Abstract
Virtually all cellular functions involve protein-protein interactions (PPIs). As an increasing number of PPIs are identified and vast amount of information accumulated, researchers are finding different ways to interrogate the data and understand the interactions in context. However, it is widely recognized that a significant portion of the data is scattered, redundant, not considered high quality, and not readily accessible to researchers in a systematic fashion. In addition, it is challenging to identify the optimal protein targets in the current PPI networks. The GeneSense server was developed to integrate gene annotation and PPI networks in an expandable architecture that incorporates selected databases with the aim to assemble, analyze, evaluate and disseminate protein-protein association information in a comprehensive and user-friendly manner. Three network models including nodenet, leafnet and loopnet are used to identify the optimal protein targets in the complex networks. GeneSense is freely available at www.biomedsense.org/genesense.php.
A cell can be viewed as an information processing system, receiving signals from its environment and its own internal state, interpreting these signals, and making appropriate cell-fate decisions1 by regulating a network of interactions among the proteins encoded by its own genes. Interaction maps rather than individual genes and proteins provide insights to protein functions and are valuable in identifying ways to fight diseases2. Large amounts of human protein-protein interactions (PPIs) have been reported by experimental techniques, manual curation of literatures, and numerous computational prediction methods3. Protein-protein associations have proven to be an instrumental approach that led to the emergence of systematic and large-scale usage scenarios for functional association networks4. Ideally, the complete set of associations is assembled into a large network that captures the up-to-date knowledge on the functional modularity and interconnectivity in the cell. For example, PPIs have been used to interpret the results of genome-wide genetic screens5, functional genomics data6,7 and elucidation of disease genes8. Such expanding knowledge base has the potential to improve the often time-consuming and cost-intensive process of biomedical analysis, and becomes a major thrust in systems biology research. However, this information is widely scattered and the rapid accumulation of data also renders it difficult to retrieve threads of information concurrently and correctly. The majority of public protein-protein interaction databases such as IntAct9, HPRD10, MINT11 and BioGRID12 archive PPI records from literature curation or direct user submissions. Databases such as PINA13, APID14, STRING4, MiMI15 and UniHI16 integrate information from these curated PPI databases to provide comprehensive sets of public PPIs. In addition, the PINA database integrates six public PPI databases, including IntAct9, MINT11, BioGRID12, DIP17, HPRD10, and MIPS Mpact18. Each of these databases has its own unique features with a large variation in architectural design and annotation. Meanwhile, these databases are heavily relied upon to facilitate studies of biological activities and formulate hypotheses on protein functions and cellular processes as a result of rapidly growing amount of public PPI data.
With the ever increasing importance of PPIs, the challenge researchers face at this point is to efficiently organize and retrieve useful information from the data, which raises the following questions: (i) Can the different data sources be integrated in order to gather a comprehensive set of information? A major imperfection across various databases is the implementation of multiple identification systems depending on the applications the individual database was designed to support or based on developer's preferences. Although some databases attempted to integrate multiple public databases, e.g., PINA, the curated information only represents partial scientific information, or focuses on a specific subset of biological characteristics. For example, the use of p53 and c-Jun in PINA whose names are TP53 and JUN in HGNC (the HUGO Gene Nomenclature Committee), respectively, does not allow for updates in future for the inconsistent names. A better way to integrate the existing PPI databases, eliminate redundancy, and prevent the compilation of inaccuracies is clearly needed. (ii) What are the methods to identify and reduce false-positive PPIs data? Suspicion has been raised about the quality and reliability of protein interaction data with the increasing size of available PPI databases. There are two distinct classes of false positives; one is biological false-positives in which the interactions can be confirmed by multiple computational methods, but the two proteins are in fact never present in the same cell or subcellular compartment at the same time; the other is technical false-positives that can occur in any experimental system19. Both computational and experimental methods for identifying PPIs generate some extent of false positives. (iii) How to identify the best associated proteins for in-depth query and research? Cellular functions are often critically dependent on the correct assembly of proteins to become functional multi-protein complexes through dynamic interactions of various components in response to signals, from internal cellular demands or a cell's external environment20. For example, the PPI network of a tumor suppressor kinase LKB1 and its 14 substrate kinases consisting of 131 proteins and 203 interactions provides hypotheses on the links and pathways critical for tumorigenesis following LKB1 deficiency13. However, it is difficult to identify the appropriate LKB1 target genes from the complex network.
To address these challenges, we developed a web-based platform called GeneSense with the following three main objectives: (i) To provide gene annotation and integrate different data sources based on HGNC in which all genes are manually curated, and the HGNC symbols and names assigned represent a standard, acceptable to be used in all publications and databases where a specific gene is discussed or referenced21. (ii) To build the PPI networks based on literature and experimental data without the false-positives. (iii) To build a user-friendly tool comprising nodenet (node network), leafnet (leaf network) and loopnet (loop network) to assist efficient identification of regulatory factors.
Results
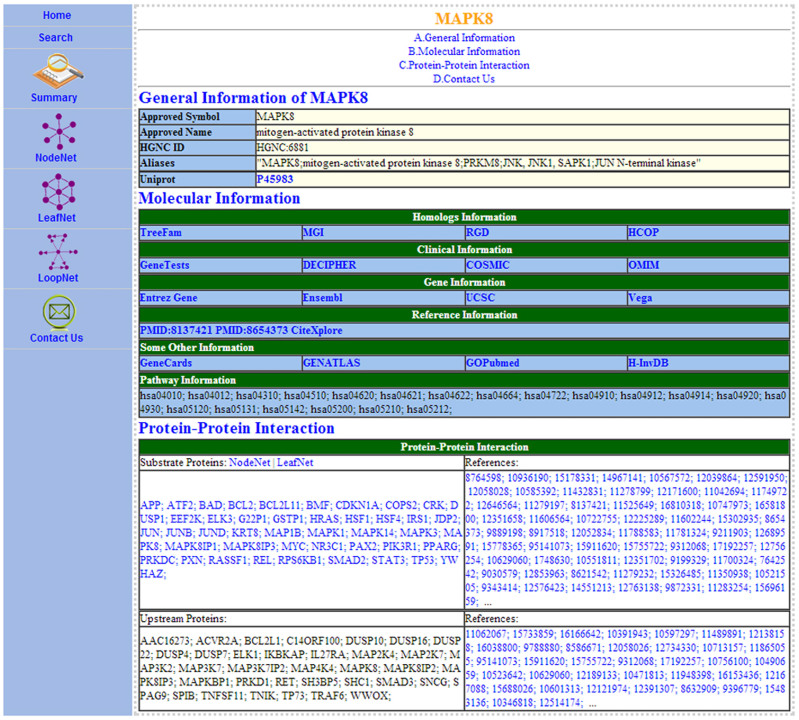
GeneSense is developed to support and integrate gene annotation and the protein level network analysis. The goal of GeneSense's team is to design a friendly, intuitive user interface and a clear presentation of the results. GeneSense requires a JavaScript-enabled browser, such as Google Chrome and Internet Explorer. It allows users to enter the database via a gene of interest using its approved symbol, alias names, approved name or descriptions. Once users submitted the gene of interest, they retrieve the gene's descriptions and are informed of similarly described genes. Subsequently, the users can choose to continue with a gene summary page (Fig. 1) or abort the process and return to the data entry page. The results page is divided into five main sections: a search button to search a new gene of interest, the gene summary (Fig. 1), the node network for the gene of interest (Fig. 2A), the leaf network for the gene of interest (Fig. 2B), and the loop network for the gene of interest (Fig. 2C). In the PPI network section, a JavaScript applet will launch and the networks will be displayed.
Figure 1. A screen shot of gene MAPK8 information summary page in GeneSense.
The table is divided into sections that show MAPK8's general information, homologs information, clinical information, gene information, reference information, pathway information, and protein-protein interactions, respectively, with links to additional information.
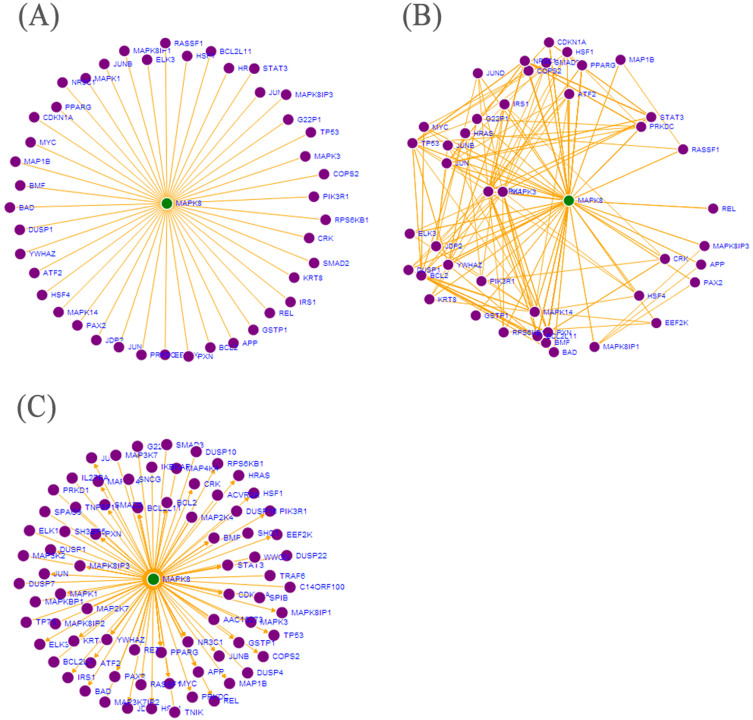
Figure 2. Network analysis of MAPK8 protein.
The green circle indicates the node protein. The purple circles indicate leaf proteins, and the orange lines indicate interactions. (A) The node network of MAPK8 protein. Fig. 3A shows the interactions of MAPK8 and 44 downstream proteins. (B) The leaf network of MAPK8 protein. The leafnet model in Fig. 3B is used to evaluate the interactions of downstream proteins. (C) The loop network of MAPK8 protein. The loopnet model in Fig. 3C shows the visualization of MAPK8 PPI network including downstream and upstream proteins that helps researchers to understand the mediated communication between interacted proteins.
Application to gene annotation
The web page for summary (Fig. 1) displays the general information of the queried gene, its homologs information, clinical information, gene information, reference information, pathway information, and protein-protein interaction information. The general information such as the approved symbol and name is mainly based on HGNC21 and implemented by Uniprot22 which provides richly and accurately annotated protein sequence knowledgebase. Biologists studying a gene in human organisms often wish to transfer functional information between species and homologs information that helps to elucidate how the gene is related to other genes in a family, such as that demonstrated in TreeFam23, MGI24, RGD25, and HCOP26. Others databases such as GeneTests27, UCSC28, CiteXplore (www.ebi.ac.uk/citexplore), GeneCards29 and pathway information are also linked to GeneSense. Gene information is based on gene definitions from HGNC21 and related links via both HGNC-curated data and mapped data provided by the external databases. A group of homology-related links, including TreeFam23, mouse genome informatics (MGI)24, rat genome databases (RGD)25, and HGNC comparison of orthology predictions (HCOP)26 are used to specify the homologs information in GeneSense. Clinical information links include GeneTests27, DECIPHER30, COSMIC31, and OMIM (http://omim.org/). Four widely used gene and genome browsers Entrez Gene32, Ensembl33, UCSC28, Vega34 are also linked in GeneSense. PubMed35 and CiteXplore (www.ebi.ac.uk/citexplore) hyperlinks are included in the references to provide active links to articles that first described the gene in question or that are particularly relevant to the nomenclature of the gene. Additional links such as GeneCards29, GENATLAS36, GOPubmed37 and H-InvDB38 are included in GeneSense based on HGNC. KEGG39 information is used for pathway analysis in GeneSense. The threads of basic protein-protein interaction information fetched from different data sources are also listed in the summary and the associated proteins can also be clicked on to retrieve the corresponding gene summary information.
Application to protein-protein interaction networks
PPI databases in GeneSense were integrated by IPI40 that mapped a variety of accession numbers from different databases, subsequently unified to HGNC accession numbers. It includes a non-redundant database based on integration of data from IntAct9, MINT11, HPRD10 and other databases, such as MEROPS41 that can be integrated by IPI. The architecture of GeneSense based on HGNC and various types of web services offers great advantages of being easily expandable with different PPI data sources. The network visualization is used to evaluate the regulatory relationship between the queried protein and associated proteins, such as the network analysis of MAPK8 gene in Fig. 2. The nodenet of MAPK8 gene (Fig. 2A) shows the interactions of MAPK8 and 44 downstream proteins. The leafnet model was further used to evaluate the interactions of downstream proteins in Fig. 2B. A regulatory network can exist under the identified post transcriptional modifications in either of two stable states (‘upstream' or ‘downstream'). The loopnet model (Fig. 2C) shows the visualization of MAPK8 PPI network, including downstream and upstream proteins that may contribute to the understanding of the mediated communication between interacted proteins. GeneSense can also be used to analyze larger complex networks of PPIs, such as the SRC PPI network (Supplementary Fig. S1).
Discussion
Most public PPI databases adopt diverse practices to annotate gene and protein-protein interaction information. These databases gather partial scientific information that is available, or focus on a specific subset of biological characteristics. The use of inconsistent names exists in these databases that often does not allow for later updates or correction of gene annotation and PPI integration from validated external sources. For example, c-Jun, of which the approved symbol name is JUN in GeneSense and HGNC, also has another synonym AP-1 in HGNC21; the use of c-Jun in PINA does not allow for straightforward update or correction for network analysis with references to a variety of external resources13,42 lacking the consideration of AP-1. Furthermore, inconsistent names also cause the use of the partial scientific information. Han et al43 found that JNK (also named MAPK8) plays a key role in the metabolic response to obesity, but Pal and coworkers' research showed that JNK1 (also named MAPK8) activation does not account for the major diet-induced effects in some other experiment44. The discrepancy exists between different researches as a result of the lack of uniform nomenclature and the asymmetric information. Genesense prevents the use of the partial scientific information by using MAPK8 to unify the inconsistent names. In GeneSense, the primary identifier for each record is the approved and updated gene symbol, which is an acronym or abbreviation of the associated gene name based on HGNC21. As a result, inconsistent names are unified and partial access to existing data is prevented. It also enables easy data tracking regardless of updates in the nomenclature of any given entry by assigning each entry to a unique ‘HGNC ID'21. On the basis of the unified gene symbol name, different databases, such as IntAct9, MINT11, HPRD10, MEROPS41 and other databases can be integrated into GeneSense.
GeneSense is also dedicated to visualization of PPI networks of the coded proteins based on HGNC, IPI and PPI databases. Visualization can be greatly enhanced by interactive presentations and animation; however, high-level abstractions may limit a developer's ability to execute fast incremental scene changes if the system lacks necessary information to avoid redundant computation. To address this problem, GeneSense cooperated with Data-Driven Documents (D3) seamlessly, which results in significantly faster page loads: twice as fast as Protovis and over three times as fast as Flash. Nodenet, leafnet and loopnet were built based on D3. The nodenet model can be useful in highlighting understudied molecular interactions of proteins. For example, the nodenet model shows the interactions of MAPK8 and downstream proteins (Fig. 2A), and it may guide the formulation of meaningful hypotheses with regard to signaling pathways critical to tumorigenesis following MAPK8 deficiency. The leafnet model helps to identify specific proteins that regulate the genes or proteins of interest by the leaf networks. The leafnet (Fig. 2B) showed that some downstream proteins such as MAPK1 and JUN having many interactions with other downstream proteins may be involved in important yet complex mechanisms in MAPK8 related signaling pathways; Some downstream proteins, such as REL and GSTP1 that do not show much interaction with other downstream proteins, may have a simple yet unique function with MAPK8. The loopnet model (Fig. 2C) can be used to assist the design of experiment that aims to distinguish between alternative mechanisms involved in the complex networks, such as the upstream protein MAP3K7 and downstream protein REL of MAPK8 can be designed to regulate MAPK8 to present bistable regulatory mechanisms in different ways. Moreover, a force-directed layout algorithm45 and D3 were applied to visualize the large complex networks in GeneSense, and they make the analysis of complex disease associated-genes relatively easy. To give another example, SRC kinase is a common signaling node in trastuzumab resistance caused by different mechanisms in HER2-positive breast cancers46. Our previous study showed that an intrinsic 40-gene set can be used to classify breast cancer subtypes and assist in optimizing therapeutic management47; however, the association between SRC and the intrinsic characteristic genes are unknown. Using GeneSense, two intrinsic genes, ESR1 and ERBB2 were identified as SRC downstream genes (Supplementary Fig. S1). Understanding the complex ways SRC interacts with its downstream genes ESR1 and ERBB2 in specific breast cancer subtypes maybe crucial for discovering and analyzing mechanisms involved in trastuzumab resistance.
In practice, GeneSense aims to frame the complicated PPI networks in precise terms and use computer simulations to derive implications about how the networks function in normal cells and in the malfunction of diseased cells supported by gene annotation. The following outcomes can be expected from using GeneSense: (i) to gain an accurate overview of genes information of interest; (ii) to build different models to highlight understudied molecular interactions of proteins coded by user-entered genes.
Methods
Distributed architecture and data sources
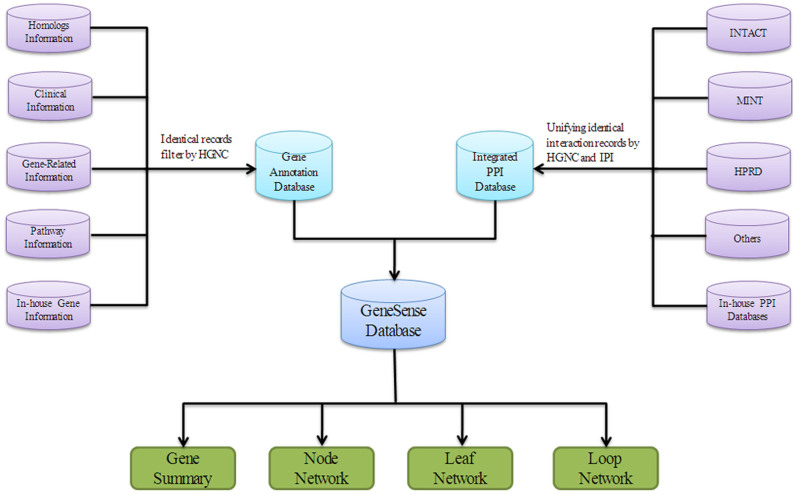
GeneSense is a web-based platform that allows users to visualize, manipulate and analyze gene information and to find the optimal gene regulatory factors by corresponding protein networks. GeneSense database contains two parts (Fig. 3): the first part consists of gene annotation and the second part consists of PPI database. The gene annotation part includes homologs information, clinical information, and gene related information from HGNC21. Pathway information from KEGG39 was also integrated to GeneSense by transferring identical records based on HGNC. PPI databases in GeneSense were integrated by international protein index (IPI)40 that mapped a variety of accession numbers from different databases, subsequently unified to HGNC accession numbers. It includes a non-redundant database based on integration of data from IntAct9, MINT11, HPRD10 and other databases, such as MEROPS41 that can be integrated by IPI. Interactions and protein information were integrated with GeneSense assuming that two proteins from different databases are the same if they have the same IPI accession. With reference to IPI, GeneSense merges results from data sources that employ different but compatible identifier systems. Unique PPI records in different databases were identified by IPI to gather a comprehensive and non-redundant protein-protein interaction dataset, and the protein names were subsequently unified based on HGNC21 to offer consistent names and non-redundant data sets of PPI information. In-house gene information and in-house PPI databases include scattered data that is not included in the existing databases, and would be integrated into GeneSense manually. When users query the genes of interest in GeneSense, the integrated information is retrieved and presented in the gene summary part, including the downstream and upstream proteins according to the post transcriptional modification events. Furthermore, among the key features of GeneSense, three different network models were developed to analyze the function of proteins coded by the retrieved genes: Node network (nodenet) is used to observe the associated downstream proteins interactions with the target proteins; leaf network (leafnet) is used to calculate the complexity of associated downstream proteins with one another and assist in the identification of probable regulatory factors; loop network (loopnet) is used to provide an overview of the upstream and downstream relationships of associated proteins with the target proteins. The architecture of GeneSense based on HGNC and various types of web services offers great advantages of being easily extendable with different PPI data sources.
Figure 3. Schematics of GeneSense platform.
Gene annotation databases (top left, purple) and PPI (top right, purple) databases are integrated into GeneSense that allows users to query interested genes and analyze the genes by pre-constructed networks, including node network, leaf network and loop network.
Network construction and implementation
In GeneSense, queried protein (node protein) is represented by the central green node, and interacted proteins (leaf proteins) are represented by purple nodes. A node can be dragged around to change the arrangement of the nodes. Edges are the connections between nodes and each edge is associated with the reference corresponding to the interactions. GeneSense adopts a number of methods to annotate protein-protein interactions. First, nodenet in GeneSense supports basic queries of PPI network for a single protein, which can be used to rapidly verify whether in-lab generated PPIs are already in the public domain or potentially being novel. Second, GeneSense provides the leafnet network model to visualize the complexity of the queried protein and its substrate proteins that can be used to find the unique or optimal substrate proteins. Third, GeneSense provides loopnet to visualize the upstream and downstream targets of the queried protein, which reveals biological events in cells at the protein-protein interaction level.
GeneSense platform runs on a Linux server and uses Data-Driven Documents (D3), an embedded domain-specific language for transforming the document object model (DOM) based on the data. The DOM combined a number of technologies, mainly, CSS for aesthetics, PHP for page content, JavaScript for interaction, SVG for vector graphics, and so on. Force-directed algorithm45 and D3 was used to generate graphs and to determine the position of each node. Each node is subject to a repulsive force from every other node, yet constrained by the edges that keep nodes connected together. It results in a flexible layout that appears inviting as it unfolds, as exemplified by the nodenet that displays the pictures of the queried protein (or node) and its interacted proteins (or leaf). Although the nodenet model appears to be a promising way to display queried protein and interacted protein datasets, it does not describe leaf-leaf relationships and their degrees of influence. The leafnode was built based on the iteration process of force-directed algorithm for each leaf. The leafnode model is constructed in such a way that high complexity corresponds to layouts in which adjacent leafs are close to each other, and in which non-adjacent leafs are well-spaced. The high complexity leafs may play a crucial role in the signaling network, while low complexity leafs may participate in the regulation of the node in a relatively simpler way. With the aim to visualize the proteins upstream or downstream, modifications to the basic nodenet model were made and the loopnet model that adds the directions to the nodenet model was built. Loopnet reflects the upstream and downstream events involved in the post transcriptional modifications.
Author Contributions
Z.C. and K.W. conceived the project, designed methods, implemented analyses and wrote the manuscript. T.Z. and J.L. analyzed data and wrote the manuscript. Z.Y., Y.W. and W.Z. assisted manuscript preparation.
Supplementary Material
Supplementary Figure S1
References
- Tyson J. J. et al. Dynamic modelling of oestrogen signalling and cell fate in breast cancer cells. Nat Rev Cancer 11, 523–532 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M. Proteomics: The interaction map. Nature 484, 271–275 (2012). [DOI] [PubMed] [Google Scholar]
- Ramirez F., Schlicker A., Assenov Y., Lengauer T. & Albrecht M. Computational analysis of human protein interaction networks. Proteomics 7, 2541–2552 (2007). [DOI] [PubMed] [Google Scholar]
- Franceschini A. et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 41, D808–815 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez O. & Zimmer R. Contextual analysis of RNAi-based functional screens using interaction networks. Bioinformatics 27, 2707–2713 (2011). [DOI] [PubMed] [Google Scholar]
- Goh W. W., Lee Y. H., Chung M. & Wong L. How advancement in biological network analysis methods empowers proteomics. Proteomics 12, 550–563 (2012). [DOI] [PubMed] [Google Scholar]
- Oppermann F. S. et al. Combination of chemical genetics and phosphoproteomics for kinase signaling analysis enables confident identification of cellular downstream targets. Mol Cell Proteomics 11, O111 012351 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I., Blom U. M., Wang P. I., Shim J. E. & Marcotte E. M. Prioritizing candidate disease genes by network-based boosting of genome-wide association data. Genome Res 21, 1109–1121 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrien S. et al. The IntAct molecular interaction database in 2012. Nucleic Acids Res 40, D841–846 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshava Prasad T. S. et al. Human Protein Reference Database--2009 update. Nucleic Acids Res 37, D767–772 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licata L. et al. MINT, the molecular interaction database: 2012 update. Nucleic Acids Res 40, D857–861 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatr-Aryamontri A. et al. The BioGRID interaction database: 2013 update. Nucleic Acids Res 41, D816–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley M. J. et al. PINA v2.0: mining interactome modules. Nucleic Acids Res 40, D862–865 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Toro J., Prieto C. & De las Rivas J. APID2NET: unified interactome graphic analyzer. Bioinformatics 23, 2495–2497 (2007). [DOI] [PubMed] [Google Scholar]
- Tarcea V. G. et al. Michigan molecular interactions r2: from interacting proteins to pathways. Nucleic Acids Res 37, D642–646 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaurasia G. et al. UniHI 4: new tools for query, analysis and visualization of the human protein-protein interactome. Nucleic Acids Res 37, D657–660 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salwinski L. et al. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res 32, D449–451 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guldener U. et al. MPact: the MIPS protein interaction resource on yeast. Nucleic Acids Res 34, D436–441 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusick M. E., Klitgord N., Vidal M. & Hill D. E. Interactome: gateway into systems biology. Hum Mol Genet 14 Spec No. 2, R171–181 (2005). [DOI] [PubMed] [Google Scholar]
- Westermarck J., Ivaska J. & Corthals G. L. Identification of Protein Interactions Involved in Cellular Signalling. Mol Cell Proteomics (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray K. A. et al. Genenames.org: the HGNC resources in 2013. Nucleic Acids Res 41, D545–552 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magrane M. & Consortium U. UniProt Knowledgebase: a hub of integrated protein data. Database (Oxford) 2011, bar009 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J. et al. TreeFam: 2008 Update. Nucleic Acids Res 36, D735–740 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bult C. J., Eppig J. T., Blake J. A., Kadin J. A. & Richardson J. E. The mouse genome database: genotypes, phenotypes, and models of human disease. Nucleic Acids Res 41, D885–891 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwinell M. R. et al. The Rat Genome Database 2009: variation, ontologies and pathways. Nucleic Acids Res 37, D744–749 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre T. A., Wright M. W., Lush M. J. & Bruford E. A. HCOP: a searchable database of human orthology predictions. Brief Bioinform 8, 2–5 (2007). [DOI] [PubMed] [Google Scholar]
- Rubinstein W. S. et al. The NIH genetic testing registry: a new, centralized database of genetic tests to enable access to comprehensive information and improve transparency. Nucleic Acids Res 41, D925–935 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer L. R. et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res 41, D64–69 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safran M. et al. GeneCards Version 3: the human gene integrator. Database (Oxford) 2010, baq020 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas M., Bragin E., Clayton S., Bevan P. & Firth H. V. Interpretation of genomic copy number variants using DECIPHER. Curr Protoc Hum Genet Chapter 8, Unit 8 14 (2012). [DOI] [PubMed] [Google Scholar]
- Forbes S. A. et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res 39, D945–950 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglott D., Ostell J., Pruitt K. D. & Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res 39, D52–57 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P. et al. Ensembl 2013. Nucleic Acids Res 41, D48–55 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilming L. G. et al. The vertebrate genome annotation (Vega) database. Nucleic Acids Res 36, D753–760 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers E. W. et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 40, D13–25 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frezal J. Genatlas database, genes and development defects. C R Acad Sci III 321, 805–817 (1998). [DOI] [PubMed] [Google Scholar]
- Doms A. & Schroeder M. GoPubMed: exploring PubMed with the Gene Ontology. Nucleic Acids Res 33, W783–786 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda J. et al. H-InvDB in 2013: an omics study platform for human functional gene and transcript discovery. Nucleic Acids Res 41, D915–919 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Kawashima S., Okuno Y. & Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res 32, D277–280 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey P. J. et al. The International Protein Index: an integrated database for proteomics experiments. Proteomics 4, 1985–1988 (2004). [DOI] [PubMed] [Google Scholar]
- Rawlings N. D., Barrett A. J. & Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res 40, D343–350 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. et al. Integrated network analysis platform for protein-protein interactions. Nat Methods 6, 75–77 (2009). [DOI] [PubMed] [Google Scholar]
- Han M. S. et al. JNK expression by macrophages promotes obesity-induced insulin resistance and inflammation. Science 339, 218–222 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal M. et al. Alteration of JNK-1 signaling in skeletal muscle fails to affect glucose homeostasis and obesity-associated insulin resistance in mice. PLoS One 8, e54247 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M. J. F. & Reingold E. M. Graph drawing by force-directed placement. Software: Practice and Experience 21, 1129–1164 (1991). [Google Scholar]
- Muthuswamy S. K. Trastuzumab resistance: all roads lead to SRC. Nat Med 17, 416–418 (2011). [DOI] [PubMed] [Google Scholar]
- Wu Y. M. et al. Exploring novel targets of basal-like breast carcinoma by comparative gene profiling and mechanism analysis. Breast Cancer Res Treat 141, 23–32 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1