Abstract
Vibrio fischeri isolated from Euprymna scolopes (Cephalopoda: Sepiolidae) was used to create twenty-four lines that were serially passaged through the non-native host E. tasmanica for 500 generations. These derived lines were characterized for biofilm formation, swarming motility, carbon source utilization, and in vitro bioluminescence. Phenotypic assays were compared between “ES” (E. scolopes) and “ET” (E. tasmanica) V. fischeri wild isolates to determine if convergent evolution was apparent between E. tasmanica evolved lines and ET V. fischeri. Ecological diversification was observed in utilization of most carbon sources examined. Convergent evolution was evident in motility, biofilm formation, and select carbon sources displaying hyperpolymorphic usage in V. fischeri. Convergence in bioluminescence (a 2.5-fold increase in brightness) was collectively evident in the derived lines relative to the ancestor. However, dramatic changes in other properties—time points and cell densities of first light emission and maximal light output and emergence of a lag phase in growth curves of derived lines suggest increased light intensity per se was not the only important factor. Convergent evolution implies gnotobiotic squid light organs subject colonizing V. fischeri to similar selection pressures. Adaptation to novel hosts appears to involve flexible microbial metabolism, establishment of biofilm and swarmer V. fischeri ecotypes, and complex changes in bioluminescence. Our data demonstrate numerous alternate fitness optima or peaks are available to V. fischeri in host adaptive landscapes, where novel host squids serve as habitat islands. Thus, V. fischeri founder flushes occur during the initiation of light organ colonization that ultimately trigger founder effect diversification.
INTRODUCTION
The Sepiolid Squid-Vibrio Mutualism
Sepiolid squids in the genera Sepiola and Euprymna form light organ mutualisms with marine bioluminescent bacteria from the genera Vibrio and Photobacterium from the family Vibrionaneae [1]. Sepiolid squids use light produced by their bacterial symbionts for a cryptic behavior termed counterillumination [2], and the light organ bacteria are in turn exposed to a nutrient-rich microcosm in the host [3]. Particularly, the mutualism between Vibrio fischeri and Euprymna has become a model for studying associations between eukaryotic hosts and bacteria, since both partners can be maintained independently of each other in the laboratory [4, 5]. Axenic juvenile Euprymna squid hatch from their eggs with sterile light organs and are quickly colonized by symbiotically competent V. fischeri present in bacterioplankton, reaching a light organ carrying capacity (104–107 colony forming units (CFUs)/light organ) within 12–24 hours [6, 7]. Sepiolid squids are nocturnal, and just before burying in sand, seed the surrounding water with symbiotic competent V. fischeri at dawn by venting 90–95% of the symbiont light organ population. By the next evening, the remaining V. fischeri inside the animal grow to repopulate the light organ to full capacity [5]. These vented symbionts in the oceanic water column serve as a source population for colonizing the next generation of squid hatchlings.
Not all strains of V. fischeri are capable of colonizing Euprymna and Sepiola hosts, as some isolates are restricted to planktonic and commensal lifestyles (non-light organ associations with animals) [6]. V. fischeri is also able to initiate light organ mutualisms with monocentrid fishes. These mutualisms are each formed with ecologically and genetically distinct V. fischeri [8], and strains indigenous to monocentrid fish hosts do not colonize sepiolid squid to the same population levels (i.e., CFUs/light organ) as “squid” isolates [9]. Additionally, Euprymna species are distributed allopatrically throughout the Indo-West Pacific Ocean, and V. fischeri colonizing this genus are host specialists, exhibiting competitive dominance, whereas strains forming mutualisms with several sympatric Sepiola species from the Mediterranean Sea [1, 9–11] are host generalists and display no competitive dominance. Prior evidence from experimentally evolved lines of V. fischeri ES114 (native to Hawaiian E. scolopes) selected through a non-native host (Australian E. tasmanica) for 500 generations exhibited a significant increase in mean fitness relative to the ancestral strain in colonizing E. tasmanica [7], yet the traits responsible for this host specificity change remain unknown. Therefore, to determine which phenotypes are subject to host selection, both ancestor and evolved clones were characterized for biofilm formation, motility, carbon source utilization, and in vitro bioluminescence. Wild V. fischeri isolates from field-caught E. tasmanica (“ET” isolates) and E. scolopes (“ES” isolates) were also compared to experimentally evolved strains to determine if the derived lines exhibited any convergent evolution relative to indigenous ET V. fischeri.
METHODS AND MATERIALS
Motility Assay
A substantial literature exists on Vibrionaceae motility, and evidence for swarming in Vibrio is prolific and well documented [12–19]. The genus Vibrio possesses a dual flagellar system [18, 20, 21]. Polar flagella are produced by cells continuously for facile locomotion such as swimming in liquid, while inducible lateral or peritrichous flagella are manufactured for more arduous navigation via swarming on solid surfaces or through viscous milieus, including 0.5% agar [18]. Polar and lateral flagella in Vibrio spp. are encoded and regulated by distinct gene sets [19]. When performing motility assays on 0.5% agar with Vibrio, swarming can be quantified by measuring the diameter of an expanding and growing bacterial mass. Since twitching motility occurs much more slowly than swarming, the diameter of the expanding bacterial mass does not represent the former. Swimming was not measured in these assays, since the locomotion we observed occurred on the surface of 0.5% agar rather than within. This was confirmed by examining the motile bacteria under a dissecting microscope. Motility assays were modified from a previously published protocol [22] to monitor swarming. All bacterial strains were grown in 18×150 mm glass test tubes containing 5 mL 70% 32 ppt seawater-tryptone (SWT: 5 g tryptone, 3 g yeast extract, 3.75 mL glycerol, 300 mL double distilled water, and 700 mL 32 ppt artificial seawater made with Instant Ocean®) [23–25] liquid media at 28°C and 225 rpm for sixteen hours. These test tubes were initially inoculated with a single colony from an SWT 1.5% agar plate. 10 µL of overnight starter cultures were used to inoculate 18×150 mm glass test tubes with 10 mL 70% 32 ppt SWT and incubated at 28°C at 225 rpm until subcultures reached 0.10 OD600 (~1.0 × 107 CFUs/mL). Bacterial cells were pelleted by 4°C centrifugation (10,000 rpm) for ten minutes in 15 mL conical tubes, discarding the supernatant afterward. Bacterial pellets were resuspended with 10 mL washes of either filter-sterilized 34 ppt artificial seawater (Instant Ocean®), minimal ribose media, or 34 ppt SWT to remove residual nutrient media from the overnight culture [26]. This centrifugation and wash procedure was performed three times. After the third centrifugation step and resuspension wash, the cell suspension was once again centrifuged for ten minutes and pelleted. The bacterial pellet was then resuspended either with filter-sterilized 34 ppt artificial seawater, minimal ribose media, or 34 ppt SWT. For each isolate or strain, 10 µL of the resuspended pellet (~105 total cells) was placed directly center onto (0.5%) swarm motility agar plates [26, 27] containing either 34 ppt artificial seawater, minimal ribose [25], or 34 ppt SWT, representing different nutrient conditions to observe motility (n=3). This was also repeated for negative control (5.0%) agar plates, where the agar concentration is sufficiently high to prevent swarming. The negative control plates also provided a baseline for comparing motility on each of the different media or nutrient conditions. All agar plates utilized in the motility assays received only a single 10 µL resuspension per isolate. Agar volume placed into each Petri dish for motility assays was measured and kept constant (20 mL). Immediately after the swarm agar plates (including negative controls) solidified, they were allowed to air dry at 25°C for 24 hours before use. The plates were incubated for 24 hours at 28°C, when diameters of swarming bacteria were measured [22, 26]. Fisher least significant differences (LSDs), Bonferroni corrected for multiple pairwise comparisons using the Dunn-Sidak method (type I experimentwise error rate α=0.05) [28], were calculated separately for the wild isolates and each evolved time point (100, 200, 300, 400, and 500 generations). Chloramphenicol resistance has no effect on motility, as V. fischeri ES114 and unevolved V. fischeri JRM200 were not significantly different from each other under all conditions. Chloramphenicol resistance is thus a neutral marker with respect to motility between V. fischeri ES114 and unevolved V. fischeri JRM200.
Microtiter Plate Biofilm Assay
Microtiter plate biofilm assay was adapted from a published methodology initially used for staphylococci [29]. Sixteen-hour V. fischeri cultures were grown in 18×150 mm glass test tubes containing 5 mL 70% SWT in a 28°C air shaker at 225 rpm. These test tubes were initially inoculated with a single colony from an SWT 1.5% agar plate. 10 µL of the overnight starter cultures were each separately inoculated into 18×150 mm glass test tubes with 10 mL of fresh 70% 32 ppt SWT and incubated in a 28°C air shaker at 225 rpm. These subcultures were grown to 0.10 OD600 (~1.0 × 107 CFUs/mL). All 0.10 OD600 subcultures were centrifuged (10,000 rpm) at 4°C, pelleted, resuspended, and washed three times as previously described with either liquid filter-sterilized 34 ppt artificial seawater, minimal ribose, or 34 ppt SWT to ensure all residual 70% 32 ppt SWT was removed. After the third centrifugation step and resuspension wash, the cell suspension was once again centrifuged for ten minutes and pelleted. The bacterial pellet was then resuspended either with filter-sterilized 34 ppt artificial seawater, minimal ribose media, or 34 ppt SWT. Cell suspensions (2 µL) were subsequently inoculated into polystyrene 96-well microtiter plates containing 200 µL of either filter-sterilized 34 ppt artificial seawater, minimal ribose media, or 34 ppt SWT (n=12). Three liquid media types were used to examine V. fischeri biofilm formation in different environments representing varying degrees of nutrient availability. Uninoculated wells remained on each of the three different liquid media microtiter plates to serve as negative controls. Plates were incubated for 18 hours at 28°C without shaking. Media in all wells were then gently removed and washed three times appropriately with either 200 µL filter-sterilized 34 ppt artificial seawater, minimal ribose, or 34 ppt SWT, removing wash liquid each time. Microtiter plates were then incubated at room temperature for 15 minutes. 200 µL of 0.2% crystal violet dye was then added to each well and incubated for 30 minutes at room temperature. After staining, crystal violet was gently removed with a pipet and the wells washed three times with either 200 µL 34 ppt artificial seawater, minimal ribose, or 34 ppt SWT, removing wash liquid after each time. Microtiter plates were allowed to incubate at room temperature for 15 minutes. 200 µL of 70% ethanol was added to all the wells in the microtiter plates and incubated at room temperature for 20 minutes. Absorbance readings were measured for all microtiter plates at 562 nm using a Bio-Tek ELx800™ microplate reader (Winooski, VT). Fisher least significant differences (LSDs), Bonferroni corrected for multiple pairwise comparisons using the Dunn-Sidak method (type I experimentwise error rate α=0.05) [28], were calculated separately for the wild isolates and each evolved time point (100, 200, 300, 400, and 500 generations). Chloramphenicol resistance has no effect on biofilm formation, as V. fischeri ES114 and unevolved V. fischeri JRM200 were not significantly different from each other under all conditions. Chloramphenicol resistance is thus a neutral marker with respect to biofilms between V. fischeri ES114 and unevolved V. fischeri JRM200.
Carbon-Source Utilization for Ninety-Five Substrates
Biolog GN2 microplates (Hayward, CA) for gram-negative bacteria were used, where substrates in each of ninety-five wells represent different carbon sources, along with one negative control well that contains only distilled water [30, 31]. Utilization of a substrate was recorded by measuring increases in well absorbance relative to the negative control at 590 nm. Manufacturer’s instructions (Biolog GN2 MicroPlate™) and protocols were used with modification. All bacterial strains were grown in 18×150 mm glass test tubes containing 5 mL 70% 32 ppt SWT at 28°C and 225 rpm for sixteen hours. These test tubes were initially inoculated with a single colony from an SWT 1.5% agar plate. 10 µL of overnight starter cultures were used to inoculate 125-mL triple-baffled culture flasks with 50 mL of fresh 70% 32 ppt SWT, which were incubated at 28°C and shaken at 225 rpm until the flask subculture reached 0.220 OD590 (~2.0 × 107 CFUs/mL). Bacterial cells from each 0.220 OD590 subculture were pelleted by 4°C centrifugation (10,000 rpm) for ten minutes and resuspended with Biolog GN/GP inoculation fluid (Product # 72101) to remove residual nutrient media from overnight cultures. To meet V. fischeri osmolar requirements, the salinity of the Biolog inoculation fluid was adjusted to 3.4% NaCl final concentration. The centrifugation and wash procedure was completed three times. After the third centrifugation step and resuspension wash, cells were once again centrifuged for ten minutes and pelleted. The bacterial pellet was then resuspended with Biolog inoculation fluid. Working V. fischeri cell suspensions for each strain were dispensed into Biolog GN2 microplates. All wells in Biolog GN2 microplates were inoculated with 150 µL of washed V. fischeri cells at 2.0 × 107 CFUs/mL suspended in Biolog GN/GP inoculation fluid. Uninoculated Biolog GN2 microplate wells containing only Biolog GN/GP inoculation fluid (adjusted to 3.4% NaCl final concentration) and no bacteria served as negative controls. All negative controls and V. fischeri strains were measured in triplicate (n=3). Microplates were incubated at 28°C for 24 hours. Afterwards, optical densities (590 nm) of the wells were measured by placing GN2 plates in a Bio-Tek ELx800™ microplate reader (Winooski, VT). Fisher least significant differences (LSDs), Bonferroni corrected for multiple pairwise comparisons using the Dunn-Sidak method (type I experimentwise error rate α=0.05) [28], were calculated separately for wild isolates and the 500-generation evolved time point.
Chloramphenicol resistance has no effect on carbon source utilization, as V. fischeri ES114 and unevolved V. fischeri JRM200 were not significantly different from each other on any carbon source. Chloramphenicol resistance is thus a neutral marker with respect to carbon source utilization between V. fischeri ES114 and unevolved V. fischeri JRM200.
Bioluminescence
Sixteen-hour V. fischeri cultures were grown in 18×150 mm glass test tubes containing 5 mL 70% SWT liquid media in a 28°C air shaker at 225 rpm. These test tubes were initially inoculated with a single colony from an SWT 1.5% agar plate. 10 µL of the overnight cultures were each separately inoculated into 18×150 mm glass test tubes with 5 mL of fresh 70% 32 ppt SWT and incubated in a 28°C air shaker at 225 rpm to obtain exponential growth subcultures at 0.5 OD600. 0.5 OD600 subcultures were used to inoculate 125-mL triple-baffled culture flasks with fresh 50 mL 34 ppt SWT to an initial 5 × 105 CFUs/mL cell density. Culture flasks were incubated at 28°C and shaken at 225 rpm for eight hours with bioluminescence readings and plate count enumeration taken every 30 minutes (n=6). Bioluminescence readings were recorded with a TD-20/20 luminometer (Turner Designs, Sunnyvale, CA). Since total bacterial luminescence is dependent on cell density and total volume, measurements were reported as relative light units (RLUs) per Log10 (CFUs/mL) per mL of liquid culture (RLUs [Log10 (CFUs/mL)]−1mL−1) over time. Cell density determinations were completed on 34 SWT 1.5% agar plates, which were incubated at 28°C for 24 hours. During these time interval experiments, V. fischeri broth cultures have nearly 100% plating efficiency [6]. Bioluminescence readings of Escherichia coli K12 MG1655 were also monitored as a negative control under the same experimental conditions with the exception that Luria Bertani (LB) broth (10 g tryptone, 5 g yeast extract, 10 g NaCl, and 1 L double distilled water) and LB 1.5% agar plates were used. There was no difference in bioluminescence between V. fischeri ES114 and unevolved V. fischeri JRM200 through all time points, showing that chloramphenicol is a neutral marker with this trait. Bioluminescence data were assessed using repeated measures analysis. No assumption was made with sphericity. The quite conservative Extreme Greenhouse-Geisser and Lower Bound corrections were applied separately to statistical main effects [32]. Fisher least significant differences (LSDs), Bonferroni corrected for multiple pairwise comparisons using the Dunn-Sidak method (type I experimentwise error rate α=0.05) [28], were calculated separately for wild isolates and the 500-generation evolved time point.
RESULTS
Motility
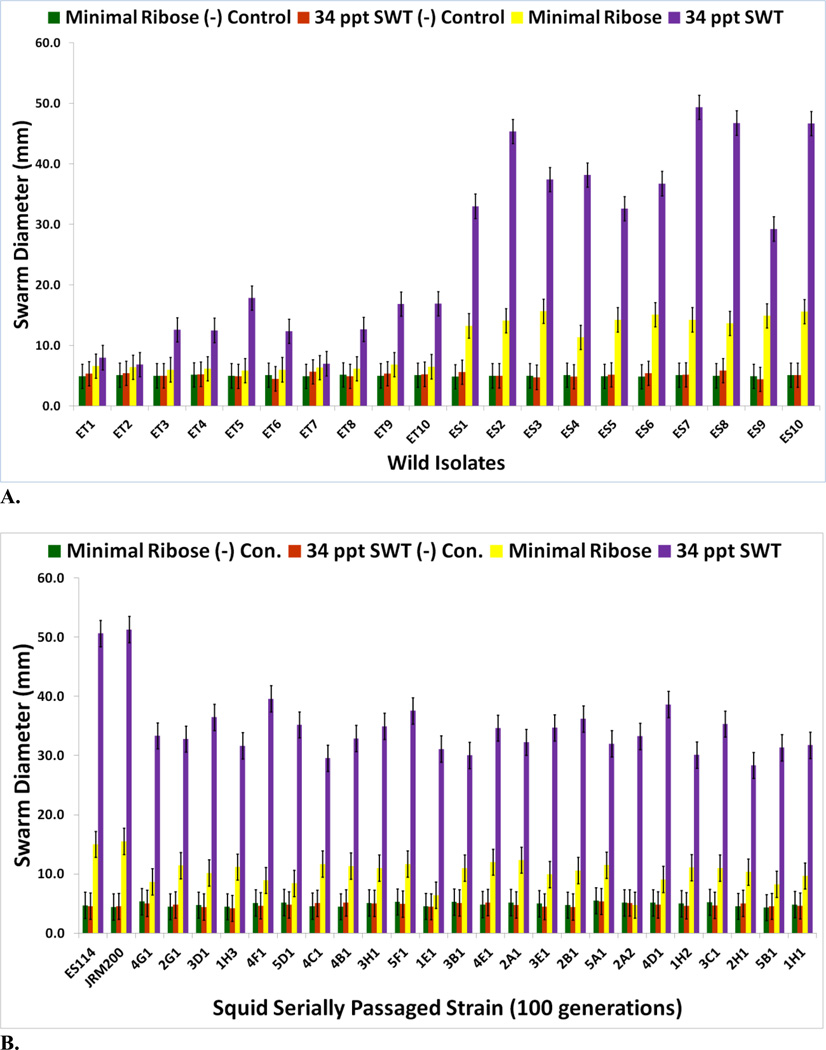
Ten ET (wild isolates from E. tasmanica) and ten ES (wild isolates from E. scolopes) V. fischeri strains were previously shown to be genetically distinct from one another [7, 8, 33]. These field isolates were randomly sampled from their Euprymna squid hosts and used as a comparison to the evolved clones for motility (Fig. 1A). ES wild type strains were more motile than ET on minimal ribose and 34 ppt SWT. V. fischeri ES114 is chloramphenicol sensitive, while V. fischeri JRM200 is a V. fischeri ES114 derivative which is chloramphenicol resistant [24]. V. fischeri JRM200 was used to create twenty-four lines that were serially transferred through novel squid host E. tasmanica for 500 generations [7] and used in this study. All 24 evolved lines improved in colonization of E. tasmanica relative to ancestor, as no lineage remained the same or became worse. V. fischeri lines serially passaged through E. tasmanica for 500 generations became less motile relative to ancestor V. fischeri ES114 and unevolved V. fischeri JRM200 (zero generations through E. tasmanica) on minimal ribose and 34 ppt SWT (Figs. 1B–1D for 100, 200, and 500 generations; supplementary Figs. S1A and S1B for 300 and 400 generations). Unlike many bacterial species, V. fischeri is capable of motility on minimal media without nutritional supplements such as casamino acids [27]. No growth was observed on 34 ppt artificial seawater 0.5% agar plates as well as on the motility negative control 5.0% agar plates. Significant decreases in motility were first observed for all lines by 100 generations (Fig. 1B) on motility SWT plates and continued to decline throughout the remainder of the selection experiment. However, by 500 generations, all derived lines still remained motile on SWT plates relative to SWT motility negative controls (Fig. 1D). Although significant decreases in motility on minimal ribose is evident by 100 generations (Fig. 1B), a marked drop was noted by 200 generations (Fig. 1C), as most of the derived lines are no longer significantly more motile than on minimal ribose motility negative control plates. Motility on minimal ribose plates was completely lost by 300 generations (Fig. S1A). Thus, the derived lines converged on ET motility on minimal ribose and 34 ppt SWT, yet significant differences (polymorphisms) were seen among the various lineages in motility as a result of adaptation to novel host E. tasmanica.
FIGURE 1.
(A) Motility assays for natural V. fischeri isolates extracted from E. tasmanica (“ET”) and E. scolopes (“ES”) squid hosts (n=3). Motility assays for V. fischeri JRM200 lines serially passaged for (B) 100, (C) 200, and (D) 500 generations through E. tasmanica (n=3). Error bars represent Fisher LSDs with Bonferroni correction for multiple pairwise comparisons (type I experimentwise error rate α=0.05). If error bars overlap between any particular pair of comparisons, they are not significantly different from each other [28].
Biolfilm Formation
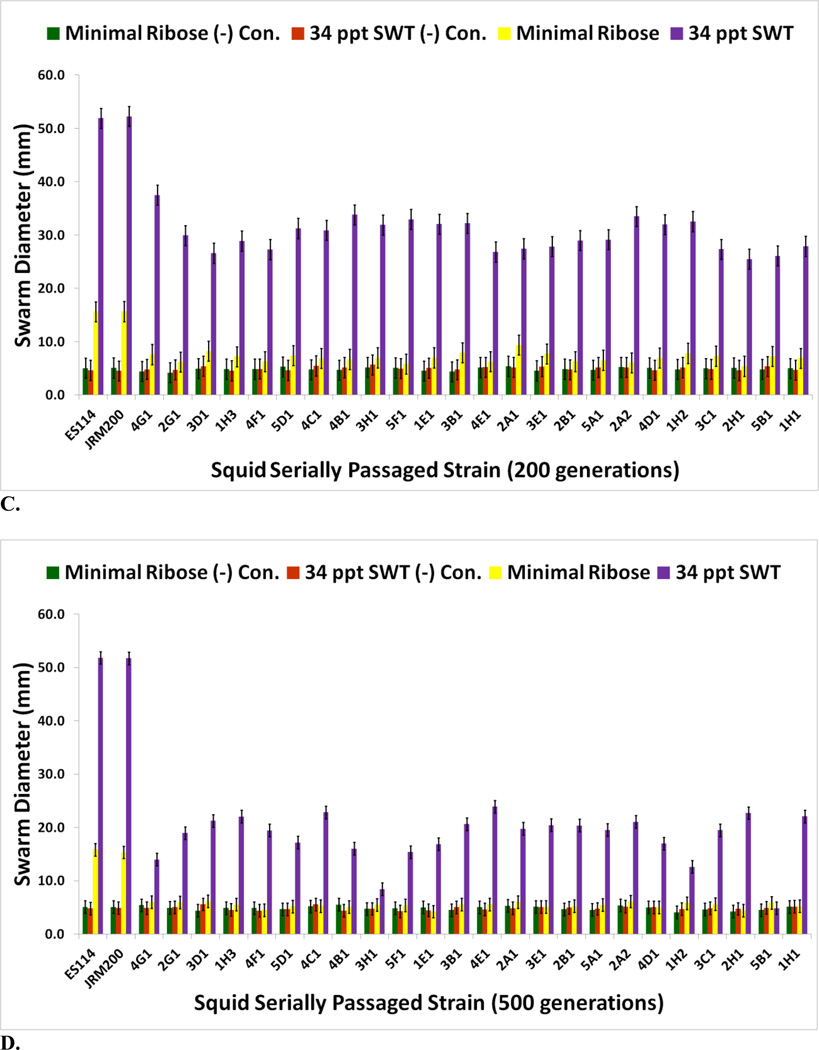
No bacterial contamination was observed in uninoculated negative control wells for 34 ppt artificial seawater, minimal ribose media, or 34 ppt SWT. Biofilm formation is higher for the ET wild isolates than the ES ones in artificial seawater, minimal ribose, and 34 ppt SWT (Fig. 2A). V. fischeri passaged through E. tasmanica increased their biofilm formation capacity (Figs. 2B–2D for 100, 400, and 500 generations; supplementary Figs. S2A and S2B for 200 and 300 generations). Relative to the loss of motility, the evolution of elevated biofilm formation was delayed, not appearing in great levels until 400 generations (Fig. 2C), when it simultaneously appeared dramatically in minimal ribose and SWT. Heightened biofilm formation in artificial seawater arose by 400 generations (Fig. 2C), as some derived lines were significantly higher than the ancestor in this medium. Interestingly, V. fischeri ES114 and unevolved V. fischeri JRM200 were initially incapable of biofilm growth in artificial seawater. Significant dissimilarity from the ancestral state in biofilm construction did not manifest in the evolved lines in 34 ppt SWT and minimal ribose until 200 and 300 generations, respectively (Figs. S2A and S2B). Thus, like motility, the relative evolutionary change in biofilm formation is nutrient dependent and convergent to ET wild variation, as “domesticated” variation created by artificial selection through novel host E. tasmanica showed a parallel biofilm response in all media types. The evolved lines have also diverged from one another in biofilm formation.
FIGURE 2.
(A) Biofilm assays for wild V. fischeri strains from E. tasmanica (“ET”) and E. scolopes (“ES”) specimens collected from the field (n=12). Biofilm assays for V. fischeri JRM200 lines serially passaged for (B) 100, (C) 400, and (D) 500 generations through E. tasmanica (n=12). Error bars represent Fisher LSDs with Bonferroni correction for multiple pairwise comparisons (type I experimentwise error rate α=0.05). If error bars overlap between any particular pair of comparisons, they are not significantly different from each other [28].
Carbon-Source Utilization for Ninety-Five Substrates
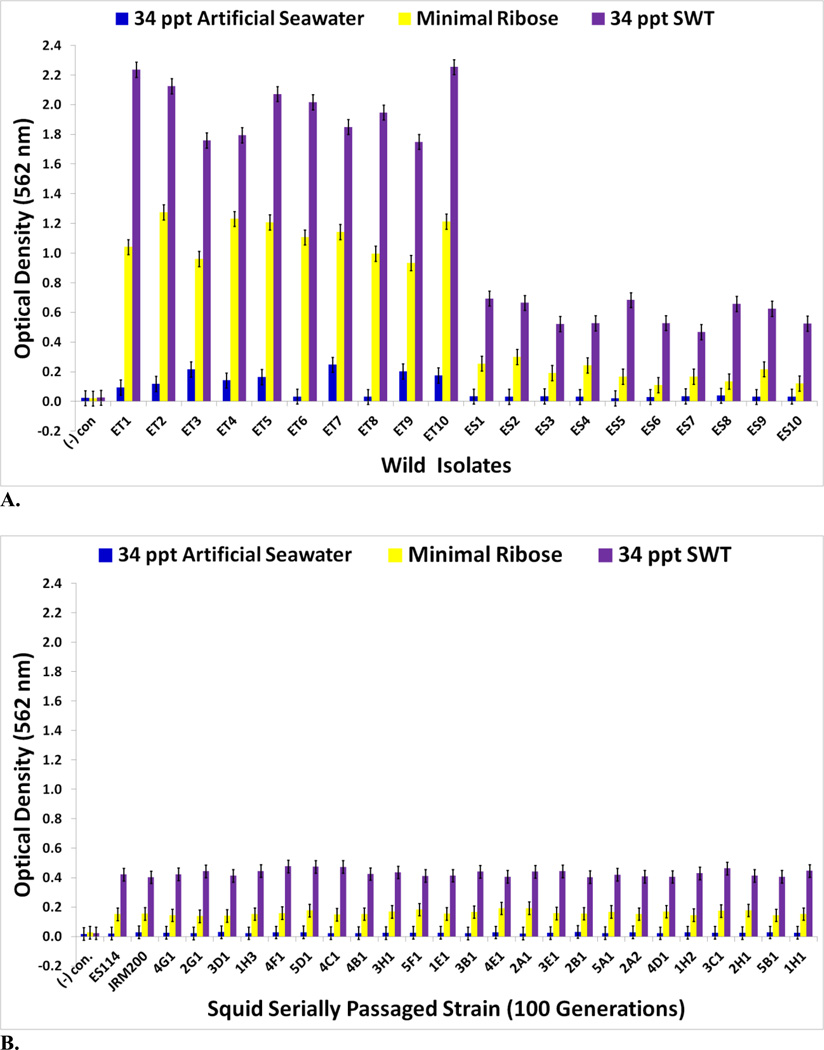
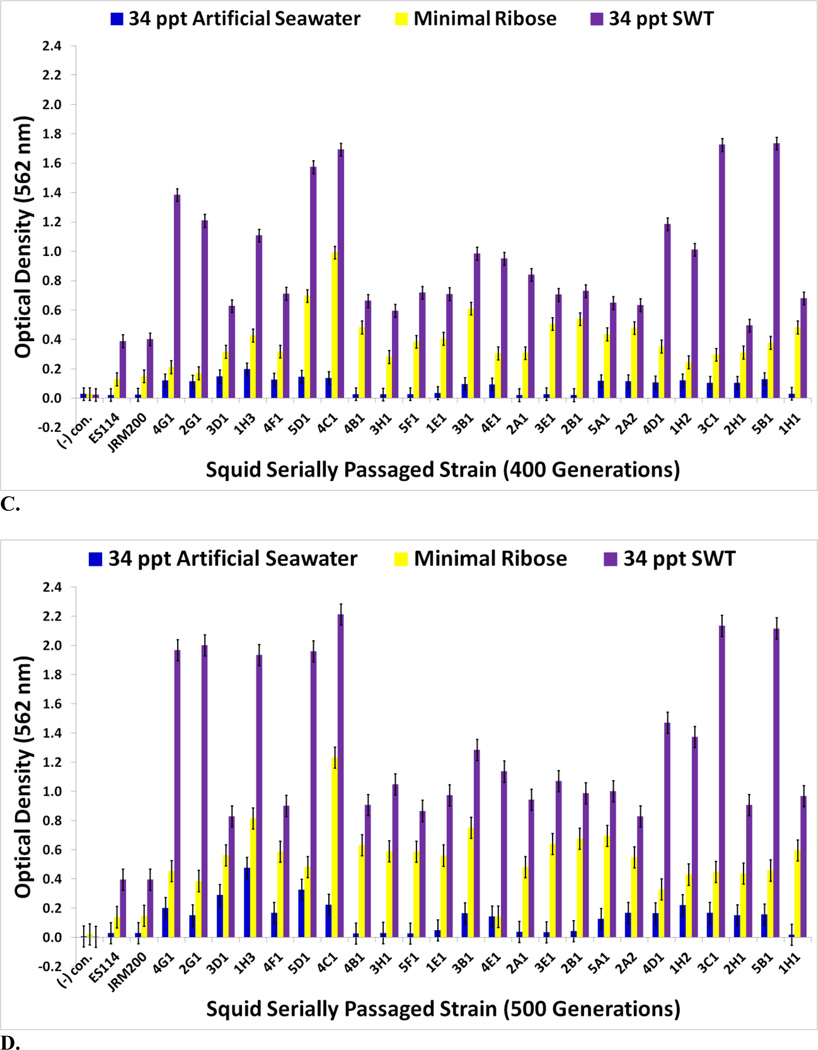
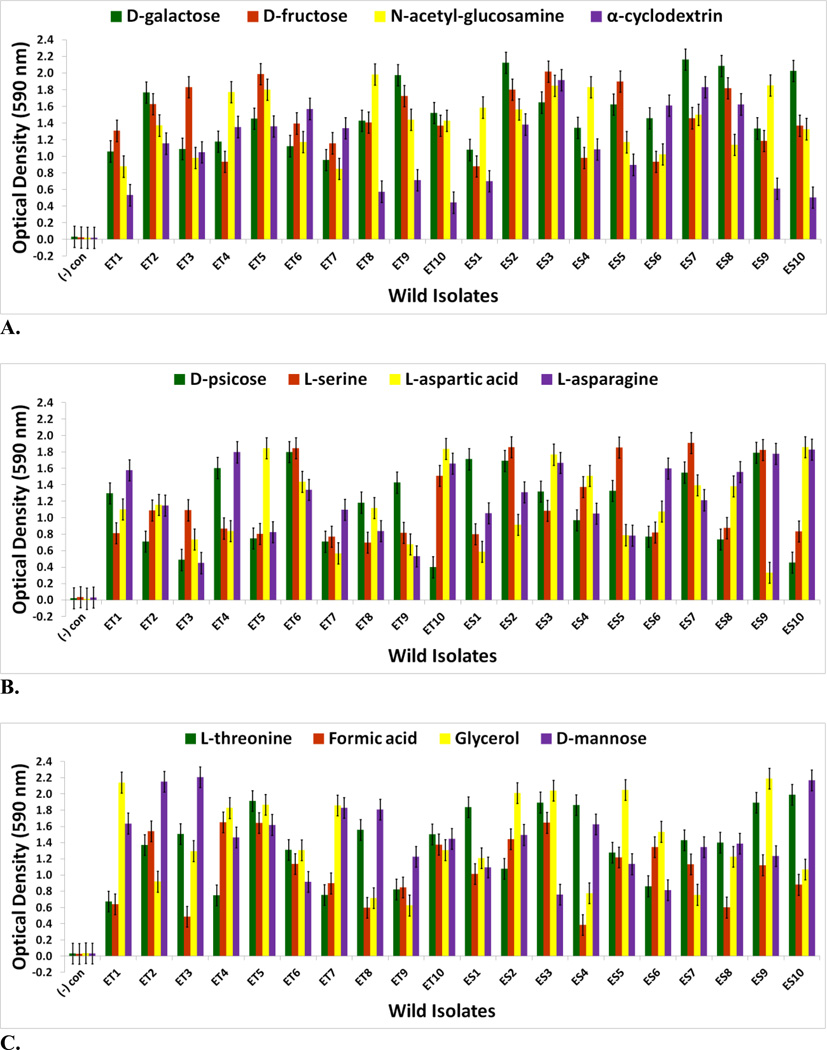
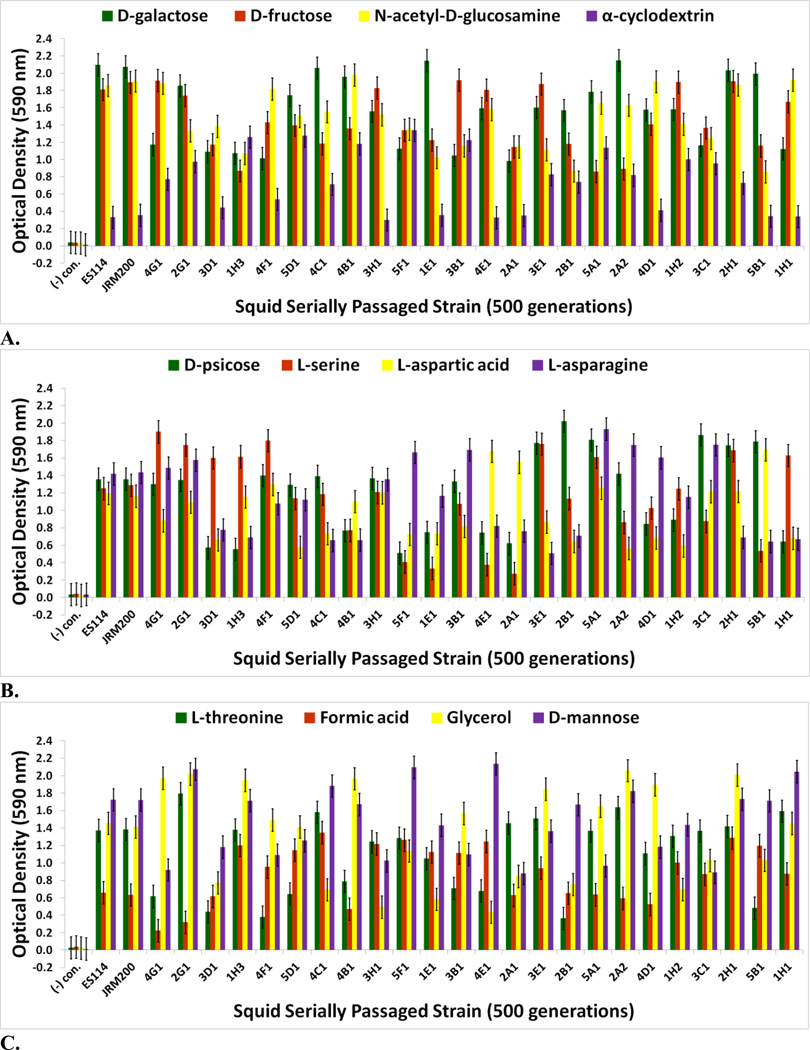
The goal of measuring carbon source utilization in this study was to identify metabolic or physiological changes in the twenty-four V. fischeri lines as results of adapting to a novel squid host environment in the light organ. Some of these changes may have been directly responsible for adaptation to the new host animal; others may have been the result of tradeoffs, epistasis, pleiotropy, or mutation accumulation [34]. No bacterial contamination was observed on the Biolog GN2 negative control plates. The Biolog GN2 microplate data for V. fischeri ES114 and V. fischeri unevolved JRM200 (supplementary Table S1) is congruent with what has been previously reported as accessible carbon sources for V. fischeri, supporting the validity of the results [35–40]. ET and ES wild isolates were heterogeneous for carbon substrate utilization (Figs. 3A–4B). Relative to ancestral metabolism, V. fischeri lines serially passaged through E. tasmanica diversified in their utilization in every substrate in the Biolog microplate initially physiologically procurable (Figs. 5A–6B). As is wild ET variation (Fig. 3A–3C), “domesticated” variation (Figs. 5A–5C) in carbon utilization from experimentally evolved V. fischeri is quite polymorphic. The precise manner of change between the derived lines and the ancestral state was carbon source dependent. For example, the various lineages remained the same or decreased on N-acetyl-D-glucosamine, while the evolutionary response with cyclodextrin was to increase or stay the same (Fig. 5A). On D-psicose (a rare sugar in nature [41]), the evolved lines either gained, lost, or were unaltered (Fig. 5B). However, no compelling convergent evolution was evident relative to wild isolates in most carbon substrates (representative examples shown in Figs. 3A–3C; Figs. 5A–5C).
FIGURE 3.
Optical density (590 nm) measurements in (A) D-galactose, D-fructose, N-acetyl-D-glucosamine, and α-cyclodextrin (B) D-psicose, L-serine, L-aspartic acid, and L-asparagine (C) L-threonine, formic acid, glycerol, and D-mannose of wild V. fischeri isolates from E. tasmanica (“ET”) and E. scolopes (“ES”) specimens collected from the field (n=3). Error bars represent Fisher LSDs with Bonferroni correction for multiple pairwise comparisons (type I experimentwise error rate α=0.05). If error bars overlap between any particular pair of comparisons, they are not significantly different from each other [28].
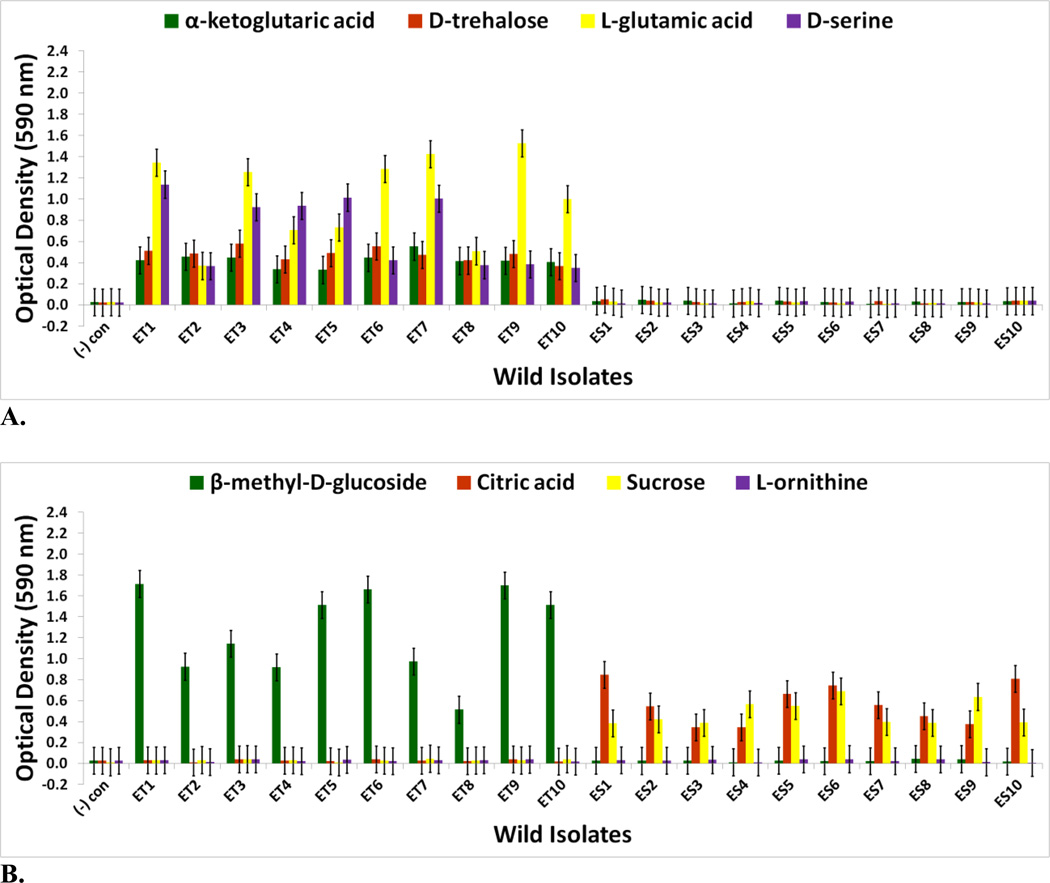
FIGURE 4.
Optical density (590 nm) measurements in (A) α-ketoglutaric acid, D-trehalose, L-glutamic acid, and D-serine (B) β-methyl-D-glucoside, citric acid, sucrose, and L-ornithine of wild V. fischeri isolates from Australian E. tasmanica (“ET”) and Hawaiian E. scolopes (“ES”) specimens collected from the field (n=3). Error bars represent Fisher LSDs with Bonferroni correction for multiple pairwise comparisons (type I experimentwise error rate α=0.05). If error bars overlap between any particular pair of comparisons, they are not significantly different from each other [28].
FIGURE 5.
Optical density (590 nm) measurements in (A) D-galactose, D-fructose, N-acetyl-D-glucosamine, and α-cyclodextrin (B) D-psicose, L-serine, L-aspartic acid, and L-asparagine (C) L-threonine, formic acid, glycerol, and D-mannose of ancestor V. fischeri ES114, unevolved JRM200, and the 24 lines evolved through the novel Australian squid host E. tasmanica for 500 generations (n=3). Error bars represent Fisher LSDs with Bonferroni correction for multiple pairwise comparisons (type I experimentwise error rate α=0.05). If error bars overlap between any particular pair of comparisons, they are not significantly different from each other [28].
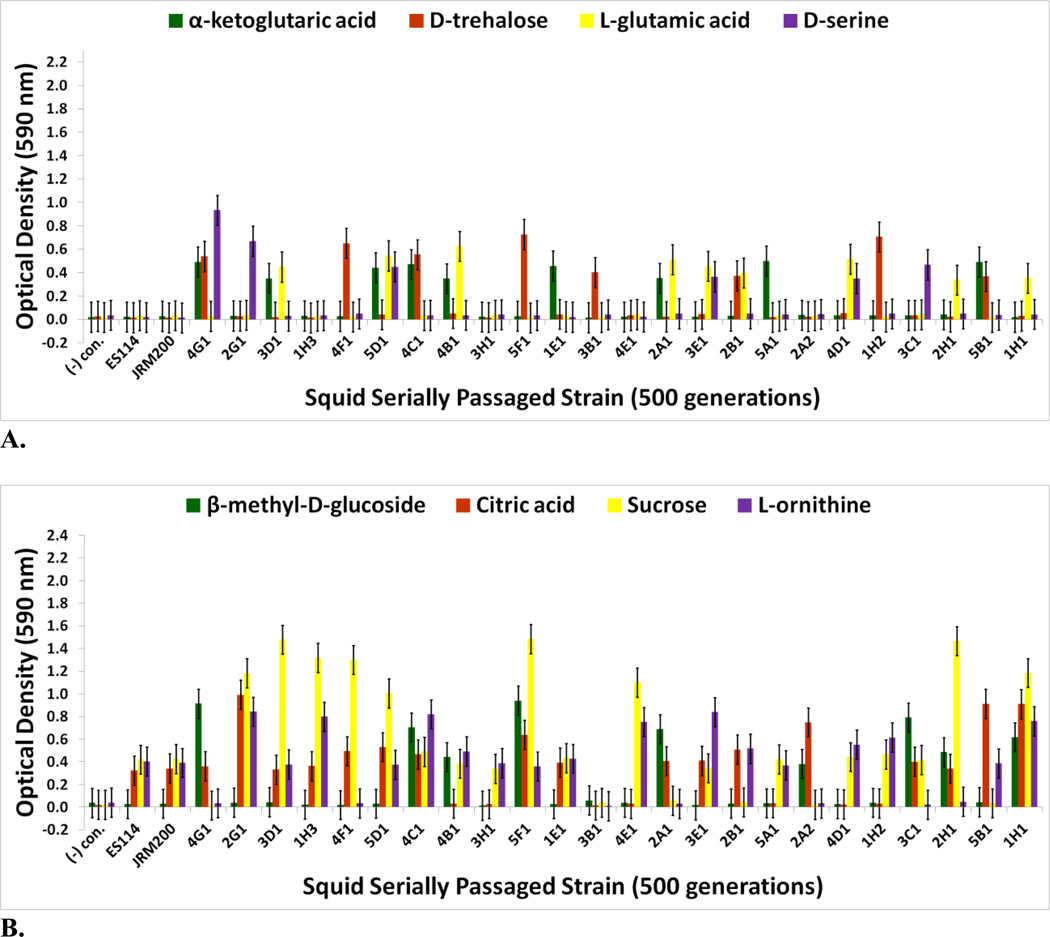
FIGURE 6.
Optical density (590 nm) measurements in (A) α-ketoglutaric acid, D-trehalose, L-glutamic acid, and D-serine (B) β-methyl-D-glucoside, citric acid, sucrose, and L-ornithine of ancestor V. fischeri ES114, unevolved JRM200, and the 24 lines evolved through the novel Australian squid host E. tasmanica for 500 generations (n=3). Error bars represent Fisher LSDs with Bonferroni correction for multiple pairwise comparisons (type I experimentwise error rate α=0.05). If error bars overlap between any particular pair of comparisons, they are not significantly different from each other [28].
Genetic evidence exists that these carbon sources play an important role for V. fischeri metabolism in its association with sepiolid squid hosts. [3, 42] L-asparaginase, chitinases (liberate N-acetyl-D-glucosamine monomers), and dextrinases are produced in the symbiont while in symbiosis with the host squid. Protein importers and permeases for L-serine, D-galactose, and D-fructose have also been found in symbiotic V. fischeri [42]. L-threonine is supplied by Euprymna squid to V. fischeri within the light organ in nutrient limiting amounts relative to other amino acids [3]. Moreover, a multitude of genes are dedicated to the V. fischeri catabolism of glycerol, formic acid, chitin, and fumaric acid while in the light organ. Fumaric acid can be made by the deamination of L-aspartic acid, a reaction carried out by aspartate ammonia-lyase, an enzyme expressed by symbiotic V. fischeri [42]. D-mannose residues coat the epithelial linings of light organs within sepiolid squid hatchlings and play a fundamental role in V. fischeri attachment to squid host eukaryotic cells through mannose-recognizing adhesins on the bacterial surface [22, 43], which potentially serve as a source for “mannose” grazing. Prolific variation in catabolism of D-mannose among the derived lines relative to precursor state was evident (Fig. 5C).
For V. fischeri, some carbon sources in the Biolog GN2 microplate exhibit the most substantial metabolic diversity (hyperpolymorphic) known for any particular substrate. They include N-acetyl-D-galactosamine, L-fucose, α-ketoglutaric acid, D-trehalose, L-glutamic acid, D-serine, β-methyl-D-glucoside, citric acid, sucrose, and L-ornithine [37–40, 44, 45]. The Biolog data for the last eight of these hyperpolymoprhic carbon sources are shown for the wild isolates (Figs. 4A & 4B). All ET and no ES strains utilize α-ketoglutaric acid, D-trehalose, L-glutamic acid, and D-serine, while no ET and all ES isolates metabolize citric acid and sucrose. The ability to utilize seven new carbon sources was gained in some V. fischeri lines serially passaged through E. tasmanica for five hundred generations—N-acetyl-D-galactosamine, L-fucose, α-ketoglutaric acid, D-trehalose, L-glutamic acid, D-serine, and β-methyl-D-glucoside, relative to forerunners V. fischeri ES114 and unevolved V. fischeri JRM200 (last five shown in Figs. 6A & 6B). Conversely, substrates such as citric acid, sucrose, and L-ornithine were lost in some lines (Fig. 6B). N-acetyl-D-galactosamine has been found to participate in cell-cell attachment through lectins, carbohydrate-binding proteins on extracellular surfaces, including bioluminescent bacteria [46]. Lectins may be involved in biofilm formation and bacterial microcolony aggregations. Previous research has hypothesized that lectins with N-acetyl-D-galactosamine specificity govern symbiosis initiation in bioluminescent bacteria with marine animals, V. fischeri included [47]. Furthermore, D-serine, citric acid, α-ketoglutaric acid, β-methyl-D-glucoside, and N-acetyl-D-galactosamine have all been listed as carbon substrates dispensable for V. cholerae, which are evolutionarily discarded and reacquired as necessary for certain niche environments [48]. Metabolic pathways can evolve modularly in function, some being especially malleable, to accommodate the niche breadth ecologically necessary for Vibrio populations under specific environmental parameters (e.g., free-living versus host-associated lifestyles). Curiously, no wild isolates from E. scolopes and E. tasmanica grew on N-acetyl-D-galactosamine, L-fucose, and L-ornithine. L-fucose residues may also serve as attachment sites for bacteria to other cells, such as fucose-sensitive hemagglutinin A (fshA) in V. cholerae [49]. In summary, ET convergence was observed with α-ketoglutaric acid, D-trehalose, L-glutamic acid, D-serine, β-methyl-D-glucoside, citric acid, and sucrose (Figs. 4A & 4B; Figs. 6A & 6B). As with motility and biofilm, evolutionary distinction is emerging among the lines in carbon source metabolism.
Bioluminescence
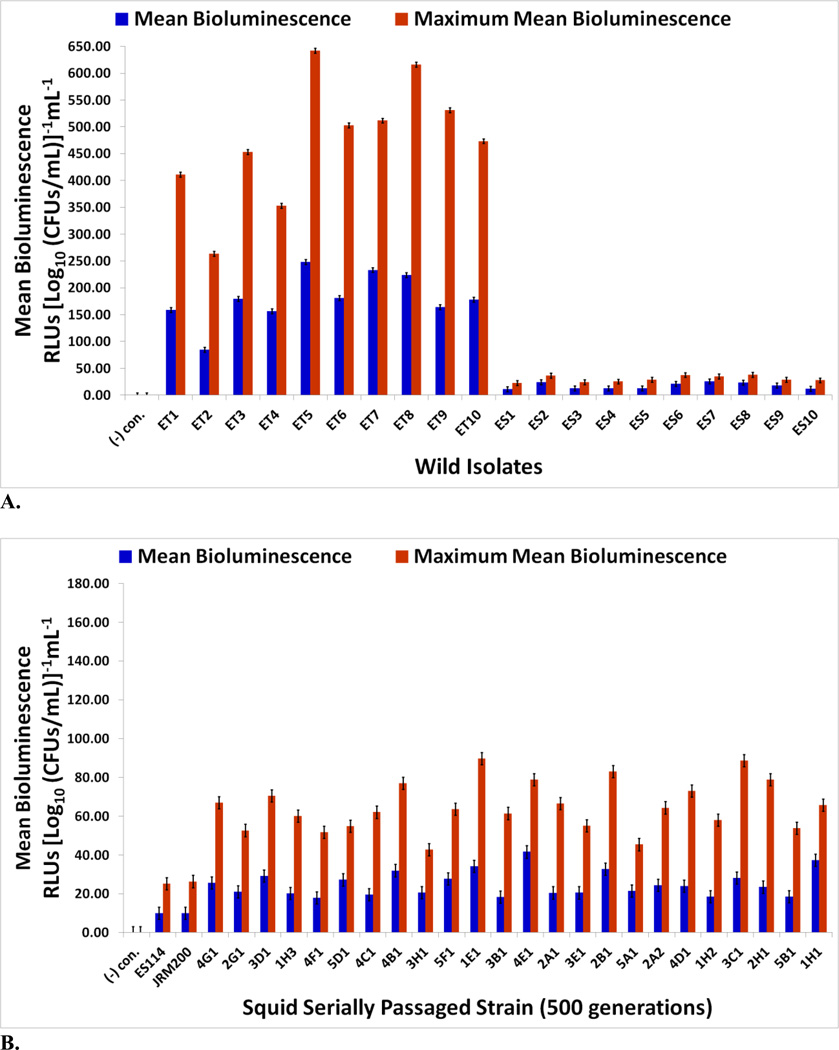
Bioluminescence in V. fischeri is necessary for sepiolid squid colonization, as lux null mutants display defects in colonization ability [6, 50, 51]. Negative control E. coli K12 MG1655 was never luminous. Mean and maximum mean bioluminescence (RLUs [Log10 (CFUs/mL)]−1mL−1) (Figs. 7A & 7B) and notable characteristics of the “growth-light” curves for the wild isolates and experimentally evolved lines were determined (Tables 1A & 1B). All ET strains are brighter than all ES ones (Fig. 7A). ET wild strains are 10.4-fold and 15.6-fold brighter (higher power) than ES strains in mean and maximum mean bioluminescence, respectively. Furthermore, ET V. fischeri produce light before ES strains (ET: 195 min versus ES: 225 min) at lower cell densities (ET: 4.80 × 107 CFUs/mL versus ES: 1.00 × 108 CFUs/mL), ET symbionts achieving total photon emission maxima higher (ET: 4,294.2 RLUs versus ES: 264.5 RLUs) and later (ET: 327 min versus ES: 294 min) at higher cell densities (ET: 1.06 × 109 CFUs/mL versus ES: 4.67 × 108 CFUs/mL) than ES isolates in the process (Table 1A). A trend in ET growth curves is the appearance of a lag phase, while this growth phase is completely lacking in ES symbiont replication. ET symbionts appear to be evolving a quorum sensing machinery to maximize light production throughout bacterial growth, including producing light earlier at lower populations, prolonging length of time cells are luminous, surging light intensity or wattage, and increasing cell density at which maximum light emission occurs to amplify total amount of light output at an instant.
FIGURE 7.
Mean and maximum mean bioluminescence for (A) wild V. fischeri strains from E. tasmanica (“ET”) and E. scolopes (“ES”) specimens collected from the field and (B) ancestor V. fischeri ES114, unevolved JRM200, and the 24 lines evolved through the novel Australian squid host E. tasmanica for 500 generations (n=6). Error bars represent Fisher LSDs with Bonferroni correction for multiple pairwise comparisons (type I experimentwise error rate α=0.05). If error bars overlap between any particular pair of comparisons, they are not significantly different from each other [28].
TABLE 1.
Notable characteristics of the “growth-light” curves (A) wild V. fischeri strains from E. tasmanica (“ET”) and E. scolopes (“ES”) specimens collected from the field and (B) ancestor V. fischeri ES114, unevolved JRM200, and the 24 lines evolved through the novel Australian squid host E. tasmanica for 500 generations (n=6). Negative control E. coli K12 MG1655 is omitted from the tables, since it produced no light. SE=standard error.
| A. | ||||||
|---|---|---|---|---|---|---|
| Strain | Time Light Emission Began (min) |
Log10 [CFUs/mL] Where Light Was First Emitted (±SE) |
Time Maximal Light Output Occurred (min) |
Maximal Light Output (RLUs) (±SE) |
Log10 [CFUs/mL] Where Maximal Light Output Occurred (±SE) |
Lag Phase (Length) |
| ET1 | 210 | 7.797 (±0.033) | 300 | 3597.9 (±29.3) | 8.754 (±0.046) | No |
| ET2 | 240 | 8.111 (±0.116) | 360 | 2464.5 (±20.9) | 9.347 (±0.127) | No |
| ET3 | 180 | 7.510 (±0.057) | 270 | 3835.1 (±23.5) | 8.464 (±0.140) | No |
| ET4 | 150 | 7.231 (±0.056) | 300 | 3072.5 (±33.4) | 8.696 (±0.053) | No |
| ET5 | 180 | 7.492 (±0.134) | 300 | 5609.8 (±38.1) | 8.737 (±0.077) | Yes (1.0 hr) |
| ET6 | 210 | 7.786 (±0.082) | 330 | 4570.8 (±25.7) | 9.088 (±0.060) | No |
| ET7 | 210 | 7.835 (±0.051) | 360 | 4809.9 (±19.9) | 9.398 (±0.146) | No |
| ET8 | 210 | 7.815 (±0.086) | 330 | 5606.6 (±39.5) | 9.096 (±0.048) | Yes (1.0 hr) |
| ET9 | 180 | 7.418 (±0.087) | 360 | 4963.8 (±29.7) | 9.338 (±0.122) | Yes (1.0 hr) |
| ET10 | 210 | 7.818 (±0.054) | 360 | 4411.4 (±25.4) | 9.317 (±0.092) | No |
| Mean | 195 (±5.8) | 7.681 (±0.056) | 327 (±5.4) | 4294.2 (±162.6) | 9.024 (±0.072) | -- |
| ES1 | 210 | 7.793 (±0.109) | 270 | 191.5 (±0.4) | 8.375 (±0.048) | No |
| ES2 | 240 | 8.152 (±0.094) | 300 | 316.1 (±5.6) | 8.716 (±0.077) | No |
| ES3 | 240 | 8.168 (±0.078) | 360 | 226.8 (±3.6) | 9.275 (±0.058) | No |
| ES4 | 270 | 8.407 (±0.093) | 330 | 229.9 (±4.1) | 9.045 (±0.093) | No |
| ES5 | 240 | 8.207 (±0.084) | 300 | 250.4 (±2.4) | 8.752 (±0.035) | No |
| ES6 | 180 | 7.542 (±0.121) | 240 | 306.9 (±2.5) | 8.170 (±0.068) | No |
| ES7 | 240 | 8.111 (±0.052) | 300 | 305.9 (±1.1) | 8.696 (±0.033) | No |
| ES8 | 240 | 8.216 (±0.059) | 330 | 345.6 (±0.9) | 9.033 (±0.071) | No |
| ES9 | 210 | 7.780 (±0.055) | 270 | 242.8 (±1.5) | 8.369 (±0.047) | No |
| ES10 | 180 | 7.628 (±0.117) | 240 | 228.6 (±2.6) | 8.254 (±0.073) | No |
| Mean | 225 (±4.8) | 8.000 (±0.032) | 294 (±6.5) | 264.5 (±8.5) | 8.669 (±0.099) | -- |
| B. | ||||||
|---|---|---|---|---|---|---|
| Strain | Time Light Emission Began (min) |
Log10 [CFUs/mL] Where Light Emission Began (±SE) |
Time Maximal Light Output Occurred (min) |
Maximal Light Output (RLUs) (±SE) |
Log10 [CFUs/mL] Where Maximal Light Output Occurred (±SE) |
Lag Phase (Length) |
| ES114 | 210 | 8.152 (±0.041) | 270 | 224.8 (±4.5) | 8.886 (±0.014) | No |
| JRM200 | 210 | 8.165 (±0.014) | 270 | 234.1 (±3.3) | 8.876 (±0.004) | No |
| Mean | 210 | 8.159 (±0.004) | 270 | 229.5 (±2.4) | 8.881 (±0.003) | -- |
| 4G1 | 150 | 6.624 (±0.006) | 270 | 514.0 (±20.8) | 7.769 (±0.045) | Yes (1.5 hr) |
| 2G1 | 180 | 7.551 (±0.005) | 360 | 481.5 (±8.4) | 9.280 (±0.008) | No |
| 3D1 | 150 | 6.869 (±0.004) | 390 | 644.5 (±8.6) | 9.246 (±0.113) | Yes (1.0 hr) |
| 1H3 | 180 | 7.595 (±0.016) | 390 | 566.3 (±8.3) | 9.539 (±0.110) | No |
| 4F1 | 180 | 7.496 (±0.041) | 330 | 461.9 (±0.1) | 9.068 (±0.053) | No |
| 5D1 | 210 | 7.817 (±0.051) | 450 | 568.0 (±8.4) | 9.770 (±0.053) | No |
| 4C1 | 150 | 6.924 (±0.009) | 330 | 536.3 (±0.7) | 8.746 (±0.025) | Yes (1.0 hr) |
| 4B1 | 180 | 7.514 (±0.046) | 300 | 663.6 (±3.1) | 8.705 (±0.044) | No |
| 3H1 | 210 | 7.801 (±0.030) | 300 | 366.0 (±9.0) | 8.705 (±0.004) | No |
| 5F1 | 150 | 7.192 (±0.004) | 360 | 583.7 (±37.2) | 9.295 (±0.084) | No |
| 1E1 | 150 | 6.897 (±0.033) | 360 | 806.3 (±13.1) | 9.058 (±0.079) | Yes (1.0 hr) |
| 3B1 | 180 | 7.702 (±0.007) | 300 | 540.8 (±0.1) | 8.916 (±0.004) | No |
| 4E1 | 150 | 7.272 (±0.012) | 390 | 749.7 (±19.1) | 9.592 (±0.064) | No |
| 2A1 | 180 | 7.491 (±0.093) | 390 | 631.9 (±1.8) | 9.597 (±0.044) | No |
| 3E1 | 180 | 7.459 (±0.049) | 330 | 490.8 (±2.7) | 9.038 (±0.020) | No |
| 2B1 | 150 | 6.896 (±0.034) | 360 | 741.6 (±19.2) | 9.008 (±0.056) | Yes (1.0 hr) |
| 5A1 | 180 | 7.544 (±0.077) | 270 | 372.2 (±11.1) | 8.344 (±0.070) | No |
| 2A2 | 180 | 7.487 (±0.068) | 330 | 575.0 (±12.0) | 9.045 (±0.045) | No |
| 4D1 | 180 | 7.501 (±0.051) | 330 | 651.5 (±16.5) | 9.022 (±0.062) | No |
| 1H2 | 180 | 7.615 (±0.007) | 330 | 518.3 (±0.7) | 9.061 (±0.015) | No |
| 3C1 | 210 | 7.807 (±0.041) | 300 | 767.5 (±11.0) | 8.732 (±0.044) | No |
| 2H1 | 180 | 7.535 (±0.014) | 300 | 684.0 (±4.7) | 8.756 (±0.001) | No |
| 5B1 | 180 | 7.578 (±0.012) | 300 | 461.0 (±0.6) | 8.702 (±0.001) | No |
| 1H1 | 180 | 7.420 (±0.005) | 390 | 622.6 (±10.4) | 9.579 (±0.007) | Yes (0.5 hr) |
| Mean | 175 (±4.0) | 7.430 (±0.038) | 340 (±9.3) | 590.4 (±24.5) | 9.033 (±0.093) | -- |
On average, mean and maximum mean bioluminescence increased 2.5-fold in the derived lines relative to the ancestor. All derived lines increased in mean and maximum mean bioluminescence as a result of adapting to novel host E. tasmanica (Fig. 7B); however, modifications of other traits suggest light intensity (i.e., power or wattage, photons emitted per cell per second) was not the only important factor influencing the evolution of bioluminescence. Population growth and quorum sensing apparatus may also have been affected. Some lineages challenged with a novel ET host developed a lag phase, a feature reminiscent of ET symbiont convergence (Tables 1A & 1B). Perhaps the ET lag phase is a cost or tradeoff associated with elevated photon production. Moreover, the time point that the evolved lines first ignited occurred earlier and at lower cell densities to ancestral V. fischeri ES114 and unevolved V. fischeri JRM200, while maximal light output occurred later at higher cell densities. Evidently, polymorphisms exist for traits affecting population growth, bioluminescence, and quorum sensing (Fig. 7B and Table 1B), as the various lineages are different from each other as well as from the ancestral state, perhaps due to differences in autoinducer sensitivity. As with motility, biofilm formation, and carbon sources demonstrating hyperpolymorphic utilization, bioluminescence displays convergent evolution to ET wild isolates yet simultaneously exhibits ecological diversification among the various lineages.
DISCUSSION
Convergent Evolution
Convergent evolution implies axenic squid light organs subject colonizing V. fischeri to similar selection pressures, and multifarious genetic changes exist to produce the same phenotypic solutions necessary for the symbionts to successfully respond to the evolutionary challenges imposed by a host [52]. Motility plays an integral role in the colonization of sepiolid squid by V. fischeri [53] and allows host-associated bacteria to reach the destination and surface desired for further colonization or attachment. Motility results suggest that adaptation to novel host E. tasmanica by V. fischeri JRM200 mandated first a decrease in motility and then a subsequent boost in biofilm formation. The latter may not have been possible until the first occurred due genetic constraints or evolutionary contingency (i.e., biofilm potential does not increase until mutations decreasing motility happens first) [54], or decreased motility may have been the more immediately important and necessary evolutionary change to adjust to a new host, with a surge in biofilm capacity coming later as a reinforcing adaptation. Interestingly enough, the realization that V. fischeri did not show statistically meaningful host adaptation to E. tasmanica until 400 generations, yet motility decreased by a significantly different amount by 100 generations [7]. Like motility, biofilm formation is also indispensable in squid colonization for V. fischeri [55, 56]. Motility and biofilms from wild symbiont populations in Euprymna squid hosts suggest convergent evolution between experimentally evolved lines in E. tasmanica and natural ET V. fischeri (Figs. 1A–1D & Figs. 2A–2D). ET V. fischeri form more prolific biofilms and swarm less than ES V. fischeri, while ES V. fischeri produce more restricted biofilms and move faster (Figs. 1A & 2A). Perhaps symbionts in E. tasmanica are selected for better host tissue attachment and elevated stress tolerance (more sticky and hardy), while those in E. scolopes are evolutionarily honed for chemotaxis (more nimble locomotion and elevated nutrient sensitivity). Biofilm formation is positively correlated in many host-associated vibrios with colonization potential, immunity avoidance, and eukaryotic cell attachment in hosts [57–59]. Biofilms are known to increase bacterial survival against environmental stress [58–61].
The artificial variation generated from the experimentally evolved lines is congruent with ET natural variation, as V. fischeri “domesticated” in E. tasmanica appear more phenotypically similar to field ET V. fischeri in motility and biofilm production and less like their original ES V. fischeri. Figure 4B suggests a threshold event or critical period was reached by 400 generations, since a sharp and sudden expanse in biofilm growth precipitously arises across many of the lineages in different media in a manner suggestive of second messenger signaling, signal transduction cascades, and quorum sensing autoinducers [62]. The decrease in motility is more progressive and continued throughout the 500 generation time period. Not only was convergent evolution observed in biofilm development and motility between the derived lines and wild isolates, but an inverse relationship was also noted. The inverse relationship between biofilm development and motility is known to have a biochemical basis within the genus Vibrio, specifically the second messenger cyclic diguanylate (c-di-GMP) [56, 59] and is a topic for future research. Generally, high intracellular concentrations of second messenger c-di-GMP correlate with increased biofilm formation, while lower quantities are associated with elevated motility (e.g., V. cholerae) [63]. Inverse relationships between motility and sessility in microbial lifestyles along a “c-di-GMP” continuum within hosts is an intriguing topic to partially explain competitive dominance and resulting tradeoffs observed in Vibrio symbionts colonizing Euprymna [9, 10]. Possibly, c-di-GMP intracellular pool levels as a “host” point of selection may be dissipated by sympatry of several Sepiola host species [1], not permitting for “c-di-GMP” ecological specialization for swarmer or biofilm ecotypes due to more homogenization of host environments in a geographical area. A scattering of few mutations affecting biofilm gene expression and c-di-GMP regulation drive convergent evolution and adaptive radiation in Pseudomonas fluorescens in structured microcosms, while simultaneously provoking astounding diversity [64, 65].
As with motility and biofilm formation, some convergent evolution appears to be occurring with carbon metabolism for substrates characteristic of evolutionary fluid allocation and redeployment in the V. fischeri physiological repertoire, when weighted against wild symbiont light organ populations from field-caught E. tasmanica and E. scolopes specimens (Figs. 4A, 4B, 6A, & 6B) [37–40, 44, 45]. Analyzing wild V. fischeri isolates from Euprymna squid hosts provides compelling evidence that citric acid and sucrose metabolic capabilities are abandoned in favor of L-glutamic acid, β-methyl-D-glucoside, D-trehalose, α-ketoglutaric acid, and D-serine for an ET lifestyle. This is a trend observed in the twenty-four lines originally retrieved from E. scolopes and experimentally adapted to novel host E. tasmanica. The functions of these carbon sources in bacteria, the genus Vibrio, and in sepiolid squid-V. fischeri mutualisms have not been fully elucidated. D-amino acids are largely restricted to peptidoglycan and teichoic acid in the bacterial cell wall [66], but D-serine could serve as an electron donor with appropriate isomerase function (e.g., mutase, racemase, or epimerase activity), as has been identified in V. cholerae [67]. β-methyl-D-glucoside may serve as an alternative nutrient for bacteria when rapidly metabolizable and more energy efficient catabolites are unavailable [68], as is true for all less preferred carbon sources—but still utilized by V. fischeri on the Biolog GN2 microplate. β-methyl-D-glucoside utilization has been implicated in chemotaxis, motility, phosphorylation by the phosphotransferase system, and rpoB mutations affecting RNA polymerase, regulatory proteins, regulatory RNA three-dimensional structures, and translation initiation/termination complexes [69]. β-methyl-D-glucoside may also be employed in membrane-derived oligosaccharide synthesis and metabolism of cellulose [69, 70], a polymer known to be a component of V. fischeri biofilms [56]. D-trehalose may also function in protection against osmotic stresses [71].
L-glutamic acid and citric acid increase V. cholerae host colonization potential in humans through the phosphotransacetylase-acetate kinase pathway (i.e., acetate switch) [72, 73], which drives ATP generation, recycles acetyl-CoA when respiration and central metabolism are backlogged, and post-translational regulation of proteins, illustrating the effect carbon sources can have especially on animal host colonization when the substrates themselves are main metabolites easily shuttled or shunted into alternate biochemical pathways (The acetate switch is present in V. fischeri [74], but acetic acid did exhibit ET convergence.) [72]. L-glutamic acid has also been reported to aid bacterial growth during iron limitation [75], which is noteworthy considering the squid light organ is low in available iron [76]. Furthermore, α-ketoglutaric acid is known to function as a substrate for the production of bacterial siderophores to assist iron acquisition in Vibrio [77]. α-ketoglutaric acid is also believed to possess value against oxidative stress [78], and the squid light organ is known to initiate a respiratory burst via innate host immunity that produces toxic oxygen species [79]. Perhaps the convergent evolution depicted in biofilm formation and these carbon sources are related, either to exploit new resources or increase resistance against unaccustomed stressors present in a novel host. Enhancing colonization and persistence in an unfamiliar host are other possibilities. How alternate carbon sources may be used by V. fischeri to manage against various host stresses (e.g., tenacious immune defenses) is a stimulating prospect for future study. For instance, amino acids may be more useful than monosaccharides for some cells against osmotic stress, while the former might be more beneficial to regulate high pH stress if nitrogen is not limiting (otherwise amino sugars such N-Acetyl-D-glucosamine could be used if available) [80]. Although not relevant to the squid light organ, carbon source is known to affect bacterial susceptibility to heavy metal toxicity, including V. fischeri [81]. Convergent evolution coupling metabolism and stress together has been described in mutualisms between invertebrate hosts and microbial symbionts [82, 83].
For bioluminescence, the derived lines displayed convergence to ET wild isolates (Figs. 7A & 7B; Tables 1A & 1B), implying light emission may be associated with biofilm formation, motility, and the convergent carbon sources. luxU links bioluminescence to biofilm formation in V. fischeri [84], and rpoN controls motility, biofilm, luminescence, and squid colonization ability [85]. Bioluminescence may be tied to biofilm formation, motility, and central metabolism through integrated circuitries involving quorum sensing and c-di-GMP second messenger signaling [86], which is consistent with the simultaneous increase in luminosity, biofilm formation, and lower motility. Relative to the ancestor, light emission occurred earlier and at a lower cell density for the derived lines, while maximal light output ensued later and at a higher cell density (Table 1B). Early quorum sensing at low cell densities and late quorum sensing at high cell densities implicate the AinS and Lux signal transduction relays in V. fischeri, respectively [87]. AinS quorum sensing has also been associated with the acetate switch [74]. Thus, evidence exists adaptation to E. tasmanica and conceivably to novel hosts in general involves selection at multiple layers of cell signaling (e.g., GGDEF, EAL, and PilZ proteins), which could account for appearance of a lag phase [14, 88]. Mutations affecting AinS quorum sensing is known to affect growth curves [89]. Previous efforts failed to find bioluminescence differences among the evolved lines within E. tasmanica, and several replicate animals were not examined within a single evolved line (e.g., using n=3 squid hosts with 3B1 instead of one animal each with 3B1, 5B1, and 4F1) [7]. At the time of study, there were insufficient E. tasmanica hatchlings available for these experiments. However, derived lines that are significantly brighter at 500 generations relative to ancestral V. fischeri ES114 and unevolved V. fischeri JRM200 have recently been observed in E. tasmanica (unpublished data). Prior investigation showed the derived lines were more dim in the ancestral host E. scolopes, giving off less photons per log10 (CFUs/mL) mL−1 [7]. E. scolopes hatchlings and adults are smaller (with concomitantly smaller light organs as well) than E. tasmanica thoughout their entire life cycles. Therefore, E. scolopes individual hosts carry one to two orders of magnitude less symbionts than E. tasmanica, regardless of the ontogenetic stage. Hence, tradeoffs may now exist in the ancestral host E. scolopes where V. fischeri cell densities and quorum sensing are mistimed for motility, biofilm formation, and bioluminescence due to adaptation of the derived lines to Australian E. tasmanica, since symbionts can no longer reach the proper population levels in the Hawaiian squid at the appropriate times of host development, as the symbiont and host life histories are no longer accurately synchronized.
Diversification
Diversifying evolution or radiation predominates over convergent evolution in morphospace when profuse phenotypes prevail to be evolutionarily fit or successful to a particular selection pressure. A multitude of mutations to attain each of these prosperous possibilities in a complex, heterogeneous environment with vacant niches provide ecological opportunity, an attribute characteristic of island evolution, including Galapagos finches, Hawaiian silversword alliance, Hawaiian honeycreepers, Anolis lizards, and cichlids in their lake habitat islands [90–92]. Aside from the carbon sources displaying convergent evolution, the remaining carbon sources metabolizable by V. fischeri wild isolates depicted a general polymorphism, a tendency also reflected in the derived lines (representative examples in Figs. 3A–3C; 5A–5C). These polymorphic carbon sources are being used distinctly and variedly, both individually and as an assortment. These results are congruent with other microbial experimental evolution studies [52, 93], including with E. coli experiments with less than 800 generations of evolution in homogenous and unstructured environments [94]. This increased “domesticated” variation may be reflective of diversifying selection in the squid light organ with incipient resource partitioning and changes in nutrient specificities toward carbon resources, perhaps even as part of cross-feeding or syntrophy between other V. fischeri ecotypes within the light organ of the same squid host individual [91, 94]. The implication of these results in nature are profound. Euprymna squid can live up to a year, which amounts to 1,500–2,000 V. fischeri generations [7]. Squid light organs are microcosms full of convoluted chasms with tremendous physical and biochemical complexity and heterogeneity [95], and a single V. fischeri clone has immense opportunity to evolve cross-feeding with either other V. fischeri subtypes or host cells within a host lifetime. The squid light organ microenvironment varies in spatial and temporal carbon source composition available to V. fischeri cells (e.g., N-acetyl-D-glucosamine at night and glycerol in the morning) [42]. Moreover, within the light organ of a single individual squid host, different V. fischeri subtypes will specialize in where they will settle and reside, with little mixing of individual V. fischeri variants within the light organ symbiont population despite the disturbance imposed by daily venting [42, 96]. In turn, this dissimilarity in the spatial localization and structured regional occupancy of the light organ by V. fischeri cells leads to differential gene expression in the symbionts, which can be a primer for the evolution of ecological differentiation.
An imperative realization is biofilm formation, motility, convergent carbon sources, and bioluminescence are also exemplifying ecological diversification. Despite the manifiestation in convergence to wild ET isolates in these traits, the evolved lines are still becoming divergent from one another. Other laboratories have also proved convergent evolution facilitates ecological diversification in separate lineages, including on other carbon sources [52, 93]. Quorum sensing and biofilms beget diversity, including the modification of population growth (e.g., lag phase changes) [97, 98]. Research with V. cholerae verifies alternate carbon sources can be incorporated or substituted into the composition of a biofilm to access new ecological niches [99, 100]. Intriguingly, bioluminescence (i.e., quorum sensing) and trans genetic regulation of the lux operon could function as a generator for V. fischeri adaptive radiation inside this symbiont’s animal hosts, as cAMP receptor protein (a transcriptional regulator) may have a continuum of graded possible effects on lux operon gene expression due to the presence of different carbon sources in various light organ microenvironments within sepiolid squids and monocentrid fishes [62]. A captivating possibility is the utilization of the sepiolid squid-Vibrio symbiosis to investigate evolvability and versatility in V. fischeri. V. fischeri possesses three biofilm gene clusters, syp, vpsII like, and the cellulose operon, yielding over 30 genes that can be independently modified to stimulate biofilm niche differentiation to exploit squid light organ microenvironments in novel ways [101]. This evolutionary process is quite analogous to the customization of the myriad bony elements in the head region of cichlids [102]. Additionally, modularity (as seen in convergent carbon sources, Figs. 4A, 4B, 6A, & 6B) increases the evolutionary diversification potential of a lineage, including for adaptive radiation [103, 104].
Squids as Host Habitat Islands
The use of biogeography theory to characterize microbial diversity has been previously applied, including mutualisms, pathogen-host interactions, and microcosms [105–110]. Axenic sepiolid squid hatchlings, first emanating from their eggs, essentially serve as sterile and mobile volcanic islands (i.e., host habitat islands) that subject their bioluminescent symbionts to severe genetic bottlenecks during colonization [7, 106, 108, 111, 112]. Only six to twelve V. fischeri cells of a 100–103 CFUs/mL seawater inoculum initiate symbiosis in an animal that ultimately reach an adult light organ carrying capacity of 108–1011 cells per host [113], providing pioneer V. fischeri colonizers with ecological release from the semi-starvation frequently encountered during the oceanic free-living phase. Such episodes of genetic bottlenecks with ensuing abundant proliferation and population recovery are called “founder flushes” [114], which provide opportunities for population movements across valleys via genetic drift to alternative adaptive peaks [90] and facilitate genetic revolutions, unique adaptations, founder effect evolution not normally possible, and novel independent evolutionary trajectories into vacant niches [e.g., island syndromes such as gigantism in the genera Geochelone and Aepyornis, nanism in Mammuthus exilis, and evolution of woody lifestyles by herbaceous plants] [90, 101, 115–118]. Founder flushes have been documented for bacteria, including V. cholerae and Helicobacter pylori, and are of significance in epidemiology [119]. Within the squid host, founder flushes lead to expanding V. fischeri niche breadth and ecological diversification (e.g., alternate metabolic utilization) from initial tight symbiont bottlenecks containing low genetic diversity, as has been observed in heterogeneous and complex microcosm environments with P. fluorescens [91, 120]. Moreover, the sepiolid squid light organ is an immensely specialized and complex structure [95], where V. fischeri populations can undergo ecological differentiation over the lifetime of the cephalopod host [7, 90, 101]. These conclusions are consistent with population genetics conducted with ES, ET, and Sepiola wild isolates of V. fischeri from sepiolid squid hosts through vast scales of space encompassing the world’s oceans and time spanning 20,000 generations of symbiont evolution [7, 33, 121, 122]. The presence of phenotypic convergent evolution and ecological diversification of V. fischeri serially passaged through E. tasmanica in motility, biofilm formation, metabolism, and bioluminescence when compared to wild isolates from this animal indicate host adaptive landscapes that have both smooth and rugged, multi-peaked topographies [123, 124], providing this bacterium with tremendous ecological opportunities to exploit a vast N-dimensional niche hypervolume space within animal hosts [125]. Innumerable evolutionary trajectories and adaptive peaks are available in the fitness landscape of a novel host. Simply put, there are many ways V. fischeri can successfully colonize a squid, permitting extensive specialization, resource partitioning, and genetically distinct ecotype subpopulations to crystallize within an animal host—in essence adaptive radiation [120, 126, 127].
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank the staff at Kewalo Marine Laboratory and the Sydney Institute of Marine Sciences for help with collecting and maintaining squids prior to shipment. F. Rivera was supported by the NMSU Minority for Access Careers program (NIH GM07667-35). W. Soto was supported by the NIH RISE program at NMSU (NIH NIGMS R25GM061222. This work was supported by NIH NIAID 1SC1AI081659, NIH NIAIDS 3SC1AI081659-02S1, and NSF IOS 074498 to M.K.N.
REFERENCES
- 1.Nishiguchi MK. Temperature affects species distribution in symbiotic populations of Vibrio spp. Applied and Environmental Microbiology. 2000;66:3550–3555. doi: 10.1128/aem.66.8.3550-3555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones BW, Nishiguchi MK. Counterillumination in the bobtail squid, Euprymna scolopes (Mollusca: Cephalopoda) Marine Biology. 2004;144:1151–1155. [Google Scholar]
- 3.Graf J, Ruby EG. Host-derived amino acids support the proliferation of symbiotic bacteria. Proceedings of the National Academy of Sciences. 1998;95:1818–1822. doi: 10.1073/pnas.95.4.1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McFall-Ngai MJ, Ruby EG. Symbiont recognition and subsequent morphogenesis as early events in an animal-bacterial mutualism. Science. 1991;254:1491–1494. doi: 10.1126/science.1962208. [DOI] [PubMed] [Google Scholar]
- 5.Nyholm SV, Nishiguchi MK. The evolutionary ecology of a sepiolid squid-Vibrio association: from cell to environment. Vie Et Milieu-Life and Environment. 2008;58:175–184. [PMC free article] [PubMed] [Google Scholar]
- 6.Ruby EG. Lessons from a cooperative, bacterial-animal association: the Vibrio fischeri-Euprymna scolopes light organ symbiosis. Annual Review of Microbiology. 1996;50:591–624. doi: 10.1146/annurev.micro.50.1.591. [DOI] [PubMed] [Google Scholar]
- 7.Soto W, Punke EB, Nishiguchi MK. Evolutionary perspectives in a mutualism of sepiolid squid and bioluminescent bacteria: combined usage of microbial experimental evolution and temporal population genetics. Evolution. 2012;66:1308–1321. doi: 10.1111/j.1558-5646.2011.01547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishiguchi MK, Nair VS. Evolution of symbiosis in the Vibrionaceae: a combined approach using molecules and physiology. International Journal of Systematics and Evolutionary Microbiology. 2003;53:2019–2026. doi: 10.1099/ijs.0.02792-0. [DOI] [PubMed] [Google Scholar]
- 9.Nishiguchi MK. Host recognition is responsible for symbiont composition in environmentally transmitted symbiosis. Microbial Ecology. 2002;44:10–18. doi: 10.1007/BF03036870. [DOI] [PubMed] [Google Scholar]
- 10.Nishiguchi MK, Ruby EG, McFall-Ngai MJ. Competitive dominance during colonization is an indicator of coevolution in an animal-bacterial symbiosis. Applied and Environmental Microbiology. 1998;64:3209–3213. doi: 10.1128/aem.64.9.3209-3213.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bello G. A key for the identification of the Mediterranean sepiolids (Mollusca: Cephalopoda) In: Boletzky Sv., editor. Mediterranean Sepiolidae. vol. 16. Monaco, Monaco: Bulletin de L’Institute oceanographiique; 1995. pp. 41–55. [Google Scholar]
- 12.O'Shea TM, Klein AH, Geszvain K, Wolfe AJ, Visick KL. Diguanylate cyclases control magnesium-dependent motility of Vibrio fischeri. Journal of Bacteriology. 2006;188:8196–8205. doi: 10.1128/JB.00728-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCarter ML. Bacterial acrobatics on a surface: swirling packs, collisions, and reversals during swarming. Journal of Bacteriology. 2010;192:3246–3248. doi: 10.1128/JB.00434-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolfe AJ, Visick KL. Get the message out: cyclic-di-GMP regulates multiple levels of flagellum-based motility. Journal of Bacteriology. 2008;190:463–475. doi: 10.1128/JB.01418-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daniels R, Vanderleyden J, Michiels J. Quorum sensing and swarming migration in bacteria. FEMS Microbiology Reviews. 2004;28:261–289. doi: 10.1016/j.femsre.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 16.Kim M-Y, Park R-Y, Choi M-H, Sun H-Y, Kim C-M, Kim S-Y, Rhee J-H, Shin S-H. Swarming differentiation of Vibrio vulnificus downregulates the expression of the vvhBA hemolysin gene via the LuxS quorum-sensing system. The Journal of Microbiology. 2006;44:226–232. [PubMed] [Google Scholar]
- 17.Trimble MJ, McCarter LL. Bis-(3'–5')-cyclic dimeric GMP-linked quorum sensing controls swarming in Vibrio parahaemolyticus. Proceedings of the National Academy of Sciences. 2011;108:18079–18084. doi: 10.1073/pnas.1113790108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCarter ML. Dual flagellar systems enable motility under different circumstances. Journal of Molecular Microbiology and Biotechnology. 2004;7:18–29. doi: 10.1159/000077866. [DOI] [PubMed] [Google Scholar]
- 19.McCarter LL. Polar flagellar motility of the Vibrionaceae. Microbiology and Molecular Biology Reviews. 2001;65:445–462. doi: 10.1128/MMBR.65.3.445-462.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merino S, Shaw JG, Tomás JM. Bacterial lateral flagella: an inducible flagella system. FEMS Microbiology Letters. 2006;263:127–135. doi: 10.1111/j.1574-6968.2006.00403.x. [DOI] [PubMed] [Google Scholar]
- 21.Merino S, Tomás JM. Lateral flagella systems. In: Jarrell KF, editor. Pili and Flagella: Current Research and Future Trends. Norfolk, UK: Caister Academic Press; 2009. [Google Scholar]
- 22.Ariyakumar DS, Nishiguchi MK. Characterization of two host-specific genes, mannose-sensitive hemagglutinin (mshA) and uridyl phosphate dehydrogenase (UDPDH) that are involved in the Vibrio fischeri-Euprymna tasmanica mutualism. FEMS Microbiology Letters. 2009;299:65–73. doi: 10.1111/j.1574-6968.2009.01732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nealson KH. Isolation, identification, and manipulation of luminous bacteria. Methods in Enzymology. 1978;57:153–165. [Google Scholar]
- 24.McCann J, Stabb EV, Millikan DS, Ruby EG. Population dynamics of Vibrio fischeri during infection of Euprymna scolopes. Applied and Environmental Microbiology. 2003;69:5928–5934. doi: 10.1128/AEM.69.10.5928-5934.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soto W, Gutierrez J, Remmenga MD, Nishiguchi MK. Salinity and temperature effects on physiological responses of Vibrio fischeri from diverse ecological niches. Microbial Ecology. 2009;57:140–150. doi: 10.1007/s00248-008-9412-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nair VS, Nishiguchi MK. Vibrio fischeri strains exhibit differential biological properties both in their free-living and symbiotic niches. Vie Et Milieu-Life and Environment. 2009;58:175–184. [Google Scholar]
- 27.Harshey RM. Bacterial motility on a surface: many ways to a common goal. Annual Review of Microbiology. 2003;57:249–273. doi: 10.1146/annurev.micro.57.030502.091014. [DOI] [PubMed] [Google Scholar]
- 28.Sokal RR, Rohlf FJ. Biometry. New York City, NY, USA: W.H. Freeman & Company; 1995. [Google Scholar]
- 29.Christensen GD, Simpson WA, Younger JJ, Baddour LM, Barrett FF, Melton DM, Beachey EH. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. Journal of Clinical Microbiology. 1985;22:996–1006. doi: 10.1128/jcm.22.6.996-1006.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bochner BR. Global phenotypic characterization of bacteria. FEMS Microbiology Letters. 2009;33:191–205. doi: 10.1111/j.1574-6976.2008.00149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bochner BR. Sleuthing out bacterial identities. Nature. 1989;339:157–158. doi: 10.1038/339157a0. [DOI] [PubMed] [Google Scholar]
- 32.Abdi H. The Greenhouse-Geisser correction. In: Salkind N, editor. Encyclopedia of Research Design. Thousand Oaks, CA, USA: SAGE Publications; 2010. [Google Scholar]
- 33.Jones BW, Lopez JE, Huttenburg J, Nishiguchi MK. Population structure between environmentally transmitted vibrios and bobtail squids using nested clade analysis. Molecular Ecology. 2006;15:4317–4329. doi: 10.1111/j.1365-294X.2006.03073.x. [DOI] [PubMed] [Google Scholar]
- 34.Elena SF, Lenski RE. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nature Reviews Genetics. 2003;4:457–469. doi: 10.1038/nrg1088. [DOI] [PubMed] [Google Scholar]
- 35.Hendrie MS, Hodgkiss W, Shewan JM. Proposal that Vibrio marinus (Russell 1891) Ford 1927 be amalgamated with Vibrio fischeri (Beijerinck 1889) Lehmann and Neumann 1896. International Journal of Systematic Bacteriology. 1971;21:217–221. [Google Scholar]
- 36.Ruby EG, Nealson KH. Symbiotic association of Photobacterium fischeri with the marine luminous fish Monocentris japonica : a model of symbiosis based on bacterial studies. Biological Bulletin. 1976;151:574–586. doi: 10.2307/1540507. [DOI] [PubMed] [Google Scholar]
- 37.Bryant TN, Lee JV, West PA, Colwell RR. Numerical classification of Vibrio and related genera. Journal of Applied Bacteriology. 1986;61:437–467. doi: 10.1111/j.1365-2672.1986.tb04308.x. [DOI] [PubMed] [Google Scholar]
- 38.Sawabe T, Sugimura I, Ohtsuka M, Nakano K, Tajima K, Ezura Y, Christen R. Vibrio halioticoli sp. nov., a non-motile alginolytic marine bacterium isolated from the gut of the abalone Haliotis discus hannai. International Journal of Systematic Bacteriology. 1998;48:573–580. doi: 10.1099/00207713-48-2-573. [DOI] [PubMed] [Google Scholar]
- 39.Alcaide E. Numerical taxonomy of Vibrionaceae isolated from cultured amberjack (Seriola dumerili) and surrounding water. Current Microbiology. 2003;46:184–189. doi: 10.1007/s00284-002-3826-2. [DOI] [PubMed] [Google Scholar]
- 40.Farmer JJ, III, Janda JM, Brenner DJ, Krieg NR, Staley JR. Vibrio. In: Brenner DJ, Krieg NR, Staley JR, editors. Bergey's Manual Systematic Bacteriology. New York, NY, USA: Springer-Verlag; 2005. pp. 494–546. [Google Scholar]
- 41.Kim H-J, Hyun E-K, Kim Y-S, Lee Y-J, Oh D-K. Characterization of an Agrobacterium tumefaciens d-psicose, 3-epimerase that converts, d-fructose to d-psicose. Applied and Environmental Microbiology. 2006;72:981–985. doi: 10.1128/AEM.72.2.981-985.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wier AM, Nyholm SV, Mandel MJ, Massengo-Tiassé RP, Schaefer AL, Koroleva I, Splinter-BonDurant S, Brown B, Manzella L, Einat Snir E, Almabrazi H, Scheetz TE, de Fatima Bonaldo M, Casavant TL, Soares MB, Cronan JE, Reed JL, Ruby EG, McFall-Ngai MJ. Transcriptional patterns in both host and bacterium underlie a daily rhythm of anatomical and metabolic change in a beneficial symbiosis. Proceedings of the National Academy of Sciences. 2010;107:2259–2264. doi: 10.1073/pnas.0909712107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Visick KL, McFall-Ngai MJ. An exclusive contract: specificity in the Vibrio fischeri-Euprymna scolopes partnership. Journal of Bacteriology. 2000;182:1779–1787. doi: 10.1128/jb.182.7.1779-1787.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noguerola I, Blanch AR. Identification of Vibrio spp. with a set of dichotomous keys. Journal of Applied Microbiology. 2008;105:175–185. doi: 10.1111/j.1365-2672.2008.03730.x. [DOI] [PubMed] [Google Scholar]
- 45.Thompson FL, Iida T, Swings J. Biodiversity of Vibrios. Microbiolology and Molecular Biology Reviews. 2004;68:403–431. doi: 10.1128/MMBR.68.3.403-431.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vydryakova GA, Kirpichenko TV, Lifant'eva AA. Formation of aggregated structures by luminescent bacteria in the presence of carbohydrates. Microbiology. 2007;2:282–284. [PubMed] [Google Scholar]
- 47.Vydryakova GA. Carbohydrate specificity of lectins from luminous bacteria. Applied Biochemistry and Microbiology. 2006;42:364–368. [Google Scholar]
- 48.Keymer DP, Miller MC, Schoolnik GK, Boehm AB. Genomic and phenotypic diversity of coastal Vibrio cholerae strains is linked to environmental factors. Applied and Environmental Microbiology. 2007;73:3705–3714. doi: 10.1128/AEM.02736-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Albert MJ, Bhuiyan NA, Talukder KA, Kaisar A, Faruque ASG, Nahar S, Faruque SM, Ansaruzzaman M, Rahman M. Phenotypic and genotypic changes in Vibrio cholerae O139 Bengal. Journal of Clinical Microbiology. 1997;35:2588–2592. doi: 10.1128/jcm.35.10.2588-2592.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soto W, Lostroh CP, Nishiguchi MK. Physiological responses to stress in the Vibrionaceae. In: Seckback J, Grube M, editors. Cooperation and Stress in Biology: Joint Ventures in Biology. vol. 17. New York, NY, USA: Springer; 2010. pp. 407–426. [Google Scholar]
- 51.Visick KL, Foster J, Doino J, McFall-Ngai MJ, Ruby EG. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. Journal of Bacteriology. 2000;182:4578–4586. doi: 10.1128/jb.182.16.4578-4586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.MacLean RC, Bell G. Divergent evolution during an experimental adaptive radiation. Proceedings of the Royal Society B: Biological Sciences. 2003;270:1645–1650. doi: 10.1098/rspb.2003.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Millikan DS, Ruby EG. Alterations in Vibrio fischeri motility correlate with a delay in symbiosis initiation and are associated with additional symbiotic colonization defects. Applied and Environmental Microbiology. 2002;68:2519–2528. doi: 10.1128/AEM.68.5.2519-2528.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Travisano M, Mongold JA, Bennett AF, Lenski RE. Experimental tests of the roles of adaptation, chance, and history in evolution. Science. 1995;267:87–90. doi: 10.1126/science.7809610. [DOI] [PubMed] [Google Scholar]
- 55.Hussa EA, Darnell CL, Visick KL. RscS functions upstream of SypG to control the syp locus and biofilm formation in Vibrio fischeri. Journal of Bacteriology. 2008;190:4576–4583. doi: 10.1128/JB.00130-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yildiz FH, Visick KL. Vibrio biofilms: so much the same yet so different. Trends in Microbiology. 2009;17:109–118. doi: 10.1016/j.tim.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McDougald D, Kjelleberg S. Adaptive responses of vibrios. In: Thompson FL, Austin B, Swings J, editors. The Biology of Vibrios. Washington, D.C., USA: ASM Press; 2006. pp. 133–155. [Google Scholar]
- 58.Chavez-Dozal AA, Nishiguchi MK. Variation in biofilm formation among symbiotic and free-living strains of Vibrio fischeri. Journal of Basic Microbiology. 2011;51:452–458. doi: 10.1002/jobm.201000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chavez-Dozal AA, Hogan D, Gorman C, Quintanal-Villalonga A, Nishiguchi MK. Multiple Vibrio fischeri genes are involved in biofilm formation and host colonization. FEMS Microbiology Letters. 2012;81:562–573. doi: 10.1111/j.1574-6941.2012.01386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Landini P. Cross-talk mechanisms in biofilm formation and responses to environmental and physiological stress in Escherichia coli. Research in Microbiology. 2009;160:259–266. doi: 10.1016/j.resmic.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 61.Chavez-Dozal AA, Gorman C, Erken M, Steinberg PD, McDougald D, Nishiguchi MK. Predation response of Vibrio fischeri biofilms to bacterivorus protists/phagotrophic protozoa. Applied and Environmental Microbiology. 2013;79:553–558. doi: 10.1128/AEM.02710-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miyashiro T, Ruby EG. Shedding light on bioluminescence regulation in Vibrio fischeri. Molecular Microbiology. 2012;84:795–806. doi: 10.1111/j.1365-2958.2012.08065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jude BA, Taylor RK. Genetics of Vibrio cholerae colonization and motility. In: Faruque SM, Nair GB, editors. Vibrio cholerae Genomics and Molecular Biology. Norfolk, United Kingdom: Caister Academic Press; 2008. pp. 67–79. [Google Scholar]
- 64.McDonald MJ, Gehrig SM, Meintjes PL, Zhang X-X, Rainey PB. Adaptive divergence in experimental populations of Pseudomonas fluorescens IV. genetic constraints guide evolutionary trajectories in a parallel adaptive radiation. Genetics. 2009;183:1041–1053. doi: 10.1534/genetics.109.107110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.MacLean RC. Adaptive radiation in microbial microcosms. Journal of Evolutionary Biology. 2005;18:1376–1386. doi: 10.1111/j.1420-9101.2005.00931.x. [DOI] [PubMed] [Google Scholar]
- 66.Madigan MT, Martinko JM. Brock Biology of Microorganisms. Upper Saddle River, NJ, USA: Pearson Prentice Hall; 2006. [Google Scholar]
- 67.Lam H, Oh D-C, Cava F, Takacs C-N, Clardy J, de Pedro MA, Matthew K, Waldor MK. D-amino acids govern stationary phase cell wall remodeling in bacteria. Science. 2009;323:1552–1155. doi: 10.1126/science.1178123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tobisch S, Stulke J, Hecker M. Regulation of the lic operon of Bacillus subtilis and characterization of potential phosphorylation sites of the licR regulator protein by site-directed mutagenesis. Journal of Bacteriology. 1999;181:4995–5003. doi: 10.1128/jb.181.16.4995-5003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Perkins AE, Nicholson WL. Uncovering new metabolic capabilities of Bacillus subtilis using phenotype profiling of rifampin-resistant rpoB mutants. Journal of Bacteriology. 2008;190:807–814. doi: 10.1128/JB.00901-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kennedy EP. Membrane-derived oligosaccharides. In: Neidhardt FC, Ingraham JL, Low KB, Magasanik B, Shaechter M, Umbarger HE, editors. Escherichia coli and Salmonella typhimurium. vol. 1. Washington, D.C., USA: ASM Press; 1987. pp. 672–679. [Google Scholar]
- 71.DeLoney CR, Bartley TM, Visick KL. Role for phosphoglucomutase in Vibrio fischeri-Euprymna scolopes symbiosis. Journal of Bacteriology. 2002;184:5121–5129. doi: 10.1128/JB.184.18.5121-5129.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Minato Y, Fassio SR, Wolfe AJ, Hase CC. Central metabolism controls transcription of a virulence gene regulator in Vibrio cholerae. Microbiology. 2013;159:792–802. doi: 10.1099/mic.0.064865-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wolfe AJ. The acetate switch. Microbiology and Molecular Biology Reviews. 2005;69:12–50. doi: 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Studer SV, Mandel MJ, Ruby EG. AinS quorum sensing regulates the Vibrio fischeri acetate switch. Journal of Bacteriology. 2008;190:5915–5923. doi: 10.1128/JB.00148-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Filiatrault MJ, Stodghill PV, Bronstein PA, Moll S, Lindeberg M, Grills G, Schweitzer P, Wang W, Schroth GP, Luo S, Khrebtukova I, Yang Y, Thannhauser T, Butcher BG, Cartinhour S, Schneider DJ. Transcriptome analysis of Pseudomonas syringae identifies new genes, noncoding RNAs, and antisense activity. Journal of Bacteriology. 2010;192:2359–2372. doi: 10.1128/JB.01445-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Graf J, Ruby EG. Novel effects of a transposon insertion in the Vibrio fischeri glnD gene: defects in iron uptake and symbiotic persistence in addition to nitrogen utilization. Molecular Microbiology. 2000;37:168–179. doi: 10.1046/j.1365-2958.2000.01984.x. [DOI] [PubMed] [Google Scholar]
- 77.Kadi N, Challis GL. Siderophore biosynthesis: a substrate specificity assay for nonribosomal peptide synthetase-independent siderophore synthetases involving trapping of acyl-adenylate intermediates with hydroxylamine. Methods in Enzymology. 2009;458:432–457. doi: 10.1016/S0076-6879(09)04817-4. [DOI] [PubMed] [Google Scholar]
- 78.Mizunoe Y, Wai SN, Takade A, Yoshida S. Restoration of culturability of starvation-stressed and low-temperature-stressed Escherichia coli O157 cells by using H2O2-degrading compounds. Archives of Microbiology. 1999;172:63–67. doi: 10.1007/s002030050741. [DOI] [PubMed] [Google Scholar]
- 79.Ruby EG, McFall-Ngai MJ. Oxygen-utilizing reactions and symbiotic colonization of the squid light organ by Vibrio fischeri. Trends in Microbiology. 1999;7:414–420. doi: 10.1016/s0966-842x(99)01588-7. [DOI] [PubMed] [Google Scholar]
- 80.Jaenicke R. Enzymes under extremes of physical conditions. Annual Review of Biophysics and Bioengineering. 1981;1:1–67. doi: 10.1146/annurev.bb.10.060181.000245. [DOI] [PubMed] [Google Scholar]
- 81.Tekerlekopoulou AG, Tsiamis G, Dermou E, Siozios S, Bourtzis K, Vayenas DV. The effect of carbon source on microbial community structure and CR(VI) reduction rate. Biotechnology and Bioengineering. 2010;107:478–487. doi: 10.1002/bit.22837. [DOI] [PubMed] [Google Scholar]
- 82.Chaston J, Goodrich-Blair H. Common trends in mutualism revealed by model associations between invertebrates and bacteria. FEMS Microbiology Reviews. 2010;34:41–58. doi: 10.1111/j.1574-6976.2009.00193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fan L, Reynolds D, Liu M, Stark M, Kjelleberg S, Webster NS, Thomas T. Functional equivalence and evolutionary convergence in complex communities of microbial sponge symbionts. Proceedings of the National Academy of Sciences. 2012;109:E1878–E1887. doi: 10.1073/pnas.1203287109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ray VA, Visick KL. LuxU connects quorum sensing to biofilm formation in Vibrio fischeri. Molecular Microbiology. 2012;86:954–970. doi: 10.1111/mmi.12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wolfe AJ, Millikan DS, Campbell JM, Visick KL. Vibrio fischeri σ54 controls motility, biofilm formation, luminescence, and colonization. Applied and Environmental Microbiology. 2004;70:2520–2524. doi: 10.1128/AEM.70.4.2520-2524.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Srivastava D, Waters CM. A tangled web: regulatory connections between quorum sensing and cyclic di-gmp. Journal of Bacteriology. 2012;194:4485–4493. doi: 10.1128/JB.00379-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lupp C, Ruby EG. Vibrio fischeri LuxS and AinS: comparative study of two signal synthases. Journal of Bacteriology. 2004;186:3873–3881. doi: 10.1128/JB.186.12.3873-3881.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Visick KL. Layers of signaling in a bacterium-host association. Journal of Bacteriology. 2005;187:3603–3606. doi: 10.1128/JB.187.11.3603-3606.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lupp C, Ruby EG. Vibrio fischeri uses two quorum-sensing systems for the regulation of early and late colonization factors. Journal of Bacteriology. 2005;187:3620–3629. doi: 10.1128/JB.187.11.3620-3629.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Whittaker RJ, Fernandez-Palacios JM. Island Biogeography: Ecology, Evolution, and Conservation. Oxford, NY, USA: Oxford University Press; 2007. [Google Scholar]
- 91.Travisano M, Rainey PB. Studies of adaptive radiation using model microbial systems. American Naturalist. 2000;156:S35–S44. doi: 10.1086/303414. [DOI] [PubMed] [Google Scholar]
- 92.Rainey PB, Travisano M. Adaptive radiation in a heterogeneous environment. Nature. 1998;394:69–72. doi: 10.1038/27900. [DOI] [PubMed] [Google Scholar]
- 93.Ostrowski EA, Woods RJ, Lenski RE. The genetic basis of parallel and divergent phenotypic responses in evolving populations of Escherichia coli. Proceedings of the Royal Society B: Biological Sciences. 2008;275:277–284. doi: 10.1098/rspb.2007.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rosenzweig RF, Sharp RR, Treves DS, Adams J. Microbial evolution in a simple unstructured environment: genetic differentiation in Escherichia coli. Genetics. 1994;137:903–917. doi: 10.1093/genetics/137.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sycuro L, Ruby E, McFall-Ngai M. Confocal microscopy of the light organ crypts in juvenile Euprymna scolopes reveals their morphological complexity and dynamic function in symbiosis. Journal of Morphology. 2006;267:555–568. doi: 10.1002/jmor.10422. [DOI] [PubMed] [Google Scholar]
- 96.Dunn AK, Millikan DS, Adin DM, Bose JL, Stabb EV. New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Applied and Environmental Microbiology. 2006;72:802–810. doi: 10.1128/AEM.72.1.802-810.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ponciano JM, La H-J, Joyce P, Forney LJ. Evolution of diversity in spatially structured Escherichia coli populations. Journal of Bacteriology. 2009;75:6047–6054. doi: 10.1128/AEM.00063-09. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Milton DL. Quorum sensing in vibrios: complexity for diversification. International Journal of Medical Microbiology. 2006;296:61–71. doi: 10.1016/j.ijmm.2006.01.044. [DOI] [PubMed] [Google Scholar]
- 99.Chatterjee SN, Chaudhuri K. Lipopolysaccharides of Vibrio cholerae I. Physical and chemical characterization. Biochimica et Biophysica Acta. 2003;1639:65–79. doi: 10.1016/j.bbadis.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 100.Sozhamannan S, Yildiz FH. Diversity and genetic basis of polysaccharide biosynthesis in Vibrio cholerae. In: Bhattacharya SK, Ramamurthy T, editors. Epidemiological and Molecular Aspects On Cholera. New York, NY, USA: Springer; 2011. [Google Scholar]
- 101.Schluter D. Ecology of Adaptive Radiation. Oxford, NY: Oxford University Press; 2000. [Google Scholar]
- 102.Galis F, Metz JAJ. Why are there so many cichlid species? Trends in Ecology and Evolution. 1998;13:1–2. doi: 10.1016/s0169-5347(97)01239-1. [DOI] [PubMed] [Google Scholar]
- 103.Yang AS. Modularity, evolvability, and adaptive radiations: a comparison of the hemi- and holometabolous insects. Evolution and Development. 2001;3:59–72. doi: 10.1046/j.1525-142x.2001.003002059.x. [DOI] [PubMed] [Google Scholar]
- 104.Rainey PB, Cooper TF. Evolution of bacterial diversity and the origins of modularity. Research in Microbiology. 2004;155:370–375. doi: 10.1016/j.resmic.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 105.Dickerson JE, Robinson JR, Robinson JV. Microcosms as islands: a test of the MacArthur-Wilson equilibrium theory. Ecology. 1985;66:966–980. [Google Scholar]
- 106.Holt RD. A biogeographical and landscape perspective on within-host infection dynamics. In: Bell CR, Brylinsky M, Johnson-Green P, editors. Proceedings of the 8th International Symposium of Microbial Ecology. Halifax, Canada: Atlantic Canada Society for Microbial Ecology; 2000. pp. 583–588. [Google Scholar]
- 107.Papke RT, Ward DM. The importance of physical isolation to microbial diversification. FEMS Microbiology Letters. 2004;48:293–303. doi: 10.1016/j.femsec.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 108.Taylor MW, Schupp PJ, De Nys R, Kjelleberg S, Steinberg PD. Biogeography of bacteria associated with the marine sponge Cymbastela concentrica. Environmental Microbiology. 2005;7:419–433. doi: 10.1111/j.1462-2920.2004.00711.x. [DOI] [PubMed] [Google Scholar]
- 109.Green J, Bohannan B. Spatial scaling of microbial biodiversity. Trends in Ecology and Evolution. 2006;21:501–507. doi: 10.1016/j.tree.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 110.Ramette A, Tiedje JM. Biogeography: an emerging cornerstone for understanding prokaryotic diversity, ecology, and evolution. Microbial Ecology. 2007;53:197–207. doi: 10.1007/s00248-005-5010-2. [DOI] [PubMed] [Google Scholar]
- 111.Kuris AM, Blaustein AR, Alio JJ. Hosts as islands. American Naturalist. 1980;116:570–586. [Google Scholar]
- 112.Van der Gast CJ. Islands shaping thought in microbial ecology. Advances in Applied Microbiology. 2008;64:167–182. doi: 10.1016/S0065-2164(08)00406-1. [DOI] [PubMed] [Google Scholar]
- 113.Wollenberg MS, Ruby EG. Population structure of Vibrio fischeri within the light organs of Euprymna scolopes from two Oahu populations. Applied and Environmental Microbiology. 2009;75:193–202. doi: 10.1128/AEM.01792-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Slatkin M. In defense of founder-flush theories of speciation. The American Naturalist. 1996;147:493–505. [Google Scholar]
- 115.Agenbroad LD. New pygmy mammoth (Mammuthus exilis) localities and radiocarbon dates from San Miguel, Santa Rosa, and Santa Cruz Islands, California. In: Weigand PW, editor. Contributions to the Geology of the Northern Channel Islands Southern California. Bakersfield, CA, USA: American Association of Petroleum Geologists, Pacific Section; 1998. pp. 169–175. [Google Scholar]
- 116.Yoshida A, Fumihito A, Yamagishi S, Tanida K. Underground temperature change throughout a year measured at a coastal dune bearing sub-fossil egg shell fragments of the elephant bird (Aepyornis) in faux-cap, southern Madagascar. Journal of the Yamashina Institute for Ornithology. 2005;36:136–140. [Google Scholar]
- 117.Templeton AR. The reality and importance of founder speciation in evolution. BioEssays. 2008;30:470–479. doi: 10.1002/bies.20745. [DOI] [PubMed] [Google Scholar]
- 118.Barton NH, Charlesworth B. Genetic revolutions, founder effects, and speciation. Annual Review of Ecology, Evolution, and Systematics. 1984;15:133–164. [Google Scholar]
- 119.Garg P, Aydanian A, Smith D, Morris J, Glenn J, Nair GB, Stine OC. Molecular epidemiology of 0139 Vibrio cholerae: Mutation, lateral gene transfer, and founder flush. Emerging Infectious Diseases. 2003;9:810–814. doi: 10.3201/eid0907.020760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.MacLean RC, Bell G. Experimental adaptive radiation in Pseudomonas. American Naturalist. 2002;160:569–581. doi: 10.1086/342816. [DOI] [PubMed] [Google Scholar]
- 121.Zamborsky DJ, Nishiguchi MK. Phylogeographical patterns among sympatric populations of sepiolid squids and their Vibrio symbionts in the Mediterranean Sea. Applied and Environmental Microbiology. 2011;77:642–649. doi: 10.1128/AEM.02105-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Soto W. Evolutionary ecology of symbionts within free-living and host environments in the sepiolid squid-Vibrio symbiosis. Las Cruces, NM, USA: New Mexico State University; 2009. [Google Scholar]
- 123.Wright S. Character change, speciation, and the higher taxa. Evolution. 1982;36:427–443. doi: 10.1111/j.1558-5646.1982.tb05065.x. [DOI] [PubMed] [Google Scholar]
- 124.Colegrave N, Buckling A. Microbial experiments on adaptive landscapes. BioEssays. 2005;27:1167–1173. doi: 10.1002/bies.20292. [DOI] [PubMed] [Google Scholar]
- 125.Hutchinson GE. Concluding remarks. Cold Spring Harbor Symposia on Quantitative Biology. 1957;22:415–427. [Google Scholar]
- 126.Habets MGJL, Rozen DE, Hoekstra RF, de Visser JGM. The effect of population structure on the adaptive radiation of microbial populations evolving in spatially structured environments. Ecology Letters. 2006;9:1041–1048. doi: 10.1111/j.1461-0248.2006.00955.x. [DOI] [PubMed] [Google Scholar]
- 127.Hunt DE, David LA, Gevers D, Preheim SP, Alm EJ, Polz MF. Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science. 2008;320:1081–1085. doi: 10.1126/science.1157890. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.