Summary
Changes to the chromatin structure accompany aging, but the molecular mechanisms underlying aging and the accompanying changes to the chromatin are unclear. Here we report a mechanism whereby altering chromatin structure regulates lifespan. We show that normal aging is accompanied by a profound loss of histone proteins from the genome. Indeed, yeast lacking the histone chaperone Asf1 or acetylation of histone H3 on lysine 56 are short lived and this appears to be at least partly due to their having decreased histone levels. Conversely, increasing the histone supply by inactivation of the Hir (histone information regulator) complex or overexpression of histones dramatically extends lifespan, via a pathway that is distinct from previously known pathways of lifespan extension. This study indicates that maintenance of the fundamental chromatin structure is critical for slowing down the aging process and reveals that increasing the histone supply extends lifespan.
Keywords: Aging, lifespan extension, chromatin, histone acetylation, transcription
Introduction
Mitotic cells divide a finite number of times (replicative lifespan) before they exhibit signs of senescence and cease replicating. The length of time that a post-mitotic or senescent cell exists in a non-dividing state, termed chronological lifespan, also has a profound influence on the physiology and longevity of an organism. Gradual alterations of biological macromolecules characterize normal aging, while adverse alterations to macromolecules are risk factors for age-related conditions and disease states. Notably, aging is the single highest risk factor for the majority of human malignancies. Despite the fundamental importance of the aging process, there are still huge gaps in our knowledge as to the biological changes that lead to aging and the molecular causes of these changes.
Several important cellular processes have been implicated in the regulation of yeast replicative lifespan, including glucose sensing, nutrient sensing, stress response, mitochondrial function and transcriptional silencing (Steinkraus et al., 2008). The conservation of the fundamental mechanisms of aging and lifespan determination across eukaryotes (Smith et al., 2007; Smith et al., 2008) enables the study of aging in budding yeast. Determining how replicative lifespan can be extended in yeast will ultimately help facilitate the development of therapeutic regimens to extend lifespan and delay the onset of age-associated disease in humans.
It is clear that the packaging of the eukaryotic genome together with histones to form the chromatin structure plays a critical role in regulating the activities of the genome (Groth et al., 2007; Li et al., 2007). Tightly packaged chromatin structure reduces access to the DNA to limit inappropriate gene expression and genomic instability, while an open chromatin structure promotes unregulated gene expression and genomic instability. Notably, increased genomic instability and inappropriate transcription are associated with increased age (Busuttil et al., 2007; Maslov and Vijg, 2009). A clear example of the influence of chromatin structure on aging is provided by the Sir2/Hst2 NAD-dependent protein and histone deacetylases that have a key role in transcriptional silencing and maintenance of chromatin structure. Inactivation of Sir2 or Hst2 shortens lifespan (Lamming et al., 2005), while introduction of an extra copy of the gene encoding Sir2 extends lifespan (Kaeberlein et al., 1999). The role of Sir2/Hst2 in yeast aging is likely related to its ability to inhibit formation of extrachromosomal rDNA circles (ERC) (Kaeberlein et al., 1999) by repressing rDNA recombination (Gottlieb and Esposito, 1989). A key substrate of the deacetylase activity of Sir2 is lysine 16 of histone H4 (Moazed, 2001). Sir2 protein levels normally decrease during aging, leading to increased levels of acetylated H4 K16 and concomitant loss of histones from specific subtelomeric regions of the genome (Dang et al., 2009). However, it is unclear how altered chromatin structure at the gene-depleted subtelomeric regions of the genome would lead to aging, or if reversed, would extend lifespan.
Chromatin is modified by the cell in a myriad of different ways. The ultimate and most profound chromatin changes are the loss of histones from the DNA, or the deposition of histones onto the DNA. The removal of histones from DNA and the incorporation of histones onto DNA are mediated by a class of proteins termed histone chaperones. Anti silencing function 1 (Asf1) is a highly conserved central chaperone of histones H3 and H4 that is required for proper regulation of gene expression, histone acetylation and the maintenance of genomic integrity (Chen et al., 2008; Ramey et al., 2004; Tyler et al., 1999). We were prompted to investigate whether yeast cells lacking Asf1 have a reduced lifespan, due to the difficulty we encountered in obtaining re-growth of asf1 mutant cells from old colonies. Our studies have led to our uncovering a causal relationship between chromatin structure and aging, while discovering a means to greatly extend yeast replicative lifespan via manipulating the chromatin structure.
Results
Yeast lacking the histone chaperone Asf1 are short-lived
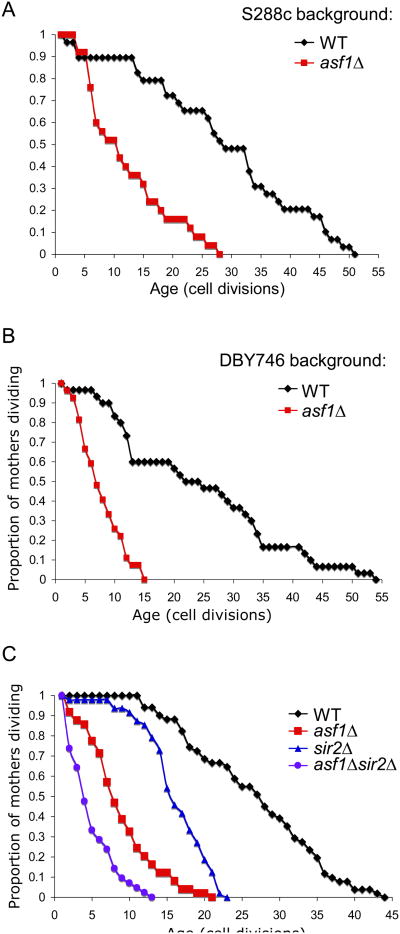
Yeast deleted for ASF1 have a very short chronological lifespan, shorter than that of yeast lacking the well known anti-aging protein Sir2 and comparable to that of yeast lacking Superoxidase dismutase 1 (Sod1) (Fig. S1). In order to determine whether yeast lacking Asf1 also have a defect in replicative lifespan we took advantage of the unique asymmetric nature of cell division in budding yeast. We found that yeast lacking Asf1 are short-lived, having a median replicative lifespan of about 7 generations in comparison to the median lifespan of about 27 generations for wild type yeast in multiple yeast backgrounds (Fig. 1A, B). While shortening of lifespan can result from several causes, asf1 mutants exhibit phenotypes characteristic of aging. Loss of transcriptional silencing occurs in telomeric regions of aged yeast (Kim et al., 1996) and asf1 mutants have transcriptional silencing defects (Le et al., 1997; Singer et al., 1998; Tyler et al., 1999). Also, yeast lacking the protein Rtt109 that functions in the same pathway as Asf1 for lifespan determination (see later) have an elevated rate of ERC formation (Han et al., 2007b), which correlates with premature aging.
Figure 1. Asf1 is required for normal lifespan.
A. Replicative lifespan of isogenic strains BY4741 (“WT”) or BY4747asf1 (“asf1”). B. Replicative lifespan of isogenic strains that were wild type (DBY746) or deleted for ASF1 (JFY022) in the DBY746 background often used for aging studies. C. Replicative aging analysis of isogenic yeast strains BY4741 (“WT”), BY4747asf1 (“asf1”), BY4747sir2 (“sir2”), and BY4747asf1sir2 (“asf1sir2”).
To investigate whether Asf1 functions in previously established pathways of aging we compared the effect of deletion of ASF1 to deletion of SIR2 on replicative lifespan. We found that asf1 mutants have a more drastic decrease in lifespan than sir2 mutants (which have a median lifespan of 15 generations) (Fig. 1C). Furthermore, yeast lacking both Asf1 and Sir2 were extremely short-lived (median lifespan of 4 to 5 generations) (Fig. 1C). These data demonstrate that Asf1 and Sir2 are functioning in non-identical pathways to promote longevity.
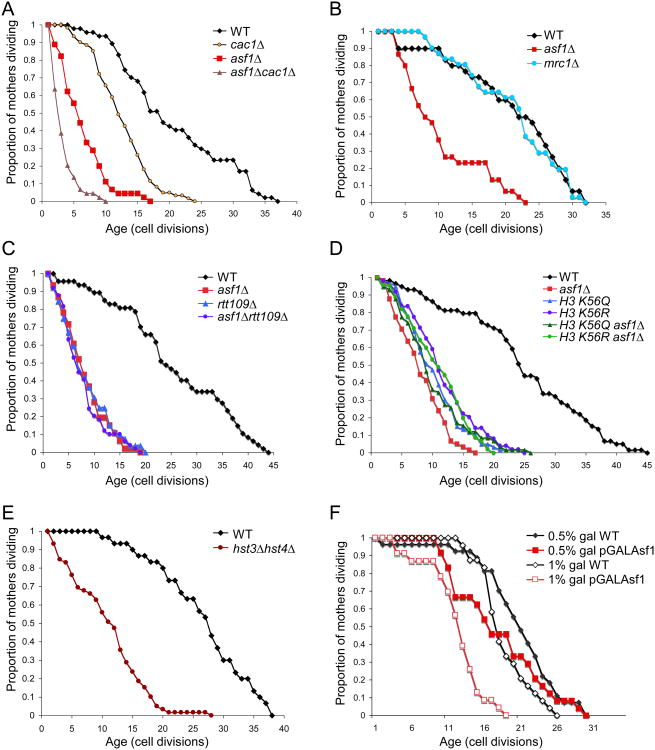
Next, we set out to determine through which molecular pathway Asf1 mediates attainment of a full replicative lifespan. Asf1 promotes replication-dependent chromatin assembly in collaboration with the histone chaperone CAF-1 (Tyler et al., 1999). Inactivation of CAF-1 via deletion of the CAC1 gene encoding the largest subunit of CAF-1 (Kaufman et al., 1997) shortens yeast replicative lifespan but to a lesser degree than deletion of ASF1 (Fig. 2A). In agreement, the double cac1asf1 mutant had a lifespan that was shorter than either single mutant (Fig. 2A), indicating that Asf1 is not solely influencing lifespan via its role in replication-dependent chromatin assembly. Genomic instability contributes to aging and Asf1 has been shown to play a role in the maintenance of genomic stability, apparently due to its role in reducing the amount of DNA damage occurring during DNA replication (Myung et al., 2003; Ramey et al., 2004; Tyler et al., 1999). Notably, yeast lacking the checkpoint protein Mrc1 that normally senses replicational stress (Robert et al., 2006) do not have a reduced lifespan even though they also experience genomic instability occurring during DNA replication (Fig. 2B). These data suggest that the short replicative lifespan of asf1 mutants is unlikely due to genomic instability during DNA replication.
Figure 2. Genetic identification of the pathway through which Asf1 regulates replicative lifespan.
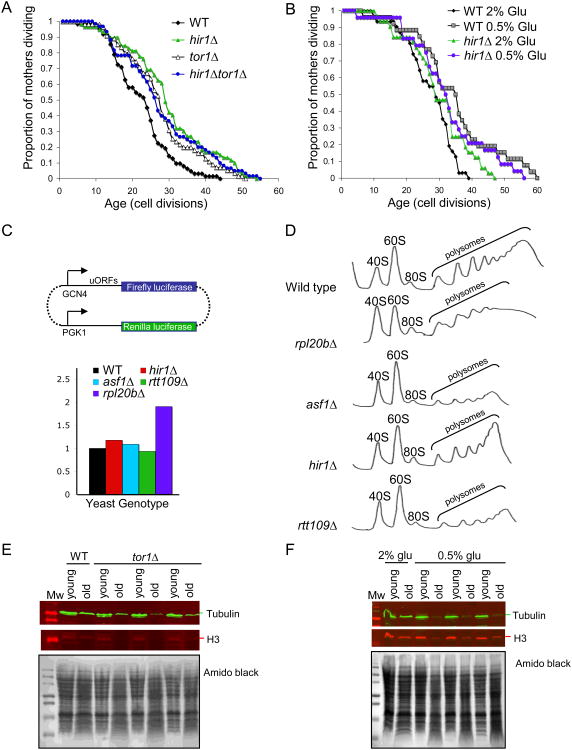
A. Replicative aging analysis of wild type W303 yeast (“WT”) or this strain deleted for ASF1 (“asf1”), CAC1 (“cac1”) or both ASF1 and CAC1 (“asf1cac1”). B. Isogenic strains that were wild type (BY4741) or deleted for ASF1 or MRC1 from the genome wide deletion collection in the S288c strain background were analyzed for replicative lifespan. C. Replicative aging analysis of yeast strains YB (“WT”), ZGY608 (“asf1”), ZGY906 (“rtt109”), and ZGY964 (“asf1rtt109”). D. Replicative aging analysis of yeast strains HMY152 (“WT”), JFY004 (“asf1”), HMY139 (“H3 K56Q”), HMY140 (“H3 K56R”), JFY005 (“H3 K56Q asf1”), and JFY006 (“H3 K56R asf1”). E. Replicative aging analysis of isogenic yeast strains BY4741 (“WT”) and YNML7 (“hst3hst4”). F. Replicative aging analysis of isogenic yeast strains SHY0015 (“WT”) and SHY0014 (“pGALAsf1”). 0.5% galactose leads to endogenous levels of Asf1, while 1% galactose leads to higher than endogenous levels of Asf1 (Zabaronick and Tyler, 2005).
The role of Asf1 in longevity is due to Asf1 facilitating acetylation of histone H3 on lysine 56
Asf1 has an additional function in helping several histone acetyl transferases (HATs) acetylate their histone substrates (Adkins et al., 2007; Fillingham et al., 2008; Han et al., 2007a). To examine whether the function of Asf1 in promoting histone acetylation by the HAT Rtt109 was responsible for normal lifespan, we compared the effect of deletion of ASF1 and RTT109 on replicative aging. The replicative lifespan was indistinguishable between strains deleted for ASF1, RTT109 or both ASF1 and RTT109 together (Fig. 2C), demonstrating that Asf1 and Rtt109 function together in the same pathway to promote achievement of a full lifespan.
Asf1 and Rtt109 together mediate acetylation of histone H3 on lysine 56 and lysine 9 (Fillingham et al., 2008; Han et al., 2007a). To determine which lysine was the critical acetylation target for anti-aging, we measured the replicative lifespan of yeast unable to acetylate K56 using a strain bearing a mutation in the sole copy of the H3 gene that leads to a lysine to arginine (K56R) substitution. Note that although these strains only have one pair of genes expressing H3/H4 (HHT2/HHF2), they produce equivalent amounts of histone proteins as a strain with both pairs of genes expressing H3/H4 (Fig. S2). The H3 K56R mutant had a reduced lifespan (Fig. 2D) (Dang et al., 2009), which was not quite as short as that of yeast with unacetylated K56 (such as the asf1 or rtt109 mutants). This is presumably because the arginine is unlikely to fully mimic the lysine for forming the proper histone-DNA contacts mediated by K56 within the nucleosome structure (Luger et al., 1997), and is consistent with the increased chromatin accessibility and reduced transcriptional silencing that occurs in yeast carrying the H3 K56R mutation (Xu et al., 2007). Nevertheless, the drastically reduced lifespan of the H3 K56R mutant clearly shows that failure to acetylate histone H3 on K56 results in a shortened lifespan (Fig. 2D). Furthermore, deletion of ASF1 did not exacerbate the effect of the H3 K56R mutation on lifespan (Fig. 2D). As such, the role of Asf1 in determining a normal lifespan is mediated via acetylation of histone H3 on K56.
Next, we examined the consequence of having too much H3 K56 acetylation on replicative lifespan. To do this, we first used a strain where the only copy of H3 had been mutated such that lysine 56 is substituted for glutamine (H3 K56Q) to mimic permanent acetylation. The H3 K56Q mutant had a greatly shortened lifespan, which was only slightly longer than that of the asf1 and rtt109 mutants (Fig. 2D) (Dang et al., 2009). To rule out the possibility that the shortened lifespan of the H3 K56Q mutant was due to the glutamine not completely mimicking acetylation of H3 K56, we examined the replicative lifespan of yeast deleted for the genes encoding the two partially redundant histone deacetylases that remove the acetyl group from H3 K56Ac; Hst3 and Hst4 (Maas et al., 2006). Deletion of HST3 and HST4 leads to a greatly elevated level of H3 K56Ac (Maas et al., 2006) and the hst3hst4 double mutant had an extremely short replicative lifespan (Fig. 2E) (Tsuchiya et al., 2006). Because it is not yet known whether deletion of HST3 and HST4 leads to alterations in the levels of other histone modifications, we took another approach to increase the level of acetylated H3 K56Ac in the yeast cells. We had previously shown that overexpression of Asf1 leads to elevated levels of H3 K56Ac (Adkins et al., 2007). Using a strain in which the endogenous ASF1 gene was under the control of the galactose inducible GAL1 promoter (Zabaronick and Tyler, 2005), we found that induction of Asf1 with 0.5% galactose leads to a 19% reduction in the median lifespan, while inducing even more Asf1 with 1% galactose leads to a 28% reduction in the median lifespan (Fig. 2F). These data demonstrate that to achieve normal lifespan it is not only important to be able to acetylate H3 on K56, but it may be equally important to be able to deacetylate H3 K56Ac. Alternatively, there may be a delicate balance or amount of acetylated H3 K56Ac that is required for achieving a normal lifespan.
Histone protein levels drop during aging
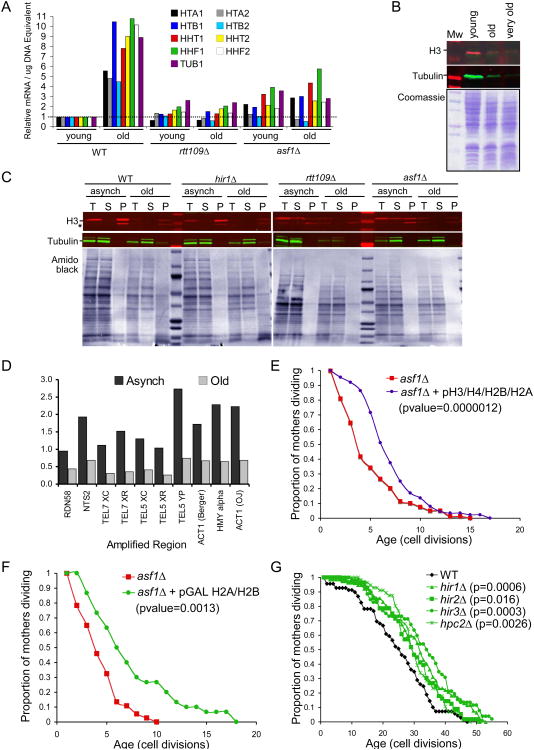
We wondered whether the short lifespan of cells that fail to acetylate histone H3 on K56 might be related to the requirement of H3 K56Ac and Asf1 for accurate expression of the cell cycle regulated histone genes (Sutton et al., 2001; Xu et al., 2005), HHT1 and HHT2 encoding H3, HHF1 and HHF2 encoding H4, HTB1 encoding H2B and HTA1 encoding H2A. Expression of these cell cycle regulated histone genes is repressed outside of S-phase by the histone information regulator (Hir) complex (Spector and Osley, 1993; Spector et al., 1997). Consistent with earlier studies, we find histone transcript levels are altered upon deletion of ASF1, RTT109 or HIR1 (Fig. S3, 3A). To investigate whether histone transcript levels change during the normal aging process, we biochemically separated young and old yeast cells. We observed a significant increase in histone transcripts in the aging population of wild type yeast (Fig. 3A, S3B, S3C). In agreement with the role of H3 K56Ac for accurate expression of the cell cycle regulated histone genes, we found that the short-lived rtt109Δ mutant also had significantly lower levels of these transcripts compared to wild type strains when the data were normalized to tubulin levels (Fig. S3A, B). Aging or deletion of ASF1 or RTT109 leads to an accumulation of cells with a G2/M DNA content (Fig. S4A). Towards understanding the possible effects of altered histone transcription on nucleosome density we decided to correct for these cell cycle defects by normalizing the transcript levels to DNA content. Accordingly, when we independently normalized the histone transcript levels to the amount of DNA per cell, histone transcript levels are slightly higher in young asf1 and rtt109 mutant cells than young wild type cells (Fig. 3A, S3C). However, although histone transcripts also increased in the asf1 and rtt109 mutant cells during the aging process, this was to a much lesser degree than in wild type cells (Fig. 3A).
Figure 3. Aging cells have reduced histone protein levels.
A. RNA levels for H3 (HHT2 and HHT1), H4 (HHF2 and HHF1), H2A (HTA1, HTA2), H2B (HTB1, HTB2) and tubulin (TUB) from strain BY4741 (“WT”) or this strain deleted for ASF1 (“asf1Δ”), or RTT109 (“rtt109Δ”). RNA levels were normalized to total DNA content, and the RNA levels were normalized to 1 for young WT RNA (data prior to normalizing to 1 are given in Suppl. Fig. 3C). Representative results are shown. B. Histone H3 protein and tubulin levels in wild type strain BY4741 were measured by western analysis of equivalent amounts of total protein extracts from biochemically separated wild type yeast. Below is shown a Coomassie stained gel of the same amounts of the same total yeast protein extracts analyzed in the western analyses. The western analyses used infrared secondary antibodies and the images were taken in the linear range of detection. C. Total (T), soluble (S) and pellet (P) proteins were isolated from strain UCC5181 (“WT”) or this strain deleted for HIR1 (“hir1Δ”), ASF1 (“asf1 Δ”), or RTT109 (“rtt109Δ”), that were growing asynchronously of mixed age or were aged via the “mother enrichment program”. The same DNA equivalents of each fraction were analyzed for histone H3 protein levels by western blotting, where non-chromatin bound histones reside in the soluble fraction and chromatinized histones reside in the pellet fraction. “*” denotes a likely proteolytic cleavage product of H3 that arises upon over-handling of the protein. Tubulin was assayed to demonstrate the efficiency of the fractionation. Below are shown amido black staining of the membranes used for the western analyses. The western analyses used infrared secondary antibodies and the images were taken in the linear range of detection. D. Cells of mixed age (“Asynch”) or following aging via the mother enrichment program (“aged”) from strain UCC5181 were subject to ChIP analysis for histone H3 occupancy at the indicated DNA regions. Data shown are the average of two independent experiments, whose results are shown individually in Suppl. Fig. S6A and B. E. Replicative lifespan of strain JFY047 and JFY048 (“asf1”) carrying the empty vector pRS426 or the 2 micron vector pFB1156 carrying the HHF1/HHT1 and HTB1/HTA1 genes. The pvalue indicates the significance of the lifespan extension. F. Replicative lifespan of strains JFY056 and JFY050 (“asf1”) carrying the empty vector pRS315 or the vector pRO689 carrying the pGAL driven HTB1/HTA1 genes. G. Replicative lifespan of isogenic strains with the indicated gene encoding subunits of the Hir complex deleted.
Given that the levels of histone transcripts increase during aging, we asked whether histone protein levels also increase during aging. We loaded equal amounts of total proteins from young, old or very old wild type yeast and determined the levels of histone H3 by quantitative western analysis. The levels of H3 and H2A protein drastically decrease during aging in wild type yeast (Fig. 3B, S4B). We see a very similar drastic decrease in histone levels during aging when we normalize to DNA content (Fig. S4C). It is possible that the increased abundance of histone transcripts during aging (Fig. 3A) is the cell's attempt to replace the histone proteins that are lost or not properly synthesized during aging. Consistent with the histone transcript analyses above, the short-lived rtt109 and asf1 mutants have significantly lower levels of total histone H3 protein than the isogenic wild type strain (Fig. S4D). Furthermore, it is clear that the total histone protein levels also decrease during aging in the short-lived rtt109 mutant and the hir1 mutant (Fig. S4E). Moreover, the relative amount of histone H3 protein in the young and old short-lived rtt109 mutant is less than that seen in the respective young and old wild type cells, while the amount of histone H3 protein in the old hir1 mutant cells is higher than that in the old wild type cells (Fig. S4E). These data are consistent with the total abundance of histone proteins decreasing during aging, and show that the short-lived asf1 and rtt109 mutants have even lower levels of histone proteins than normal.
To determine whether the decrease in histone protein levels in the short-lived asf1 and rtt109 mutants and during the normal aging process reflects a loss of free histones or chromatinized histones, we performed chromatin fractionations. We analyzed the same DNA equivalents of proteins for the total, supernatant (free proteins) and pellet (chromatin proteins) fractions to enable direct comparison of the distribution of the histones between the free and chromatinized forms. In order to isolate sufficient numbers of aged cells for this analysis, we utilized a new system developed by Dan Gottschling's lab, termed the “mother enrichment program” (Lindstrom and Gottschling, 2009). Coupled with the traditional biochemical isolation of old cells, this system enabled us to easily isolate much older cells, that had reached approximately 50% of the maximum lifespan for each strain. As before, we observe a greatly reduced total amount of histones during aging (Fig. 3C). Furthermore, we find that in both young and old wild type and hir1 cells, the vast majority of histone H3 is chromatinized (Fig. 3C). Quite unexpectedly, we find that in the asf1 and rtt109 short-lived mutants, a significant amount of the histones are free, even in the mixed age (asynch) population (Fig. 3C). This may reflect a defect in chromatin assembly in the absence of Rtt109 and Asf1 (Li et al., 2008). To provide further evidence for reduced histone occupancy on the DNA during aging, we performed chromatin immunoprecipitation (ChIP) analysis of histone H3. Again, we utilized the mother enrichment program (Lindstrom and Gottschling, 2009) to isolate old cells (median 30 generations) for this analysis. At all areas examined, including the active Actin gene and the silent HML locus, the rDNA and telomere proximal loci, we observed a 50-75% reduction in histone occupancy in the aged population in comparison to the mixed age asynchronous culture (Fig. 3D, S4F).
Supplying extra histone proteins increases longevity
We hypothesized that the increase in histone transcript levels that normally occurs during aging serves to protect cells from premature aging, while the reduced histone expression in the short-lived mutants is a cause of their shortened lifespan. To test this hypothesis, we investigated whether ectopic expression of extra histones could extend the lifespan of the short-lived asf1 mutants. A high copy number plasmid encoding all four core histones extended the median lifespan of asf1 mutants by 65% (Fig. 3E). Overexpression of the H2A/H2B pair of histones from a galactose inducible promoter also significantly increased the lifespan of asf1 mutants (Fig. 3F). Overexpression of H3/H4 did not significantly extend the lifespan of the asf1 mutant (Fig. S5A,B), which is not too surprising given that these cells are lacking the major H3/H4 histone chaperone. These data show that the shortened lifespan of the asf1 mutant can be partly reversed by supplying additional histones.
Given that the reduced level of histone proteins in the asf1 mutant is partly responsible for the shortened lifespan of this strain, we predicted that supplying higher levels than normal of histone proteins would extend the lifespan of wild type yeast. To test this prediction, we analyzed the replicative lifespan of the hir1 mutant that has significantly higher histone protein levels in the old cells as compared to wild type old cell (Fig. 3G). In addition, we examined the other three components of the Hir complex; Hir2, Hir3, Hpc2 (Green et al., 2005). Inactivation of any component of the Hir complex extended the median replicative lifespan by 25-35% (Fig. 3G). This extent of lifespan extension is comparable to that achieved by other known individual manipulations that result in lifespan extension. In summary, mutants with low histone protein levels such as rtt109 and asf1 mutants are short-lived, while mutants with high histone protein levels such as hir1, hir2, hir3 and hpc2 mutants have extended lifespan.
Increased histone supply appears to extend replicative lifespan via a pathway that is distinct from other lifespan extension pathways
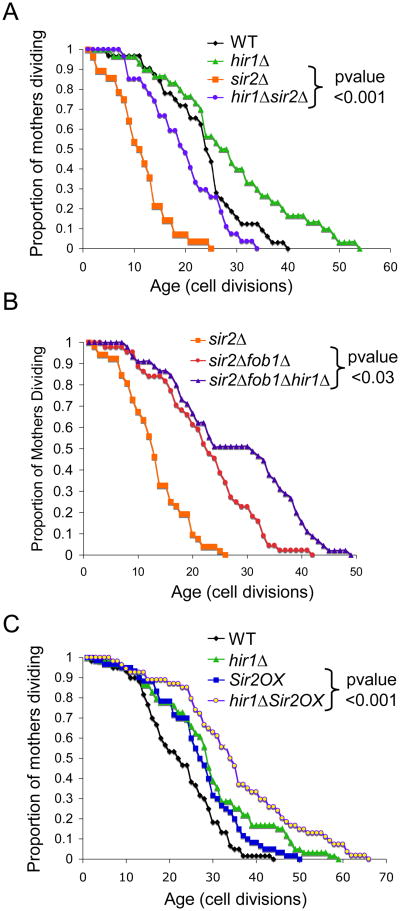
To determine how the histone supply pathway of longevity relates to other known pathways of lifespan extension, we performed epistasis analyses. Deletion of HIR1 significantly extends the lifespan of the short-lived sir2 mutant (Fig. 4A), indicating that the extra histone supply pathway does not require the Sir2 histone deacetylase to achieve lifespan extension. The short lifespan of sir2 mutants is due to the accumulation of ERCs because it is suppressed by deletion of FOB1 (Defossez et al., 1999) (Fig. 4B), where FOB1 encodes a replication fork protein whose deletion prevents ERC formation (Defossez et al., 1999). Notably, additional deletion of HIR1 from the sir2fob1 double mutant led to a significant extension in lifespan beyond that of the sir2fob1 double mutant (Fig. 4B), indicating that extra histone supply is unlikely to extend lifespan via reducing ERC formation. An extra copy of the gene encoding Sir2 (Sir2 OX) extends lifespan (Kaeberlein et al., 1999) (Fig. 4C), mediated via reduced ERC accumulation because it is blocked by FOB1 deletion (Imai et al., 2000). Deletion of HIR1 leads to a greater degree of lifespan extension than overexpression of Sir2 (Fig. 4C), while deletion of HIR1 and Sir2 overexpression together significantly increased lifespan when compared to Sir2 overexpression alone (Fig. 4C). Collectively, these data indicate that extra histone supply is unlikely to extend lifespan via the identical pathways by which ERC accumulation and Sir2 influence lifespan.
Figure 4. Increased histone supply does not extend lifespan by the same pathways as Sir2 or ERCs.
A. Replicative lifespan of isogenic strains with the indicated genes deleted or containing an extra copy of Sir2 (Sir2OX). B. Replicative lifespan of isogenic strains with the indicated genes deleted. C. Replicative lifespan of isogenic strains with the indicated genes deleted and/or containing an extra copy of Sir2 (Sir2OX).
Lifespan extension due to caloric restriction (CR) is likely mediated through the highly conserved nutrient-responsive target of rapamycin (Tor1) kinase (Kaeberlein et al., 2005). To investigate the relationship between the lifespan extension due to increased histone supply and the lifespan extension due to Tor1 deletion, we combined the hir1 and tor1 mutants. The hir1 mutant extended lifespan more the tor1 mutant (Fig. 5A). However, the double hir1tor1 mutant had a lifespan that was intermediate between that of the tor1 mutant and the hir1 mutant (Fig. 5A). CR of yeast is achieved by growth in 0.5% glucose in contrast to the 2% glucose that is used for standard yeast growth. Although individually CR or HIR1 deletion extends lifespan, CR of the hir1 mutant led to a lifespan extension that was intermediate between that resulting from the individual manipulations, not additive (Fig. 5B). These results suggest two possibilities: either (i) that deletion of TOR1/CR and deletion of HIR1 function through the same pathway to extend lifespan or alternatively, (ii) that HIR1 deletion cannot extend lifespan in the context of a tor1 mutant/CR.
Figure 5. Epistasis analysis with the Tor1 pathway or CR.
A. Replicative lifespan of isogenic strains with the indicated genes deleted. B. Replicative lifespan of isogenic strains with or without HIR1 deleted, grown on either 0.5% glucose to induce CR or on 2% glucose. C. Schematic of the dual luciferase reporter plasmid used to measure Gcn4 translation levels. The ratio of the signal from the GCN4-firefly luciferase to renilla luciferase is shown for isogenic strains from the genome wide deletion collection in the S288c strain background. D. Polysome profiles of the indicated isogenic strains from the genome wide deletion collection in the S288c strain background. The position of the 60S, 40S, 80S ribosome and the polysomes are indicated. E. Histone H3 protein and tubulin levels were measured by western analysis of equivalent amounts of total protein extracts from biochemically separated young and old wild type and tor1 mutant yeast. Below is shown the amido black stained membrane to show equivalent total protein loading. The western analyses used infrared secondary antibodies and the images were taken in the linear range of detection. F. As for E, but with wild type yeast grown in normal conditions (2% glucose) or calorie restricted conditions (0.5% glucose).
Depletion of the 60S ribosomal subunits extends lifespan (Steffen et al., 2008). Genetic evidence places CR, tor1 inhibition and depletion of the 60S subunits in a longevity pathway that is partially dependent on upregulated translation of the transcription factor Gcn4 that promotes stress resistance (Steffen et al., 2008). To investigate whether the histone supply pathway extends lifespan via the same pathways that are targeted by deletion of TOR1 and CR, we measured Gcn4 translation and the abundance of 60S ribosomal subunits in the mutants that influence histone supply. Using a dual luciferase reporter construct to measure Gcn4 translation, we observed increased Gcn4 levels upon deletion of the gene encoding the 60S subunit Rpl20b (Fig. 5C) as observed previously (Steffer et al., 2008). However, the isogenic short-lived rtt109 and asf1 mutants and the long-lived hir1 mutant did not have significant alterations in Gcn4 protein synthesis (Fig. 5C), suggesting that 60S ribosomal subunit abundance is not drastically altered in these strains.
To examine directly whether the abundance of the 60S ribosomal subunit was altered in the mutants that effect histone supply, we performed polysome profiling. The rpl20b mutant has a lower amount of 60S ribosomal subunit as compared to wild type (Fig. 5D, Fig. S5C) as seen previously (Steffer et al., 2008). We note that the amount and size of the polysomes was lower in the rpbl20b mutant compared to wild type indicating reduced amounts of translation in general and each polysome peak was a doublet although the relevance of this is unclear (Fig. 5D). The short-lived asf1 and rtt109 mutants had less polysomes than the wild type, indicating that less translation in general is occurring. However, no significant change in the 60S to 40S ratio was observed with the short-lived asf1 and rtt109 mutants or the long-lived hir1 mutant, as compared to the wild type strain (Fig. 5D). Taken together, these data indicate that extra histone supply is not influencing lifespan via the same pathways of altered protein synthesis that are influenced by inhibition of Tor1 to extend lifespan.
To further investigate the relationship between lifespan extension by extra histone supply and lifespan extension due to deletion of TOR1 or DR, we examined total histone protein levels during these manipulations. Surprisingly, we found that biochemically isolated old cells from tor1 mutants or during DR have significantly less histone proteins than old wild type yeast cells (Fig. 5E,F, Fig. S5D). These data indicate that the pathway by which DR and TOR1 deletion leads to extended lifespan is not a consequence of increased histone supply. These data also raise the possibility that deletion of HIR1 does not further extend the lifespan of tor1 mutants or during DR because HIR1 deletion may not be sufficient to raise the histone levels significantly during these manipulations. Taken together, our analyses suggest that HIR1 deletion leads to lifespan extension via a pathway that is not shared with other known mechanisms of lifespan extension.
Ectopic expression of histones H3 and H4 profoundly increases replicative lifespan
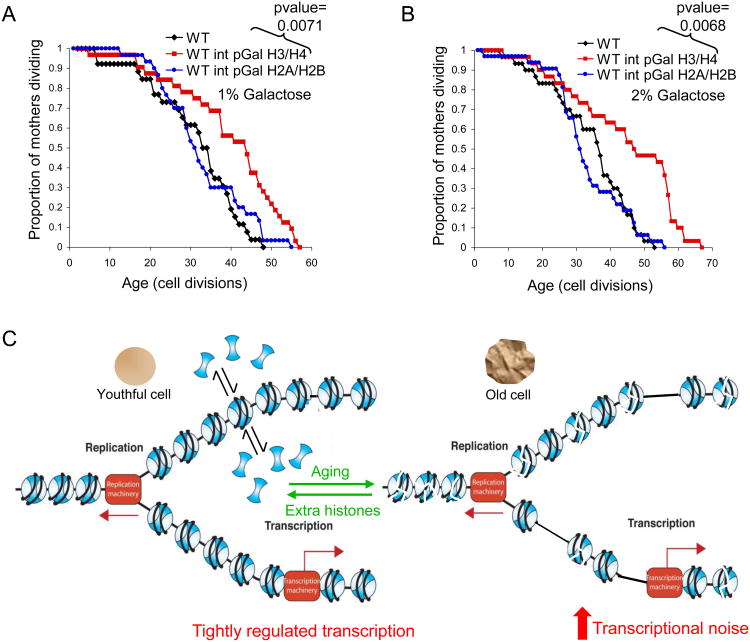
To determine which of the core histones impart anti-aging properties, we individually integrated extra copies of the genes encoding H3/H4 or H2A/H2B under the control of the pGAL1/10 promoters. Overexpression of integrated genes encoding H3/H4, but not H2A/H2B extended the median lifespan of wild type cells by 30% (Fig. 6A, Fig. S6A). Higher levels of H3/H4 induction, via addition of more galactose to more strongly induce the promoters controlling H3/H4, resulted in lifespan extension of up to 50% (Fig. 6B). Noteworthy, this degree of yeast replicative lifespan extension is far greater than that usually attained by most other single manipulations in yeast. Expression of H3/H4 in these conditions leads to approximately a 50% increase in the level of total histone proteins (Fig. S6B,C). Overexpression of histones H3/H4 in these conditions did not lead to significant differences in the polysome profile compared to wild type cells (data not shown) indicating that excess histones are not drastically influencing protein synthesis or ribosomal subunit composition. Also, no significant changes in the cell cycle resulted from overexpression of H3/H4 (Fig. S6D) Importantly, the expression of extra histone proteins does not lead to resistance to the oxidative damage that results from treatment with hydrogen peroxide, replication stress due to treatment with hydroxyurea or DNA damage due to treatment with the alkylating agent methylmethane sulfonate (Figs. S6E,F). As such, the extended lifespan that results from extra histone supply is not due to their protecting the genome from genomic instability.
Figure 6. Overexpression of histones H3/H4 extends lifespan.
A. Replicative lifespan of isogenic strains that are wild type (“WT”) or carrying an extra copy of either HHT2/HHF2 or HTB1/HTA1 driven from the galactose inducible pGAL1/10 divergent promoter integrated into the genome grown on 1% galactose and B. on 2% galactose. C. Model for lifespan extension by increasing histone supply.
Discussion
In this study we show that aging is accompanied by a profound loss of histone proteins. This loss of histones is causal for aging because significant lifespan extension is achieved by ectopically increasing histone expression in wild type yeast, via a pathway that is distinct from other known lifespan extension pathways.
Age dependent changes in chromatin structure
The drastic decrease in the level of histone proteins on chromatin during the normal aging process in yeast (Fig. 3B-D) is consistent with the loss of transcriptional silencing that occurs in wild type yeast during aging (Kim et al., 1996). More recently, an age-dependent increase in levels of histone H4 K16Ac at the X elements within subtelomeric regions of yeast chromosomes has been shown to occur due to a progressive loss of the Sir2 deacetylase protein during aging (Dang et al., 2009). This particular histone modification is unique in that it prevents higher order packaging between adjacent nucleosomes (Shogren-Knaak et al., 2006), implying that increased H4 K16ac during aging would lead to a more open chromatin structure in aged cells. Indeed, when reporter genes were inserted into the X elements within subtelomeric regions, the age dependent increase in H4 K16Ac levels led to increased expression of these reporter genes during aging (Dang et al., 2009). However the mechanistic relevance of opening up the chromatin structure of the X elements of subtelomeric regions on aging is not clear.
Many other previous studies have also revealed transcriptional dysregulation occurring during the aging process and it is clear that at least some of these transcriptional changes are due to age-dependent alterations in the chromatin structure. For example, DNA methylation, which is known to lead to a tighter packaging of the chromatin structure (Fuks, 2005), diminishes throughout the lifespan of mouse liver cells with consequent expression of previously repressed genes (Mays-Hoopes et al., 1983; Singhal et al., 1987). Age-related mRNA changes have also been documented extensively in yeast (Yiu et al., 2008). Similarly, microarray studies of global messenger RNAs (mRNAs) in aging animals and in mutants considered to affect the aging process have found many transcriptional changes (Zahn and Kim, 2007). Furthermore, the finding that stochastic differences in gene expression between individuals can influence lifespan in C. elegans points to a causal role for epigenetic changes during aging (Rea et al., 2005).
How does extra histone supply extend lifespan?
Our model for how interventions that increase histone supply result in lifespan extension proposes that the resulting enlarged soluble pool of free histones facilitates histone exchange via stimulating the ongoing equilibrium of dynamic chromatin assembly and disassembly (Fig. 6C). The resulting higher rate of histone exchange would accelerate the rate of removal of post-translationally modified and damaged histone proteins from the DNA, and would additionally enable sporadically occurring nucleosome free regions in the genome to be repackaged into chromatin. This in turn would reduce inappropriate access of proteins to the genome during aging, which would otherwise lead to the increased transcription and genomic instability – each of which are frequent characteristics of increased age and conditions that exhibit accelerated aging phenotypes.
If the chromatin is packaged into to a tighter structure by overexpressing histones, one would predict that the increased histone supply would restore the transcriptional silencing that is normally lost during aging (Kim et al., 1996). Indeed, hir1 mutants, which have elevated histone levels, have enhanced transcriptional silencing as compared to wild type cells (Kaufman et al., 1998; Smith et al., 1999). By analogy, inactivation of the histone deacetylase Rpd3 also leads to elevated histone levels, extending lifespan in some yeast backgrounds and increasing transcriptional silencing (Bernstein et al., 2000; Kim et al., 1999; Rogina et al., 2002; Smith et al., 1999). Although the increased transcriptional silencing that occurs upon inactivation of Rpd3 and Hir1 provides a marker for altered chromatin structure, it is thought that the influence of Rpd3 on the aging process is most likely mediated through influencing the expression of euchromatin genes (Frankel and Rogina, 2005; Kim et al., 1999). Indeed, inactivation of Rpd3 and the concomitant increase in histone levels results in significant down-regulation of 40% of genes located within 20kb of the telomeres (Bernstein et al., 2000). This is reminiscent of the induction of 40% of genes located within 20kb of the telomeres that occurs upon depletion of histone H4 (Wyrick et al., 1999).
It is clear from the increased silencing in the hir mutants and rpd3 mutants that supplying extra histone proteins results in a tighter chromatin structure, and we anticipate that this will be the case not only at the silent regions but also at other regions of the genome. We propose a model where the expression of euchromatic genes increases inappropriately during aging due to the deterioration of the chromatin structure with age and that the mis-expression of genes contributes to the aging process. By extension, we propose that our manipulations that result in increased histone supply with age result in a tighter chromatin structure, maintaining the regulated state of these genes, leading to lifespan extension (Fig. 6C). The next challenge is to identify the specific transcript(s) that are down regulated by the formation of a tighter chromatin structure that lead to lifespan extension.
Relevance to lifespan extension in higher eukaryotes and via other pathways
The ultimate goal of this field is to achieve lifespan and healthspan extension in humans. As such, it is critically important to consider the potential relevance of our results in the yeast system to the situation in higher eukaryotes. Consistent with the reduction in histone proteins that we observed in aged yeast, we have found that levels of histone H3 protein decrease in mitotically active mouse tissues during aging (unpublished data), suggesting that histone protein loss may also occur in mammals during aging. This could be the cause of the elevated transcript levels that are characteristic during the aging of renewable tissues in mice (Warren et al., 2007). By examining the published transcriptome analyses, it is apparent that at least one regimen of lifespan extension in C. elegans is accompanied by increased histone expression (McColl et al., 2008). Furthermore, calorie restriction in mice is accompanied by increased histone expression (Barger et al., 2008). It will be important to determine whether the increased histone transcript levels during these lifespan regimens in higher eukaryotes are mirrored in a higher histone protein level. Indeed, we find increased histone transcript levels of 4 of the 6 cell cycle regulated histone genes in our long-lived tor1 yeast mutants (Fig. S3A), while histone protein levels are decreased in the old tor1 mutants and during CR (Fig. 5E & 5F), suggesting that they are trying to compensate for the depletion of histone proteins by transcribing more histones. Presumably the lower level of histone proteins in the yeast tor1 mutants and during CR (Fig. 5E & 5F) is a reflection of the greatly decreased degree of protein synthesis that occurs during nutritional stress and in the absence of Tor1. Our model leads us to ask why deletion of TOR1 or CR extends lifespan if they have lower histone protein levels than normal in their old cells? Presumably the pathway by which deletion of TOR1 and CR extends lifespan, which is largely unknown, can function without the need for extra histones to repress the key aging transcript(s) that are inappropriately generated during aging. This may be due to the fact that the reduced protein synthesis of these key aging transcripts during CR or Tor1 inactivation circumvents a requirement for their transcriptional repression in old cells for extended lifespan. Given the conservation of the mechanistic basis of aging, it will be necessary to identify these key aging transcripts to better understand and potentially ameliorate the age-related rise of human disease.
Experimental Procedures
Yeast Strains and Media
Yeast strains are described in Supplemental Table I. Replicative lifespan assays were performed on YPD with 2% glucose or YP with 1 % raffinose and 0.5%, 1 % or 2% galactose as indicated. Plasm ids expressing histones were maintained by growth in synthetic complete media lacking the appropriate amino acid and supplemented with raffinose and galactose for strains carrying galactose inducible promoter fusions.
Lifespan Analysis
Replicative lifespan of virgin mothers cells was determined as described previously (Kennedy et al., 1994). The number of mother cells analyzed for each Figure is itemized in Suppl. Table 2. Yeast cells were kept overnight at 12°C to slow cell division. Yeast were initially grown on YP glycerol plates to eliminate yeast cells lacking mitochondria. For yeast with extended lifespan, the Wilcoxon Rank-Sum Test was used to determine statistical significance. The Wilcoxon.test function was used in the R software version 2.80 and p values less than the significance cut off of 0.05 are given in the figures and in Suppl. Table 2.
Isolation of Young and Old Yeast Cells
Old yeast cells were isolated as described previously by using EZ Link Sulfo-NHL-LC-LC-Biotin (Pierce) to label cell surface proteins and MagnaBind Streptavidin beads (Thermo Scientific) (Lin et al., 2001). The old cells generated by this protocol had divided an average 8 times, as determined by calcafluor staining. The “very old” cells were isolated by two sequential rounds of this affinity purification, and had divided an average of 15 times, as determined by calcafluor staining.
For isolating older cells, we utilized the MEP as described in (Lindstrom and Gottschling, 2009). For the chromatin fractionations, strains UC5181 and DTY011 were grown for a total of 36 hours in the presence of estradiol, DT009 and DT010 for 16 hours. For ChIP experiments in galactose, UC5181 was grown for 72 hours total.
Reverse Transcriptase Real-time PCR
RNA was analyzed by real-time RT-PCR analysis using a Roche Light Cycler and SYBR green detection. Primer sequences are given in the supplemental methods.
Quantitative Western Blotting Analysis, ChIP and Chromatin Fractionations
Total protein extracts were isolated by boiling in Llaemli buffer prior to loading gels. Anti-rat and anti-rabbit secondary antibodies were used that fluoresced at 800nm and 700nm respectively. Membranes were scanned with a LiCor Odyssey scanning system and their software was used to quantitate the bands, using the average density method. The methods for ChIP were published previously (Williams et al., 2008) and are detailed in the supplemental methods section. The chromatin fractionation was performed using a variation of the method published in (Frei and Gasser, 2000), and is detailed in the supplemental methods section.
Gcn4 Reporter Assay and Polysome Profiling
The analysis of Gcn4 translation used a dual reporter assay as previously described (Steffen et al., 2008). Briefly, a reporter plasm id pVW31 containing the GCN4 promoter fused to firefly luciferase and Renilla luciferase under the control of a constitutive promoter was used. Cells were grown overnight in synthetic media lacking uracil to maintain the plasmid. Before lysing, cells were grown 2 hrs in YPD to minimize GCN4 expression due to nutrient stress. The Promega Dual-Luciferase Reporter Assay was used along with a Luminoskan Ascent to monitor expression. Firefly luciferase was normalized to Renilla luciferase activity. For polysome profiling, cells were grown in 75 mL of YPD to an OD of 0.8. Cultures were centrifuged and cooled with ice-cold YPD containing 100 μg/ml cycloheximide. After centrifuging again, cells were rinsed with ice-cold ddH2O containing 100 μg/ml cycloheximide and polysome profiles were measured as previously described (Steffen et al., 2008).
Supplementary Material
Acknowledgments
We thank Candice Wike for assistance and Brian Kennedy for sharing data prior to publication. We are grateful to William Feser for aiding in the statistical analysis. We thank Dan Gottschling, Brian Kennedy, Michael Grunstein, Allain Verreault, Fred Winston, Valter Longo, David Botstein, David Toczyski, Zhigou Zhang, Rohinton Kamakaka and Namrita Dhillon for kindly providing strains and plasmids. This work was supported by NIH grant GM64475 (to JKT), T32-GM08370 (to JF) and University of Colorado Cancer Center Aging Initiative seed grant funding to JKT.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adkins MW, Carson JJ, English CM, Ramey CJ, Tyler JK. The histone chaperone anti-silencing function 1 stimulates the acetylation of newly synthesized histone H3 in S-phase. J Biol Chem. 2007;282:1334–1340. doi: 10.1074/jbc.M608025200. [DOI] [PubMed] [Google Scholar]
- Barger JL, Kayo T, Pugh TD, Prolla TA, Weindruch R. Short-term consumption of a resveratrol-containing nutraceutical mixture mimics gene expression of long-term caloric restriction in mouse heart. Exp Gerontol. 2008;43:859–866. doi: 10.1016/j.exger.2008.06.013. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Tong JK, Schreiber SL. Genomewide studies of histone deacetylase function in yeast. Proc Natl Acad Sci U S A. 2000;97:13708–13713. doi: 10.1073/pnas.250477697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busuttil R, Bahar R, Vijg J. Genome dynamics and transcriptional deregulation in aging. Neuroscience. 2007;145:1341–1347. doi: 10.1016/j.neuroscience.2006.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Carson JJ, Feser J, Tamburini B, Zabaronick S, Linger J, Tyler JK. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell. 2008;134:231–243. doi: 10.1016/j.cell.2008.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang W, Steffen KK, Perry R, Dorsey JA, Johnson FB, Shilatifard A, Kaeberlein M, Kennedy BK, Berger SL. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defossez PA, Prusty R, Kaeberlein M, Lin SJ, Ferrigno P, Silver PA, Keil RL, Guarente L. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol Cell. 1999;3:447–455. doi: 10.1016/s1097-2765(00)80472-4. [DOI] [PubMed] [Google Scholar]
- Fillingham J, Recht J, Silva AC, Suter B, Emili A, Stagljar I, Krogan NJ, Allis CD, Keogh MC, Greenblatt JF. Chaperone Control of the Activity and Specificity of the Histone H3 Acetyltransferase Rtt109. Mol Cell Biol. 2008 doi: 10.1128/MCB.00182-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel S, Rogina B. Drosophila longevity is not affected by heterochromatin-mediated gene silencing. Aging Cell. 2005;4:53–56. doi: 10.1111/j.1474-9726.2005.00143.x. [DOI] [PubMed] [Google Scholar]
- Frei C, Gasser SM. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev. 2000;14:81–96. [PMC free article] [PubMed] [Google Scholar]
- Fuks F. DNA methylation and histone modifications: teaming up to silence genes. Curr Opin Genet Dev. 2005;15:490–495. doi: 10.1016/j.gde.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Gottlieb S, Esposito RE. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 1989;56:771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- Green EM, Antczak AJ, Bailey AO, Franco AA, Wu KJ, Yates JR, 3rd, Kaufman PD. Replication-independent histone deposition by the HIR complex and Asf1. Curr Biol. 2005;15:2044–2049. doi: 10.1016/j.cub.2005.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- Han J, Zhou H, Horazdovsky B, Zhang K, Xu RM, Zhang Z. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science. 2007a;315:653–655. doi: 10.1126/science.1133234. [DOI] [PubMed] [Google Scholar]
- Han J, Zhou H, Li Z, Xu RM, Zhang Z. Acetylation of lysine 56 of histone H3 catalyzed by RTT109 and regulated by ASF1 is required for replisome integrity. J Biol Chem. 2007b;282:28587–28596. doi: 10.1074/jbc.M702496200. [DOI] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW, 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- Kaufman PD, Cohen JL, Osley MA. Hir proteins are required for position-dependent gene silencing in Saccharomyces cerevisiae in the absence of chromatin assembly factor I. Mol Cell Biol. 1998;18:4793–4806. doi: 10.1128/mcb.18.8.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman PD, Kobayashi R, Stillman B. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- Kennedy BK, Austriaco NR, Jr, Guarente L. Daughter cells of Saccharomyces cerevisiae from old mothers display a reduced life span. J Cell Biol. 1994;127:1985–1993. doi: 10.1083/jcb.127.6.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Benguria A, Lai CY, Jazwinski SM. Modulation of life-span by histone deacetylase genes in Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:3125–3136. doi: 10.1091/mbc.10.10.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Villeponteau B, Jazwinski SM. Effect of replicative age on transcriptional silencing near telomeres in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1996;219:370–376. doi: 10.1006/bbrc.1996.0240. [DOI] [PubMed] [Google Scholar]
- Lamming DW, Latorre-Esteves M, Medvedik O, Wong SN, Tsang FA, Wang C, Lin SJ, Sinclair DA. HST2 mediates SIR2-independent life-span extension by calorie restriction. Science. 2005;309:1861–1864. doi: 10.1126/science.1113611. [DOI] [PubMed] [Google Scholar]
- Le S, Davis C, Konopka JB, Sternglanz R. Two new S-phase-specific genes from Saccharomyces cerevisiae. Yeast. 1997;13:1029–1042. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1029::AID-YEA160>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, Verreault A, Zhang Z. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell. 2008;134:244–255. doi: 10.1016/j.cell.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SS, Manchester JK, Gordon JI. Enhanced gluconeogenesis and increased energy storage as hallmarks of aging in Saccharomyces cerevisiae. J Biol Chem. 2001;276:36000–36007. doi: 10.1074/jbc.M103509200. [DOI] [PubMed] [Google Scholar]
- Lindstrom DL, Gottschling DE. The mother enrichment program a genetic system for facile replicative life span analysis in Saccharomyces cerevisiae. Genetics. 2009;183:413–422. 411SI–413SI. doi: 10.1534/genetics.109.106229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Maas NL, Miller KM, DeFazio LG, Toczyski DP. Cell cycle and checkpoint regulation of histone H3 K56 acetylation by Hst3 and Hst4. Mol Cell. 2006;23:109–119. doi: 10.1016/j.molcel.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Maslov AY, Vijg J. Genome instability, cancer and aging. Biochim Biophys Acta. 2009;1790:963–969. doi: 10.1016/j.bbagen.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mays-Hoopes LL, Brown A, Huang RC. Methylation and rearrangement of mouse intracisternal a particle genes in development, aging, and myeloma. Mol Cell Biol. 1983;3:1371–1380. doi: 10.1128/mcb.3.8.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McColl G, Killilea DW, Hubbard AE, Vantipalli MC, Melov S, Lithgow GJ. Pharmacogenetic analysis of lithium-induced delayed aging in Caenorhabditis elegans. J Biol Chem. 2008;283:350–357. doi: 10.1074/jbc.M705028200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D. Enzymatic activities of Sir2 and chromatin silencing. Curr Opin Cell Biol. 2001;13:232–238. doi: 10.1016/s0955-0674(00)00202-7. [DOI] [PubMed] [Google Scholar]
- Myung K, Pennaneach V, Kats ES, Kolodner RD. Saccharomyces cerevisiae chromatin-assembly factors that act during DNA replication function in the maintenance of genome stability. Proc Natl Acad Sci U S A. 2003;100:6640–6645. doi: 10.1073/pnas.1232239100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramey CJ, Howar S, Adkins M, Linger J, Spicer J, Tyler JK. Activation of the DNA damage checkpoint in yeast lacking the histone chaperone anti-silencing function 1. Mol Cell Biol. 2004;24:10313–10327. doi: 10.1128/MCB.24.23.10313-10327.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea SL, Wu D, Cypser JR, Vaupel JW, Johnson TE. A stress-sensitive reporter predicts longevity in isogenic populations of Caenorhabditis elegans. Nat Genet. 2005;37:894–898. doi: 10.1038/ng1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert T, Dervins D, Fabre F, Gangloff S. Mrc1 and Srs2 are major actors in the regulation of spontaneous crossover. EMBO J. 2006;25:2837–2846. doi: 10.1038/sj.emboj.7601158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogina B, Helfand SL, Frankel S. Longevity regulation by Drosophila Rpd3 deacetylase and caloric restriction. Science. 2002;298:1745. doi: 10.1126/science.1078986. [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- Singer MS, Kahana A, Wolf AJ, Meisinger LL, Peterson SE, Goggin C, Mahowald M, Gottschling DE. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics. 1998;150:613–632. doi: 10.1093/genetics/150.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal RP, Mays-Hoopes LL, Eichhorn GL. DNA methylation in aging of mice. Mech Ageing Dev. 1987;41:199–210. doi: 10.1016/0047-6374(87)90040-6. [DOI] [PubMed] [Google Scholar]
- Smith ED, Kennedy BK, Kaeberlein M. Genome-wide identification of conserved longevity genes in yeast and worms. Mech Ageing Dev. 2007;128:106–111. doi: 10.1016/j.mad.2006.11.017. [DOI] [PubMed] [Google Scholar]
- Smith ED, Tsuchiya M, Fox LA, Dang N, Hu D, Kerr EO, Johnston ED, Tchao BN, Pak DN, Welton KL, et al. Quantitative evidence for conserved longevity pathways between divergent eukaryotic species. Genome Res. 2008;18:564–570. doi: 10.1101/gr.074724.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Caputo E, Boeke JD. A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol Cell Biol. 1999;19:3184–3197. doi: 10.1128/mcb.19.4.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector MS, Osley MA. The HIR4-1 mutation defines a new class of histone regulatory genes in Saccharomyces cerevisiae. Genetics. 1993;135:25–34. doi: 10.1093/genetics/135.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector MS, Raff A, DeSilva H, Lee K, Osley MA. Hir1p and Hir2p function as transcriptional corepressors to regulate histone gene transcription in the Saccharomyces cerevisiae cell cycle. Mol Cell Biol. 1997;17:545–552. doi: 10.1128/mcb.17.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen KK, MacKay VL, Kerr EO, Tsuchiya M, Hu D, Fox LA, Dang N, Johnston ED, Oakes JA, Tchao BN, et al. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell. 2008;133:292–302. doi: 10.1016/j.cell.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinkraus KA, Kaeberlein M, Kennedy BK. Replicative aging in yeast: the means to the end. Annu Rev Cell Dev Biol. 2008;24:29–54. doi: 10.1146/annurev.cellbio.23.090506.123509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton A, Bucaria J, Osley MA, Sternglanz R. Yeast asf1 protein is required for cell cycle regulation of histone gene transcription. Genetics. 2001;158:587–596. doi: 10.1093/genetics/158.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya M, Dang N, Kerr EO, Hu D, Steffen KK, Oakes JA, Kennedy BK, Kaeberlein M. Sirtuin-independent effects of nicotinamide on lifespan extension from calorie restriction in yeast. Aging Cell. 2006;5:505–514. doi: 10.1111/j.1474-9726.2006.00240.x. [DOI] [PubMed] [Google Scholar]
- Tyler JK, Adams CR, Chen SR, Kobayashi R, Kamakaka RT, Kadonaga JT. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- Warren LA, Rossi DJ, Schiebinger GR, Weissman IL, Kim SK, Quake SR. Transcriptional instability is not a universal attribute of aging. Aging Cell. 2007;6:775–782. doi: 10.1111/j.1474-9726.2007.00337.x. [DOI] [PubMed] [Google Scholar]
- Williams SK, Truong D, Tyler JK. Acetylation in the globular core of histone H3 on lysine-56 promotes chromatin disassembly during transcriptional activation. Proc Natl Acad Sci U S A. 2008;105:9000–9005. doi: 10.1073/pnas.0800057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrick JJ, Holstege FC, Jennings EG, Causton HC, Shore D, Grunstein M, Lander ES, Young RA. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature. 1999;402:418–421. doi: 10.1038/46567. [DOI] [PubMed] [Google Scholar]
- Xu F, Zhang K, Grunstein M. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell. 2005;121:375–385. doi: 10.1016/j.cell.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Xu F, Zhang Q, Zhang K, Xie W, Grunstein M. Sir2 deacetylates histone H3 lysine 56 to regulate telomeric heterochromatin structure in yeast. Mol Cell. 2007;27:890–900. doi: 10.1016/j.molcel.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yiu G, McCord A, Wise A, Jindal R, Hardee J, Kuo A, Shimogawa MY, Cahoon L, Wu M, Kloke J, et al. Pathways change in expression during replicative aging in Saccharomyces cerevisiae. J Gerontol A Biol Sci Med Sci. 2008;63:21–34. doi: 10.1093/gerona/63.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabaronick SR, Tyler JK. The histone chaperone anti-silencing function 1 is a global regulator of transcription independent of passage through S phase. Mol Cell Biol. 2005;25:652–660. doi: 10.1128/MCB.25.2.652-660.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn JM, Kim SK. Systems biology of aging in four species. Curr Opin Biotechnol. 2007;18:355–359. doi: 10.1016/j.copbio.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.