Abstract
The underlying pathology of schizophrenia (SZ) is likely as heterogeneous as its symptomatology. A variety of cortical and subcortical regions, including the prefrontal cortex, have been implicated in its pathology, and a number of genes have been identified as risk factors for disease development. We used in situ hybridization (ISH) to examine the expression of 58 genes in the dorsolateral prefrontal cortex (DLPFC, comprised of Brodmann areas 9 and 46) from 19 individuals with a premorbid diagnosis of SZ and 33 control individuals. Genes were selected based on: (1) previous identification as risk factors for SZ; (2) cell type markers or (3) laminar markers. Cell density and staining intensity were compared in the DLPFC, as well as separately in Brodmann areas 9 and 46. The expression patterns of a variety of genes, many of which are associated with the GABAergic system, were altered in SZ when compared with controls. Additional genes, including C8orf79 and NR4A2, showed alterations in cell density or staining intensity between the groups, highlighting the need for additional studies. Alterations were, with only a few exceptions, limited to Brodmann area 9, suggesting regional specificity of pathology in the DLPFC. Our results agree with previous studies on the GABAergic involvement in SZ, and suggest that areas 9 and 46 may be differentially affected in the disease. This study also highlights additional genes that may be altered in SZ, and indicates that these potentially interesting genes can be identified by ISH and high-throughput image analysis techniques.
Keywords: gene expression, in situ hybridization, prefrontal cortex, schizophrenia
Introduction
Schizophrenia (SZ) is a complex psychiatric disorder characterized by disorganized thought processes. It can present with a variety of positive and negative symptoms, including hallucinations, delusions, deficits in speech and behavior, blunted affect, and decline in speech and motivation.1, 2 Changes have also been noted in a variety of memory and executive functions.3, 4, 5, 6, 7 Owing to the heterogeneous nature of the clinical symptoms, attempts to define a single underlying pathogenomic lesion have been unsuccessful. Various cortical and subcortical abnormalities have been associated with SZ,8, 9, 10, 11, 12, 13, 14, 15 as well as alterations at the level of morphology,16, 17 neurotransmitter systems18, 19 and neurophysiology.20, 21, 22, 23 Additionally, polymorphisms in multiple genes have been identified as risk factors for disease development through genome-wide association studies.24, 25
The dorsolateral prefrontal cortex (DLPFC) has been identified as one region that may be altered in SZ.9, 13, 14, 15, 26 It has been shown to be crucial for verbal memory and fluency as well as working memory, processes that are altered in SZ.27, 28, 29 A variety of changes have been noted in the DLPFC of schizophrenic patients, including alterations in overall cell density, in the number of specific receptors, changes in gene expression, as well as alterations found using fMRI during specific tasks.13, 14, 15, 30, 31
In this study, we used in situ hybridization (ISH) to examine the expression patterns of 58 genes in the DLPFC of individuals who had been diagnosed with SZ prior to death, or individuals with no history of neuropsychiatric illnesses. The genes examined segregate into those previously implicated as risk factors for the development of SZ, cell type markers (most of which are markers for GABAergic interneurons) or genes that show enhanced expression in a given layer or layers of the cortex. In order to determine if there was any regional specificity to expression patterns, distribution of gene expression was assessed in Brodmann areas 9 or 46, as well as for a combination of the two areas (hereafter referred to as ‘DLPFC'). We assessed the density of cells expressing each gene and the intensity of staining within each cell to develop a better understanding of how gene expression may be altered in SZ compared with controls.
Materials and methods
Case selection
Materials from individuals that met DMS-IV criteria for a premorbid diagnosis of SZ were compared with control individuals with no history of neurologic or psychiatric illnesses. Tissue from the DLPFC of the right hemisphere was dissected at the NIMH (Section on Neuropathology, Clinical Brain Disorders Branch, GCAP, IRP), frozen in isopentane, stored at −80 °C and shipped to the Allen Institute on dry ice. Post-mortem interval, pH, cause of death, handedness and information on whether the subjects were smokers or had alcohol or antipsychotics in their tissues at the time of death were collected from the source. Prior to use in the study, each case underwent analysis of tissue quality, RNA quality (RIN) and verification of the presence of the regions of interest (ROIs); only those cases that met the inclusion criteria were included in the study, resulting in 19 individuals with a diagnosis of SZ and 33 control individuals. A complete list of inclusionary criteria can be found in Supplementary Table 1. With the exception of pH and age, no significant differences were found between the two groups with regard to PMI, RIN values or the presence of alcohol post-mortem (Table 1). Differences in pH have been ascribed to the use of medication and the resulting buildup of lactic acid,32 though other factors may contribute to the difference. As has also been previously noted in literature,33 the schizophrenic group had a higher incidence of smoking than the control group. However, without longitudinal data to assess how long or how often a subject had smoked, or the types and number of cigarettes smoked per day, smoking was not used in the models for data analysis. Evidence of antipsychotic use at the time of death was not seen in any of the controls, but was documented in 75% of the SZ cases. Owing to the fact that nearly all SZ subjects had evidence of antipsychotic treatment at the time of death, and the likelihood that the remaining SZ subjects were on antipsychotic medication at some point prior to death, no attempt was made to assess whether gene expression levels correlated with antipsychotic treatment.
Table 1. Demographics.
| Control (mean±s.d.) | Schizophrenia (mean±s.d.) | Test value, P-value | |
|---|---|---|---|
| Gender (M/F; % male) | 24/9 (72.7%) | 14/5 (73.7%) | χ2=0.006, df=1, NS |
| Age (years) | 37.21±9.61 (20–49) | 43.74±8.59 (22–57) | t=2.52, df=41.22, P<0.01 |
| PMI (h) | 29.94±13.74 (9–68) | 31.13±13.05 (7–51) | t=0.311, df=39.03, NS |
| pH | 6.43±0.27 (5.9–7.0) | 6.23±0.12 (6.0–6.4) | t=−3.60, df=46.56, P<0.001 |
| RIN | 6.95±0.73 (5.6–8.4) | 6.70±0.43 (6.0–7.4) | t=−1.59, df=49.96, NS |
| Smoking (Y/N/NA; % yes) | 9/32/1 (27.7%) | 16/3/0 (84.21%) | χ2=15.714, df=2, P<0.001 |
Once selected for inclusion in the study, the tissue blocks were sectioned at 25 μm on a cryostat. Every 20th section was stained for Nissl substance, and the location of areas 9 and 46 were confirmed and identified when present. Three series of 60 consecutive sections, interspersed by the Nissl-stained slides, were identified as containing the ROIs, and were labeled by ISH for expression of a panel of genes (see below). This yielded three stained sections for each probe, with an interval of no less than 1.5 mm between each section.
ISH and gene selection
The use of a high-throughput ISH platform to generate data on gene expression patterns in human tissue has been previously described.34, 35 Probes were generated against the human genes of interest and their performance validated in a pilot study. All probes were chosen based on their ability to show specific expression during image analysis/informatics processing. All slides were scanned using an Aperio system (Aperio Technologies, Vista, CA, USA), resulting in digital images with a resolution of 1 μm per pixel. Images were then assessed for quality control, and images that failed quality control metrics were eliminated from the study. All images that passed the quality control metrics are freely available at http://human.brain-map.org/ish/search.
Fifty-eight study genes (Supplementary Table 2) were selected based on their inclusion into one or more of the three categories: approximately half of the genes studied were selected as genes previously associated with SZ; ∼30% of the genes represented specific cell subtypes (primarily interneurons) and almost 40% were used as laminar markers. Genes included in the SZ-associated group were selected based on previous association and expression studies, or are binding partners or in pathways of SZ candidate genes as identified by the gene list provided by Schizophrenia Forum (http://www.schizophreniaforum.org/). SZ-associated genes that are not listed as laminar or cell type markers tended to show pan-cortical labeling patterns.
Image analysis
The presence of areas 9 and 46 were verified by cytoarchitectural analysis, using images from Nissl-stained sections. Areas 9 and 46, corresponding to areas FDm/FDp and FDΔ respectively, were delineated as described by von Economo and Koskinas.36 Area 9 was identified by a relatively cell-sparse layer, 5b, with clumps of von Economo cells present in layers 3 and 5. Area 46 was identified by a relatively uniform distribution of pyramidal cells throughout layer 5 and absence of a discernible layer 5b, whereas von Economo cells were rare or absent. The locations of areas 9 and 46, which often occurred in the same section, were transferred from images of Nissl-stained sections to images of matched ISH-stained sections. The ROI, extending from the pia to just beyond the gray matter/white matter boundary, were taken from domains of the cortex in which the plane of section was perpendicular to the cortical surface. Artifacts such as blood vessels, tears in the tissue or debris were avoided while defining ROIs. A minimum of 5-mm linear cortical length was delineated as ROI on each section; for image analysis, each ROI was analyzed at the scanned resolution of 1 μm2 per pixel (Supplementary Figure 1).
Once ROIs were assigned, they were assessed individually with informatics algorithms34, 35, 37, 38 designed to assess two different parameters: cell density (reported as cells mm−2), and the average staining intensity for each stained cell in the ROI. These assessments were made for the entirety of the ROI (‘all'), as well as for 100 equal subdivisions parallel to the pia and white matter, with the first subdivision at the pia, and the 100th subdivision lying just under the gray matter/white matter border. The subdivisions were scaled such that in all cases exactly 100 would be applied, regardless of the depth of the cortex. This partitioning allowed analysis of the pattern of distribution across the cortex in a comparable way for all individual ROI, independent of the thickness of the cortex, in a manner similar to the methods developed by Zilles and colleagues.39
Statistical analysis
JMP 8.0.2 (SAS Institute, Cary, NC, USA) and R40 statistical programs were used to analyze the data. Histograms for each factor of interest were produced for each gene and area, and outliers were identified as those data points that were greater than the upper quartile +1.5 * (interquartile range) or less than the lower quartile −1.5 * (interquartile range). Diagnosis was covaried with pH, age and PMI for all analyses, and compared via an analysis of covariance (ANCOVA). Cell density and average staining intensity were compared between controls and subjects with SZ for areas 9 and 46, and a combination of the two (‘DLPFC'), and were compared for both the entirety of the ROIs and for individual layers. Every gene/area combination was tested simultaneously across the layers. P-values were corrected across the layers for multiple hypothesis testing using the Benjamini–Hochberg method41 in R. Statistical significance was set at α=0.05.
Results
In this study, we examined the expression of a variety of genes in areas 9 and 46, as well as in the two areas combined (‘DLPFC'). High-throughput image analysis techniques allowed us to analyze thousands of ROIs across 52 individuals and 58 genes. Within each area, ROIs were identified and 100 sublayers were assessed to ascertain gene expression patterns across the depth of the cortex. The expression patterns of laminar marker genes were used to identify which sublayers were contained within the various cortical layers. Cell density and average staining intensity of the reaction product were independently assessed for each gene and compared between control and SZ groups (Supplementary Figure 1). Significant differences were found in 22 of the 58 genes examined; of these, only 5 showed significant differences in area 46. The remaining differences were noted in the DLPFC as a whole or specifically in area 9.
SZ-associated genes
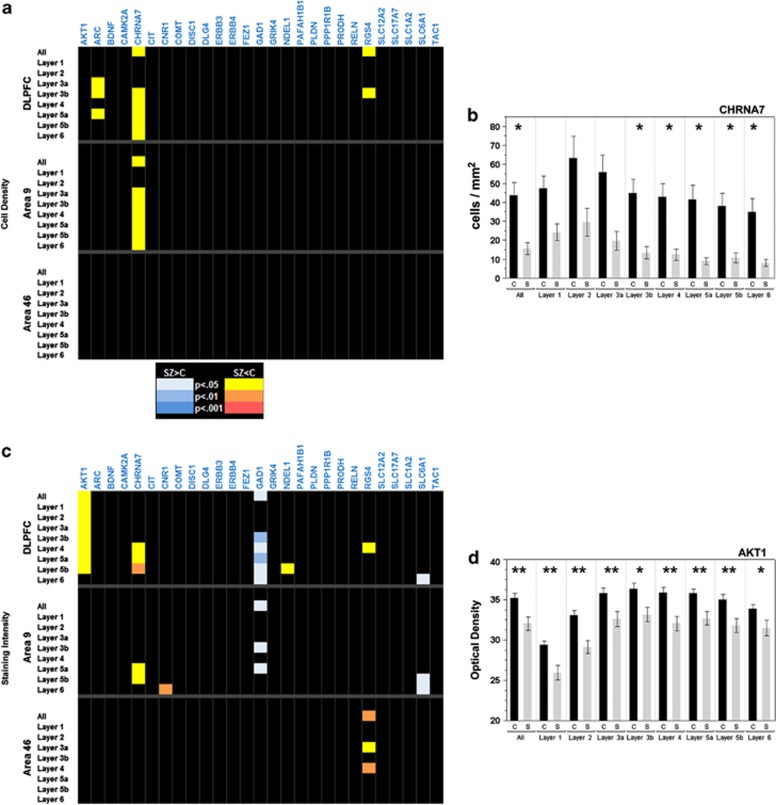
Of the 27 genes chosen for their association with SZ, only 3 (11.1%) showed significant differences in density within the DLPFC. When areas 9 and 46 were examined independently, only one gene, CHRNA7, retained significant differences, and only in area 9 (Figure 1a). The significantly lower densities displayed by ARC and RGS4 in the DLPFC did not survive analysis in individual areas, though the trend for lower densities persisted. The most robust differences were found for the gene CHRNA7, which showed decreased densities compared with controls in both DLPFC and area 9 alone (Figure 1b).
Figure 1.
(a–d) P-value charts for cell density (a), and staining intensity of SZ-associated genes (c). Genes are listed across the top of the graph, while the areas analyzed and their respective layers are on the left. Corrected P-values are denoted by color for each layer as shown in the color key between the two images. Black, NS. (b) Cell density measurements by layer for CHRNA7 in the DLPFC. (d) Staining intensity measurements for AKT1 in the DLPFC. Controls: black bars; SZ: gray bars. Bars represent mean values±s.e.m. for each layer. *P<0.05; **P<0.01.
Changes in staining intensity were found for seven of the SZ-associated genes (25.9%, Figure 1c). The significant differences found for AKT1- and NDEL1- expressing cells were in DLPFC as a whole, but not in individual areas. CHRNA7-expressing cells, which had shown decreased densities, also showed changes in intensity; these changes were found in the DLPFC, but were limited to area 9 when the two regions were examined independently, similar to the finding for density. Decreased expression of CNR1 was limited to layer 6 of area 9. Only RGS4-expressing cells showed significant intensity alterations in area 46. In general, intensity was decreased in SZ when compared with controls, with the most consistent and significant changes found in AKT-expressing cells (Figure 1d). Within the group of SZ-associated genes, only the interneuron markers GAD1 and SLC6A1 had increased expression in SZ. This intensity-specific increase was consistent with other interneuron marker genes (see below).
Interneuron markers
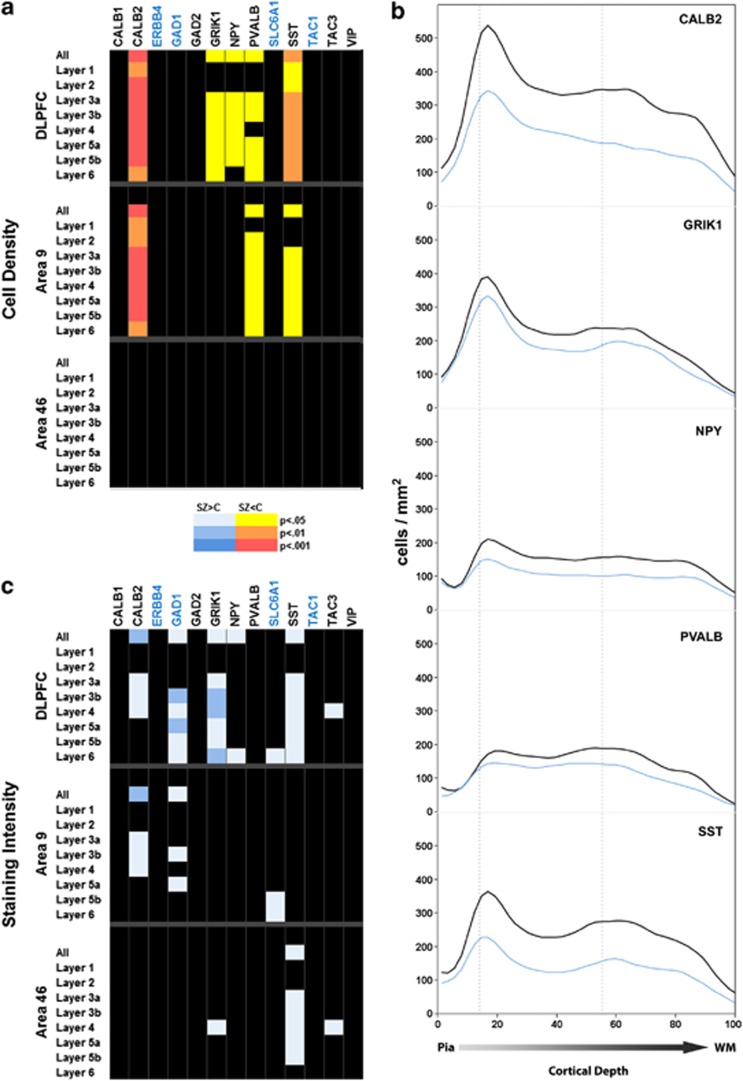
Of the 13 genes chosen to examine interneurons, 5 (38.5%) showed significant differences in density measurements in the DLPFC between controls and SZ subjects (Figure 2a). For these genes, differences were found throughout the depth of the cortex, though less often for layers 1 and 2. Although there was a trend for decreased density in both areas examined, three of these genes showed significant differences only in area 9. The most robust of the density decreases were found for the genes CALB2 and SST, which encode calretinin and somatostatin, respectively. Decreases in densities were also detected in PVALB-, GRIK1- and NPY-expressing cells (Figure 2b). Density was not universally decreased across interneuron markers, however. Density showed no significant alteration for the pan-interneuron markers GAD1, GAD2 or SLC6A1, suggesting that the overall density of interneurons remains unchanged in SZ. Additionally, significant differences were not detected for CALB1- and VIP-expressing interneurons, or for additional markers such as ERBB4 and the tachykinins (TAC1 and TAC3). Although not significant, the trend for several other interneuron markers such as TAC1 and TAC3 was for decreased density in SZ patients, implying a broader decrease in this category of genes.
Figure 2.
P-value charts for cell density (a), and staining intensity (c) of interneuron marker genes. Genes are listed across the top of the graph (SZ-associated genes are in blue), whereas the areas analyzed and their respective layers are on the left. Corrected P-values are denoted by color for each layer as shown in the color key between the two charts. Black, NS. (b) Cortical expression pattern for the cell density of interneuron markers in the DLPFC for the five interneuron markers that showed significant differences. Black lines: control values; blue lines: SZ subjects.
Staining intensity was also altered in the DLPFC for 7 of the 13 (53.8%) interneuron markers, most notably for CALB2, GAD1, GRIK1 and SST (Figure 2c). In contrast to the general finding of more significant differences in area 9, GRIK1-, SST- and TAC3-expressing cells were altered only in area 46. These differences, together with the decreased densities for RGS4-expressing cells noted above and the decreased density of PCP4 in layer 4 noted below, are the only differences that reached significance between the two groups in area 46; alterations in staining intensity for interneuron markers account for two-thirds of the changes seen in area 46. In all instances, unlike the intensity decreases found for the SZ-associated genes, the direction of change was for an increase in interneuron-staining intensity. A possible explanation for this discrepancy is that unlike the SZ-associated genes, decreased expression in this set of genes resulted in the complete loss of detectable staining for a subset of cells that express these genes at low levels. While the density of stained cells would therefore decrease, the average staining intensity of remaining cells would increase due to the remaining higher-expressing cells.
Laminar markers
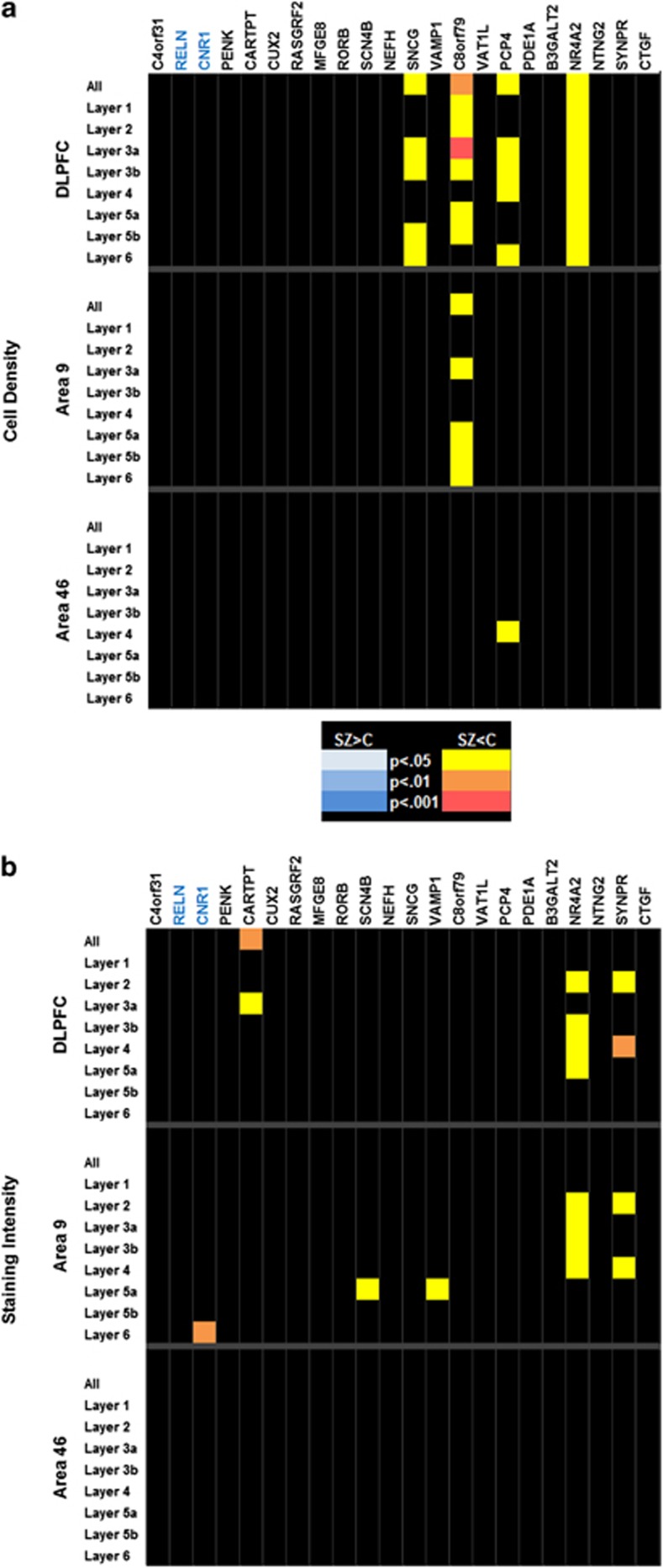
Twenty-two of the genes selected for this study were chosen as markers for specific layers of the cortex. Whereas each of the markers had been chosen because it showed enhanced expression in one or more layers, lighter labeling was also often seen at lower levels or at decreased densities in other layers of the cortex. Of these, 4 (18.2%) showed significant changes in density (Figure 3a), with the changes occurring throughout the depth of the cortex as opposed to only the layer for which the gene was preferentially expressed. One of these genes, C8orf79 was significantly different in area 9, whereas PCP4 showed differences only in layer 4 of area 46.
Figure 3.
(a–d) P-value charts for cell density and staining intensity of laminar marker genes. No significant differences were found in area 46. Genes are listed across the top of the graph (SZ-associated genes are in blue), with genes that are preferentially expressed in layer 1 to the left, and those preferentially expressed in layer 6 to the right. The areas analyzed and their respective layers are on the left. Corrected P-values are denoted by color for each layer as shown in the color key between the charts. Black, NS.
Significant differences in intensity were found in the DLPFC for three genes: CARTPT, NR4A2, and SYNPR (Figure 3b). Changes in intensity of the latter two genes were also seen in area 9, but not in area 46. Layer specific changes in intensity were also found for CNR1, SNCG, SCN4B and VAMP1 in area 9 only, whereas no changes were found for any gene in area 46.
Remaining cell type markers
Probes directed against oligodendrocytes (MBP) or astrocytes (SLC1A2) showed no significant differences in either density or intensity. CLDN5 expression, used as a marker of vascular endothelium, showed significant decreases in intensity in the DLPFC, but these changes were limited to area 9 when the areas were examined independently (data not shown).
Discussion
Reports of differences in cell densities within the prefrontal cortex have varied widely in the SZ literature. Some studies have found loss of specific subgroups of cells42, 43 or decreases in cell density.44 Other studies have reported increased density in specific layers of the cortex9, 14, 15 or alterations in the underlying white matter.45, 46 In this study, we have found no evidence of decreases in the overall density of cells in SZ as compared to controls. Although specific subgroups of interneurons showed significantly lower cell densities in disease, the densities for GAD1, GAD2 and SLC6A1 suggest that there is no significant decline in interneurons as a whole. It is also important to note that the findings from this study may be attributable to confounding factors, including increased tobacco use, antipsychotic medication and/or environmental factors such as social isolation or a lack of intellectual stimulation.
In this study, we examined the expression of a variety of genes of several types. Consistent with previous reports, we have shown that a variety of genes expressed in interneurons show alterations in SZ. Disruption of the GABAergic system of the prefrontal cortex is well accepted as being a component of SZ pathology. Akbarian et al.47 initially reported a decrease in the density of GAD1-expressing neurons in the DLPFC of subjects with SZ. Volk et al.48 reported a similar decrease in GAD1-expressing cells in area 9 and in the density of the GABA transporter SLC6A1-expressing cells in areas 9 and 10 of the prefrontal cortex,49 although the expression per cell for each of these genes did not differ. Both of these studies concluded that the decrease in density was a result of expression levels falling below detectable level, rather than loss of interneurons. In our study, the density of cells identified by pan-interneuron markers (i.e., GAD1, GAD2 and SLC6A2) showed no differences, suggesting that the overall population of GABAergic interneurons in the DLPFC is not changed between the two groups.
However, we did find alterations in the expression patterns of multiple genes that are markers of interneurons, particularly in Brodmann area 9. Specifically, decreased densities were seen in cells expressing a variety of markers for subsets of interneurons: CALB2, SST and PVALB. Interestingly, these genes do not all highlight the same subset of interneurons, and in many cases, markers for genes in the same subset were unaffected. For example, while SST-expressing cells showed robust changes in both density and intensity, CALB1 showed no detectable changes. Although it is unclear to what degree these two populations may overlap in the DLPFC, it has been shown that in the temporal cortex, over 80% of cells that stained for somatostatin also stain for calbindin.50 In that study, somatostatin did not colocalize with calretinin or parvalbumin, both of which show changes in our study. The most plausible explanation is consistent with previous studies showing that the expression of specific genes is decreased below the level of detection in a subset of cells, resulting in what appears to be cell loss. This is further substantiated in light of a failure to identify a loss of interneurons overall with GAD1, GAD2 or SLC1A2. It is possible that the effects we are observing are due to a shift in the proportion of interneurons displaying specific markers; however, we did not find a compensatory increase in any of the markers examined. It should be noted that while the tyramide protocol employed here utilizes amplification, and is therefore inherently less linear of a reaction than what is observed with autoradiography, the relative relationship between expression levels and staining intensity remains applicable. As with all labeling procedures, a threshold for detection exists such that lack of labeling cannot be easily dismissed as a complete absence of expression, but merely failure of the expression levels to reach detection threshold.
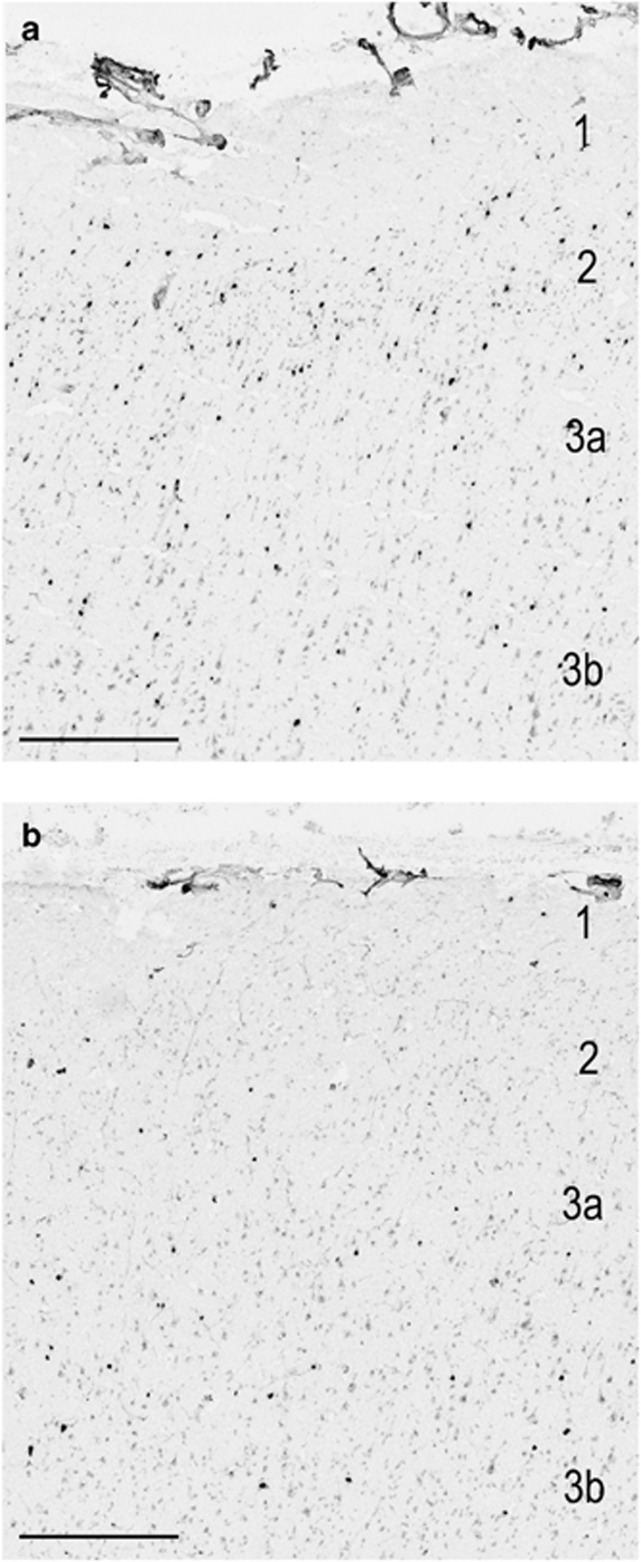
Interestingly, our most robust finding involved a decrease in the density of cells expressing CALB2 (Figure 4). Although numerous immunohistochemical studies have shown no decrease in the number of cells that express calretinin,51, 52, 53, 54, 55, 56, 57 one quantitative PCR study showed a significant decrease in the amount of RNA for CALB2 when examining subjects with SZ,58 similar to our results, although an additional study using RT-PCR failed to recapitulate these results.59 It is therefore possible that although the CALB2 mRNA expression levels may fall to below the detection levels for ISH, even low-expressing cells are capable of producing enough calretinin to be indistinguishable at the protein level.
Figure 4.
The decrease in the density of cells stained for CALB2 in area 9 is apparent in the supragranular layers of the cortex when control tissue (a), is compared with tissue from an individual with SZ (b). Size bar=400 μm.
It is also worth noting that one of the genes showing the most robust differences was CHRNA7, which encodes the α7 nicotinic acetylcholine receptor. Interestingly, CHRNA7 has been localized to a subset of interneurons, especially in the striatum and the hippocampus,60 and studies have suggested that CHRNA7 expression is decreased in a variety of brain regions in SZ.61, 62 Additionally, a number of studies have identified various SNPs in CHRNA7 as possible risk factors for disease development,63, 64, 65 although earlier studies reported no evidence for linkage.66, 67 Some of the markers identified, however, are located within or near a region of the gene that is duplicated in a chimeric gene, CHRFAM7A. Studies have shown that this chimeric gene may also be implicated in SZ,68, 69 making it unclear to what extent CHRNA7 itself is affected. The probe that was used in this study had been specifically designed to have no overlap with the chimeric gene, and our results can therefore be attributed specifically to changes in CHRNA7 alone.
An intriguing finding from our study is that within the DLPFC, almost all of the significant changes were specific to area 9, extending the results of previous studies13, 14, 15 that suggest regional specificity in the disease. Similarly, a recent study70 using weighted-gene coexpression analysis also found only minimal changes in area 46, whereas additional Brodmann areas (21, 32 and 38) showed significant differences between SZ subjects and controls. Although false negative results due to an insufficient N for area 46 are possible, power analysis on our data set suggests that when significance was reached in area 9, the number of samples required to reach significance in area 46 was significantly greater (t=−4.07, df=70, P=0.0001). For genes that failed to reach significance in area 9, there was no such difference. These analyses, together with other studies, suggest that the failure to find significance in area 46 may, at least in part, reflect regional differences in pathology.
The results found with interneuron markers and CHRNA7 support the use of a high-throughput, cellular resolution platform for identifying changes associated with gene expression level in diseased individuals and the use of image analysis to detect changes that may not be evident with other methodologies. While the laminar markers were not expected to have robust changes between the groups, some showed interesting results that could justify additional studies. Of note is the expression of C8orf79 and NR4A2. C8orf79 encodes a putative methyltransferase, which is interesting in light of the recent focus on epigenetic factors in SZ, as well as proposed involvement of other methyltransferases such as COMT71 and DNMT172, 73, 74 (DNA methyltransferase-1). NR4A2 is a nuclear receptor protein that can act as a transcription factor. The mouse homolog, Nurr1, has been shown to influence the expression of several genes involved in brain development, especially within the dopaminergic system.75, 76, 77, 78 Haplotype effects of NR4A2 have been proposed in SZ,79, 80 although multiple studies have failed to find a direct linkage between NR4A2 alleles and SZ.81, 82 Large-scale ISH studies such as the one presented here can shed light on specifically where and to what degree expression levels of both known and novel genes are altered in human disease.
Acknowledgments
We would like to thank the families of the deceased for the donations of brain tissue, and their time and effort devoted to the consent process and interviews, and the staff of the Offices of the Chief Medical Examiner of District of Columbia and Northern Virginia for their assistance. We would also like to thank the Allen Institute founders, PG Allen and J Allen, for their vision, encouragement and support.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
Supplementary Material
References
- American Psychiatric Association, American Psychiatric Association. Task Force on DSM-IV Diagnostic and Statistical Manual of Mental Disorders: DSM-IV4th edn.American Psychiatric Association: Washington, DC; 886.1994. xxvii. [Google Scholar]
- Jablensky A. The diagnostic concept of schizophrenia: its history, evolution, and future prospects. Dialogues Clin Neurosci. 2010;12:271–287. doi: 10.31887/DCNS.2010.12.3/ajablensky. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry JD, Rendell PG, Kliegel M, Altgassen M. Prospective memory in schizophrenia: primary or secondary impairment. Schizophr Res. 2007;95:179–185. doi: 10.1016/j.schres.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Nestor PG, Niznikiewicz M, McCarley RW. Distinct contribution of working memory and social comprehension failures in neuropsychological impairment in schizophrenia. J Nerv Ment Dis. 2010;198:206–212. doi: 10.1097/NMD.0b013e3181d14143. [DOI] [PubMed] [Google Scholar]
- Schwartz BL, Rosse RB, Deutsch SI. Toward a neuropsychology of memory in schizophrenia. Psychopharmacol Bull. 1992;28:341–351. [PubMed] [Google Scholar]
- Tek C, Gold J, Blaxton T, Wilk C, McMahon RP, Buchanan RW. Visual perceptual and working memory impairments in schizophrenia. Arch Gen Psychiatry. 2002;59:146–153. doi: 10.1001/archpsyc.59.2.146. [DOI] [PubMed] [Google Scholar]
- Horan WP, Braff DL, Nuechterlein KH, Sugar CA, Cadenhead KS, Calkins ME, et al. Verbal working memory impairments in individuals with schizophrenia and their first-degree relatives: findings from the Consortium on the Genetics of Schizophrenia. Schizophr Res. 2008;103:218–228. doi: 10.1016/j.schres.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danos P, Baumann B, Kramer A, Bernstein HG, Stauch R, Krell D, et al. Volumes of association thalamic nuclei in schizophrenia: a postmortem study. Schizophr Res. 2003;60:141–155. doi: 10.1016/s0920-9964(02)00307-9. [DOI] [PubMed] [Google Scholar]
- Goldman-Rakic PS, Selemon LD. Functional and anatomical aspects of prefrontal pathology in schizophrenia. Schizophr Bull. 1997;23:437–458. doi: 10.1093/schbul/23.3.437. [DOI] [PubMed] [Google Scholar]
- Jones EG. Cortical development and thalamic pathology in schizophrenia. Schizophr Bull. 1997;23:483–501. doi: 10.1093/schbul/23.3.483. [DOI] [PubMed] [Google Scholar]
- Popken GJ, Bunney WE, Potkin SG, Jones EG. Subnucleus-specific loss of neurons in medial thalamus of schizophrenics. Proc Natl Acad Sci USA. 2000;97:9276–9280. doi: 10.1073/pnas.150243397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selemon LD. Increased cortical neuronal density in schizophrenia. Am J Psychiatry. 2004;161:1564. doi: 10.1176/appi.ajp.161.9.1564. [DOI] [PubMed] [Google Scholar]
- Selemon LD, Mrzljak J, Kleinman JE, Herman MM, Goldman-Rakic PS. Regional specificity in the neuropathologic substrates of schizophrenia: a morphometric analysis of Broca's area 44 and area 9. Arch Gen Psychiatry. 2003;60:69–77. doi: 10.1001/archpsyc.60.1.69. [DOI] [PubMed] [Google Scholar]
- Selemon LD, Rajkowska G, Goldman-Rakic PS.Abnormally high neuronal density in the schizophrenic cortex. A morphometric analysis of prefrontal area 9 and occipital area 17 Arch Gen Psychiatry 199552805–818.discussion 819-820. [DOI] [PubMed] [Google Scholar]
- Selemon LD, Rajkowska G, Goldman-Rakic PS. Elevated neuronal density in prefrontal area 46 in brains from schizophrenic patients: application of a three-dimensional, stereologic counting method. J Comp Neurol. 1998;392:402–412. [PubMed] [Google Scholar]
- Weinberger DR. On localizing schizophrenic neuropathology. Schizophr Bull. 1997;23:537–540. doi: 10.1093/schbul/23.3.537. [DOI] [PubMed] [Google Scholar]
- Weinberger DR, Wagner RL, Wyatt RJ. Neuropathological studies of schizophrenia: a selective review. Schizophr Bull. 1983;9:193–212. doi: 10.1093/schbul/9.2.193. [DOI] [PubMed] [Google Scholar]
- Brigman JL, Padukiewicz KE, Sutherland ML, Rothblat LA. Executive functions in the heterozygous reeler mouse model of schizophrenia. Behav Neurosci. 2006;120:984–988. doi: 10.1037/0735-7044.120.4.984. [DOI] [PubMed] [Google Scholar]
- Enomoto T, Tse MT, Floresco SB. Reducing prefrontal gamma-aminobutyric acid activity induces cognitive, behavioral, and dopaminergic abnormalities that resemble schizophrenia. Biol Psychiatry. 2011;69:432–441. doi: 10.1016/j.biopsych.2010.09.038. [DOI] [PubMed] [Google Scholar]
- Barr MS, Farzan F, Rusjan PM, Chen R, Fitzgerald PB, Daskalakis ZJ. Potentiation of gamma oscillatory activity through repetitive transcranial magnetic stimulation of the dorsolateral prefrontal cortex. Neuropsychopharmacology. 2009;34:2359–2367. doi: 10.1038/npp.2009.79. [DOI] [PubMed] [Google Scholar]
- Basar-Eroglu C, Brand A, Hildebrandt H, Karolina Kedzior K, Mathes B, Schmiedt C. Working memory related gamma oscillations in schizophrenia patients. Int J Psychophysiol. 2007;64:39–45. doi: 10.1016/j.ijpsycho.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Basar-Eroglu C, Mathes B, Brand A, Schmiedt-Fehr C. Occipital gamma response to auditory stimulation in patients with schizophrenia. Int J Psychophysiol. 2011;79:3–8. doi: 10.1016/j.ijpsycho.2010.10.011. [DOI] [PubMed] [Google Scholar]
- Basar-Eroglu C, Schmiedt-Fehr C, Marbach S, Brand A, Mathes B. Altered oscillatory alpha and theta networks in schizophrenia. Brain Res. 2008;1235:143–152. doi: 10.1016/j.brainres.2008.06.114. [DOI] [PubMed] [Google Scholar]
- Kleinman JE, Law AJ, Lipska BK, Hyde TM, Ellis JK, Harrison PJ, et al. Genetic neuropathology of schizophrenia: new approaches to an old question and new uses for postmortem human brains. Biol Psychiatry. 2011;69:140–145. doi: 10.1016/j.biopsych.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Sanders AR, Gejman PV. Genome-wide approaches to schizophrenia. Brain Res Bull. 2010;83:93–102. doi: 10.1016/j.brainresbull.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones LB. Recent cytoarchitechtonic changes in the prefrontal cortex of schizophrenics. Front Biosci\ 2001;6:E148–E153. doi: 10.2741/jones. [DOI] [PubMed] [Google Scholar]
- Forbes NF, Carrick LA, McIntosh AM, Lawrie SM. Working memory in schizophrenia: a meta-analysis. Psychol Med. 2009;39:889–905. doi: 10.1017/S0033291708004558. [DOI] [PubMed] [Google Scholar]
- Heinrichs RW, Zakzanis KK. Neurocognitive deficit in schizophrenia: a quantitative review of the evidence. Neuropsychology. 1998;12:426–445. doi: 10.1037//0894-4105.12.3.426. [DOI] [PubMed] [Google Scholar]
- Lee J, Park S. Working memory impairments in schizophrenia: a meta-analysis. J Abnorm Psychol. 2005;114:599–611. doi: 10.1037/0021-843X.114.4.599. [DOI] [PubMed] [Google Scholar]
- Lee J, Folley BS, Gore J, Park S. Origins of spatial working memory deficits in schizophrenia: an event-related FMRI and near-infrared spectroscopy study. PloS ONE. 2008;3:e1760. doi: 10.1371/journal.pone.0001760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mexal S, Berger R, Logel J, Ross RG, Freedman R, Leonard S. Differential regulation of alpha7 nicotinic receptor gene (CHRNA7) expression in schizophrenic smokers. J Mol Neurosci. 2010;40:185–195. doi: 10.1007/s12031-009-9233-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halim ND, Lipska BK, Hyde TM, Deep-Soboslay A, Saylor EM, Herman MM, et al. Increased lactate levels and reduced pH in postmortem brains of schizophrenics: medication confounds. J Neurosci Methods. 2008;169:208–213. doi: 10.1016/j.jneumeth.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JR, Hatsukami DK, Mitchell JE, Dahlgren LA. Prevalence of smoking among psychiatric outpatients. Am J Psychiatry. 1986;143:993–997. doi: 10.1176/ajp.143.8.993. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Zeng H, Shen EH, Hohmann JG, Oh SW, Bernard A, Royall JJ, et al. Large-scale cellular-resolution gene profiling in human neocortex reveals species-specific molecular signatures. Cell. 2012;149:483–496. doi: 10.1016/j.cell.2012.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Economo C, Koskinas GN. Atlas of Cytoarchitectonics of the Adult Human Cerebral Cortex. S Karger AG: Basel; 2008. [Google Scholar]
- Ng L, Pathak SD, Kuan C, Lau C, Dong H, Sodt A, et al. Neuroinformatics for genome-wide 3D gene expression mapping in the mouse brain. IEEE/ACM Trans Comput Biol Bioinform. 2007;4:382–393. doi: 10.1109/tcbb.2007.1035. [DOI] [PubMed] [Google Scholar]
- Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleicher A, Palomero-Gallagher N, Morosan P, Eickhoff SB, Kowalski T, de Vos K, et al. Quantitative architectural analysis: a new approach to cortical mapping. Anat Embryol (Berl) 2005;210:373–386. doi: 10.1007/s00429-005-0028-2. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- Akbarian S, Kim JJ, Potkin SG, Hetrick WP, Bunney WE, Jones EG. Maldistribution of interstitial neurons in prefrontal white matter of the brains of schizophrenic patients. Arch Gen Psychiatry. 1996;53:425–436. doi: 10.1001/archpsyc.1996.01830050061010. [DOI] [PubMed] [Google Scholar]
- Benes FM, McSparren J, Bird ED, SanGiovanni JP, Vincent SL. Deficits in small interneurons in prefrontal and cingulate cortices of schizophrenic and schizoaffective patients. Arch Gen Psychiatry. 1991;48:996–1001. doi: 10.1001/archpsyc.1991.01810350036005. [DOI] [PubMed] [Google Scholar]
- Benes FM, Davidson J, Bird ED. Quantitative cytoarchitectural studies of the cerebral cortex of schizophrenics. Arch Gen Psychiatry. 1986;43:31–35. doi: 10.1001/archpsyc.1986.01800010033004. [DOI] [PubMed] [Google Scholar]
- Davis KL, Stewart DG, Friedman JI, Buchsbaum M, Harvey PD, Hof PR, et al. White matter changes in schizophrenia: evidence for myelin-related dysfunction. Arch Gen Psychiatry. 2003;60:443–456. doi: 10.1001/archpsyc.60.5.443. [DOI] [PubMed] [Google Scholar]
- Hoistad M, Segal D, Takahashi N, Sakurai T, Buxbaum JD, Hof PR. Linking white and gray matter in schizophrenia: oligodendrocyte and neuron pathology in the prefrontal cortex. Front Neuroanat. 2009;3:9. doi: 10.3389/neuro.05.009.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbarian S, Kim JJ, Potkin SG, Hagman JO, Tafazzoli A, Bunney WE, et al. Gene expression for glutamic acid decarboxylase is reduced without loss of neurons in prefrontal cortex of schizophrenics. Arch Gen Psychiatry. 1995;52:258–266. doi: 10.1001/archpsyc.1995.03950160008002. [DOI] [PubMed] [Google Scholar]
- Volk DW, Austin MC, Pierri JN, Sampson AR, Lewis DA. Decreased glutamic acid decarboxylase67 messenger RNA expression in a subset of prefrontal cortical gamma-aminobutyric acid neurons in subjects with schizophrenia. Arch Gen Psychiatry. 2000;57:237–245. doi: 10.1001/archpsyc.57.3.237. [DOI] [PubMed] [Google Scholar]
- Volk D, Austin M, Pierri J, Sampson A, Lewis D. GABA transporter-1 mRNA in the prefrontal cortex in schizophrenia: decreased expression in a subset of neurons. Am J Psychiatry. 2001;158:256–265. doi: 10.1176/appi.ajp.158.2.256. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Albo MC, Elston GN, DeFelipe J. The human temporal cortex: characterization of neurons expressing nitric oxide synthase, neuropeptides and calcium-binding proteins, and their glutamate receptor subunit profiles. Cereb Cortex. 2001;11:1170–1181. doi: 10.1093/cercor/11.12.1170. [DOI] [PubMed] [Google Scholar]
- Beasley CL, Zhang ZJ, Patten I, Reynolds GP. Selective deficits in prefrontal cortical GABAergic neurons in schizophrenia defined by the presence of calcium-binding proteins. Biol Psychiatry. 2002;52:708–715. doi: 10.1016/s0006-3223(02)01360-4. [DOI] [PubMed] [Google Scholar]
- Cotter D, Landau S, Beasley C, Stevenson R, Chana G, MacMillan L, et al. The density and spatial distribution of GABAergic neurons, labelled using calcium binding proteins, in the anterior cingulate cortex in major depressive disorder, bipolar disorder, and schizophrenia. Biol Psychiatry. 2002;51:377–386. doi: 10.1016/s0006-3223(01)01243-4. [DOI] [PubMed] [Google Scholar]
- Daviss SR, Lewis DA. Local circuit neurons of the prefrontal cortex in schizophrenia: selective increase in the density of calbindin-immunoreactive neurons. Psychiatry Res. 1995;59:81–96. doi: 10.1016/0165-1781(95)02720-3. [DOI] [PubMed] [Google Scholar]
- Reynolds GP, Abdul-Monim Z, Neill JC, Zhang ZJ. Calcium binding protein markers of GABA deficits in schizophrenia—postmortem studies and animal models. Neurotox Res. 2004;6:57–61. doi: 10.1007/BF03033297. [DOI] [PubMed] [Google Scholar]
- Sakai T, Oshima A, Nozaki Y, Ida I, Haga C, Akiyama H, et al. Changes in density of calcium-binding-protein-immunoreactive GABAergic neurons in prefrontal cortex in schizophrenia and bipolar disorder. Neuropathology. 2008;28:143–150. doi: 10.1111/j.1440-1789.2007.00867.x. [DOI] [PubMed] [Google Scholar]
- Tooney PA, Chahl LA. Neurons expressing calcium-binding proteins in the prefrontal cortex in schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry. 2004;28:273–278. doi: 10.1016/j.pnpbp.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Sun J, Reynolds GP. A selective reduction in the relative density of parvalbumin-immunoreactive neurons in the hippocampus in schizophrenia patients. Chin Med J (Engl) 2002;115:819–823. [PubMed] [Google Scholar]
- Fung SJ, Webster MJ, Sivagnanasundaram S, Duncan C, Elashoff M, Weickert CS. Expression of interneuron markers in the dorsolateral prefrontal cortex of the developing human and in schizophrenia. Am J Psychiatry. 2010;167:1479–1488. doi: 10.1176/appi.ajp.2010.09060784. [DOI] [PubMed] [Google Scholar]
- Hashimoto T, Bazmi HH, Mirnics K, Wu Q, Sampson AR, Lewis DA. Conserved regional patterns of GABA-related transcript expression in the neocortex of subjects with schizophrenia. Am J Psychiatry. 2008;165:479–489. doi: 10.1176/appi.ajp.2007.07081223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams CE, Stitzel JA, Collins AC, Freedman R. Alpha7-nicotinic receptor expression and the anatomical organization of hippocampal interneurons. Brain Res. 2001;922:180–190. doi: 10.1016/s0006-8993(01)03115-8. [DOI] [PubMed] [Google Scholar]
- Freedman R, Hall M, Adler LE, Leonard S. Evidence in postmortem brain tissue for decreased numbers of hippocampal nicotinic receptors in schizophrenia. Biol Psychiatry. 1995;38:22–33. doi: 10.1016/0006-3223(94)00252-X. [DOI] [PubMed] [Google Scholar]
- Guan ZZ, Zhang X, Blennow K, Nordberg A. Decreased protein level of nicotinic receptor alpha7 subunit in the frontal cortex from schizophrenic brain. Neuroreport. 1999;10:1779–1782. doi: 10.1097/00001756-199906030-00028. [DOI] [PubMed] [Google Scholar]
- Freedman R, Leonard S, Gault JM, Hopkins J, Cloninger CR, Kaufmann CA, et al. Linkage disequilibrium for schizophrenia at the chromosome 15q13-14 locus of the alpha7-nicotinic acetylcholine receptor subunit gene (CHRNA7) Am J Med Genet. 2001;105:20–22. [PubMed] [Google Scholar]
- Riley BP, Makoff A, Mogudi-Carter M, Jenkins T, Williamson R, Collier D, et al. Haplotype transmission disequilibrium and evidence for linkage of the CHRNA7 gene region to schizophrenia in Southern African Bantu families. Am J Med Genet. 2000;96:196–201. doi: 10.1002/(sici)1096-8628(20000403)96:2<196::aid-ajmg15>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Stassen HH, Bridler R, Hagele S, Hergersberg M, Mehmann B, Schinzel A, et al. Schizophrenia and smoking: evidence for a common neurobiological basis. Am J Med Genet. 2000;96:173–177. [PubMed] [Google Scholar]
- Curtis L, Blouin JL, Radhakrishna U, Gehrig C, Lasseter VK, Wolyniec P, et al. No evidence for linkage between schizophrenia and markers at chromosome 15q13-14. Am J Med Genet. 1999;88:109–112. doi: 10.1002/(sici)1096-8628(19990416)88:2<109::aid-ajmg1>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Neves-Pereira M, Bassett AS, Honer WG, Lang D, King NA, Kennedy JL. No evidence for linkage of the CHRNA7 gene region in Canadian schizophrenia families. Am J Med Genet. 1998;81:361–363. doi: 10.1002/(sici)1096-8628(19980907)81:5<361::aid-ajmg3>3.0.co;2-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempster EL, Toulopoulou T, McDonald C, Bramon E, Walshe M, Wickham H, et al. Episodic memory performance predicted by the 2 bp deletion in exon 6 of the "alpha 7-like" nicotinic receptor subunit gene. Am J Psychiatry. 2006;163:1832–1834. doi: 10.1176/ajp.2006.163.10.1832. [DOI] [PubMed] [Google Scholar]
- Sinkus ML, Lee MJ, Gault J, Logel J, Short M, Freedman R, et al. A 2-base pair deletion polymorphism in the partial duplication of the alpha7 nicotinic acetylcholine gene (CHRFAM7A) on chromosome 15q14 is associated with schizophrenia. Brain Res. 2009;1291:1–11. doi: 10.1016/j.brainres.2009.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussos P, Katsel P, Davis KL, Siever LJ, Haroutunian VA. A System-level transcriptomic analysis of schizophrenia using postmortem brain tissue samples. Arch Gen Psychiatry. 2012;69:1–11. doi: 10.1001/archgenpsychiatry.2012.704. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Weickert CS, Beltaifa S, Kolachana B, Chen J, Hyde TM, et al. Catechol O-methyltransferase (COMT) mRNA expression in the dorsolateral prefrontal cortex of patients with schizophrenia. Neuropsychopharmacology. 2003;28:1521–1530. doi: 10.1038/sj.npp.1300218. [DOI] [PubMed] [Google Scholar]
- Costa E, Dong E, Grayson DR, Guidotti A, Ruzicka W, Veldic M. Reviewing the role of DNA (cytosine-5) methyltransferase overexpression in the cortical GABAergic dysfunction associated with psychosis vulnerability. Epigenetics. 2007;2:29–36. doi: 10.4161/epi.2.1.4063. [DOI] [PubMed] [Google Scholar]
- Veldic M, Guidotti A, Maloku E, Davis JM, Costa E. In psychosis, cortical interneurons overexpress DNA-methyltransferase 1. Proc Natl Acad Sci USA. 2005;102:2152–2157. doi: 10.1073/pnas.0409665102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldic M, Kadriu B, Maloku E, Agis-Balboa RC, Guidotti A, Davis JM, et al. Epigenetic mechanisms expressed in basal ganglia GABAergic neurons differentiate schizophrenia from bipolar disorder. Schizophr Res. 2007;91:51–61. doi: 10.1016/j.schres.2006.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eells JB, Lipska BK, Yeung SK, Misler JA, Nikodem VM. Nurr1-null heterozygous mice have reduced mesolimbic and mesocortical dopamine levels and increased stress-induced locomotor activity. Behav Brain Res. 2002;136:267–275. doi: 10.1016/s0166-4328(02)00185-7. [DOI] [PubMed] [Google Scholar]
- Eells JB, Misler JA, Nikodem VM. Early postnatal isolation reduces dopamine levels, elevates dopamine turnover and specifically disrupts prepulse inhibition in Nurr1-null heterozygous mice. Neuroscience. 2006;140:1117–1126. doi: 10.1016/j.neuroscience.2005.12.065. [DOI] [PubMed] [Google Scholar]
- Galleguillos D, Fuentealba JA, Gomez LM, Saver M, Gomez A, Nash K, et al. Nurr1 regulates RET expression in dopamine neurons of adult rat midbrain. J Neurochem. 2010;114:1158–1167. doi: 10.1111/j.1471-4159.2010.06841.x. [DOI] [PubMed] [Google Scholar]
- Moore TM, Brown T, Cade M, Eells JB. Alterations in amphetamine-stimulated dopamine overflow due to the Nurr1-null heterozygous genotype and postweaning isolation. Synapse. 2008;62:764–774. doi: 10.1002/syn.20550. [DOI] [PubMed] [Google Scholar]
- Chen YH, Tsai MT, Shaw CK, Chen CH. Mutation analysis of the human NR4A2 gene, an essential gene for midbrain dopaminergic neurogenesis, in schizophrenic patients. Am J Med Genet. 2001;105:753–757. doi: 10.1002/ajmg.10036. [DOI] [PubMed] [Google Scholar]
- Xing G, Zhang L, Russell S, Post R. Reduction of dopamine-related transcription factors Nurr1 and NGFI-B in the prefrontal cortex in schizophrenia and bipolar disorders. Schizophr Res. 2006;84:36–56. doi: 10.1016/j.schres.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Carmine A, Buervenich S, Galter D, Jonsson EG, Sedvall GC, Farde L, et al. NURR1 promoter polymorphisms: Parkinson's disease, schizophrenia, and personality traits. Am J Med Genet B Neuropsychiatr Genet. 2003;120B:51–57. doi: 10.1002/ajmg.b.20033. [DOI] [PubMed] [Google Scholar]
- Ruano D, Macedo A, Dourado A, Soares MJ, Valente J, Coelho I, et al. NR4A2 and schizophrenia: lack of association in a Portuguese/Brazilian study. Am J Med Genet B Neuropsychiatr Genet. 2004;128B:41–45. doi: 10.1002/ajmg.b.30031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.