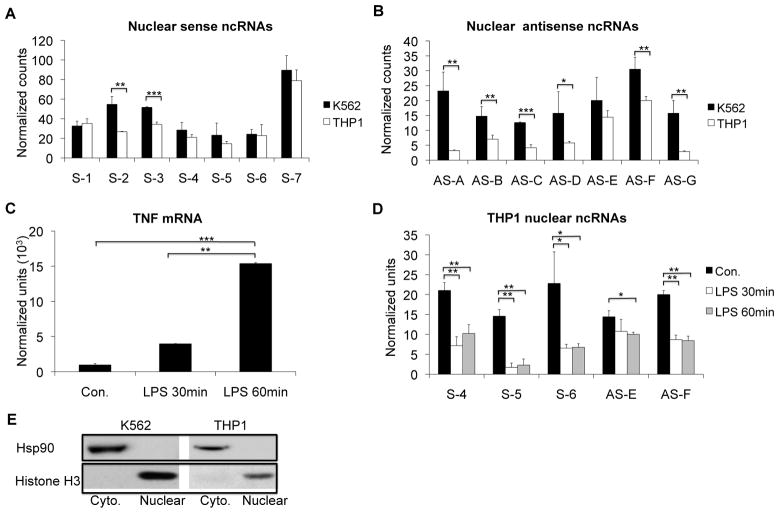
Knowing that the ncRNAs were chromatin-associated, we hypothesized that the peaks of RNA at the 3′ end of LTA and the 5′ end of TNF represented “spill” from adjacent LTA and TNF mRNAs (Figure 1). We therefore purified nuclear RNA to eliminate most mRNAs and were still able to detect signal across the region using nCounter, without the peaks near the ends of LTA and TNF (Figure 3A and 3B). We noted that the repressed K562 nuclear RNA preparations had higher levels of the ncRNAs than the competent THP1 nuclear RNA preparations (Figure 3A and 3B). When THP1 cells were stimulated with LPS, there was a rapid increase in TNF message (Figure 3C), as expected. There was a concomitant decline of sense and antisense ncRNAs in the −300 region of TNF promoter (Figure 3D).
Figure 3. ncRNA levels are inversely related to TNF mRNA expression.
NanoString nCounter analysis indicated that there were more nuclear (A) sense and (B) antisense ncRNAs in K562 than THP1 cells. N=2. C) TNF mRNA levels greatly increased after LPS stimulation of THP1 cells. N=3. D) LPS stimulation led to decreased ncRNAs in THP1 cells. N=2. Cells were treated with 1μg/ml LPS for 30 or 60 minutes and NanoString technology was used to measure nuclear ncRNAs and total TNF message. Error bars in panels denote SE. One asterisk (p<0.05); two asterisks (p<0.01), three asterisks (p<0.001). E) The purity of the cell fractions for the nuclear RNA analyses were confirmed by Western blot. HSP90 was used as a marker for cytoplasm and Histone H3 was used as a marker for nuclear material.
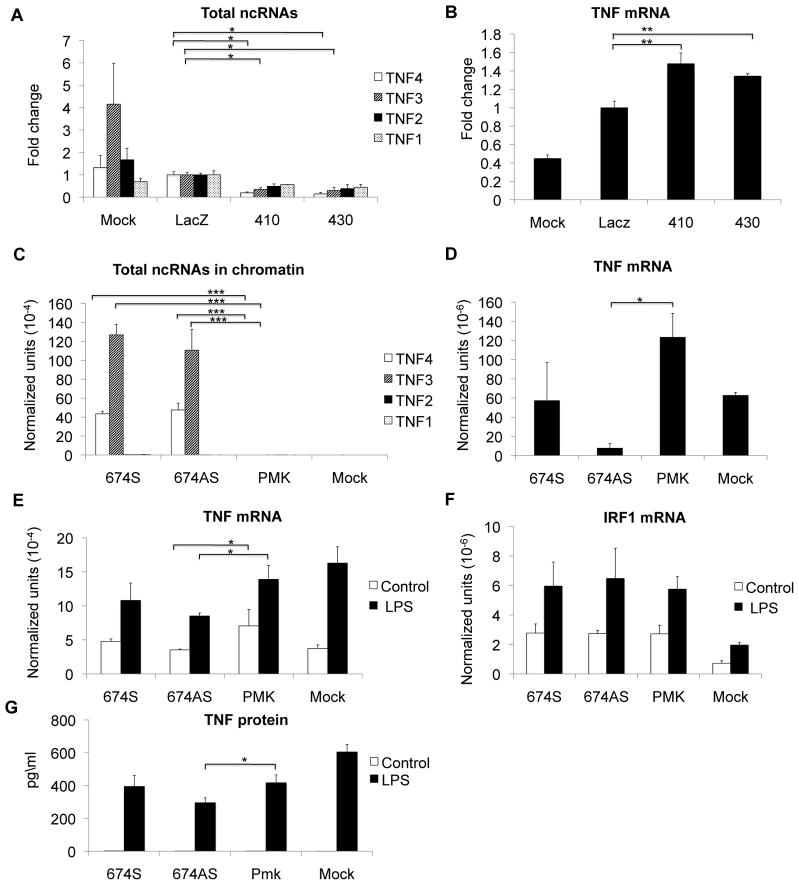
To determine whether these ncRNAs function in the regulation of TNF expression, we designed a set of phosphorothioate oligonucleotides to knock down these ncRNAs. They had variable efficacy de-repressing TNF mRNA and variable efficacy at knocking down the ncRNAs (Supplemental Figure S2). We selected a pair of oligonucleotides (410, 430) that successfully targeted the ncRNA, de-repressed TNF expression and were near the site where we had previously identified histone marks of repression (Figure 1A). In the repressed K562 cell type, the 410 and 430 oligonucleotides effectively knocked down the ncRNAs and de-repressed the TNF message (Figure 4A and 4B). Although the level of TNF mRNA is quite low in these cells, the depletion of the ncRNAs clearly led to increased spliced message.
Figure 4. The ncRNA regulation of TNF mRNA expression.
Levels of ncRNAs were modulated to examine the effect. A) K562 cells were transfected with the indicated 20bp phosphorothioate oligonucleotides which knock down either the sense (410) or the antisense (430) transcripts. The ncRNAs were measured directly using qRT-PCR with random primers, demonstrating appropriate knockdown. N=5 B) Both sense and antisense ncRNA knockdown increased TNF mRNA in K562 cells. N=5. C) K562 cells were pretreated with 10ng/ml PMA for 3 days to induce low levels of TNF expression and then transfected with the 674 ncRNA sense (S), antisense (AS) over-expression constructs or the empty vector alone (PMK). These exogenously transcribed ncRNAs successfully targeted chromatin. N=3. D) ncRNA over-expression decreased the TNF mRNA level in low dose PMA-treated K562 cells. N=3. E) The effect of ncRNA over-expression was examined in THP1 cells. Both basal and LPS-induced TNF expression was diminished in the ncRNA-transected cells compared to vector (PMK) alone. N=4. F) As a specificity control, we measured IRF1 transcript abundance from the same cultures. No effect was seen. N=4. G) THP1 cells were transfected with the over-expression constructs or the control vector. Cells were stimulated with LPS and supernatants collected. A TNF ELISA was used to quantitate the TNF protein production (n=2). Error bars in panels denote SE. One asterisk (p<0.05); two asterisks (p<0.01), three asterisks (p<0.001). Asterisks refer to the difference between the 410 or 430 and LacZ.
To further investigate the role of the ncRNAs, we made ncRNA over-expression constructs of 674bp in both the sense and antisense orientation (Figure 1A). Because K562 cells are already repressed cells, we treated the cells with low dose PMA to induce low levels of TNF mRNA. We found that transfection of ncRNAs did successfully target chromatin (Figure 4C). Transfection of antisense ncRNA led to significantly decreased TNF mRNA levels with transfection of sense ncRNA having a more modest effect (Figure 4D). We additionally examined the effects in THP1 cells, a monocyte line competent for expression. We did not observe any effect of knockdown of the ncRNAs (data not shown), probably because the ncRNA levels are already so low in THP1 cells (Figure 3D). Over-expression of the ncRNAs was associated with repression of TNF mRNA both in resting and stimulated THP1 cells (Figure 4E). To demonstrate the specificity of the effect, we examined IRF1, a transcription factor induced by LPS. Transfection of the ncRNAs had no effect on IRF1 transcript abundance in THP1 cells (Figure 4F). Furthermore, the effect of the ncRNAs was demonstrable at the level of protein production (Figure 4G). Therefore, knockdown of the ncRNAs de-repressed expression of TNF mRNA and over-expression led to repression of TNF mRNA. These studies suggested that these ncRNAs are mechanistically involved in repression of TNF.
Chromatin characteristics at the site of the ncRNAs
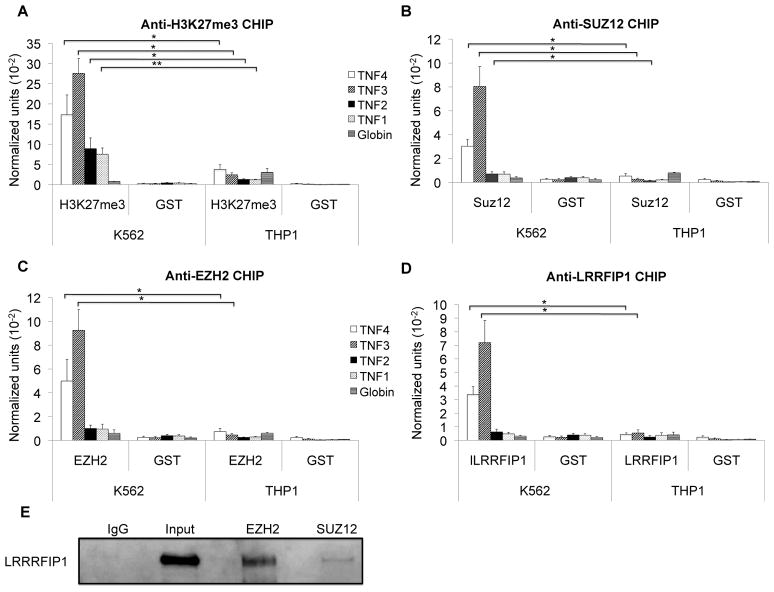
To better understand the chromatin environment at the site of the ncRNAs, we first examined histone modifications. One epigenetic mark of particular interest is tri-methylation of lysine 27 on histone H3 (H3K27me3). EZH2 (Enhancer of Zeste Homolog 2) and SUZ12 (Suppressor of Zeste 12 Homolog) are two important polycomb protein family members that have been implicated in ncRNA-mediated repression (72–74). EZH2 methylates H3K27 and SUZ12 regulates methyltransferase activity at both H3K9 and H3K27. We utilized ChIP assays to investigate chromatin marks of repression as well as the presence of EZH2 and SUZ12. We also examined a known repressor of TNF, LRRFIP1, which was of interest because of its reported RNA-binding function. As expected, H3K27me3, EZH2, SUZ12, and LRRFIP1 were all present on the TNF promoter in repressed K562 cells but not competent THP1 cells (Figure 5). They appeared to occupy a similar genomic space with a peak of amplification centered in the TNF3 amplimer site. These data suggested that a complex of proteins resided on the repressed promoter and that the repressed state was associated with H3K27me3. To determine if LRRFIP1 might directly interact with EZH2 and SUZ12, non-crosslinked co-immunoprecipitation was performed, demonstrating that EZH2 and SUZ12 both interacted with LRRFIP1 (Figure 5E).
Figure 5. Chromatin characteristics upstream of TNF.
Chromatin marks were compared in K562 and THP1 cell lines. ChIP assays showed that A) H3K27me3, B) EZH2, and C) Suz12 were increased in K562 cells compared to THP1 cells. N=5. D) LRRFIP1 was also found to bind to the same region of TNF promoter in K562 cells. N=5. Error bars in all panels denote SE. * indicates the P value of K562 compared with THP1 cells for the same antibody. One asterisk (p<0.05); two asterisks (p<0.01), three asterisks (p<0.001). E) Co-immunoprecipitation analysis of the interaction of LRRFIP1 with SUZ12 and EZH2 in K562 cells. The lysates were precipitated with anti-IgG, EZH2 or SUZ12 and analyzed by western blot with LRRFIP1 antibodies. This is one representative western blot from three experiments.
ncRNAs interact with LRRFIP1 and are required for LRRFIP1 binding to chromatin
LRRFIP1 has been previously described as an RNA-binding protein (71) and we hypothesized that it might interact with chromatin via an RNA-mediated process. LRRFIP1 exists as two major RNA splice variants, which in turn lead to two very different proteins (Supplemental Figure S3A). We made a full-length exon 2-start LRRFIP1 construct that expressed the 160kD LRRFIP1 protein and we made a truncated LRRFIP1 construct that was predicted to lack the entire DNA-binding region and half of the RNA-binding domain (Supplemental Figure S3A). Using a biosensor approach with 32bp oligonucleotides centered on the LRRFIP1 binding site at −308 or an off-target sequence, we examined binding to different nucleic acid structures. We found that the full length LRRFIP1 bound to dsRNA, ssRNA, DNA: RNA, dsDNA and ssDNA, but it had higher affinity for dsRNA, ssRNA, and DNA: RNA compared with dsDNA (Table 1). It recognized a dsRNA off-target structure (actin) similarly. In contrast, the truncated LRRFIP1 protein and GST did not exhibit any binding over background (data not shown).
Table I.
Biosensor analysis of LRRFIP1 binding
Average of 5 experiments for TNF oligonucleotides
| ka (1/MS) | kd (1/S) | KD (M) | P value (Compared to dsRNA) | |
|---|---|---|---|---|
| dsRNA | 1.62E+04 (±4.09E+3) | 4.36E-04 (±1.3E-4) | 3.97E-08 (±2.73E-08) | |
| dsDNA | 1.25E+04 (±9.19E+3) | 2.39E-03 (±9.1E-4) | 2.46E-07 (±8.90E-08) | <0.0005 |
| DNA-RNA | 2.01E+04 (±1.10E+4) | 1.07E-03 (±3.60E-4) | 5.74E-08 (±1.82E-08) | =0.245 |
| ssDNA | 4.80 E+04 (±3.60E+3) | 4.44E-03 (±1.78E-3) | 1.85E-07 (±1.21E-07) | <0.05 |
| ssRNA | 1.31E+04 (±8.05E+3) | 9.18E-04 (±5.2E-4) | 7.00E-08 (±5.81E-08) | =0.1881 |
| Off target dsRNA | 1.91E+04 | 9.72E-04 | 5.08E-08 |
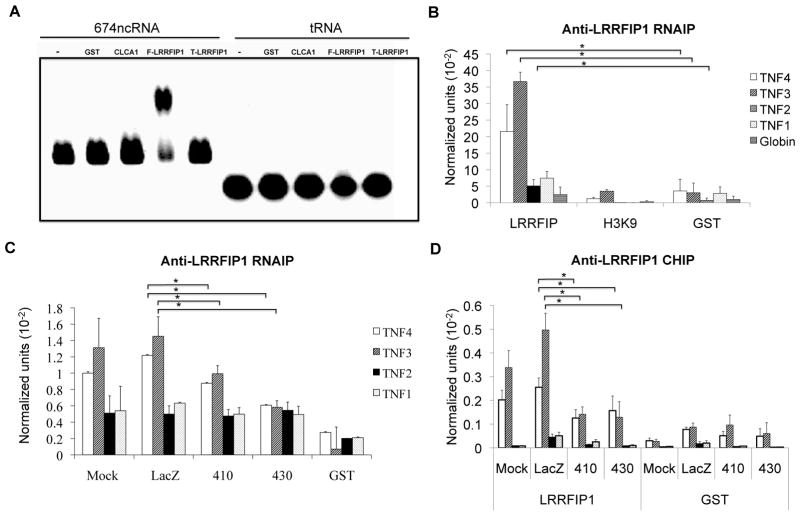
To further confirm direct RNA binding, an RNA binding assay was performed (Figure 6A). An RNA species of 674bp was produced by in vitro transcription (Figure 1A) and was incubated with several different proteins including LRRFIP1. The free RNA appeared as a low smear, while the LRRFIP1-RNA complex appeared as a shifted, higher band, confirming direct RNA binding. There was no binding to tRNA. We further examined the structural requirements of RNA binding of LRRFIP1 (Supplemental Figure S3). Longer RNAs bound LRRFIP1 better than shorter species. Double-stranded RNA of all sizes bound LRRFIP1 better than single stranded RNA. Not all RNAs bound LFFRIP1 equally (Supplemental Figure S3F), but there did not seem to be a strict sequence requirements as sense and antisense were equally capable of binding. MFold (75) was used to predict the secondary structure of the 674 antisense ncRNA and predicted a highly folded arrangement that would lead to extensive dsRNA structures even when transcribed as a single stranded species (Supplemental Figure S3H). Thus, this ncRNA would have the structural requirements that have been identified as ideal for LRRFIP1 binding.
Figure 6. LRRFIP1 interactions.
LRRFIP1 is an RNA-binding protein implicated in TNF repression. A) LRRFIP1 bound ncRNAs in vitro. The 674 ncRNA species or yeast tRNA were incubated with purified full length LRRFIP1 (F-LRRFIP1) or truncated LRRFIP1 protein (T-LRRFIP1). GST and CLCA1 protein were used as negative controls. The free RNA is indicated as the (-) lane. Only full length LRRFIP1 bound to the 674 ncRNA. LRRFIP1 bound tRNAs very poorly. This is representative of three experiments. B) LRRFIP1 bound ncRNAs in vivo. An RNA IP was performed with LRRFIP1, H3K9me3 or GST antibodies using K562 nuclei. The recovered RNA was reverse transcribed and amplified with the indicated upstream primer/probe pairs. LRRFIP1 immunoprecipitation brought down the ncRNAs but not globin. N=3. C) K562 cells were transfected with the indicated 20bp phosphorothioate oligonucleotides which knock down either the sense or the antisense transcripts. Immunoprecipitaton of LRRFIP1 recovered less ncRNA in the knockdown cells. N=3. D) LRRFIP1 binding to the TNF promoter was diminished in K562 after transfection of the 430 oligo (which knocked down anti-sense ncRNA) or the 410-transfected cells (which knocked down sense ncRNA). A GST ChIP is included as a negative control. N=5. Error bars in panels denote SE. One asterisk (p<0.05); two asterisks (p<0.01), three asterisks (p<0.001). All asterisks refer to the difference between the 410 or 430 and LacZ.
Therefore, LRRFIP1 was found to bind dsRNA>ssRNA by both biosensor and RNA binding analyses. To investigate LRRFIP1 binding to ncRNA in vivo, we performed an RNA-IP using K562 nuclei. ncRNAs were detected by qRT-PCR from LRRFIP1 immunoprecipitated nuclear RNA but not with H3K9me3 nor GST negative control antibodies (Figure 6B). Globin mRNA was also not recognized, thereby demonstrating specificity. These analyses demonstrated LRRFIP1 binding to RNA by in vitro and in vivo.
We wished to determine whether LRRFIP1 binding to chromatin was dependent on the ncRNAs. Transfection of both the reverse oligonucleotide 410 (targeting sense ncRNA) and forward oligonucleotide 430 (targeting antisense ncRNA) led to diminished LRRFIP1 binding to the upstream ncRNAs in K562 nuclei, with antisense targeting having slightly more of an effect. Mock transfected cells and LacZ oligonucleotide-transfected cells were comparable and immunoprecipitation with a GST antibody demonstrated background levels of signal (Figure 6C). These data demonstrated that the 410 and 430 oligonucleotides interfere with LRRFIP1 binding to the chromatin-associated ncRNA, presumably by depleting the ncRNAs. Having demonstrated that the 410 and 430 oligonucleotides diminished ncRNA interactions with LRRFIP1, we investigated whether that was sufficient to disrupt LRRFIP1 binding to chromatin. Transfection of the two phosphorothioate oligonucleotides led to decreased binding of LRRFIP1 to chromatin in a ChIP assay (Figure 6D). Therefore, the ncRNAs are critical for LRRFIP1 targeting.
Over-expression of ncRNAs can drive repressive chromatin
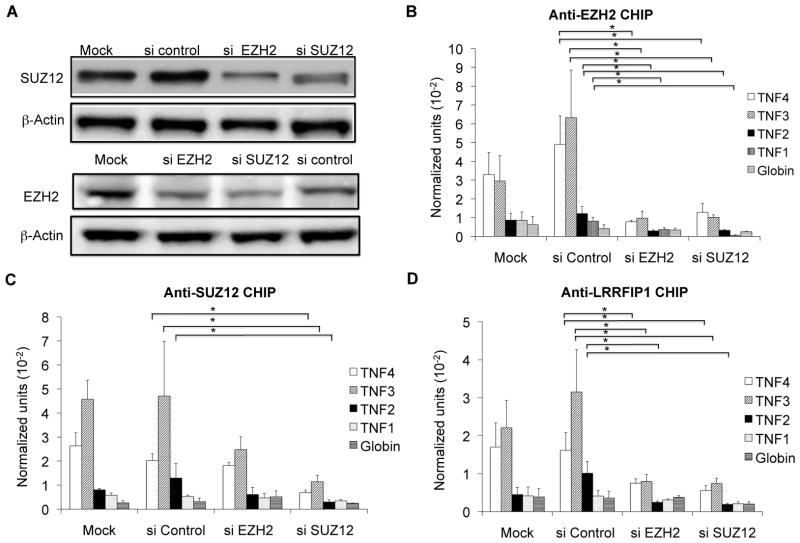
We had demonstrated that over-expression of the ncRNA repressed TNF message levels (Figure 4). We hypothesized that over-expression of the ncRNA could also drive marks of repression. The same over-expression constructs were utilized as before. We were restricted to using the TNF2 probe due to the high level of exogenous DNA after transfection. In this assay, we found increased levels of H3K27me3, EZH2, and SUZ12 in antisense 674-transfected cells compared to vector-transfected cells (PMK) (Figure 7). For the 674 sense transfection, only EZH2 binding was found to be significantly increased. Over-expression of the ncRNA was sufficient to induce marks of repression but the role of LRRFIP1 was still not clear. To identify reciprocal relationships between LRRFIP1, EZH2, and SUZ12, we knocked down EZH2 and SUZ12 (Figure 8). Knocking down either EZH2 or SUZ12 compromised binding of EZH2, SUZ12 and LRRFIP1 to chromatin (Figure 8B–8D), demonstrating that the protein-protein interactions seen by co-immunoprecipitation are functionally significant and impact recruitment to chromatin.
Figure 7. Over-expression of ncRNAs induced repressor recruitment to the TNF promoter.
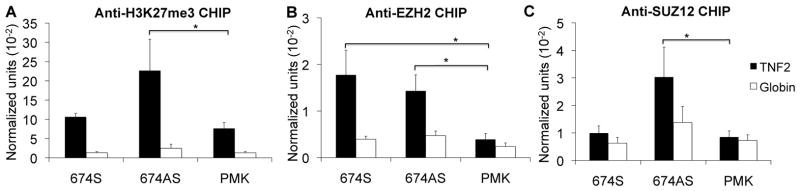
The effect of ncRNAs on chromatin conformation was examined. K562 cells were treated with 10ng/ml PMA for 3 days to induce low levels of message and then transfected with the 674 ncRNA sense or antisense over-expression constructs or the vector (PMK). CHIP assays were utilized to investigate A) H3K27me3, B) EZH2, and C) SUZ12 binding at the TNF promoter. Over-expression of the ncRNA led to increased marks of repression. N=4. Error bars in panels denote SE. One asterisk (p<0.05); two asterisks (p<0.01), three asterisks (p<0.001).
Figure 8. Knockdown of SUZ12 and EZH2 impairs LRRFIP1 binding.
Functional interactions of LRRFIP1, EZH2, and SUZ12 were examined. A) A western blot was used to determine the SUZ12 and EZH2 protein levels in the K562 cells transfected with siRNA-control, siRNA-EZH2 and siRNA-SUZ12. Knockdowns were effective. This is representative of three experiments. B–D) ChIP assays demonstrated that knockdown of SUZ12 or EZH2 affected their own binding and that of LRRFIP1 at the TNF promoter. N=3. Error bars in panels denote SE. One asterisk (p<0.05); two asterisks (p<0.01), three asterisks (p<0.001).
LRRFIP1 regulation of TNF mRNA
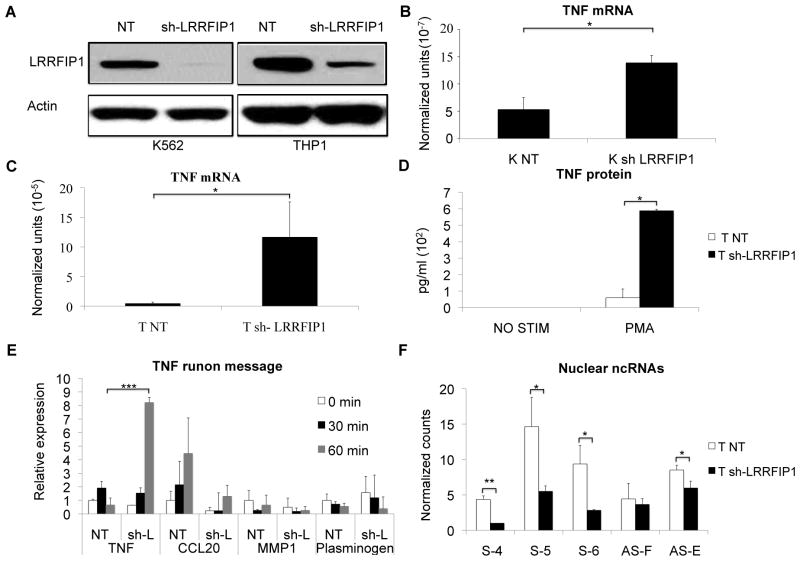
Our data suggested that the ncRNAs, especially antisense ncRNAs, are involved in LRRFIP1 binding to the promoter as a transcriptional repressor. To confirm the function of LRRFIP1, we created stably-transfected LRRFIP1 knockdown cells with short hairpin RNAs (shRNAs) in K562 and THP1 cells (Figure 9A). We found that TNF mRNA was significantly increased in both the K562 and THP1 LRRFIP1 knock-down cells (Figure 9B and 9C) and TNF protein was also increased in the LRRFIP1 knockdown THP1 cells after stimulation (Figure 9D), consistent with its role as a repressor. We used PMA as an acute stimulus in THP1 cells and also found knockdown of LRRFIP1 was associated with increased transcription as defined by a modified run-on assay (Figure 9E). Other genes, both induced or not by PMA, did not exhibit increased transcription on the LRRFIP1 knockdown cells. We also performed CHIP assays to define the effects of LRRFIP1 knock-down on chromatin marks of repressors, however, no changes were observed in H3K27me3, EZH2, SUZ12 in both LRRFIP1 knock-down K562 and THP1 cells (data not shown). Therefore, LRRFIP1 binding to chromatin is dependent on EZH2 and SUZ12 but EZH2 and SUZ12 binding is not dependent on LRRFIP1. However, we found that knock-down of LRRFIP1 resulted in diminished ncRNA abundance, especially around the LRRFIP binding site (Figure 9F). This suggests that the stability and function of ncRNA might depend on LRRFIP1 binding.
Figure 9. LRRFIP1 regulation of TNF mRNA.
LRRFIP1 was knocked down to examine the effect on transcription. A) THP1 or K562 cells were transfected with LRRFIP1 short hairpin RNA (sh-LRRFIP1) or non-target control shRNA (NT) and puromycin was used to establish the stable LRRFIP1 knockdown cell lines. The Western blot demonstrates effective knockdown of LRRFIP1. B) LRRFIP1 knockdown increased resting TNF mRNA level in K562 cells and C) THP1cells. N=3. D) TNF protein was markedly increased in PMA-stimulated-LRRFIP1 knockdown THP1 cells compared to the control cells. 10ng/ml PMA was used to treat the cells for 6 hours. N=3. E) Nascent transcripts were measured after 100ng/ml PMA treatment for 0, 30, 60 minutes in THP1 cells. LRRFIP1 knockdown cells expressed higher levels of TNF message, but this effect was limited to TNF. NT= Non-target shRNA, sh-L=LRRFIP1 knockdown. CCL20 is PMA-inducible, MMP1 and plasminogen are not PMA-inducible. N=3. F) LRRFIP1 knockdown altered the ncRNA abundance at the LRRFIP1 binding region. N=2. NanoString nCounter technology was used to detect the ncRNAs. Error bars in panels denote SE. One asterisk (p<0.05); two asterisks (p<0.01), three asterisks (p<0.001).
Discussion
Our data have defined a novel transcriptional regulatory pathway for the TNF gene. We first observed that the intergenic ncRNAs were tightly associated with chromatin and were of diverse lengths. K562 cells, repressed for TNF expression, had more abundant ncRNAs than THP1 cells, which are competent for expression of TNF protein. We used both knock-down and over-expression strategies and demonstrated that the more abundant the ncRNAs, the less TNF was expressed. The effects were small suggesting that this mechanism may represent a fine tuning strategy for transcriptional regulation. Alternatively, the knock-down strategy and the over-expression strategy may not have led to as effective targeting as the normal endogenous pathway, although we did show that the over-expressed ncRNA localized to chromatin. We believe the phenomenon is biologically relevant because effects on protein production were significant. We then pursued a strategy to define the mechanism of the ncRNA effect. We hypothesized that LRRFIP1, a known RNA-binding protein, which we had demonstrated acted as a repressor of TNF, might interact directly with the ncRNAs (37, 71). Our data demonstrated that not only did LRRFIP1 interact with the ncRNAs in vitro but that the interaction in vivo was required for localization of LRRFIP1 to the chromatin. Localization was also dependent on EZH2 and SUZ12.
LRRFIP1 (also known as TRIP, GCF2, and FLAP) was originally identified as a GC-rich binding protein that repressed epidermal growth factor receptor expression and platelet-derived growth factor expression (76–78). It has also been described as a transcriptional repressor in other settings, a tumor suppressor, a platelet regulator, an early responder to foreign nucleic acids, and as a β-catenin cofactor (71, 79–87). It exists as two major isoforms, a long 160kD isoform transcribed starting from exon 2 that includes RNA- and DNA-binding motifs and a shorter 120kD isoform transcribed from exon 1 that includes part of the RNA-binding motif but not the DNA-binding motif. The diverse functional descriptions may be in part due to isoform-specific effects. There is a homolog designated as LRRFIP2 that shares sequence homology only at the 5′ end (85). LRRFIP1 has no other homologs and no other defined motifs other than potential phosphorylation sites and a nuclear localization domain (88).
In our study, we demonstrated that LRRFIP1 interacts with the ncRNAs in a sequence-independent fashion. The sense and antisense sequences both bound LRRFIP1 and off-target dsRNA also bound, supporting a model where the structure is more important than the sequence. Double-stranded RNA bound more effectively than single stranded RNA and longer species were also favored. These qualities are both found in the ncRNA upstream of TNF and we speculate that the other chromatin sites of LRRFIp1 binding share similar structural qualities. We noted some asymmetry in the effects of the ncRNAs with knock down of antisense ncRNA slightly more effective at altering LRRFIP1 binding. Over-expression of antisense RNA also seemed to have a more robust effect on chromatin marks of repression and TNF mRNA abundance. It is not known whether the functions of the sense and antisense are different and further studies will better define the structural requirement of the ncRNA for LRRFIP1 binding.
There are now numerous examples of ncRNAs regulating transcription and their functional complexity, pervasive nature, and structural characteristics are just beginning to be understood. Transcription start sites, enhancers, and CpG islands all produce short transcripts of uncertain function. The ncRNAs described here were found to be strongly associated with chromatin, a population of RNAs initially described in 1975 (89). Several models have been posited to explain the role of ncRNAs in transcriptional regulation. 1) The ncRNA can serve as a decoy that titrates DNA-binding proteins away from genomic target sequences (90, 91). 2) The ncRNA can serve as a structural scaffold to facilitate interactions of multiple proteins (92–94). 3) Guide RNAs operating in cis or trans localize regulatory proteins and provide locus specificity (50, 95, 96). Our data clearly support the binding of LRRFIP1 to the RNA and via the RNA to the chromatin, consistent with a model where the RNA provides locus specificity in cis. Our data are also consistent with a scaffolding function with EZH2, SUZ12 and LRRFIP1 assembling on an RNA tether to the region. These data suggest a model where repressed cells express ncRNAs in a region where polycomb proteins bind. Once bound, LRRFIP1 is recruited to the site where it participates in the repressive complex. It is anchored to the site by the ncRNA. This model represents a hybrid of scenarios two and three above.
This study identified a novel transcriptional regulatory pathway. We presume that this pathway is not unique to TNF, although our studies did not extend beyond the one gene. Instead, we pursued a detailed analysis of one gene as an archetype. TNF is a relevant subject because of its importance in human disease states. Dissecting the mechanisms underlying the regulatory functions of ncRNAs is just beginning. This study adds to that fundamental knowledge by examining a gene with highly dynamic expression and by identifying the role of LRRFIP1 in ncRNA function.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the support of the protein core facility.
Footnotes
This study was funded by 1RO1 AI 51323, R21 AI090914, and the Wallace Chair of Pediatrics.
References
- 1.Beutler B, Brown T. A CAT reporter construct allows ultrasensitive estimation of TNF synthesis and suggests that the TNF gene has been silenced in non-macrophage cell lines. J Clin Invest. 1991;87:1336–1344. doi: 10.1172/JCI115137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cerami A, Beutler B. Cachectin: the dark side of tumor necrosis factor. Cold Spring Harbor symposia on quantitative biology. 1986;51(Pt 1):625–629. doi: 10.1101/sqb.1986.051.01.074. [DOI] [PubMed] [Google Scholar]
- 3.Cerami A, Beutler B. The role of cachectin/TNF in endotoxic shock and cachexia. Immunol Today. 1988;9:28–31. doi: 10.1016/0167-5699(88)91353-9. [DOI] [PubMed] [Google Scholar]
- 4.Gardam MA, Keystone EC, Menzies R, Manners S, Skamene E, Long R, Vinh DC. Anti-tumour necrosis factor agents and tuberculosis risk: mechanisms of action and clinical management. The Lancet infectious diseases. 2003;3:148–155. doi: 10.1016/s1473-3099(03)00545-0. [DOI] [PubMed] [Google Scholar]
- 5.Jacobs M, Togbe D, Fremond C, Samarina A, Allie N, Botha T, Carlos D, Parida SK, Grivennikov S, Nedospasov S, Monteiro A, Le Bert M, Quesniaux V, Ryffel B. Tumor necrosis factor is critical to control tuberculosis infection. Microbes and infection / Institut Pasteur. 2007;9:623–628. doi: 10.1016/j.micinf.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 6.Roach DR, Bean AG, Demangel C, France MP, Briscoe H, Britton WJ. TNF regulates chemokine induction essential for cell recruitment, granuloma formation, and clearance of mycobacterial infection. J Immunol. 2002;168:4620–4627. doi: 10.4049/jimmunol.168.9.4620. [DOI] [PubMed] [Google Scholar]
- 7.Bean AG, Roach DR, Briscoe H, France MP, Korner H, Sedgwick JD, Britton WJ. Structural deficiencies in granuloma formation in TNF gene-targeted mice underlie the heightened susceptibility to aerosol Mycobacterium tuberculosis infection, which is not compensated for by lymphotoxin. J Immunol. 1999;162:3504–3511. [PubMed] [Google Scholar]
- 8.Marino MW, Dunn A, Grail D, Inglese M, Noguchi Y, Richards E, Jungbluth A, Wada H, Moore M, Williamson B, Basu S, Old LJ. Characterization of tumor necrosis factor-deficient mice. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:8093–8098. doi: 10.1073/pnas.94.15.8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Butler DM, Malfait AM, Mason LJ, Warden PJ, Kollias G, Maini RN, Feldmann M, Brennan FM. DBA/1 mice expressing the human TNF-alpha transgene develop a severe, erosive arthritis: characterization of the cytokine cascade and cellular composition. J Immunol. 1997;159:2867–2876. [PubMed] [Google Scholar]
- 10.Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-a production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 11.Dal Canto RA, Shaw MK, Nolan GP, Steinman L, Fathman CG. Local delivery of TNF by retrovirus-transduced T lymphocytes exacerbates experimental autoimmune encephalomyelitis. Clinical Immunology. 1999;90:10–14. doi: 10.1006/clim.1998.4653. [DOI] [PubMed] [Google Scholar]
- 12.Douni E, Akassoglou K, Alexopoulou L, Georgopoulos S, Haralambous S, Hill S, Kassiotis G, Kontoyiannis D, Pasparakis M, Plows D, Probert L, Kollias G. Transgenic and knockout analyses of the role of TNF in immune regulation and disease pathogenesis. J Inflamm. 1995;47:27–38. [PubMed] [Google Scholar]
- 13.Kontoyiannis D, Pasparakis M, Pizarro TT, Cominelli F, Kollias G. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU- rich elements: implications for joint and gut-associated immunopathologies. Immunity. 1999;10:387–398. doi: 10.1016/s1074-7613(00)80038-2. [DOI] [PubMed] [Google Scholar]
- 14.Willuweit A, Sass G, Schoneberg A, Eisel U, Tiegs G, Clauss M. Chronic inflammation and protection from acute hepatitis in transgenic mice expressing TNF in endothelial cells. J Immunol. 2001;167:3944–3952. doi: 10.4049/jimmunol.167.7.3944. [DOI] [PubMed] [Google Scholar]
- 15.Armstrong L, Godinho SI, Uppington KM, Whittington HA, Millar AB. Contribution of TNF-alpha converting enzyme and proteinase-3 to TNF-alpha processing in human alveolar macrophages. American journal of respiratory cell and molecular biology. 2006;34:219–225. doi: 10.1165/rcmb.2005-0087OC. [DOI] [PubMed] [Google Scholar]
- 16.Buxade M, Parra JL, Rousseau S, Shpiro N, Marquez R, Morrice N, Bain J, Espel E, Proud CG. The Mnks are novel components in the control of TNF alpha biosynthesis and phosphorylate and regulate hnRNP A1. Immunity. 2005;23:177–189. doi: 10.1016/j.immuni.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 17.Yang Y, Chang JF, Parnes JR, Fathman CG. T cell receptor (TCR) engagement leads to activation-induced splicing of tumor necrosis factor (TNF) nuclear pre-mRNA. J Exp Med. 1998;188:247–254. doi: 10.1084/jem.188.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osman F, Jarrous N, Ben-Asouli Y, Kaempfer R. A cis-acting element in the 3′-untranslated region of human TNF-alpha mRNA renders splicing dependent on the activation of protein kinase PKR. Genes Dev. 1999;13:3280–3293. doi: 10.1101/gad.13.24.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sullivan KE, Reddy AB, Dietzmann K, Suriano AR, Kocieda VP, Stewart M, Bhatia M. Epigenetic regulation of tumor necrosis factor alpha. Mol Cell Biol. 2007;27:5147–5160. doi: 10.1128/MCB.02429-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahlknecht U, Will J, Varin A, Hoelzer D, Herbein G. Histone deacetylase 3, a class I histone deacetylase, suppresses MAPK11-mediated activating transcription factor-2 activation and represses TNF gene expression. J Immunol. 2004;173:3979–3990. doi: 10.4049/jimmunol.173.6.3979. [DOI] [PubMed] [Google Scholar]
- 21.Sullivan KE, Reddy AB, Dietzmann K, Suriano AR, Kocieda VP, Stewart M, Bhatia M. Epigenetic regulation of tumor necrosis factor alpha. Mol Cell Biol. 2007;27:5147–5160. doi: 10.1128/MCB.02429-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wessels I, Rosenkranz E, Ventura Ferreira M, Neuss S, Zenke M, Rink L, Uciechowski P. Activation of IL-1beta and TNFalpha genes is mediated by the establishment of permissive chromatin structures during monopoiesis. Immunobiology. 2013;218:860–868. doi: 10.1016/j.imbio.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 23.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pham TH, Langmann S, Schwarzfischer L, El Chartouni C, Lichtinger M, Klug M, Krause SW, Rehli M. CCAAT enhancer-binding protein beta regulates constitutive gene expression during late stages of monocyte to macrophage differentiation. J Biol Chem. 2007;282:21924–21933. doi: 10.1074/jbc.M611618200. [DOI] [PubMed] [Google Scholar]
- 25.Pham TH, Benner C, Lichtinger M, Schwarzfischer L, Hu Y, Andreesen R, Chen W, Rehli M. Dynamic epigenetic enhancer signatures reveal key transcription factors associated with monocytic differentiation states. Blood. 2012;119:e161–171. doi: 10.1182/blood-2012-01-402453. [DOI] [PubMed] [Google Scholar]
- 26.Pham TH, Minderjahn J, Schmidl C, Hoffmeister H, Schmidhofer S, Chen W, Langst G, Benner C, Rehli M. Mechanisms of in vivo binding site selection of the hematopoietic master transcription factor PU.1. Nuc Acids Res. 2013;41:6391–6402. doi: 10.1093/nar/gkt355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barthel R, Tsytsykova AV, Barczak AK, Tsai EY, Dascher CC, Brenner MB, Goldfeld AE. Regulation of tumor necrosis factor alpha gene expression by mycobacteria involves the assembly of a unique enhanceosome dependent on the coactivator proteins CBP/p300. Mol Cell Biol. 2003;23:526–533. doi: 10.1128/MCB.23.2.526-533.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai EY, Falvo JV, Tsytsykova AV, Barczak AK, Reimold AM, Glimcher LH, Fenton MJ, Gordon DC, Dunn IF, Goldfeld AE. A lipopolysaccharide-specific enhancer complex involving Ets, Elk-1, Sp1, and CREB binding protein and p300 is recruited to the tumor necrosis factor alpha promoter in vivo. Mol Cell Biol. 2000;20:6084–6094. doi: 10.1128/mcb.20.16.6084-6094.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuprash DV, I, Udalova A, Turetskaya RL, Kwiatkowski D, Rice NR, Nedospasov SA. Similarities and differences between human and murine TNF promoters in their response to lipopolysaccharide. J Immunol. 1999;162:4045–4052. [PubMed] [Google Scholar]
- 30.Lai WS, Carballo E, Strum JR, Kennington EA, Phillips RS, Blackshear PJ. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol Cell Biol. 1999;19:4311–4323. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 32.Lopez de Silanes I, Galban S, Martindale JL, Yang X, Mazan-Mamczarz K, Indig FE, Falco G, Zhan M, Gorospe M. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol Cell Biol. 2005;25:9520–9531. doi: 10.1128/MCB.25.21.9520-9531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ronkina N, Menon MB, Schwermann J, Tiedje C, Hitti E, Kotlyarov A, Gaestel M. MAPKAP kinases MK2 and MK3 in inflammation: complex regulation of TNF biosynthesis via expression and phosphorylation of tristetraprolin. Biochemical pharmacology. 2010;80:1915–1920. doi: 10.1016/j.bcp.2010.06.021. [DOI] [PubMed] [Google Scholar]
- 34.Loflin P, Chen CY, Shyu AB. Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes & Development. 1999;13:1884–1897. doi: 10.1101/gad.13.14.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Min H, Turck CW, Nikolic JM, Black DL. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes & Development. 1997;11:1023–1036. doi: 10.1101/gad.11.8.1023. [DOI] [PubMed] [Google Scholar]
- 36.Zhang W, Wagner BJ, Ehrenman K, Schaefer AW, DeMaria CT, Crater D, DeHaven K, Long L, Brewer G. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol Cell Biol. 1993;13:7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suriano AR, Sanford AN, Kim N, Oh M, Kennedy S, Henderson MJ, Dietzmann K, Sullivan KE. GCF2/LRRFIP1 represses tumor necrosis factor alpha expression. Mol Cell Biol. 2005;25:9073–9081. doi: 10.1128/MCB.25.20.9073-9081.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sullivan KE, Suriano A, Dietzmann K, Lin J, Goldman D, Petri MA. The TNFalpha locus is altered in monocytes from patients with systemic lupus erythematosus. Clin Immunol. 2007;123:74–81. doi: 10.1016/j.clim.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes & Development. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ponting CP. The functional repertoires of metazoan genomes. Nature reviews Genetics. 2008;9:689–698. doi: 10.1038/nrg2413. [DOI] [PubMed] [Google Scholar]
- 41.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H, Chalk AM, Chiu KP, Choudhary V, Christoffels A, Clutterbuck DR, Crowe ML, Dalla E, Dalrymple BP, de Bono B, Della Gatta G, di Bernardo D, Down T, Engstrom P, Fagiolini M, Faulkner G, Fletcher CF, Fukushima T, Furuno M, Futaki S, Gariboldi M, Georgii-Hemming P, Gingeras TR, Gojobori T, Green RE, Gustincich S, Harbers M, Hayashi Y, Hensch TK, Hirokawa N, Hill D, Huminiecki L, Iacono M, Ikeo K, Iwama A, Ishikawa T, Jakt M, Kanapin A, Katoh M, Kawasawa Y, Kelso J, Kitamura H, Kitano H, Kollias G, Krishnan SP, Kruger A, Kummerfeld SK, Kurochkin IV, Lareau LF, Lazarevic D, Lipovich L, Liu J, Liuni S, McWilliam S, Madan Babu M, Madera M, Marchionni L, Matsuda H, Matsuzawa S, Miki H, Mignone F, Miyake S, Morris K, Mottagui-Tabar S, Mulder N, Nakano N, Nakauchi H, Ng P, Nilsson R, Nishiguchi S, Nishikawa S, Nori F, Ohara O, Okazaki Y, Orlando V, Pang KC, Pavan WJ, Pavesi G, Pesole G, Petrovsky N, Piazza S, Reed J, Reid JF, Ring BZ, Ringwald M, Rost B, Ruan Y, Salzberg SL, Sandelin A, Schneider C, Schonbach C, Sekiguchi K, Semple CA, Seno S, Sessa L, Sheng Y, Shibata Y, Shimada H, Shimada K, Silva D, Sinclair B, Sperling S, Stupka E, Sugiura K, Sultana R, Takenaka Y, Taki K, Tammoja K, Tan SL, Tang S, Taylor MS, Tegner J, Teichmann SA, Ueda HR, van Nimwegen E, Verardo R, Wei CL, Yagi K, Yamanishi H, Zabarovsky E, Zhu S, Zimmer A, Hide W, Bult C, Grimmond SM, Teasdale RD, Liu ET, Brusic V, Quackenbush J, Wahlestedt C, Mattick JS, Hume DA, Kai C, Sasaki D, Tomaru Y, Fukuda S, Kanamori-Katayama M, Suzuki M, Aoki J, Arakawa T, Iida J, Imamura K, Itoh M, Kato T, Kawaji H, Kawagashira N, Kawashima T, Kojima M, Kondo S, Konno H, Nakano K, Ninomiya N, Nishio T, Okada M, Plessy C, Shibata K, Shiraki T, Suzuki S, Tagami M, Waki K, Watahiki A, Okamura-Oho Y, Suzuki H, Kawai J, Hayashizaki Y. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 43.Xu Z, Wei W, Gagneur J, Clauder-Munster S, Smolik M, Huber W, Steinmetz LM. Antisense expression increases gene expression variability and locus interdependency. Mol Syst Biol. 2011;7:468. doi: 10.1038/msb.2011.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ogawa Y, Sun BK, Lee JT. Intersection of the RNA interference and X-inactivation pathways. Science. 2008;320:1336–1341. doi: 10.1126/science.1157676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Splinter E, de Wit E, Nora EP, Klous P, van de Werken HJ, Zhu Y, Kaaij LJ, van Ijcken W, Gribnau J, Heard E, de Laat W. The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes & Development. 2011;25:1371–1383. doi: 10.1101/gad.633311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, Byron M, Monks B, Henry-Bezy M, Lawrence JB, O’Neill LA, Moore MJ, Caffrey DR, Fitzgerald KA. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341:789–792. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peng X, Gralinski L, Ferris MT, Frieman MB, Thomas MJ, Proll S, Korth MJ, Tisoncik JR, Heise M, Luo S, Schroth GP, Tumpey TM, Li C, Kawaoka Y, Baric RS, Katze MG. Integrative deep sequencing of the mouse lung transcriptome reveals differential expression of diverse classes of small RNAs in response to respiratory virus infection. MBio. 2011:2. doi: 10.1128/mBio.00198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, Johnston D, Kim GE, Spitale RC, Flynn RA, Zheng GX, Aiyer S, Raj A, Rinn JL, Chang HY, Khavari PA. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Molecular Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Redrup L, Branco MR, Perdeaux ER, Krueger C, Lewis A, Santos F, Nagano T, Cobb BS, Fraser P, Reik W. The long noncoding RNA Kcnq1ot1 organises a lineage-specific nuclear domain for epigenetic gene silencing. Development (Cambridge, England) 2009;136:525–530. doi: 10.1242/dev.031328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kanduri C. Functional insights into long antisense noncoding RNA Kcnq1ot1 mediated bidirectional silencing. RNA Biol. 2008;5:208–211. doi: 10.4161/rna.7113. [DOI] [PubMed] [Google Scholar]
- 54.Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 55.Suzuki K, Juelich T, Lim H, Ishida T, Watanebe T, Cooper DA, Rao S, Kelleher AD. Closed chromatin architecture is induced by an RNA duplex targeting the HIV-1 promoter region. J Biol Chem. 2008;283:23353–23363. doi: 10.1074/jbc.M709651200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ting AH, Schuebel KE, Herman JG, Baylin SB. Short double-stranded RNA induces transcriptional gene silencing in human cancer cells in the absence of DNA methylation. Nat Genet. 2005;37:906–910. doi: 10.1038/ng1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weinberg MS, Villeneuve LM, Ehsani A, Amarzguioui M, Aagaard L, Chen ZX, Riggs AD, Rossi JJ, Morris KV. The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells. RNA (New York, NY. 2006;12:256–262. doi: 10.1261/rna.2235106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Janowski BA, Huffman KE, Schwartz JC, Ram R, Nordsell R, Shames DS, Minna JD, Corey DR. Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nature structural & molecular biology. 2006;13:787–792. doi: 10.1038/nsmb1140. [DOI] [PubMed] [Google Scholar]
- 59.Han J, Kim D, Morris KV. Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells. Proc Natl Acad Sci U S A. 2007;104:12422–12427. doi: 10.1073/pnas.0701635104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nature structural & molecular biology. 2006;13:793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- 61.Lee JY, Sullivan KE. Gamma interferon and lipopolysaccharide interact at the level of transcription to induce tumor necrosis factor alpha expression. Infection and immunity. 2001;69:2847–2852. doi: 10.1128/IAI.69.5.2847-2852.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mapendano CK, Lykke-Andersen S, Kjems J, Bertrand E, Jensen TH. Crosstalk between mRNA 3′ end processing and transcription initiation. Molecular Cell. 2010;40:410–422. doi: 10.1016/j.molcel.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 64.Seidl CI, Stricker SH, Barlow DP. The imprinted Air ncRNA is an atypical RNAPII transcript that evades splicing and escapes nuclear export. The EMBO journal. 2006;25:3565–3575. doi: 10.1038/sj.emboj.7601245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bochnig P, Reuter R, Bringmann P, Luhrmann R. A monoclonal antibody against 2,2,7-trimethylguanosine that reacts with intact, class U, small nuclear ribonucleoproteins as well as with 7-methylguanosine-capped RNAs. European journal of biochemistry / FEBS. 1987;168:461–467. doi: 10.1111/j.1432-1033.1987.tb13439.x. [DOI] [PubMed] [Google Scholar]
- 66.Patrone G, Puppo F, Cusano R, Scaranari M, Ceccherini I, Puliti A, Ravazzolo R. Nuclear run-on assay using biotin labeling, magnetic bead capture and analysis by fluorescence-based RT-PCR. BioTechniques. 2000;29:1012–1014. 1016–1017. doi: 10.2144/00295st02. [DOI] [PubMed] [Google Scholar]
- 67.Bendak K, Loughlin FE, Cheung V, O’Connell MR, Crossley M, Mackay JP. A rapid method for assessing the RNA-binding potential of a protein. Nuc Acids Res. 2012;40:e105. doi: 10.1093/nar/gks285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee JY, Kim NA, Sanford A, Sullivan KE. Histone acetylation and chromatin conformation are regulated separately at the TNF alpha promoter in monocytes and macrophages. Journal of Leukocyte Biology. 2003;73:862–871. doi: 10.1189/jlb.1202618. [DOI] [PubMed] [Google Scholar]
- 69.Garrett S, Dietzmann-Maurer K, Song L, Sullivan KE. Polarization of primary human monocytes by IFN-gamma induces chromatin changes and recruits RNA Pol II to the TNF-alpha promoter. J Immunol. 2008;180:5257–5266. doi: 10.4049/jimmunol.180.8.5257. [DOI] [PubMed] [Google Scholar]
- 70.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wilson SA, Brown EC, Kingsman AJ, Kingsman SM. TRIP: a novel double stranded RNA binding protein which interacts with the leucine rich repeat of flightless I. Nucleic Acids Res. 1998;26:3460–3467. doi: 10.1093/nar/26.15.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev. 2004;14:155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 73.Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 74.Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, Bollen M, Esteller M, Di Croce L, de Launoit Y, Fuks F. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 75.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nuc Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Santiago FS, Khachigian LM. Ets-1 stimulates platelet-derived growth factor A-chain gene transcription and vascular smooth muscle cell growth via cooperative interactions with Sp1. Circ Res. 2004;95:479–487. doi: 10.1161/01.RES.0000141135.36279.67. [DOI] [PubMed] [Google Scholar]
- 77.Khachigian LM, Santiago FS, Rafty LA, Chan OL, Delbridge GJ, Bobik A, Collins T, Johnson AC. GC factor 2 represses platelet-derived growth factor A-chain gene transcription and is itself induced by arterial injury. Circulation Research. 1999;84:1258–1267. doi: 10.1161/01.res.84.11.1258. [DOI] [PubMed] [Google Scholar]
- 78.Reed AL, Yamazaki H, Kaufman JD, Rubinstein Y, Murphy B, Johnson AC. Molecular cloning and characterization of a transcription regulator with homology to GC-binding factor. J Biol Chem. 1998;273:21594–21602. doi: 10.1074/jbc.273.34.21594. [DOI] [PubMed] [Google Scholar]
- 79.Soler G, Nusbaum S, Varet B, Macintyre EA, Vekemans M, Romana SP, Radford-Weiss I. LRRFIP1, a new FGFR1 partner gene associated with 8p11 myeloproliferative syndrome. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2009;23:1359–1361. doi: 10.1038/leu.2009.79. [DOI] [PubMed] [Google Scholar]
- 80.Lee YH, Stallcup MR. Interplay of Fli-I and FLAP1 for regulation of beta-catenin dependent transcription. Nuc Acids Res. 2006;34:5052–5059. doi: 10.1093/nar/gkl652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ohtsuka H, Oikawa M, Ariake K, Rikiyama T, Motoi F, Katayose Y, Unno M, Johnson AC. GC-binding factor 2 interacts with dishevelled and regulates Wnt signaling pathways in human carcinoma cell lines. International journal of cancer Journal international du cancer. 2011;129:1599–1610. doi: 10.1002/ijc.25837. [DOI] [PubMed] [Google Scholar]
- 82.Goodall AH, Burns P, Salles I, Macaulay IC, Jones CI, Ardissino D, de Bono B, Bray SL, Deckmyn H, Dudbridge F, Fitzgerald DJ, Garner SF, Gusnanto A, Koch K, Langford C, O’Connor MN, Rice CM, Stemple D, Stephens J, Trip MD, Zwaginga JJ, Samani NJ, Watkins NA, Maguire PB, Ouwehand WH. Transcription profiling in human platelets reveals LRRFIP1 as a novel protein regulating platelet function. Blood. 2010;116:4646–4656. doi: 10.1182/blood-2010-04-280925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang P, An H, Liu X, Wen M, Zheng Y, Rui Y, Cao X. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a beta-catenin-dependent pathway. Nature Immunology. 2010;11:487–494. doi: 10.1038/ni.1876. [DOI] [PubMed] [Google Scholar]
- 84.Li Y, Li W, Yang Y, Lu Y, He C, Hu G, Liu H, Chen J, He J, Yu H. MicroRNA-21 targets LRRFIP1 and contributes to VM-26 resistance in glioblastoma multiforme. Brain research. 2009;1286:13–18. doi: 10.1016/j.brainres.2009.06.053. [DOI] [PubMed] [Google Scholar]
- 85.Dai P, Jeong SY, Yu Y, Leng T, Wu W, Xie L, Chen X. Modulation of TLR signaling by multiple MyD88-interacting partners including leucine-rich repeat Fli-I-interacting proteins. J Immunol. 2009;182:3450–3460. doi: 10.4049/jimmunol.0802260. [DOI] [PubMed] [Google Scholar]
- 86.Rikiyama T, Curtis J, Oikawa M, Zimonjic DB, Popescu N, Murphy BA, Wilson MA, Johnson AC. GCF2: expression and molecular analysis of repression. Biochim Biophys Acta. 2003;1629:15–25. doi: 10.1016/s0167-4781(03)00156-8. [DOI] [PubMed] [Google Scholar]
- 87.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 88.Arakawa R, Bagashev A, Song L, Maurer K, Sullivan KE. Characterization of LRRFIP1. Biochem Cell Biol. 2010;88:899–906. doi: 10.1139/O10-014. [DOI] [PubMed] [Google Scholar]
- 89.Paul J, Duerksen JD. Chromatin-associated RNA content of heterochromatin and euchromatin. Molecular and cellular biochemistry. 1975;9:9–16. doi: 10.1007/BF01731728. [DOI] [PubMed] [Google Scholar]
- 90.Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, Kong B, Langerod A, Borresen-Dale AL, Kim SK, van de Vijver M, Sukumar S, Whitfield ML, Kellis M, Xiong Y, Wong DJ, Chang HY. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nature Genetics. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Molecular Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 94.Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, Attardi LD, Regev A, Lander ES, Jacks T, Rinn JL. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Simon MD, Wang CI, Kharchenko PV, West JA, Chapman BA, Alekseyenko AA, Borowsky ML, Kuroda MI, Kingston RE. The genomic binding sites of a noncoding RNA. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:20497–20502. doi: 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.