Abstract
The symptoms of ataxia-telangiectasia (A-T) include a progressive neurodegeneration caused by ATM protein deficiency. We previously found that nuclear accumulation of histone deacetylase-4, HDAC4, contributes to this degeneration; we now report that increased histone H3K27 trimethylation (H3K27me3) mediated by polycomb repressive complex 2 (PRC2) also plays an important role in the A-T phenotype. Enhancer of zeste homolog 2 (EZH2), a core catalytic component of PRC2, is a new ATM kinase target, and ATM-mediated S734 phosphorylation of EZH2 reduces protein stability. Thus, PRC2 formation is elevated along with H3K27me3in ATM deficiency. ChIP-sequencing shows a significant increase in H3K27me3 ‘marks’ and a dramatic shift in their location. The change of H3K27me3 chromatin-binding pattern is directly related to cell cycle re-entry and cell death of ATM-deficient neurons. Lentiviral knockdown of EZH2 rescues Purkinje cell degeneration and behavioral abnormalities in Atm−/− mice, demonstrating that EZH2 hyperactivity is another key factor in A-T neurodegeneration.
Ataxia-telangiectasia (A-T) is a multisystem disease characterized by neurodegeneration in the central nervous system (CNS)1–4. The earliest and most profound neuropathology involves the Purkinje and granule cells of the cerebellum. A-T is caused by mutation of the A-T mutated (ATM) gene, which is ubiquitously expressed throughout development and encodes a serine/threonine protein kinase of the phosphatidylinositol-3 kinase-related kinase (PIKK) family5. The best-known function of ATM is to ensure the integrity of the genome. After DNA damage, ATM activates cell cycle checkpoints, arresting the cycle until DNA repair is complete6. Its role in maintaining the health and survival of neurons is complex. Consistent with its function in DNA damage repair7, ATM protects post-mitotic neurons from degeneration by suppressing the cell cycle8–10. But ATM is also required for neuronal vesicle trafficking11 and maintaining HDAC4 (histone deacetylase 4) in the cytoplasm12. Both of these processes are crucial for neuronal viability and yet neither is directly induced by DNA damage12. These later observations hint at a complex web of nerve cell phenotypes that are dependent on ATM function and suggest that the mechanisms behind the neurodegeneration in A-T are multifaceted and still incompletely defined.
Our finding of ectopic nuclear HDAC4 in ATM deficiency and the subsequent genome-wide alterations in the acetylation marks on histones H3 and H4 led us to explore other aspects of the histone code. Polycomb group (PcG) family proteins are transcriptional repressors that epigenetically modify chromatin and participate in the establishment and maintenance of many cell fates13–17. PcG proteins participate in both stem cell self-renewal and tumorigenesis as well as in brain development18,19. In mammals, there are two distinct polycomb protein complexes: polycomb repressive complex 1 (PRC1) and PRC220. The PRC2 complex consists of EZH2 (enhancer of zeste homolog 2), SUZ12 (suppressor of zeste 12) and EED (embryonic ectoderm development)21. EZH2 catalyzes the addition of methyl groups to histone H3 at Lys 27 (H3K27) in target gene promoters, leading to epigenetic silencing. EZH2 plays a role in X-chromosome inactivation, germ line development, cell fate decisions and stem cell pluripotency14,15,22,23. EZH2 is overexpressed in aggressive solid tumors and its overexpression has been implicated in cancer progression and metastases15,24,25; its knockdown leads to decreased proliferation of cancer cells and a delay in the G2/M transition. During development and stem cell maturation, CDK1/EZH2 or CDK2/EZH2 interactions integrate developmental cues and cell cycle activity to control self-renewal or differentiation15,25. EZH2 also regulates the balance between self-renewal and differentiation of cortical neurons26,27.
Histone modification is a major form of epigenetic gene regulation that plays a critical role in many neuronal processes28,29. Spurred by our finding of dramatic HDAC4-mediated histone acetylation changes in Atm-deficient neurons, we asked whether EZH2-mediated H3K27 trimethylation (H3K27me3) is also affected by a defect of ATM function. We now report that in the cerebella of A-T patients and in A-T mouse models, the levels of H3K27 trimethylation are elevated due to the loss ATM-dependent EZH2 phosphorylation. This phosphorylation normally de-stabilizes EZH2 thus reducing its association with the PRC2 complex and subsequent H3K27 methylation. ChIP-sequencing using H3K27me3 antibody reveals genome-wide differences in this histone mark between wild type and Atm−/−. These differences help to drive the CNS symptoms of A-T; knockdown of EZH2 prevents cell cycle reentry and neuronal death in Atm-deficient neurons and improves behavior of Atm−/− mice. We propose a model of A-T in which EZH2-mediated hypermethylation of H3K27me3 is a crucial step leading to the degeneration of the Atm-deficient neurons.
Results
Elevated H3K27me3 in A-T brains
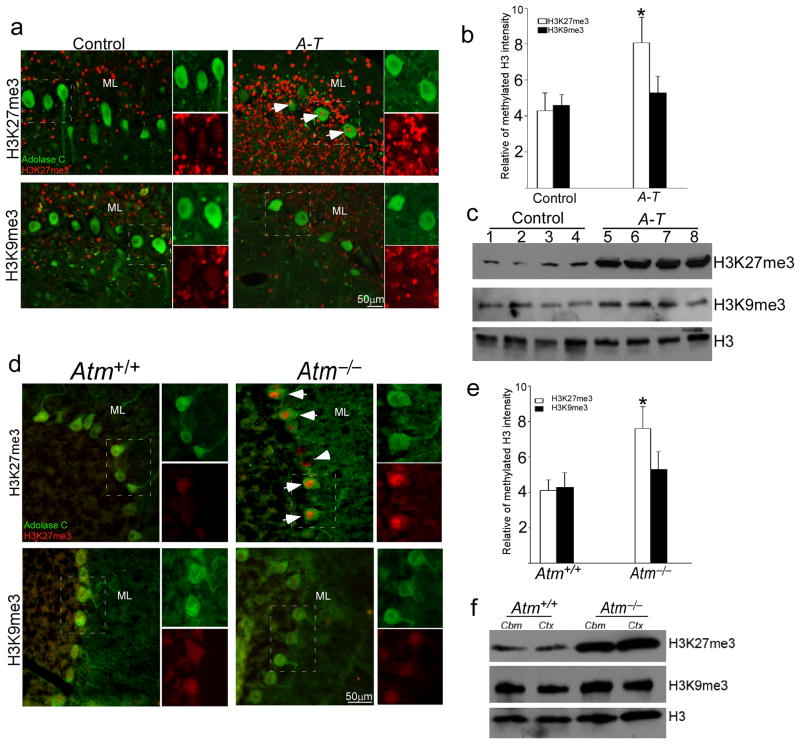
EZH2-mediated H3K27 trimethylation is a key activity of the polycomb epigenetic system. It affects many processes including proliferation, tumorigenesis as well as brain development15,22,30. We became interested in EZH2 as a possible player in ataxia-telangiectasia (A-T) because of its roles of in cell cycle regulation. ATM-deficient Purkinje and granule cells both reenter the cell cycle before dying in human A-T and in its Atm−/− mouse model10,31. We speculated that if EZH2 were involved, the disease process in the A-T nervous system might derive in part from the over-activation of PRC2. As H3K27 is the main target of EZH2, first, we examined H3K27me3 in human cerebellar cortex. Purkinje and granule cell nuclei contained low levels of H3K27me3 in healthy control cerebellar sections (Fig. 1a, top and Fig. 1b). Yet, as predicted by our hypothesis, both neuronal types in sections from A-T patients showed enhanced nuclear H3K27me3 staining; H3K9me3 was unaffected (Fig. 1a, bottom and Fig. 1b). Western blots of tissue extracts validated the immunostaining – elevated levels of H3K27me3 in A-T compared to control (Fig. 1c). We next repeated these observations in Atm−/− mice. Immunostaining for H3K27me3 (Fig. 1d, upper and Fig. 1e), but not H3K9me3 (Fig. 1d, lower and Fig. 1e), was substantially increased in Atm−/− Purkinje cell nuclei. We also found increased H3K27me3 in mouse Atm−/− neocortex and hippocampus (Fig. 1f; Supplementary Fig. 1a). As expected, an increased interaction of H3K27me3 with EZH2 was also found in ATM-deficiency (Supplementary Fig. 1b–c). Thus, there is a close association between elevated H3K27me3 and Atm-deficiency in both humans and mice.
Figure 1. Hypermethylation of H3K27 in human A-T and mouse Atm−/− cerebellum.
a) Paraffin sections of human cerebellar cortex from individuals who died with no known brain disease or with a diagnosis of A-T were immunostained with H3K27me3 and H3K9me3 antibodies (red). Aldolase C immunostaining (green) was used as a marker of Purkinje cells. The white arrows indicate labeled neurons. Scale bar, 50 μm.
b) The relative intensity of H3K27me3 immunostaining illustrated in panel a). * = p < 0.05 by Student’s t-test. Error bars represent SEM.
c) The protein level of H3K27me3 increases in A-T cerebellum. Nuclear protein extracts from frozen cerebellar cortex were probed on western blots with H3K27me3 and H3K9me3 antibodies. Total histone 3 (H3) was used as a loading control. Full-length blots are presented in Supplementary Figure 12. d) Ten micron cryostat sections of wild type (left panels) and Atm−/− (right panels) mouse cerebellum were immunostained with H3K27me3 and H3k9me3 (red). Aldolase C immunostaining (green) was used as a marker of Purkinje cells. The white arrows indicate labeled Purkinje neurons. Scale bar, 50 μm.
e) The relative intensity of H3K27me3 immunostaining illustrated in panel d). * = p < 0.05 by Student’s t-test. Error bar represents SEM.
f) The protein level of H3K27me3 increases dramatically in Atm−/− mouse cerebellum. Nuclear protein extracts from adult mouse cerebellar cortex were probed on western blots with H3K27me3 and H3K9me3 antibodies. H3 was used as a loading control. Full-length blots are presented in Supplementary Figure 12.
EZH2 is an ATM target
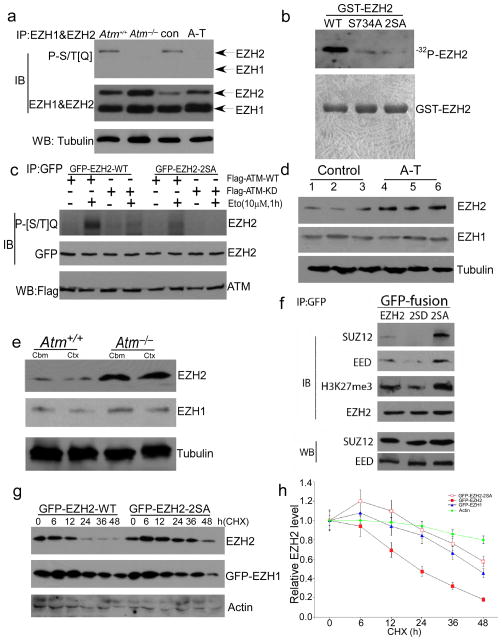
The remarkable change in the levels of H3K27me3 in A-T brain suggested that EZH2 might be a novel ATM target. Analysis of the EZH2 protein sequence predicts that two serines, S652 and S734, are highly probable sites of ATM phosphorylation. We could find no comparably favorable sites in the EZH1 histone methyltransferase, or in other members (e.g., Suz12) of the polycomb repressor complex 2 (PRC2) to which EZH2 belongs. To determine whether these two EZH2 sites are used as ATM targets in vivo, we prepared cerebellar extracts from control and A-T patients as well as from wild type and Atm−/− mice and performed immunoprecipitation with EZH2 or EZH1 antibodies. Western blots of the immunoprecipitates were then probed with an antibody against a phosphorylated serine or threonine in the context of a following glutamine residue (P[S/T]Q), the canonical ATM/ATR target site32,33. A strong P[S/T]Q signal was found on the EZH2 band in control samples but not in A-T or Atm−/− extracts (Fig. 2a). We found no P[S/T]Q signal in either genotype using EZH1 immunoprecipitates (Fig. 2a). We verified that S734 is the predominant ATM site on EZH2 by performing ATM in vitro kinase assays using GST-EZH2 fusion proteins as substrates (Fig. 2b). The phosphorylation signal was nearly lost when the EZH2-S734A mutant was used as substrate; the 2SA (S652A/S734A) double mutation blocked the phosphorylation signal entirely. This indicates S734 is the major ATM phosphorylation site, but S652 may serve as an alternate kinase target. To confirm whether ATM-mediated EZH2 phosphorylation responds to DNA damage in vivo, GFP-EZH2 wild type or the non-phosphorylatable 2SA mutant was overexpressed in human A-T fibroblasts with Flag-ATM wild type or the kinase dead (KD) mutant. A strong P[S/T]Q signal was found with etoposide treatment when GFP-EZH2-WT was expressed with Flag-ATM-WT (Fig. 2c). Expression of either the EZH2-2SA mutant (lane 6) or the ATM-KD mutant (lane 3) significantly reduced the response.
Figure 2. ATM-mediated EZH2 phosphorylation prevents PRC2 formation and H3K27 methylation.
a) Protein extracts from mouse and frozen human cerebellar cortex were immunoprecipitated with the EZH2 and EZH1, and blotted with the [S/T]Q or EZH2 and EZH1 antibodies. Tubulin was used as a loading control. Full-length blots are presented in Supplementary Figure 12.
b) ) In vitro ATM kinase confirmed EZH2 as a novel ATM target. N2a cell extract (100 μg) was immunoprecipitated with the 2C1 anti-ATM antibody. Kinase reactions (upper panel) were performed in the presence of purified GST-tagged EZH2, GST-tagged EZH2S734A and GST-tagged EZH2S652&734A (2SA) (1 μg of each) as indicated. Coomassie blue staining (lower panel) was used to demonstrate total protein load. Full-length blots are presented in Supplementary Figure 12.
c) GFP-EZH2 and Flag-ATM were transfected into human A-T fibroblast cells. Lysates were immunoprecipitated with anti-GFP antibody and blotted with phosphor-[S/T]Q antibody plus anti-GFP. Flag antibody was used to test expression of Flag-ATM-WT and KD. Full-length blots are presented in Supplementary Figure 12.
d) The protein level of EZH2 increases dramatically in A-T cerebellum. Nuclear protein extracts from frozen cerebellar cortex were probed on western blots with EZH2 and EZH1 antibodies. Tubulin was loading control. Full-length blots are presented in Supplementary Figure 12.
e) The protein level of EZH2 increases dramatically in Atm−/− mouse cerebellum. Nuclear protein extracts from adult mouse cerebellar cortex were probed on western blots with EZH2 and EZH1 antibodies. Full-length blots are presented in Supplementary Figure 12.
f) PRC2 formation and H3K27 methylation is depended on ATM-mediated EZH2 phosphorylation. GFP-EZH2 WT and phosphorylation mutant (2SA) and phosphorylation mimic (2SD) were overexpressed in N2a cells. PRC2 complex were evaluated by examination of SUZ12, EED and H3K27me3 from EZH2 immunoprecipitates. Full-length blots are presented in Supplementary Figure 12.
g) ATM-dependent EZH2 phosphorylation is critical to EZH2 degradation. GFP-EZH2 WT and the non-phosphorylatable mutant (2SA) were overexpressed in N2a cells followed by treatment with CHX for different times. GFP-EZH1 was control. Full-length blots are presented in Supplementary Figure 12.
h) Quantification of three repetitions of the experiment illustrated in panel g). Data represent the mean ± SD.
ATM phosphorylation of EZH2 blocks its assembly into the PRC2 complex
In mature neurons, the levels of EZH2 are normally very low26, a finding that we verified in wild type mouse and human disease-free cerebella (Fig. 2d–e and Supplementary Fig. 2a). In A-T cerebellum and Atm−/− cerebellum by contrast, we found that levels of EZH2 were significantly higher (Fig. 2d–e). Elevated levels of EZH2 were also found in other Atm−/− brain regions such as neocortex and hippocampus (Fig. 2e, Supplementary Fig. 2b) suggesting that this effect may be a general one. The enhanced levels of EZH2 protein in ATM-deficient neurons were likely not a result of increased transcription; by reverse transcription PCR (RT-PCR), EZH2 mRNA levels were nearly equal in the two genotypes (Supplementary Fig. 2c–d).
As a core protein of the polycomb repressive complex 2 (PRC2), the function and enzymatic activity of EZH2 relies on its physical interaction with the other PRC2 proteins. When we overexpressed EZH2in N2a cells, we found that, compared to wild type EZH2, the association with EED and Suz12 was stronger with the non-phosphorylatable 2SA mutant and weaker with the 2SD phosphomimetic mutant (Fig. 2f). To assess whether the stability of EZH2 affects its ability to participate in PRC2 complex formation, we used the protein synthesis inhibitor cycloheximide to block new protein synthesis, then followed the degradation of the existing pools of protein. After overexpression in N2a cells, wild type EZH2 had a protein half-life of 17.3 hrs (Fig. 2g–h). The half-life of the phosphomimetic mutant (2SD) was comparable (not shown). By contrast, the non-phosphorylatable 2SA mutant proved remarkably stable (t½ ~ 2 days; Fig. 2g–h). In all cases, stability was directly correlated with increased EZH2 binding to H3K27me3 (Fig. 2f). Endogenous EZH2 protein in human A-T fibroblasts behaved similarly – more stable after cycloheximide treatment than EZH2 in control fibroblasts (Supplementary Fig. 3a–b). Next we tested whether inhibiting protease activity with ALLN blocked the degradation of non-phosphorylatable EZH2. When given 24 hours after transfection of GFP-EZH2, protease inhibition blocked the degradation of the 2SA EZH2 mutant (Supplementary Fig. 3c–d). These data suggest that PRC2 complex formation is enhanced in ATM deficiency and that this protects EZH2 from degradation.
H3K27me3 chromatin-binding correlates with down-regulated transcription
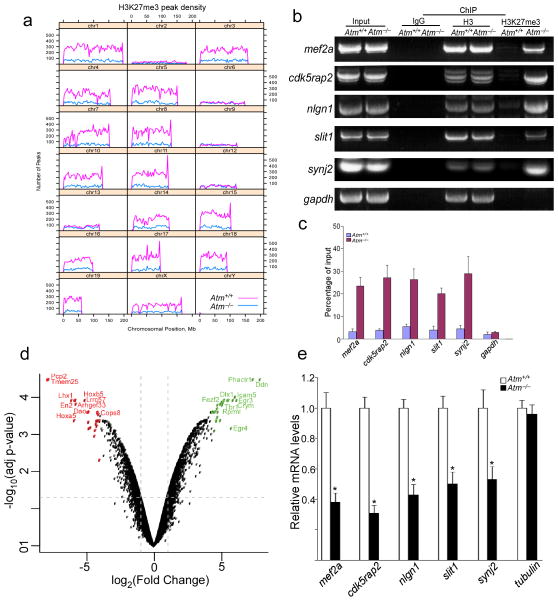
The substantially increased levels of H3K27me3 in ATM-deficient neurons led us to ask whether the evidence of altered chromatin remodeling was related to the elevated levels of EZH2. We thus performed H3K27me3 chromatin immunoprecipitation (ChIP) from Atm−/− cerebellar cortex, and found a substantial (4-fold) increase in the number of H3K27me3 binding sites in Atm−/− compared to wild type chromatin (Fig. 3a, Supplementary Fig. 4a). In addition, this increase was accompanied by a substantial shift in the pattern of binding; just 5% of the sites identified in wild type chromatin overlapped with the sites found in Atm−/−. Most of the Atm−/− binding sites (93%) were not represented in the wild type; and of the total wild type sites identified only 23% were found in Atm−/− as well. Despite these local shifts in discrete binding sites at the genomic level, the increased in H3K27me3 density was uniformly distributed over genome (Supplementary Fig. 4b). Analyzed by gene function, a large proportion of H3K27me3 targets are in categories that include neuronal function and cell cycle regulation (Table S1), but the peaks in the different genotypes did not show a striking difference in gene ontology (GO – Table S1). Among genes identified in this analysis were Slit1, Synj2, and Nlgn1 – genes with important function in synaptic development and maintenance. The H3K27me3 marking of each of these genes was confirmed by qPCR (Fig. 3b–c). We also found H3K27me3 enrichment in promoter areas of cell cycle regulatory genes cdkn2a and cdkn2b in Atm−/− samples compared to wild type (Supplementary Fig. 5a–b). All ChIP-qPCR assays displayed increased enrichment of H3K27me3 within the Atm−/− peak predicted by sequencing and peak finding, confirming our ChIPseq analysis.
Figure 3. Shifted H3K27me3 chromatin-binging pattern is related to downregulated neuronal genes in Atm−/− mouse cerebellum.
a) An illustration of H3K27me3 ChIP-seq peak distribution. The numbers of peaks per bin (1% of genome) are plotted by chromosomal position, comparing Atm+/+ (red) with Atm−/− (blue).
b) Semi-quantitative RT-PCRs from product of H3K27me3 ChIP show increased enrichment of H3K27me3 binding to promoter regions of mef2a, slit1, synj2, nlgn1 and cdk5rap2 in Atm−/− cerebellum. Full-length gel are presented in Supplementary Figure 12.
c) Following total H3 ChIP or H3K27me3 ChIP from wild-type and Atm−/− cerebellar cortex, quantitative real-time PCR analysis was performed for the presence of specific neuronal target genes (* = p < 0.01). gapdh was used as a control.
d) Microarray results are summarized in a volcano plot of log2 fold change (Atm+/+ - Atm−/−) versus log10 of the p value adjusted for multiple measurements. The top 25 downregulated (red) and upregulated (green) genes are colored, with the top 10 genes labeled. Dashed lines indicate 2-fold up or down (vertical) and p = 0.05 (horizontal).
e) Quantitative real-time polymerase chain reaction (PCR) was performed with specific primers for target genes. Total RNA was extracted from mouse cerebellar cortex. Data represent the mean ± SD of three independent experiments.
We next performed microarray analysis on RNA samples from wild type and Atm−/− cerebellar cortex. Topping the list of down-regulated genes were several associated with Purkinje and granule cells, such as Purkinje cell protein 2 (pcp2 – also known as L7) and gamma-amino butyric acid A receptor subunit alpha 6 (gabra6), engrailed-1 and -2 (en1, en2) (Fig. 3d). Moreover, several neuronal genes described above based on enrichment of H3K27me3 in their promoters’ regions – mef2a, cdk5rap2, slit1, synj2, and nlgn1 – are among the down-regulated genes identified by expression array (Table S2–3). The GO analysis of the down regulated genes also highlighted developmental processes (Table S4). To assess the direct effect of enrichment of H3K27me3 in the promoter regions of these genes, we examined mRNA expression in wild type and Atm−/− cerebella. Consistent with microarray data, we found a significant reduction of gene expression in Atm−/− samples (Fig. 3e). Although consistent with the ChIP data, we note that the loss could be due in part to the death of these cell types as well as to reduced gene expression.
EZH2 has a direct effect on postmitotic neurons
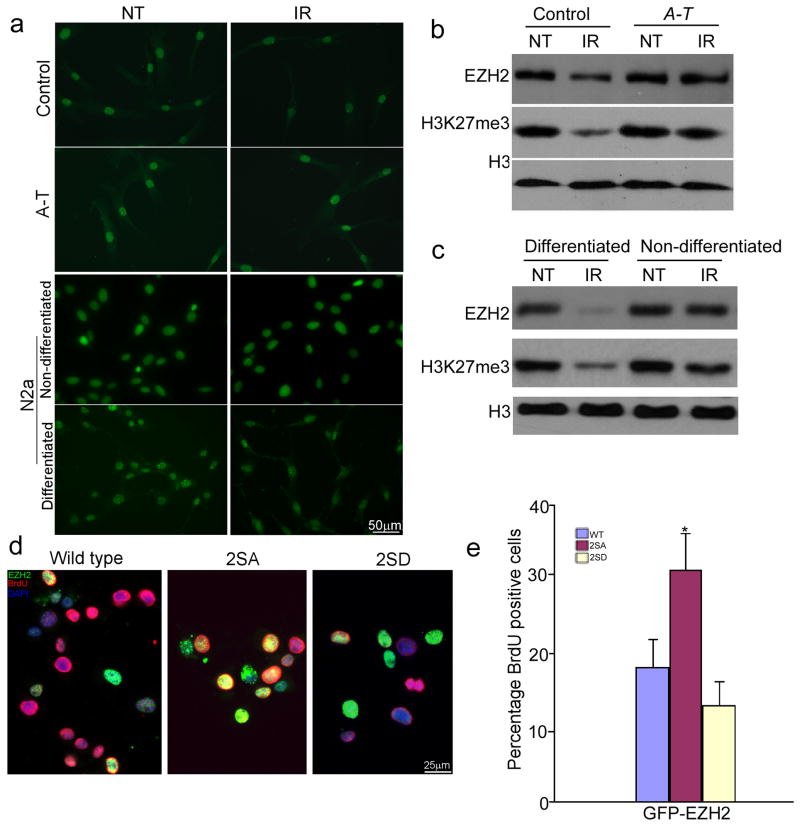
Our previous work on HDAC4 had led us to conclude that the ATM-dependent effects on histone acetylation were independent of DNA damage. By contrast, ATM-mediated EZH2 phosphorylation responds robustly to DNA damage (Fig. 2c). To pursue this difference we treated normal human fibroblasts or differentiated rodent N2a cells with ionizing radiation (IR). In control fibroblasts we saw a reduction of EZH2 and H3K27me3 (Fig. 4a–c), while in A-T fibroblast lines IR had virtually no effect. Log phase N2a cells responded in a manner similar to ATM-deficient fibroblasts (Fig. 4a and c), suggesting that the ATM effect was more pronounced in differentiated, non-mitotic cells.
Figure 4. ATM-dependent EZH2 phosphorylation affects neuronal cell cycle and DNA damage response.
a) Human control and A-T fibroblast and N2a cells (non-differentiated and differentiated) were immunostained with H3K27me3 with or without γ–radiation (10 Gy) treatment. Scale bar, 50 μm.
b–c) Protein extracts from control and A-T fibroblast (b) and N2a cells (c) with or without γ–radiation (IR) were probed on western blots with the antibodies indicated at left. Full-length blots are presented in Supplementary Figure 12.
d) ATM-dependent EZH2 phosphorylation affected BrdU incorporation. N2a cells were transfected with GFP-EZH2 WT (wild type), 2SA and 2SD at Day1. Differentiation media was added at Day3. BrdU (Red) was added to the medium to assess cell cycle activity at Day5. Scale bar = 25 μm.
e) Quantification of BrdU incorporation illustrated in panel d). The bars represent the percentage of BrdU/GFP double positive cells to total GFP positive cells. Each bar represents the average of three independent experiments; error bars denote SEM (* = p < 0.01).
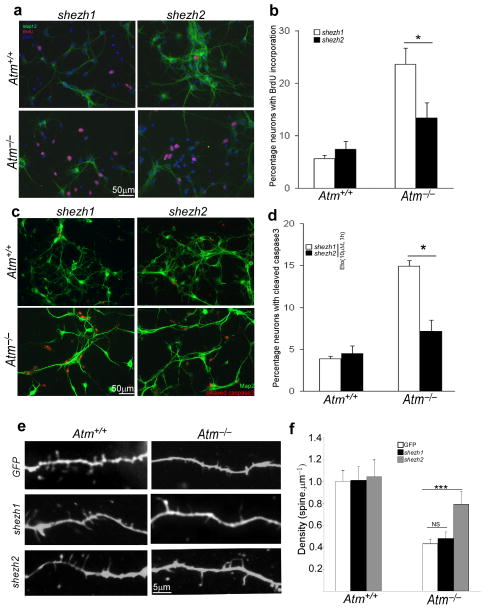
We next asked whether ATM-mediated EZH2 phosphorylation was involved in the ability of ATM to regulate the neuronal cell cycle. In differentiated N2a cells, 2SA-EZH2, but not wild type or 2SD-EZH2, significantly increased the level of BrdU incorporation (Fig. 4d–e). We found the same effect of an acute reduction of EZH2 in primary ATM-deficient neurons by using EZH2 shRNA (shezh2) lentiviral particles. The efficiency of viral infection was identified by examining EZH2 and H3K27me3 level (Supplementary Fig. 6a). As expected, knockdown of EZH2 significantly blocked BrdU incorporation (Fig. 5a–b). There was no effect from EZH1 shRNA (shezh1) infection. Knocking down EZH2 also prevented etoposide-induced activation of the cell death pathway in Atm−/− primary neurons (Fig. 5c–d), consistent with the defective DNA damage response found in A-T brain34.
Figure 5. Knockdown of EZH2 prevents cell death and cell cycle reentry in Atm−/− neurons.
a) E16.5 cortical neurons were infected at DIV3 with either control lentivirus (shezh1) or virus encoding shezh2. On DIV6, BrdU was added to the medium to assess cell cycle activity (red) in Map2-positve neurons (green). Scale bar, 50 μm.
b) Map2/BrdU double-labeled cells were counted in replicate cultures, their numbers are displayed as a percent of the total number of Map2-positive cells (* = p < 0.05).
c) E16.5 cortical neurons were infected at DIV3 with either control lentivirus (shezh1) or virus encoding shezh2. On DIV7, after treatment with etoposide for 3 hours, cells were fixed and stained with cleaved caspase-3 antibody. Scale bar, 50 μm.
d) Cell death was quantified by counting the number of activated caspase-3 immunostained cells and expressing these numbers as a percentage of the total Map2-positive cells (*, p < 0.05).
e) E16.5 cortical neurons were infected at DIV3 with either control lentivirus (shezh1) or virus encoding shezh2. On DIV14, cells were fixed and stained with V-GLUT and Map2 antibodies. Scale bar = 5 μM.
f) Quantification of mean spine density for cultured neurons; genotype and lentiviral infection as indicated (error bars = SEM). Sample numbers: nAtm+/+&GFP = 20, nAtm−/−&GFP = 21, nAtm+/+&shezh1 = 26, nAtm−/−&shezh1 = 23, nAtm+/+&shezh2 = 28, and nAtm−/−&shezh2=33. *** = p < 0.01, NS = p > 0.05 by Mann-Whitney test.
This reversal of the A-T cell cycle phenotype suggests that for neurons to keep their cell cycle in check they must maintain a constitutive ATM activity that keeps the levels of EZH2 low. We found that this suppression of EZH2-dependent histone methylation is also important for maintaining the proper structure of the neuronal dendrite. We measured spine density in cultured E16.5 neocortical neurons from wild type and Atm−/− brains. Mutant cortical neurons have a markedly reduced number of spines along a given length of dendrite (as previously reported for cerebellar Purkinje cells35). Knockdown of EZH2, however, partially rescued this effect and led to an increase of spine density in Atm−/− primary neurons (Fig. 5e–f). For wild type neurons, no dramatic changes occurred after shezh2 infection. The improved Atm−/− dendritic structure achieved by decreasing EZH2 was associated with changes in mRNA for specific neuronal genes. Using qPCR after shezh2 infection, we found significant increase of neuronal genes such slit1, synj2 and nlgn1 (Supplementary Fig. 6b and c). By contrast, the mRNA levels of cell cycle regulatory genes such as pcna and cyclin A were decreased (Supplementary Fig. 6b and d).
Knocking down Ezh2 prevents neurodegenerative process of Atm−/− mice
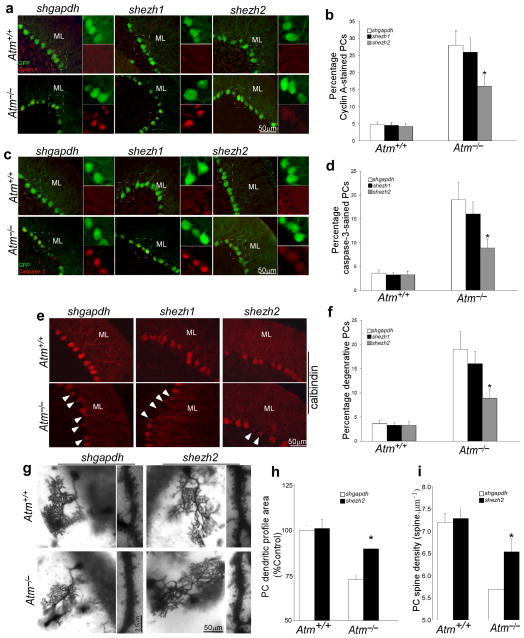
Previous studies have documented a degenerative process in AtmtmlBal mutant mouse cerebellum10,12,31. To examine the effect of ezh2 knockdown on this process, we surgically delivered shezh2-GFP expressing lentivirus into live adult mouse cerebellum. Three to four weeks after virus delivery, the efficiency of the EZH2 shRNAs was evaluated both by GFP expression (Supplementary Fig. 7a) and by the level of EZH2 immunoreactivity (Supplementary Fig. 7b). We found that the elevated levels of EZH2 in Atm−/− cerebellum were reduced after shezh2 infection; the same virus had little effect in wild type brains. Additionally, immunostaining for H3K27me3 revealed a dramatic reduction in cerebellar neurons of shezh2-injected Atm−/− mice (Supplementary Fig. 7b). We also monitored cell cycle re-entry and the initiation of cell death in these preparations. We found that in Atm−/− cerebellar GFP-positive neurons, reduction of EZH2 and H3K27me3 not only reduced the expression of cell cycle proteins such as cyclin A (Fig. 6a–b, red) and PCNA (Supplementary Fig. 8a), it also partially inhibited the activation of caspase-3 (Fig. 6c–d, red). The Purkinje cell marker calbindin also revealed a substantial attenuation of Atm−/− Purkinje cell atrophy after injection of shezh2 (Fig. 6e–f).
Figure 6. Knockdown of EZH2 prevents neurodegeneration of Atm−/− mice.
a) Representative images of cyclin A- and GFP-stained PCs show the effects of knockdown of ezh2. Scale bar, 50 μm.
b) Quantification of the cyclin A staining shown in panel a). Each bar represents the average of three independent experiments. Error bar = SEM.
c) Representative images of cleaved caspase-3- and GFP-stained PCs show the effects of knockdown of ezh2.
d) Quantification of the staining shown in panel c). Each bar represents the average of three independent experiments. Error bar = SEM.
e) Representative images of calbindin-stained PCs show the effects of knockdown of ezh2. The white arrows mark degenerating PCs with atrophy. Scale bar = 50 μm.
f) Quantification of the staining shown in panel e). Each bar represents the average of three independent experiments. Error bar = SEM.
g) Representative Golgi-stained images of the dendritic arbors and distal spine density of PCs show the effects of lentiviral delivery of shezh2 on degenerative progression in Atm−/− cerebellum.
h–i) Quantitative assessments of dendritic profiles and density of PCs shown in panel g). There were significant differences in PCs dendritic profile areas (ANOVA; F(2,14) = 3.6; p< 0.005) and spine density (ANOVA: F(2,14) = 18.7; p< 0.01) between wild type and Atm−/− mice. The dendritic profiles (F(2,14) = 6.7; p< 0.005) and spine density (F(2,14) = 13.6; p< 0.01) of Purkinje cells in Atm−/− mice with shezh2 infection was significantly increased. The counted PCs cell numbers: nAtm+/+&shgdph = 26, nAtm−/−&shgdpah = 29, nAtm+/+&shezh2 = 27, nAtm−/−&shezh2 = 31.
Each bar represents the average of three independent experiments. Error bar =SEM.
We next sought to determine if these in vivo improvements in the molecular profile of the Atm−/− cerebellar neurons was reflected in their cytological properties. While a previous study found no evidence of Purkinje cell loss in the AtmtmlAwb allele of Atm36, by applying the optical fractionator cell counting technique, we found a slight reduction in PCs counts in Atmtm1Bal homozygous mice (Table S5). This difference is too small, however, to serve as a reference for lentiviral shezh2 rescue. We therefore analyzed the structural properties of Purkinje cells in a 2mm diameter area surrounding the injection site of the shezh2-encoding lentivirus and found several important changes. First, by H&E staining we noted a reduction in the number of Purkinje cells with the large cytoplasmic vacuoles described previously31,10 (Supplementary Fig. 8b). In addition, when we stained mouse cerebellum using the rapid Golgi stain (Supplementary Fig. 9) we noted changes in both the dendrites and spine density of the Purkinje cells in the infected areas. Although there was no significant difference in Purkinje cell density between wild type and Atm−/− cerebella with or without shezh2 viral infection, we found that the cross sectional Purkinje cell dendritic profile area was reduced in untreated Atm−/− mice compared to wild type (Fig. 6g and h; Table S6). This shrinkage was reversed in the areas surrounding the injection and the difference between injected and uninjected brains was significant. Furthermore, knockdown of ezh2 significantly increased the spine density of Purkinje cells in Atm−/− mice in contrast to controls (Fig. 6g and i).
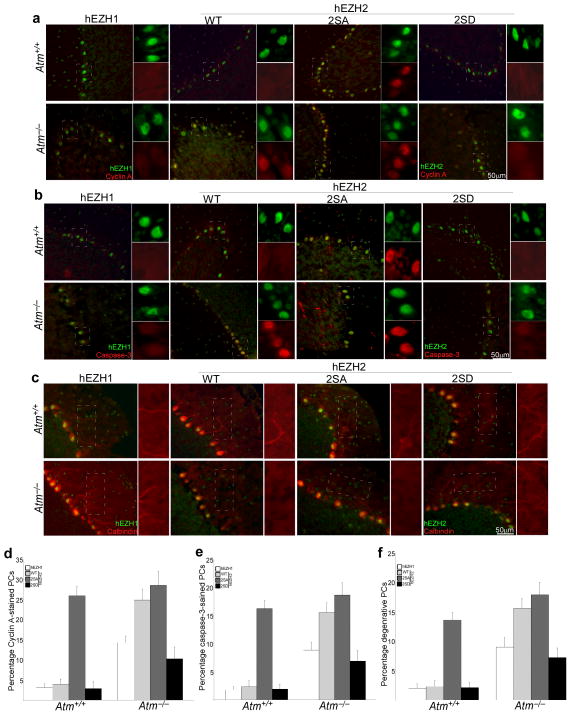
To verify the direct effect of loss of ATM-mediated EZH2 phosphorylation on the degenerative process of Purkinje cells in ATM-deficiency, we co-injected lentiviral particles encoding human wild type EZH2, a non-phosphorylatable mutant (2SA, which has the S652A, S734A mutations) or a phosphorylation mimic mutant (2SD, which has the S652D, S734D mutations) together with ezh2 shRNA (which is specific for the mouse Ezh2 message) into the cerebella of wild-type and Atm−/− adult mice. One week after injection of the two lentiviruses, gene transfer was monitored by immunohistochemistry using a human-specific EZH2 antibody (Fig. 7 and Supplementary Fig. S10). In infected Atm−/− cerebellar neurons, EZH2 phosphorylation mimic mutant (2SD) prevented cell cycle reentry (Fig. 7a and d) and caspase-3 activation (Fig. 7b and e) as well as Purkinje cell atrophy (Fig. 7c and f). By contrast, EZH2 non-phosphorylatable mutant (2SA) resulted in cell cycle reentry and caspase-3 activation as well as Purkinje cell atrophy in both Atm−/− and wild-type mice. As expected, wild type human EZH2 directly lead to cyclin A and caspase-3 re-appearance in the Atm−/− mutant but not in the wild type, however, overexpression of unmodified human EZH1 had little effect on either wild-type or Atm−/− neurons.
Figure 7. Effect of ATM-dependent EZH2 phosphorylation on the degeneration of Purkinje cell.
a–b) Representative images of Cyclin A- and cleaved caspase3-stained Purkinje cells show the effects of lentiviral delivery of different EZH2 mutants on degenerative progression in Atm−/− mouse cerebellum. Brain sections collected 1 week after injection were stained with Cyclin A and cleaved caspase3 (Red) and compared with ectopic EZH2 positive neurons (green). Non-phosphorylatable mutant = 2SA; phosphorylation mimic mutant = 2SD.
c) Representative images of calbindin-stained Purkinje cells show the effects of lentiviral delivery of different EZH2 mutants on Purkinje cell atrophy in Atm−/− mouse cerebellum. Brain sections collected 1 week after injection were stained with calbindin (Red) and compared with ectopic EZH2 positive neurons (green).
d–f) Quantification of the staining shown in panel a–c). Each bar represents the average of three independent experiments. Error bar = SEM.
Knocking down Ezh2 attenuates behavioral abnormalities of Atm−/− mice
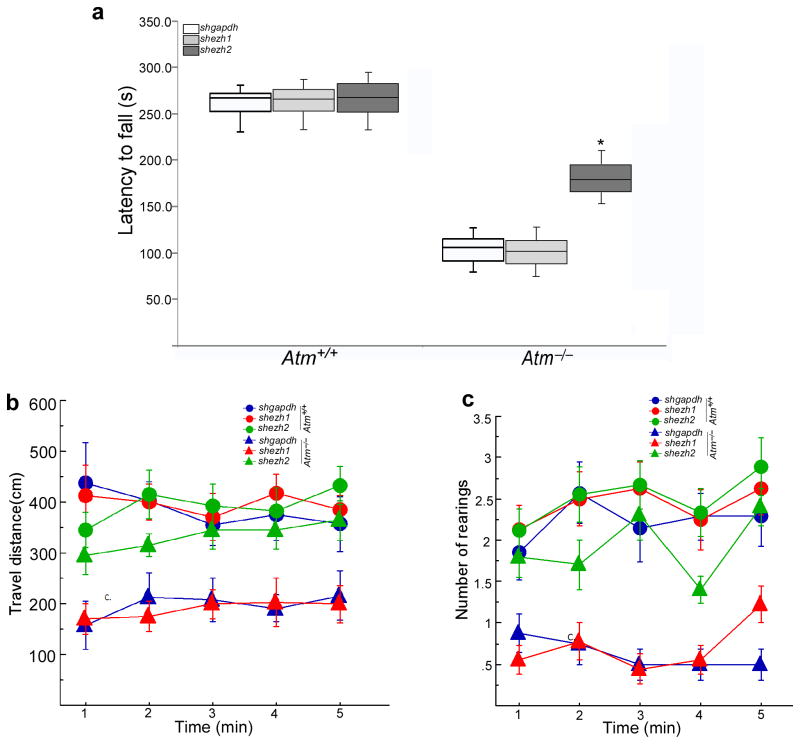
Although Atm mutant mice lack the profound ataxia found in human A-T, they do have reduced motor coordination compared to wild type mice12,37–39. Therefore, to test the role of enhanced EZH2 stability in this phenotype, we injected shezh2-encoding lentiviral particles into mouse cerebellum at postnatal day 18 (P18) – an age at which the areas reached by viral infection should be greater than in the adult. Three weeks after intracerebellar infusions, when transgene expression should be maximal, we subjected animals to rota-rod testing and measured locomotor activity in an open field. In the rota-rod test, the shezh2-injected Atm−/− mice showed a significant delay in falling (p < 0.05) compared to Atm−/− mutants infused with shezh1 or shgapdh (Fig. 8a). Furthermore, spontaneous open field activity (measured as distance travelled) was significantly greater in shezh2-injected Atm−/− mice, and approached wild type values (Fig. 8b). A secondary parameter of ataxic behavior, rearing, was also different dependent on genotype and treatment. Wild type mice reared more often (p < 0.05), compared with mutants receiving virus encoding shezh1 or shgapdh. However, shezh2-treated Atm−/− mice showed a reversal of this deficiency (Fig. 8c).
Figure 8. Knockdown of EZH2 reverses neurological behavioral abnormalities of Atm−/− mice.
a) Rota-rod testing demonstrated thatshezh2-injected Atm−/− mice had longer latency-to-fall times. Time measured to remain on the rota-rod was analyzed by a two-way ANOVA, and found to reveal a significant genotype x treatment interaction (F(2,45) = 23.1, p< 0.0001). A marked main effect of genotype was also observed (F(1,45) = 656.4, p< 0.0001), reflecting the ataxia in Atm−/− mice.
b–c) Locomotor activity and rearing in the open field were measured across one-minute bins for a total of five consecutive minutes. (b) For distance travelled, a two-way ANOVA with repeated measures revealed significant effects of genotype (F(1,45) = 37.8, p< 0.0001), treatment (F(2,45) = 3.9, p< 0.027) and a genotype x treatment interaction (F(2,45) = 3.7, p < 0.05). The latter was supported by a clear separation of values in Atm−/− mice between those given shezh2 and those administered shezh1 and shgapdh. (c) For rearing, significant main effects (Group: F(1,45) = 172.4, p< 0.0001; Treatment: F(2,45) = 25.0, p< 0.0001) and an interaction (F(2,45) = 13.8, p < 0.0001) were similarly observed, once again contributed to by the deviation to normal wild type numbers by shezh2-infused Atm−/− mice.
Discussion
We have previously shown that nuclear accumulation of HDAC4 plays a critical role in A-T neurodegeneration12. In the current work we have uncovered a role for EZH2-mediated histone methylation in the A-T neurodegenerative process. In A-T and Atm−/− mouse brain, histone H3K27 trimethylation showed a significant increase. Further, in the absence of ATM the histone methyltransferase, EZH2, is stabilized, is more readily incorporated into the PRC2 complex and displays enhanced binding to H3K27. The elevation in H3K27me3 was quantified by a genome-wide analysis that represents the first unbiased, high-resolution mapping of H3K27me3 occupancy in the ATM-deficient brain. The increase in chromatin sites where H3K27me3 associates in Atm−/− brain is correlated with a shift in the expression of a variety of cell cycle and neurotrophic genes. The result is an EZH2-induced increase in neuronal process degeneration, cell cycle reentry, suppression of neuronal genes, and induction of the apoptotic process. The physiological relevance of these changes in the epigenome is underlined by our finding that knockdown of EZH2 can partially rescue the behavioral disabilities found in Atm−/− mice, a finding with immediate therapeutic interest. In the aggregate, therefore, our data define a pathological role for elevated EZH2-mediated H3K27me3 in the etiology of A-T.
EZH2 was discovered based on its functions during embryogenesis, yet the current work highlights its importance to signaling in the adult brain. Other kinases have also been shown to modulate adult EZH2 activity including AKT, CDK1, CDK2 and p38 kinase15,25,40,41. Adding ATM to this list further integrates the pattern of histone methylation into the signaling activity of a cell by providing evidence that the levels and location of H3K27me3 can respond to DNA damage and oxidative stress. This can be seen directly in the alteration of EZH2 levels observed after ionizing radiation or etoposide treatment. We note that while Cdk1-mediated EZH2 phosphorylation increases H3K27 methylation15,30, AKT-mediated EZH2 phosphorylation on serine 21 prevents it41. This suggests that EZH2 regulation in response to a change in cellular state is complex and bi-directional.
ATM deficiency leads to enhanced histone methylation, which is generally associated with a more compact form of chromatin. The observation that DNA damage, which activates ATM, leads to a substantial reduction in histone methylation suggests that activated ATM, by accelerating the degradation of EZH2, would be expected to reduce chromatin condensation. This would lead to a more open chromatin structure that could aid in the access of the various DNA repair proteins to the injured double helix and suggests a new and previously unrecognized way in which ATM helps in the DNA repair process. We note that this concept is fully consistent with work showing slower rates of DNA repair in heterochromatin compared with euchromatin42. Previous studies indicate that EZH2 plays a role in determining cancer cell response during the DNA damage response43; two additional studies demonstrate that EZH2 contributes to cancer progression44,45. Thus our data suggest that ATM-mediated degradation of EZH2 during the DNA damage response is likely to serve a protective function in brain.
Note that as ATM activity increases not only would histone methylation decrease, but because HDAC4 would be less likely to move to the nucleus, the levels of histone deacetylation should decrease12. These two changes would act in consort with one another; both changes in the histone code would favor a more open chromatin configuration and thus more efficient DNA repair. Additional insight into the consequences of the ATM-dependent changes in the histone code can be achieved by comparing the H3K27me3 sites defined in this study with the nuclear HDAC4 chromatin binding we reported previously12 (Supplementary Fig. 11). The locations of the two ‘marks’ overlap at only ~2% of the sites in the wild-type. In ATM-deficiency, however, the overlap increases to more than 5%. The dual epigenetic marks would be predicted to induce gene silencing in the Atm mutant. The H3K27me3-HDAC4 intersection peaks also appear to be particularly enriched in areas likely to contain cis-acting regulatory sequences. This can be seen most easily by comparing the distribution of these areas with those defined by randomly selected peak sets. Therefore, the H3K27me3-HDAC4 intersections are more likely to select marks within a reasonable cis-acting distance of a gene and are therefore more likely to regulate adjacent gene expression. It should be noted that while it may be important in determining the A-T phenotype, this overlapping subset of marks comprises only a small fraction of the total, suggesting that the two systems – methylation and acetylation – have largely independent functions.
In conclusion, our study presents evidence to suggest an expanded epigenetic regulatory model for the neurodegenerative process in A-T. ATM-deficiency leads to abnormal alteration in EZH2-dependent polycomb epigenetic system. We propose that changes in the status of cell cycle and neuronal survival gene expression in A-T brain is not only dependent on the consequences of HDAC4 nuclear accumulation; it also relies on PRC2 complex-mediated H3K27me3 changes. Identification of a role for ATM in the polycomb silencing system expands our understanding of the dynamic changes underlying histone modifications in the A-T neurodegenerative processes and suggests novel therapeutic targets for treatment.
Materials and Methods
Antibodies and chemical regents
Antibodies against EZH1, EZH2, EED and SUZ12, Aldolase C, calbindin, H3K27me3, alpha Tublin, BrdU and cleaved caspase3 were obtained from Abcam. Antibodies against EZH2, H3K27me3 and H3K9me3, total histone 3 and phospho-S/T[Q] were obtained from Cell Signaling. Antibodies against cyclin A and cyclin D1 and β–actin were from Santa Cruz. Antibodies specific against human EZH1 and EZH2 were obtained from Abcam. Secondary antibodies used for immunocytochemistry were as follows: Alexa 488-labeled chicken anti-mouse or anti-rabbit; Alexa 594-labeled donkey anti-mouse or anti-rabbit (Invitrogen, Eugene, OR); all were used at a dilution of 1:500. DAPI (4′,6′-diamidino-2-phenylindol) was used as a nuclear counter stain at 1 μg/ml.
Constructs and Plasmids
myc-EZH2 and GST-EZH2 were provided by Dr. Mien-chie Hung (The University of Texas M.D. Anderson Cancer Center, Houston, TX) and Haojie Huang (Stony Brook University Medical Center, Stony Brook, NY). Site-direct mutation of GFP-EZH2 on S652&734/A andS652&734/D and GST-EZH2 on S734A and S652&734/A were performed by Quik Change Mutagenesis Kit (Stratagene, La Jolla, CA).
Human subjects
Human autopsy tissue from four individuals diagnosed with A-T and four age-matched controls was obtained from UCLA Medical Center and Children’s Hospital Los Angeles. Brain tissue was routinely fixed, paraffin embedded, and sectioned at 10 μm. All A-T cases were confirmed both clinically and by post-mortem examination. The average age of the subjects was 21 years for the A-T patients and 25 years for non-A-T controls. The non-A-T controls were free of any known CNS neurological diseases at the time of death. Human frozen tissue was secured from 4 additional individuals diagnosed with A-T and four age-matched controls; samples were obtained from NICHD Brain and Tissue Bank of Developmental Disorders at the University of Maryland, Baltimore, MD.
Animals
AtmtmBal/+ mice were a gift from Dr. Yang Xu. Timed pregnancies were established from these matings; the date of appearance of a vaginal plug was considered embryonic day 0.5. Embryos were taken at embryonic day 16.5 (E16.5) for either cortical cultures or histology. Animals were allowed to survive for 7 days before sacrifice. All animal procedures were carried out in accordance with Rutgers University IACUC standards. The animal facilities at Rutgers University are fully AAALAC accredited.
Human fibroblast and N2a cell culture
All human control and A-T primary fibroblast cell lines described in this paper were purchased from the Coriell Cell Repository. Fibroblasts were cultured in fibroblast medium (Dulbecco’s modified Eagle’s medium [DMEM] supplemented with 15% fetal bovine serum [FBS; Hyclone], 1 mM glutamine [Invitrogen], 1% nonessential amino acids [Invitrogen], and penicillin/streptomycin [Invitrogen]). N2a cells were cultured in DMEM medium supplemented with 10% FBS. To differentiate N2a cells, 24 hours after seeding, culture medium was replaced by DMEM with 1 mM dbcAMP and 0.25% FBS. Cells were kept in this differentiation medium for 3–7 days before use.
Primary Neuronal Cultures
Embryonic cortical neurons were isolated by standard procedures. For ATM-deficient cultures, all embryos from an AtmtmBal/+ x Atmtm1Bal/+ mating were harvested and treated separately then retrospectively genotyped by PCR. Isolated E16.5 embryonic cerebral cortices were treated with 0.25% Trypsin-EDTA and dissociated into single cells by gentle trituration. Cells were suspended in Neurobasal medium supplemented with B27 and 2 mM glutamine, then plated on coverslips or dishes coated with poly-L-Lysine (5 mg/mL). All cultures were grown for a minimum of 5 days in vitro (DIV) before any treatment. All experiments were performed on a minimum of three cultures from three separate embryos; each condition was examined in triplicate. All transfections were carried out using Lipofectamine 2000 after 4–5 DIV. Live imaging of GFP was performed on a heated stage in a controlled 5% CO2 atmosphere at 7–10 DIV.
Immunohistochemistry
For DAB/bright field staining, all paraffin-embedded human sections were deparaffinized inxylene and then rehydrated through graded ethanol to water. The sections were pretreated in 0.3% hydrogen peroxide in methanol for 30 min to remove endogenous peroxidase activity, rinsedin Tris-buffered saline (TBS), and then treated with 0.1 M citrate buffer in a microwave at sufficient power to keep the solution at 100°C for 20 min. Sections were cooled in the same buffer at room temperature (RT) for 30 min and rinsed in TBS. Slides were incubated in 10% goat serum in PBS blocking solution for1 h at RT, after which primary antibody was applied to the sections that were then incubated at 4°C overnight. The sections were washed three times in TBS before applying the secondary antibody (Vector Laboratories). Secondary antibody was applied for 1 h at RT. Afterwards, sections were rinsed three times in TBS. Rinsed sections were then incubated in Vectastain ABC Elitereagent for 1 h and developed using diaminobenzidine, according to the protocol of the manufacturer (Vector Laboratories). The sections were counterstained with hematoxylin, and after dehydration all sections were mounted in Permount under a glass coverslip. Control sections were subjected to the identical staining procedure, except for the omission of the primary antibody.
Immunofluorescence
All paraffin-embedded human sections were deparaffinized inxylene and then rehydrated through graded ethanol to water and PBS. For the mouse, cryostat sections were first rinsed in PBS, Subsequent steps were identical for paraffin and cryostat material. After rehydration, sections were treated in antigen unmasking solution (low pH) for 30 min at 100°C. After the slides had cooled in buffer for 30 min at room temperature, slides were rinsed in PBS. Then sections were incubated in 10% goatserum in PBS to block nonspecific binding for 1 h at room temperature. All primary antibodies were diluted in PBS containing 0.5% Triton X-100 and 5% goat serum and incubated with sections overnight at 4°C. After rinsing in PBS, they were incubated for 1 h with secondary antibody, which was conjugated with various fluorescent labeled secondary antibodies. All sections were mounted in ProLong® Gold antifade reagent with DAPI (Invitrogen) under a glass coverslip. All experiments were conducted in triplicates.
ChIP-quantitative real-time PCR
After immunoprecipitation, recovered chromatin fragments were analyzed by real-time PCR using primer pairs specific for 150–250-base-pair (bp) segments corresponding to mouse gene promoter regions (regions upstream of the start codon, near the first exon). Real-time PCR was performed using EXPRESS SYBR® GreenER™ qPCRSupermix Universal (Invitrogen) on ABI Prism® 7900HT Sequence Detection System (Applied Biosystems). The relative quantities of immunoprecipitated DNA fragments were calculated by comparison with a standard curve generated by serial dilutions of input DNA. Data were derived from three independent amplifications. All PCR primer sequences can be found in Table S7.
RNA extraction and reverse transcription PCR
RNA was prepared using PureLink micro-to-midi total RNA purification system (Invitrogen). Semi-quantitative RT-PCR was performed with Superscript III one-step RT-PCR system with platinum Taq High Fidelity (Invitrogen). For quantitative Real-time PCR, cDNA was prepared by using oligonucleotide (dT), random primers, and a Thermo Reverse Transcription kit (Signal way Biotechnology). All qPCRs were performed using EXPRESS SYBR® GreenER™ qPCRSupermix Universal (Invitrogen) on ABI Prism® 7900HT Sequence Detection System (Applied Biosystems). The mRNA level of tubulin or 36B4 was used as an internal control. All RT-PCR primers are shown on Table S8.
Western blotting and immunoprecipitation
For Western blots, protein extracts from mouse tissue or primary cultures were separated with SDS–PAGE. For immunoprecipitation, protein extracts were subsequently incubated for overnight at 4°C with the primary antibodies and Protein G Dynabeads (Invitrogen). The precipitates were washed twice with a lysis buffer and then were resolved on SDS-PAGE gel.
In vitro kinase assay
N2A cells overexpressing wild type Flag-ATM were lysed in TNN buffer and immunoprecipitated with protein G Dynabeads beads (20 μl) and anti-Flag antibody (Sigma). Kinase reactions were performed using precipitated ATM on beads (~ 15 μl) with either GST-tagged EZH2, the GST-tagged EZH2S734A and GST-tagged EZH2S652&734A purified from bacteria (~ 1 μg protein) in a kinase buffer (50 mM Hepes at pH 7.4, 150 mM NaCl, 6 mM MgCl2, 4 mM MnCl2, 10% glycerol, 1 mM dithiothreitol, 0.1 mM NaOV and 15 μ Ci of [γ-32P] ATP] and incubated at 30 °C for 30 min. Proteins were separated by SDS–PAGE, transferred to nitrocellulose membranes and visualized by autoradiography.
BrdU incorporation
For BrdU labeling, 10 μm BrdU was added to the Neurobasal media at DIV6. After labeling for 24 hours, the primary neurons were then fixed, and DNA was hydrolyzed by exposing the cells to 2N HCl for 10 minutes. Specimens were then neutralizing in 0.1 M sodium borate (pH 8.6) for 10 minutes and then rinsed extensively in PBS (3X) for 45 min before treatment with blocking reagent. Non-specific antibody binding was blocked by exposing the fixed cells to 5% normal goat serum in 0.1% Triton-X for 1 hour before application of the BrdU primary antibody. Total cells number was based on DAPI-positive nuclei. Counts were made of BrdU-positive, DAPI-positive and double-positive cells.
Chromatin immunoprecipitation (ChIP)
ChIP was performed with mouse cerebellar cortex fixed with 4% paraformaldehyde (PFA) solution and stored at −80 °C before use. Brains were cut into small (~1 mm3) pieces and chemically cross-linked by 1% PFA in 1xDPBS solution for 15 min at room temperature, then homogenized, resuspended, lysed in TNN lysis buffers with RNAse, followed by digestion of chromatin DNA with micrococal nuclease (NEB). TNN lysates were then incubated with nuclease 10–15 min at 37°C (1 μl, 2000U) plus 1 ml lysate plus 10 μl 0.5 M CaCl2). The reaction was stopped with 100 μl 0.5 M EDTA on ice, then sonicated to shear crosslinked DNA. We used a Branson S450 digital Sonifier at 20% power and delivered ten 30-s pulses (60-s pause between pulses) during which all samples were immersed in an ice bath. The resulting whole-cell extract was incubated overnight at 4 °C with 30 μl of Dynal Protein G magnetic beads that had been preincubated with 5 μg of the appropriate antibody. Beads were washed five times with TNN lysis buffer and once with 1xChIP buffer. Bound complexes were eluted from the beads by heating to 65 °C. Whole-cell extract DNA (reserved from the sonication step) was treated to reverse crosslinks. Immunoprecipitated DNA and whole-cell extract DNA were then purified by treatment with 50 mM NaCl and proteinase K for 2 hours at 65 °C with rotation. Purified DNA samples were normalized and subjected to PCR analysis. Antibodies used for pull downs were: anti-H3K27me3 and anti-histone3, mouse and rabbit IgG were from Cell Signaling. After immunoprecipitation, recovered chromatin fragments were subjected to semi-quantitative PCR or real-time PCR for 32–40 cycles, using primer pairs specific for 150–500 bp segments as described in the text.
ChIP sequencing analysis
ChIP sequencing libraries were constructed from anti-H3k27me3-immunoprecipitated chromatin using the SOLiD™ DNA fragment library kit (Invitrogen) following the standard ChIP-Seq protocol. Libraries containing Atm+/+ or Atm−/− samples were bar-coded, applied to beads with emulsion PCR, enriched, and sequenced using the SOLiD™ System v4 at the Centre for Applied Genomics at SickKids Hospital, Toronto. A sample prepared without immunoprecipitation from Atm+/+ was used as the input control. Sequenced 50-nt reads and quality strings were aligned to the mouse genome (mm9) using Bowtie 0.12.546 to identify the single, best-quality match location. Results were converted to BAM format using Samtools. Peaks were selected, comparing Atm+/+ or Atm−/− against the input control, using Find Peaks 4.047. Resulting bed-formatted files were imported into the ChIPpeakAnno package48 in R/BioConductor for analysis. Peak tracks were created and visualized using the UCSC Genome Browser where subsets of peaks were chosen for qPCR validation.
Microarray Analysis
RNA was hybridized with the Illumina WG-6 arrays at the Sanford Burnham microarray core facility. Samples from three independent animals were run for each genotype (Atm+/+ and Atm−/−). Data were extracted using GenomeStudio and imported into R/BioConductor using the lumi package and then fit to a linear model using the limma package.
shRNA Knockdown in primary neurons
The hairpin sequences for MISSION® shRNA Lentiviral Transduction Particles including shezh1, shezh2, shgapdh (Table S9) were from Sigma. After 3 days in culture, primary neurons were infected using a multiplicity of infection (moi) between 5 and 10 to provide an efficiency of infection above 70%. Samples were collected 3–5 days later and different assays were performed.
Lentivirus production and cerebellar injections
Lentiviral preparation and cerebellar microinjection were conducted as previous described12. Briefly, premade iLenti™ constructs including a set of ezh1 siRNA-GFP and a set of ezh2 siRNA-GFP were from Applied Biological Material Inc. (Table S10). Human EZH2 and EZH1 lentiviral constructs were from GeneCopoeia. Site-direct mutation of lentiviral EZH2 to 2SA (S652A/S734A) and 2SD (S652D/S734D) were performed by QuikChange Mutagenesis Kit (Stratagene, La Jolla, CA). The high lentiviral particle titers were prepared with Lenti-Pac™ Expression Packaging Kits (GeneCopoeia) in HEK 293T cells and purified by Ultra-Pure Lentivirus purification Kits (Applied Biological Material Inc. Vancouver). Stereotaxic intra-cerebellar infusions were delivered to wild type and Atm−/− mice (8 weeks of age) under isoflurane anesthesia. Mice were positioned in a Kopf stereotaxic apparatus and burr holes drilled into the skull. A 5 μl Hamilton syringe fitted with a 33-gauge needle was lowered into the cerebellum for vector delivery. The coordinates of the burr hole and ventral location for infusion were: −7.2 mm from bregma, 1.0 mm lateral from the midline, and 3 mm ventral. For each cerebellum, slow infusion over 15 minutes of 3–5 μl of lentiviral particles (1–5×109 TU/ml), was performed; the needle was withdrawn 5 min after completing infusion. For knocking down endogenous EZH2 and overexpressing human EZH2, we delivered shezh2: human EZH1,2 lentiviruses as 1:1 ratio in 2-month old mice. After surgery, animals were injected subcutaneously with 0.3 ml pre-warmed saline to avoid dehydration, and were allowed 7 days recovery prior to tissue collection. For behavioral experiments, lentiviruses were injected into mouse cerebellum at postnatal day 18 (P18), and 3 weeks later, trained for tests of motor behavior.
Rota-rod test
The rota-rod consisted of a felt-covered steel cylinder fitted to a rotating pump with variable speed settings. Age-matched animals were initially trained for two days to walk on the rota-rod, with each day consisting of two 5-minute training trials (inter-trial interval: 2 hr), with the rota-rod speed adjusted to accelerate from 6 rpm to 40 rpm over a 5 min period each day. After 1 week, mice were returned for testing using the rota-rod adjusted to 10 and 16 rpm over a 5 min test period. Latency to fall off was measured in seconds using a stopwatch by an observer blind to experimental treatment.
Open-Field Test
The open-field test was conducted as previously described12,49. Briefly, a four-sided chamber was used as the open field (63 × 57 × 28 cm). The floor of the arena was divided into a grid of 30 squares. After placing a mouse at the center of the arena, locomotor activity was measured over a 5-minute period using an overhead camera connected to a videotape recorder. Mouse movement behavior was digitally tracked using video-tracking software (Spontaneous Motor Activity Recording and Tracking (SMART), San Diego instruments, San Diego, CA) by running the videotape recording through a Dell computer containing a frame grabber. In addition to obtaining a measure of distance travelled (measured in cm), an observer blind to experimental treatment scored animals for rearing behavior. A rear was defined as an animal raising both front paws off the floor and extending the head and torso upward. Both rearing numbers and distance travelled were divided into one-minute bins across the 5-minute test. This was conducted to determine whether changes in behavior between the various groups were time dependent relative to initial exposure to the apparatus.
Stereological analysis of Purkinje cells
Golgi neurohistological staining was performed following a modification of classical Golgi procedure described previously50. Briefly, mice were killed by cervical dislocation, and the freshly dissected brain was processed with FD Rapid GolgiStain kit™ by FD NeuroTechnologies, Inc. (Ellicott City, MD), and counterstained with cresyl violet. To examine morphological alteration of Purkinje neurons, six randomly selected cerebellar sagittal cryosections (60 μm) through the entire cerebellum were observed. The numbers of Purkinje cells per cerebellum were estimated with the optical fractionator technique on a Leica microscope using Stereologer software (IPLab Scanalytics Software). Purkinje cells were counted and analyzed according to the parameters of the optical dissector described as below. The optical fractionator probe randomly selected locations within each contour to place a 3-dimensional (250×250×10 μm) counting box within the defined tissue. Statistical comparisons between different groups were made using a one-factor analysis of variance (ANOVA). Post-hoc comparisons were made using the Bonferroni-Dunn test.
Statistical analyses
All non-behavioral data are presented as the means ± SEM of a minimum of three replicates. With the exception of the behavioral data, we evaluated statistical differences by Student’s t test. Behavioral data were analyzed by two-way ANOVAs, with repeated measures included for the open field data. Post hoc testing used Neuman-Keuls or Bonferroni t-tests where appropriate. Effects were considered significant at p < 0.05. No statistical methods were used to predetermine sample size, but sample sizes were similar to those described in previous related studies12. No randomization of samples was performed. Data distribution was assumed to be normal but this was not formally tested. No blinding was done. The number of samples or cells examined in each analysis is shown in the legends.
Supplementary Material
Acknowledgments
We thank Dr. Yang Xu (UCSD, San Diego) for providing AtmtmBal mutant strain. We thank Dr. Richard Gatti (UCLA, Los Angles) for sharing human A-T paraffin-fixed samples. Human frozen tissue was obtained from NICHD Brain and Tissue Bank of Developmental Disorders at the University of Maryland, Baltimore, MD. Special thanks to Dr. Rishen Ma (Mount Sinai School of Medicine) for the help in stereological analysis of Purkinje cells. We thank Drs. Michael B. Kastan (Duke Univeristy), Haojie Huang (SUNY at Stony Brook) and Mien-chie Hung (The University of Texas M.D. Anderson Cancer Center, Houston) for providing the Flag-ATM, myc-EZH2 and GST-EZH2 plasmids. The long-term support of the A-T Children’s Project is gratefully acknowledged as is the support of Rutgers University. This work was also funded by grants from the NIH: NS20591 and NS71022 to K.H.; RC1 CA147187 to R.P.H. and MH60706 and NIEHS P30 ES05022 to A.K.
Footnotes
Contributions
J.L. and K.H. designed the experiments, analyzed data and wrote the manuscript. J.L. carried out biochemistry experiments and ChIP-qPCR and semi-quantitative PCR. M.S and R.P.H. developed, carried out and analyzed data for ChIP-seq analyses. J.L., E.M. and A.K. carried out the mouse cerebellar lentiviral injections and behavioral tests.
Accession codes
The resulting H3K27me3 ChIP sequence files and quality scores are available from the NIH SRA, accession number SRA051384. Microarray data are available at NIH GEO accession GSE50951.
References
- 1.Sedgwick RP, Boder E. Progressive ataxia in childhood with particular reference to ataxia-telangiectasia. Neurology. 1960;10:705–715. doi: 10.1212/wnl.10.7.705. [DOI] [PubMed] [Google Scholar]
- 2.Boder E. Ataxia-telangiectasia: some historic, clinical and pathologic observations. Birth Defects Orig Artic Ser. 1975;11:255–270. [PubMed] [Google Scholar]
- 3.Lavin MF, Shiloh Y. Ataxia-telangiectasia: a multifaceted genetic disorder associated with defective signal transduction. Curr Opin Immunol. 1996;8:459–464. doi: 10.1016/s0952-7915(96)80030-6. [DOI] [PubMed] [Google Scholar]
- 4.Biton S, Barzilai A, Shiloh Y. The neurological phenotype of ataxia-telangiectasia: solving a persistent puzzle. DNA Repair (Amst) 2008;7:1028–1038. doi: 10.1016/j.dnarep.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 5.Savitsky K, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 6.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 7.Dar I, Biton S, Shiloh Y, Barzilai A. Analysis of the ataxia telangiectasia mutated-mediated DNA damage response in murine cerebellar neurons. J Neurosci. 2006;26:7767–7774. doi: 10.1523/JNEUROSCI.2055-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Y, Herrup K. Loss of neuronal cell cycle control in ataxia-telangiectasia: a unified disease mechanism. J Neurosci. 2005;25:2522–2529. doi: 10.1523/JNEUROSCI.4946-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rimkus SA, et al. Mutations in String/CDC25 inhibit cell cycle re-entry and neurodegeneration in a Drosophila model of Ataxia telangiectasia. Genes Dev. 2008;22:1205–1220. doi: 10.1101/gad.1639608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J, Chen J, Vinters HV, Gatti RA, Herrup K. Stable brain ATM message and residual kinase-active ATM protein in ataxia-telangiectasia. J Neurosci. 31:7568–7577. doi: 10.1523/JNEUROSCI.0778-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, Han YR, Plummer MR, Herrup K. Cytoplasmic ATM in neurons modulates synaptic function. Curr Biol. 2009;19:2091–2096. doi: 10.1016/j.cub.2009.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J, et al. Nuclear accumulation of HDAC4 in ATM deficiency promotes neurodegeneration in ataxia telangiectasia. Nat Med. 2012;18:783–790. doi: 10.1038/nm.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev. 2004;14:155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 14.Juan AH, et al. Polycomb EZH2 controls self-renewal and safeguards the transcriptional identity of skeletal muscle stem cells. Genes Dev. 2011;25:789–794. doi: 10.1101/gad.2027911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei Y, et al. CDK1-dependent phosphorylation of EZH2 suppresses methylation of H3K27 and promotes osteogenic differentiation of human mesenchymal stem cells. Nat Cell Biol. 2011;13:87–94. doi: 10.1038/ncb2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Francis NJ, Follmer NE, Simon MD, Aghia G, Butler JD. Polycomb proteins remain bound to chromatin and DNA during DNA replication in vitro. Cell. 2009;137:110–122. doi: 10.1016/j.cell.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Surface LE, Thornton SR, Boyer LA. Polycomb group proteins set the stage for early lineage commitment. Cell Stem Cell. 7:288–298. doi: 10.1016/j.stem.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 18.Hirabayashi Y, et al. Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron. 2009;63:600–613. doi: 10.1016/j.neuron.2009.08.021. [DOI] [PubMed] [Google Scholar]
- 19.Sauvageau M, Sauvageau G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 7:299–313. doi: 10.1016/j.stem.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 22.Bracken AP, et al. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su IH, et al. Polycomb group protein ezh2 controls actin polymerization and cell signaling. Cell. 2005;121:425–436. doi: 10.1016/j.cell.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 24.Varambally S, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–1699. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeng X, Chen S, Huang H. Phosphorylation of EZH2 by CDK1 and CDK2: a possible regulatory mechanism of transmission of the H3K27me3 epigenetic mark through cell divisions. Cell Cycle. 2011;10:579–583. doi: 10.4161/cc.10.4.14722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pereira JD, et al. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc Natl Acad Sci U S A. 2010;107:15957–15962. doi: 10.1073/pnas.1002530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohn F, et al. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 28.Chouliaras L, et al. Epigenetic regulation in the pathophysiology of Alzheimer’s disease. Prog Neurobiol. 2010;90:498–510. doi: 10.1016/j.pneurobio.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 29.Fagiolini M, Jensen CL, Champagne FA. Epigenetic influences on brain development and plasticity. Curr Opin Neurobiol. 2009;19:207–212. doi: 10.1016/j.conb.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen S, et al. Cyclin-dependent kinases regulate epigenetic gene silencing through phosphorylation of EZH2. Nat Cell Biol. 2010;12:1108–1114. doi: 10.1038/ncb2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuljis RO, Xu Y, Aguila MC, Baltimore D. Degeneration of neurons, synapses, and neuropil and glial activation in a murine Atm knockout model of ataxia-telangiectasia. Proc Natl Acad Sci U S A. 1997;94:12688–12693. doi: 10.1073/pnas.94.23.12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian B, Yang Q, Mao Z. Phosphorylation of ATM by Cdk5 mediates DNA damage signalling and regulates neuronal death. Nat Cell Biol. 2009;11:211–218. doi: 10.1038/ncb1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Demonacos C, et al. A new effector pathway links ATM kinase with the DNA damage response. Nat Cell Biol. 2004;6:968–976. doi: 10.1038/ncb1170. [DOI] [PubMed] [Google Scholar]
- 34.Biton S, et al. Nuclear ataxia-telangiectasia mutated (ATM) mediates the cellular response to DNA double strand breaks in human neuron-like cells. J Biol Chem. 2006;281:17482–17491. doi: 10.1074/jbc.M601895200. [DOI] [PubMed] [Google Scholar]
- 35.Chen P, et al. Oxidative stress is responsible for deficient survival and dendritogenesis in purkinje neurons from ataxia-telangiectasia mutated mutant mice. J Neurosci. 2003;23:11453–11460. doi: 10.1523/JNEUROSCI.23-36-11453.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barlow C, et al. ATM is a cytoplasmic protein in mouse brain required to prevent lysosomal accumulation. Proc Natl Acad Sci U S A. 2000;97:871–876. doi: 10.1073/pnas.97.2.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barlow C, et al. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 1996;86:159–171. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- 38.Gueven N, et al. Dramatic extension of tumor latency and correction of neurobehavioral phenotype in Atm-mutant mice with a nitroxide antioxidant. Free Radic Biol Med. 2006;41:992–1000. doi: 10.1016/j.freeradbiomed.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 39.Browne SE, et al. Treatment with a catalytic antioxidant corrects the neurobehavioral defect in ataxia-telangiectasia mice. Free Radic Biol Med. 2004;36:938–942. doi: 10.1016/j.freeradbiomed.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 40.Kaneko S, et al. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev. 2010;24:2615–2620. doi: 10.1101/gad.1983810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cha TL, et al. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science. 2005;310:306–310. doi: 10.1126/science.1118947. [DOI] [PubMed] [Google Scholar]
- 42.Goodarzi AA, Jeggo P, Lobrich M. The influence of heterochromatin on DNA double strand break repair: Getting the strong, silent type to relax. DNA Repair (Amst) 2010;9:1273–1282. doi: 10.1016/j.dnarep.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 43.Chou DM, et al. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci U S A. 2010;107:18475–18480. doi: 10.1073/pnas.1012946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stefansson OA, Esteller M. EZH2-mediated epigenetic repression of DNA repair in promoting breast tumor initiating cells. Breast Cancer Res. 2011;13:309. doi: 10.1186/bcr2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeidler M, et al. The Polycomb group protein EZH2 impairs DNA repair in breast epithelial cells. Neoplasia. 2005;7:1011–1019. doi: 10.1593/neo.05472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fejes AP, et al. FindPeaks 3.1: a tool for identifying areas of enrichment from massively parallel short-read sequencing technology. Bioinformatics. 2008;24:1729–1730. doi: 10.1093/bioinformatics/btn305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu LJ, et al. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics. 2010;11:237. doi: 10.1186/1471-2105-11-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cooper JF, Kusnecov AW. Methylmercuric chloride induces activation of neuronal stress circuitry and alters exploratory behavior in the mouse. Neuroscience. 2007;148:1048–1064. doi: 10.1016/j.neuroscience.2007.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Friedland DR, Los JG, Ryugo DK. A modified Golgi staining protocol for use in the human brain stem and cerebellum. J Neurosci Methods. 2006;150:90–95. doi: 10.1016/j.jneumeth.2005.06.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.