Abstract
We propose nonparametric inference methods on the mean difference between two correlated functional processes. We compare methods that (1) incorporate different levels of smoothing of the mean and covariance; (2) preserve the sampling design; and (3) use parametric and nonparametric estimation of the mean functions. We apply our method to estimating the mean difference between average normalized δ power of sleep electroencephalograms for 51 subjects with severe sleep apnea and 51 matched controls in the first 4 h after sleep onset. We obtain data from the Sleep Heart Health Study, the largest community cohort study of sleep. Although methods are applied to a single case study, they can be applied to a large number of studies that have correlated functional data.
Keywords: EEG, sleep, penalized splines, measurement error, spectrogram
1. Introduction
We propose nonparametric methods for estimating the mean difference and the associated variability between two correlated functional processes. There is a vast literature on estimating parametric fixed effects with correlated residuals, which led to two separate ‘schools’ of thought in statistics: estimating equations and covariance modeling. In short, estimating equations have focused on using a working covariance matrix to obtain unbiased estimators of the mean. The empirical covariance matrix is then used in a sandwich formula to obtain corrected covariance estimators; see [1–3] for more details. Covariance modeling can be carried out either explicitly using parametric, parametric mixtures, or nonparametric methods, or implicitly using random effects; see [4–8] for more details. Nonparametric smoothing with correlated residuals has a similar long history, with most papers being inspired and applied to smoothing of time series data [9–13].
The literature on functional data analysis also contains some papers on comparing the means of two functional processes. In particular, Benko et al. [14] provided theoretical arguments for using bootstrap tests for assessing the equality of means, eigenfunctions, and eigenvalues of the covariance functions for the two sample problem. Hall and Van Keilegom [15] used bootstrap for the hypothesis testing of the equality of distributions of two independent samples of curves. Zhang et al. [16] proposed L2-based and bootstrap-based statistics for testing the equality of two mean curves when curves are independent and observed without noise. Authors have also published several Bayesian approaches for inference on the mean difference in the presence of complex correlation structures, including [17–22].
The main novelty of our paper is that we propose simple, easy-to-implement, bootstrap-based methods for inference on the difference in the means of two functional processes that exhibit complex correlation patterns. In Section 1.1, we introduce our motivating example obtained from a matching case–control study, where subjects with severe sleep disrupted breathing (SDB) were matched to controls. The correlation between cases and controls is induced by matching. There are many other examples where correlated functional data appear naturally: (1) longitudinal observations of images or functions; (2) replication experiments; and (3) multilevel sampling experiments.
This is a new and important problem, as many new medical and public health studies contain nonindependent samples of functions. Our primary aim is to estimate the difference in means between two correlated functional samples together with its associated variability. Our secondary aim is to test whether and where the difference in means is statistically different from zero. We provide three easy-to-use, fast, and statistically principled techniques that address our primary and secondary aims. These techniques contain variations, adaptations, and refinements of ideas sprinkled throughout the literature and are easy to implement, computationally fast, scalable, and adaptable to increasingly complex designs.
Our methods and discussion will be general, but we consider a motivating example from the Sleep Heart Health Study (SHHS) [23], the largest community cohort study of sleep.
1.1. Short description of the data and problem
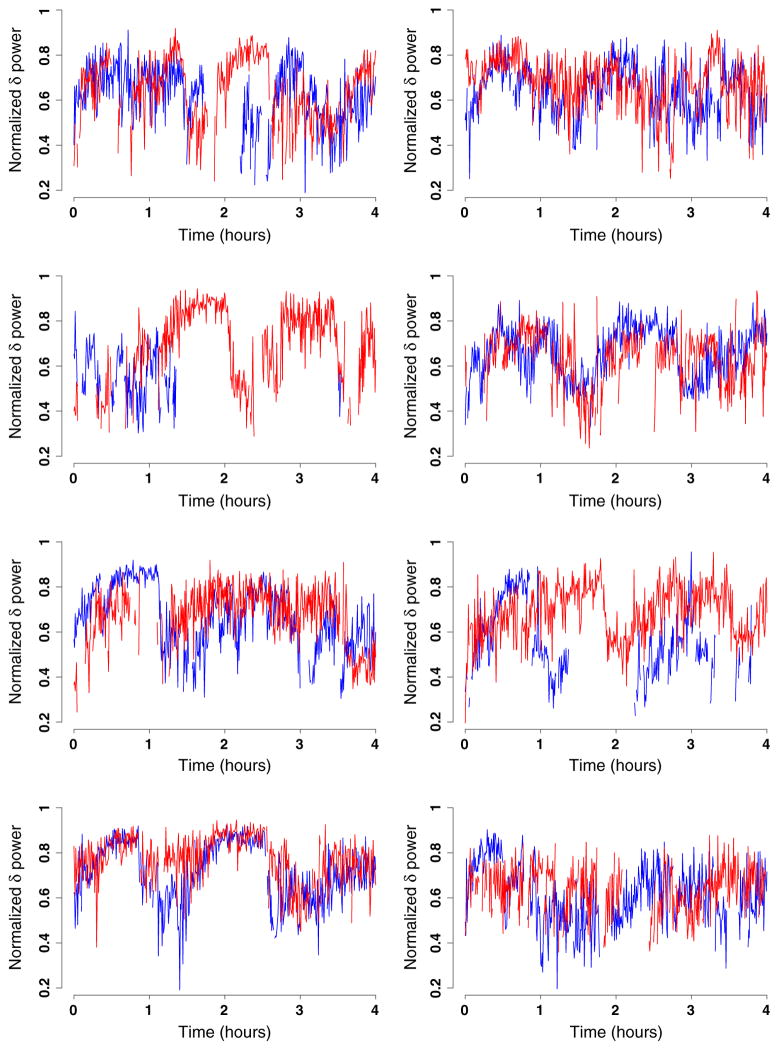
The SHHS collected in-home polysomnogram data on thousands of subjects at multiple visits. Two-channel electroencephalograph (EEG) data were collected as part of the polysomnogram at a frequency of 125 Hz or 125 observations per second. Thus, for each subject, visit, and EEG channel, a total of 3:6 millions observations were collected for a typical 8-h sleep interval. Here we focus on modeling a particular characteristic of the spectrum of the EEG data, the proportion of δ power. For more details on the definition and interpretation of δ power, see, for example, [24–26]. For our purpose, it is sufficient to know that percent δ power is a summary measure of the spectral representation of the EEG signal; in this paper, we use percent δ power calculated in 30-s intervals. Figure 1 displays the sleep EEG proportion of δ power in each of the 30-s intervals of the first 4 h after sleep onset for eight matched pairs of subjects. Each panel displays a matched pair with the red lines corresponding to subjects with SDB and the blue lines corresponding to their matched controls. The x-axis represents time in hours since sleep onset, and the y-axis represents the estimated proportion of δ power. We show observations in adjacent 30-s intervals with missing observations indicating wake periods.
Figure 1.
Normalized δ power for the first 4 h after sleep onset for eight matched pairs of controls (blue) and subjects with sleep apnea (red).
We obtained a total of 51 matched pairs with the use of propensity score matching [27]. We identified subjects with severe SDB as those with a respiratory disturbance index greater than 30 events/hour. We identified subjects without SDB as those with a respiratory disturbance index smaller than five events per hour. Other exclusion criteria included prevalent cardiovascular disease, hypertension, chronic obstructive pulmonary disease, asthma, coronary heart disease, history of stroke, and current smoking. We utilized propensity score matching to balance the groups on demographic factors and to minimize confounding. SDB subjects were matched with no-SDB subjects on the factors of age, BMI, race, and sex. Race and sex were exactly matched, whereas age and BMI were matched using the nearest neighbor Mahalanobis technique with a caliper of 0.10. The resultant match was 51 pairs that met the strict inclusion criteria outlined previously and exhibiting very low standardized biases, a vast improvement on the imbalance of BMI between diseased and nondiseased groups of past studies [28].
Inspection of the eight pairs displayed in Figure 1 reveals several notable features of the data. First, there is large within/between-subject as well as within-group variability. Second, there are no readily recognizable patterns within groups. Third, and more conspicuous, functions are correlated within groups because of the matching process. Fourth, missing data patterns are subject specific with the proportion of missing observations varying dramatically across subjects; note, for example, that more than 50% of data are missing for the healthy subject in the third plot. Thus, simply taking the within-group differences would be inefficient by throwing away data.
1.2. Short description of challenges
Data of the type shown in Figure 1 have many of the complexities encountered in similar applications: missing observations, correlated functions, complex dependence structures, and noise. The problem we are interested in is estimating the difference in the mean functions corresponding to subjects with sleep apnea (red lines in Figure 1) and matched controls (blue lines in Figure 1).
To better understand the problem and the various assumptions, it helps to provide a reasonable statistical framework. The data in our study are pairs of functions {YiA(t), YiC(t)}, where i denotes subject, t = t1, …, tT = 480 denotes the time measured in 30-s intervals from sleep onset, A stands for apneic, and C stands for control. For each subject, some of the observations might be missing. We write both processes as
| (1) |
and we are interested in estimators of d(t) = μA(t) − μC(t) and their associated variability. Many functional data papers concerned with estimating d(·) [14–16] assume that ViA(·) and ViC(·) to be independent, an unreasonable assumption in our and other contexts. Thus, the main challenge is to estimate the function d(·) when the residual processes ViA(·) and ViC(·) have complex covariance structures and are correlated. In most cases, assuming a parametric covariance function, such as working independence, autoregressive, or exchangeable, would badly misfit the observed functional covariance. Taking mixtures of such families tends to fail equally badly because of the complex nature of functional data. A secondary challenge is that making a priori parametric assumptions about either the mean functions or the difference between them would likely be misleading.
To address these issues, we propose three strategies. The first strategy uses nonparametric estimators of the mean functions based on penalized splines [29, 30] under the independence assumption. The variability of the difference function estimate is then obtained via a nonparametric bootstrap of pairs. We call this the ‘nonparametric estimation using nonparametric bootstrap’, and we describe it in details in Section 2.1. The second strategy uses the same nonparametric estimators of the mean functions. The procedure then relies on modeling and smoothing the error processes, ViA(·) and ViC(·), using multilevel functional techniques [26]. Thus, instead of a nonparametric bootstrap of pairs, we simulate data from the joint distribution of the error processes. We call this the ‘nonparametric estimation using parametric bootstrap’ because it uses parametric simulations from the functional distributions. We describe the method in Section 2.2. The third strategy uses parametric estimation of the mean functions where the number of degrees of freedom (df) is fixed a priori. Nonparametric bootstrap of pairs is then used to estimate estimators variability. We describe the method in Section 2.3.
2. Functional bootstrap
Because subjects are matched, it is reasonable to assume that the processes ViA(t) and ViC(t) are correlated. In this section, we propose two bootstrap methods that preserve the pair-specific correlation. The first approach employs a fully nonparametric bootstrap, whereas the second combines elements of nonparametric modeling of covariance operators and parametric simulations from the induced mixed effects model.
Both methods use estimators of the mean function under the independence assumption. We start by describing two smooth estimators of μA(t); the estimator for μC(t) is obtained similarly. The first estimator, denoted μ˜A(t), is obtained by using penalized spline smoothing of all pairs {t, YiA(t)} under the independence assumption, that is assuming that ViA(t) is a mean zero, uncorrelated, homoscedastic process. The second estimator, denoted by μ̂A(t), is obtained by using penalized spline smoothing of {t, Ȳ·A(t)}, where for all t. Penalized splines are one of the most successful and practical automatic smoothing techniques; we refer here to the excellent monographs [30, 31]. A penalized spline approach represents the mean function as μA(t) = BA(t)βA, where BA(t) is a low-rank spline basis obtained by fixing the number and location of knots and achieves smoothing by imposing that the spline coefficients are random with a distribution βA ~ N(0, DA). The penalty matrix DA is intrinsically related to the choice of spline basis, BA(t), and typically depends on one smoothing parameter that is estimated from the data. In this paper, we use thin-plate splines with 20 knots positioned at the empirical quantiles of the observed time points [30, Chapter 13.4]. We used the function spm implemented in the implemented in R[32] package SemiPar[33].
There are some important points to make before we proceed. First, note that Ȳ·A(t) is a consistent estimator of μA(t). Second, obtaining μ˜A(t) is more computationally expensive than obtaining μ̂A(t) as it requires smoothing of I T pairs compared with only T pairs. This is especially important when the number of subjects, I, is large and one uses the nonparametric bootstrap to estimate the variability of mean estimators. However, both estimators can be used in most applications. We will show that they provide almost identical results in our application and in simulations. The intuition for this result is quite simple: the mean of local means is the local mean.
2.1. Nonparametric estimation using nonparametric bootstrap
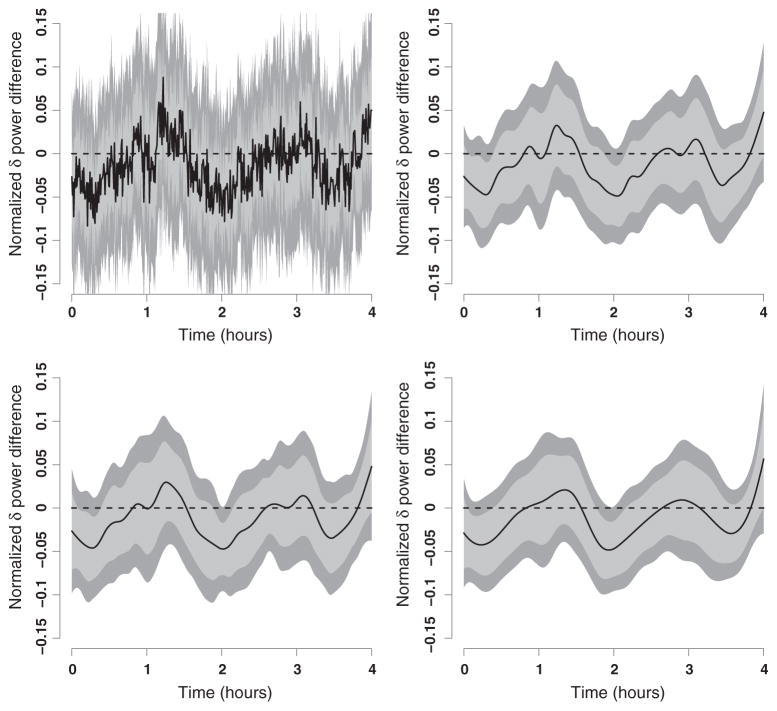
We applied the bootstrap methods to the 51 matched pairs of controls and subjects with sleep apnea. The top-left panel in Figure 2 displays the average normalized δ power for the 51 subjects with severe sleep apnea (red) and 51 matched controls (blue). Raw means are depicted as dots, whereas penalized spline smoothers of the raw means are depicted as lines. Similar curves could be shown using a penalized spline smoother of the entire data set, but the results are indistinguishable from the ones shown. We used B = 1000 nonparametric bootstrap samples of matched pairs, and we repeated the penalized spline fitting of the raw means described previously; the total computation time was 27 min (Dual Core Processor 3 GHz, 32 Gb RAM PC). This created B bootstrap estimators d̂b(t) = μ̂A,b(t) − μ̂C,b(t) of d(t), b = 1, …, B. The top-right panel in Figure 2 displays the bootstrap estimator of mean differences as a solid black line. The estimator d̂B(t) tends to be negative during most of the 4-h interval, which suggests that subjects with severe sleep apnea tend to have lower normalized δ power. However, d̂B(t) is far from being a flat line indicating that the difference is more pronounced during certain intervals.
Figure 2.
Top-left panel: average normalized δ power for 51 subjects with severe sleep apnea (red) and 51 matched controls (blue). Raw means are depicted as dots, whereas penalized spline smoothers of the raw means are depicted as lines. Top-right panel: estimated mean difference between average normalized δ power of subjects with apnea and controls. The pointwise 95% confidence intervals (shaded gray area) are obtained by nonparametric bootstrapping of pairs of subjects, estimating the mean of the sleep apneic and control groups using penalized splines and taking the difference between the estimated means of the groups. Bottom-left panel: similar to the top-right panel, obtained using nonparametric bootstrap of pairs but with a nonparametric smooth estimates of the entire data set for each group. Bottom-right panel: shows the results in the top-right panel as a percent of the range of the mean normalized δ power of controls.
To better visualize where these differences are likely to occur, we construct 95% pointwise confidence intervals based on the bootstrap samples. At time point t, a 95% bootstrap confidence interval for dB(t) is [q̂B,0.025(t), q̂B,0.975(t)], where q̂B,p(t) is the p-quantile of the bootstrap sample d̂b(t), b = 1, …, B. Because the distribution is symmetric, we chose instead to use d̂B(t) ± 2 ŝB(t), where ŝB(t) is the estimated standard deviation of the bootstrap sample d̂b(t), b = 1, …, B. Here we focus on point-wise confidence intervals, but we will discuss joint confidence intervals in Section 3. The top-right panel in Figure 2 also displays the 95% pointwise confidence intervals as a shaded gray area. Statistically significant differences between normalized δ power of subjects with sleep apnea and controls can be detected between minutes 4 and 23 with the largest difference around minute 18 and between minutes 113 and 129 with the largest difference around minute 122. These findings seem to agree with the observed variability in the top-left panel of Figure 2.
We have conducted the same analysis using d˜(t) instead of d̂(t). Recall that d˜(t) is based on nonparametric smooth estimates of the entire data set, whereas d̂(t) is based on nonparametric smooth estimates of the empirical means. The bottom-left panel in Figure 2 displays the same information for d˜(t) as the top-right panel in the same figure. This indicates that results are practically indistinguishable with the two methods. We prefer using d̂(t), because it is much faster to calculate.
The raw difference between normalized δ power displayed in the top-right panel in Figure 2 provides valuable information but does not directly quantify the relative size of observed differences. To provide that, the bottom-right panel in Figure 2 displays the difference between normalized δ power between the two groups as a percentage of the range of the estimated mean functions of controls. More precisely, we plot 100d̂(t) = {maxt μ̂C(t) − mint μ̂C(t)} and its associated variability.
An alternative approach would be to bootstrap the pair differences YiA(t) − YiC(t) = d(t) + ViA(t) − ViC(t). However, calculating the difference YiA(t) − YiC(t) can only be performed when both YiA(t) and YiC(t) are observed. Thus, missing data in either process would compound the problem and would lead to serious efficiency losses. When data are not missing, this approach provides similar results to the ones obtained with the method described previously.
2.2. Nonparametric estimation using parametric bootstrap
In this section, we will continue to use the smooth estimates of the mean functions under the independence assumption. The main difference is in how we estimate the variability of these estimators when the distribution of error processes ViA(t) and ViC(t) is unknown. We start by noting that the pairing of subjects induces within-pair correlation. We account for this by defining a multilevel functional model for both processes as described by Di et al. [26]
| (2) |
where Xi(t) is a functional process with smooth covariance operator KX(·, ·), UiA(t) and UiC(t) are functional processes with the same smooth covariance operator KU(·, ·), εiA(t) and εiC(t) are independent mean zero variance random variables, and Xi(t), UiA(t), UiC(t), εiA(t), and εiC(t) are assumed mutually independent within and between pairs. The role of the process Xi(t) is to account for the within-pair correlation, as cov{YiA(t), YiC(s)} = KX(t, s). The processes UiA(t) and UiA(t) are assumed to share the same covariance operator, a reasonable assumption in this context. However, this assumption is not necessary and may be relaxed in other applications. Both KX(·, ·) and KU(·, ·) are left unspecified, are assumed to be smooth, and are estimated from the data.
Here we proceed in two stages. First, we obtain WiA(t) = YiA(t) − μ̂A(t) = ViA(t) + {μA(t) − μ̂A(t)} and WiC(t) = YiC(t) − μ̂C(t) = ViC(t) + {μC(t) − μ̂C(t)}, where μ̂A(t) and μ̂C(t) are the nonparametric smooth estimates of μA(t) and μC(t) described in Section 2.1. Note that the covariance operators of the W(·) and V(·) are identical and supi |WiA(t) − ViA(t)| ≤ |μA(t) − μ̂A(t)|. Thus, we can use the observed W(·) process to estimate the covariance operators of V(·).
Second, we use multilevel functional principal component analysis (MFPCA) [26] to obtain the parsimonious bases that capture most of the functional variability of the space spanned by Xi(t) and Uij(t) j = A, C, respectively. MFPCA is based on the spectral decomposition of the within-visit and between-visit functional variability covariance operators. We summarize here the main components of this methodology. Denote by and for j ≠ k the total and between covariance operator corresponding to the observed process, Wij(·), respectively. Denote by KX(t, s) = cov{Xi(t), Xi(s)} the covariance operator of the Xi(·) process and by KU(t, s) = cov{Uij(s), Uij(t)} the covariance operator of the Uij(·) process. By definition, for j ≠ k. Moreover, and , where δts is equal to 1 when t = s and 0 otherwise. Thus, KX(s, t) can be estimated using a method of moments estimator of , say . For t ≠ s, a method of moments estimator of , say K̂U(s, t), can be used to estimate KU(s, t). To estimate K̂U(t, t), one predicts KU(t, t) using a bivariate thin-plate spline smoother of K̂U(s, t) for s ≠ t. Staniswalis and Lee [34] proposed this method for nonparametric longitudinal data analysis and showed that this works well for MFPCA [24, 26, 35].
Once consistent estimators of KX(s, t) and KU(s, t) are available, the spectral decomposition and functional regression proceed as in the single-level case. More precisely, Mercer’s theorem [36, Chapter 4] provides the following convenient spectral decompositions , where are the ordered eigenvalues and are the associated orthonormal eigenfunctions of KX(·, ·) in the L2 norm. Similarly, , where are the ordered eigenvalues and are the associated orthonormal eigenfunctions of KU(·, ·) in the L2 norm. The Karhunen–Loève decomposition [37, 38] provides the following infinite decompositions and , where are the principal component scores with E(ξik) = E(ζijl) = 0, . The zero-correlation assumption between the Xi(·) and Uij(·) processes is ensured by the assumption that cov(ξi, ζijl) = 0. These properties hold for every i, j, k, and l. For simplicity, we will refer to and as the levels 1 and 2 eigenfunctions and eigenvalues, respectively.
Given these developments, we propose to parametrically simulate functional residuals from the model
| (3) |
where , k = 1, …, K, ζiAl, , l = 1, …, L, and εiA(t), are mutually independent. A parametric bootstrap is then obtained by calculating and , where and are obtained by simulation from model (3) and b = 1, …, B. This could replace the nonparametric bootstrap described in Section 2.1; the methods for obtaining the variability of estimators remain the same.
Simulating from model (3) is easy once MFPCA is used to estimate the eigenfunctions and eigenvalues. The only technical point is deciding what values of K and L to use in practice. In general, the particular choice does not influence the confidence intervals provided that K and L are large enough. To show that this is, indeed, the case in our application, we considered four different choices from reasonable to extreme. Figure 3 displays the 95% confidence intervals obtained using four different choices. The top-left panel displays results for K and L chosen such that 90% of the variability described by KX(·, ·) and KU(·, ·) is explained. We also used the standard estimator of the variability, σε. The top-right panel displays results for the case when the explained variability is increased to 99% with the same estimator of σε. The left-bottom panel displays results for the case when the explained variability is 99% with a conservative estimator of σε, that is, an estimator that is roughly 20% larger than the standard estimator. The right-bottom panel shows results for the case when the explained variability is 99.95% at both levels with the same conservative estimator of σε.
Figure 3.
Mean difference and 95% pointwise confidence intervals via the parametric bootstrap. Top-left panel: results for K = 36 and L = 26 chosen such that 90% of the variability described by KX(·, ·) and KU(·, ·) is explained. We also used the standard estimator of the variability, σε. Top-right panel: explained variability is increased to 95% (K = 49, L = 40) with the same estimator of σε. Left-bottom panel: explained variability is 95% with an estimator that is roughly 20% larger than the standard estimator. Right-bottom panel: explained variability is 99% (K = 76, L = 69) at both levels with the same conservative estimator of σε.
For these data, we conclude that the choices of K, L, and estimator of σε have a minimal impact on the inference about the mean function. Alternatively, one could consider estimating K and L using restricted likelihood ratio testing in an associated mixed model [35], cross validation [39], or variance explained [26]. In our data example, for a wide range of choices of K and L, the confidence intervals do not change too much and are shorter than those obtained from a fully nonparametric bootstrap; compare Figures 2 and 3. The fact that the confidence intervals are shorter does not automatically make them better. It could be that the longer fully nonparametric bootstrap intervals are necessary to achieve the nominal coverage level. We investigate this further in Section 4.
2.3. Parametric estimation using nonparametric bootstrap
Both previous methods rely on nonparametric smoothing of the group-specific mean functions. A simple alternative is to parameterize the mean functions, estimate the means under the independence assumption, and use the nonparametric bootstrap of pairs described in Section 2.1 to estimate variability. Such a method is especially useful in the case when prior information about the shape of the mean functions exists before data analysis is conducted.
The top-left panel in Figure 4 displays the estimator of the mean difference based on the assumption that both means have 1 df per hour (or 4 df for the 4-h period). The other three panels display the estimators based on the assumptions that both means have 2, 3, and 4 df per hour, which correspond to 8, 12, and 16 total df, respectively. As the number of df increases, the function becomes wigglier while its variability increases. Indeed, the average pointwise standard deviation increases from 0.0124 for the 1 df per hour fit to 0.0204, or 64.5%, for the 4 df per hour fit; for more details, see Section 3.
Figure 4.
Mean difference estimators between subjects with apnea and controls using thin-plate regression splines with unpenalized coefficients. The number of degrees of freedom is fixed (not estimated) and is set equal to the following: 1 degree of freedom per hour (top-left panel); 2 degrees of freedom per hour (top-right panel); 3 degrees of freedom per hour (bottom-left panel); and 4 degrees of freedom per hour (bottom-right panel).
The cases shown here are between two extremes. At one extreme is the model with 1/4 df per hour, which would correspond to fitting a constant mean both to cases and controls. At the other extreme is the model with 120 df per hour, which would fit a different mean to every time point. Both these cases are important in themselves, and we provide their results in Section 3.
3. Pointwise and joint confidence intervals
We now proceed with our application. Single testing for differences in means of two processes can be stated as
When testing for one t, then there are standard methods to preserve the α level of the test; for example, using normal or bootstrap approximations of the null distribution. These are called pointwise confidence intervals and are depicted as light gray bands in figures throughout this paper. Multiple testing for differences at all locations can be stated as
All three methods described in this paper produce samples from the joint distributions of an estimator of the difference function d(t) = μA(t) − μC(t).
Given these samples, there is a simple way to produce joint confidence intervals. Assume that we have a T × B dimensional matrix S that stores the samples from the target distribution. Each row contains one sample of length T and corresponds to a particular estimator of d(·). The column mean, d̄(t), over all samples is an estimator of the mean function, whereas the covariance ΣS = cov(S) is a T × T dimensional matrix. With enough samples, the sampling variability in d̄(t) and ΣS can be ignored. To obtain the joint confidence intervals, we use the following easy-to-implement algorithm:
Simulate dn(t) from a multivariate N {d̄(t), ΣS}
Calculate xn = maxt {|dn(t) − d̄(t)|/σ(t)}, where σ2(t) is the tth diagonal element of ΣS
Repeat for n = 1, …, N and obtain q1−α the 1 − α empirical quantile of the sample {xn : n = 1, …, N}
Obtain the joint confidence intervals d̄(t) ± q1−ασ(t)
To the best of our knowledge, this is the first time such an approach is proposed for functional data, although the original idea has been around for some time; see, for example, its description in the context of scatter plot smoothing using penalized splines [30]. Note that the normal approximation in step 1 of the algorithm is not necessary, and the bootstrap samples can be used directly to obtain the joint confidence intervals. More precisely, assume that db(t) is an estimator of the difference between the mean functions at time point t for bootstrap number b. Then pointwise estimators for the mean and the standard deviation of the mean are and , respectively. Then we can easily construct the random variable realizations Mb = maxt |db(t) − d̄(t)|/s̄(t), the maximum over the entire range of t values of the standardized mean realizations. Then if q1−α is the 1 − α quantile of Mb, b = 1, …, B, then a 1 − α% joint confidence interval for the difference in means will take the form d̄(t) ± q1−α s̄(t). A similar discussion would hold in a Bayesian context for posterior simulations using MCMC from the joint distribution of d(t) given the data. The only reason for using a normal approximation, as we do, is to reduce computation time in cases when bootstrap or MCMC are computationally expensive.
This was the algorithm used to obtain the joint confidence intervals in Figure 5; note the dark gray bands extending the light gray areas. The interpretation for the pointwise confidence intervals shown in light gray is that, at each location in repeated samples, the true mean function will be covered by the shaded light gray interval 100(1 − α)% of the time. The interpretation for the joint confidence intervals is that, at all locations in repeated samples, the true mean function will be covered by the shown dark or light gray areas 100(1 − α)% of the time. One could think of the dark gray band extensions as the correction for multiple comparisons that takes into account the observed correlation between test statistics.
Figure 5.
Pointwise (light gray) and joint (dark extensions) 95% confidence intervals for the difference in mean of normalized δ power between subjects with sleep apnea and matched controls as a function of time from sleep onset. Top left: t-testing without smoothing the mean functions. Top right: nonparametric estimation using non-parametric bootstrap. Bottom left: nonparametric estimation using parametric bootstrap. Bottom right: parametric estimation with 3 degrees of freedom per hour using nonparametric bootstrap.
Figure 5 displays the estimated mean difference together with the 95% pointwise (light gray) and joint (dark gray extensions) under various estimation scenarios. The top-left panel corresponds to the case when no smoothing of the mean function is used. Although the method is wasteful and results are extreme, this is the most popular approach and has been used extensively in genomics, where it is called ‘pointwise testing’, and imaging, where it is termed ‘voxelwise testing’. The top-right panel displays the results for ‘nonparametric estimation using nonparametric bootstrap’ (NE/NB), the bottom-left panel displays the results for ‘nonparametric estimation using parametric bootstrap’ (NE/PB), and the bottom-right panel displays the results for ‘parametric estimation using nonparametric bootstrap’ (PE/NB) with 3 df per hour or 12 total df.
The four plots display some obvious differences, but they convey the same general message: there is no statistically significant difference between the normalized δ power in the first 4 h after sleep onset between subjects with apnea and controls in this data set. Moreover, if a difference exists, then it is more pronounced around minutes 20 and 120 after sleep onset. The distinction between point-wise and joint confidence intervals as well as between pointwise and joint tests for mean differences is not just academic. Indeed, ignoring this distinction would lead to fundamentally different conclusions from analyzing the same data set. For example, using pointwise confidence intervals would lead to the conclusion that there is a statistically significant difference between individuals with sleep apnea and controls.
These findings should not be disappointing. Indeed, this study could be used to generate simple, plausible, and easy-to-test hypotheses that could be analyzed in other studies. We hypothesize that there is a difference between average normalized δ power of individuals with sleep apnea and controls, and this difference is localized between minutes 5 and 20 and between minutes 110 and 125. In fact, if this information were available, a standard t-test for difference in the [5, 20]-minute period would have a p-value of 0.0190, whereas in the [110, 125] minute would have a p-value of 0.0037 with the use of a two-sided t-test. These tests are invalid after conducting 480 other tests to identify regions of large differences. However, they provide extremely interesting findings that could become more focused hypotheses for future studies. Note that a two-sided t-test for the difference in the mean over all time points and subjects between individuals with apnea and controls has a p-value of 0.2, indicating that there is not enough statistical evidence to reject the null of no difference. We also intend to use the average δ power between minutes [5, 20] and [110, 125] as potential health biomarkers in our future studies.
We now quantify more precisely the observed differences between the three procedures we applied to the SHHS data set. In particular, we report the average estimated standard deviation across all periods, , where σ̂t is an estimator of the variability of the estimator d̂(t) for a particular method. Visually, σ̄ is a measure of width of the pointwise confidence intervals depicted as light gray areas. We also report the 0.95 quantile, q0.95, of the distribution used for multiple test corrections. This quantile will depend on the method of estimation of the covariance of test statistics. Note that the average length of the joint confidence intervals shown as dark gray extensions is 2q0.95σ̄.
Table I displays results for the three methods discussed in this paper and compares them with the results obtained using pointwise t-testing without smoothing the mean function. This is considered to be the reference procedure and is labeled ‘PE/NB (120 df/h)’ because it is equivalent to using a parametric estimation with 1 df for every time point. Rows 2 and 3, labeled NE/NB and NE/PB, are the methods introduced in Sections 2.1 and 2.2, respectively. The last four rows correspond to the PE/NB method described in Section 2.3 using from 4 to 1 df per hour.
Table I.
Average estimated standard deviation, , of the mean estimator, d̂(t), 95% quantile used to correct for multiple testing, q0.95, average length of joint confidence intervals, 2q0.953σ̄, and percent reduction in average length of confidence intervals compared with t-testing without smoothing of the mean function, labeled PE/NB (120 df/h).
| Method | σ̄ | q0.95 | q0.95σ̄ | % reduction |
|---|---|---|---|---|
| PE/NB (120 df/h) | 0.0286 | 3.79 | 0.108 | Reference |
| NE/NB | 0.0204 | 3.15 | 0.064 | 41 |
| NE/PB | 0.0204 | 3.17 | 0.064 | 41 |
| PE/NB (4 df/h) | 0.0204 | 3.06 | 0.062 | 43 |
| PE/NB (3 df/h) | 0.0191 | 3.05 | 0.058 | 46 |
| PE/NB (2 df/h) | 0.0177 | 2.96 | 0.052 | 51 |
| PE/NB (1 df/h) | 0.0124 | 2.59 | 0.032 | 70 |
For example, the percent reduction in average length was calculated for method NE/NB as 100(0.108 − 0.064)/0.108 = 41%. The label NE/NB is short for nonparametric estimation using nonparametric bootstrap described in Section 2.1. The label NE/PB is short for nonparametric estimation using parametric bootstrap as described in Section 2.2. The label PE/NB is short for parametric estimation using nonparametric bootstrap as described in Section 2.3. After the label PE/NB, the number of degrees of freedom per hour for parametric estimation is provided within brackets.
The results now quantify our findings. In particular, they indicate that not smoothing the mean is wasteful. Indeed, nonparametrically smoothing the mean across time reduces the average estimated standard deviation of the mean from 0.0286 to 0.0204, or roughly 30%. Moreover, the quantile used for multiple corrections, also decreases from 3.79 to 3.15, or roughly 17%. This decrease is likely due to the increased correlation after smoothing. For reference, the Bonferonni correction quantile for 480 two-sided tests with a family-wide error rate of 0.05 is 3.88. Thus, the average length of the joint 95% confidence intervals decreased from 0.108 to 0.064, which is a 100(0.108 − 0.064)/0.108 = 41% reduction. Parametric smoothing further reduces the average length of the joint confidence intervals. Indeed, average length is increasing as a function of df from 0.032, for 4 df per hour, to 0.062, for 4 df per hour. However, one should not conclude that a smaller number of df is better, as the estimator of the mean function is shrunk towards zero; see Figure 3 for more details.
We conclude that the reduction in average length of confidence intervals can be quite dramatic using very simple smoothing methods. In practice, if little information is available before conducting the analysis, it makes sense to use nonparametric smoothing of the mean. However, if some information is available, then it may make sense to use that information to commit to a particular smoothing method. For example, in future studies of differences in normalized sleep δ power, one can start by assuming that the functions have 3 df per hour. If in doubt, it is probably better to allow for more rather than less df.
4. Simulations
Here we investigate the performance of the observed methods in a simulation study. For all settings, our results are based on simulation of 200 data sets from model (1), where μd(t) is detailed in the following text and Vid(t) =Xi(t) + Uid(t) + εid(t), for i = 1, …, I and d = A, C indicating whether the ith curve is from the case (A) or control (C) group. We set and consider curves sampled at T = 100 points. We consider many scenarios that combine various choices:
Number of subjects: (a) I = 30, (b) I = 50, (c) I = 100, and (d) I = 200;
Sample design: (a) equally spaced time points in [0, 1] and (b) unequally spaced time points in [0, 1] obtained by deleting at random observations that are equally spaced;
Group mean function: (M1) μA(t) = μC(t) = sin(tπ), (M2) μA(t) = 0.5(1 − t)2, μC(t) = 0.1(t + 1)2, (M3) μA(t) = 3t2/2 + t3 − 1.5t; μC(t) = −5(t2− t)/3 + 0.2.
-
Variance processes: (CV1) ; (CV2) , where GP denotes a Gaussian process with mean 0, variance , and Matern autocorrelation function ρU(t). The Matern autocorrelation function is defined as
(4) where ϕ and κ are unknown parameters and Kκ is the modified Bessel function of order κ. We set and the parameters of the Matern correlation function, ρU(t), equal to κ = 5 and ϕ = 0.07. Where appropriately, for k = 1, …, K, l = 1, …, L and , for d = A, C. We set K = 2 and L = 3, for k = 1, 2 and l = 1, 2, 3. We used Legendre polynomials for the process X, in particular ; and we used Fourier basis functions for the process U, in particular , and .
We used inferential methods described in Section 2: nonparametric estimation using nonparametric bootstrap (NE/NB), nonparametric estimation using parametric bootstrap (NE/PB), and parametric estimation using nonparametric bootstrap (PE/NB) using a different number of df. For PE/NB, we used 4 (small), 7 (moderate), and 22 (large) number of df. We obtained pointwise and joint confidence intervals and compared methods in terms of actual coverage and length of the corresponding confidence intervals.
We calculate integrated actual coverage (IAC) for pointwise confidence intervals as , where CIP(t) is the pointwise confidence interval at time t, the expectation is taken with respect to the distribution of the confidence intervals, and 1{·;} denotes the indicator function. In repeated samples, we estimate IACP as , where B is the number of samples, T is the number of grid points, and are the pointwise confidence intervals obtained in the bth simulation. Similarly, we calculate IAC for joint confidence intervals as IACJ= E[1{d(t) ∈ CIJ(t)}: for every t ∈ [0, 1]], where CIJ(t) is the joint confidence interval at time t and the expectation is taken with respect to the distribution of the confidence intervals. In repeated samples, we estimate IACJ as : for every t ∈ [0, 1]}/B, where B is the number of samples and are the joint confidence intervals obtained in the bth simulation.
One can interpret IACP as the average coverage probability of pointwise confidence intervals (light gray areas throughout this paper) across grid points t. In contrast, IACJ is the probability that the entire function is covered by the joint confidence intervals (dark gray extensions throughout this paper). The performance of both intervals is important, although only joint confidence intervals and IACJ are directly related to answering the scientifically important questions: (1) ‘Is there statistical evidence of difference between cases and controls?’ and (2) ‘If there is statistical evidence of difference, then where is this evidence localized and how can it be quantified?’
We presented results for sample sizes ranging from 30 to 200, with and without missing data. For the missing data scenarios, we removed at random 30% of the complete set of observations per subject. We consider the case when the two group mean functions are equal and investigate the confidence intervals for different covariance structures. Tables II and III provide the IAC and expected length of the various pointwise 90% and 95% confidence intervals. We compare results with the t-test method based on empirical mean estimates that do not take into account smoothing. Because there are 100 observations per function, this method is a particular case of parametric estimation with nonparametric bootstrap, where the mean function is estimated using 100 df. Thus, we denote this method PE/NB (100 df).
Table II.
Estimates of the integrated actual coverage of the pointwise (1 − α)100% confidence intervals obtained with NB/NE, PB/NE, and NB/PE with various degrees of freedom for the fit.
| 1 − α | I | CV | NB/PE (100) | NB/NE | PB/NE | NB/PE (4) | NB/PE (7) | NB/PE (22) |
|---|---|---|---|---|---|---|---|---|
| 0.90 | 30 | CV1 | 0.88 (0.89) | 0.88 (0.89) | 0.87 (0.90) | 0.88 (0.89) | 0.88 (0.89) | 0.88 (0.88) |
| CV2 | 0.88 (0.88) | 0.87 (0.88) | 0.89 (0.89) | 0.87 (0.87) | 0.87 (0.88) | 0.88 (0.88) | ||
| 50 | CV1 | 0.88 (0.89) | 0.88 (0.89) | 0.89 (0.90) | 0.89 (0.89) | 0.88 (0.88) | 0.88 (0.88) | |
| CV2 | 0.88 (0.89) | 0.88 (0.90) | 0.90 (0.90) | 0.86 (0.89) | 0.87 (0.89) | 0.88 (0.89) | ||
| 100 | CV1 | 0.88 (0.90) | 0.89 (0.92) | 0.90 (0.89) | 0.88 (0.92) | 0.88 (0.91) | 0.88 (0.91) | |
| CV2 | 0.89 (0.89) | 0.89 (0.90) | 0.89 (0.89) | 0.89 (0.87) | 0.88 (0.89) | 0.89 (0.89) | ||
| 200 | CV1 | 0.91 (0.90) | 0.91 (0.89) | 0.91 (0.89) | 0.91 (0.89) | 0.91 (0.89) | 0.91 (0.89) | |
| CV2 | 0.89 (0.90) | 0.89 (0.89) | 0.91 (0.89) | 0.91 (0.88) | 0.90 (0.88) | 0.89 (0.89) | ||
| 0.95 | 30 | CV1 | 0.93 (0.94) | 0.93 (0.94) | 0.93 (0.94) | 0.93 (0.93) | 0.93 (0.94) | 0.93 (0.93) |
| CV2 | 0.93 (0.94) | 0.93 (0.94) | 0.94 (0.94) | 0.93 (0.94) | 0.93 (0.93) | 0.93 (0.93) | ||
| 50 | CV1 | 0.94 (0.94) | 0.94 (0.94) | 0.94 (0.95) | 0.95 (0.94) | 0.94 (0.94) | 0.94 (0.94) | |
| CV2 | 0.93 (0.94) | 0.93 (0.95) | 0.95 (0.95) | 0.92 (0.95) | 0.92 (0.94) | 0.93 (0.94) | ||
| 100 | CV1 | 0.94 (0.95) | 0.95 (0.96) | 0.95 (0.94) | 0.94 (0.96) | 0.95 (0.96) | 0.94 (0.96) | |
| CV2 | 0.94 (0.95) | 0.94 (0.95) | 0.94 (0.94) | 0.94 (0.93) | 0.94 (0.94) | 0.94 (0.94) | ||
| 200 | CV1 | 0.95 (0.95) | 0.95 (0.95) | 0.95 (0.94) | 0.96 (0.95) | 0.96 (0.95) | 0.95 (0.95) | |
| CV2 | 0.94 (0.95) | 0.95 (0.94) | 0.95 (0.94) | 0.95 (0.93) | 0.95 (0.94) | 0.94 (0.94) |
We present results for the two types of covariance structures for the case that data are observed completely (incompletely).
Table III.
Estimates of the integrated expected length of the pointwise (1 − α)100% confidence intervals obtained with NB/NE, PB/NE, and NB/PE with various degrees of freedom (provided between brackets) for the fit.
| 1 − α | I | CV | NB/PE (100) | NB/NE | PB/NE | NB/PE (4) | NB/PE (7) | NB/PE (22) |
|---|---|---|---|---|---|---|---|---|
| 30 | CV1 | 1.11 (1.46) | 1.08 (1.07) | 1.11 (1.11) | 1.00 (1.01) | 1.08 (1.11) | 1.09 (1.17) | |
| CV2 | 0.87 (1.18) | 0.69 (0.69) | 0.70 (0.69) | 0.49 (0.51) | 0.62 (0.65) | 0.80 (0.88) | ||
| 50 | CV1 | 0.87 (1.13) | 0.85 (0.84) | 0.85 (0.86) | 0.78 (0.78) | 0.85 (0.86) | 0.85 (0.91) | |
| CV2 | 0.68 (0.91) | 0.53 (0.54) | 0.55 (0.54) | 0.38 (0.40) | 0.48 (0.51) | 0.62 (0.69) | ||
| 100 | CV1 | 0.62 (0.80) | 0.60 (0.60) | 0.61 (0.60) | 0.56 (0.56) | 0.61 (0.62) | 0.61 (0.65) | |
| CV2 | 0.48 (0.65) | 0.38 (0.39) | 0.39 (0.39) | 0.27 (0.28) | 0.35 (0.36) | 0.45 (0.49) | ||
| 200 | CV1 | 0.44 (0.57) | 0.43 (0.43) | 0.43 (0.43) | 0.40 (0.40) | 0.43 (0.44) | 0.43 (0.46) | |
| CV2 | 0.34 (0.46) | 0.27 (0.28) | 0.27 (0.27) | 0.19 (0.20) | 0.25 (0.26) | 0.32 (0.35) | ||
| 0.95 | 30 | CV1 | 1.33(1.74) | 1.28 (1.28) | 1.32 (1.32) | 1.19 (1.20) | 1.29 (1.32) | 1.30 (1.40) |
| CV2 | 1.03 (1.41) | 0.82 (0.82) | 0.83 (0.82) | 0.58 (0.61) | 0.74 (0.78) | 0.95 (1.05) | ||
| 50 | CV1 | 1.04 (1.34) | 1.01 (1.00) | 1.02 (1.03) | 0.93 (0.94) | 1.01 (1.03) | 1.02 (1.09) | |
| CV2 | 0.81 (1.09) | 0.64 (0.65) | 0.65 (0.64) | 0.45 (0.47) | 0.58 (0.61) | 0.74 (0.82) | ||
| 100 | CV1 | 0.74 (0.96) | 0.72 (0.72) | 0.72 (0.72) | 0.67 (0.67) | 0.72 (0.74) | 0.73 (0.78) | |
| CV2 | 0.58 (0.77) | 0.45 (0.46) | 0.46 (0.46) | 0.32 (0.34) | 0.41 (0.43) | 0.53 (0.58) | ||
| 200 | CV1 | 0.53 (0.68) | 0.51 (0.51) | 0.51 (0.51) | 0.47 (0.47) | 0.51 (0.52) | 0.51 (0.55) | |
| CV2 | 0.41 (0.55) | 0.32 (0.33) | 0.33 (0.33) | 0.23 (0.24) | 0.29 (0.31) | 0.38 (0.41) |
We present results for the two types of covariance structures for the case that data are observed completely (incompletely).
Tables III and V indicate that confidence intervals that do not account for the smoothness of the mean function tend to be unnecessarily wide; compare columns labeled PE/NB (100 df) with all other columns. The problem is even more serious when data are missing; compare results shown within brackets. The nonparametric estimation of the mean function with parametric or nonparametric bootstrap yields relatively similar confidence intervals, with respect to both coverage and length; compare results in columns labeled NE/NB and NE/PB in Tables II–V. Parametric estimation using nonparametric bootstrap (labeled PE/NB) tend to have good coverage probability especially when the number of df of the fit is in a neighborhood of the true number of df. Our simulations seem to indicate that there is a wide range of number of df that provide reasonable results. This matches our experience that as long as the main features of the mean functions are captured, the coverage probabilities are remarkably robust to the choice of df. Thus, in general, it seems reasonable to choose a number of df that is likely to exceed the complexity of the functions. However, the length of PE/NB confidence intervals increases slowly with the number of df of the fit and becomes extreme when the maximum complexity of the model is reached. Thus, PE/NB could be recommended in situations where previous information about the expected complexity of the mean function exists or in cases where one expects to have very noisy empirical mean estimators. When the PE/NB strategy is employed, the number of df has to be chosen a priori; hunting for a number of df that provides some statistically significant results should be viewed as scientific cheating. The NE/NB and NE/PB methods provide a reasonable compromise for those situations when the scientist knows very little about the expected shape of the mean functions. They tend to trade some of the length of the confidence interval for the ‘peace of mind’ provided by automatic smoothing.
Table V.
Estimates of the integrated expected length of the joint (1 − α)100% confidence intervals obtained with with NB/NE, PB/NE, and NB/PE with various degrees of freedom (provided between brackets) for the fit.
| 1 − α | I | CV | NB/PE (100) | NB/NE | PB/NE | NB/PE (4) | NB/PE (7) | NB/PE (22) |
|---|---|---|---|---|---|---|---|---|
| 30 | CV1 | 1.78 (2.74) | 1.53 (1.57) | 1.58 (1.59) | 1.38 (1.41) | 1.54 (1.63) | 1.58 (1.90) | |
| CV2 | 1.63 (2.30) | 1.14 (1.14) | 1.16 (1.14) | 0.72 (0.75) | 1.00 (1.04) | 1.41 (1.57) | ||
| 50 | CV1 | 1.40 (2.12) | 1.21 (1.23) | 1.22 (1.23) | 1.09 (1.10) | 1.21 (1.27) | 1.25 (1.47) | |
| CV2 | 1.28 (1.79) | 0.89 (0.90) | 0.91 (0.89) | 0.56 (0.59) | 0.78 (0.82) | 1.11 (1.23) | ||
| 100 | CV1 | 1.00 (1.52) | 0.86 (0.88) | 0.87 (0.86) | 0.78 (0.79) | 0.87 (0.91) | 0.89 (1.06) | |
| CV2 | 0.92 (1.27) | 0.63 (0.64) | 0.64 (0.64) | 0.40 (0.42) | 0.56 (0.59) | 0.79 (0.88) | ||
| 200 | CV1 | 0.71 (1.07) | 0.61 (0.62) | 0.61 (0.61) | 0.55 (0.56) | 0.61 (0.65) | 0.63 (0.75) | |
| CV2 | 0.65 (0.91) | 0.45 (0.46) | 0.46 (0.45) | 0.28 (0.30) | 0.40 (0.42) | 0.56 (0.62) | ||
| 0.95 | 30 | CV1 | 1.96 (2.95) | 1.72 (1.75) | 1.77 (1.78) | 1.56 (1.59) | 1.73 (1.82) | 1.77 (2.09) |
| CV2 | 1.75 (2.45) | 1.24 (1.25) | 1.27 (1.24) | 0.80 (0.83) | 1.09 (1.14) | 1.53 (1.70) | ||
| 50 | CV1 | 1.54 (2.28) | 1.35 (1.37) | 1.37 (1.38) | 1.22 (1.24) | 1.36 (1.42) | 1.39 (1.63) | |
| CV2 | 1.38 (1.91) | 0.97 (0.98) | 0.99 (0.98) | 0.62 (0.65) | 0.85 (0.90) | 1.20 (1.33) | ||
| 100 | CV1 | 1.10 (1.63) | 0.97 (0.98) | 0.97 (0.97) | 0.87 (0.89) | 0.97 (1.02) | 0.99 (1.17) | |
| CV2 | 0.98 (1.36) | 0.69 (0.70) | 0.70 (0.70) | 0.44 (0.47) | 0.61 (0.64) | 0.86 (0.95) | ||
| 200 | CV1 | 0.78 (1.15) | 0.68 (0.69) | 0.69 (0.69) | 0.62 (0.63) | 0.69 (0.72) | 0.70 (0.82) | |
| CV2 | 0.70 (0.96) | 0.49 (0.50) | 0.50 (0.50) | 0.32 (0.33) | 0.43 (0.46) | 0.61 (0.68) |
We present results for the two types of covariance structures for the case that data are observed completely (incompletely).
Tables IV and V provide the IAC and expected length of the joint 90% and 95% confidence intervals, respectively. Although both NE/NB and NE/PB performed similarly, NE/PB required careful covariance modeling in the CV2 scenario. In this case, the covariance decays slowly, and a large number of eigenfunctions had to be retained to ensure at least 99% variance explained. Although, in practice, more liberal thresholds, such as 90% or 95%, are used to model functional data, we argue that higher thresholds should be used for obtaining close to nominal coverage probabilities in most scenarios. As a final point, missing data are handled well by the three methods, at least when data are missing at random.
Table IV.
Estimates of the integrated actual coverage of the joint (1 − α)100% confidence intervals obtained with NB/NE, PB/NE, and NB/PE with various degrees of freedom (provided between brackets) for the fit.
| 1 − α | I | CV | NB/PE (100) | NB/NE | PB/NE | NB/PE (4) | NB/PE (7) | NB/PE (22) |
|---|---|---|---|---|---|---|---|---|
| 30 | CV1 | 0.82 (0.76) | 0.86 (0.84) | 0.84 (0.83) | 0.85 (0.85) | 0.84 (0.82) | 0.84 (0.82) | |
| CV2 | 0.68 (0.72) | 0.77 (0.82) | 0.82 (0.75) | 0.84 (0.86) | 0.78 (0.80) | 0.72 (0.76) | ||
| 50 | CV1 | 0.84 (0.82) | 0.86 (0.88) | 0.84 (0.85) | 0.86 (0.88) | 0.88 (0.88) | 0.86 (0.82) | |
| CV2 | 0.80 (0.79) | 0.85 (0.92) | 0.86 (0.84) | 0.86 (0.87) | 0.84 (0.84) | 0.85 (0.86) | ||
| 100 | CV1 | 0.88 (0.88) | 0.88 (0.92) | 0.89 (0.82) | 0.88 (0.92) | 0.87 (0.92) | 0.88 (0.90) | |
| CV2 | 0.84 (0.81) | 0.86 (0.92) | 0.90 (0.86) | 0.86 (0.82) | 0.88 (0.88) | 0.86 (0.88) | ||
| 200 | CV1 | 0.91 (0.88) | 0.90 (0.91) | 0.88 (0.84) | 0.89 (0.87) | 0.92 (0.88) | 0.89 (0.88) | |
| CV2 | 0.88 (0.85) | 0.90 (0.91) | 0.89 (0.86) | 0.90 (0.86) | 0.90 (0.84) | 0.88 (0.86) | ||
| 0.95 | 30 | CV1 | 0.89 (0.84) | 0.90 (0.89) | 0.86 (0.90) | 0.91 (0.90) | 0.90 (0.88) | 0.90 (0.88) |
| CV2 | 0.80 (0.85) | 0.84 (0.90) | 0.92 (0.88) | 0.90 (0.92) | 0.86 (0.86) | 0.82 (0.88) | ||
| 50 | CV1 | 0.90 (0.91) | 0.92 (0.94) | 0.91 (0.90) | 0.92 (0.93) | 0.92 (0.92) | 0.92 (0.92) | |
| CV2 | 0.88 (0.90) | 0.92 (0.95) | 0.94 (0.91) | 0.92 (0.94) | 0.94 (0.91) | 0.90 (0.93) | ||
| 100 | CV1 | 0.92 (0.95) | 0.94 (0.98) | 0.96 (0.92) | 0.95 (0.96) | 0.93 (0.98) | 0.94 (0.96) | |
| CV2 | 0.92 (0.88) | 0.92 (0.94) | 0.94 (0.92) | 0.92 (0.92) | 0.92 (0.94) | 0.91 (0.94) | ||
| 200 | CV1 | 0.94 (0.90) | 0.94 (0.95) | 0.94 (0.91) | 0.93 (0.96) | 0.94 (0.94) | 0.94 (0.94) | |
| CV2 | 0.95 (0.90) | 0.95 (0.95) | 0.94 (0.92) | 0.94 (0.92) | 0.95 (0.91) | 0.94 (0.92) |
We present results for the two types of covariance structures for the case that data are observed completely (incompletely).
We have also conducted extensive additional simulation studies to investigate the robustness of our results to departures from the assumption of normality of both the scores and the errors. We investigated mixture of normal distributions for the scores and double exponential and t-distributions for the errors. We report results in the attached web supplement and indicate the excellent robustness of our approach to these types of departures from normality.
5. More general models
So far, we have considered a very specific model, where the observed subject-specific functions have a natural structure with two levels of functional stochastic variability. However, the idea of fitting the population level parameters under the independence assumption and then bootstrapping subjects to estimate their variability is very general. Consider, for example, data models of the form
where η(t, Xij) is the population-level mean of the functional process Yij(t), Xij is a vector of covariates that may depend only on the cluster, i, or on the cluster and observation within subject, i and j, and Vij is a residual process that may have a complex correlation structure. We assume that Vij and Vi′j′ are independent for every i ≠ i′, but we do not specify any covariance structure. Greven et al. [40] considered a particular case of this model, where they observed functional data at multiple visits over time and Xij = Tij, the time of the j th visit for the ith subject. There are many important particular cases of η(t, Xij): (1) η(t, Xij) = Xijγ, which is the standard parametric linear regression; (2) η(t, Xij) = μ(t) + Xij γ, where μ(t) is modeled either parametrically or nonparametrically; and (3) η(t, Xij) = μA(t)I{i ∈ A} − μC(t)I{i ∈ C }, where μA(t) and μC(t) are mean functions of groups labeled A and C, respectively, and I {· } is the indicator function.
The approach we propose for this type of problem is simple: bootstrap the subjects and obtain estimators of η(t, Xij) under the assumption of independence, that is, under the assumption that Vij (t) are independent identically distributed zero-mean homoscedastic random variables. We propose to conduct inference about η(t, Xij) using the empirical bootstrap distribution obtained from the collection of bootstrap estimators, η̂b(t, Xij), for b = 1, …, B. The performance of the bootstrap needs to be assessed in many different applications, although we consider this approach to be simple and promising.
6. Discussion
In this paper, we provide simple and fast methods for testing if and where the means of two correlated functional processes are different. We contend that the importance of multiple testing is established beyond a reasonable doubt, widely acknowledged, and almost universally ignored. Here we provide testing methods based on joint pointwise confidence intervals that take into account the following: (1) the size and complexity of the data; (2) the sampling mechanisms; and (3) the large number of hypotheses being tested. We conclude that for formal hypothesis testing, joint bounds should be used; for biomarker discovery followed by validation, pointwise confidence intervals can also be used as an exploratory tool.
We conclude that nonparametric estimation using nonparametric bootstrap that respects the data correlation structure is a powerful, simple, and practical method for making inference about the fixed effects of longitudinal functional models. In our case study, a bootstrap of pairs is necessary to account for the sampling mechanism induced by matching. Nonparametric estimation using parametric bootstrap based on liberal choices of the number of eigenvectors is a viable alternative. This approach is slightly more computationally intensive but provides an excellent platform for generalization to more complex models. Parametric estimation using nonparametric bootstrap is a simple methodology that is especially appealing when prior information about the shape of the mean functions is available.
A potential limitation of the bootstrap is that it is conditional, that is, the estimated eigenfunctions and eigenvalues are fixed after the initial estimation step. One way around the problem could be to bootstrap the clusters nonparametrically, apply a parametric bootstrap, and then combine results. Although this may sound complicated, it could actually be carried out very fast. We will investigate such approaches in the future. Also, we do not consider here the theoretical properties of the bootstrap and rely instead on simulations. Another limitation of the bootstrap is when the number of subjects is small or very small. Indeed, having three to five replicates or subjects with millions of observations is quite common. A reasonable question in this context is ‘What should one do when the number of subjects is very small, say 3?’ This is a very difficult question without a standard answer. To illustrate that, consider the case when one observes scalar variables. For example, assume that the long-term observed systolic blood pressure (SBP) for three subjects was 120, 125, and 140, respectively. The mean estimator is m̂ = 128.33 with a standard deviation (using division by n = 3 not n − 1 = 2) of the mean estimator ŝ = 4.91. With the t-distribution with 2 df approximation of the t-statistic distribution used, a 95% confidence interval for the mean SBP of the population would be m̂ ± 4.30 * ŝ, or (107.22, 149.44). Of course, this interval is obtained under the assumption that SBP are independent and identically distributed normal variables, which is a big assumption when there are only three observations. Using 10, 000 bootstraps, we obtained m̂b = 128.27 and ŝb = 4.88. These estimators are very close to the standard ones, as expected. However, a 95% confidence intervals based on the empirical quantiles of the bootstrap distribution is [120, 1403, that is, the interval between the smallest and largest observation in the sample. This interval is much shorter than the one based on the t-distribution approximation. Moreover, all (1 − α)% confidence interval based on the empirical quantiles of the bootstrap are equal to [120, 1403 for every α < 0.05. Thus, we do not recommend using the empirical quantiles of the bootstrap samples to construct confidence intervals in cases with small and very small sample sizes. However, it is quite clear that the mean and standard deviation estimators are very close. Thus, construction of confidence intervals and their properties will depend heavily on the true and assumed distributions of the t-statistic, a problem that cannot be typically resolved by observing three data points. Assuming a t-distribution with 2 df will be incorrect for most applications but will probably result in decent coverage probability in nonexotic examples. These suggestions do not guarantee nominal coverage of confidence intervals. We are not aware of any method that could guarantee it when sample sizes are small or very small in the absence of strong assumptions.
A simple alternative to the bootstrap procedures introduced in this paper is a permutation test that would permute the ‘case’ and ‘control’ labels within matched pairs. This could be an excellent avenue of future research.
Supplementary Material
Acknowledgments
Crainiceanu’s research was supported by award number R01NS060910 from the National Institute Of Neurological Disorders And Stroke. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute Of Neurological Disorders And Stroke or the National Institutes of Health. Staicu’s research was sponsored by the U.S. National Science Foundation grant number DMS 1007466.
Footnotes
Supporting information may be found in the online version of this article.
References
- 1.Hansen LP. Large sample properties of generalized method of moments estimators. Econometrica. 1982;50:1029–1054. [Google Scholar]
- 2.Hardin J, Hilbe J. Generalized Estimating Equations. Chapman & Hall/CRC; Boca Raton, FL, USA: 2003. [Google Scholar]
- 3.Liang KY, Zeger S. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73(1):13–22. [Google Scholar]
- 4.Demidenko E. Mixed Models: Theory and Applications. John Wiley & Sons; Hoboken, New Jersey: 2004. [Google Scholar]
- 5.McCulloch C, Shayle R, Searle S, Neuhaus J. Generalized, Linear, and Mixed Models. John Wiley & Sons; Hoboken, New Jersey: 2008. [Google Scholar]
- 6.Qu A, Lindsay BG, Li B. Improving generalised estimating equations using quadratic inference functions. Biometrika. 2000;87:823–836. [Google Scholar]
- 7.Verbeke G, Molenberghs G. Linear Mixed Models for Longitudinal Data. Springer Verlag; New York: 2000. [Google Scholar]
- 8.Yin G. Bayesian generalized method of moments. Bayesian Analysis. 2009;4(1):1–17. [Google Scholar]
- 9.Beran J, Feng Y. Local polynomial estimation with FARIMA-GARCH error process. Bernoulli. 2001;7:733–750. [Google Scholar]
- 10.Currie I, Durban M. Flexible smoothing with PUsplines: a unified approach. Statistical Modelling. 2002;2:333–349. [Google Scholar]
- 11.Krivobokova T, Kauermann G. A note on penalized splines with correlated errors. Journal of the American Statistical Association. 2007;102(480):1328–1337. [Google Scholar]
- 12.Ray B, Tsay R. Bandwidth selection for kernel regression with long-range dependent errors. Biometrika. 1997;84:791–802. [Google Scholar]
- 13.Wang Y. Smoothing spline models with correlated random errors. Journal of the American Statistical Association. 1998;93:341–348. [Google Scholar]
- 14.Benko M, Härdle W, Kneip A. Common functional principal components. Annals of Statistics. 2009;37:1–34. [Google Scholar]
- 15.Hall P, Van Keilegom I. Two sample tests in functional data analysis, starting from discrete data. Statistica Sinica. 2007;17:1511–1531. [Google Scholar]
- 16.Zhang C, Peng H, Zhang JT. Two sample inference in functional linear models. Communications in Statistics - Theory and Methods. 2010;39:559–578. [Google Scholar]
- 17.Behseta S, Kass RE. Testing equality of two functions using bars. Statistics in Medicine. 2005;24:3523–3534. doi: 10.1002/sim.2195. [DOI] [PubMed] [Google Scholar]
- 18.Behseta S, Kass RE, Moorman DE, Olson CR. Testing equality of several functions: analysis of single-unit firing rate curves across multiple experimental conditions. Statistics in Medicine. 2007;26:3958–3975. doi: 10.1002/sim.2940. [DOI] [PubMed] [Google Scholar]
- 19.Morris JS, Vanucci M, Brown PJ, Carroll RJ. Wavelet-based nonparametric modeling of hierarchical functions in colon carcinogenesis. Journal of the American Statistical Association. 2003;98:573–583. [Google Scholar]
- 20.Morris JS, Carroll RJ. Wavelet-based functional mixed models. Journal of the Royal Statistical Society, B. 2006;68:179–199. doi: 10.1111/j.1467-9868.2006.00539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morris JS, Brown PJ, Herrick RC, Baggerly KA, Coombes KR. Bayesian analysis of mass spectrometry data using wavelet based functional mixed models. Biometrics. 2008;12:479–489. doi: 10.1111/j.1541-0420.2007.00895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morris JS, Baladandauthapani V, Herrick RC, Sanna PP, Gutstein HG. Automated analysis of quantitative image data using isomorphic functional mixed models, with application to proteomic data. Annals of Applied Statistics. 2011;5(2A):894–923. doi: 10.1214/10-aoas407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quan SF, Howard BV, Iber C, Kiley JP, Nieto FJ, O’Connor GT, Rapoport DM, Redline S, Robbins J, Samet JM, Wahl PW. The Sleep Heart Health Study: design, rationale, and methods. Sleep. 1997;20:1077–1085. [PubMed] [Google Scholar]
- 24.Crainiceanu CM, Staicu AM, Di CZ. Generalized multilevel functional regression. Journal of the American Statistical Association. 2009;104:1550–1561. doi: 10.1198/jasa.2009.tm08564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crainiceanu C, Caffo B, Di CZ, Punjabi N. Nonparametric signal extraction and measurement error in the analysis of electroencephalographic activity during sleep. Journal of the American Statistical Association. 2009;104(486):541–555. doi: 10.1198/jasa.2009.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Di CZ, Crainiceanu CM, Caffo BS, Punjabi NM. Multilevel functional principal component analysis. Annals of Applied Statistics. 2009;3(1):458–488. doi: 10.1214/08-AOAS206SUPP. Online access 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swihart BJ, Caffo BS, Crainiceanu CM, Punjabi NM. Collection of Biostatistics Research Archive (COBRA) Johns Hopkins University, Dept. of Biostatistics Working Papers; Nov, 2009. Modeling multilevel sleep transitional data via poisson log-linear multilevel models. Working Paper 212. (Available from: http://www.bepress.com/jhubiostat/paper212) [Google Scholar]
- 28.Swihart BJ, Caffo BS, Bandeen-Roche K, Punjabi NM. Characterizing sleep structure using the hypnogram. Journal of Clinical Sleep Medicine. 2008;4(4):349–355. [PMC free article] [PubMed] [Google Scholar]
- 29.O’Sullivan F. A statistical perspective on ill-posed inverse problems (with discussion) Statistical Science. 1986;1:505–527. [Google Scholar]
- 30.Ruppert D, Wand MP, Carroll RJ. Semiparametric Regression. Cambridge University Press; Cambridge, UK: 2003. [Google Scholar]
- 31.Wood S. Generalized Additive Models. An Introduction with R. Chapman & Hall/CRC; Boca Raton, FL, USA: 2006. [Google Scholar]
- 32.R Development Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2010. (Available from: http://www.R-project.org/) [Google Scholar]
- 33.Wand M, Coull B, French J, Ganguli B, Kammann E, Staudenmayer J, Zanobetti A. Semipar 1.0 r package [Google Scholar]
- 34.Staniswalis J, Lee J. Nonparametric regression analysis of longitudinal data. Journal of the American Statistical Association. 1998;93(444):1403–1418. [Google Scholar]
- 35.Staicu A-M, Crainiceanu CM, Carroll RJ. Fast methods for spatially correlated multilevel functional data. Biostatistics. 2010;11(2):177–194. doi: 10.1093/biostatistics/kxp058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Indritz J. Methods in Analysis. Macmillan & Collier-Macmillan; New York: 1963. [Google Scholar]
- 37.Karhunen K. Über lineare Methoden in der Wahrscheinlichkeitsrechnung. Annales Academiæ Scientiarum Fennicæ, Series A1: Mathematica-Physica, Suomalainen Tiedeakatemia. 1947;37:3–79. [Google Scholar]
- 38.Loève M. Functions Aleatoire de Second Ordre. Comptes Rendus de l’Académie des Sciences. 1945:220. [Google Scholar]
- 39.Zhou L, Huang JZ, Martinez JG, Maity A, Baladandayuthapani V, Carroll RJ. Reduced rank mixed effects models for spatially correlated hierarchical functional data. Journal of the American Statistical Association. 2010;105(489):390–400. doi: 10.1198/jasa.2010.tm08737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Greven S, Crainiceanu C, Caffo B, Reich D. Longitudinal functional principal component analysis. Electronic Journal of Statistics. 2010;4:1022–1054. doi: 10.1214/10-EJS575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.