Abstract
Digital PCR is a new technology that enables detection and quantification of cancer DNA molecules from peripheral blood. Using this technique, we identified mutant PIK3CA DNA in circulating plasma tumor DNA (ptDNA) from a patient with concurrent early stage breast cancer and non-small cell lung cancer. The patient underwent successful resection of both her breast and lung cancers, and using standard Sanger sequencing the breast cancer was shown to harbor the identical PIK3CA mutation identified in peripheral blood. This case report highlights potential applications and concerns that can arise with the use of ptDNA in clinical oncology practice.
Keywords: plasma tumor DNA, breast cancer, lung cancer, PIK3CA, digital PCR
Introduction
PIK3CA encodes the p110α component of phosphatidylinositol 3-kinase (PI3K) and has been shown to harbor somatic mutations in ~ 36% of breast cancers and up to 3% of non-small cell lung cancers. PIK3CA is one of the most frequently mutated oncogenes in human cancers (1) and currently there is intense interest in developing PI3 kinase inhibitors that could serve as targeted therapies for cancers that have these mutations. Three hot-spot mutations have been discovered within two exons (Exon 9: E542K and E545K and Exon 20: H1047R) and account for 80–90% of all PIK3CA mutations in human cancers. Lung cancers contain a broader range of PIK3CA mutations with E545K Exon 9 mutations representing the majority at approximately 30% frequency (2). Although the prognostic significance of PIK3CA mutations remains controversial, hotspot mutations have been shown to confer oncogenic features and resistance to some chemotherapies (3, 4). We and others have demonstrated that in metastatic breast cancer patients tumors that harbor PIK3CA mutations will release their DNA such that the mutations can also be isolated from blood by analyzing circulating cell-free plasma derived tumor DNA (ptDNA) using various methods (5–7). Although there is likely to be a lower amount of ptDNA in early stage cancers compared to advanced disease due to less tumor burden, patient tumor PIK3CA status may be determined by a simple blood draw even in this setting depending on the sensitivity of the assay used.
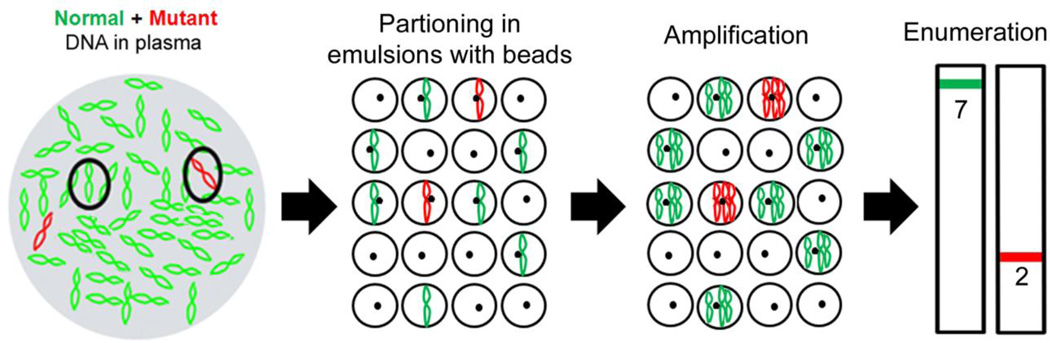
There are a number of techniques for detecting ptDNA which employ mechanisms for separating out the smaller variant or mutant fraction that is ptDNA from the larger circulating cell-free DNA population derived from normal cells. BEAMing is a digital polymerase chain reaction (PCR) platform that stands for its primary components: Beads, Emulsion, Amplification and Magnetics and uses emulsion PCR technology to separately amplify individual molecules and then quantify specific variants using flow cytometry (8, 9) (Figure 1). BEAMing has been shown to detect and quantify somatic mutations found in ptDNA with a high level of sensitivity and specificity (7, 9, 10). In this case report, we describe one of our patients from an ongoing prospective study who presented with early-stage synchronous primary breast and non-small cell lung cancers. This afforded us the unique opportunity to examine which tumor harbored the PIK3CA mutation identified in the patient’s ptDNA. Importantly, as ptDNA analysis becomes more widespread as a means of “liquid biopsy” to detect cancer mutations, this case report demonstrates the need for vigilance when ascribing a mutation found in blood to a patient’s known tumor. Thus, the possibility of occult malignancies should be considered when a mutation identified in ptDNA does not necessarily align with the diagnosed tumor type.
Figure 1. Schematic of BEAMing.
DNA is diluted and partitioned into emulsions with magnetic beads such that single or no DNA template molecules are present within each emulsion. Emulsions also contain primers, thermostabile DNA polymerase, dNTPs and other PCR reagents. Emulsions are then amplified massively in parallel and then the emulsions are burst open followed by hybridization to mutant and wild type specific fluorescent probes. Amplified DNA on beads are then quantified using flow cytometry to determine the number of mutant and wild type DNA molecules present.
Materials and Methods
Case Report
A 67 year-old African American female non-smoker was incidentally noted by imaging to have a 2 cm right sided breast mass in the axillary tail and a 1 cm mass in the upper lobe of her right lung. The patient underwent a biopsy of the axillary tail mass and was found to have a poorly differentiated adenocarcinoma consistent with a primary breast cancer. A review of the imaging studies suggested that, while the lung lesion could represent metastatic breast cancer, it was more consistent with the diagnosis of a synchronous primary lung cancer. The patient underwent a right lumpectomy with sentinel lymph node biopsy and a right upper lobectomy with thoracic lymphadenectomy for curative intent.
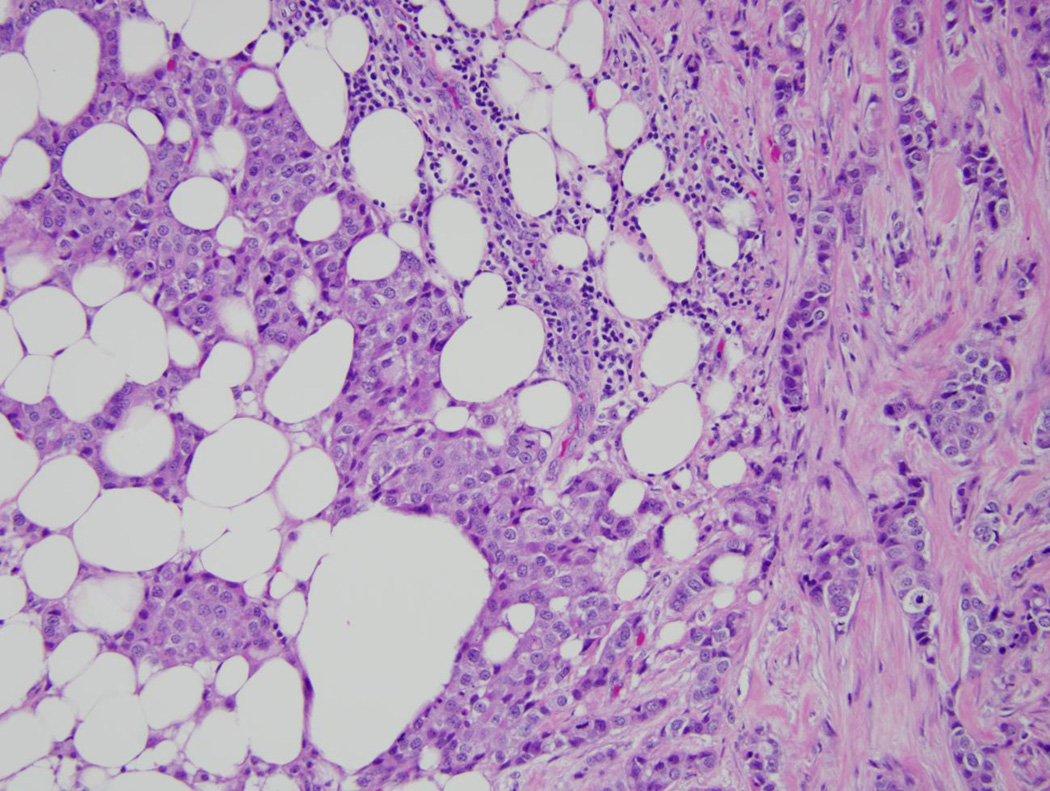
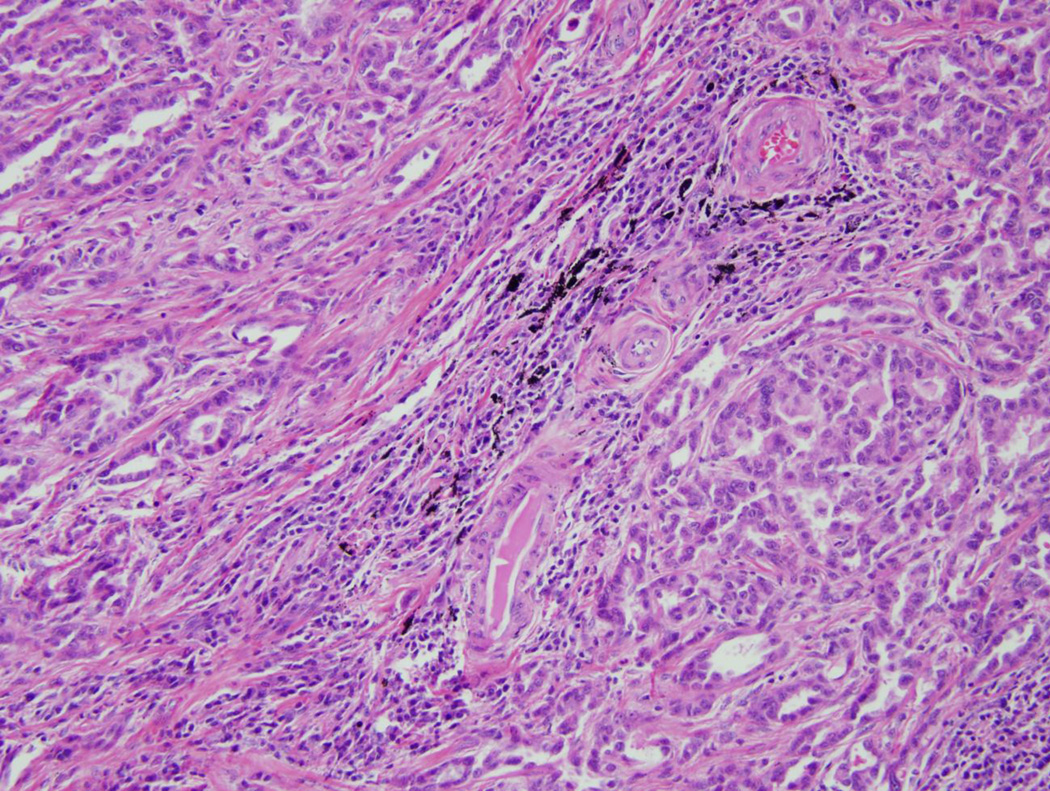
Pathologic review of the breast tumor demonstrated a 1.5cm, grade II, estrogen receptor/progesterone receptor positive, HER2 negative infiltrating mammary carcinoma (Figure 2A) with ductal and lobular features. Sentinel lymph node was negative. Her lobectomy specimen contained a 1.3 cm adenocarcinoma of mixed histology (Figure 2B) and was diagnosed as a non-small cell lung cancer which invaded the visceral pleura and had two positive lymph nodes. Thus she was concurrently staged with Stage IA breast and Stage IIA non-small cell lung cancer. The lung cancer was sent for mutational analysis and was found to be KRAS and EGFR wild type, and no ALK gene rearrangements were detected by fluorescence in situ hybridization (FISH).
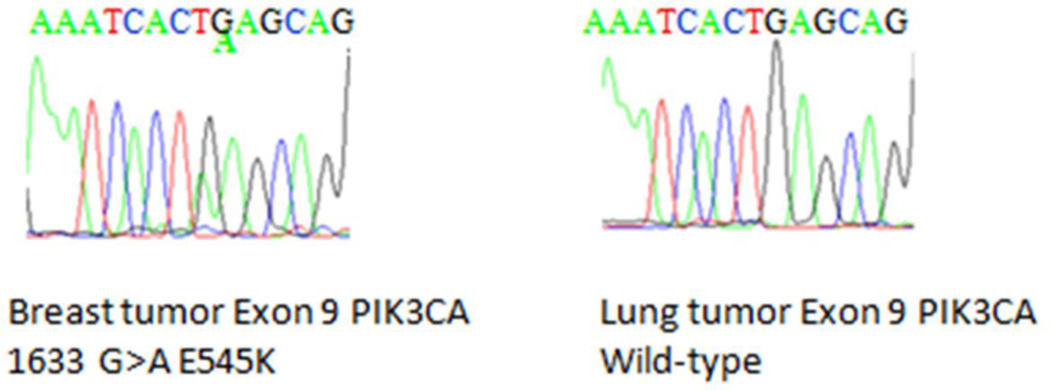
Figure 2. Pathology and sequencing analysis of tumor specimens.
FFPE samples were prepared for routine hematoxylin and eosin staining and photographed under 20× magnification. In addition, genomic DNA (gDNA) was extracted from serial sections obtained from FFPE tumor blocks and then used for Sanger sequencing. Shown are representative sections from the patient’s A: breast cancer, and B: lung cancer. C: PIK3CA Exon 9 Sequencing results are shown for breast cancer gDNA harboring the mutation (left panel) and lung cancer gDNA that is wild type for PIK3CA (right panel).
The patient received four cycles of adjuvant pemetrexed/cisplatin for her lung cancer. She then received radiation in order to complete local therapy for her breast cancer and began adjuvant hormonal therapy with an aromatase inhibitor with the expectation of undergoing five years of treatment. Follow-up for greater than two years has not revealed any recurrent disease.
Methods
The patient was consented to enroll in a prospective Johns Hopkins IRB-approved repository study. Ten milliliters (10ml) of blood was collected at the time of study entry preoperatively and subsequently approximately two weeks after surgery. The patient’s blood samples were prepared within two hours of collection and the plasma component was isolated. Genomic DNA was extracted and purified from plasma via the QIAamp Circulating Nucleic Acid kit (Qiagen). This purified circulating cell-free DNA containing ptDNA was then subjected to BEAMing (10). The plasma sample was analyzed for three PIK3CA “hotspot” mutations with three mutation specific probes: PIK3CA Exon 9 1624 G>A E542K, PIK3CA Exon 9 1633 G>A E545K and PIK3CA Exon 20 3140 A>G H1047R. The result of the BEAMing assay is reported as the percentage or fraction of mutant DNA alleles to wild-type DNA alleles.
Archival tumor specimens were collected and underwent pathology evaluation of the hematoxylin and eosin-stained slides to identify areas of tumor versus normal tissue. DNA was extracted from formalin-fixed paraffin embedded (FFPE) unstained slides (10 µm) using the Zymo pen and Pinpoint solution (Zymo Research) per the manufacturer’s protocol. Genomic DNA was isolated and purified via the QIAamp DNA FFPE tissue kit (Qiagen) according to the manufacturer’s protocol. Primers for PIK3CA Exon 9 and Exon 20 were designed and used to amplify the two exons for each of the lung and breast tumor samples. PIK3CA Exon 9 and Exon 20 were also amplified via PCR for the matched normal tissue controls for the breast and lung cancer specimens. The amplified segments (tumor and normal from each breast and lung specimen) were sent for Sanger sequencing using nested sequencing primers (see Supplemental Table 1 for all primer sequences). Investigators performing the FFPE sequencing assays were blinded to the BEAMing results.
Results
We sought to determine if this patient had detectable mutant PIK3CA in her pre-operative blood sample, and if so whether this mutant PIK3CA was still detectable after definitive surgery. BEAMing of the pre-operative plasma sample demonstrated a PIK3CA Exon 9 E545K mutation at 0.0759% of the total cell-free circulating DNA. The post-operative blood sample taken at 14 days post-surgery was negative for the PIK3CA E545K mutation by BEAMing. To determine whether the PIK3CA mutation was derived from the breast cancer, lung cancer or both, we next performed standard sequencing using tissues derived from these FFPE samples. Sanger sequencing of PIK3CA Exon 9 and Exon 20 was performed on genomic DNA extracted from normal and cancerous lung and breast tissues. This identified a PIK3CA Exon 9 E545K mutation in the breast tumor specimen but not in the normal breast tissue confirming the somatic nature of the mutation (Figure 2C). In contrast, both tumor and normal lung tissue demonstrated only wild type PIK3CA sequence.
Discussion
With the advent of digital PCR technologies such as BEAMing, the ability to detect and quantify cancer DNA molecules using peripheral blood has become a reality. Our own studies and those of others have demonstrated that ptDNA can be reliably detected in the blood of metastatic cancer patients using a number of technologies (5, 6, 11–13). Interestingly, a recent study also suggests that ptDNA can be detected in early stage breast cancer patients using whole genome amplification and SNP array analysis of blood, though the nature of this platform precludes its use as a quantitative tool (14). In contrast, BEAMing only detected ptDNA in a limited number of early stage colon cancer patients (10); however, BEAMing has not been addressed in a rigorous study for early stage breast cancers. We are currently conducting a study to address this question and the current report demonstrates that we can indeed detect ptDNA in the pre-operative blood from this patient as evidenced by the positive result for the PIK3CA E545K mutation by BEAMing.
The PIK3CA ptDNA result in this case presented an unusual situation since the patient had two synchronous primary cancers, breast and lung, both of which have been reported to contain PIK3CA mutations. While sequencing of these tumors demonstrated that the breast cancer harbored the PIK3CA mutation responsible for the same mutation found in ptDNA, this case highlights a potential pitfall in finding hotspot mutations in the blood of cancer patients unless verified with the primary tumor DNA. One could picture clinical situations where mutations found in the blood are not present in the primary tumor and this could reflect genetic heterogeneity of the primary tumor, or in this case other synchronous cancers that are either occult or not yet clinically evident. Thus, although the sensitivity of BEAMing for ptDNA is a technologic achievement, the specificity of these mutations for identifying the actual cancer in question will need to be verified in future studies. These results also emphasize many possible uses for ptDNA particularly in the setting of multiple primary tumors. Although EGFR and KRAS mutational analyses and the EML4-ALK rearrangement were negative for the patient’s lung cancer, this lung cancer likely harbors other tumor-specific mutations which could be identified using peripheral blood. Thus, the use of multiple markers that are specific for a given tumor type may afford the ability to follow response to therapies independently for each tumor and increase the sensitivity in the adjuvant setting. Given our understanding of the evolution of tumor mutational status, having several unique alterations would also ensure that we can follow tumor burden over time as genetic instability could lead to loss of one or more of these personalized markers. The use of peripheral blood for this analysis offers added appeal for a noninvasive, easy and quantitative approach for measuring tumor burden based upon molecular genotypes.
This case report demonstrates that BEAMing can detect ptDNA in the pre-operative setting in an early stage 1.5 cm node negative breast cancer patient. In addition we demonstrate that this ptDNA was undetectable post-surgery. Although the relevance of non-detectable ptDNA post-surgery is unclear, one could hypothesize that if ptDNA reflects tumor burden it could also correlate with residual micrometastatic disease and therefore have prognostic implications. Although the sensitivity of BEAMing is currently reported as 1:5,000 to 1:10,000 mutant per wild type molecules, clinically the sensitivity is currently dictated by the amount of circulating DNA that can be assayed. Of note, newer technologies including droplet digital PCR, a next generation digital PCR technology that is capable of increased numbers of emulsions and partitioning of DNA molecules, may afford higher sensitivities for detecting ptDNA. Further definitive prospective trials are needed to address the clinical relevance of the detection of ptDNA in early stage patients and the ability to potentially utilize ptDNA detection post-surgery as a prognostic and predictive marker.
Supplementary Material
Acknowledgments
This work was supported by: The Avon Foundation, the Komen Foundation, the Pearl M. Stetler Fellowship, NIH CA088843, CA009071, CA121937 and the Breast Cancer Research Foundation.
B.H.P. is a paid consultant for GlaxoSmithKline and Novartis. B.H.P. is a paid member of the scientific advisory boards of Horizon Discovery, LTD and Loxo Oncology. Under separate licensing agreements between Horizon Discovery, LTD and The Johns Hopkins University, B.H.P. is entitled to a share of royalties received by the University on sales of products. The terms of this arrangement are being managed by the Johns Hopkins University in accordance with its conflict of interest policies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure of Potential Conflicts of Interest
All other authors declare no potential conflicts.
References
- 1.Karakas B, Bachman KE, Park BH. Mutation of the PIK3CA oncogene in human cancers. Br J Cancer. 2006;94:455–459. doi: 10.1038/sj.bjc.6602970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Catalogue of Somatic Mutations in Cancer (COSMIC) Cambridge, UK: Welcome Trust Genome Campus; 2011. Distribution of Somatic Mutations in PIK3CA. [Google Scholar]
- 3.Gustin JP, Karakas B, Weiss MB, Abukhdeir AM, Lauring J, Garay JP, Cosgrove D, Tamaki A, Konishi H, Konishi Y, Mohseni M, Wang G, Rosen DM, Denmeade SR, Higgins MJ, Vitolo MI, Bachman KE, Park BH. Knockin of mutant PIK3CA activates multiple oncogenic pathways. Proc Natl Acad Sci U S A. 2009;106:2835–2840. doi: 10.1073/pnas.0813351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Isakoff SJ, Engelman JA, Irie HY, Luo J, Brachmann SM, Pearline RV, Cantley LC, Brugge JS. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res. 2005;65:10992–11000. doi: 10.1158/0008-5472.CAN-05-2612. [DOI] [PubMed] [Google Scholar]
- 5.Board RE, Wardley AM, Dixon JM, Armstrong AC, Howell S, Renshaw L, Donald E, Greystoke A, Ranson M, Hughes A, Dive C. Detection of PIK3CA mutations in circulating free DNA in patients with breast cancer. Breast Cancer Res Treat. 2010;120:461–467. doi: 10.1007/s10549-010-0747-9. [DOI] [PubMed] [Google Scholar]
- 6.Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, Dunning MJ, Gale D, Forshew T, Mahler-Araujo B, Rajan S, Humphray S, Becq J, Halsall D, Wallis M, Bentley D, Caldas C, Rosenfeld N. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368:1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 7.Higgins MJ, Jelovac D, Barnathan E, Blair B, Slater S, Powers P, Zorzi J, Jeter SC, Oliver GR, Fetting J, Emens L, Riley C, Stearns V, Diehl F, Angenendt P, Huang P, Cope L, Argani P, Murphy KM, Bachman KE, Greshock J, Wolff AC, Park BH. Detection of tumor PIK3CA status in metastatic breast cancer using peripheral blood. Clin Cancer Res. 2012;18:3462–3469. doi: 10.1158/1078-0432.CCR-11-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dressman D, Yan H, Traverso G, Kinzler KW, Vogelstein B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc Natl Acad Sci U S A. 2003;100:8817–8822. doi: 10.1073/pnas.1133470100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, Kinzler KW, Vogelstein B, Diaz LA., Jr Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diehl F, Li M, Dressman D, He Y, Shen D, Szabo S, Diaz LA, Jr, Goodman SN, David KA, Juhl H, Kinzler KW, Vogelstein B. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci U S A. 2005;102:16368–16373. doi: 10.1073/pnas.0507904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higgins MJ, Jelovac D, Barnathan E, Blair B, Slater S, Powers P, Zorzi J, Jeter SC, Oliver GR, Fetting J, Emens L, Riley C, Stearns V, Diehl F, Angenendt P, Huang P, Cope L, Argani P, Murphy KM, Bachman KE, Greshock J, Wolff AC, Park BH. Detection of Tumor PIK3CA Status in Metastatic Breast Cancer Using Peripheral Blood. Clin Cancer Res. 2012 doi: 10.1158/1078-0432.CCR-11-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leary RJ, Sausen M, Kinde I, Papadopoulos N, Carpten JD, Craig D, O'Shaughnessy J, Kinzler KW, Parmigiani G, Vogelstein B, Diaz LA, Jr, Velculescu VE. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci Transl Med. 2012;4:162ra154. doi: 10.1126/scitranslmed.3004742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, Valtorta E, Schiavo R, Buscarino M, Siravegna G, Bencardino K, Cercek A, Chen CT, Veronese S, Zanon C, Sartore-Bianchi A, Gambacorta M, Gallicchio M, Vakiani E, Boscaro V, Medico E, Weiser M, Siena S, Di Nicolantonio F, Solit D, Bardelli A. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–536. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaw JA, Page K, Blighe K, Hava N, Guttery D, Ward B, Brown J, Ruangpratheep C, Stebbing J, Payne R, Palmieri C, Cleator S, Walker RA, Coombes RC. Genomic analysis of circulating cell-free DNA infers breast cancer dormancy. Genome Res. 2012;22:220–231. doi: 10.1101/gr.123497.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.