Abstract
The G12/13 class of heterotrimeric G proteins, comprising the α-subunits Gα12 and Gα13, regulates multiple aspects of cellular behavior, including proliferation and cytoskeletal rearrangements. Although guanine nucleotide exchange factors for the monomeric G protein Rho (RhoGEFs) are well characterized as effectors of this G protein class, a variety of other downstream targets has been reported. To identify Gα12 determinants that mediate specific protein interactions, we used a structural and evolutionary comparison between the G12/13, Gs, Gi, and Gq classes to identify “class-distinctive” residues in Gα12 and Gα13. Mutation of these residues in Gα12 to their deduced ancestral forms revealed a subset necessary for activation of serum response element (SRE)–mediated transcription, a G12/13-stimulated pathway implicated in cell proliferative signaling. Unexpectedly, this subset of Gα12 mutants showed impaired binding to heat-shock protein 90 (Hsp90) while retaining binding to RhoGEFs. Corresponding mutants of Gα13 exhibited robust SRE activation, suggesting a Gα12-specific mechanism, and inhibition of Hsp90 by geldanamycin or small interfering RNA–mediated lowering of Hsp90 levels resulted in greater downregulation of Gα12 than Gα13 signaling in SRE activation experiments. Furthermore, the Drosophila G12/13 homolog Concertina was unable to signal to SRE in mammalian cells, and Gα12:Concertina chimeras revealed Gα12-specific determinants of SRE activation within the switch regions and a C-terminal region. These findings identify Gα12 determinants of SRE activation, implicate Gα12:Hsp90 interaction in this signaling mechanism, and illuminate structural features that arose during evolution of Gα12 and Gα13 to allow bifurcated mechanisms of signaling to a common cell proliferative pathway.
Introduction
The ability to perceive and respond to information from the environment is a fundamental property of living cells. G protein–coupled receptors (GPCRs) are integral membrane proteins that evolved to detect a wide range of extracellular signals, including neurotransmitters, hormones, odorants, and light. On activation, GPCRs undergo a conformational change that stimulates heterotrimeric G proteins, intracellular signaling molecules that consist of a GTP-binding α-subunit that exists in 1:1:1 stoichiometry with a β- and a γ-subunit. This heterotrimer is inactive when Gα is GDP-bound and GPCR activation causes Gα to exchange GDP for GTP, dissociate from the Gβγ heterodimer, and activate downstream effector proteins (Oldham and Hamm, 2008). Gα proteins are categorized into four classes based on amino acid sequence: Gs, Gi, Gq, and G12/13. Although each class signals to distinct effector proteins, Gα proteins share a common fold; thus, it is widely accepted that Gα–effector interactions are dictated by class-specific amino acids that create permissive binding surfaces (Cabrera-Vera et al., 2003). Understanding the principles that govern Gα–effector interaction is essential to deciphering the role of specific signaling pathways.
The G12/13 class of Gα proteins is involved in signaling networks that regulate cell migration, cytoskeletal rearrangements, adhesion, apoptosis, and growth (Juneja and Casey, 2009; Suzuki et al., 2009). Consistent with their role as signaling hubs in these pathways, overexpressed or constitutively activated Gα12 and Gα13 are potent stimulators of oncogenic transformation (Chan et al., 1993; Jiang et al., 1993; Xu et al., 1994). Some of these responses appear to involve transcriptional activation of growth-promoting genes governed by promoters harboring the serum response element (SRE) (Vara Prasad et al., 1994; Hill et al., 1995; Fromm et al., 1997). Other responses point to cell migration: endogenous Gα12 levels correlate with the degree of metastatic invasiveness in breast cancer tissue samples (Kelly et al., 2006a), and androgen-insensitive, invasive prostate tumor cells exhibit higher Gα12 levels than less aggressive prostate cancers (Kelly et al., 2006b). Despite 65% amino acid identity, Gα12 and Gα13 have nonoverlapping roles in signaling, embryonic development, and disease physiology. Mice lacking Gα13 exhibit embryonic lethality with impairment in vascular development (Offermanns et al., 1997), whereas mice lacking Gα12 appear to develop normally (Gu et al., 2002). However, in mice that are haplo-insufficient for Gα13, at least one wild-type allele of Gα12 is required to avoid lethality (Worzfeld et al., 2008), suggesting that Gα12 function during development is not eclipsed by Gα13. Also, studies of mouse embryonic fibroblasts lacking Gα12 and Gα13 reveal that both proteins are necessary for orientation of the microtubule-organizing center (Goulimari et al., 2008).
The mechanisms through which Gα12 and Gα13 stimulate cell proliferation are not well understood. The best characterized effector proteins are Rho-specific guanine nucleotide exchange factors (RhoGEFs) that interact with Gα12 and Gα13 via RGS (regulator of G protein signaling) homology domains; these include p115RhoGEF, leukemia-associated RhoGEF (LARG), and PDZ-RhoGEF (Sternweis et al., 2007). In addition to regulating cytoskeletal events such as contraction, this G12/13-RhoGEF-Rho axis also stimulates transcription through the SRE, a promoter element of the c-fos protooncogene (Fromm et al., 1997; Bhattacharyya and Wedegaertner, 2000; Shi et al., 2000). This signaling event is mediated by the transcriptional activator serum response factor, which itself requires Rho-mediated nuclear translocation of its cofactor, myocardin-related transcription factor-A (MRTF-A) (Wang et al., 2002). Although Gα12 and Gα13 signal through several common binding partners, numerous studies provide evidence of selectivity within the G12/13 class (Kelly et al., 2007). For example, activity of heat-shock protein 90 (Hsp90) is required for Gα12, but not Gα13, to stimulate cellular transformation (Vaiskunaite et al., 2001). Hsp90 is a molecular chaperone that forms a homodimer in the cytoplasm, and hydrolysis of ATP at N-terminal binding pockets within this dimer facilitates engagement and stabilization of a wide variety of Hsp90 client proteins, many of which are implicated in cancer progression (Samant et al., 2012). Hsp90 binds specifically to Gα12 within the G12/13 class, but the structural features that mediate this interaction are unknown.
Recent studies used a taxonomic comparison of Gα proteins to identify changes in key residues that contributed to the evolutionary diversification of the four classes (Friedman et al., 2009; Temple et al., 2010). In the present study, we mutated “class-distinctive” residues in Gα12 to ancestral forms to dissect their roles in distinct signaling pathways. Our results revealed a subset of these class-distinctive mutations in Gα12 that disrupted both SRE activation and Hsp90 binding. Gα13 variants harboring identical mutations displayed robust SRE activation, suggesting a Gα12-specific mechanism. In addition, the Drosophila G12/13 homolog Concertina was unable to drive SRE signaling when expressed in mammalian cells, and subsequently we used this protein as a platform to identify key determinants of growth signaling in the switch regions and C-terminal region of Gα12. These findings define Gα12-specific structural determinants of SRE activation and implicate Hsp90 binding as a requirement for this Gα12-mediated pathway.
Materials and Methods
Materials and DNA Constructs.
Glutathione-S-transferase (GST) fusions of the N-terminal 252 amino acids of p115RhoGEF and the region spanning Leu320 to Arg606 of LARG were provided by Tohru Kozasa (University of Illinois, Chicago) and have been described previously (Meigs et al., 2005). A GST fusion of the C-terminal 107 amino acids of Hsp90-α was provided by Tatyana Voyno-Yasenetskaya (University of Illinois, Chicago), and enhanced green fluorescent protein (EGFP)–fused MRTF-A was a gift from Christopher Mack (University of North Carolina, Chapel Hill). All point mutants of Gα12, Gα13, and Concertina were engineered by oligonucleotide-directed mutagenesis using the QuikChange II system (Agilent Technologies, Englewood, CO), with the following exception to the manufacturer’s instructions. Each initial amplification reaction was divided into equal halves, with each receiving one of two mutagenic oligonucleotides for the first two polymerase chain reaction (PCR) cycles. These half-reactions were then combined and subjected to 15 additional PCR cycles. Concertina mutants in the internal ribosomal entry site (IRES) vector pLL-5.5 (Uetrecht and Bear, 2009) were engineered by first excising the Concertina cDNA from pLL-5.5 using ApaI and EcoRI, subcloning into a truncated pGEX-2T vector (GE Healthcare, Little Chalfont, Buckinghamshire, UK) at these sites and then using QuikChange II reagents to introduce codon substitutions. Mutants were subcloned into pLL-5.5 and then confirmed by sequencing. To introduce myc-Gα12QL to pLL-5.5, we excised the Concertina cDNA from pLL-5.5 by digesting with ClaI, blunting 5′-overhangs with Klenow and digesting with EcoRI. Next, myc-Gα12QL cDNA was excised from pcDNA3.1 (Life Technologies, Grand Island, NY), using EcoRI and the blunt end-generating PmeI, and was ligated into pLL-5.5. All DNA constructs expressed in Drosophila cell culture were subcloned into a metallothionein promoter, pmtA vector backbone (Life Technologies), and a myc epitope tag was introduced N-terminally after the initiator Met using KOD Xtreme Hot Start Polymerase (EMD Millipore, Billerica, MA). All Gα12/Concertina and Gα13/Concertina chimeras were engineered by a two-step PCR procedure, in which desired regions of the mutationally activated Gα12 (Q229L), Gα13 (Q226L), and Concertina (Q303L) cDNAs were amplified so that each initial product contained an additional nine base pairs that overlapped with the product amplified from the other cDNA. This created junctions in which 18 base pairs formed, allowing the initial PCR products (two for chimeras 1, 2, 3, and 4, three for all other chimeras) to be combined with end primers in a second round of PCR cycling to generate a full chimeric product. For N-terminal tagging of chimeras 3, 4, 5, and 4-sub, the coding sequence was amplified by PCR with the myc epitope tag EQKLISEEDL encoded in the forward primer immediately downstream of the initiator methionine, and each product was subcloned into pcDNA3.1+ (Life Technologies). All constructs were verified by sequencing.
Expression and Immobilization of GST Fusion Proteins.
The GST fusion constructs were transformed into BL21(Gold)-DE3 cells (Agilent Technologies), liquid cultures were grown at 37°C under 75 µg/ml ampicillin selection to OD600 of 0.5–0.7, and recombinant protein expression was induced by 0.5 mM isopropyl-β-D-thiogalactopyranoside (Fisher Scientific, Pittsburgh, PA). After 3 hours, the cells were lysed on ice using 0.32 mg/ml lysozyme (MP Biomedicals, Santa Ana, CA), and GST fusion proteins were bound to glutathione Sepharose 4B (GE Healthcare) as described previously (Meigs et al., 2005). After three washes in 50 mM Tris pH 7.7 supplemented with 1 mM EDTA, 1 mM dithiothreitol, and 150 mM NaCl, samples were snap-frozen and stored at −80°C.
Preparation of Detergent-Soluble Extracts Harboring Gα Mutants.
Human embryonic kidney cells (HEK293) were grown in Dulbecco’s modified Eagle’s medium (Mediatech, Manassas, VA) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT), penicillin, and streptomycin. For myc-Gα12QL and each of the class-distinctive mutants, 7.0 µg of plasmid DNA was transfected into a 10-cm dish of HEK293 cells at approximate 90% confluence, using Lipofectamine 2000 (Invitrogen) in accordance with the manufacturer’s instructions. After 36–40 hours, cells were scraped from dishes, washed twice with phosphate-buffered saline (PBS), and solubilized in lysis buffer [50 mM HEPES pH 7.5, 1 mM EDTA, 3 mM dithiothreitol, 10 mM MgSO4, 1% (w/v) polyoxyethylene-10-lauryl ether] containing the protease inhibitors 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride (1.67 mM), leupeptin (2.1 µM), pepstatin (1.45 µM), Nα-tosyl-l-lysine chloromethyl ketone (58 µM), tosyl-l-phenylalanyl-chloromethane (61 µM), and phenylmethylsulfonyl fluoride (267 µM). Samples were centrifuged at 80,000g for 1 hour, and supernatants were snap-frozen and stored at −80°C.
Trypsin Protection Assays.
HEK293 cells grown in 10-cm dishes were transfected with various Gα12 constructs using Lipofectamine 2000, and tryptic digestions were performed as a modification of the procedure of Kozasa and Gilman (1995). Briefly, cells were lysed in 50 mM HEPES, pH 7.5, 1 mM EDTA, 3 mM dithiothreitol, and 1% polyoxyethylene-10-lauryl ether containing the same protease inhibitors as lysis buffer (see preceding) but at 2-fold lower concentration. Samples were cleared by centrifugation at 80,000g for 1 hour, and supernatants were diluted 20-fold in volume using 50 mM HEPES pH 7.5, 1 mM EDTA, 3 mM dithiothreitol, and 10 mM MgSO4. Samples were digested with 10 µg/ml TPCK-treated trypsin (New England Biolabs, Ipswich, MA) for 20 minutes at 30°C, and proteolysis was terminated by addition of 100 µg/ml lima bean trypsin inhibitor (Worthington, Lakewood, NJ). Proteins were precipitated by addition of 20% trichloroacetic acid and 0.8 mg/ml sodium deoxycholate, washed with acetone, dried, and then analyzed by SDS-PAGE and immunoblotting using J169 antisera specific to the Gα12 C terminus (Kozasa and Gilman, 1995), provided by Tohru Kozasa (University of Illinois, Chicago).
Protein Interaction Assays.
Extracts from transfected HEK293 cells were diluted in lysis buffer (see preceding description) lacking polyoxyethylene-10-lauryl ether, using sufficient volume to dilute this detergent to 0.05% (w/v). Next, Sepharose-bound GST fusion proteins were added and continuously inverted for 2 hours at 4°C. A percentage of the diluted extract was set aside as starting material (i.e., load) before Sepharose addition. Samples were centrifuged at 1,300g, and pellets were washed extensively and subjected to SDS-PAGE and immunoblot analysis using an antibody specific to either the Gα12 N terminus (Santa Cruz Biotechnology, Santa Cruz, CA), Gα13 (EMD Millipore), or the myc epitope tag (EMD Millipore), followed by alkaline phosphatase conjugated secondary antibodies (Promega, Madison, WI). For each Gα protein, the Gaussian intensity value was determined for the ∼44-kDa band in the precipitated material and divided by the Gaussian intensity of the corresponding ∼44 kDa-band in the load to normalize Gα variants for different expression levels in cells.
RhoA Activity Assays.
Pull-downs using the Rho-binding domain (RBD) of Rhotekin were performed as previously described (Ren et al., 1999) with the following minor modifications. Cells were washed with ice-cold Tris buffered saline (pH 7.6) with 2 mM MgCl2 and lysed in buffer A (50 mM Tris pH 7.6, 500 mM NaCl, 1% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate, 0.5 mM MgCl2, 200 µM orthovanadate, 10 µg/ml aprotinin, 10 µg/ml leupeptin, 1 mM phenylmethylsulfonyl fluoride). Lysates were sonicated briefly, clarified by centrifugation, and equalized for total volume, as well as protein concentration, and then incubated with 30 µg of GST-RBD beads prepared as previously described (Guilluy et al., 2011). Samples were washed with buffer B (50 mM Tris pH 7.6, 150 mM NaCl, 1% Triton X-100, 0.5 mM MgCl2, 200 µM orthovanadate, and protease inhibitors as described) three times before SDS-PAGE. Active RhoA, total RhoA, Gα12, and actin levels were determined by immunoblotting with anti-RhoA (polyclonal antibody 67B9; Cell Signaling Technology, Danvers, MA), anti-Gα12 (Santa Cruz Biotechnology), and anti-actin (clone C4; EMD Millipore). Total RhoA and Gα12 levels were similarly determined using an aliquot of whole cell lysate.
Reporter Gene Assays and RNA Interference.
SRE-luciferase plasmid was provided by Channing Der (University of North Carolina, Chapel Hill). HEK293 cells grown to approximately 80% confluence in 12-well plates were transfected with 0.2 µg of SRE-luciferase and 0.02 µg pRL-TK harboring the cDNA for Renilla luciferase (Promega), plus 1.0 µg plasmid encoding a variant of myc-Gα12QL, Gα13QL, or Concertina. Plasmid mixtures were combined with Lipofectamine 2000 in Opti-MEM reduced serum medium (Life Technologies) according to the manufacturer’s instructions. For RNA interference experiments, small interfering RNA (siRNA) specific to Hsp90-α (ON-TARGETplus SMARTpool; Thermo Scientific Dharmacon, Pittsburgh, PA) was reconstituted in 1× siRNA buffer (Thermo Scientific Dharmacon), and 60 pmol per sample was combined with the plasmids described above and cotransfected using Lipofectamine 2000 into 12-well plates of HEK293 cells. Assays for SRE activation were performed as described previously (Meigs et al., 2005). Briefly, cells were washed with phosphate-buffered saline and lysed in 1× passive lysis buffer (Promega), and lysates were analyzed using a Dual-luciferase assay system and GloMax 20/20 luminometer (Promega). Light output from firefly luciferase activity was divided by output from Renilla luciferase activity to normalize for variations in transfection efficiency.
Visualization of MRTF-A Localization.
Subcellular distribution of EGFP-tagged MRTF-A was assayed as previously reported (Hinson et al., 2007; Medlin et al., 2010) with minor modifications. HEK293 cells grown on glass coverslips to approximately 50% confluence were transfected with plasmids encoding EGFP-MRTF-A and variants of Gα12 using Lipofectamine 2000 (Life Technologies). After 36 hours, cells were washed in PBS and fixed in 4% paraformaldehyde for 10 minutes, rinsed three times in PBS, and mounted on 4′,6-diamidino-2-phenylindole–Fluoromount-G (Southern Biotech, Birmingham, AL). Cells were visualized on a TE-2000S (Nikon, Melville, NY) equipped with a SPOT monochrome digital camera (Diagnostic Instruments, Sterling Heights, MI). Individual cells were examined for 4′,6-diamidino-2-phenylindole fluorescence to define the nuclear compartment, followed by green fluorescence to visualize EGFP-MRTF-A distribution.
Drosophila Cell Culture and Imaging.
S2 cells were obtained and cultured as previously described (Rogers and Rogers, 2008). Cells were maintained in SF900 media (Life Technologies) and were transfected with 2 mg/ml DNA using the Amaxa nucleofector system (Lonza, Basel, Switzerland) with KitV and program G-30. Cells were plated onto ConA-coated coverslips in petri dishes and induced with 1 mM CuSO4 for 2.5 hours. Cells were prepared for imaging as previously described (Rogers and Rogers, 2008). Probes used were 1:400 diluted anti-myc (9E10) antibody (DSHB, Iowa City, IA) and 1:100 diluted Alexa Fluor 488 phalloidin (Life Technologies). Cells were imaged using an Eclipse Ti-E (Nikon).
Data Analysis and Statistics.
Immunoblot results were quantified using a Kodak Gel Logic 100 imaging system equipped with Molecular Imaging 5.X software (Carestream Health, New Haven, CT) to calculate Gaussian fit for each protein band. For graphical data presentation, error bars represent either ± range or ± S.E.M., as indicated in the figure legends (Figs. 1–7). Student’s t test with unequal variance was used to compare the means of two data sets with one measurement variable, and the Kruskal–Wallis test was used for one-way analysis of variance. A P value < 0.05 was considered significant.
Fig. 1.
Construction, expression, and solubilization of class-distinctive Gα12 mutants. (A) Amino acid sequence of Gα12 is shown. Each class-distinctive residue, indicated by an oval, was mutated to its nonclass-distinctive form (above right of ovals) in myc-tagged, constitutively active Gα12. Shaded areas indicate the switch I, II, and III regions, and the dashed box indicates the site of the activating Q229L mutation. (B) The indicated mutant constructs were expressed in HEK293 cells, from which detergent-soluble extracts were subjected to immunoblot analysis as described in Materials and Methods. All mutants were single amino acid substitutions except the double-mutant Q232E/Q234K (QE/QK). Extracts from cells transfected with the Q229L variant of myc-tagged Gα12 (12QL) and empty pcDNA3.1 (vector) were analyzed in parallel.
Fig. 7.
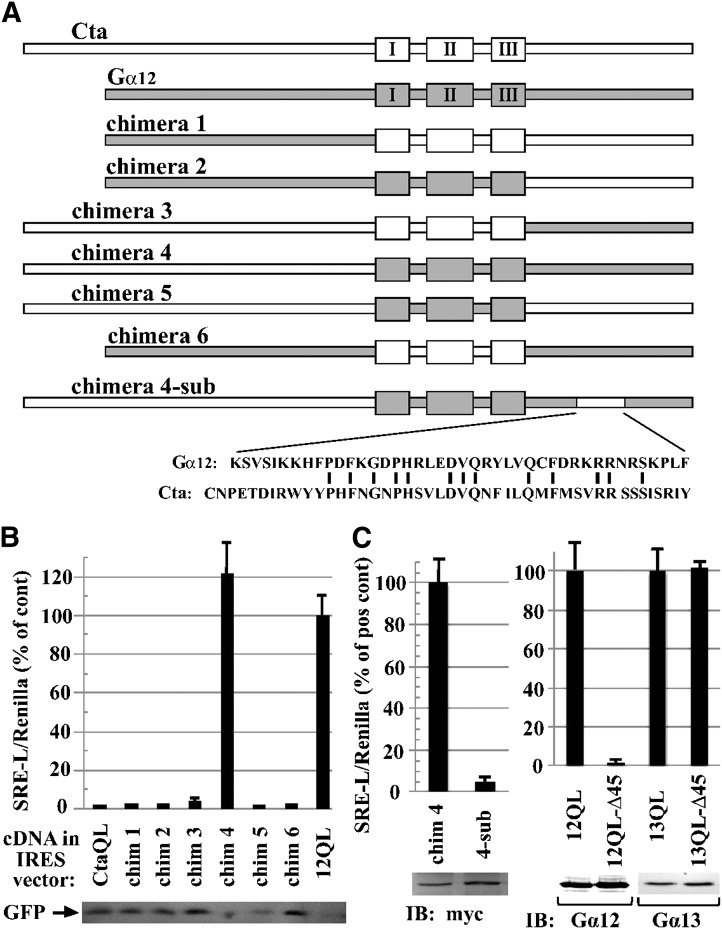
Identification of additional determinants of SRE activation in Gα12. (A) Schematic of Gα12/Concertina chimeras is shown. Chimeras were engineered as described in Materials and Methods, with the switch regions demarcated by the N-terminal boundary of the switch I region (Cys276 of Concertina, Ala202 of Gα12) and the C-terminal boundary of the switch III region (Arg342 of Concertina, Arg267 of Gα12). An activating Gln-to-Leu mutation was engineered in the switch II region of each construct. For chimera 4-sub, the indicated 45-residue Concertina region was introduced to chimera 4 in place of the indicated 42-residue Gα12 region. (B) Stimulation of SRE-mediated transcription in HEK293 cells (see Materials and Methods) by Gα12/Concertina chimeras 1–6 is shown. SRE activation is displayed as percent of the positive control myc-Gα12QL (12QL) in the same experiment. Values are mean ± range for three or more independent trials per chimera, with only the mean displayed for samples with values ≤3% of positive control. Immunoblot levels of GFP, coexpressed in the IRES vector harboring each chimera, are shown (lower panel). (C) Effects of C-terminal Concertina substitutions within chimera 4 and G12/13 proteins are shown. N-terminally myc-tagged chimera 4 and chimera 4-sub were tested for SRE activation, and mean ± range is shown for three independent experiments. Lysates were examined by immunoblot (IB) analysis using anti-myc antibody; results from a representative experiment are shown. SRE activation assays for mutants harboring the aforementioned 45-residue Concertina region at the C terminus of myc-Gα12QL (12QL-Δ45) and Gα13QL (13QL-Δ45) are presented in comparison with positive controls myc-Gα12QL and Gα13QL, respectively, and results are shown as mean ± range for three independent experiments. Comparative expression levels of the two Gα12 constructs, and the two Gα13 constructs, were determined by immunoblot analysis using anti-Gα12 (Santa Cruz Biotechnology) and anti-Gα13 (EMD Millipore) antibodies, and images from a representative experiment are shown.
Results
Identification and Mutation of Class-Distinctive Residues within Gα12.
Each Gα class is thought to signal to its downstream effectors via specialized contact surfaces that are ultimately determined by primary sequence. Therefore, we hypothesized that these effector-binding surfaces harbor unique, “class-distinctive” amino acids that confer specificity for downstream signaling partners. To identify G12/13 class-distinctive residues, we conducted a structural alignment of all four classes of mammalian G protein α-subunits: Gs, Gi, Gq, and G12/13 as described by Temple et al. (2010) and identified 19 class-distinctive residues in the G12/13 class. Of these residues, 16 were located either in Gα12 alone or in both Gα12 and Gα13 and were selected for further study. Through ancestral reconstruction analyses, we determined that the “nonclass-distinctive” counterpart, represented by a residue invariant among the other three mammalian Gα classes, most frequently represents the ancestral amino acid value (Temple et al., 2010). We engineered substitutions within a myc-tagged, constitutively active variant (Q229L) of Gα12, termed myc-Gα12QL, with each mutation converting a class-distinctive residue to its nonclass-distinctive form (Fig. 1A). Each mutant was engineered as a single substitution, with the exception of Q232E/Q234K in which two Gln residues were changed in the same construct, yielding a total of 15 Gα12 variants. When expressed in HEK293 cells, nearly all class-distinctive mutants were detected at levels comparable to unaltered myc-Gα12QL (Fig. 1B). Endogenous Gα12 migrated slightly faster on gels than the myc-tagged variants and was not detectable (Fig. 1B, vector) except when immunoblots were developed for relatively long times.
Class-Distinctive Gα12 Mutants Selectively Uncoupled from SRE Activation and Hsp90 Binding.
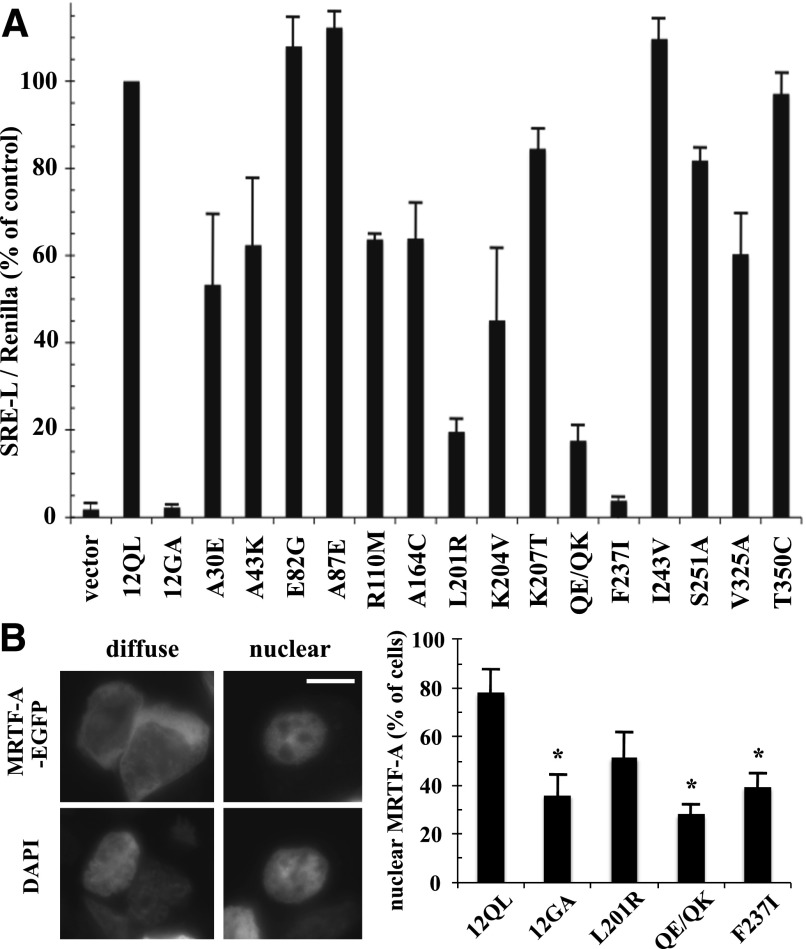
We examined the Gα12 class-distinctive mutants for activation of SRE-mediated transcription, a Rho-dependent cell growth pathway that is responsive to Gα12 and Gα13 (Fromm et al., 1997; Bhattacharyya and Wedegaertner, 2000). As shown in Fig. 2A, most of these mutants displayed normal or moderately impaired ability to stimulate a SRE-luciferase reporter construct (>50% of control, nonmutated myc-Gα12QL); however, the mutant L201R and the double-mutant Q232E/Q234K showed severe impairment of SRE stimulation (<20% of control), whereas the mutant F237I showed near-complete loss of SRE activation that was comparable to a constitutively inactive mutant (G228A) of Gα12 that is unable to release GDP (Miller et al., 1988). We further tested these SRE-uncoupled Gα12 mutants for an additional readout in this signaling pathway, nuclear translocation of MRTF-A/megakaryoblastic leukemia 1. This trafficking is mediated by activated Gα12 and Gα13 and requires downstream Rho activation, which participates in mediating MRTF-A nuclear import and activity by decreasing the availability of the MRTF-A binding partner, G-actin (Evelyn et al., 2007; Medjkane et al., 2009). As shown in Fig. 2B, cells expressing EGFP-tagged MRTF-A and cotransfected with myc-Gα12QL showed a higher percentage of cells with exclusively nuclear EGFP-MRTF-A staining than cells cotransfected with the inactive (G228A) myc-Gα12. Although the SRE-uncoupled mutants of myc-Gα12QL (L201R, Q232E/Q234K, F237I) showed varied results in this assay, all exhibited a lower percentage of cells with nuclear EGFP-MRTF-A localization than the positive control myc-Gα12QL, suggesting the impaired ability of these mutants to signal to SRE involves a mechanism upstream of MRTF-A transport to the nucleus.
Fig. 2.
Identification of class-distinctive mutants of Gα12 impaired in SRE-mediated transcriptional activation. (A) HEK293 cells were transfected with the plasmids SRE-luciferase (0.2 µg) and pRL-TK (0.02 µg) plus 1 µg of plasmid encoding each indicated class-distinctive mutant of myc-Gα12QL (x-axis). Firefly luciferase activity values were normalized for Renilla luciferase activity values and presented as a percent of positive control myc-Gα12QL (12QL) within the same experiment. Empty pcDNA3.1 (vector) and Gα12 harboring the inactivating G228A mutation (12GA) were examined as negative controls. Data presented are the mean ± range of two independent experiments per myc-Gα12QL variant, with three or more experiments performed for mutants that showed greater than 50% impairment of SRE stimulation. (B) HEK293 cells grown in six-well plates were cotransfected with 1.0 µg of EGFP-MRTF-A plasmid and 1.0 µg of each indicated Gα12 construct. Cells (50 per transfection) were scored for either diffuse or exclusively nuclear localization of the EGFP signal, using 4′,6-diamidino-2-phenylindole (DAPI) costaining to define the boundaries of the nucleus. Examples are shown in the left panels (two cells exhibiting diffuse EGFP-MRTF-A staining) and right panels (single cell exhibiting nuclear staining of EGFP-MRTF-A). Scale bar is 10 µm. The column graph shows results compiled from three independent trials, presented as mean ± S.E.M. Significance of the difference in mutant values compared with positive control (12QL) was determined by Student’s t test (*P < 0.05).
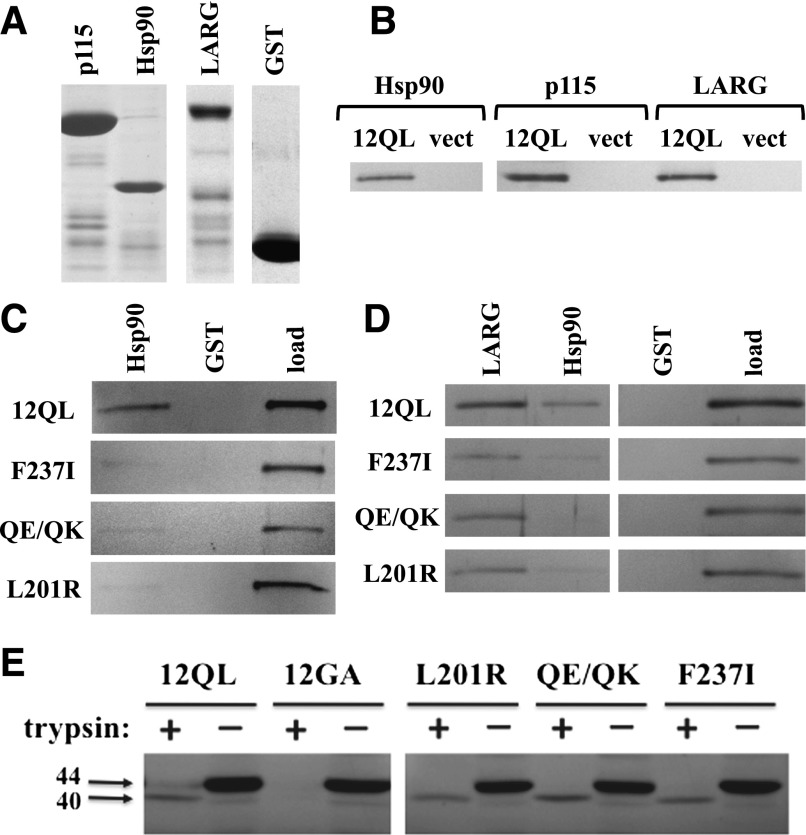
In parallel to the SRE activation studies shown in Fig. 2A, we examined whether mutation of class-distinctive residues in Gα12 to their nonclass-distinctive forms disrupted interaction with specific effector proteins. Myc-tagged Gα12QL and its mutants were expressed in HEK293 cells and tested for the ability to bind GST fusions of p115RhoGEF, LARG, and Hsp90 (Fig. 3A). As shown in Fig. 3B, each of these proteins was able to precipitate myc-Gα12QL from cell extracts. Among the class-distinctive mutants, L201R, F237I, and Q232E/Q234K were most severely impaired in binding Hsp90, with a precipitate-to-load ratio between 0 and 20% of the value for myc-Gα12QL (Fig. 3C; Table 1). This indicated that the subset of Gα12 class-distinctive residues most indispensable for stimulating SRE-mediated transcription were also the most important in Gα12–Hsp90 interaction. These mutations that attenuated SRE signaling and Hsp90 binding did not globally affect Gα12 interaction with downstream targets, as all three mutants bound strongly to LARG and p115RhoGEF (Fig. 3D; Table 1). Furthermore, these mutations did not disrupt Gα12 structural conformation. As shown in Fig. 3E, the mutants L201R, F237I, and Q232E/Q234K yielded a trypsin-protected 40-kDa fragment comparable to the fragment generated from nonmutated myc-Gα12QL, whereas the inactive G228A variant of myc-Gα12 was fully digested. Therefore, it appears these Gα12 mutants retain fundamental properties of guanine nucleotide binding, and their impairment in SRE stimulation and Hsp90 binding appears not to be caused by failed conformational activation or other, nonspecific disruptions of protein folding or effector binding.
Fig. 3.
Effector binding and conformational activation of SRE-uncoupled class-distinctive Gα12 mutants. (A) GST fusions of the N-terminal 252 amino acids of p115RhoGEF (p115), the C-terminal 107 residues of cytoplasmic Hsp90, and the region spanning Leu320 to Arg606 of LARG were analyzed by SDS-PAGE and Coomassie blue staining. Unmodified GST was analyzed in parallel. (B) Coprecipitation experiments using these GST fusion proteins were performed on detergent extracts from HEK293 cells expressing myc-tagged, constitutively active Gα12 (12QL) or empty pcDNA3.1 (vect). (C) Coprecipitations of indicated Gα12 mutants by immobilized Hsp90 are shown. For each panel, the left and center lanes harbor samples in which Sepharose-bound GST-Hsp90 or GST (indicated at top) were used to precipitate fractions from HEK293 extracts harboring myc-Gα12QL (12QL) or the mutants indicated at left. The right lane of each panel (load) harbors 5% of the HEK293 extract set aside before coprecipitation, and band intensities at ∼44 kDa were quantified and used to normalize the coprecipitated bands for Gα12 variants indicated here, as well as others (Table 1). A representative of three independent experiments is shown. (D) Coprecipitation of myc-Gα12QL and its indicated mutants by multiple effectors are shown. The indicated GST fusions of LARG and Hsp90, plus GST with no adduct (GST) are indicated, along with the load for each sample. Data shown are a representative of three independent experiments. (E) Trypsin protection assays were performed on the indicated class-distinctive mutants, plus constitutively activated (12QL) and inactivated (12GA) myc-Gα12. HEK293 cells grown in 10-cm plates were transfected with 10 µg of plasmid DNA, and after 40 hours, cell lysates were subjected to tryptic proteolysis as described in Materials and Methods. Molecular weights are indicated at left in kilodaltons. Results shown are one representative of three independent experiments.
TABLE 1.
Coprecipitation of Gα12 class-distinctive mutants by downstream effectors
For each class-distinctive mutant of myc-Gα12QL (left column), precipitation by each GST-fused effector protein (top row) and starting material (i.e., load) were quantified as described in Materials and Methods to determine a precipitate:load ratio. Categories are as follows: ++++, ratio >70% of positive control myc-Gα12QL; +++, 40–70% of control; ++, 20–40% of control; +, 0–20% of control; −, no detectable coprecipitation. Each sample is the result of two trials, except for samples scoring ++, +, or –, which were analyzed in at least three trials.
| p115 | LARG | Hsp90 | |
|---|---|---|---|
| A30E | ++++ | ++++ | ++/+++ |
| A43K | +++ | +++ | +++ |
| E82G | ++++ | ++++ | ++++ |
| A87E | ++++ | ++++ | ++++ |
| R110M | ++++ | ++++ | ++ |
| A164C | ++/+++ | +++ | ++++ |
| L201R | ++++ | +++ | −/+ |
| K204V | +++ | +++ | ++/+++ |
| K207T | ++++ | ++++ | ++ |
| QE/QK | +++ | ++++ | −/+ |
| F237I | ++/++++a | ++++ | + |
| I243V | ++/+++ | +++ | ++++ |
| S251A | ++ | ++++ | +++ |
| V325A | +++ | +++/++++ | +++ |
| T350C | ++ | ++ | ++ |
Variation spanning more than two categories.
Phylogenetic Substitutions of Class-Distinctive Gα12 Residues Have Differential Effects on SRE Signaling.
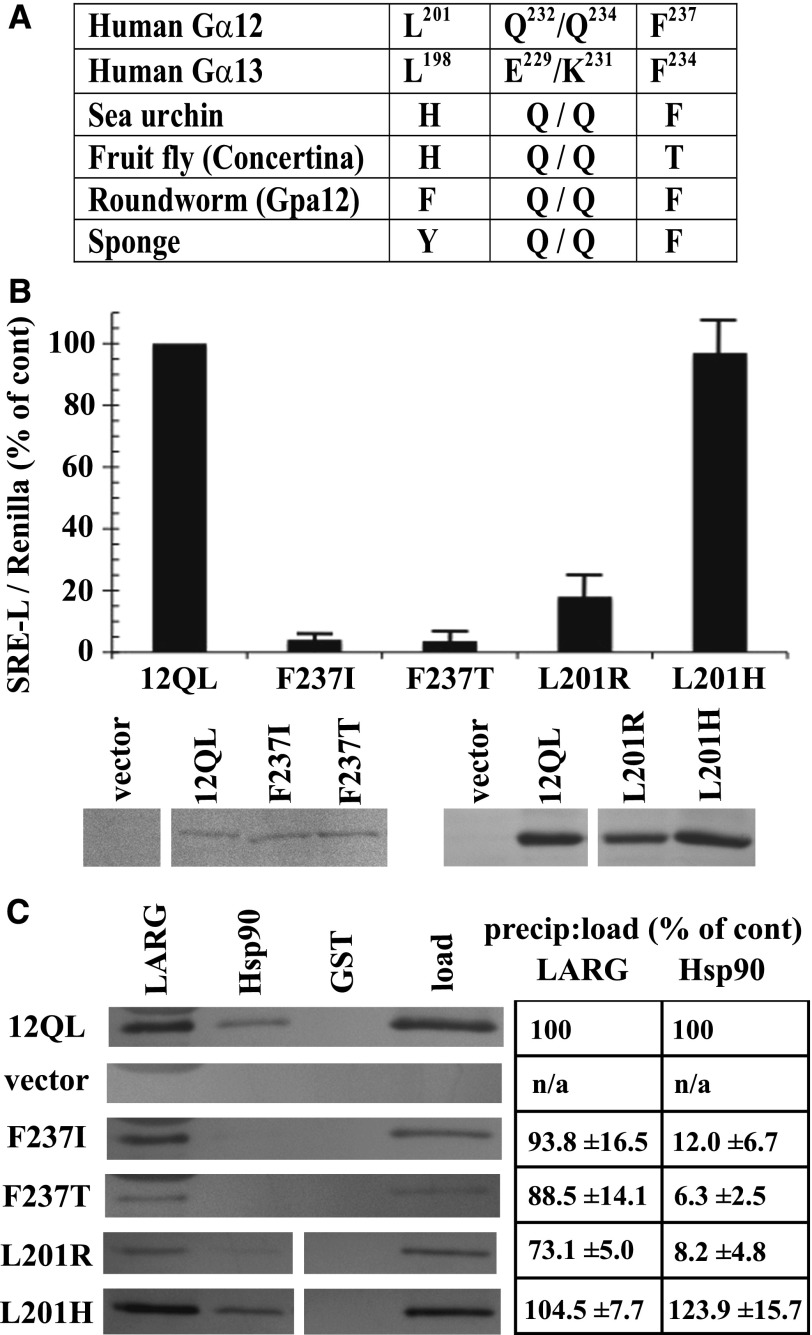
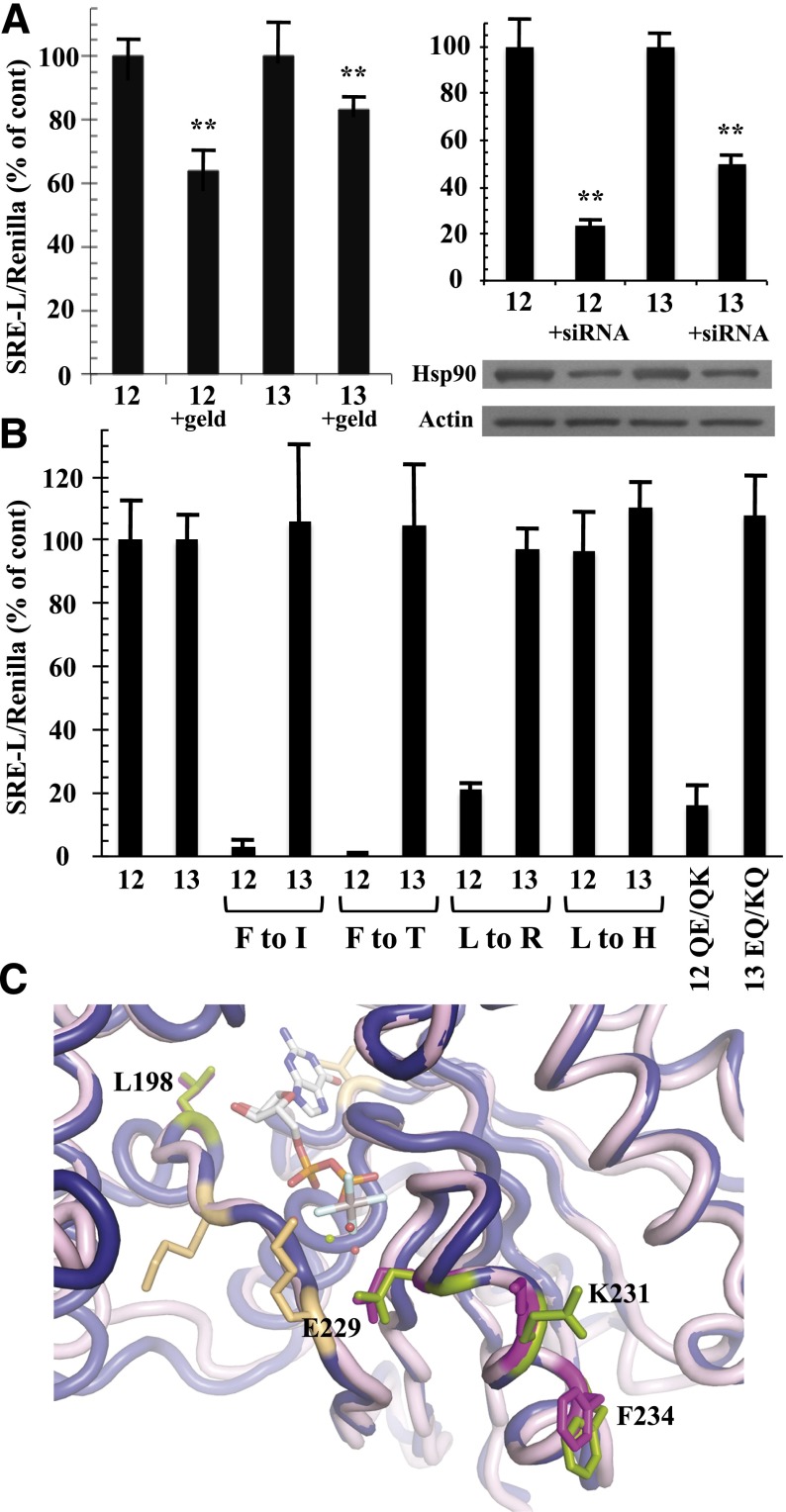
In addition to comparing the different mammalian Gα classes to identify residues that confer signaling specificity, we determined the phylogenetic timeline of G12/13 class-distinctive residues by comparing divergent sequences from sea sponge, roundworm, fruit fly, sea urchin, and mouse or human (Temple et al., 2010). Evolutionary emergence of the Leu residue immediately upstream of the switch I region (Leu201 in Gα12; Leu198 in Gα13) was relatively recent for the G12/13 class because a His residue occupies this position in the sea urchin and Drosophila homologs, and the Caenorhabditis elegans and sea sponge G12/13 proteins harbor other residues (Fig. 4A). In contrast, Phe237 in the switch II region of Gα12 is highly conserved in G12/13 proteins of all the taxa examined (Fig. 4A) except Drosophila, in which Thr occupies this position. To assess the structural and functional consequences of these evolutionary changes within the G12/13 class, we mutated Leu201 and Phe237 in myc-Gα12QL to the corresponding Drosophila residues His and Thr, respectively. The L201H mutant (i.e., mammal → fly) exhibited normal SRE activation and binding to Hsp90, in sharp contrast to L201R (i.e., class-distinctive → nonclass-distinctive) that caused impairment of both functions (Fig. 4, B and C). Surprisingly, the F237T mutant (mammal → fly) in myc-Gα12QL showed near-complete loss of SRE stimulation (Fig. 4B), phenocopying the Phe237 mutation to Ile (class-distinctive → nonclass-distinctive), and both F237I and F237T mutants were markedly impaired in Hsp90 binding (Fig. 4C). For all variants of residues Leu201 and Phe237 that we engineered, binding of Gα12 to LARG was unperturbed or slightly diminished (Fig. 4C). In some experiments, protein levels of mutants L201R and F237I were lower than the positive control myc-Gα12QL; therefore, to ascertain that impaired SRE signaling was not due merely to lowered expression of these mutants, we examined a series of cell samples transfected with decreasing amounts of myc-Gα12QL plasmid (Fig. 4B). Mutants F237I and L201R showed impaired SRE activation compared with myc-Gα12QL expressed at similar levels. From these data, we conclude that Leu201 and Phe237 in mammalian Gα12 were critical components of its evolved ability to stimulate SRE and bind Hsp90, but the corresponding residues in the Drosophila G12/13 homolog Concertina represent different stages of this process. The His residue upstream of the switch I region in Concertina is effectively a G12/13 class-distinctive residue (i.e., facilitated the same signaling properties as Leu201 in Gα12) and therefore might be an intermediate to fixation of the Leu residue in mammalian G12/13 proteins. On the other hand, the Thr residue in the switch II region of Concertina fails to act as a G12/13 class-distinctive residue; its introduction to Gα12 disrupted both SRE-mediated signaling and Hsp90 binding. It remains to be determined whether this Thr residue confers a unique signaling property in Concertina despite the conservation of Phe at this position in the G12/13 class from sea sponge to humans. To our knowledge, these results provide the first instance of a signaling function being disrupted in a mammalian G12/13 protein by substitution of corresponding sequence from an evolutionary homolog in the same class.
Fig. 4.
Ancestral and Concertina-specific substitutions of Gα12 residues involved in SRE activation and Hsp90 binding. (A) An evolutionary profile of selected class-distinctive G12/13 residues is shown, with position in the primary amino acid sequence shown in superscript for Gα12 and Gα13 (Temple et al., 2010). (B) The indicated substitutions in myc-Gα12QL (x-axis) were examined for SRE-luciferase activation as described in Materials and Methods, and results are presented as a percent of positive control myc-Gα12QL (12QL). Panels beneath this graph show expression levels of the indicated mutants, matched with one of several different samples of myc-Gα12QL, as determined by immunoblot analysis of HEK293 cell lysates using anti-Gα12 antibody. (C) Effects of the indicated substitutions within Gα12 on coprecipitation by immobilized LARG and Hsp90 are shown, with lysates from cells transfected with myc-Gα12QL and empty vector analyzed in parallel. Panel at left shows blot images representative of three independent experiments, and panel at right shows precipitate-to-load ratios as a percent of the same ratio determined for myc-Gα12QL (set at 100%). Quantitative data are presented as mean ± S.E.M.
Structural Determinants of Hsp90 Interaction Are Selectively Required for Gα12 in G12/13-Mediated SRE Activation.
Gα13, like Gα12, is a potent stimulator of SRE-mediated transcriptional activation (Fromm et al., 1997), and activated mutants of both Gα12 and Gα13 are blocked in SRE activation by the Clostridium botulinum C3 exoenzyme, a specific inhibitor of Rho activity (Shi et al., 2000). To assess the requirement for Hsp90 function in Gα12 and Gα13 signaling, we used a constitutively active (Q226L) variant of Gα13 along with constitutively active Gα12 and examined effects of the Hsp90 inhibitor geldanamycin on SRE activation. Hydrolysis of ATP is critical for Hsp90 in forming a “clamped” complex with client proteins, and geldanamycin disrupts this biologic function of Hsp90 by competing for ATP binding to the N-terminal domain (Peterson and Blagg, 2009). We found geldanamycin to partially block Gα12-mediated activation of SRE-luciferase in HEK293 cells, whereas this compound showed significantly lower efficacy in blocking Gα13 stimulation of this reporter (Fig. 5A). This selectivity of geldanamycin on Gα12 signaling within the G12/13 class agreed with previous findings in NIH3T3 fibroblasts (Vaiskunaite et al., 2001). In addition, we tested the ability of Gα12 and Gα13 to stimulate SRE-luciferase in HEK293 cells cotransfected with siRNA targeting human Hsp90-α. These experiments yielded results similar to the effects of geldanamycin; this RNA interference caused significantly greater downregulation of Gα12-mediated SRE activation in comparison with the Gα13-driven response (Fig. 5A). To determine whether the SRE-uncoupled mutants define a mechanism specific to Gα12 within the G12/13 family, we engineered constitutively active Gα13 to harbor substitutions of Leu198 and Phe234 to their nonclass-distinctive forms (mutants L198R and F234I) and their Concertina-specific forms (mutants L198H and F234T). In striking contrast to Gα12, these substitutions in Gα13 caused no disruption in SRE signaling (Fig. 5B). We also examined the role of Gα13 residues that correspond to the class-distinctive Gln232/Gln234 pair found in Gα12. As described already herein (Figs. 2 and 3), the Q232E/Q234K mutant converts Gln residues within the Gα12 switch II region to their nonclass-distinctive forms, disrupting SRE stimulation and Hsp90 binding. Paradoxically, these nonclass-distinctive Glu and Lys residues are present in Gα13, despite conservation of the Gln/Gln pair in the G12/13 class for all nonmammalian taxa examined (Fig. 4A). We hypothesized that “reversion” from Gln/Gln to the ancestral Glu/Lys was necessary for Gα13 to evolve its distinct mechanism of growth signaling, and therefore we mutated Glu229 and Lys231 in Gα13 to a pair of Gln residues and tested this E229Q/K231Q mutant in SRE-luciferase assays. Surprisingly, these substitutions caused no disruption of SRE activation triggered by constitutively active Gα13 (Fig. 5B), suggesting that Glu229 and Lys231 of Gα13 are uninvolved in its stimulation of SRE-mediated transcription, whereas Gln residues at these positions in Gα12 are critical for its mechanism of activating the same response. A structural comparison of Gα12 to Gα13 in the switch II region reveals similarities and differences in backbone and side-chain atoms at the Q232E/Q234K positions (Fig. 5C). In Gα12, the Q232E substitution alters only the charge of the side-chain, whereas the Q234K substitution changes both size and charge. Taken as a whole, these findings suggest that the effector proteins stimulated by Gα12 and Gα13 in the pathway(s) leading to SRE-mediated transcription are not identical and that interaction with functional Hsp90 is a specific requirement of Gα12 in this signaling response.
Fig. 5.
Differential effects of Hsp90 inhibition and SRE-uncoupling mutations in Gα12 and Gα13. (A) Effects of geldanamycin (left graph) and Hsp90-specific siRNA (right graph) on Gα12- and Gα13-mediated SRE activation are shown. HEK293 cells grown in 12-well plates were transfected with 0.2 µg of SRE-luciferase and 0.02 µg of pRL-TK, plus 1 µg of a constitutively active variant of either Gα12 (12) or Gα13 (13). For the left graph, cells were serum-starved for 24 hours, and then 1 µg/ml geldanamycin (geld) was added for 6 hours. For the right graph, siRNA specific to cytoplasmic Hsp90-α was cotransfected with the above plasmids as described in Materials and Methods. Luminometry assays were performed as described in Fig. 2, and each G12/13 control sample (geldanamycin absent, or siRNA absent) was set at 100%. For siRNA experiments, cell lysates were subjected to immunoblot analysis, using antibodies specific to Hsp90-α (Santa Cruz Biotechnology) and actin (clone C4; Millipore), and representative blots are shown. Results presented in each graph are the mean of three or more independent experiments, with error bars representing ± S.E.M. Effects of geldanamycin (left graph) and siRNA (right graph) were analyzed by one-way analysis of variance for significant difference in disruption of the Gα12 response compared with disruption of the Gα13 response (**P < 0.001). (B) HEK293 cells were transfected with SRE-luciferase and pRL-TK as described above plus 1 µg of each Gα plasmid. All constructs encoded the constitutively active form of the Gα protein (12 or 13), and substitutions of class-distinctive residues are indicated below the x-axis (e.g., F to I). Additional mutants were tested: QE/QK, Q232E/Q234K substitutions in constitutively active Gα12; EQ/KQ, E229Q/K231Q substitutions in constitutively active Gα13. Data are presented as mean ± range and are the result of three or more independent experiments per construct. (C) Rendering of Gα12 (PDB ID 1ZCA; Kreutz et al., 2006) is shown in blue cartoon and Gα13 (PDB ID 3AB3; Hajicek et al., 2011) in light pink cartoon with the bound nucleotide displayed as sticks and colored by atom type with carbons in white. For Gα13, side chains of Leu198, Glu229, Lys231, and Phe234 are displayed as magenta sticks and numbered. Corresponding Gα12 side chains of Leu201, Gln232, Gln234, and Phe237 are shown as green sticks and are not numbered in this diagram. Figure was rendered in The PyMOL Molecular Graphics System, Version 1.5.0.1 Schrödinger, LLC.
Class-Distinctive Mutants of Gα12 Uncouple SRE Activation from Rho-Mediated Signaling.
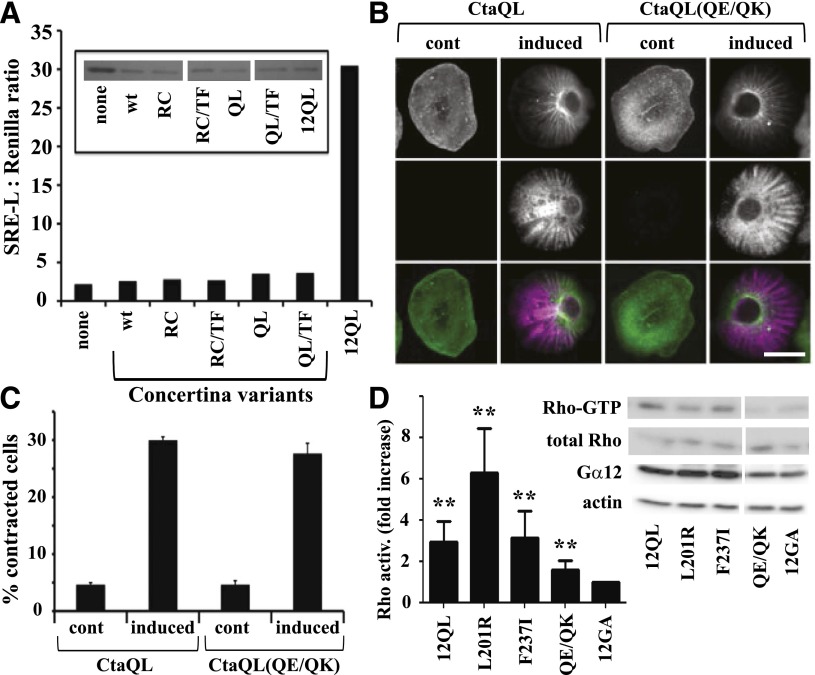
Our results, shown in Fig. 4, revealed a single amino acid substitution (F237T; mammal → fly) that uncoupled Gα12 from SRE activation, and this finding compelled us to examine whether the Drosophila Gα12 homolog Concertina could stimulate the SRE pathway when expressed in mammalian cells. Concertina is required for cell shape changes and migration during gastrulation (Parks and Wieschaus, 1991), but its growth signaling properties are not known. Because Concertina-specific antibodies were not available, we subcloned both wild-type and constitutively active Concertina (both Q303L and R277C variants) into the mammalian cell expression vector pLL5.5 (Uetrecht and Bear, 2009) harboring an IRES to allow translation from a bicistronic mRNA, along with green fluorescent protein (GFP). Neither wild-type nor constitutively active Concertina (Q303L or R277C) stimulated SRE-luciferase when expressed ectopically in HEK293 cells, despite the expression of GFP (Fig. 6A). We were not able to obtain a definitive result in experiments testing ability of the Q303L variant of Concertina to trigger a cytoskeletal response in HEK293 cells (data not shown); however, this Q303L variant was functional in its normal cellular context as an activator of contractility in S2 cells (Fig. 6, B and C), as was the R277C variant (data not shown). As a result of the acute disruption of Gα12-mediated SRE signaling observed when Phe237 was converted to the Concertina-specific Thr, we engineered the converse mutant by replacing Thr311 of Concertina with the Gα12-specific Phe residue. However, this T311F substitution did not confer on Concertina the ability to activate SRE-mediated transcription in HEK293 cells (Fig. 6A).
Fig. 6.
Differential effects of Gα12 and Concertina on SRE-mediated transcription and cytoskeletal rearrangements. (A) Effect of Concertina variants on SRE-mediated transcriptional activation. HEK293 cells were transfected with a GFP-encoding IRES vector harboring myc-Gα12QL (12QL), no additional coding sequence (none), or Concertina variants with the following designations: wt, wild-type; RC, R277C; QL, Q303L; TF, T311F. For each sample, luciferase assays were performed (graph) and GFP levels were determined by immunoblot analysis (inset) to allow indirect measure of expression of Concertina variants. Data shown are a representative of three independent experiments. (B) Effect of Concertina and its Q306E/Q308K variant on cell contraction. S2 cells expressing inducible myc-tagged, constitutively active Concertina (CtaQL) or the same protein harboring Q306E/Q308K substitutions (QE/QK) were induced with 50 µM copper sulfate and 24 hours later scored for contractility as described in Materials and Methods. Cells were stained with phalloidin (top row) and anti-myc antibody (middle row), and images were merged (bottom row). Scale bar is 10 µm, and images are from a representative of three independent experiments. (C) Compiled quantitative data from experiments described in (B) are shown, with bars indicating S.E.M. (D) HEK293 cells were assayed for basal RhoA activity 36–42 hours after transfection with constitutively active (12QL) or inactive (12GA) myc-tagged Gα12, or the indicated mutants of myc-Gα12QL. Blots were developed with SuperSignal West Pico chemiluminescent substrate (Pierce, Rockford, IL) and Kodak Biomax film, and representative blots of active RhoA (Rho-GTP) precipitated by GST-RBD versus total RhoA in cell lysates are shown, along with blots of Gα12 and actin in the same experiment. RhoA activity is the ratio of active RhoA to total RhoA signal, with densitometry (NIH ImageJ software) used to quantify results from seven independent experiments. Results are graphed as the -fold increase over cells expressing inactive, GDP-bound Gα12 (12GA). Graphical data represent mean ± S.E.M.; **P < 0.001 as calculated through analysis of variance using the Kruskal–Wallis test.
Of the class-distinctive Gα12 residues we identified as critical for SRE activation, only the switch II region Gln/Gln pair is conserved between Gα12 and Concertina. To determine whether these Gln residues are important in Concertina-driven cytoskeletal rearrangements, which is a signaling pathway known to use the RhoGEF-Rho axis (Barrett et al., 1997), we tested a Q306E/Q308K mutant of constitutively active Concertina (Q303L) for the ability to stimulate S2 cell contraction. This Concertina mutant showed no loss of efficacy in triggering contractility of S2 cells (Fig. 6, B and C), suggesting that these Gln residues are not involved in the Gα-driven mechanism that mediates these Rho-dependent cytoskeletal changes. We also directly examined the SRE-uncoupled Gα12 mutants (L201R, F237I, and Q232E/Q234K) for Rho activation, by using RBD assays to selectively precipitate GTP-bound RhoA from HEK293 cell lysates. As shown in Fig. 6D, the F237I and L201R mutants showed activation of Rho comparable to positive control myc-Gα12QL. The Q232E/Q234K mutant of myc-Gα12QL triggered a low, although significant, activation of Rho in comparison with inactive, GDP-bound myc-Gα12, despite the robust cytoskeletal response triggered in S2 cells by Concertina harboring the same substitutions.
Gα12 Switch Regions and C Terminus Harbor Independent Determinants of SRE Activation.
Because Concertina failed to stimulate SRE-mediated transcription in HEK293 cells, we used this Drosophila Gα12 homolog as a “blank canvas” in these mammalian cells for identifying regions of Gα12 that mediate SRE activation. We engineered a set of Gα12/Concertina chimeras, designated numbers 1–6, in which a domain encompassing the three switch regions and the flanking N- and C-terminal domains were interchanged in all possible combinations (Fig. 7A). Each chimera was engineered to harbor an activating Gln-to-Leu mutation within the switch II region and was subcloned into the IRES vector pLL-5.5 to allow normalization of expression through GFP immunoblotting (Uetrecht and Bear, 2009). Strikingly, the only construct that stimulated SRE signaling was chimera 4, which harbors the N-terminal 275 residues of Concertina and an additional 178 residues comprising the switch regions and C terminus of Gα12 (Fig. 7B). Whereas this chimera showed robust SRE activation equal to or greater than myc-Gα12QL, the other chimeras exhibited no SRE stimulation, with readings comparable to mutationally activated Concertina or the IRES vector expressing GFP only (Fig. 7B). GFP levels in cell lysates were similar, suggesting that the bicistronic mRNAs encoding all six chimeras were comparable in levels and translation rate (Fig. 7B). These results indicated the switch regions and C-terminal domain of Gα12 harbor determinants that are independently required for SRE activation in HEK293 cells because replacement of either domain with Concertina sequence abrogated this response. To define determinants within the Gα12 C terminus critical for this signaling function, we aligned the Gα12 and Concertina C-terminal domains and found two regions of close homology, separated by a divergent region spanning Lys304 to Phe345 in Gα12 and Cys379 to Tyr423 in Concertina (Fig. 7A). To determine whether this region harbors Gα12-specific determinants of SRE activation, we engineered a “sub-chimera” (designated chimera 4-sub) in which this Gα12 region in chimera 4 was replaced by Concertina sequence. Because N-terminal epitope-tagging of Concertina did not disrupt its ability to stimulate S2 cell contraction (Fig. 6, B and C), we engineered an N-terminal myc tag in chimeras 4 and 4-sub and similarly tagged the other chimeras that harbored the Concertina N-terminal domain (chimeras 3 and 5). As shown in Fig. 7C, chimera 4-sub displayed near-complete loss of SRE signaling, even though immunoblots showed similar expression levels to chimera 4. Myc-tagged chimeras 3, 4, and 5 generated essentially the same SRE-luciferase readouts as their untagged forms (data not shown). We also generated a variant of myc-Gα12QL, designated Δ45, harboring a substitution that introduced the same C-terminal 45-residue Concertina sequence found in chimera 4-sub, and as predicted, this mutation caused a complete loss of SRE activation (Fig. 7C). The corresponding Δ45 mutant of Gα13QL, in which its region spanning Gln301 to Tyr343 was replaced by the aforementioned Concertina sequence, exhibited robust SRE stimulation (Fig. 7C). These findings bolster our results shown in Fig. 5, suggesting that Gα12 and Gα13 use different structural features and effector binding events in stimulating this cell proliferative pathway. Taken as a whole, our results define specific residues in the switch regions, as well as a region near the C terminus, that differ between Gα12 and its Drosophila homolog and are required for SRE signaling by Gα12, but not Gα13.
Discussion
To illuminate the structural features of Gα12 and Gα13 that mediate their signaling functions, a useful approach has been to test mutants for disruption of cellular responses or binding to downstream targets (Jones and Gutkind, 1998; Adarichev et al., 2003; Nakamura et al., 2004; Vazquez-Prado et al., 2004; Grabocka and Wedegaertner, 2005). Such studies have revealed Gα12 determinants of binding to effectors that include RhoGEFs, protein phosphatase-2A, and polycystin-1 (Meigs et al., 2005; Zhu et al., 2007; Yu et al., 2011). Our current study was guided by a method that identified “class-distinctive” residues within each Gα protein and deduced the ancestral, “nonclass-distinctive” residue at each position (Temple et al., 2010). We engineered a panel of Gα12 mutants by converting each class-distinctive residue to its nonclass-distinctive counterpart; these substitutions were intended to render features of Gα12 as ancestral-like conformations that might lack ability to engage specific targets. These mutants revealed Gα12 residues required for binding to Hsp90 but not RhoGEFs: Leu201 just upstream of the switch I region and Phe237 and Gln232/Gln234 within the switch II region. Unexpectedly, these mutations matched the subset of class-distinctive substitutions we identified as disruptive to Gα12 stimulation of SRE-mediated transcription. The effects of geldanamycin, which inhibits ATP binding to the Hsp90 homodimer and would be predicted to disrupt its functional interaction with client proteins that include Gα12, suggest a relationship between Gα12-Hsp90 interaction and SRE activation, and this hypothesis is further supported by our results after siRNA-mediated targeting of Hsp90 expression (Fig. 5). However, the incomplete effect of geldanamycin on Gα12-stimulated SRE signaling leaves open the possibility that the aforementioned class-distinctive mutations disrupt SRE signaling by hindering Gα12 interaction with other, unexamined binding partners in addition to Hsp90.
Gα12 signaling to SRE requires activation of Rho (Fromm et al., 1997). Because the SRE-uncoupled Gα12 mutants exhibited normal binding to RhoGEFs, as well as robust Rho activation for mutants L201R and F237I, these class-distinctive Gα12 residues may mediate engagement of an effector pathway(s) required in addition to the canonical RhoGEF-Rho pathway for SRE activation. Results for the Q232E/Q234K mutant of Gα12 were less clear; these substitutions disrupted SRE activation and also diminished Rho activation, even though RhoGEF binding remained intact. It is possible this mutation disrupts SRE signaling by partially uncoupling Gα12 from the RhoGEF-Rho signaling pathway. However, our finding that Concertina harboring the same mutation triggered normal Rho-mediated contractility in S2 cells (Fig. 6) suggests that this conserved Gln/Gln pair is not required for the ancient G12/13-RhoGEF-Rho axis that mediates cell shape changes in organisms such as Drosophila and C. elegans (Barrett et al., 1997; Yau et al., 2003). Our overall results suggest the L201R and F237I mutants are more decisive than the Q232E/Q234K mutant in selectively uncoupling Gα12 from Hsp90 without perturbing Gα12-RhoGEF interaction. It is possible this Gln/Gln pair plays a Gα12-specific role in mediating Gα binding to a target protein (e.g., Hsp90) that must be engaged for RhoGEF-Rho signaling to commence. The Drosophila Gα12 homolog Concertina may lack this requirement, as its Q306E/Q308K mutant was unimpeded in triggering cytoskeletal rearrangements. Taken as a whole, our results suggest the class-distinctive residues essential for Gα12-specific SRE activation either mediate a Rho-independent signaling pathway or participate in an effector binding event that facilitates coupling of Gα12 to the RhoGEF-Rho pathway.
Both Gα12 and Gα13 stimulate SRE-mediated transcription via mechanisms that are sensitive to Rho inhibition (Fromm et al., 1997; Bhattacharyya and Wedegaertner, 2000; Shi et al., 2000). However, Hsp90 perturbation preferentially hindered SRE activation by Gα12 in comparison with Gα13 (Fig. 5), suggesting that nonredundant signaling mechanisms evolved within the G12/13 class. Geldanamycin inhibits thrombin-mediated signaling through protease-activated receptor 1 in mouse neuroblasts but fails to inhibit LPA-mediated signaling (Pai and Cunningham, 2002), and because the thrombin receptor preferentially couples to Gα12 whereas LPA signaling uses Gα13 (Yamaguchi et al., 2003), this finding implicates Hsp90 in a Gα12-specific role. Furthermore, our mutations of residues Leu201 and Phe237 disrupted Gα12 but not Gα13 in stimulating SRE-mediated transcription, despite conservation of these amino acids in both proteins. Another apparent Gα12 determinant of SRE activation, the Gln232/Gln234 pair, is conserved throughout taxa harboring the G12/13 class, whereas the Gs, Gi, and Gq classes use Glu/Lys at these positions. Therefore, it is intriguing that Gα13 “reverted” to the ancestral Glu/Lys pair during its evolution and that the Gln/Gln pair is critical for Gα12 in SRE activation, whereas Gα13 utilizes a signaling mechanism unaffected by Glu/Lys mutation to Gln/Gln. The functional significance of this Glu/Lys motif in Gα13 remains to be determined. Substitution of this Glu residue in Gα13 (Glu229) for a positively charged residue disrupted its RhoGEF binding, SRE activation, and recruitment of p115RhoGEF to the plasma membrane. Conversely, mutation of Glu229 to Ala did not disrupt RhoGEF binding by Gα13, and effects on SRE signaling were not reported (Grabocka and Wedegaertner, 2005). Disruption of SRE signaling by charge reversal at this site may be due solely to interference with RhoGEF engagement, whereas E229Q substitution in our study allowed Gα13 to retain SRE activation and presumably functional RhoGEF binding.
Because Gα12 and Gα13 both stimulate SRE-mediated transcription, albeit through nonredundant mechanisms, the inability of the closely related Concertina to activate this pathway in HEK293 cells was surprising. It is possible that requisite downstream effector proteins in mammalian cells are not engaged by Concertina or that this fly protein fails to fold properly in these cells. However, our data from Gα12/Concertina chimeras (Fig. 7) demonstrate that the N-terminal 275 amino acids, as well as a 45-residue C-terminal region of Concertina, can fold correctly and facilitate robust Gα12- or Gα13-mediated signaling in HEK293 cells. These results suggest that key growth-signaling properties, including mechanisms potentially involved in oncogenic transformation, evolved in the lineage that yielded mammalian G12/13 proteins after divergence from Concertina. Moreover, because Concertina activates cytoskeletal rearrangements in Drosophila through a RhoGEF (DRhoGEF2) homologous to the G12/13-coupled mammalian RhoGEFs (Barrett et al., 1997), the striking difference in Gα12 and Concertina signaling to SRE in HEK293 cells bolsters the hypothesis that Gα12 stimulation of this response requires additional effector(s) in these cells besides the canonical RhoGEF-Rho axis. These findings compelled us to interchange residues between Gα12 and Concertina, revealing F237T mutation in Gα12 as disruptive to SRE activation in HEK293 cells, whereas replacement of the Gα12 residue Leu201 with the Concertina residue His had no apparent effect. These results suggest Concertina harbors some, not all, of the structural features necessary for SRE activation in mammalian cells and that such characteristics were partially in place in a common ancestor of Concertina and the mammalian G12/13 proteins. Furthermore, Concertina provided a uniquely useful “foil” for Gα12 in our study, due to its ∼55% identical amino acid sequence and shared signaling properties in regulating Rho-mediated cytoskeletal rearrangements. Our experiments utilizing Gα12/Concertina chimeras revealed both the domain encompassing the switch regions and a span near the Gα12 C terminus as harboring independent determinants of SRE activation. Furthermore, our studies using a corresponding Gα13/Concertina chimera suggested that the same C-terminal span in Gα13 is not involved in its signaling toward SRE activation. Future comparison of G12/13 proteins with these and other chimeras, in assays of effector binding and cellular signaling readouts, may reveal structural features and mechanisms used by Gα12 and Gα13 to activate proliferation, migration, and other responses.
A question that arises from our findings is: what advantage was gained by Gα12 and Gα13 evolving different mechanisms for stimulating SRE-mediated transcription? In considering this question, it is noteworthy that similar examples have been reported in the G12/13 class. Both Gα12 and Gα13 stimulate Na+/H+ exchange across the plasma membrane; however, Gα12 requires protein kinase C, whereas Gα13 uses a pathway independent of this kinase (Dhanasekaran et al., 1994). Gα13 stimulates activity of the Na+/H+ exchangers NHE-1, NHE-2, and NHE-3, but Gα12 stimulates only the latter two proteins while inhibiting NHE-1 (Lin et al., 1996). Also, Gα12-mediated regulation of tight junctions and paracellular permeability occurs via a Src-dependent mechanism, whereas the Gα13-mediated response does not require Src (Meyer et al., 2003; Donato et al., 2009). In another example, recruitment of p115RhoGEF to the inner face of the plasma membrane by activated Gα13 is dependent on RhoA, whereas recruitment by activated Gα12 is refractory to RhoA inhibition (Bhattacharyya et al., 2009). One clue regarding different Hsp90 requirements of the G12/13 proteins has emerged from studies of lipid rafts; Gα12, but not Gα13, localizes to these sphingolipid- and cholesterol-enriched areas, and targeting of Gα12 to lipid rafts is sensitive to Hsp90 inhibition (Waheed and Jones, 2002). The different subcellular locations of Gα12 and Gα13 may facilitate differences in signaling inputs, downstream effectors, and regulators of their respective signaling properties, so that a binding partner such as Hsp90 may be critical for one G12/13 protein but not the other in driving a common downstream response such as SRE activation.
In addition to illuminating target sites for inhibiting Gα12-mediated signaling to SRE-regulated genes, our findings validate the “class-distinctive” methodology (Temple et al., 2010) as a technique that could be extended to other Gα classes to dissect the roles of different binding partners. With the increasing list of proteins that interact with Gα12, Gα13, or other Gα proteins, class-distinctive mutants should be useful for identifying connections between specific Gα-effector interactions and cellular responses mediated by different classes of G proteins.
Acknowledgments
The authors thank Tohru Kozasa, Tatyana Voyno-Yasenetskaya, Channing Der, and Christopher Mack for providing reagents, and Dan Kaplan, Pat Casey, and Aisha Chow for helpful discussions.
Abbreviations
- EGFP
enhanced green fluorescent protein
- GFP
green fluorescent protein
- GPCR
G protein–coupled receptor
- GST
glutathione-S-transferase
- HEK
human embryonic kidney
- Hsp90
heat-shock protein 90
- IRES
internal ribosomal entry site
- LARG
leukemia-associated RhoGEF
- MRTF
myocardin-related transcription factor
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- RBD
Rho-binding domain
- RhoGEF
guanine nucleotide exchange factor for Rho
- siRNA
small interfering RNA
- SRE
serum response element
Authorship Contributions
Participated in research design: Montgomery, Temple, Peters, Tolbert, Rogers, Jones, Meigs.
Conducted experiments: Montgomery, Peters, Tolbert, Smolski, Hamilton, Tagliatela, Rogers, Meigs.
Contributed new reagents or analytic tools: Montgomery, Peters, Booker, Martin, Hamilton, Tagliatela, Rogers, Meigs.
Performed data analysis: Montgomery, Temple, Peters, Tolbert, Rogers, Jones, Meigs.
Wrote or contributed to the writing of the manuscript: Montgomery, Temple, Peters, Tolbert, Rogers, Jones, Meigs.
Footnotes
This work was supported by awards from the North Carolina Biotechnology Center [Grant BRG1229]; National Institutes of Health National Institute of General Medical Sciences [Grants R01-GM081645, R01-GM029860, and R01-GM65989]; the Department of Energy [Grant DE-FG02-05er15671]; the National Science Foundation [Grants MCB0718202 and MCB0723515]; the Arnold and Mabel Beckman Foundation; and the Lineberger Comprehensive Cancer Center.
References
- Adarichev VA, Vaiskunaite R, Niu J, Balyasnikova IV, Voyno-Yasenetskaya TA. (2003) Gα13-mediated transformation and apoptosis are permissively dependent on basal ERK activity. Am J Physiol Cell Physiol 285:C922–C934 [DOI] [PubMed] [Google Scholar]
- Barrett K, Leptin M, Settleman J. (1997) The Rho GTPase and a putative RhoGEF mediate a signaling pathway for the cell shape changes in Drosophila gastrulation. Cell 91:905–915 [DOI] [PubMed] [Google Scholar]
- Bhattacharyya R, Banerjee J, Khalili K, Wedegaertner PB. (2009) Differences in Galpha12- and Galpha13-mediated plasma membrane recruitment of p115-RhoGEF. Cell Signal 21:996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya R, Wedegaertner PB. (2000) Galpha 13 requires palmitoylation for plasma membrane localization, Rho-dependent signaling, and promotion of p115-RhoGEF membrane binding. J Biol Chem 275:14992–14999 [DOI] [PubMed] [Google Scholar]
- Cabrera-Vera TM, Vanhauwe J, Thomas TO, Medkova M, Preininger A, Mazzoni MR, Hamm HE. (2003) Insights into G protein structure, function, and regulation. Endocr Rev 24:765–781 [DOI] [PubMed] [Google Scholar]
- Chan AM, Fleming TP, McGovern ES, Chedid M, Miki T, Aaronson SA. (1993) Expression cDNA cloning of a transforming gene encoding the wild-type Gα12 gene product. Mol Cell Biol 13:762–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanasekaran N, Prasad MV, Wadsworth SJ, Dermott JM, van Rossum G. (1994) Protein kinase C-dependent and -independent activation of Na+/H+ exchanger by Gα12 class of G proteins. J Biol Chem 269:11802–11806 [PubMed] [Google Scholar]
- Donato R, Wood SA, Saunders I, Gundsambuu B, Yan Mak K, Abbott CA, Powell BC. (2009) Regulation of epithelial apical junctions and barrier function by Galpha13. Biochim Biophys Acta 1793:1228–1235 [DOI] [PubMed] [Google Scholar]
- Evelyn CR, Wade SM, Wang Q, Wu M, Iñiguez-Lluhí JA, Merajver SD, Neubig RR. (2007) CCG-1423: a small-molecule inhibitor of RhoA transcriptional signaling. Mol Cancer Ther 6:2249–2260 [DOI] [PubMed] [Google Scholar]
- Friedman EJ, Temple BRS, Hicks SN, Sondek J, Jones CD, Jones AM. (2009) Prediction of protein-protein interfaces on G-protein β subunits reveals a novel phospholipase C β2 binding domain. J Mol Biol 392:1044–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm C, Coso OA, Montaner S, Xu N, Gutkind JS. (1997) The small GTP-binding protein Rho links G protein-coupled receptors and Galpha12 to the serum response element and to cellular transformation. Proc Natl Acad Sci USA 94:10098–10103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulimari P, Knieling H, Engel U, Grosse R. (2008) LARG and mDia1 link Galpha12/13 to cell polarity and microtubule dynamics. Mol Biol Cell 19:30–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabocka E, Wedegaertner PB. (2005) Functional consequences of Gα13 mutations that disrupt interaction with p115RhoGEF. Oncogene 24:2155–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu JL, Müller S, Mancino V, Offermanns S, Simon MI. (2002) Interaction of Gα(12) with G α(13) and G α(q) signaling pathways. Proc Natl Acad Sci USA 99:9352–9357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilluy C, Swaminathan V, Garcia-Mata R, O’Brien ET, Superfine R, Burridge K. (2011) The Rho GEFs LARG and GEF-H1 regulate the mechanical response to force on integrins. Nat Cell Biol 13:722–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajicek N, Kukimoto-Niino M, Mishima-Tsumagari C, Chow CR, Shirouzu M, Terada T, Patel M, Yokoyama S, Kozasa T. (2011) Identification of critical residues in G(α)13 for stimulation of p115RhoGEF activity and the structure of the G(α)13-p115RhoGEF regulator of G protein signaling homology (RH) domain complex. J Biol Chem 286:20625–20636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill CS, Wynne J, Treisman R. (1995) The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell 81:1159–1170 [DOI] [PubMed] [Google Scholar]
- Hinson JS, Medlin MD, Lockman K, Taylor JM, Mack CP. (2007) Smooth muscle cell-specific transcription is regulated by nuclear localization of the myocardin-related transcription factors. Am J Physiol Heart Circ Physiol 292:H1170–H1180 [DOI] [PubMed] [Google Scholar]
- Jiang H, Wu D, Simon MI. (1993) The transforming activity of activated Gα12. FEBS Lett 330:319–322 [DOI] [PubMed] [Google Scholar]
- Jones TL, Gutkind JS. (1998) Galpha12 requires acylation for its transforming activity. Biochemistry 37:3196–3202 [DOI] [PubMed] [Google Scholar]
- Juneja J, Casey PJ. (2009) Role of G12 proteins in oncogenesis and metastasis. Br J Pharmacol 158:32–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly P, Casey PJ, Meigs TE. (2007) Biologic functions of the G12 subfamily of heterotrimeric G proteins: growth, migration, and metastasis. Biochemistry 46:6677–6687 [DOI] [PubMed] [Google Scholar]
- Kelly P, Moeller BJ, Juneja J, Booden MA, Der CJ, Daaka Y, Dewhirst MW, Fields TA, Casey PJ. (2006a) The G12 family of heterotrimeric G proteins promotes breast cancer invasion and metastasis. Proc Natl Acad Sci USA 103:8173–8178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly P, Stemmle LN, Madden JF, Fields TA, Daaka Y, Casey PJ. (2006b) A role for the G12 family of heterotrimeric G proteins in prostate cancer invasion. J Biol Chem 281:26483–26490 [DOI] [PubMed] [Google Scholar]
- Kozasa T, Gilman AG. (1995) Purification of recombinant G proteins from Sf9 cells by hexahistidine tagging of associated subunits: characterization of α 12 and inhibition of adenylyl cyclase by α z. J Biol Chem 270:1734–1741 [DOI] [PubMed] [Google Scholar]
- Kreutz B, Yau DM, Nance MR, Tanabe S, Tesmer JJG, Kozasa T. (2006) A new approach to producing functional G α subunits yields the activated and deactivated structures of Gα(12/13) proteins. Biochemistry 45:167–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Voyno-Yasenetskaya TA, Hooley R, Lin CY, Orlowski J, Barber DL. (1996) Galpha12 differentially regulates Na+-H+ exchanger isoforms. J Biol Chem 271:22604–22610 [DOI] [PubMed] [Google Scholar]
- Medjkane S, Perez-Sanchez C, Gaggioli C, Sahai E, Treisman R. (2009) Myocardin-related transcription factors and SRF are required for cytoskeletal dynamics and experimental metastasis. Nat Cell Biol 11:257–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medlin MD, Staus DP, Dubash AD, Taylor JM, Mack CP. (2010) Sphingosine 1-phosphate receptor 2 signals though leukemia-associated RhoGEF (LARG), to promote smooth muscle cell differentiation. Arterioscler Thromb Vasc Biol 30:1779–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meigs TE, Juneja J, DeMarco CT, Stemmle LN, Kaplan DD, Casey PJ. (2005) Selective uncoupling of Gα12 from Rho-mediated signaling. J Biol Chem 280:18049–18055 [DOI] [PubMed] [Google Scholar]
- Meyer TN, Hunt J, Schwesinger C, Denker BM. (2003) Galpha12 regulates epithelial cell junctions through Src tyrosine kinases. Am J Physiol Cell Physiol 285:C1281–C1293 [DOI] [PubMed] [Google Scholar]
- Miller RT, Masters SB, Sullivan KA, Beiderman B, Bourne HR. (1988) A mutation that prevents GTP-dependent activation of the α chain of Gs. Nature 334:712–715 [DOI] [PubMed] [Google Scholar]
- Nakamura S, Kreutz B, Tanabe S, Suzuki N, Kozasa T. (2004) Critical role of lysine 204 in switch I region of Galpha13 for regulation of p115RhoGEF and leukemia-associated RhoGEF. Mol Pharmacol 66:1029–1034 [DOI] [PubMed] [Google Scholar]
- Offermanns S, Mancino V, Revel JP, Simon MI. (1997) Vascular system defects and impaired cell chemokinesis as a result of Galpha13 deficiency. Science 275:533–536 [DOI] [PubMed] [Google Scholar]
- Oldham WM, Hamm HE. (2008) Heterotrimeric G protein activation by G-protein-coupled receptors. Nat Rev Mol Cell Biol 9:60–71 [DOI] [PubMed] [Google Scholar]
- Pai KS, Cunningham DD. (2002) Geldanamycin specifically modulates thrombin-mediated morphological changes in mouse neuroblasts. J Neurochem 80:715–718 [DOI] [PubMed] [Google Scholar]
- Parks S, Wieschaus E. (1991) The Drosophila gastrulation gene concertina encodes a Gα-like protein. Cell 64:447–458 [DOI] [PubMed] [Google Scholar]
- Peterson LB, Blagg BS. (2009) To fold or not to fold: modulation and consequences of Hsp90 inhibition. Future Med Chem 1:267–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren XD, Kiosses WB, Schwartz MA. (1999) Regulation of the small GTP-binding protein Rho by cell adhesion and the cytoskeleton. EMBO J 18:578–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SL, Rogers GC. (2008) Culture of Drosophila S2 cells and their use for RNAi-mediated loss-of-function studies and immunofluorescence microscopy. Nat Protoc 3:606–611 [DOI] [PubMed] [Google Scholar]
- Samant RS, Clarke PA, Workman P. (2012) The expanding proteome of the molecular chaperone HSP90. Cell Cycle 11:1301–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CS, Sinnarajah S, Cho H, Kozasa T, Kehrl JH. (2000) G13α-mediated PYK2 activation. PYK2 is a mediator of G13α-induced serum response element-dependent transcription. J Biol Chem 275:24470–24476 [DOI] [PubMed] [Google Scholar]
- Sternweis PC, Carter AM, Chen Z, Danesh SM, Hsiung YF, Singer WD. (2007) Regulation of Rho guanine nucleotide exchange factors by G proteins. Adv Protein Chem 74:189–228 [DOI] [PubMed] [Google Scholar]
- Suzuki N, Hajicek N, Kozasa T. (2009) Regulation and physiological functions of G12/13-mediated signaling pathways. Neurosignals 17:55–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple BRS, Jones CD, Jones AM. (2010) Evolution of a signaling nexus constrained by protein interfaces and conformational states. PLOS Comput Biol 6:e1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetrecht AC, Bear JE. (2009) Golgi polarity does not correlate with speed or persistence of freely migrating fibroblasts. Eur J Cell Biol 88:711–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaiskunaite R, Kozasa T, Voyno-Yasenetskaya TA. (2001) Interaction between the Gα subunit of heterotrimeric G(12) protein and Hsp90 is required for Gα(12) signaling. J Biol Chem 276:46088–46093 [DOI] [PubMed] [Google Scholar]
- Vara Prasad MV, Shore SK, Dhanasekaran N. (1994) Activated mutant of G alpha 13 induces Egr-1, c-fos, and transformation in NIH 3T3 cells. Oncogene 9:2425–2429 [PubMed] [Google Scholar]
- Vázquez-Prado J, Miyazaki H, Castellone MD, Teramoto H, Gutkind JS. (2004) Chimeric G α i2/G α 13 proteins reveal the structural requirements for the binding and activation of the RGS-like (RGL)-containing Rho guanine nucleotide exchange factors (GEFs) by G α 13. J Biol Chem 279:54283–54290 [DOI] [PubMed] [Google Scholar]
- Waheed AA, Jones TLZ. (2002) Hsp90 interactions and acylation target the G protein Galpha 12 but not Galpha 13 to lipid rafts. J Biol Chem 277:32409–32412 [DOI] [PubMed] [Google Scholar]
- Wang DZ, Li S, Hockemeyer D, Sutherland L, Wang Z, Schratt G, Richardson JA, Nordheim A, Olson EN. (2002). Potentiation of serum response factor activity by a family of myocardin-related transcription factors. Proc Natl Acad Sci USA 99:14855–14860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worzfeld T, Wettschureck N, Offermanns S. (2008) G(12)/G(13)-mediated signalling in mammalian physiology and disease. Trends Pharmacol Sci 29:582–589 [DOI] [PubMed] [Google Scholar]
- Xu N, Voyno-Yasenetskaya T, Gutkind JS. (1994) Potent transforming activity of the G13 α subunit defines a novel family of oncogenes. Biochem Biophys Res Commun 201:603–609 [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Katoh H, Negishi M. (2003) N-terminal short sequences of α subunits of the G12 family determine selective coupling to receptors. J Biol Chem 278:14936–14939 [DOI] [PubMed] [Google Scholar]
- Yau DM, Yokoyama N, Goshima Y, Siddiqui ZK, Siddiqui SS, Kozasa T. (2003) Identification and molecular characterization of the Gα12-Rho guanine nucleotide exchange factor pathway in Caenorhabditis elegans. Proc Natl Acad Sci USA 100:14748–14753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Ritchie BJ, Su X, Zhou J, Meigs TE, Denker BM. (2011) Identification of polycystin-1 and Gα12 binding regions necessary for regulation of apoptosis. Cell Signal 23:213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D, Tate RI, Ruediger R, Meigs TE, Denker BM. (2007) Domains necessary for Galpha12 binding and stimulation of protein phosphatase-2A (PP2A): Is Galpha12 a novel regulatory subunit of PP2A? Mol Pharmacol 71:1268–1276 [DOI] [PubMed] [Google Scholar]