We thank the Editor for the opportunity to respond to the letter from Belgard et al.1 In their letter, these authors consider that the issue of ethnic population stratification may have negatively impacted the findings in our original manuscript.2 We agree that population stratification is an important issue that needs to be accounted for in such analyses.
We wrote to Dr Belgard who kindly provided the 19 single-nucleotide polymorphisms (SNPs) used in their analysis.1 These 19 SNPs were derived from the 30 SNPs provided in our original article. Of these 19 SNPs, the number of SNPs with positive weights exceeded the number of SNPs with negative weights, including the second most negative weighted SNP, rs12317962, on KCNMB4, which would bias the classifier score. Our original analyses included a total of 237 SNPs. In order to address the issue of ethnic population stratification, we downloaded data from the 1000 genome cohort,3 including Central European (CEU), Finnish (FIN), Great British (GBR) and Iberian Spanish (IBS) populations.
In their analysis using 19 SNPs, Belgard et al. indicated that in Finns (non-autism spectrum disorder (ASD)), our classifier had a higher chance of classifying individuals as ASD compared with CEU (non-ASD) individuals. They concluded that our classifier might be better at separating between European subpopulations than cases from controls. In order to examine this in detail, we tested our classifier performance in correctly identifying control individuals from the CEU, FIN, GBR and IBS control populations. As not all SNPs were available across all data sets, we retrained the classifier using the common SNPs on our training set and then applied the classifier on unseen validation data from the FIN, GBR and IBS control cohorts. Comparing these ethnic European subpopulations, we found that greater differences in classifier score between these populations occurred when only part of the classifier was used (a difference as high as 25% was observed between the FIN and GBR groups). However, using the full classifier, the effects of ethnic population contributed to <6% of the total difference in classifier score. We also provide the full 237 SNPs relevant to our classifier (Table 1). The full code used in the generation of the classifier has been made available on the Autism Genetic Resource Exchange (AGRE) website (http://agre.org), together with testing of the classifier on other ASD data sets.
Table 1. List of all 237 SNPs for ASD classifier in the CEU Cohort, 2 organised from highest to lowest median weightings.
| SNP | Weight lower | Weight median | Weight upper | Gene no. | Gene symbol |
|---|---|---|---|---|---|
| rs968122 | 1.5465 | 1.5555 | 1.5645 | 27345 | KCNMB4 |
| rs876619 | 0.9476 | 1.2092 | 1.4708 | 2775 | GNAO1 |
| rs11020772 | 0.8553 | 0.8641 | 0.8729 | 2915 | GRM5 |
| rs9288685 | 0.5856 | 0.5998 | 0.614 | 3635 | INPP5D |
| rs10193128 | 0.5836 | 0.5946 | 0.6056 | 3635 | INPP5D |
| rs7842798 | 0.5298 | 0.5386 | 0.5474 | 114 | ADCY8 |
| rs3773540 | 0.5125 | 0.5208 | 0.5291 | 55799 | CACNA2D3 |
| rs1818106 | 0.5002 | 0.5161 | 0.532 | 80310 | PDGFD |
| rs2384061 | 0.4195 | 0.4306 | 0.4417 | 109 | ADCY3 |
| rs12582971 | 0.3983 | 0.4295 | 0.4607 | 5288 | PIK3C2G |
| rs10409541 | 0.4067 | 0.4189 | 0.4311 | 773 | CACNA1A |
| rs2300497 | 0.3782 | 0.3889 | 0.3996 | 801 | CALM1 |
| rs7562445 | 0.3741 | 0.3843 | 0.3945 | 2066 | ERBB4 |
| rs7313997 | 0.3382 | 0.3567 | 0.3752 | 5801 | PTPRR |
| rs2239118 | 0.3348 | 0.3552 | 0.3756 | 775 | CACNA1C |
| rs4688054 | 0.1801 | 0.3476 | 0.515 | 2932 | GSK3B |
| rs10823195 | 0.2597 | 0.3445 | 0.4294 | 1763 | DNA2 |
| rs9798267 | 0.2759 | 0.3388 | 0.4017 | 84083 | ZRANB3 |
| rs1075354 | 0.4236 | 0.3177 | 0.6402 | 55799 | CACNA2D3 |
| rs1942052 | 0.2641 | 0.3088 | 0.3535 | 130013 | ACMSD |
| rs4696443 | 0.2525 | 0.3047 | 0.3569 | 23321 | TRIM2 |
| rs243196 | 0.2402 | 0.2976 | 0.3549 | 1112 | FOXN3 |
| rs16929470 | 0.1854 | 0.2712 | 0.3571 | 775 | CACNA1C |
| rs7580690 | 0.1647 | 0.2248 | 0.285 | 83439 | TCF7L1 |
| rs7145618 | 0.1515 | 0.2238 | 0.296 | 5528 | PPP2R5C |
| rs3770132 | 0.1514 | 0.2093 | 0.2673 | 3676 | ITGA4 |
| rs3790095 | 0.1215 | 0.2017 | 0.2819 | 2775 | GNAO1 |
| rs1013459 | 0.1417 | 0.1969 | 0.2522 | 2774 | GNAL |
| rs11001056 | 0.1519 | 0.1891 | 0.2263 | 5592 | PRKG1 |
| rs10952662 | 0.148 | 0.1868 | 0.2257 | 26047 | CNTNAP2 |
| rs7756516 | 0.152 | 0.1853 | 0.2186 | 3120 | HLA-DQB2 |
| rs8054767 | 0.1322 | 0.1803 | 0.2284 | 5579 | PRKCB |
| rs2239028 | 0.1121 | 0.1763 | 0.2405 | 775 | CACNA1C |
| rs3935743 | 0.0969 | 0.1737 | 0.2505 | 5336 | PLCG2 |
| rs1928168 | 0.0657 | 0.099 | 0.1322 | 401237 | LINC00340 |
| rs7100765 | 0.0434 | 0.0935 | 0.1436 | 5593 | PRKG2 |
| rs1369450 | 0.0563 | 0.0924 | 0.1285 | 114 | ADCY8 |
| rs1040336 | −0.0615 | 0.091 | 0.2435 | 2272 | FHIT |
| rs10407144 | 0.0434 | 0.0872 | 0.131 | 773 | CACNA1A |
| rs10794197 | 0.045 | 0.0869 | 0.1287 | 1488 | CTBP2 |
| rs3734464 | 0.0247 | 0.0868 | 0.149 | 5071 | PARK2 |
| rs7864216 | −0.0072 | 0.0863 | 0.1798 | 9630 | GNA14 |
| rs4254056 | 0.0432 | 0.0846 | 0.126 | 338751 | OR52L1 |
| rs988920 | 0.0453 | 0.0842 | 0.1232 | 9229 | DLGAP1 |
| rs12393998 | 0.0536 | 0.0839 | 0.1142 | 8450 | CUL4B |
| rs872794 | 0.0413 | 0.0813 | 0.1213 | 3778 | KCNMA1 |
| rs2503220 | −0.0527 | 0.0806 | 0.214 | 5142 | PDE4B |
| rs10468681 | 0.0356 | 0.08 | 0.1243 | 2774 | GNAL |
| rs7258489 | 0.0428 | 0.079 | 0.1152 | 808 | CALM3 |
| rs153968 | 0.0379 | 0.0765 | 0.115 | 5144 | PDE4D |
| rs944761 | 0.0361 | 0.076 | 0.1159 | 9568 | GABBR2 |
| rs2161630 | 0.0232 | 0.0754 | 0.1276 | 10725 | NFAT5 |
| rs7097311 | 0.0294 | 0.0703 | 0.1111 | 5593 | PRKG2 |
| rs2088747 | −0.0137 | 0.0693 | 0.1522 | 11060 | WWP2 |
| rs9832697 | −0.0766 | 0.0689 | 0.2144 | KCNMB2 | |
| rs7731023 | 0.0343 | 0.0683 | 0.1023 | 6502 | SKP2 |
| rs7120612 | 0.0224 | 0.0659 | 0.1094 | 390055 | OR52A6 |
| rs2033655 | 0.0277 | 0.0647 | 0.1017 | 109 | ADCY3 |
| rs1453541 | −0.1057 | 0.0354 | 0.1766 | 219983 | OR4D6 |
| rs3746821 | −0.0262 | 0.0335 | 0.0932 | 958 | CD40 |
| rs220740 | −0.0085 | 0.0332 | 0.0749 | 10846 | PDE10A |
| rs2299679 | −0.014 | 0.0331 | 0.0801 | 5332 | PLCB4 |
| rs887387 | −0.0028 | 0.0317 | 0.0662 | 489 | ATP2A3 |
| rs7174459 | −0.0092 | 0.0288 | 0.0669 | 4735 | NEDD5 |
| rs884399 | −0.0073 | 0.0281 | 0.0634 | 5581 | PRKCE |
| rs5021051 | −0.0146 | 0.027 | 0.0686 | 2895 | GRID2 |
| rs2903813 | −0.0208 | 0.0252 | 0.0711 | 3315 | HSPB1 |
| rs1062935 | −0.0207 | 0.0245 | 0.0697 | 57521 | RPTOR |
| rs9347553 | −0.0154 | 0.0228 | 0.0609 | 5071 | PARK2 |
| rs11072416 | −0.0259 | 0.0222 | 0.0703 | 6263 | RYR3 |
| rs4553343 | −0.0304 | 0.0204 | 0.0712 | 2977 | GUCY1A2 |
| rs7146234 | −0.0132 | 0.0202 | 0.0535 | 5495 | PPM1A |
| rs848282 | −0.0191 | 0.0172 | 0.0536 | 55120 | FANCL |
| rs7962764 | −0.0495 | 0.0126 | 0.0748 | 5801 | PTPRR |
| rs12726519 | −0.0377 | 0.0098 | 0.0572 | 5321 | PLA2G4A |
| rs718949 | −0.0303 | 0.0093 | 0.0489 | 1488 | CTBP2 |
| rs1954787 | −0.0264 | 0.0089 | 0.0441 | 2900 | GRIK4 |
| rs2238079 | −0.0283 | 0.0084 | 0.045 | 775 | CACNA1C |
| rs1337420 | −0.0398 | 0.008 | 0.0558 | 2898 | GRIK2 |
| rs917948 | −0.0553 | 0.0075 | 0.0704 | 5536 | PPP5C |
| rs3817222 | −0.1848 | 0.0055 | 0.1957 | 4660 | PPP1R12B |
| rs17531147 | −0.0612 | 0.003 | 0.0672 | 55970 | GNG12 |
| rs11048476 | −0.0801 | −0.0384 | 0.0033 | 3709 | ITPR2 |
| rs4145903 | −0.0762 | −0.0395 | −0.0028 | 783 | CACNB2 |
| rs10505029 | −0.1011 | −0.0404 | 0.0203 | 51366 | UBR5 |
| rs1122838 | −0.1213 | −0.0408 | 0.0396 | 9630 | GNA14 |
| rs1993477 | −0.0818 | −0.0434 | −0.0049 | 51366 | UBR5 |
| rs2179871 | −0.0912 | −0.0454 | 0.0005 | 10369 | CACNG2 |
| rs10740244 | −0.0892 | −0.0467 | −0.0041 | 5592 | PRKG1 |
| rs2503220 | −0.1151 | −0.0472 | 0.0207 | 5142 | PDE4B |
| rs1065657 | −0.0838 | −0.0488 | −0.0139 | 51465 | UBE2J1 |
| rs12714137 | −0.1234 | −0.0528 | 0.0179 | 83439 | TCF7L1 |
| rs7176475 | −0.1275 | −0.0537 | 0.0201 | 123746 | PLA2G4E |
| rs1937671 | −0.0953 | −0.0545 | −0.0138 | 5592 | PRKG1 |
| rs7079293 | −0.0902 | −0.0549 | −0.0196 | 10581 | SORBS2 |
| rs1003854 | −0.1288 | −0.0551 | 0.0187 | 326 | AIRE |
| rs919741 | −0.0962 | −0.0565 | −0.0169 | 815 | CAMK2A |
| rs750438 | −0.1075 | −0.0574 | −0.0074 | 11184 | MAP4K1 |
| rs6139034 | −0.0997 | −0.0576 | −0.0154 | 3704 | ITPA |
| rs1554606 | −0.1087 | −0.0599 | −0.0111 | 6018 | IL6 |
| rs7108524 | −0.0938 | −0.0603 | −0.0267 | 81286 | OR51E3 |
| rs1002424 | −0.1023 | −0.0626 | −0.0229 | 5562 | PRKAA1 |
| rs2239316 | −0.1033 | −0.0631 | −0.0228 | 1387 | CREBBP |
| rs5030949 | −0.157 | −0.0653 | 0.0264 | 3098 | HK1 |
| rs17682073 | −0.1006 | −0.066 | −0.0315 | 6262 | RYR2 |
| rs1872902 | −0.1108 | −0.0665 | −0.0221 | 80310 | PDGFD |
| rs11602535 | −0.166 | −0.1236 | −0.0812 | 219981 | OR5A2 |
| rs11644436 | −0.1733 | −0.1253 | −0.0774 | 5336 | PLCG2 |
| rs10762342 | −0.1909 | −0.1283 | −0.0658 | 5592 | PRKG1 |
| rs11583646 | −0.2023 | −0.1311 | −0.0599 | 6262 | RYR2 |
| rs6118611 | −0.1819 | −0.1321 | −0.0822 | 5332 | PLCB4 |
| rs2587891 | −0.1722 | −0.1322 | −0.0922 | 2775 | GNA01 |
| rs4651343 | −0.1739 | −0.1333 | −0.0926 | 5321 | PLA2G4A |
| rs1659506 | −0.1761 | −0.1363 | −0.0966 | 23295 | MGRN1 |
| rs2271986 | −0.1968 | −0.1367 | −0.0767 | 4842 | NOS1 |
| rs2302898 | −0.1775 | −0.1375 | −0.0975 | 10381 | TUBB3 |
| rs6971999 | −0.2088 | −0.1425 | −0.0763 | 26212 | OR2F2 |
| rs2272197 | −0.1896 | −0.1485 | −0.1073 | 4216 | MAP3K4 |
| rs4947963 | −0.1867 | −0.1493 | −0.1119 | 1956 | EGFR |
| rs7536307 | −0.1876 | −0.1507 | −0.1138 | 26289 | AK5 |
| rs12462609 | −0.2085 | −0.151 | −0.0936 | 773 | CACNA1A |
| rs1517521 | −0.2925 | −0.152 | −0.0114 | 23180 | RFTN1 |
| rs8063461 | −0.1865 | −0.1534 | −0.1203 | 7249 | TSC2 |
| rs888817 | −0.1937 | −0.1604 | −0.1272 | 5924 | RASGRF2 |
| rs922445 | −0.2435 | −0.1659 | −0.0883 | 2775 | GNAO1 |
| rs339408 | −0.203 | −0.167 | −0.131 | 9322 | TRIP10 |
| rs7512378 | −0.2068 | −0.1691 | −0.1314 | 55811 | ADCY10 |
| rs7870040 | −0.2408 | −0.1892 | −0.1376 | 774 | CACNA1B |
| rs3904668 | −0.2423 | −0.2069 | −0.1715 | 29993 | PACS1N1 |
| rs12716928 | −0.2784 | −0.2073 | −0.1362 | 5336 | PLCG2 |
Abbreviations: ASD, autism spectrum disorder; CEU, Central European; SNP, single-nucleotide polymorphism.
Weight indicates the contribution of each SNP to ASD clinical status. The lower and upper weights represent the 95% confidence intervals (CIs) of the distribution of weights for each SNP.
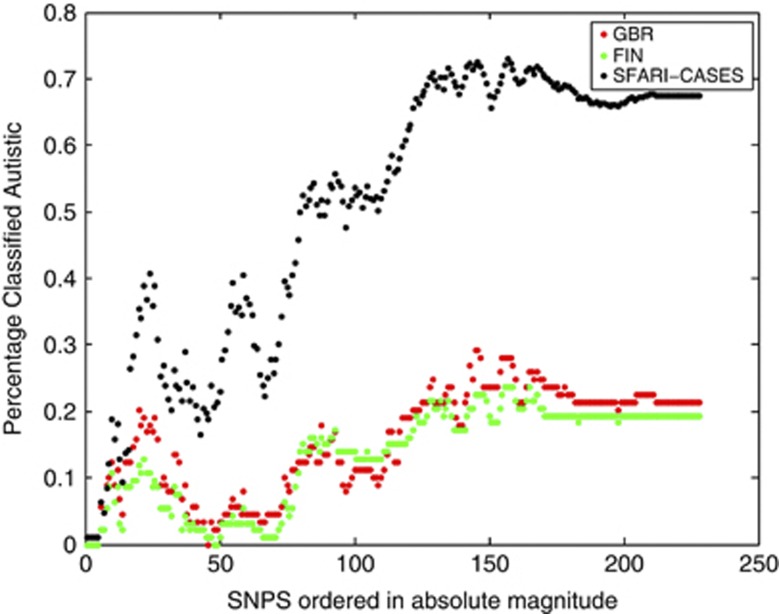
Using our SNPs, we then examined their predictive accuracy in classifying control individuals from the FIN and GBR (non-ASD) populations, as well as SFARI (Simons Foundation Autism Research Initiative) ASD probands (the independent validation sample in our paper). We plotted the percentage of individuals classified as ASD against the number of SNPs used in the classifier, with SNPs ordered by absolute magnitude of their weightings. As can be seen in Figure 1, while population stratification may have an influence at lower SNP numbers with regard to differences in classifier accuracy between populations, such an effect is diminished as a greater number of SNPs are included. The separation in percentage classified as ASD between the SFARI/ASD and the FIN/GBR groups occurred with increasing gradient between 50 and 100 SNPs, whereas at >150 SNPs the separation between these groups plateaus. This is to be expected, as these SNPs have the smallest weightings within the classifier. Therefore, in keeping with Belgard et al's analysis, we show that at low SNP numbers, population effects may influence classification accuracy, but these effects are of second order to the ASD signal as the number of SNPs increases.
Figure 1.
Percentage of individuals classified as ASD as a function of the number of single-nucleotide polymorphisms (SNPs) ordered in decreasing absolute magnitude. Significant variance was observed at smaller number of SNPs (not plotted). Note the gradient differential between SFARI cases versus FIN and GBR between SNPs 80 and 150. ASD, autism spectrum disorder; SNPs, single-nucleotide polymorphisms; SFARI-CASES, Simons Foundation Autism Research Initiative ASD probands; population samples from the 1000 genome cohort3: GBR, Great British; FIN, Finnish.
Using the classifier, as described above, we tested its accuracy in correctly classifying controls (non-ASD) within individual European cohorts. We achieved accuracies (that is, correct classification as non-ASD) of 82% for the FIN, 78% for GBR and 67% for the Spanish cohorts. In addition, to determine classifier performance confidence intervals, we performed a bootstrap analysis (1000 permutations were undertaken; 80% of the data was used to train a classifier to predict the remaining 20%) on all white non-hispanic populations, including all available populations (that is, SFARI and Autism Genetic Resource Exchange probands, and WTBC, CEU, FIN, GBR and IBS Controls). Diagnostic accuracy for ASD was 66.0% (90% CI: 61.5–71.9), with a sensitivity of 63.4% (90% CI: 54.3–75.9) and specificity of 67.2% (90% CI: 59.5–74.3). This equates to a positive likelihood ratio of 1.9 (90% CI: 1.3–3.0).
In our paper, we reported positive and negative predictive accuracies that were 70.8% and 71.8%, respectively.2 Based on a population prevalence of 1:88 cases of ASD in the US population,4 this equates to a positive predictive value (that is, precision) of 2.8% and a negative predictive value of 99.5%. This suggests that the classifier is not suitable as a general screening method, rather it should only be considered in high-risk populations where the base rate of ASD is high and produces acceptable positive and negative predictive values.
In conclusion, we demonstrate that the SNPs in our classifier show some ability to non-randomly distinguish between ASD and controls and that our results are not merely explained by population stratification as demonstrated in our analyses in independent cohorts of individuals of European ancestry. Further work on such approaches is needed in order to validate these findings, for example, prospective studies that examine children at risk for ASD (such as families with an affected member).
Acknowledgments
We thank Dr Belgard for his helpful suggestions and feedback on further analyses in the preparation of this letter.
A patent application has been filed by The University of Melbourne.
References
- Belgard TG, Jankovic I, Lowe JK, Geschwind DH.Mol Psychiatrydoi: 10.1038/mp.2013.34(e-pub ahead of print). [DOI] [PMC free article] [PubMed]
- Skafidas E, Testa R, Zantomio D, Chana G, Everall IP, Pantelis C.Mol Psychiatrydoi: 10.1038/mp.2012.126(e-pub ahead of print). [DOI] [PMC free article] [PubMed]
- Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, et al. Nature. 2012. pp. 56–65. [DOI] [PMC free article] [PubMed]
- Centers for Disease Control and Prevention Surveillance Summaries. 2012. pp. 1–19.