Summary
A number of inducible yeast genes are targeted to the nuclear periphery upon transcriptional activation. However, when repressed again, the INO1 and GAL1 genes remain at the nuclear periphery for multiple generations. Retention at the nuclear periphery represents a novel type of transcriptional memory; the peripherally localized, recently repressed state of GAL1 is activated more rapidly than the nucleoplasmically localized long-term repressed state of GAL1. This rapid reactivation involves localization at the nuclear periphery, the SWI/SNF chromatin remodeling complex, the histone variant H2A.Z and the Gal1 protein itself. Here, I review what we have learned about this type of transcriptional memory in yeast, what remains to be resolved and the challenges associated with understanding such epigenetic phenomena.
Gene recruitment to the nuclear periphery
DNA is spatially organized within the nucleus. It has long been appreciated that certain parts of the genome localize at the nuclear periphery and associate with the nuclear envelope [1–4]. Peripheral localization has been proposed to promote transcriptional repression. Much of the DNA at the nuclear periphery is transcriptionally repressed; heterochromatin, centromeres, telomeres and chromatin insulators (which can block enhancer function) localize at the nuclear periphery [5]. Proximity to the nuclear envelope in Saccharomyces cerevisiae [6] and association with the mammalian nuclear lamina [7,8] promotes transcriptional silencing of some genes.
Recent work, however, shows that peripheral localization is not always repressive. Artificially tethering a reporter gene to the nuclear envelope that lacks any silencing elements to the nuclear envelope does not promote repression [6]. Furthermore, certain inducible yeast genes are targeted rapidly to the nuclear periphery upon activation [9–11••]. A number of additional inducible genes have now been shown to undergo recruitment to the nuclear periphery in yeast [12–15] and peripheral localization correlates with a physical association with the NPC [10,12,16]. Peripheral targeting promotes transcription [9,13,17,18] and may be important for post-transcriptional events such as mRNA processing, mRNA export or translation [10,12].
The mechanism of gene recruitment
Physical interaction of genes with the NPC in vivo has been observed using different methods [10,12,16,19]. Furthermore, targeting of genes to the nuclear periphery requires NPC components, the SAGA histone acetyltransferase complex, the transcription-mRNA export complex TREX2 and the export receptor Mex67 [14,18–20]. These observations suggest that relocalization of genes to the nuclear periphery represents a “tethering” of genes to the NPC. Consistent with this interpretation, in mutant strains lacking in which NPCs cluster together into plaques, there is a significant increase in the colocalization of the HXK1 gene with clustered NPCs upon transcriptional activation [13]. However, the fraction of cells in which HXK1 colocalized with clustered NPCs (17% of the cells in the population) is still much lower than the fraction of the cells in which HXK1 is localized at the nuclear periphery (>85% of the cells in the population). Furthermore, it is unclear if the ~150 NPCs in the typical haploid nucleus in yeast would be able to accommodate both a large number of transcriptionally active genes (as suggested by the NPC association data in refs 11, 12 and 16) and nucleo-cytoplasmic transport without interference. The current model for gene recruitment to the NPC, on which I will build for the purposes of this review, is consistent with both the in vivo physical interaction of genes with the NPC and the genetic requirements for various NPC and NPC-associated proteins for gene recruitment. However, a direct physical interaction between genes and the NPC has not been demonstrated and it remains possible that the site to which genes are recruited is actually the nuclear envelope, a platform of the filamentous Mlp/Tpr proteins or other, unidentified structures at the nuclear periphery.
Gene recruitment of at least one gene to the nuclear periphery also requires the 3′ UTR [13]. Furthermore, the association of certain genes with NPC components, as measured by chromatin immunoprecipitation, is sensitive to RNase treatment [12]. The connection between the NPC, RNA elements important for mRNA export and the peripheral relocalization of genes suggested that relocalization might represent a consequence of transcription. In other words, relocalization of genes to the nuclear periphery might be mediated by a bridging interaction between chromosomal DNA, the mRNA, export factors and the NPC. If so, then relocalization to the nuclear periphery might simply represent an emergent property of highly expressed genes, rather than a specific targeting event.
Several results argue against this interpretation. From experiments using a temperature-sensitive allele of RNA polymerase to block transcription, it became clear that transcription is not essential for either the interaction of GAL1 with the NPC [16] or for relocalization of INO1 to the nuclear periphery [11••]. Furthermore, association of the Mex67 mRNA export receptor with the GAL2 gene was independent of RNA, suggesting that the export receptor can interact with DNA or chromatin directly [14]. Also, both INO1 and GAL1 remain at the nuclear periphery for generations after transcription is repressed (see below; ref. [11••]). Finally, we have recently identified an 8 basepair element in the INO1 promoter that functions as a “DNA zip code” to target an ectopic locus to the nuclear periphery [18]. Therefore, although the mRNA may play important roles in the localization of genes with the NPC after transcription is initiated (see below), transcription-independent mechanisms of gene relocalization occur and relocalization to the nuclear periphery represents an active and specific targeting mechanism.
Transcriptional memory at the nuclear periphery
In addition to a role in transcriptional activation, we have identified a role for peripheral targeting in the rapid reactivation of recently repressed genes. When cells are shifted from activating to repressing conditions, both the INO1 and GAL1 genes remain at the nuclear periphery through multiple cell divisions [11••]. Retention of GAL1 at the nuclear periphery is very stable, lasting greater than seven generations. This suggested that peripheral localization might represent a novel epigenetic state that reflects previous transcription. We hypothesized that this recently repressed “memory” state might be functionally different from the long-term repressed state. Indeed, we found that the reactivation of the GAL1 gene, even after seven generations of repression, was much faster than the initial activation of the GAL1 gene [11••]. This suggested that cells have cellular and molecular mechanisms to mark previously expressed genes and to promote their reactivation, a phenomenon I call adaptive transcriptional memory.
Concurrent work independently demonstrated that previous transcription of GAL1, GAL7 and GAL10 promoted their rapid reactivation [21••]. Rapid reactivation of GAL1 required as little as one hour of activation and persisted through cell division. The rapid reactivation of the GAL1 gene was not due to the lingering association of RNA polymerase II, the TATA binding protein (which binds upstream of preinitiation complex formation) or coactivators such as Mediator (which interacts with RNA Polymerase II to promote transcription), the SWI/SNF chromatin remodeling complex or the SAGA histone acetyltransferase [21••]. Therefore, recently repressed GAL1 is not primed through a lingering association with a stalled RNA polymerase II, but through faster recruitment of RNA polymerase II upon reactivation.
Defects in mRNA processing can also lead to post-transcriptional targeting of genes to the nuclear periphery. GAL1 promoter-driven reporter genes that produce mRNAs that are improperly polyadenylated remain associated with post-transcriptional RNA “dots” at the nuclear periphery [22]. Localization at the nuclear periphery is maintained for approximately 1 hour after transcription is repressed [22,24•], increases with defects in mRNA processing [22] and requires both an mRNA export factor (TREX 2; ref. [23•]) and the nuclear exosome, an ribonuclease that degrades aberrant mRNAs in the nucleus [24•]. In mutants lacking the THO complex (which coordinates mRNA processing with export), an intermediate in the mRNA processing pathway accumulates associated chromatin [25•]. This intermediate also appears as RNA dots, requires the nuclear exosome for its formation and leads to the relocalization of genes to the nuclear pore complex [25•]. It is unclear how this mechanism of peripheral retention relates to the epigenetic peripheral retention observed for the endogenous, wild type GAL1 gene.
The mechanism of transcriptional memory
Although both INO1 and GAL1 are retained at the nuclear periphery after transcriptional repression, the mechanism of GAL1 transcriptional memory is easier to study because it lasts longer [11••]. Recently repressed GAL1 is both rapidly reactivated and localized at the nuclear periphery. The mechanism of reactivation of recently repressed GAL1 is different from the mechanism of activation of long-term repressed GAL1 (see below; refs. [11••,21••]). Therefore, I propose that the cell marks recently repressed genes and that this alters their mechanism of transcriptional activation.
Many epigenetic phenomena require post-translational histone modifications or alterations in nucleosome composition [26–29]. Adaptive transcriptional memory requires two chromatin-based mechanisms: the histone variant H2A.Z [11••] and the SWI/SNF chromatin remodeling complex [21••] play essential roles in either the establishment of transcriptional memory or the rapid reactivation of the memory state.
H2A.Z nucleosomes are found in two regions of the genome. They are found in subtelomeric regions, where they are important for insulating euchromatin from the spread of heterochromatin [30]. They are also found in the promoters of many eukaryotic genes, where they play a role in promoting transcriptional activation [31–36]. We identified a third role for H2A.Z nucleosomes: H2A.Z is essential both for the retention of INO1 and GAL1 at the nuclear periphery after repression and for the rapid reactivation of these genes [11••]. Importantly, the initial targeting of INO1 or GAL1 to the nuclear periphery and the initial activation of these genes was not dependent on H2A.Z. Thus, H2A.Z plays a specific, essential role in the peripheral retention and reactivation of certain recently repressed genes.
The reactivation of GAL1 does not require histone modifications that are associated with transcriptional activation [21••]. This is important because previous work had shown that immediately after GAL1 transcription is repressed, the trimethylation of histone H3 lysine 4 that is associated with transcription persists [37]. However, the reactivation of GAL1 does require the SWI/SNF chromatin-remodeling complex. In the absence of the Swi2 protein, the activation and reactivation of GAL1 are indistinguishable and slow. The initial activation of GAL1 was normal in the swi2Δ mutant, indicating that, like H2A.Z, the role of SWI/SNF was specific to the reactivation of the memory state.
The role of SWI/SNF appears to be to counteract the repressive effects of either Isw1 or Isw2 remodeling complexes. The requirement for SWI/SNF in GAL1 reactivation was bypassed in strains lacking either Isw1 of Isw2 [21••]. Isw1 and Isw2 are the catalytic subunits of two related chromatin-remodeling complexes implicated in transcriptional repression. When the swi2Δ mutation was combined with either isw1Δ or isw2Δ, the double mutant exhibited normal transcriptional memory, reactivating GAL1 more rapidly.
Both the SWI/SNF chromatin remodeling complex and the histone variant H2A.Z are required for the rapid reactivation of GAL1. The major difference between the studies that implicated H2A.Z and that implicated SWI/SNF in transcriptional memory is the particular regime of repression before reactivation. Kundu et al. (2007) analyzed the effect of SWI/SNF on GAL1 reactivation after one hour of repression, whereas our work has focused on the effect of H2A.Z on GAL1 reactivation after 12 hours of repression. It remains to be determined how these regimes are related.
The role of DNA localization in transcriptional memory
The correlation of rapid reactivation with peripheral localization suggests that localization may be important for transcriptional memory. Loss of H2A.Z causes both a dramatic decrease in the reactivation rate and loss of peripheral localization of INO1 and GAL1 after repression. Therefore, peripheral localization is coupled to reactivation and both are dependent on H2A.Z. Intriguingly, H2A.Z physically associates with the NPC-associated protein Nup2 [38]. Tethering silenced genes to the NPC creates a boundary that prevents the spreading of silencing [39]. Furthermore, loss of Nup2 causes a spread of silencing from telomeres, similar to the effect of loss of H2A.Z [30,38]. Therefore, perhaps the memory state and boundary activity represent mechanistically related chromatin states that both require H2A.Z nucleosomes and interaction with the NPC.
Artificially tethering the INO1 gene to the nuclear periphery leads to more rapid transcriptional activation [9,11••]. This effect may represent a type of “artificial memory”. Like genuine transcriptional memory, tethering does not affect the ultimate steady-state activation of INO1 [11••]. However, unlike genuine memory, tethering to the nuclear envelope induces rapid activation in the absence of previous transcription.
The mechanism of epigenetic inheritance
SWI/SNF is required for rapid reactivation of GAL1. H2A.Z is required for rapid reactivation of both GAL1 and INO1 and for their retention at the nuclear periphery after repression. These results raised the possibility that transcriptional memory might represent a self-perpetuating chromatin state and that this might explain the epigenetic inheritance of transcriptional memory. In other words, a change in the chromatin state that reflects the history of the gene might “bookmark” a gene and, if it persists until DNA replication, this bookmark might somehow promote its own inheritance. The concept that chromatin might promote its own inheritance to produce stable epigenetic changes in gene expression has been broadly endorsed and has even been proposed as a modern definition of epigenetics [40]. However, the perpetuation of epigenetic states through chromatin alone, without input from trans-acting factors, is still controversial and has been greeted with skepticism [41].
To directly test the hypothesis that chromatin alone is the source of epigenetic inheritance of GAL transcriptional memory, a recent study used heterokaryon analysis, in which cells fuse, but nuclei do not [42••]. They showed that rapid reactivation of GAL1 could be transferred from one cell to another via cytoplasmic mixing. Therefore, GAL transcriptional memory is a cytoplasmically inherited phenomenon. The cytoplasmic factor was identified as the Gal1 protein itself; a gal1Δ mutant is unable to rapidly reactivate the GAL7 gene. Furthermore, constitutive expression of GAL1 was sufficient to confer rapid reactivation of GAL7 [42••]. GAL1 encodes the galactokinase enzyme. Galactokinase is 70% identical to the galactose sensor Gal3, which senses galactose and inactivates the Gal80 transcriptional repressor [43]. Therefore, Gal1 may directly inhibit Gal80, priming the GAL genes for reactivation. The ~100 fold activation of the GAL1 gene, coupled with an extremely long half-life, provides a plausible mechanism for the inheritance of transcriptional memory for > 7 generations [11••,42••]. Thus, although GAL transcriptional memory requires chromatin-based mechanisms, the inheritance of memory is mediated by a cytoplasmic factor.
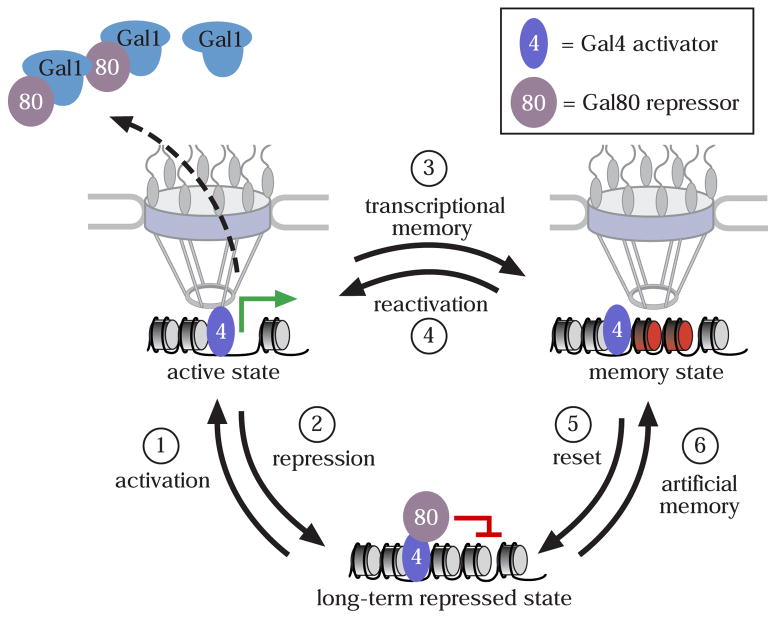
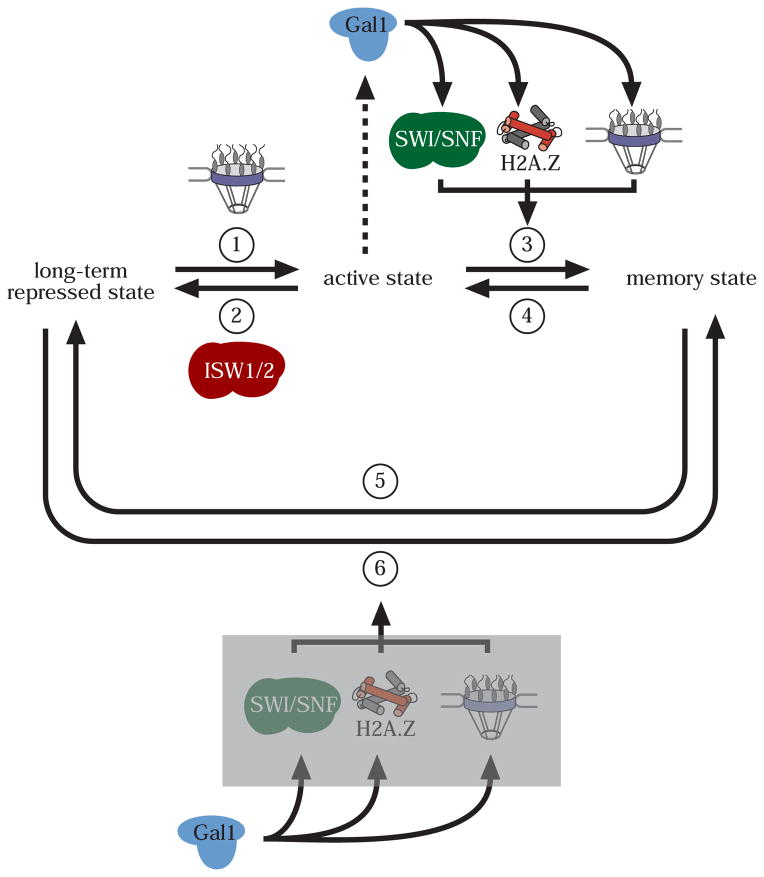
Based on what we have learned from the regulation of the GAL genes, I propose a model for adaptive transcriptional memory (Figure 1). For genes that utilize this type of transcriptional memory, three qualitatively distinct states can exist: a long-term repressed state, an active state and an epigenetic memory state. Each state is distinguished from the other two by its subnuclear localization, its transcriptional activity and its mechanism of regulation (Figures 1 & 2). Furthermore, I propose that it is possible to convert between any state and any other state. Conversion from the active state to the memory state requires a cytoplasmic effector (Gal1 in the case of the GAL genes). I propose that the cytoplasmic effector functions upstream of peripheral localization, SWI/SNF and H2A.Z (Figure 2). The long-term repressed state can also be converted directly into the memory state (i.e. artificial memory) by expression of the cytoplasmic regulator (Figure 2). I propose that, like genuine memory, this pathway also requires peripheral targeting, SWI/SNF or H2A.Z (Figure 2).
Figure 1. Three distinct states of the GAL genes.
Initial activation of the GAL1, GAL2, GAL7 and GAL10 genes (step 1) through the Gal3 galactose sensor, leads to relocalization to the nuclear periphery through association with the NPC and the production of the Gal1 protein. Gal1 binds the repressor Gal80 and inhibits its function. After repressing transcription (via the glucose repression system), the genes remain at the nuclear periphery in a memory state that can be rapidly reactivated. The establishment of the memory state requires chromatin changes (indicated as red nucleosomes). Eventually, transcriptional memory is lost (reset, step 5). The memory state differs from the long-term repressed state in its localization, its chromatin requirements for activation and its rate of reactivation.
Figure 2. Regulation of transcriptional memory.
The states and the interconversion between them are the same as in Figure 1. Conversion between the three states is regulated by the production of a cytoplasmic regulator (the Gal1 protein in the case of the GAL genes). The cytoplasmic regulator functions upstream of the SWI/SNF chromatin remodeling complex, the H2A.Z histone variant and peripheral localization (indicated as the NPC) to promote transcriptional memory (step 3). Because we cannot order these events relative to each other currently, I represent them as parallel inputs. SWI/SNF is necessary to counteract the conversion of the active state into the long-term repressed state by the ISW1 and ISW2 complexes. The memory state can be generated directly from the long-term repressed state by expression of the cytoplasmic regulator (artificial memory; step 6). Grey box: possible role for SWI/SNF, H2A.Z and peripheral localization in the generation of artificial memory.
Two observations indicate that the memory state is qualitatively different from the long-term repressed state and that reactivation occurs by a novel mechanism that is not simply due to titration of a negative regulator. First, the long-term repressed state and the memory state localize to different parts of the nucleus (Figure 1). Second, the requirement for H2A.Z and SWI/SNF in rapid reactivation of recently repressed INO1 and GAL1 is specific; loss of these proteins has no significant effect on the activation of long-term repressed INO1 and GAL1 [11••,21••]. This, too, is not simply due to a change in the rate-limiting step because SWI/SNF and H2A.Z are not required for the rapid activation of GAL1 when cells are shifted from non-repressing raffinose medium to activating galactose medium (our unpublished data; ref. [21••]). Finally, if the tethering of INO1 truly generates artificial memory and if cytoplasmic regulators are a general feature of transcriptional memory, this suggests that their role can be bypassed by tethering INO1 to the nuclear envelope. If so, then the transient expression of a cytoplasmic regulator (like Gal1) generates a transcriptional memory through downstream chromatin modifications mediated by peripheral localization, SWI/SNF and H2A.Z (Figures 1 & 2).
Future directions
Future work will integrate what we have learned to create a more coherent model of transcriptional memory and to answer a number of important questions. How do cytoplasmic factors, chromatin-based changes and gene localization work together to create a marked promoter that can be activated more rapidly? What cytoplasmic factors, if any, are involved in INO1 transcriptional memory? How many genes utilize this type of short-term memory? What are the selective advantages of this system? Finally, do metazoan cells utilize this type of transcriptional memory? SWI/SNF and H2A.Z are conserved among eukaryotes and peripheral targeting of genes has been observed in flies [44•] and mammalian cells [45•]. If metazoan cells utilize adaptive transcriptional memory, it might provide a mechanism by which environmental or physiological inputs can produce short-term to long-term changes in gene expression.
Acknowledgments
I thank the members of the lab, as well as Andreas Matouschek, Rick Gaber, Jonathan Widom, Sandy Westerheide and Jacklyn Jansen for their helpful comments on the manuscript. JHB is supported by grant GM080484 from the National Institute of General Medical Sciences and the Baldwin Fund for Biomedical Research at Northwestern University.
References and recommended reading
Papers of particular interest, published within the period of review have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Rabl C. Über Zeillteilung. In: Gegenbaur C, editor. Morphol Jahrbuch. Vol. 10 1885. pp. 214–330. [Google Scholar]
- 2.Boveri T. Die blastermerenkerne von Ascaris megalocephala und die theorie der chromosomenindividualität. Arch Zellforschung. 1909;3:181. [Google Scholar]
- 3.Marshall WF, Dernburg AF, Harmon B, Agard DA, Sedat JW. Specific interactions of chromatin with the nuclear envelope: positional determination within the nucleus in Drosophila melanogaster. Mol Biol Cell. 1996;7:825–842. doi: 10.1091/mbc.7.5.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gasser SM. Visualizing chromatin dynamics in interphase nuclei. Science. 2002;296:1412–1416. doi: 10.1126/science.1067703. [DOI] [PubMed] [Google Scholar]
- 5.Cockell M, Gasser SM. Nuclear compartments and gene regulation. Curr Opin Genet Dev. 1999;9:199–205. doi: 10.1016/S0959-437X(99)80030-6. [DOI] [PubMed] [Google Scholar]
- 6.Andrulis ED, Neiman AM, Zappulla DC, Sternglanz R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature. 1998;394:592–595. doi: 10.1038/29100. [DOI] [PubMed] [Google Scholar]
- 7.Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- 8.Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brickner JH, Walter P. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2004;2:e342. doi: 10.1371/journal.pbio.0020342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–439. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- ••11.Brickner DG, Cajigas IC, Fondufe-Mittendorf Y, Ahmed S, Lee P-C, Widom J, Brickner JH. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 2007;5:e81. doi: 10.1371/journal.pbio.0050081. This paper describes the discovery that INO1 and GAL1 remain at the nuclear periphery after repression and that this promotes more rapid reactivation. The authors also show that the histone variant H2A.Z is essential for both retention at the nuclear periphery and rapid reactivation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casolari JM, Brown CR, Drubin DA, Rando OJ, Silver PA. Developmentally induced changes in transcriptional program alter spatial organization across chromosomes. Genes Dev. 2005;19:1188–1198. doi: 10.1101/gad.1307205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, Gasser SM. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 2006;441:774–778. doi: 10.1038/nature04845. [DOI] [PubMed] [Google Scholar]
- 14.Dieppois G, Iglesias N, Stutz F. Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Mol Cell Biol. 2006;26:7858–7870. doi: 10.1128/MCB.00870-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarma NJ, Haley TM, Barbara KE, Buford TD, Willis KA, Santangelo GM. Glucose-responsive regulators of gene expression in Saccharomyces cerevisiae function at the nuclear periphery via a reverse recruitment mechanism. Genetics. 2007;175:1127–1135. doi: 10.1534/genetics.106.068932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmid M, Arib G, Laemmli C, Nishikawa J, Durussel T, Laemmli UK. Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol Cell. 2006;21:379–391. doi: 10.1016/j.molcel.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 17.Menon BB, Sarma NJ, Pasula S, Deminoff SJ, Willis KA, Barbara KE, Andrews B, Santangelo GM. Reverse recruitment: the Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation. Proc Nat’l Acad Sci U S A. 2005;102:5749–5754. doi: 10.1073/pnas.0501768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahmed S, Brickner DG, McDonough M, Light WH, Volpe T, Brickner JH. DNA zip codes: an ancient mechanism for targeting genes to the nuclear periphery. doi: 10.1038/ncb2011. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luthra R, Kerr SC, Harreman MT, Apponi LH, Fasken MB, Ramineni S, Chaurasia S, Valentini SR, Corbett AH. Actively transcribed GAL genes can be physically linked to the nuclear pore by the SAGA chromatin modifying complex. J Biol Chem. 2007;282:3042–3049. doi: 10.1074/jbc.M608741200. [DOI] [PubMed] [Google Scholar]
- 20.Cabal GG, Genovesio A, Rodriguez-Navarro S, Zimmer C, Gadal O, Lesne A, Buc H, Feuerbach-Fournier F, Olivo-Marin JC, Hurt EC, et al. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 2006;441:770–773. doi: 10.1038/nature04752. [DOI] [PubMed] [Google Scholar]
- ••21.Kundu S, Horn PJ, Peterson CL. SWI/SNF is required for transcriptional memory at the yeast GAL gene cluster. Genes Dev. 2007;21:997–1004. doi: 10.1101/gad.1506607. This paper describes the discovery that GAL1, GAL2, GAL7 and GAL10 are more rapidly activated if they have previously been expressed. This transcriptional memory persists through cell division. Memory does not require histone acetylation or methylation but does require the SWI/SNF chromatin-remodeling complex. SWI/SNF functions to overcome ISW1/2-mediated repression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abruzzi KC, Belostotsky DA, Chekanova JA, Dower K, Rosbash M. 3′-end formation signals modulate the association of genes with the nuclear periphery as well as mRNP dot formation. EMBO J. 2006;25:4253–4262. doi: 10.1038/sj.emboj.7601305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •23.Chekanova JA, Abruzzi KC, Rosbash M, Belostotsky DA. Sus1, Sac3, and Thp1 mediate post-transcriptional tethering of active genes to the nuclear rim as well as to non-nascent mRNP. RNA. 2008;14:66–77. doi: 10.1261/rna.764108. This paper shows that proteins involved in mRNA export are essential for the accumulation of mRNA “dots” in association with GAL1 promoter-driven reporter genes and for their post-transcriptional localization at the nuclear periphery. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •24.Vodala S, Abruzzi KC, Rosbash M. The nuclear exosome and adenylation regulate posttranscriptional tethering of yeast GAL genes to the nuclear periphery. Mol Cell. 2008;31:104–113. doi: 10.1016/j.molcel.2008.05.015. Using both GAL1 promoter-driven reporter genes and the endogenous GAL1 gene, this paper argues that the nuclear exosome normally generates unpolyadenylated RNAs that accumulate in gene-associated “dots”. These RNA dots and the associated reporter genes are localized at the nuclear periphery. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •25.Rougemaille M, Dieppois G, Kisseleva-Romanova E, Kanth Gudipati R, Lemoine S, Blugeon C, Boulay J, Heick Jensen T, Stutz F, Devaux F, et al. THO/Sub2p functions to coordinate 3′-end processing with gene-nuclear pore association. Cell. 2008;135:308–321. doi: 10.1016/j.cell.2008.08.005. This paper describes the accumulation of a stalled mRNA processing/export intermediate in mutants lacking THO components. This intermediate, which may correspond to RNA “dots” observed in refs [22–24] is associated with chromatin and interacts with the NPC. The accumulation of this intermediate and its targeting to the NPC requires the nuclear exosome. [DOI] [PubMed] [Google Scholar]
- 26.Andersen AA, Panning B. Epigenetic gene regulation by noncoding RNAs. Curr Opin Cell Biol. 2003;15:281–289. doi: 10.1016/s0955-0674(03)00041-3. [DOI] [PubMed] [Google Scholar]
- 27.Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- 28.Lawrence RJ, Earley K, Pontes O, Silva M, Chen ZJ, Neves N, Viegas W, Pikaard CS. A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol Cell. 2004;13:599–609. doi: 10.1016/s1097-2765(04)00064-4. [DOI] [PubMed] [Google Scholar]
- 29.Henikoff S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat Rev Genet. 2008;9:15–26. doi: 10.1038/nrg2206. [DOI] [PubMed] [Google Scholar]
- 30.Meneghini MD, Wu M, Madhani HD. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 2003;112:725–736. doi: 10.1016/s0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- 31.Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang H, Roberts DN, Cairns BR. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123:219–231. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guillemette B, Bataille AR, Gevry N, Adam M, Blanchette M, Robert F, Gaudreau L. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 2005;3:e384. doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 35.Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whittle CM, McClinic KN, Ercan S, Zhang X, Green RD, Kelly WG, Lieb JD. The genomic distribution and function of histone variant HTZ-1 during C. elegans embryogenesis. PLoS Genet. 2008;4:e1000187. doi: 10.1371/journal.pgen.1000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 38.Dilworth DJ, Tackett AJ, Rogers RS, Yi EC, Christmas RH, Smith JJ, Siegel AF, Chait BT, Wozniak RW, Aitchison JD. The mobile nucleoporin Nup2p and chromatin-bound Prp20p function in endogenous NPC-mediated transcriptional control. J Cell Biol. 2005;171:955–965. doi: 10.1083/jcb.200509061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ishii K, Arib G, Lin C, Van Houwe G, Laemmli UK. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell. 2002;109:551–562. doi: 10.1016/s0092-8674(02)00756-0. [DOI] [PubMed] [Google Scholar]
- 40.Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 41.Ptashne M. On the use of the word ‘epigenetic’. Curr Biol. 2007;17:R233–236. doi: 10.1016/j.cub.2007.02.030. [DOI] [PubMed] [Google Scholar]
- ••42.Zacharioudakis I, Gligoris T, Tzamarias D. A yeast catabolic enzyme controls transcriptional memory. Curr Biol. 2007;17:2041–2046. doi: 10.1016/j.cub.2007.10.044. Using heterokaryon analysis, the authors show that GAL gene transcriptional memory is conferred by a cytoplasmic factor. They identify the Gal1 protein as the cytoplasmic factor. Constitutive expression of Gal1 leads to more rapid transcriptional activation of GAL7, suggesting that this is sufficient to generate artificial memory. [DOI] [PubMed] [Google Scholar]
- 43.Peng G, Hopper JE. Gene activation by interaction of an inhibitor with a cytoplasmic signaling protein. Proc Nat’l Acad Sci U S A. 2002;99:8548–8553. doi: 10.1073/pnas.142100099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •44.Kurshakova MM, Krasnov AN, Kopytova DV, Shidlovskii YV, Nikolenko JV, Nabirochkina EN, Spehner D, Schultz P, Tora L, Georgieva SG. SAGA and a novel Drosophila export complex anchor efficient transcription and mRNA export to NPC. EMBO J. 2007;26:4956–4965. doi: 10.1038/sj.emboj.7601901. The hsp70 locus in Drosophila localizes at the nuclear periphery, in association with the nuclear pore complex (NPC). Full transcription and the localization at the nuclear periphery requires the same NPC components that have been shown to function in gene recruitment to the nuclear periphery in yeast. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •45.Brown CR, Kennedy CJ, Delmar VA, Forbes DJ, Silver PA. Global histone acetylation induces functional genomic reorganization at mammalian nuclear pore complexes. Genes Dev. 2008;22:627–639. doi: 10.1101/gad.1632708. In mammalian cells, certain genes associate with the NPC and relocalize from the nucleoplasm to the nuclear periphery upon treatment with histone deacetylase inhibitors. This suggests that peripheral targeting of active genes can also occur in mammalian cells. [DOI] [PMC free article] [PubMed] [Google Scholar]