Abstract
Gene amplifications in the 17q chromosomal region are observed frequently in breast cancers. An integrative bioinformatics analysis of this region nominated the MAP3K 3 gene as a potential therapeutic target in breast cancer. This gene encodes mitogen-activated protein kinase kinase kinase 3 (MAP3K3/MEKK3), which has not yet been reported to be associated with cancer-causing genetic aberrations. We found that MAP3K3 was amplified in approximately 8–20% of breast cancers. Knockdown of MAP3K3 expression significantly inhibited cell proliferation and colony formation in MAP3K3-amplified breast cancer cell lines MCF-7 and MDA-MB-361 but not in MAP3K3 non-amplified breast cancer cells. Knockdown of MAP3K3 expression in MAP3K3-amplified breast cancer cells sensitized breast cancer cells to apoptotic induction by TNFα and TRAIL, as well as doxorubicin, VP-16 and fluorouracil, three commonly used chemotherapeutic drugs for treating breast cancer. In addition, ectopic expression of MAP3K3, in collaboration with Ras, induced colony formation in both primary mouse embryonic fibroblasts and immortalized human breast epithelial cells (MCF-10A). Combined, these results suggest that MAP3K3 contributes to breast carcinogenesis and may endow resistance of breast cancer cells to cytotoxic chemotherapy. Therefore, MAP3K3 may be a valuable therapeutic target in patients with MAP3K3-amplified breast cancers, and blocking MAP3K3 kinase activity with a small molecule inhibitor may sensitize MAP3K3-amplified breast cancer cells to chemotherapy.
Keywords: MAP3K3, oncogene, breast cancer, chemo-resistance
Introduction
Activation of oncogenes through DNA amplification and over-expression plays important roles in cancer initiation and progression. Classical chromosomal analysis and large-scale genome projects have revealed a number of amplified genes in breast cancers, including HER2, CCND1, PAK1 and RPS6KB1, but only a few have been demonstrated to be causally involved and therapeutically important in human breast cancers [1–3]. The long arm of chromosome 17 is a frequent site of cancer-associated genetic anomalies. Amplification of several genomic regions in 17q, such as 17q23 and 17q12 amplicons, has been identified in breast cancers [4]. This cytogenetic anomaly is strongly associated with poor prognosis and is a significant predictor of relapse in breast cancers [5–7]. These amplifications are typically discontinuous and complex in structure, suggesting that multiple oncogenes in this chromosomal segment may be co-selected during breast carcinogenesis. To discover potential novel oncogenes in this chromosome region, we leveraged a concept signature (ConSig) analysis to quickly identify cancer-relevant genes from thousands of candidates [8]. This analysis is based on the fact that cancer-causal genes tend to share characteristic molecular concepts (signature concepts), including cancer-related gene ontologies, pathways, protein interactions and gene domains. Such ‘signatures’ of molecular concepts can be used to nominate biologically meaningful cancer-relevant genes from a large number of candidate genes, by computing their strength of association with those cancer signature concepts. We applied this ConSig analysis to prioritize the amplified gene list on Chr17q, nominated based on a matched DNA copy number and gene expression datasets for breast cancers. The analysis revealed MAP3K3 as a potential new oncogene in breast cancer.
The MAP3K3 gene in 17q encodes mitogen-activated protein kinase kinase kinase 3 (MAP3K3/MEKK3), a serine/threonine protein kinase within the MAP3K family. Members of this protein family play important roles in mouse embryonic development and in growth factor and cytokine-induced signal transduction pathways [9–13]. However, the role of MAP3K3 in human cancers is poorly understood. In this study, we confirmed the deregulation of MAP3K3 in human breast cancer cell lines and tumour tissue specimens, and further explored the role of this gene in breast tumourigenesis as well as in the response of breast cancer cells to cytotoxic chemo-drugs. Our data provide compelling evidence that MAP3K3 has a critical role in breast tumourigenesis and may be an important therapeutic target.
Materials and methods
Cell lines, tissue specimens, expression vectors and antibodies
Mammary epithelial cell line MCF-10A and human breast cancer cell lines MCF-7, MDA-MB-361, MDA-MB-231, MDA-MB-435, MDA-MB-468 and SK-BR-3 were purchased from the American Type Culture Collection (Manassas, VA, USA) and maintained in the suggested medium with 10% fetal calf serum (FCS). MDA-MB-453 cells were kindly provided by Dr Ana M. Gonzalez-Angulo (MD Anderson Cancer Center). The retroviral expression vectors for RAS and Myc were provided by Dr Scott W Lowe. The retrovirus packing vector Pegpam 3e and RDF vectors were obtained from Dr Gianpietro Dotti. The PLC-ECO plasmid was provided by Dr Biao Zheng. The retroviral expression vector for MEKK3 was constructed by subcloning the MEKK3 into the pBabepuro vector. The antibodies for MAP3K3 (MEKK3; 611103), Vimentin (550513) and mouse (554002) were from BD Biosciences Pharmingen (San Diego, CA, USA). The antibodies for ICAM1 (4915S), mouse (7076S), rabbit (7074S) and PARP (9532S) were from Cell Signalling (Danvers, MA, USA). The antibody against β-Actin was from Sigma (St. Louis, MO, USA).
Integrative analysis of public copy number datasets for breast cancers
Agilent 244A two-channel array CGH datasets of breast cancers were compiled from the Gene Expression Omnibus (GSE20393; http://www.ncbi.nlm.nih.gov/geo). The differential ratio between the processed testing channel signal and the processed reference channel signal was calculated, after which the resulting relative DNA copy number data were log2-transformed, reflecting the DNA copy number difference between the testing and reference samples. Copy number data were segmented by the circular binary segmentation (CBS) algorithm [14]. Genomic loci with log2 relative copy number ≥ 0.75 were defined as amplification. To reveal potential drug targets from chromosome 17, we first identified all genes on this chromosome with genomic amplifications in > 10% of breast cancers. To reveal genes with gene expression primarily affected by copy number, we extracted matched gene expression data from GSE16534 (Affymetrix HuEx1.0 array) and correlated with the copy number data from GSE20393 through Pearson’s correlation analysis (153 samples have matched copy number and gene expression data). The candidate genes (n = 107) with increased gene expression correlating with copy number (R > 0.5) were then ranked with a ConSig score that revealed the most biologically meaningful genes underlying cancer. The ConSig score used in this study is available at: http://consig.cagenome.org (release 2). In addition, we also analysed an Affymetrix SNP 6.0 array dataset for 503 breast tumours from the Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov/). Normalized ’level 3’ data from TCGA were directly applied in the analysis.
Meta-analysis of public gene expression datasets for breast cancers
For correlation analysis of MAP3K3 with ICAM1 and vimentin, we compiled nine public breast tumour expression profiling datasets (Loi, GEO:GSE6532; Wang, GEO:GSE2034; Desmedt, GEO:GSE7390; Miller, GEO:GSE3494; Schmidt, GEO:GSE 11121; Zhang, GEO:GSE12093; Minn, GEO:GSE2603 and Chin, GEO:GSE5327; http://cancer.lbl.gov/breastcancer/data.php), including 1340 patients in total. Genes within each dataset were first normalized to standard deviations (SDs) from the median.
Fluorescence in situ hybridization
The BAC containing the MAP3K3 locus at chromosome 17q23 (RP11-51 F16) was purchased from Invitrogen. This BAC and a plasmid containing chromosome 17 centromeric sequence (pZ17-14) were fluorescently labelled with spectrum red and spectrum green, respectively (Vysis, Downers Grove, IL, USA), by the nick translation method. The map position of the BAC clone was confirmed on normal human metaphase spreads and then the labelled BAC probe was hybridized, together with a 17 centromere probe, to metaphase or interphase spreads of MCF-7 and MDA-MB-361 cell lines and interphase nuclei of touch preparations from primary breast tumour specimens. The slides were counterstained with DAPI, and the images were captured using the Quips Pathvysion System (Applied Imaging, Santa Clara, CA, USA). To determine the amplification status, 200 individual interphase nuclei were analysed for each cell line and primary tumour specimens. The criterion for amplification was > 5% of tumour nuclei displaying increased MAP3K3 copy number relative to the chromosome 17 centromeric probe signals and ploidy of the tumour cells.
MAP3K3 knockdown in breast cancer cell lines
A pSUPER-retro vector was used to generate shRNA plasmids for human MAP3K3. The following target sequences were selected: 5′-AACATGATTGTTCACCGGGAC-3′ (sh-MAP3K3-13) and 5′-AAGTACCTGGAATGCAGAGTA-3′ (sh-MAP3K3-15). The sh-control sequence was 5′-CGTCTTTTCGGACTTAGAGAG-3′. The authenticity of these plasmids was confirmed by DNA sequencing. The resulting pSU-PER vectors were then transfected into the packaging cell line HEK293T with retrovirus packaging vector Pegpam 3e and RDF, using FuGENE 6 (Roche) to make retroviruses expressing small hairpin RNA against MAP3K3. The medium containing retroviruses was collected 48 h after transfection. Two other lentiviruses with small hairpin RNAs against MAP3K3 (TRCN0000002306 and TRCN0000002307) were obtained from Sigma. Breast cancer cells were incubated with virus-containing medium in the presence of 4 mg/ml polybrene. Stable cell lines were established after 6 days of puromycin (2 µg/ml) selection.
Establishment of stable MEF cell lines expressing RAS, Myc or MAP3K3
The pBabe or pWZL retroviral empty vector or vectors for K-RASV12D, c-Myc or MAP3K3 were co-transfected with retrovirus packing vector Pegpam 3e and PLC-ECO into HEK293T cells to obtain retroviral supernatant. Viral supernatant were collected after 48 and 72 h. Wild-type early passage MEFs were incubated with virus-containing medium in the presence of 4 mg/ml polybrene (Sigma). Stable cell lines were established after 6 days of puromycin (2 µg/ml) and/or hygromycin (500 µg/ml) selection.
Results
Integrative analyses of public genomic datasets reveal MAP3K3 as a potential oncogene in breast cancers
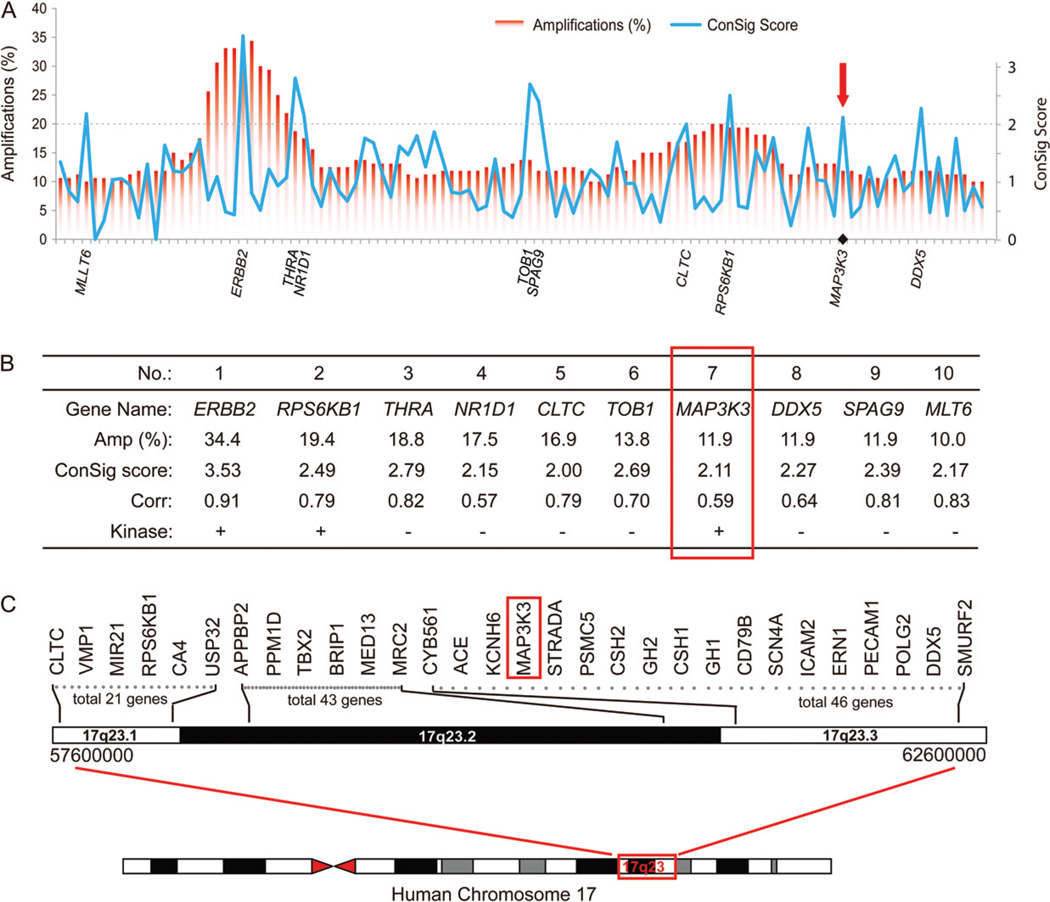
To identify new oncogenes from the chromosome 17q region, we compiled copy number and matched gene expression datasets for 153 breast cancers from Gene Expression Omnibus (GSE20393 and GSE16534) [15]. Based on these data, we identified 107 genes in chromosome 17q that displayed frequent genomic amplifications and correlated gene expression changes. To determine the potential roles of these candidate genes in tumourigenesis, we leveraged the concept signature (ConSig) analysis developed in our previous study [8]. Ranking these 107 genes by ConSig score revealed 10 leading candidates, among which three encode druggable kinases (ERBB2, RPS6KB1 and MAP3K3) (Figure 1A, B). ERBB2 and RPS6KB1 are well-known oncogenes and primary targets of genomic amplifications in breast tumours. The third candidate, MAP3K3, encodes a serine/threonine protein kinase and is located in an amplified region discrete from the ERBB2 and RPS6KB1 amplicons (Figure 1). This protein kinase regulates a number of important signalling pathways, including NF-κB, ERK and JNK signalling [9–13]; therefore, it has potential to be a new therapeutic target for breast cancer.
Figure 1.
Integrative analyses of public genomics data reveal MAP3K3 as a potential oncogene target in breast cancers. (A) ConSig score revealing primary oncogene targets of genomic amplifications in 17q. The x axis shows 107 candidate genes sorted by their physical locations in the chromosome. Red bars show the frequency of genomic amplification for each candidate gene. ConSig scores are shown in the blue line chart, with the spikes indicating biologically important genes. The top 10 candidate genes benchmarked by ConSig score are labelled on the x axis, with the cut-off shown by the dashed line. (B) List of the top 10 candidate genes by integrative analysis of public genomic datasets. (C) A physical map of the chromosome 17q23 region, with MAP3K3 indicated in red.
Detection of MAP3K3 amplification and over-expression in breast cancer cell lines and primary tumour specimens
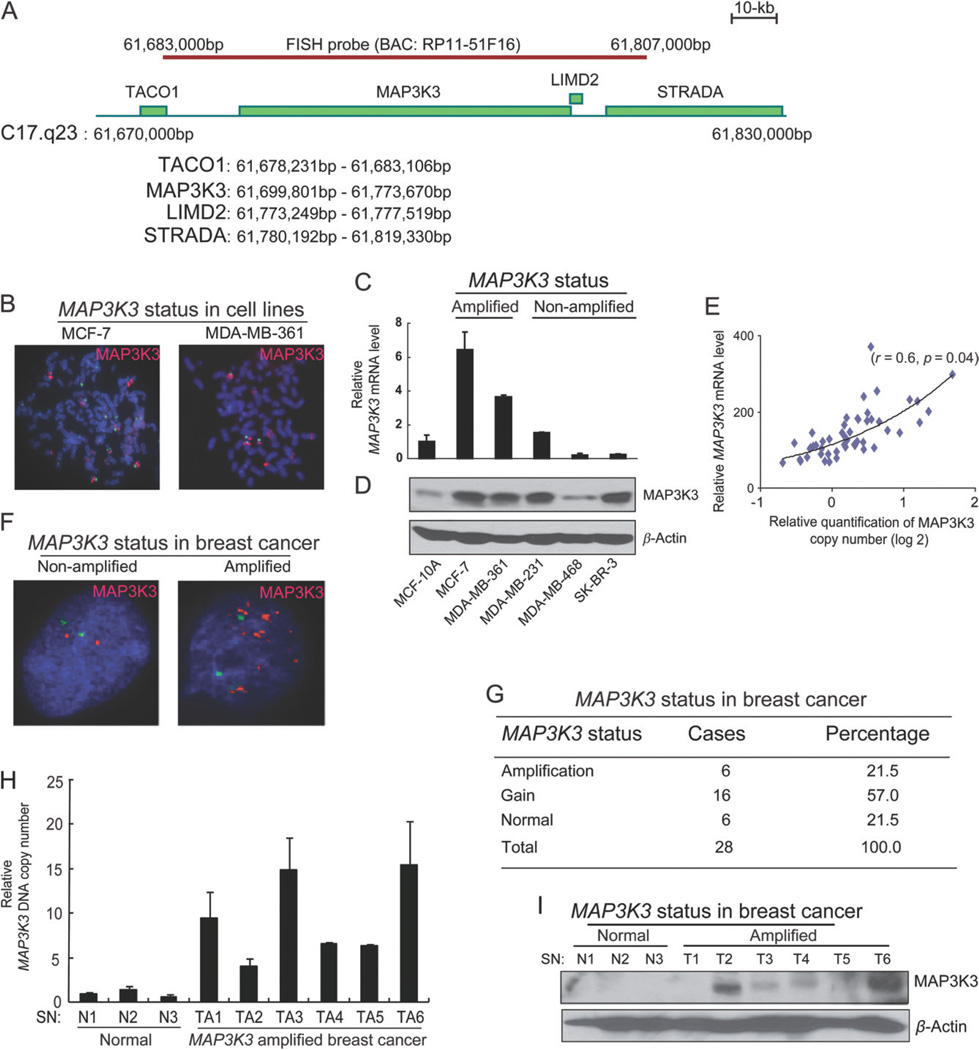
To experimentally confirm genomic amplification at the MAP3K3 locus in breast cancer cells, we first performed fluorescence in situ hybridization (FISH) analysis on five human breast cancer cell lines (MCF-7, MDA-MB-361, MDA-MB-231, MDA-MB-468 and SK-BR-3) as well as a non-transformed breast epithelial cell line (MCF-10A). We used a BAC probe from the 17q23 chromosomal DNA segment (RP11-51 F16), which covers a 123 kb region containing two known full-length genes, MAP3K3 and LIMD2 (LIM domain-containing protein 2) (Figure 2A), the latter of which has no known function. The FISH probe can specifically recognize the MAP3K3 locus in MAP3K3 non-amplified cell line MDA-MB-435 (see supplementary material, Figure S1A). The analysis of the hybridization patterns on metaphase and interphase chromosomes revealed 10 copies of this locus in MCF-7 cells, 16 copies in MDA-MB-361 and two copies in the other cell lines (after the data were normalized to the number of chromosome 17 centrosomes) (Figure 2B and data not shown). The FISH data suggest that MAP3K3 is amplified in a subset of human breast cancer cell lines.
Figure 2.
MAP3K3 is amplified and over-expressed in a subset of breast cancers. (A) A schematic presentation of the BAC (RP11-51F16) FISH probe and the genes located within this probe. (B) FISH analysis of the indicated cell lines using the BAC probe (red) and a chromosome 17 centromere probe (pZ17-14, green). (C) Quantitative real-time RT–PCR showing MAP3K3 transcript levels, which were normalized to GAPDH and represented as fold-change relative to MCF-10A. Error bars represent SD from triplicate samples. (D) Immunoblotting analysis for MAP3K3 in the indicated cell lines. (E) The mRNA level of MAP3K3 positively correlates with its gene copy number in 51 breast cancer cell lines. (F) Representative images of FISH analysis of MAP3K3 in two primary breast cancers. (G) Summary of MAP3K3 status in 28 primary breast cancer specimens analysed by FISH. (H) The analyses of MAP3K3 DNA copy numbers by quantitative genomic PCR in primary breast cancers. (I) Immunoblotting analysis for MAP3K3 in normal breast tissue and breast cancers with MAP3K3 amplification.
Next, we examined whether changes of the DNA copy number in the locus in these breast cancer cell lines corresponded to an increase at the MAP3K3 transcript and protein levels. Quantitative reverse transcriptase–polymerase chain reactions (qRT–PCR) revealed that the MAP3K3 transcript level was much higher in MCF-7 and MDA-MB-361 cells than in MCF-10A control (Figure 2C). Western blotting revealed that the MAP3K3 protein level was also higher in MCF-7 and MDA-MB-361 than in MCF-10A (Figure 2D). In addition, increased MAP3K3 protein levels were also observed in MDA-MB-231 and SK-BR3, which bear no MAP3K3 amplification (Figure 2D), suggesting that MAP3K3 protein levels can be up-regulated in some human breast cancer cell lines by a gene amplification-independent mechanism. Figure S1B (see supplementary material) summarizes the MAP3K3 amplification status, MAP3K3 protein expression level and other important breast cancer biomarkers in the above analysed breast cancer cell lines. Furthermore, we analysed the MAP3K3 gene copy number and expression in 51 breast cancer cell lines in previously published datasets [16]. The MAP3K3 mRNA levels correlated positively with its gene copy numbers in these breast cancer cell lines (Figure 2E). Interestingly, the luminal subtype exhibited a significantly higher MAP3K3 mRNA level and gene copy numbers than the basal A subtype (see supplementary material, Figure S1C, D). Collectively, these results suggest that the MAP3K3 gene is amplified and/or over-expressed in a subset of human breast cancer cell lines and that the MAP3K3 protein level is often up-regulated in these breast cancer cell lines.
We next sought to experimentally validate genomic amplification and over-expression of MAP3K3 in primary human breast tumours. We performed FISH analysis using the same probe (RP11-51 F16) on 28 tumour samples of invasive ductal carcinoma of the breast. Amplification (more than six copies) of this locus was observed in six cases and gain (three to six copies) was observed in 16 specimens (Figure 2F, G). Then we used quantitative genomic PCR analysis to confirm MAP3K3 copy number expansion in this locus. Six amplified cases showed higher levels of MAP3K3 genomic DNA than all three normal breast specimens (Figure 2H). Next, we investigated whether the MAP3K3 protein was also increased in primary human breast cancer tissue samples with MAP3K3 amplification. The MAP3K3 protein levels in the samples of six breast tumour tissues that displayed 17q23 amplification, based on FISH data, were compared with that in the three samples of normal breast tissues. Four of these six tumour samples showed increased MAP3K3 protein levels compared to the control normal breast tissues (Figure 2I). Together, these data demonstrate that MAP3K3 is amplified and over-expressed in a subset of human primary breast cancers. Of note, in this relatively small cohort of cases, we did not find a correlation of MAP3K3 amplification with a specific subtype (ER+, HER2+ or triple-negative; data not shown). Furthermore, Oncomine analysis did not detect a correlation of MAP3K3 over-expression with ER, PR or HER2 status (see supplementary material, Figure S2).
Of note, in the above copy number dataset of 160 breast tumours, besides genomic amplifications in 19 cases (12%), we also observed rare unbalanced breakpoints at the MAP3K3 locus in two cases. Further interrogation of the copy number data of 503 breast tumours from the Cancer Genome Atlas (TCGA) revealed unbalanced breakpoints at the MAP3K3 locus in eight cases, in addition to the genomic amplifications in 8% of tumours (see supplementary material, Figure S3A). Copy number data for 51 breast cancer cell lines revealed a similar unbalanced breakpoint at the MAP3K3 locus in the luminal breast cancer cell line MDA-MB-453 (see supplementary material, Figure S3A). To test whether the unbalanced breakpoint in MDA-MB-453 was associated with a gene rearrangement, we analysed paired-end transcriptome sequencing data for this cell line and detected a MYO15B–MAP3K3 fusion transcript, which was verified by reverse-transcription PCR and capillary sequencing (see supplementary material, Figure S3B, C).
MAP3K3 amplification/over-expression promotes the growth of breast cancer cells and MAP3K3 is a potential therapeutic target in breast cancer
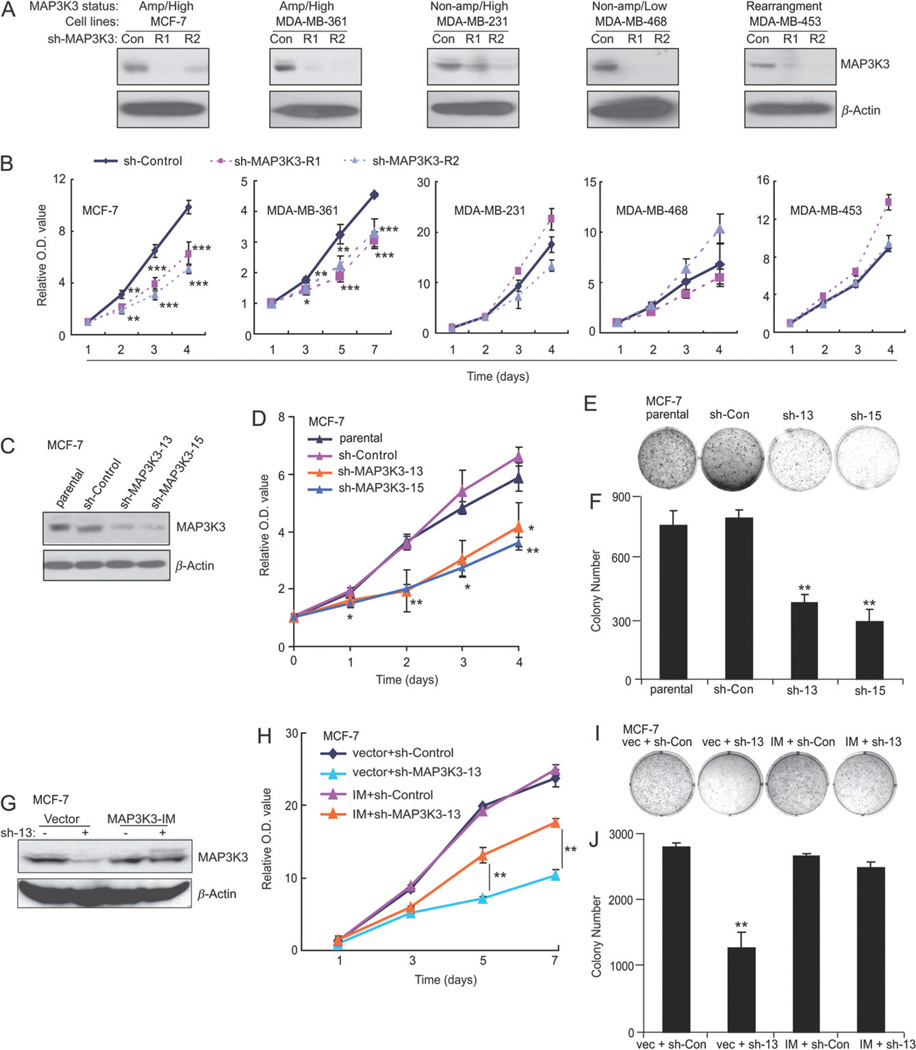
After discovering frequent MAP3K3 amplification and over-expression in human breast tumours, we next determined whether MAP3K3 amplification/over-expression was required for breast cancer cell proliferation and could be a potential therapeutic target in breast cancer. We assessed the effect of MAP3K3 stable knockdown in five cell lines with different MAP3K3 amplification/over-expression status (amplification/over-expression, MCF-7 and MDA-MB-361; non-amplification/over-expression, MDA-MB-231; non-amplification/low expression, MDA-MB-468; MAP 3K3 rearranged, MDA-MB-453). Two lentivirus-mediated shRNAs successfully knocked down MAP3K3 expression in all tested five cell lines (Figure 3A). Knockdown MAP3K3 expression in MAP3K3 amplified cell lines (MCF-7 and MDA-MB-361) significantly inhibited cell proliferation. However, knockdown MAP3K3 expression in MAP3K3 non-amplified cell lines (MDA-MB-231, MDA-MB-468 and MDA-MB-453) showed no inhibitory effect on cell proliferation. These data suggest that MAP3K3 is required for breast cancer proliferation in MAP3K3-amplified breast cancers. In addition, lentivirus-mediated knockdown of MAP3K3 showed that MAP3K3 is also required for colony formation of MCF-7 but not MDA-MB-231 cells (see supplementary material, Figure S4A–D), further supporting that MAP3K3 is required for breast cancer malignancy in MAP3K3-amplified breast cancers.
Figure 3.
MAP3K3 knockdown causes a decreased cell proliferation and colony formation potential in MCF-7 and MDA-MB-361 cells. (A) Immunoblotting analysis showing MAP3K3 knockdown in MCF-7, MDA-MB-361, MDA-MB-231, MDA-MB-468 and MDA-MB-453 cells by lentivirus-mediated expression of shRNAs. (B) Cell growth curve assay showing a proliferation defect in MAP3K3 knockdown MCF-7 and MDA-MB-361 cells. The cell counting kit-8 (CCK-8) was used to measure relative cell numbers. Error bars represent SDs of six samples. (C) Immunoblotting analysis showing MAP3K3 knockdown in MCF-7 cells infected with retroviruses stably expressing the indicated shRNAs. (D) Cell growth curve assay showing a proliferation defect in MAP3K3 knockdown cells. Error bars represent the standard error of the mean (SEM) from three independent experiments. (E) Colony assay showing reduced colony formation in MAP3K3-knockdown cells (day 14). (F) Quantification of mean colony numbers in (E). Error bars represent SD from triplicate samples. (G) Over-expression of MAP3K3-IM rescues MAP3K3 expression in cells stably expressing sh-MAP3K3-13. (H) MAP3K3-IM-mediated rescue of the above proliferation defect. Error bars represent SD from triplicate samples. (I) MAP3K3-IM-mediated rescue of the colony formation defect caused by MAP3K3 knockdown. (J) Quantification of mean colony numbers in (I). Error bars represent SD from triplicate samples. *p < 0.05, **p < 0.01, sh-MAP3K3 versus sh-Control, t-test. Results from (D, E, H and I) are representative of three independent experiments.
To further confirm these results, we transduced MCF-7 cells with two other retrovirus-expressing MAP3K3 short-hairpin RNAs (sh-MAP3K3) to generate cell lines with a stable MAP3K3 knockdown. As expected, two sh-MAP3K3 sequences (# 13 and 15) resulted in a significant decrease in MAP3K3 protein levels in MCF-7 cells compared to the levels in cells with sh-control (Figure 3C). We then tested the effect of MAP3K3 knockdown on MCF-7 proliferation. Both stable MAP3K3 knockdown cell lines displayed a significant proliferation defect compared to the parental and sh-control cell lines, as we observed in lentivirus-mediated knockdown of MAP3K3 expression (Figure 3D). Consistent with the above result, in a soft agar colony-formation assay, the two knockdown cell lines also formed significantly fewer colonies than parental or sh-control cells (Figure 3E, F).
To confirm that this effect was not caused by off-target effects of shRNA, MCF-7 cells with stable expression of sh-MAP3K3-13 were transfected with a MAP3K3 shRNA-immune expression vector (MAP3K3-IM), which forced the expression of a full-length MAP3K3 that contained a mutated sh-MAP3K3 target sequence. MAP3K3-IM successfully restored the MAP3K3 protein levels (Figure 3G). As expected, MAP3K3-IM reversed both the proliferation defect (Figure 3H) and colony formation deficiency (Figure 3I, J) caused by sh-MAP3K3. Therefore, we conclude that MAP3K3 plays a critical role in MCF-7 cell proliferation and colony formation.
To confirm that the above results are not cell line-dependent, we tested the effect of MAP3K3 knockdown in another MAP3K3-amplified cell line, MDA-MB-361. We first confirmed the knockdown effect of both sh-MAP3K3 sequences on the expression of MAP3K3 in this cell line (see supplementary material, Figure S4E). As expected, MAP3K3 knockdown also inhibited cell proliferation and colony formation in MDA-MB-361 cells (see supplementary material, Figure S4F–H). Together, these experiments suggest that amplification and over-expression of MAP3K3 promote breast cancer growth, and that the inhibition of MAP3K3 may be helpful in the treatment of breast cancer that carries MAP3K3 amplification.
MAP3K3 mediates cell survival in human breast cancer cells, and contributes to the resistance of breast cancer cells to TNFα-, TRAIL-, doxorubicin-, VP-16- and fluorouracil (5-FU)-induced apoptosis
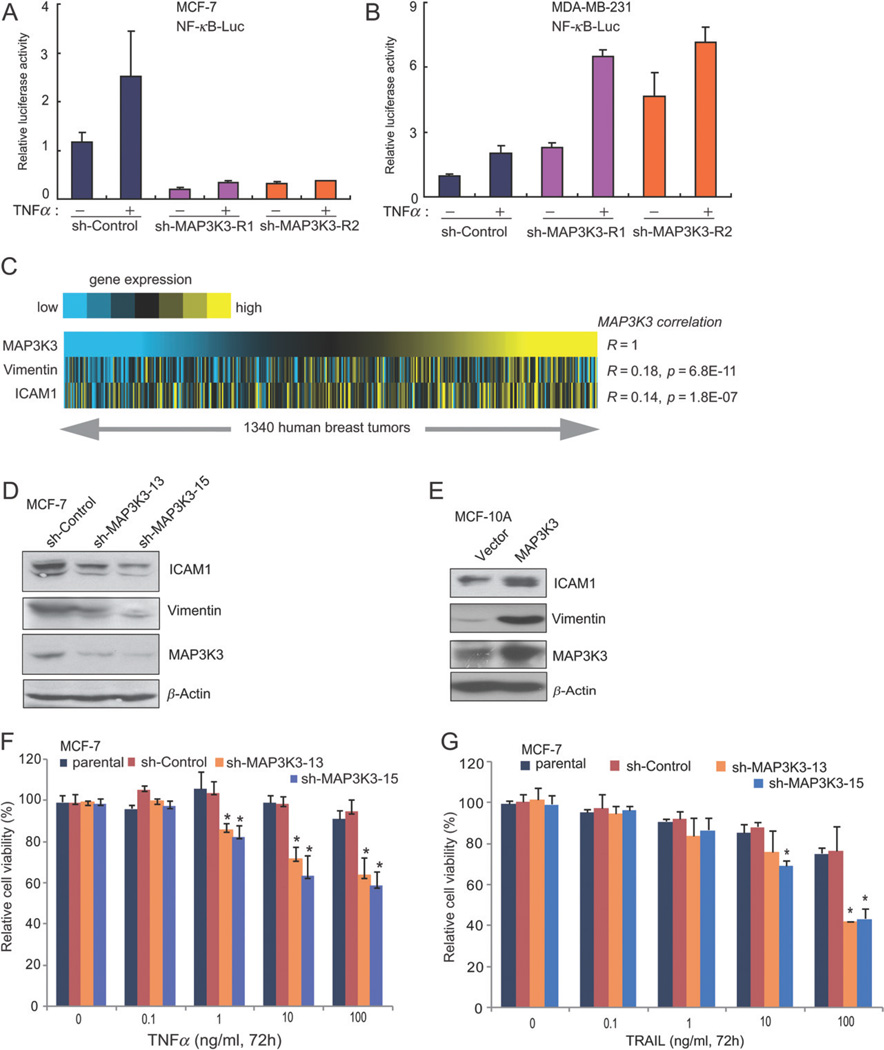
Previous studies in non-breast cell lines demonstrate that MAP3K3 mediates cytokine-induced NF-κB activation by phosphorylating and activating IKK, leading to phosphorylation and degradation of IκBα, releasing its binding partner RelA for nuclear translocation and transcriptional activation of anti-apoptosis targets [12,17–19]. Increased NF-κB activity is frequently present in human cancers, including breast tumours [20–22], and may enhance tumour cell survival [23,24]. Here, we found that knockdown expression of MAP3K3 in MCF-7 but not in MDA-MB-231 cells inhibited basal and TNFα-induced NF-κB activity (Figure 4A, B), suggesting that MAP3K3 is required for basal and TNFα-induced NF-κB activity in MAP3K3-amplified but not in non-amplified breast cancer cells. Furthermore, we found that expression of both NF-κB target genes (ICAM1 and vimentin) correlated with the expression of MAP3K3 across 1340 human breast cancers from nine public breast tumour expression profiling datasets (Figure 4C). ICAM1 and vimentin have been shown to play important roles in invasion and epithelial–mesenchymal transition and over-expression of these two genes correlated with poor breast cancer outcome in breast cancer patients [25–27]. Consistently, knockdown of MAP3K3 expression in MCF-7 cells resulted in decreased protein levels of ICAM1 and vimentin (Figure 4D). Conversely, over-expression of MAP3K3 in MCF-10A cells up-regulated the expression of ICAM1 and vimentin (Figure 4E). Next, we tested the effect of MAP3K3 knockdown in human breast cancer cells on TNFα-mediated NF-κB activation and apoptotic induction. In our assay, TNFα induced a dose-dependent loss of cell viability in MCF-7 cells stably expressing sh-MAP3K3, but not in the parental or sh-control MCF-7 cells (Figure 4F). We also compared the sh-MAP3K3 MCF-7 cells versus parental and sh-control MCF-7 cells for their response to TRAIL-induced apoptosis. Consistent with a recent report, TRAIL induced a stronger apoptosis in sh-MAP3K3 cells than that in parental and sh-control cells [28] (Figure 4G).
Figure 4.
MAP3K3 regulates NF-κB targets and knockdown of MAP3K3 expression enhances TNFα- and TRAIL-induced apoptosis in MCF-7 cells. (A) Knockdown of MAP3K3 expression in MCF-7 cells inhibited basal and TNFα-induced NF-κB activity. NF-κB-dependent luciferase reporter and control Renilla luciferase reporter vectors were transfected into control and sh-MAP3K3 MCF-7 cells; 36 h after transfection, cells were left untreated or treated with TNFα for 16 h before luciferase reporter assays were performed. (B) The effect of knockdown MAP3K3 expression on NF-κB activity in MDA-MB-231 cells. (C) Heat map showing positive correlation of ICAM1 and Vimentin expression with MAP3K3 expression across 1340 breast tumours (yellow, high expression; R, Spearman’s correlation). (D) Immunoblotting analysis showing decreased levels of ICAM1 and Vimentin in MCF-7 cells with MAP3K3 knockdown. (E) Immunoblotting analysis showing the increased levels of ICAM1 and Vimentin in MCF-10A cells stably over-expressing MAP3K3. Results from (D, E) are representative of three independent experiments. (F, G) Quantification of cell numbers (by CCK-8) in cell cultures treated with the indicated concentrations of TNFα (F) or TRAIL (G). Error bars represent SEM from three independent experiments.
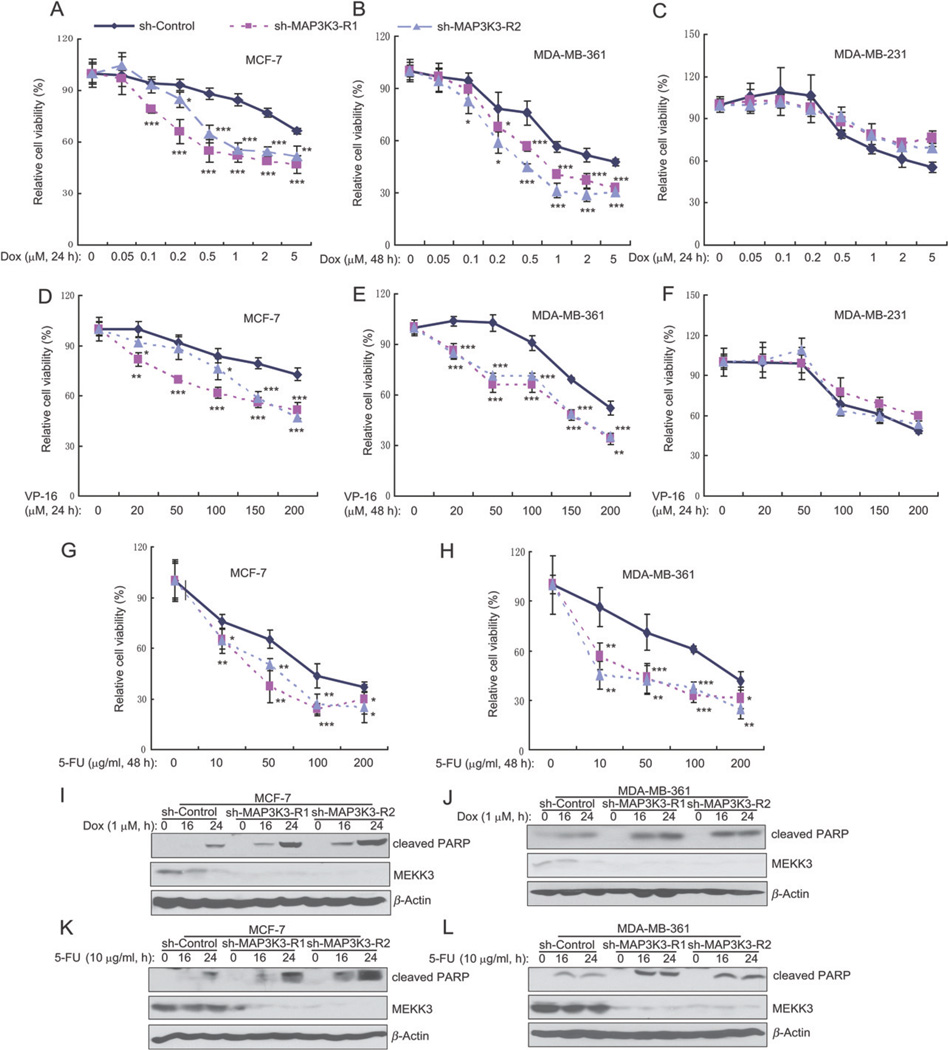
The above findings that MAP3K3 blockade sensitizes breast cancer cells to apoptosis-inducing cytokines suggest that MAP3K3 inhibition might also sensitize breast cancer cells to apoptotic induction by chemotherapeutic agents. Therefore, we tested the effect of MAP3K3 knockdown on the survival of breast cancer cells treated with doxorubicin, VP-16 and 5-FU, three chemotherapeutic drugs commonly used in the clinic to treat breast cancers. Two MAP3K3-amplified cell lines (MCF-7 and MDA-MB-361), but not three MAP3K3-non-amplified cell lines (MDA-MB-231, MDA-MB-468 and MDA-MB-453), were more sensitive to doxorubicin-induced cell death when MAP3K3 expression was knocked down in the cells (Figure 5A–C; see also supplementary material, Figure S5A, B). Similarly, MAP3K3 knockdown in two MAP3K3-amplified cell lines (MCF-7 and MDA-MB-361), but not in three MAP3K3-non-amplified cell lines (MDA-MB-231, MDA-MB-468 and MDA-MB-453), made cells more sensitive to VP-16-induced cell death (Figure 5D–F; see also supplementary material, Figure S5C, D). Knockdown of MAP3K3 expression in MCF-7 and MDA-MB-361 cells also made cells more sensitive to 5-FU-induced cell death (Figure 5G, H). Consistent with these results, doxorubicin and 5-FU induced increased amounts of cleavage of PARP in MAP3K3 knockdown cells compared to the control cells (Figure 6I–L). Together, our data suggest that knockdown of MAP3K3 enhances apoptotic induction by TNFα, TRAIL, doxorubicin, VP-16 and 5-FU, and that MAP3K3 amplification and over-expression contribute to the resistance of breast cancer cells to chemotherapy.
Figure 5.
Knockdown of MAP3K3 expression enhances doxorubicin-, VP16- and 5-FU-induced apoptosis in MCF-7 and MDA-MB-361 cells. (A–C) Quantification of cell numbers (by CCK-8) of MCF-7 (A), MDA-MB-361 (B) and MDA-MB-231 (C). Cultured cells were treated with the indicated concentrations of doxorubicin. (D–F) Quantification of cell numbers (by CCK-8) of MCF-7 (D), MDA-MB-361 (E) and MDA-MB-231 (F). Cultured cells were treated with the indicated concentrations of VP-16. (G, H) Quantification of cell numbers (by CCK-8) of MCF-7 (G) and MDA-MB-361 (H). Cultured cells were treated with the indicated concentrations of 5-FU. (I, J) Immunoblotting analysis showing the level of cleaved PARP in MAP3K3 stable knockdown cells treated with doxorubicin at the indicated time points. (K, L) Immunoblotting analysis showing the level of cleaved PARP in MAP3K3 stable knockdown cells treated with 5-FU at the indicated time points.
Figure 6.
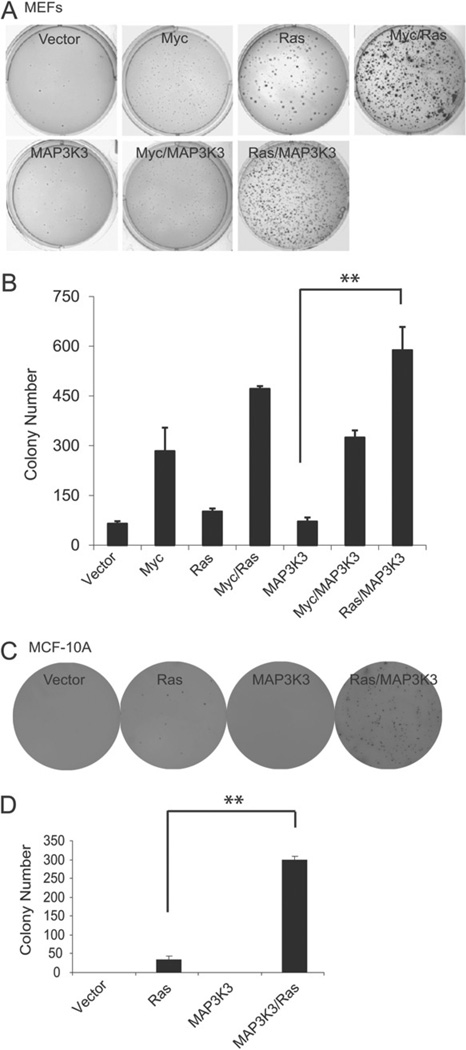
MAP3K3 promotes mammalian cell transformation. (A) Colony formation assay of MEFs stably transfected with retroviruses expressing the indicated genes. (B) Quantification of the data from colony assay in (A). Error bars represent SD from triplicate samples. (C) Colony formation assay of MCF-10A cells stably transfected with vectors expressing the indicated genes. (D) Quantification of the results from formation colony assay in (C). Error bars represent SD from triplicate samples. **p < 0.01, t-test.
MAP3K3 promotes transformation of breast cells
Finally, we also determined whether amplified MAP3K3 was a transforming oncogene. We first transduced primary mouse embryonic fibroblasts (MEFs) with a retrovirus carrying MAP3K3. MAP3K3 overexpression alone did not transform MEFs, as judged by colony formation in soft agar. Over-expression of K-RASV12D (a constitutively activated version of RAS) or c-Myc resulted in the formation of some or a few colones, respectively; and co-transfection of both resulted in the formation of many colonies (Figure 6A, B), as expected from the demonstrated collaborative effect of these two genes in transformation. While MAP3K3 did not increase the potential of c-Myc to transform MEFs, co-over-expression of MAP3K3 and K-RASV12D led to colony numbers comparable to those in the dishes co-transfected with K-RASV12D and c-Myc (Figure 6A, B). This indicates a significant transforming potential of MAP3K3 in cells that have an activated Ras signalling, but not in cells with c-Myc activation. Next we examined whether MAP3K3 over-expression could transform immortalized mammary epithelial cells (MCF-10A). Indeed, MAP3K3 over-expression also caused the transformation of MCF-10A cells in collaboration with K-RASV12D (Figure 6C, D). These results indicate that MAP3K3 has oncogenic properties.
Discussion
In this study, we applied an integrative bioinformatics approach to reveal potential oncogene targets on chromosome 17q, one of the most severely altered chromosomal regions in breast cancers. This approach utilized the concept signature analysis that we developed in our previous study to reveal the most biologically relevant genes from this genomic region [8,14]. Applying this approach to public genomic datasets uncovered that MAP3K3, encoding a serine/threonine protein kinase, was altered by genomic amplification in 8–12% of primary breast cancers. To verify this finding, we performed FISH analysis on 28 invasive breast cancer tissues and confirmed MAK3K3 amplification in six cases.
Our knockdown experiments demonstrate that MAP3K3 is also required for the growth of breast cancer cells harbouring MAP3K3 amplification. This finding suggests that MAP3K3 is a potential therapeutic target in MAP3K3-amplified breast cancers. The importance of targeting of MAP3K3 in breast cancer is further supported by our results exhibiting that MAP3K3 conferred the resistance of breast cancer cells to apoptotic induction by doxorubicin, VP16 and 5-FU (three commonly used chemotherapeutic agents in the clinic). Our data are consistent with previous observations that forced over-expression of MAP3K3 induced resistance to doxorubicin [18].
Besides its potential significance in breast cancer treatment, MAP3K3 also appears to be a transforming oncogene, based on soft-agar assays. It is interesting that MAP3K3 collaborated with Ras, but not c-Myc, to transform cells. Perhaps MAP3K3 and c-Myc are in the same oncogenic pathway, distinct from Ras signalling. Indeed, NF-κB has been suggested to up-regulate Myc expression [29]. This finding provides additional support for NF-κB in mediating MAP3K3 function in breast cancer. Of note, the mechanism by which MAP3K3 is involved in breast tumourigenesis is probably unrelated to ER- or HER2-mediated signalling pathways. This is supported by the observation that the amplification/over-expression status of MAP3K3 in our 28 breast cancer cases and in public expression datasets was not affected by the status of the established clinically used tumour markers, including ER, PR or HER2 (see supplementary material, Figure S2).
In conclusion, our data provide compelling evidence that MAP3K3 is amplified and over-expressed in a subset of breast cancers and that its amplification and over-expression contribute to breast cancer formation. MAP3K3 amplification and over-expression may also contribute to the resistance of breast cancer cells to apoptosis-inducing agents, including commonly used chemotherapeutic drugs. Therefore, MAP3K3 may be a valuable therapeutic target in a subset of breast cancers, and targeting MAP3K3 may sensitize breast cancer cells with MAP3K3 amplification to existing chemotherapy.
Supplementary Material
Acknowledgements
We thank Kristine Yang for editing our manuscript. We are grateful to Drs Ana M Gonzalez-Angulo, Scott W Lowe, Gianpietro Dotti and Biao Zheng for providing cell lines and plasmids described in this paper. This study was supported by the University Cancer Foundation via the Institutional Research Grant Program at the University of Texas MD Anderson Cancer Center (to HZ), the Fleming-Davenport Award from the Texas Medical Center (to HZ), the Virginia and LE Simmons Family Foundation Collaborative Research Fund (to JY), a developmental grant from the Baylor Breast Center NIH Specialized Program of Research Excellence (Grant No. P50 CA58183, to JY), NIH-NINDS (Grant No. 1R01NS072420, to JY) and the Orchid Cancer Appeal (to YJL, Grant No. CDMRP W81XWH-12-1-0166 to XW, and Grant No. W81XWH-12-1-0167 to RS).
Footnotes
Author contributions
YF, NG, XW and WS conceived and carried out experiments, performed data analysis and wrote the manuscript; RM, WB, PZ, SV, LA, XNL, EL, SGH and HZ performed experiments and data analysis; RS, JWG, CKO, JY and HZ proposed experiments, oversaw performance and reviewed the manuscript; XW and CJC performed the bioinformatics analysis; JSY, YJZ, YJL and PHR performed FISH and analysed the data; and JY and HZ planned experiments, reviewed all data and wrote the manuscript. All authors were involved in writing the paper and had final approval of the submitted version.
SUPPLEMENTARY MATERIAL ON THE INTERNET
The following supplementary material may be found in the online version of this article:
Supplementary materials and methods
Table S1. Reverse transcription PCR primers specific to MYO15B–MAP3K3 fusion.
Figure S1. MAP3K3 is amplified and over-expressed in breast cancer cell lines.
Figure S2. MAP3K3 is not associated with ER, PR or HER2 in human breast cancers.
Figure S3. Identification of MYO15B–MAP3K3 gene fusion in the MDA-MB-453 breast carcinoma cell line.
Figure S4. The effect of knockdown of MAP3K3 expression on cell proliferation and colony formation in MCF-7, MDA-MB-361 and MDA-MB-231 breast cancer cells.
Figure S5. The effect of knockdown of MAP3K3 expression on Dox- and VP16-induced cell death in MDA-MB-468 and MDA-MB-453 cells.
References
- 1.Slamon DJ, Clark GM, Wong SG, et al. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 2.Ong CC, Jubb AM, Haverty PM, et al. Targeting p21-activated kinase 1 (PAK1) to induce apoptosis of tumor cells. Proc Natl Acad Sci USA. 2011;108:7177–7182. doi: 10.1073/pnas.1103350108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li J, Yang Y, Peng Y, et al. Oncogenic properties of PPM1D located within a breast cancer amplification epicenter at 17q23. Nat Genet. 2002;31:133–134. doi: 10.1038/ng888. [DOI] [PubMed] [Google Scholar]
- 4.Kallioniemi A, Kallioniemi OP, Piper J, et al. Detection and mapping of amplified DNA sequences in breast cancer by comparative genomic hybridization. Proc Natl Acad Sci USA. 1994;91:2156–2160. doi: 10.1073/pnas.91.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Isola JJ, Kallioniemi OP, Chu LW, et al. Genetic aberrations detected by comparative genomic hybridization predict outcome in node-negative breast cancer. Am J Pathol. 1995;147:905–911. [PMC free article] [PubMed] [Google Scholar]
- 6.Plummer SJ, Paris MJ, Myles J, et al. Four regions of allelic imbalance on 17q12-qter associated with high-grade breast tumors. Genes Chromosomes Cancer. 1997;20:354–362. [PubMed] [Google Scholar]
- 7.Hermsen MA, Baak JP, Meijer GA, et al. Genetic analysis of 53 lymph node-negative breast carcinomas by CGH and relation to clinical, pathological, morphometric, and DNA cytometric prognostic factors. J Pathol. 1998;186:356–362. doi: 10.1002/(SICI)1096-9896(199812)186:4<356::AID-PATH196>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 8.Wang XS, Prensner JR, Chen G, et al. An integrative approach to reveal driver gene fusions from paired-end sequencing data in cancer. Nat Biotechnol. 2009;27:1005–1011. doi: 10.1038/nbt.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blank JL, Gerwins P, Elliott EM, et al. Molecular cloning of mitogen-activated protein/ERK kinase kinases (MEKK) 2 and 3. Regulation of sequential phosphorylation pathways involving mitogen-activated protein kinase and c-Jun kinase. J Biol Chem. 1996;271:5361–5368. doi: 10.1074/jbc.271.10.5361. [DOI] [PubMed] [Google Scholar]
- 10.Chao TH, Hayashi M, Tapping RI, et al. MEKK3 directly regulates MEK5 activity as part of the big mitogen-activated protein kinase 1 (BMK1) signaling pathway. J Biol Chem. 1999;274:36035–36038. doi: 10.1074/jbc.274.51.36035. [DOI] [PubMed] [Google Scholar]
- 11.Yang J, Boerm M, McCarty M, et al. Mekk3 is essential for early embryonic cardiovascular development. Nat Genet. 2000;24:309–313. doi: 10.1038/73550. [DOI] [PubMed] [Google Scholar]
- 12.Yang J, Lin Y, Guo Z, et al. The essential role of MEKK3 in TNF-induced NF-κB activation. Nat Immunol. 2001;2:620–624. doi: 10.1038/89769. [DOI] [PubMed] [Google Scholar]
- 13.Xu BE, Stippec S, Lenertz L, et al. WNK1 activates ERK5 by an MEKK2/3-dependent mechanism. J Biol Chem. 2004;279:7826–7831. doi: 10.1074/jbc.M313465200. [DOI] [PubMed] [Google Scholar]
- 14.Wang XS, Shankar S, Dhanasekaran SM, et al. Characterization of KRAS rearrangements in metastatic prostate cancer. Cancer Discov. 2011;1:35–43. doi: 10.1158/2159-8274.CD-10-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kan Z, Jaiswal BS, Stinson J, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 16.Heiser LM, Sadanandam A, Kuo WL, et al. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc Natl Acad Sci USA. 2012;109:2724–2729. doi: 10.1073/pnas.1018854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Q, Yang J, Lin Y, et al. Differential regulation of interleukin 1 receptor and Toll-like receptor signaling by MEKK3. Nat Immunol. 2004;5:98–103. doi: 10.1038/ni1014. [DOI] [PubMed] [Google Scholar]
- 18.Samanta AK, Huang HJ, Bast RC, Jr, et al. Overexpression of MEKK3 confers resistance to apoptosis through activation of NFκB. J Biol Chem. 2004;279:7576–7583. doi: 10.1074/jbc.M311659200. [DOI] [PubMed] [Google Scholar]
- 19.Abbasi S, Su B, Kellems RE, et al. The essential role of MEKK3 signaling in angiotensin II-induced calcineurin/nuclear factor of activated T-cells activation. J Biol Chem. 2005;280:36737–36746. doi: 10.1074/jbc.M506493200. [DOI] [PubMed] [Google Scholar]
- 20.Karin M. Nuclear factor-κB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 21.Karin M. NF-κB and cancer: mechanisms and targets. Mol Carcinog. 2006;45:355–361. doi: 10.1002/mc.20217. [DOI] [PubMed] [Google Scholar]
- 22.Lilienbaum A, Duc Dodon M, Alexandre C, et al. Effect of human T-cell leukemia virus type I tax protein on activation of the human vimentin gene. J Virol. 1990;64:256–263. doi: 10.1128/jvi.64.1.256-263.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grivennikov SI, Karin M. Inflammation and oncogenesis: a vicious connection. Curr Opin Genet Dev. 2010;20:65–71. doi: 10.1016/j.gde.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lilienbaum A, Paulin D. Activation of the human vimentin gene by the Tax human T-cell leukemia virus. I. Mechanisms of regulation by the NF-κB transcription factor. J Biol Chem. 1993;268:2180–2188. [PubMed] [Google Scholar]
- 25.Marui N, Offermann MK, Swerlick R, et al. Vascular cell adhesion molecule-1 (VCAM-1) gene transcription and expression are regulated through an antioxidant-sensitive mechanism in human vascular endothelial cells. J Clin Invest. 1993;92:1866–1874. doi: 10.1172/JCI116778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Creighton CJ, Li X, Landis M, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci USA. 2009;106:13820–13825. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bewick M, Conlon M, Lee H, et al. Evaluation of sICAM-1, sVCAM-1, and sE-Selectin levels in patients with metastatic breast cancer receiving high-dose chemotherapy. Stem Cells Dev. 2004;13:281–294. doi: 10.1089/154732804323099217. [DOI] [PubMed] [Google Scholar]
- 28.Guo SY, Liu SG, Liu L, et al. RNAi silencing of the MEKK3 gene promotes TRAIL-induced apoptosis in MCF-7 cells and suppresses the transcriptional activity of NF-κB. Oncol Rep. 2012;27:441–446. doi: 10.3892/or.2011.1509. [DOI] [PubMed] [Google Scholar]
- 29.Romashkova JA, Makarov SS. NF-κB is a target of AKT in anti-apoptotic PDGF signalling. Nature. 1999;401:86–90. doi: 10.1038/43474. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.