Abstract
Previous studies in hepatocyte-derived cell lines and the whole liver established that the aryl hydrocarbon receptor (AhR) can disrupt G1-phase cell cycle progression following exposure to persistent AhR agonists, such as TCDD (dioxin, 2,3,7,8-tetrachlorodibenzo-p-dioxin). Growth arrest was attributed to inhibition of G1-phase cyclin-dependent kinase 2 (CDK2) activity. The present study examined the effect of TCDD exposure on liver regeneration following 70% partial hepatectomy in mice lacking the Cip/Kip inhibitors p21Cip1 or p27Kip1 responsible for regulating CDK2 activity. Assessment of the regenerative process in wild-type, p21Cip1 knockout, and p27Kip1 knockout mice confirmed that TCDD-induced inhibition of liver regeneration is entirely dependent on p21Cip1 expression. Compared with wild-type mice, the absence of p21Cip1 expression completely abrogated the TCDD inhibition, and accelerated hepatocyte progression through G1 phase during the regenerative process. Analysis of the transcriptional response determined that increased p21Cip1 expression during liver regeneration involved an AhR-dependent mechanism. Chromatin immunoprecipitation studies revealed that p21Cip1 induction required AhR binding to the newly characterized nonconsensus xenobiotic response element, in conjunction with the tumor suppressor protein Kruppel-like factor 6 functioning as an AhR binding partner. The evidence also suggests that AhR functionality following partial hepatectomy is dependent on a p21Cip1-regulated signaling process, intimately linking AhR biology to the G1-phase cell cycle program.
Introduction
The liver is the only solid organ known to be capable of regenerating organ mass in response to damage (Huang and Elferink, 2005). This is important given the role of the liver in metabolism of both endogenous and exogenous compounds. One such example is the metabolism of exogenous aromatic hydrocarbon compounds by enzymes of the cytochrome P450 system (Denison and Nagy, 2003). Central to both the regenerative and metabolic functions of the liver is the ligand-activated basic helix-loop-helix transcription factor of the Per-aryl hydrocarbon receptor nuclear translocator (Arnt)-Sim family, known as the aryl hydrocarbon receptor (AhR) (Fukunaga et al., 1995). The AhR is activated by numerous polycyclic and halogenated aromatic hydrocarbons, including the prototypical ligand TCDD (dioxin, 2,3,7,8-tetrachlorodibenzo-p-dioxin) (Denison and Nagy, 2003). In humans, TCDD is known to cause a number of deleterious health effects, including liver toxicity, immunotoxicity, dermal toxicity, tumor promotion, and developmental abnormalities (Safe, 1986). These effects of TCDD are all mediated by the AhR (Poland and Knutson, 1982; Nebert et al., 1993; Fernandez-Salguero et al., 1996).
In the absence of a ligand, the AhR exists in the cytosol bound by chaperones, including heat-shock proteins (Hankinson, 1995) and an immunophilin-like protein (Whitlock, 1999). Upon ligand binding, the AhR translocates to the nucleus, dissociates from the chaperones, and interacts with DNA-binding partners to regulate target gene expression. Canonically, the nuclear AhR heterodimerizes with the Arnt (Probst et al., 1993; Reisz-Porszasz et al., 1994) and binds to a core recognition motif (5′-GCGTG-3′) termed the xenobiotic response element (XRE) (Watson and Hankinson, 1992; Swanson et al., 1995). CYP1A1 represents the prototypical XRE-regulated AhR target gene (for review, see Denison et al., 2011). Recent studies, however, have revealed that the TCDD-activated AhR also forms a transcriptionally active heterodimeric complex with the Kruppel-like factor 6 (KLF6) protein bound to a nonconsensus XRE (NC-XRE) (Huang and Elferink, 2012; Wilson et al., 2013).
It is now well accepted that the AhR plays an important role in cell cycle progression (Ma and Whitlock, 1996; Kolluri et al., 1999; Abdelrahim et al., 2003; Denison et al., 2011). Indeed, studies have shown that TCDD exposure leads to G1 cell cycle arrest in cell lines that express a functional AhR, but not in cell lines lacking AhR expression (Gottlicher and Wiebel, 1991; Weiss et al., 1996). Previous work has also shown that the cell cycle effects depend on a direct interaction between the AhR and the active (hypophosphorylated) form of the retinoblastoma tumor suppressor protein (Rb) (Ge and Elferink, 1998; Puga et al., 2000; Elferink et al., 2001; Marlowe et al., 2004). One way the AhR–Rb interaction regulates cell cycle progression is by repressing E2F transcription factor (E2F)–dependent gene expression, thus preventing entry into S-phase (Puga et al., 2000; Marlowe et al., 2004). Separate from its role as a repressor complex, the AhR–Rb interaction also promotes gene expression consistent with Rb functioning as a transcriptional coactivator, where sustained receptor activity in 5L rat hepatoma cells leads to induction of the G1-phase cyclin-dependent kinase inhibitor p27Kip1 (Kolluri et al., 1999; Levine-Fridman et al., 2004; Huang and Elferink, 2005). p27Kip1, as well as p21Cip1 and the less well studied p57Kip2, are members of the Cip/Kip family of cyclin-dependent kinase inhibitors targeting cyclin-dependent kinase 2 (CDK2) activity (Sherr and Roberts, 1995). We have previously shown that AhR activation by TCDD suppresses cell proliferation during partial hepatectomy–induced liver regeneration (Mitchell et al., 2006). The in vivo studies showed that CDK2 activity was diminished in the regenerating liver in mice pretreated with TCDD. It is noteworthy that the decrease in CDK2 activity was associated with increased binding of the p21Cip1 and p27Kip1 inhibitors to the CDK2–cyclin E complex. The receptor’s role in hepatic cell cycle control is more complicated, however. Abdelrahim et al. (2003) showed that the AhR can both inhibit (MCF7 cells) and promote (HepG2 cells) cell growth, which was underscored by significant changes in the expression of several G1-phase regulatory proteins in HepG2 cells. Likewise, whereas TCDD suppresses liver growth in hepatectomized mice, it promotes cell proliferation in animals treated with a hepatomitogen (Mitchell et al., 2010), despite suppression of CDK2 activity.
Given the role for p21Cip1 and p27Kip1 in G1-phase cell cycle control, we set out to determine if these proteins are functionally required for the TCDD-induced inhibition of liver regeneration in hepatectomized animals. The present study examined the regenerative response following 70% partial hepatectomy in mice lacking either p21Cip1 or p27Kip1. The data reveal that p21Cip1, rather than p27Kip1, is essential for the TCDD-induced growth arrest, and that p21Cip1 induction by TCDD involves the recently described novel AhR-KLF6 complex binding to an NC-XRE in the p21Cip1 promoter.
Materials and Methods
Animals.
C57Bl/6 [wild-type (WT)], p21Cip1 knockout (KO), and p27Kip1 KO mice were purchased from The Jackson Laboratories (Bar Harbor, ME). These animals were maintained on a 12-hour light/dark cycle, in a temperature-controlled facility in the University of Texas Medical Branch (UTMB) animal resource center, with food and water ad libitum. Female mice 8–10 weeks old were used for our experiments. Animals were euthanized via isoflurane overdose followed by cervical dislocation. All experiments were conducted in accordance with approved Institutional Animal Care and Use Committee procedures.
Chemicals.
As reported previously (Mitchell et al., 2010), TCDD was purchased from Cerilliant (Round Rock, TX) and dissolved in anisole, followed by dilution in peanut oil to 2 mg/ml. TCDD was administered via oral gavage at 20 μg TCDD/kg body weight. Control animals received similar amounts of anisole dissolved in peanut oil.
Partial Hepatectomy.
Partial hepatectomy (PH) or sham surgery was performed as previously described (Mitchell et al., 2006). Briefly, mice were anesthetized via isoflurane inhalation and PH was performed, resecting ∼70% of the liver (as previously described by Higgins and Anderson, 1931). Sham surgery involved anesthetization followed by opening the abdominal cavity and gentle manipulation of the liver tissue without resection. Mice were allowed to recover and were then euthanized at indicated times by isoflurane inhalation followed by cervical dislocation. All animal handling and surgical procedures were performed in strict compliance with an approved UTMB Institutional Animal Care and Use Committee protocol.
BrdU Incorporation.
To measure cell proliferation following partial hepatectomy, we performed BrdU (5-bromo-2′-deoxyuridine) incorporation studies as previously described (Mitchell et al., 2006). Briefly, 50 mg/kg BrdU (Sigma-Aldrich, St. Louis, MO) was administered intraperitoneally 2 hours before euthanization. Tissues were then resected and fixed in 10% buffered formalin for 18 hours. For continuous labeling studies, mice were provided with drinking water containing 0.8 mg/ml BrdU (bottles shielded from UV light) immediately following surgery until mice were sacrificed, at which time the tissue was removed and fixed in 10% buffered formalin for 18 hours. Fixed tissues were sectioned, processed, and stained by the UTMB Histopathology Core Facility. Briefly, sections were stained for BrdU incorporation using a biotinylated anti-BrdU antibody (Invitrogen, Carlsbad, CA). These sections were then incubated with avidin-conjugated horseradish peroxidase and 3,3′-diaminobenzidine. For the analysis of proliferation, the numbers of brown stained (BrdU-positive) nuclei were counted in six randomly selected low-power fields per animal and expressed as a percentage of the total number of nuclei.
Immunoprecipitation/Western Blotting.
Western blots were conducted as previously described (Mitchell et al., 2006). Briefly, fresh-frozen liver tissue was homogenized using a polytron in TGH buffer (50 mM HEPES pH 7.4, 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA pH 8.0, 1% Triton X-100, 10% glycerol) supplemented with 1 mM phenylmethanesulfonyl fluoride, 10 mM NaF, 1 mM Na3VO4, and 5 µg/ml protease inhibitors (p8340; Sigma-Aldrich). Protein concentrations were determined using the DC protein assay kit (Bio-Rad Laboratories, Inc., Hercules, CA). For immunoprecipitation experiments, 50 µg of total protein from whole-cell lysates was incubated with 2 μg anti-CDK2 antibody (sc-163 or sc-163g; Santa Cruz Biotechnology, Dallas, TX) for 4 hours at 4°C. Protein A or protein G beads were then added and incubated 1 hour at 4°C. Beads were washed five times in NETN buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 0.5% Nonidet P-40, 1 mM EDTA) and resuspended in 2× SDS loading buffer, then boiled for 10 minutes at 100°C and fractionated on SDS gel as detailed later.
For Western blots, proteins were resolved on SDS-polyacrylamide (10%) gel and transferred to polyvinylidene difluoride membranes (Amersham Hybond-LFP; GE Healthcare Life Sciences, Piscataway, NJ). Membranes were blocked for 1 hour at room temperature (RT) in 5% dry milk in Tris-buffered saline, 0.1% Tween 20 (v/v) (TBST), and incubated overnight at 4°C with primary antibody in 5% milk in TBST. Membranes were washed three times with TBST, 10 minutes each, and incubated at RT for 1 hour with secondary antibody in 5% milk in TBST. Finally, membranes were washed one time with TBST for 15 minutes and visualized on the Typhoon Trio Variable Mode Imager (GE Healthcare Life Sciences). Primary antibodies used include CDK2 (sc-163), p21Cip1 (sc-6246), p27Kip1 (sc-776), cyclin A (sc-751), and cyclin E (sc-247; Santa Cruz Biotechnology). Secondary antibodies were Cy3/Cy5 anti-mouse or anti-rabbit (GE Healthcare Life Sciences). Blot analysis and densitometry were performed using ImageQuant software (GE Healthcare Life Sciences).
Kinase Assay.
Kinase assays were conducted as previously described (Mitchell et al., 2006). Briefly, 500 μg of total protein was subjected to CDK2 immunoprecipitation as described earlier, except it was washed for four times in NETN (rather than five times), one time in kinase assay buffer (see Mitchell et al., 2006), resuspended in 15 μl of kinase assay buffer supplemented with 10 μg of histone H1 and 5 μCi [32P]ATP (3000 Ci/mmol), and incubated for 40 minutes at RT. The reaction was stopped with 15 μl of 2× SDS loading buffer, and reaction products were resolved on 15% SDS-polyacrylamide gel and analyzed by autoradiography.
Real-Time Polymerase Chain Reaction.
RNA was isolated from total liver tissue using TRIzol (Life Technologies Corp., Carlsbad, CA) according to the manufacturer’s recommendations. SuperScript II (Invitrogen) was used for first-strand cDNA synthesis using 1 µg of total RNA according to the manufacturer’s recommendations. Two microliters of this cDNA was used as a template for the polymerase chain reactions (PCRs). Ten times PCR buffer (Fischer, Waltham, MA) was diluted to 1× with dH2O, forward and reverse primers (Table 1) were added at 0.2 µM final concentration, and 10 U/per reaction of Taq DNA polymerase was added to make a master mix. cDNA was warmed to 95°C before adding the master mix (hot-start). Cycling parameters were 95°C for 45 seconds, 68°C for 30 seconds, and 72°C for 90 seconds. This sequence was used to amplify the target sequence for the indicated number of cycles, and a final extension at 72°C for 6 minutes was used. Reaction products were resolved on a 0.8% agarose ethidium bromide gel. Gels were visualized on a Typhoon Trio variable mode imager (GE Healthcare Life Sciences), and densitometry was analyzed with ImageQuant software.
TABLE 1.
PCR primers
| Gene | Forward | Reverse |
|---|---|---|
| p21Cip1 qRT-PCR | TGTCTTGCACTCTGGTGTCTGAG | CAATCTGCGCTTGGAGTGATAG |
| p27Kip1 qRT-PCR | TCCAGGGATGAGGAAGCG | CTCCACAGTGCCAGCGTTC |
| p21Cip1 Transcript 1 | GGAGCATGAATGGAGACAGAGACC | ACCAGAGTGCAAGACAGCGACAAG |
| p21Cip1 Transcript 2 | AGCAGCCGAGAGGTGAGC | ACCAGAGTGCAAGACAGCGACAAG |
| GAPDH | ACGGCAAATTCAACGGCACAGTCA | CATTGGGGGTAGGAACACGGAA |
| p21Cip1 ChIP | GTTAGTCCTTCCCACAGTTGGT | CGTCGAGCTGCCTCCTTAT |
| CYP1A1 qRT-PCR | GCCTAACTCTTCCCTGGATGC | TCAATGAGGCTGTCTGTGATGTC |
GAPDH, glyceraldehyde 3-phosphate dehydrogenase; qRT-PCR, quantitative real-time PCR.
Quantitative Real-Time PCR.
Total RNA was isolated from fresh liver tissue using TRIzol according to the manufacturer’s recommendations. Quantitative real-time PCR was performed by the Real-Time PCR core facility at the University of Texas Medical Branch as previously described (Mitchell et al., 2006). Briefly, RT-PCR was performed using 20 ng of total RNA for target gene detection. Target gene expression (2-CT) was calculated, following normalization of the triplicate threshold cycle value to 18S rRNA.
Chromatin Immunoprecipitation.
Chromatin immunoprecipitation (ChIP) experiments were performed as previously described (Huang and Elferink, 2012; Wilson et al., 2013). Briefly, C57Bl/6 mice were treated in vivo with 20 μg TCDD per kilogram of body weight, or an equivalent amount of vehicle via oral gavage. Mice were then euthanized by isoflurane inhalation and cervical dislocation. Whole livers were resected and the gall bladder removed, then rinsed in 1× phosphate-buffered saline. Livers were diced into fine pieces and cross-linked in 1% formaldehyde for 10 minutes at RT. Cross-linking was stopped by addition of 500 mM glycine. Tissue was pelleted via centrifugation at 3200g for 5 minutes at 4°C. Pellet was resuspended in 6 ml of cold 1× phosphate-buffered saline and homogenized using a dounce homogenizer with a tight pestle (Wheaton, Millville, NJ). The homogenate was pelleted at 3200g for 5 minutes at 4°C and resuspended in 5 ml of cell lysis buffer (150 mM NaCl, 25 mM Tris pH 7.5, 5 mM EDTA, 1% Triton X-100, 0.1% SDS, 0.5% deoxycholate) supplemented with protease inhibitor cocktail (p8340; Sigma-Aldrich), and homogenized in a dounce homogenizer with a tight pestle. Homogenates were incubated on ice for 15 minutes and centrifuged at 3200g for 5 minutes at 4°C. The samples were processed for ChIP using the Active Motif ChIP-IT Express Enzymatic Kit (Active Motif, Carlsbad, CA). Protein DNA complexes were immunoprecipitated with the following antibodies: mouse monoclonal anti-AhR (ab2769; Abcam, Cambridge, MA), goat polyclonal anti-KLF6 (SC-20885; Santa Cruz Biotechnology), mouse monoclonal anti-histone H3 (ab10799; Abcam), or rabbit monoclonal IgG isotype control (#3900; Cell Signaling Technologies, Danvers, MA). Immunoprecipitated DNA was purified by phenol-chloroform extraction and ethanol precipitation. Cyp1a1 and p21Cip1 promoter sequences were amplified by PCR using primers flanking the XRE and NC-XRE sequences, respectively (Table 1). PCR products were resolved on a 5% polyacrylamide Tris/borate/EDTA gel and stained with 1× SYBR Green for 30 minutes in the dark. Gels were rinsed with dH2O and imaged on a Typhoon Trio Variable Mode Imager. Band intensity was measured using ImageQuant software.
Statistical Analysis.
Data were analyzed using Prism 6 (GraphPad Software, La Jolla, CA). Animal numbers for surgery experiments were based on a preliminary power analysis. Statistical significance of P < 0.05 was used for all experiments. Unless otherwise indicated, significance was determined by Holm–Sidak t test.
Results
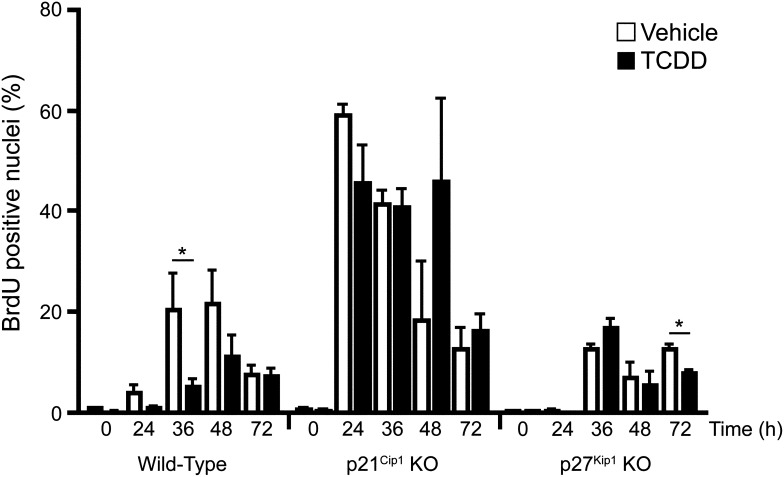
We have previously shown that persistent AhR activation by TCDD suppresses liver regeneration in WT mice following 70% PH, concomitant with a decrease in CDK2 activity (Mitchell et al., 2006). To test the hypothesis that this antiproliferative response was due to the p21Cip1 and/or p27Kip1 CDK inhibitor(s), we monitored hepatocyte proliferation in regenerating livers in WT, p21Cip1, and p27Kip1 knockout mice following TCDD treatment (Fig. 1). Commitment to DNA synthesis (S-phase) was measured using BrdU incorporation at various times after the PH. Substantial BrdU incorporation was first detected in WT mice at 36 hours post-PH in keeping with a precisely regulated temporal program (Weglarz and Sandgren, 2000). Consistent with our previous findings (Mitchell et al., 2006), a significant decrease in BrdU-positive nuclei is detected in livers from TCDD pretreated WT mice. Extensive BrdU incorporation occurred 12 hours sooner in the hepatectomized p21Cip1 KO mice, and, significantly, the antiproliferative property associated with TCDD treatment was absent in the p21Cip1-null background, suggesting that p21Cip1 plays a critical role in regulating normal liver regeneration. In contrast, BrdU incorporation in p27Kip1 KO mice was modest in both vehicle and TCDD-treated WT mice, although peak DNA synthesis matched the temporal process observed in WT mice. TCDD-induced inhibition of DNA synthesis was reproducibly detected in the p27Kip1 KO mouse livers, but only 72 hours postsurgery. These findings suggest that p21Cip1, rather than p27Kip1, plays a critical role in regulating passage through G1 phase during liver regeneration, and that loss of p21Cip1 function both hastens entry into S-phase and abrogates the AhR-mediated TCDD-induced growth arrest. Indeed, Albrecht and coworkers (1998) have previously demonstrated that loss of p21Cip1 accelerated hepatocyte progression through the G1 phase after PH. BrdU incorporation was minimal (<4%) in sham-operated mice that were pretreated with either vehicle or TCDD (data not shown).
Fig. 1.
Analysis of hepatocyte proliferation during liver regeneration in vehicle or TCDD-treated wild-type, p27Kip1 KO mice, or p21Cip1 KO mice. Mice were treated for 24 hours with vehicle or TCDD prior to PH (t = 0 hours). Animals were pulsed with BrdU 2 hours before being euthanized at indicated time points. BrdU-positive nuclei were counted in six random immunohistochemical fields (>300 cells/field) in liver tissue from each mouse. Data represent the average percent positive nuclei from three animals per treatment group for each genotype, and are representative of three separate experiments. Data are plotted as the mean ± S.E.M. *Significant difference between the vehicle and TCDD-treated groups (P < 0.05).
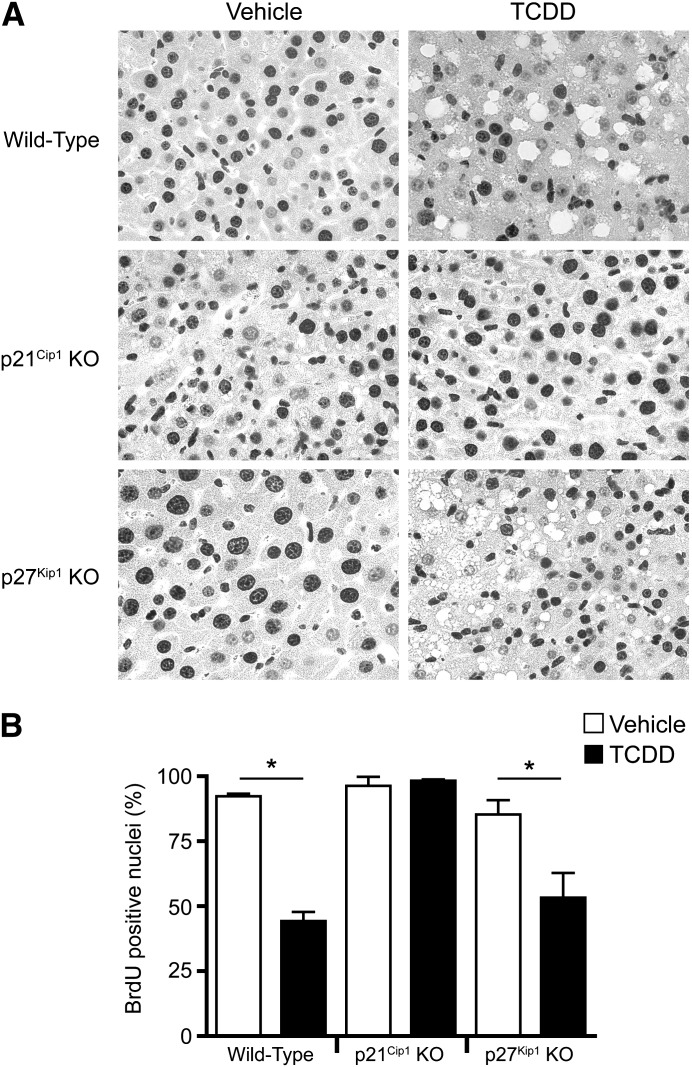
To examine the cumulative effect of p21Cip1 or p27Kip1 loss on liver regeneration, we performed a continuous BrdU labeling study over a 120-hour period following partial liver resection in WT, p21Cip1 KO, and p27Kip1 KO mice (Fig. 2). The immunohistochemical data show that, over the course of 5 days, DNA synthesis occurred in 85–100% of the hepatic nuclei in vehicle-treated mice irrespective of the genotype. In contrast, TCDD pretreatment suppressed liver cell proliferation in the WT and p27Kip1 KO mice by about 50–60%, whereas the p21Cip1 KO mouse livers were completely resistant to the TCDD effect. These data support the BrdU pulse labeling results (Fig. 1). We also observed hydropic degeneration in the hepatectomized livers from TCDD-treated WT and p27Kip1 KO mice that was undetectable in the p21Cip1 KO mouse livers. The precise molecular basis for this difference is unknown, but is suggestive of a link between p21Cip1 activity and the TCDD-induced pathology detected in the liver sections.
Fig. 2.
TCDD-induced suppression of liver regeneration is specifically abrogated in p21Cip1 KO mice. (Top) Representative sections of cumulative BrdU incorporation during liver regeneration in wild-type, p21Cip1 KO, or p27Kip1 KO mice. Mice were treated for 24 hours with vehicle or TCDD prior to PH surgery, and provided BrdU in the drinking water (0.8 mg BrdU/ml, shielded from light) for a period of 5 days post-PH. Liver tissue was processed and stained for immunohistochemistry, and BrdU-positive cells identified as dark brown (3,3-diaminobenzidine–stained) nuclei in the photomicrographs. (Bottom) Cumulative BrdU incorporation was quantified as described in Fig. 1. Data represent the average percent positive nuclei from three animals per treatment group and are representative of three separate experiments. Data are plotted as the mean ± S.E.M. *Significant difference between the vehicle and TCDD-treated groups (P < 0.05).
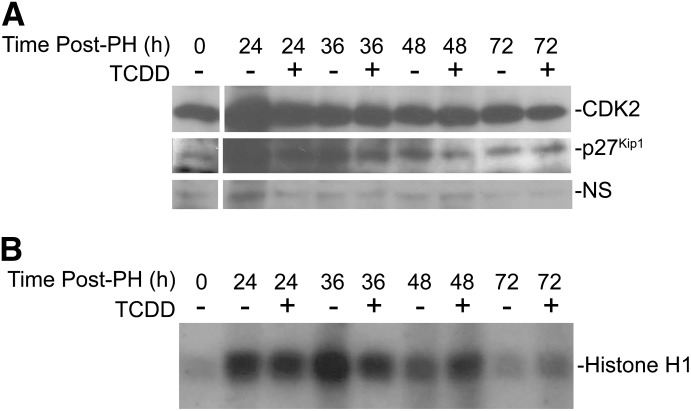
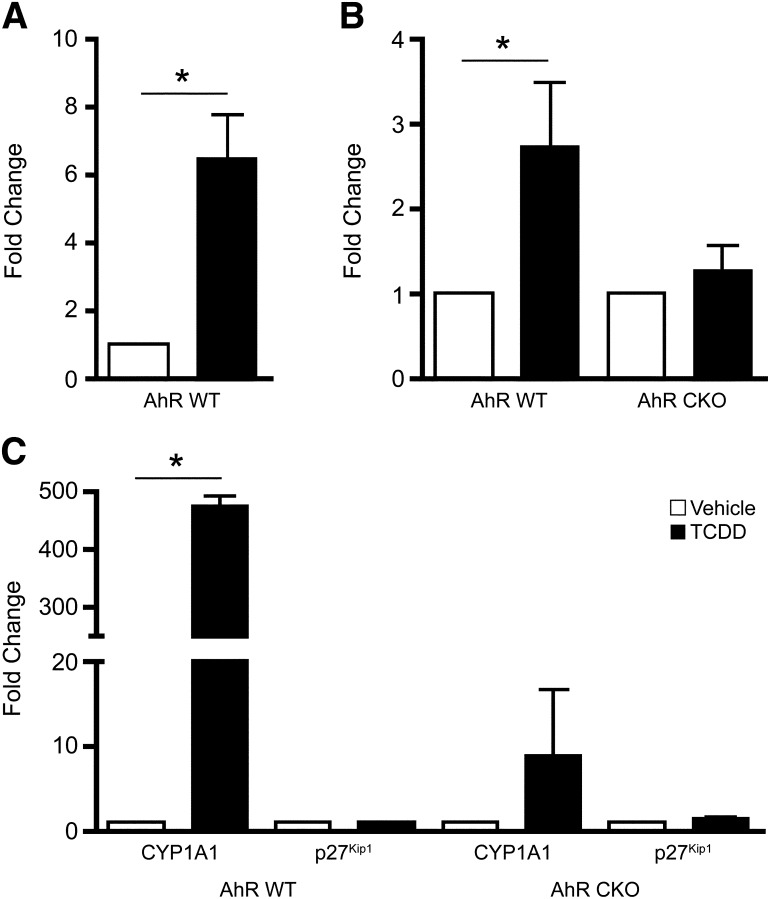
Given the finding that p21Cip1 KO mouse livers entered S-phase prematurely and were refractory to the TCDD effects, we examined CDK2 expression and function in these livers. CDK2 protein levels transiently increased during the first 24 hours post-PH, but remained largely unaltered during the subsequent 48 hours of liver regeneration and showed no TCDD dependency (Fig. 3A). Consistent with observations made in WT mice (Albrecht et al., 1998), coimmunoprecipitation of the p27Kip1 protein in the p21Cip1-null background revealed that CDK2 binding increased marginally following PH. Moreover, formation of the p27Kip1/CDK2 complex did not display a TCDD dependency. Assessment of CDK2 activity in p21Cip1 KO mice readily detected enhanced kinase activity by 24 hours post-PH (Fig. 3B), without the pronounced TCDD-induced inhibition seen in WT mice (Mitchell et al., 2006). Whereas CDK2 activity is first evident at 36 hours post-PH in WT mice (Albrecht et al., 1998; Mitchell et al., 2006), detecting CDK2 activity by 24 hours in the p21Cip1 KO mice underscores the accelerated commitment to DNA synthesis. Therefore, despite the presence of p27Kip1, the lack of p21Cip1 resulted in premature upregulation of CDK2 activity and loss of TCDD-induced growth arrest. Under normal physiologic conditions, p21Cip1 expression is largely undetectable in the quiescent liver, although PH triggers p21Cip1 gene expression (Albrecht et al., 1998). In contrast, p21Cip1 expression is inducible in quiescent mouse livers following a TCDD treatment of 24 hours (Fig. 4A). In fact, significant hepatic p21Cip1 mRNA induction was evident within 2 hours of TCDD treatment in WT mice, but not in the hepatocyte-specific AhR conditional KO (CKO) mice lacking receptor protein expression in the liver parenchyma (Fig. 4B), suggesting that p21Cip1 is indeed an AhR target gene. A comparable observation was made for the prototypical AhR target gene, Cyp1a1, exhibiting robust induction in WT livers that was markedly attenuated in the CKO mouse liver (Fig. 4C). The residual (≈2%) Cyp1a1 induction observed is attributed to AhR activity in the nonparenchymal cells (e.g., stellate cells, endothelial cells, and Kupffer cells) that retained AhR expression in this CKO model. In contrast, p27Kip1 mRNA levels did not change following TCDD exposure, indicating that this is not a TCDD-responsive murine gene in vivo, despite being TCDD-responsive in the 5L rat hepatoma cell line (Kolluri et al., 1999).
Fig. 3.
CDK2 expression and activity in the p21Cip1 KO mice. (A) p21Cip1 KO mice were treated with vehicle or TCDD for 24 hours prior to PH, and euthanized at the indicated times postsurgery. Livers were harvested and homogenized followed by immunoprecipitation with anti-CDK2 antibody. Immunoprecipitates were fractionated by SDS-PAGE and probed sequentially for CDK2 and p27Kip1 by Western blotting. Blots are representative of at least three separate experiments, and a nonspecific (NS) band routinely detected on the CDK2 blots was used as a loading control. (B) CDK2 kinase assays were performed on liver homogenates from mice treated as in A. Homogenates were immunoprecipitated with an anti-CDK2 antibody and protein A/G beads. Bead-bound CDK2 was incubated with histone H1 and 32P-γATP and radiolabeled histone H1 visualized by autoradiography.
Fig. 4.
AhR-dependent expression of p21Cip1 expression in the murine liver. (A) Wild-type mice were treated with vehicle or TCDD by gavage 24 hours prior to being euthanized and total RNA prepared from the liver. Quantitative real-time PCR was performed on the RNA to measure the p21Cip1 mRNA level. Data represent the average value for RNA isolated from five independent experiments for each treatment. Data are plotted as the mean ± S.E.M. *Significant difference between vehicle and TCDD-treated animals (P < 0.01). (B) Wild-type and AhR CKO mice were treated with vehicle or TCDD by gavage 2 hours prior to sacrifice. p21Cip1 mRNA levels were measured by quantitative real-time PCR on total RNA. Data are representative of six wild-type animals per treatment and four AhR CKO animals per treatment. Data are plotted as the mean ± S.E.M. *Significant difference between the vehicle and TCDD-treated animals (P < 0.05). (C) Quantitative real-time PCR on total RNA isolated in B was performed to measure CYP1A1 and p27Kip1 expression.
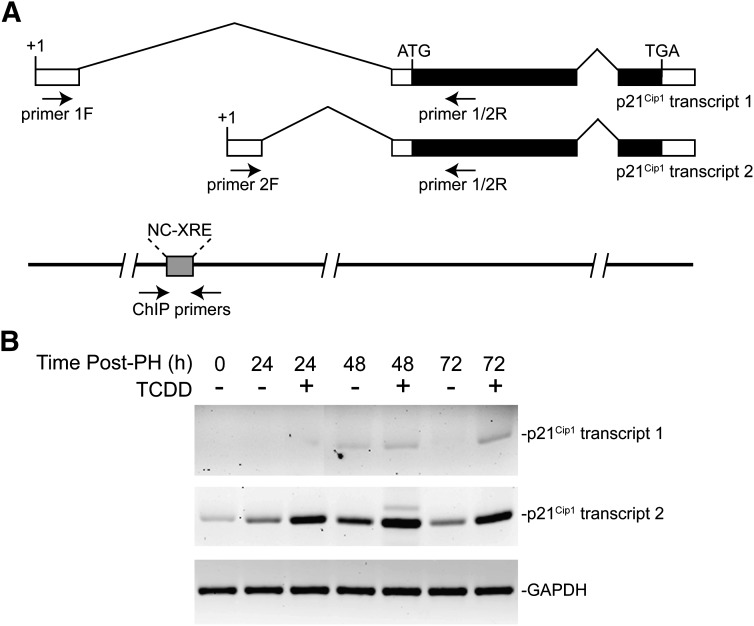
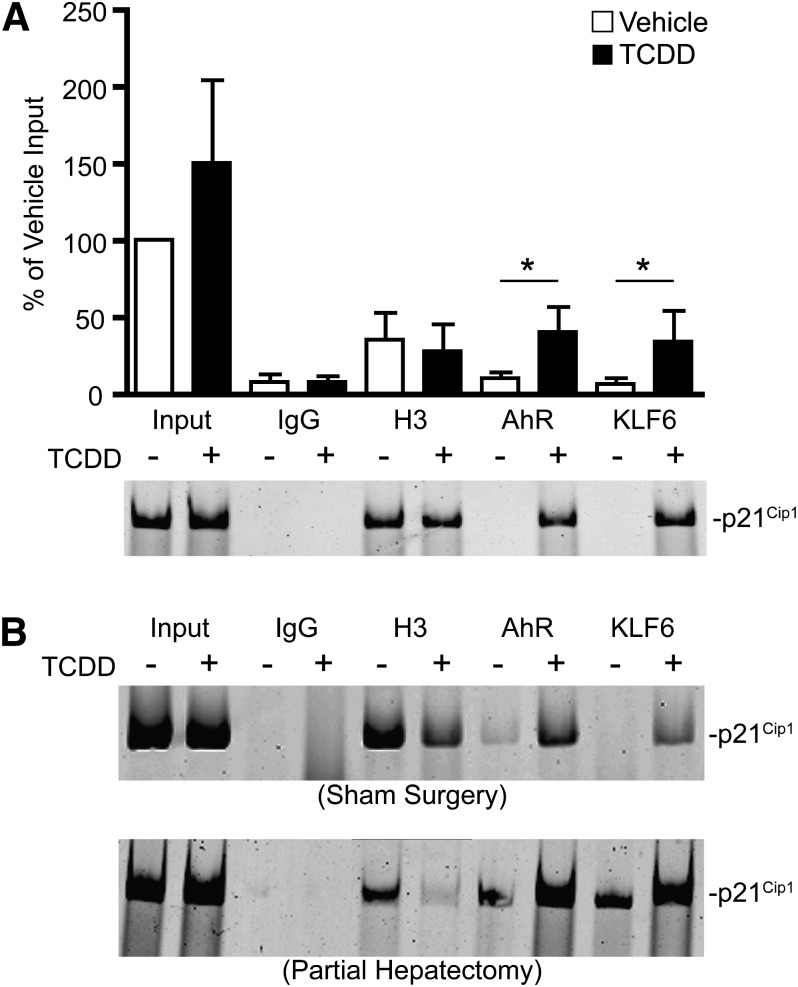
To verify that p21Cip1 is an AhR target gene, we performed ChIP assays. However, prior to assaying for AhR binding to the p21Cip1 promoter in vivo, it was first necessary to ascertain which of two alternate promoters (Gartel et al., 2004) conferred TCDD-inducible p21Cip1 expression in the liver. A third alternate p21Cip1 transcript has been described previously (Huppi et al., 1994), but is not expressed in the liver (Gartel et al., 2004). The genomic context for the two transcripts expressed in the liver is shown in Fig. 5A. Transcription is under the control of two distinct promoters, where transcripts 1 and 2 differ in their 5′-untranslated region, but encode the same p21Cip1 protein. Real-time PCR on total RNA using variant-specific primers revealed that the downstream promoter is by far the dominant promoter during liver regeneration as well as in response to TCDD treatment (Fig. 5B). Our laboratory recently documented that the NC-XRE is a functional target site for a novel interaction involving the tumor suppressor KLF6 and the AhR (Wilson et al., 2013). It is noteworthy that the downstream promoter of p21Cip1 contains an NC-XRE. Given the previous finding that p21Cip1 is a KLF6 target gene (Narla et al., 2001, 2007), ChIP assays concentrated on the NC-XRE–containing promoter region. ChIP experiments were performed on mouse liver tissue obtained from animals treated with vehicle or TCDD via oral gavage. PCR amplification of the genomic region encompassing the NC-XRE revealed that both the AhR and KLF6 proteins were recruited to the promoter within 2 hours following TCDD exposure (Fig. 6A). Analysis on quantitated replicate experiments confirmed that AhR and KLF6 DNA binding is significantly increased by TCDD treatment, consistent with the rapid increase in p21Cip1 mRNA (Fig. 4B). DNA binding by the AhR-KLF6 complex to the p21Cip1 promoter persists for at least 24 hours following TCDD exposure (Fig. 6B, sham surgery). Significantly, PH—in the absence of TCDD—also induced recruitment of the AhR and KLF6 to the p21Cip1 promoter within 2 hours (Fig. 6B, partial hepatectomy), consistent with the transient increase of transcript 2 following PH (Fig. 5B). TCDD pretreatment potentiated AhR-KLF6 binding to the p21Cip1 promoter in hepatectomized livers, in keeping with the enhanced mRNA expression.
Fig. 5.
Differential expression of the p21Cip1 locus in the mouse genome. (A) Diagrammatic representation of the p21Cip1 locus encoding liver-expressed transcripts. The diagram also identifies the location of the NC-XRE and the variant-specific primers used for real-time PCR and ChIP analysis. (B) Mice were treated with vehicle or TCDD for 24 hours prior to PH surgery, and euthanized at the indicated times after surgery. Primers specific for transcripts 1, transcript 2, and GAPDH (glyceraldehyde 3-phosphate dehydrogenase) were used in real-time PCR and the products visualized by agarose gel electrophoresis. Data are representative of three separate experiments.
Fig. 6.
ChIP analysis on the p21Cip1 transcript 2 promoter encompassing the NC-XRE. (A) Mice were treated with vehicle or TCDD via gavage for 2 hours. Livers were excised and processed for ChIP on the AhR and KLF6. IgG and histone H3 antibodies were used as negative and positive controls, respectively. PCR products were fractionated by 6% PAGE and stained with SYBR Green, and quantitation was presented as a percentage of the input DNA. The data shown are representative of four independent experiments. Data are plotted as the mean ± S.E.M. *Significant difference between vehicle and TCDD treatments via one-sided ratio-paired t tests (P < 0.05). (B) Mice were treated with vehicle or TCDD by gavage for 24 hours prior to sham surgery (top) or PH (bottom). Mice were euthanized 2 hours after surgery, and livers were processed for ChIP. Gels are representative of three separate experiments.
These data indicate that PH triggers AhR activation in the absence of an exogenous agonist such as TCDD. The evidence for spontaneous AhR activation following PH resulting in p21Cip1 expression (Fig. 6) is further supported by an increase in CYP1A1 protein expression observed during the first 48 hours following PH (Fig. 7). This represents a transient induction of the Cyp1a1 gene attributed to an endogenous signaling mechanism activating the AhR that was previously observed during liver regeneration (Mitchell et al., 2006). The surprising observation that this Cyp1a1 induction is attenuated in the p21Cip1 KO mice following PH implies that AhR functionality is dependent on a p21Cip1-regulated process absent in the null mice.
Fig. 7.
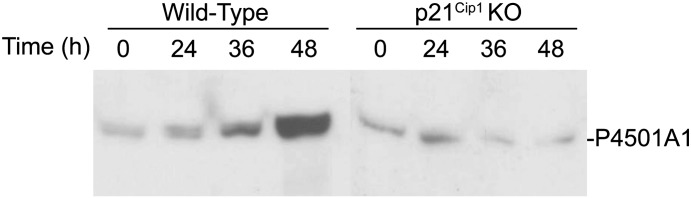
AhR activation is disrupted in p21Cip1 KO mice. Wild-type and p21Cip1 KO mice were subjected to PH and euthanized at the indicated times. Livers were excised and processed for Western blot analysis to detect CYP1A1 protein levels. Blot is representative of three experiments.
Discussion
Current understanding of the signaling events that regulate liver regeneration after two-thirds PH is incomplete. Although 70% PH induces most of the remaining hepatocytes to replicate at least once before exiting the cell cycle, we have shown that TCDD treatment can disrupt the normal regenerative process following PH, with fully half of the hepatocytes in the liver remnant failing to proliferate (Mitchell et al., 2006). The study also demonstrated that CDK2 activity, rather than CDK4 activity, was inhibited by TCDD treatment. Hence, the work reported here focused on the CDK2 inhibitors, p21Cip1 and p27Kip1, rather than the INK4 inhibitors known to suppress CDK4 activity (Sherr and Roberts, 1995). Moreover, TCDD-induced growth arrest in the 5L rat hepatoma cell line was attributed to p27Kip1 function (Kolluri et al., 1999; Levine-Fridman et al., 2004). However, the present finding using KO mice demonstrated that, in vivo, p21Cip1 rather than p27Kip1 conferred the TCDD-induced inhibition in liver regeneration. BrdU incorporation studies revealed that loss of p27Kip1 did not substantially alter the TCDD-induced inhibition in hepatocyte proliferation observed in wild-type mice (Fig. 2). In contrast, the absence of p21Cip1 expression completely abrogated the TCDD effect seen in WT mice. Accordingly, CDK2 activity in the p21Cip1 KO mouse liver was unaffected by TCDD treatment (Fig. 3B), juxtaposing the earlier findings in WT mice where TCDD treatment inhibited CDK2 activity (Mitchell et al., 2006). In keeping with other studies (Albrecht et al., 1998), PH increased CDK2 activity in p21Cip1 KO mice at 24 hours, substantially earlier than the peak of CDK2 activity normally observed at 36 hours post-PH (Mitchell et al., 2006). This is congruent with the accelerated commitment to S-phase measured by BrdU incorporation (Fig. 1). Collectively, the results indicate that TCDD-induced growth arrest in the regenerating liver is absolutely dependent on p21Cip1 activity, and that p27Kip1 does not confer functional redundancy. The finding that p21Cip1, rather than p27Kip1, is required for the TCDD effect in vivo also bolsters the recognition that studies examining cell cycle control in tissue culture systems cannot automatically be extrapolated to liver regeneration in vivo (Loyer et al., 1994; Kren and Steer, 1996).
p21Cip1 is the founding member of the Cip/Kip family of cyclin-dependent kinase inhibitors (Gartel et al., 1996). Expression of p21Cip1 is controlled mostly at the transcriptional level by both p53-dependent and -independent mechanisms (Gartel and Tyner, 1999). Although the p21Cip1 genomic locus is organized differently in mice and humans, both produce multiple transcripts utilizing different promoters (Nozell and Chen, 2002; Gartel et al., 2004). In mice, expression of the classic transcript (transcript 2) is p53-independent (Gartel et al., 2004). This transcript appears to be the dominant transcript upregulated following PH and exposure to TCDD (Fig. 5B). It is noteworthy that the promoter regulating transcript 2 contains an NC-XRE. We recently showed that the NC-XRE is an AhR DNA-binding site. AhR-mediated regulation of gene expression via the NC-XRE occurs through a mechanism involving the novel binding partner, KLF6 (Huang and Elferink, 2012; Wilson et al., 2013). The basis for the modest increase in transcript 1 detected after PH and TCDD treatment is uncertain (Fig. 5B). As such, further studies are required to explore whether this increased expression is due to direct transcriptional regulation of the upstream promoter or a secondary consequence of transcriptional activation at the downstream promoter. Albrecht and coworkers demonstrated that, in contrast to p27Kip1 expression, which changed little during liver regeneration, p21Cip1 expression, although undetectable in the quiescent liver, was dramatically induced within 3 hours following PH (Albrecht et al., 1997, 1998). Our data show that p21Cip1 transcript levels transiently peaked at 48 hours post-PH, declining thereafter. Induction of p21Cip1 by TCDD is detectable at the mRNA level within 2 hours (Fig. 4B), rapidly reaching a sustained level exceeding the increase observed following PH alone (Fig. 5B). The lack of induction in the AhR CKO mouse liver suggests that p21Cip1 is an AhR target gene. Accordingly, p21Cip1 induction is concomitant with recruitment of the AhR and KLF6 to the transcript 2 promoter region harboring the NC-XRE (Fig. 6). KLF6 was first implicated in regulating p53-independent p21Cip1 expression in human prostate cancer (Narla et al., 2001). More recently, studies in transgenic mice overexpressing KLF6 in the liver showed marked p21Cip1 expression and diminished hepatocyte proliferation (Narla et al., 2007). These authors concluded that KLF6 was a critical regulator of hepatocyte proliferation and liver size in vivo, through a mechanism dependent in large part on p21Cip1 expression. Remarkably, the phenotype in the transgenic KLF6 mice closely resembled that observed in p21Cip1 transgenic mice overexpressing the kinase inhibitor in hepatocytes (Wu et al., 1996). Moreover, liver regeneration in the p21Cip1 transgenic mice was markedly attenuated following PH (0.5–14.5% of normal). Collectively, these data suggest that the increase in p21Cip1 expression during hepatocyte proliferation following PH contributes to the normal temporal process associated with liver regeneration; prolonged and elevated p21Cip1 expression in the transgenic mice, however, impedes hepatocyte proliferation. We conclude that TCDD-treated, hepatectomized mice similarly show enhanced induction of p21Cip1 expression through an AhR-KLF6–dependent transcriptional response resulting in growth arrest.
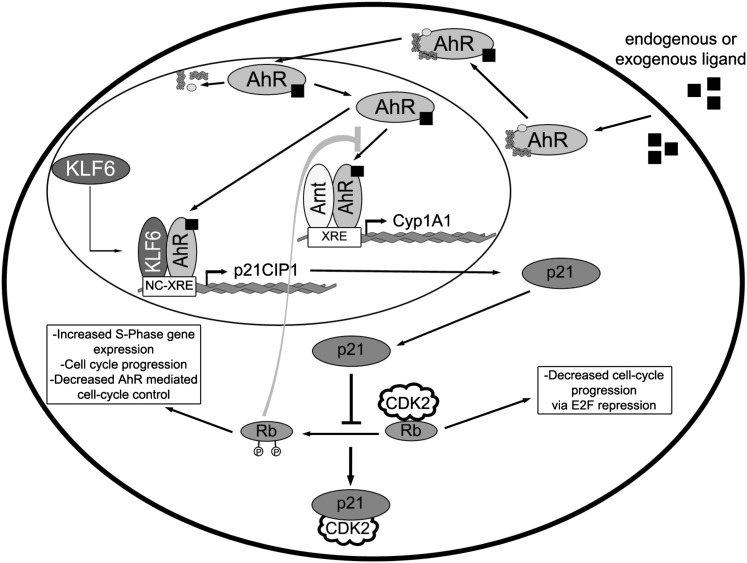
Hyperphosphorylation and concomitant inactivation of the Rb protein is considered to be the major function of the G1- and S-phase CDKs (Sherr and Roberts, 2004). Rb protein inactivation triggers its release from E2F, allowing for the transcriptional activation of S-phase genes, thus resulting in cell cycle progression. Therefore, circumstances that interfere with CDK activation inhibit cell proliferation. Albrecht et al. (1998) showed that Rb protein hyperphosphorylation and loss of E2F binding occurred sooner (36 versus 48 hours) in partially hepatectomized p21Cip1 KO mice as compared with wild-type mice, consistent with the shortened G1 interval. Given our previous observation that the active (hypophosphorylated) Rb protein also binds to the AhR and contributes to AhR transcriptional activity (Ge and Elferink, 1998; Elferink et al., 2001; Levine-Fridman et al., 2004), it is reasonable to speculate that processes which facilitate Rb protein inactivation may also disrupt AhR activity. This prediction is borne out by the failure of PH to increase CYP1A1 expression in the p21Cip1 KO mouse liver, a response normally observed in the regenerating liver of wild-type mice (Fig. 7). Based on the data we have collected and the findings presented elsewhere, we propose the following model (Fig. 8). Liver regeneration following PH results in AhR activation as evidenced by AhR recruitment to the p21Cip1 promoter and CYP1A1 induction, which involves AhR binding to the NC-XRE and XRE, respectively. Receptor binding to the XRE is in concert with the Arnt protein, whereas NC-XRE binding depends on partnering with KLF6. Under normal physiologic conditions, p21Cip1 expression is increased to temporally regulate G1-phase CDK activity and Rb protein inactivation. The absence of p21Cip1 hastens passage through the G1 phase of the cell cycle by inactivating the Rb protein prematurely. Conversely, prolonged overexpression of p21Cip1 due to TCDD treatment inhibits G1-phase kinase activity and delays Rb protein inactivation. Under normal physiologic conditions, the consequent increase in CYP1A1 activity is presumed to facilitate the metabolic clearance of endogenous AhR agonists (Chang and Puga, 1998) resulting in attenuated AhR transcriptional activity, thus restoring balance to the signaling processes. However, because TCDD is refractory to metabolic clearance of the aforementioned endogenous agonists by CYP1A1, AhR activity persists, thereby resulting in prolonged p21Cip1 expression and cell cycle arrest. Conversely, since the Rb protein functions as an AhR coactivator in Cyp1a1 gene expression (Ge and Elferink, 1998; Elferink et al., 2001; Levine-Fridman et al., 2004), the absence of p21Cip1 is expected to hasten Rb protein inactivation to promote cell cycle progression and thus impair Cyp1a1 induction.
Fig. 8.
AhR signal integration during liver regeneration. Activation of the AhR in response to an exogenous agonist or endogenous cue stimulates nuclear translocation and chaperone dissociation. The liganded AhR partners with either the Arnt protein or KLF6, followed by binding to the XRE and NC-XRE, respectively. Under normal physiologic conditions, the AhR-Arnt heterodimer induces Cyp1a1 expression and subsequent metabolic turnover of the AhR agonist to prevent sustained AhR activation. Concomitantly, the AhR-KLF6 heterodimer induces p21Cip1 expression to modulate CDK2 activity and affect the rate of Rb protein inactivation. Since the hypophosphorylated (active) Rb protein contributes to AhR-Arnt–dependent Cyp1a1 expression, the level of p21Cip1 expression indirectly affects Cyp1a1 expression. This cross-talk imputes G1-phase cell cycle progression with a delicate mechanism to fine tune AhR activity and subsequent p21Cip1 expression.
In conclusion, we have shown that TCDD-induced suppression of liver regeneration depends on induction of the p21Cip1 gene involving recruitment of the AhR–KLF6 complex to the NC-XRE present in the downstream promoter. Moreover, p21Cip1, rather than p27Kip1, confers the AhR-mediated attenuation of liver regeneration, and absence of p21Cip1 abrogates the TCDD-induced effect on liver regeneration. The data also attest to integrated signaling of multiple AhR target genes regulated by distinct DNA recognition sites.
Acknowledgments
The authors thank Dr. Tod Harper for assistance with the p21 transcript experiments. The authors also thank the UTMB Histopathology core for preparing and staining liver tissue, and Deborah Prusak of the UTMB Molecular Genomics core for running the quantitative real-time PCR experiments.
Abbreviations
- AhR
aryl hydrocarbon receptor
- Arnt
aryl hydrocarbon receptor nuclear translocator
- BrdU
5-bromo-2′-deoxyuridine
- CDK2
cyclin-dependent kinase 2
- ChIP
chromatin immunoprecipitation
- CKO
conditional KO
- E2F
E2F transcription factors
- KLF6
Kruppel-like factor 6
- KO
knockout
- NC-XRE
nonconsensus xenobiotic response element
- PCR
polymerase chain reaction
- PH
partial hepatectomy
- Rb
retinoblastoma tumor suppressor protein
- RT
room temperature
- TBST
Tris-buffered saline/Tween 20
- TCDD
dioxin, 2,3,7,8-tetrachlorodibenzo-p-dioxin
- UTMB
University of Texas Medical Branch
- WT
wild-type
- XRE
xenobiotic response element
Authorship Contributions
Participated in research design: Jackson, Li, Mitchell, Elferink.
Conducted experiments: Jackson, Li, Mitchell, Joshi.
Performed data analysis: Jackson, Li, Mitchell, Joshi, Elferink.
Wrote or contributed to the writing of the manuscript: Jackson, Elferink.
Footnotes
This work was supported by the National Institutes of Health National Institute of Environmental Health Sciences [Grants R01-ES07800, P30-ES006676, and T32-ES007254].
References
- Abdelrahim M, Smith R, 3rd, Safe S. (2003) Aryl hydrocarbon receptor gene silencing with small inhibitory RNA differentially modulates Ah-responsiveness in MCF-7 and HepG2 cancer cells. Mol Pharmacol 63:1373–1381 [DOI] [PubMed] [Google Scholar]
- Albrecht JH, Meyer AH, Hu MY. (1997) Regulation of cyclin-dependent kinase inhibitor p21(WAF1/Cip1/Sdi1) gene expression in hepatic regeneration. Hepatology 25:557–563 [DOI] [PubMed] [Google Scholar]
- Albrecht JH, Poon RY, Ahonen CL, Rieland BM, Deng C, Crary GS. (1998) Involvement of p21 and p27 in the regulation of CDK activity and cell cycle progression in the regenerating liver. Oncogene 16:2141–2150 [DOI] [PubMed] [Google Scholar]
- Chang CY, Puga A. (1998) Constitutive activation of the aromatic hydrocarbon receptor. Mol Cell Biol 18:525–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison MS, Nagy SR. (2003) Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu Rev Pharmacol Toxicol 43:309–334 [DOI] [PubMed] [Google Scholar]
- Denison MS, Soshilov AA, He G, DeGroot DE, Zhao B. (2011) Exactly the same but different: promiscuity and diversity in the molecular mechanisms of action of the aryl hydrocarbon (dioxin) receptor. Toxicol Sci 124:1–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elferink CJ, Ge NL, Levine A. (2001) Maximal aryl hydrocarbon receptor activity depends on an interaction with the retinoblastoma protein. Mol Pharmacol 59:664–673 [DOI] [PubMed] [Google Scholar]
- Fernandez-Salguero PM, Hilbert DM, Rudikoff S, Ward JM, Gonzalez FJ. (1996) Aryl-hydrocarbon receptor-deficient mice are resistant to 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced toxicity. Toxicol Appl Pharmacol 140:173–179 [DOI] [PubMed] [Google Scholar]
- Fukunaga BN, Probst MR, Reisz-Porszasz S, Hankinson O. (1995) Identification of functional domains of the aryl hydrocarbon receptor. J Biol Chem 270:29270–29278 [DOI] [PubMed] [Google Scholar]
- Gartel AL, Radhakrishnan SK, Serfas MS, Kwon YH, Tyner AL. (2004) A novel p21WAF1/CIP1 transcript is highly dependent on p53 for its basal expression in mouse tissues. Oncogene 23:8154–8157 [DOI] [PubMed] [Google Scholar]
- Gartel AL, Serfas MS, Gartel M, Goufman E, Wu GS, el-Deiry WS, Tyner AL. (1996) p21 (WAF1/CIP1) expression is induced in newly nondividing cells in diverse epithelia and during differentiation of the Caco-2 intestinal cell line. Exp Cell Res 227:171–181 [DOI] [PubMed] [Google Scholar]
- Gartel AL, Tyner AL. (1999) Transcriptional regulation of the p21((WAF1/CIP1)) gene. Exp Cell Res 246:280–289 [DOI] [PubMed] [Google Scholar]
- Ge N-L, Elferink CJ. (1998) A direct interaction between the aryl hydrocarbon receptor and retinoblastoma protein. Linking dioxin signaling to the cell cycle. J Biol Chem 273:22708–22713 [DOI] [PubMed] [Google Scholar]
- Göttlicher M, Wiebel FJ. (1991) 2,3,7,8-Tetrachlorodibenzo-p-dioxin causes unbalanced growth in 5L rat hepatoma cells. Toxicol Appl Pharmacol 111:496–503 [DOI] [PubMed] [Google Scholar]
- Hankinson O. (1995) The aryl hydrocarbon receptor complex. Annu Rev Pharmacol Toxicol 35:307–340 [DOI] [PubMed] [Google Scholar]
- Higgins GM, Anderson RM. (1931) Experimental pathology of the liver: restoration of the liver of the white rat following partial surgical removal. Arch Pathol 12:186–202 [Google Scholar]
- Huang G, Elferink CJ. (2005) Multiple mechanisms are involved in Ah receptor-mediated cell cycle arrest. Mol Pharmacol 67:88–96 [DOI] [PubMed] [Google Scholar]
- Huang G, Elferink CJ. (2012) A novel nonconsensus xenobiotic response element capable of mediating aryl hydrocarbon receptor-dependent gene expression. Mol Pharmacol 81:338–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppi K, Siwarski D, Dosik J, Michieli P, Chedid M, Reed S, Mock B, Givol D, Mushinski JF. (1994) Molecular cloning, sequencing, chromosomal localization and expression of mouse p21 (Waf1). Oncogene 9:3017–3020 [PubMed] [Google Scholar]
- Kolluri SK, Weiss C, Koff A, Göttlicher M. (1999) p27(Kip1) induction and inhibition of proliferation by the intracellular Ah receptor in developing thymus and hepatoma cells. Genes Dev 13:1742–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kren BT, Steer CJ. (1996) Posttranscriptional regulation of gene expression in liver regeneration: role of mRNA stability. FASEB J 10:559–573 [DOI] [PubMed] [Google Scholar]
- Levine-Fridman A, Chen L, Elferink CJ. (2004) Cytochrome P4501A1 promotes G1 phase cell cycle progression by controlling aryl hydrocarbon receptor activity. Mol Pharmacol 65:461–469 [DOI] [PubMed] [Google Scholar]
- Loyer P, Glaise D, Cariou S, Baffet G, Meijer L, Guguen-Guillouzo C. (1994) Expression and activation of cdks (1 and 2) and cyclins in the cell cycle progression during liver regeneration. J Biol Chem 269:2491–2500 [PubMed] [Google Scholar]
- Ma Q, Whitlock JP., Jr (1996) The aromatic hydrocarbon receptor modulates the Hepa 1c1c7 cell cycle and differentiated state independently of dioxin. Mol Cell Biol 16:2144–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlowe JL, Knudsen ES, Schwemberger S, Puga A. (2004) The aryl hydrocarbon receptor displaces p300 from E2F-dependent promoters and represses S phase-specific gene expression. J Biol Chem 279:29013–29022 [DOI] [PubMed] [Google Scholar]
- Mitchell KA, Lockhart CA, Huang G, Elferink CJ. (2006) Sustained aryl hydrocarbon receptor activity attenuates liver regeneration. Mol Pharmacol 70:163–170 [DOI] [PubMed] [Google Scholar]
- Mitchell KA, Wilson SR, Elferink CJ. (2010) The activated aryl hydrocarbon receptor synergizes mitogen-induced murine liver hyperplasia. Toxicology 276:103–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narla G, Heath KE, Reeves HL, Li D, Giono LE, Kimmelman AC, Glucksman MJ, Narla J, Eng FJ, Chan AM, et al. (2001) KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science 294:2563–2566 [DOI] [PubMed] [Google Scholar]
- Narla G, Kremer-Tal S, Matsumoto N, Zhao X, Yao S, Kelley K, Tarocchi M, Friedman SL. (2007) In vivo regulation of p21 by the Kruppel-like factor 6 tumor-suppressor gene in mouse liver and human hepatocellular carcinoma. Oncogene 26:4428–4434 [DOI] [PubMed] [Google Scholar]
- Nebert DW, Puga A, Vasiliou V. (1993) Role of the Ah receptor and the dioxin-inducible [Ah] gene battery in toxicity, cancer, and signal transduction. Ann N Y Acad Sci 685:624–640 [DOI] [PubMed] [Google Scholar]
- Nozell S, Chen X. (2002) p21B, a variant of p21(Waf1/Cip1), is induced by the p53 family. Oncogene 21:1285–1294 [DOI] [PubMed] [Google Scholar]
- Poland A, Knutson JC. (1982) 2,3,7,8-tetrachlorodibenzo-p-dioxin and related halogenated aromatic hydrocarbons: examination of the mechanism of toxicity. Annu Rev Pharmacol Toxicol 22:517–554 [DOI] [PubMed] [Google Scholar]
- Probst MR, Reisz-Porszasz S, Agbunag RV, Ong MS, Hankinson O. (1993) Role of the aryl hydrocarbon receptor nuclear translocator protein in aryl hydrocarbon (dioxin) receptor action. Mol Pharmacol 44:511–518 [PubMed] [Google Scholar]
- Puga A, Barnes SJ, Dalton TP, Chang Cy, Knudsen ES, Maier MA. (2000) Aromatic hydrocarbon receptor interaction with the retinoblastoma protein potentiates repression of E2F-dependent transcription and cell cycle arrest. J Biol Chem 275:2943–2950 [DOI] [PubMed] [Google Scholar]
- Reisz-Porszasz S, Probst MR, Fukunaga BN, Hankinson O. (1994) Identification of functional domains of the aryl hydrocarbon receptor nuclear translocator protein (ARNT). Mol Cell Biol 14:6075–6086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safe SH. (1986) Comparative toxicology and mechanism of action of polychlorinated dibenzo-p-dioxins and dibenzofurans. Annu Rev Pharmacol Toxicol 26:371–399 [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. (1995) Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev 9:1149–1163 [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. (2004) Living with or without cyclins and cyclin-dependent kinases. Genes Dev 18:2699–2711 [DOI] [PubMed] [Google Scholar]
- Swanson HI, Chan WK, Bradfield CA. (1995) DNA binding specificities and pairing rules of the Ah receptor, ARNT, and SIM proteins. J Biol Chem 270:26292–26302 [DOI] [PubMed] [Google Scholar]
- Watson AJ, Hankinson O. (1992) Dioxin- and Ah receptor-dependent protein binding to xenobiotic responsive elements and G-rich DNA studied by in vivo footprinting. J Biol Chem 267:6874–6878 [PubMed] [Google Scholar]
- Weglarz TC, Sandgren EP. (2000) Timing of hepatocyte entry into DNA synthesis after partial hepatectomy is cell autonomous. Proc Natl Acad Sci USA 97:12595–12600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss C, Kolluri SK, Kiefer F, Göttlicher M. (1996) Complementation of Ah receptor deficiency in hepatoma cells: negative feedback regulation and cell cycle control by the Ah receptor. Exp Cell Res 226:154–163 [DOI] [PubMed] [Google Scholar]
- Whitlock JP., Jr (1999) Induction of cytochrome P4501A1. Annu Rev Pharmacol Toxicol 39:103–125 [DOI] [PubMed] [Google Scholar]
- Wilson SR, Joshi AD, Elferink CJ. (2013) The tumor suppressor Kruppel-like factor 6 is a novel aryl hydrocarbon receptor DNA binding partner. J Pharmacol Exp Ther 345:419–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Wade M, Krall L, Grisham J, Xiong Y, Van Dyke T. (1996) Targeted in vivo expression of the cyclin-dependent kinase inhibitor p21 halts hepatocyte cell-cycle progression, postnatal liver development and regeneration. Genes Dev 10:245–260 [DOI] [PubMed] [Google Scholar]