Abstract
The RNA interference (RNAi) pathway in animal cells can be harnessed to silence gene expression with artificial small interfering RNAs (siRNAs) or transgenes that express small hairpin RNAs (shRNAs). The transgene-expressing shRNA approach has been adapted into large-scale resources for genome-wide loss-of-function screens, whereas focused studies on a narrow set of genes can be achieved by using individual shRNA constructs from these resources. Although current shRNA repositories generally work, they might fail in certain situations and therefore necessitate other alternatives. We detail here a new highly-accessible and rational design of custom shRNAs that utilizes a refined backbone configuration termed the ‘Organic’ shRNA (OshR) platform. The OshR platform is ‘organic’ because it conforms more naturally to the endogenous vertebrate miRNAs by maintaining specific bulges and incorporating strategic mismatches to insure the desired guide strand is produced while reducing the accumulation of passenger strands that might contribute to off-target effects. We also demonstrate that the reliability of the OshR platform for gene silencing is increased when sequences target the 3' UnTranslated Region (3'UTR) of a gene. We further compare the OshR platform with the current and emerging shRNA designs, and propose that the OshR platform is a novel approach that can allow investigators to generate custom and effective shRNAs for individual gene functional studies.
Keywords: RNA interference, shRNA, gene silencing
INTRODUCTION
RNA interference (RNAi) is a broad genetic phenomenon in animals where gene expression can be interfered by small RNAs, such as microRNAs (miRNAs), endogenous small interfering RNAs (endo-siRNAs) and Piwi-interacting RNAs (piRNAs). Together, these pathways can be considered as different “arms” of the RNAi phenomenon in animals (1). Because RNAi is a natural and robust gene silencing process in animal cells, investigators can harness these pathways to silence the expression of nearly any gene of interest. The power and versatility of RNAi stems from the relative ease in synthesizing and introducing small RNAs with enormous variety of different sequences that efficiently incorporate into the RNA Induced Silencing Complex (RISC) as Guide strands (2). Guide strands direct the RISC to specific transcripts with complementarity to the guide, and RISC can trigger catalytic turnover of the targeted transcripts. The harnessing of RNAi has revolutionized many areas of biology because it has enabled highly specific reverse genetics analyses in animal cultured cells and has become an essential tool in studying diseases from cancer to diabetes to neurological disorders (3–5).
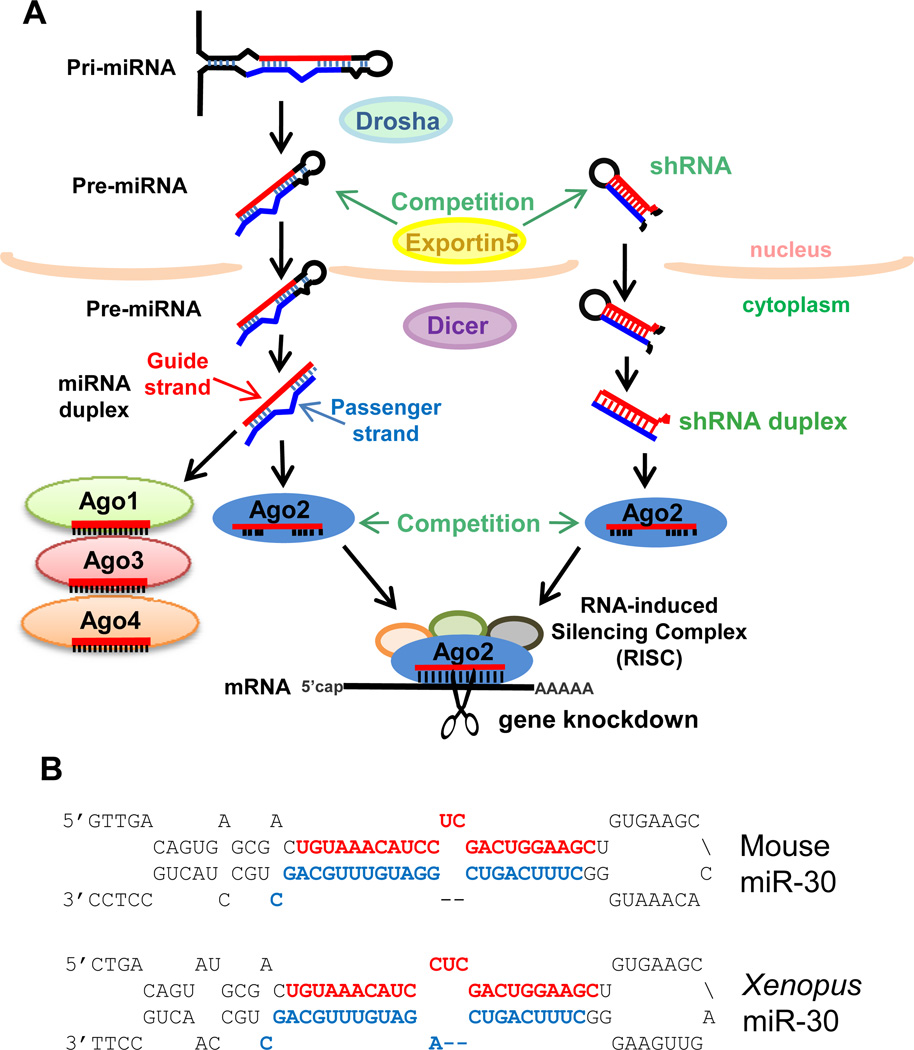
In most vertebrate cells, one of the main biological functions of the RNAi pathway is to regulate the expression of the transcriptome through miRNAs (Figure 1). Genome-encoded miRNA genes are transcribed by RNA-Polymerase (Pol) II as a long primary transcript (pri-miRNA) which is processed into a small hairpin-shaped precursor (pre-miRNA) by the Drosha enzyme, and then further processed by the Dicer enzyme into a small duplexed RNA (2). This small RNA duplex is then sampled on both ends of the duplex for ease in unwinding the RNA ends according to the thermodynamics of RNA base-pairing (6, 7). Typically, the RNA strand whose 5' start is at the duplex end that is easier to unwind will predominantly accumulate as the ‘Guide’ strand, while the other ‘Passenger’ strand will either be discarded or accumulate only modestly. Thus, a single-stranded guide RNA such as a mature miRNA sequence then becomes incorporated into an Argonaute protein that is the core effector protein of the RISC.
Figure 1. Exogenous RNAi by shRNAs may compete with the microRNA pathway.
A) Schematic of the biogenesis pathway for miRNAs which requires Drosha and Dicer to sequentially and specifically cut the miRNA and shRNA precursor into a small RNA duplex. The Guide strand will be incorporated into the Argonaute (Ago1– 4) proteins and the Ago2-RISC targets mRNA degradation. shRNAs may compete with miRNAs for specific limiting factors like Ago2 and Exportin5. B) The secondary structure for the priand pre-miRNA-30 from mouse, top, and Xenopus, bottom, with the miR-30-5p noted in red and the miR-30-3p noted in blue. There is conservation in the mechanism that directs greater accumulation of miR-30-5p as the Guide strand (red) in both mouse (10 fold to 300 fold, (55)) and frog (3 fold, (56)).
The RISC loaded with a small RNA can bind transcripts with as few as 7–8nt of complementarity with the ‘seed’ sequence in the 5' end of the guide RNA to trigger inhibition of translation and promote mRNA destabilization (8). However, complete complementarity through the entire ~22nt length of the guide RNA can trigger catalytic cleavage of the mRNA and lead to more robust gene silencing. Although seed pairing is one of the best understood modes for predicting and observing miRNA-mediated silencing, there are other less understood mechanisms that can contribute to gene silencing events such as extensive 3’ end pairing between an miRNA and target transcript (9) and non-canonical interactions between Argonaute/miRNA complexes and endogenous transcripts (10, 11).
Investigators can harness RNAi by programming the RISC with two types of triggers: chemically synthesized small interfering RNA (siRNA) or vector-driven transgenes that express a short hairpin RNA (shRNA). siRNAs were the original molecule that the Tuschl lab used to demonstrate that RNAi could silencing genes in vertebrate cells (12, 13), and although they are still extensively used (14), siRNAs have the restrictions of relatively higher cost and more transient gene knockdown effects when the siRNA is diluted from cell division and RNA turnover (15). Since recombinant DNA technology is inexpensive and highly scalable, the next evolution was the shRNA: a plasmid that expressed a short transcript driven by RNA Pol III type of promoters to make a small fold back structure which would be short enough not to trigger the undesired innate immunity response (16–19). These original shRNAs were reasonably successful in silencing gene expression in mammalian cultured cells while avoiding innate immunity toxic effects because they likely mimicked the endogenous miRNAs and entered the RISC through the same miRNA biogenesis factors.
However, the original shRNA design was tested when miRNA strand biogenesis was not yet fully understood. Now we know that the Drosha and Dicer enzymes work sequentially and measure intimately the length of the shRNA’s stem base and loop to know where to cut to yield a particular duplex (20). If the register for Drosha and Dicer cutting is unclear, inadvertent small RNA duplexes are generated that display thermodynamic properties which cause loading of an undesired passenger strand instead of a guide strand (21). The majority of pri-miRNA and pre-miRNA precursors contain various mismatched bulges in the secondary structure that drive the predominant accumulation of one guide strand, either from the 5' or 3' arm (2, 22, 23). However, there are some miRNA genes whose secondary structures are much more evenly double-stranded and have sequence features that allow for similar accumulation of the miRNA-5p and miRNA-3p to denote strands from both arms (24–28).
A second generation shRNA platform took more lessons from nature by borrowing structural elements from the mammalian miR-30 backbone, such as an RNA Pol II promoter, and parts of the endogenous stem base and loop features (29–31). This platform has revolutionized reverse genetic studies in mammalian cells because shRNA libraries have been scaled up to genome-wide screens for functional discovery of biological processes, to name a few like the DNA damage response gene, oncogenesis, or viral responses (32–35). The 2nd generation shRNA platform has even been co-opted back into invertebrates like Drosophila (3, 36). Although the RNAi machinery in these organisms is receptive to siRNA generation from long double-stranded RNA triggers (37, 38), it appears that germline gene silencing is more effective with shRNAs.
Nevertheless, there is continuing interest to improve the efficacy and reliability of shRNA design and deployment (21). When researchers target individual genes with shRNAs, they must obtain a panel of multiple shRNA constructs in order to sort out variable efficacies as well as mitigate potential off-targeting effects from each individual shRNA (39, 40). In some instances however, an entire panel of shRNAs can fail. It is unclear if failure is due to competition with the endogenous miRNA pathway (41–44), unpredictable guide versus passenger strand production (21, 45), secondary structures, or RNA binding proteins that occlude the RNAi machinery from accessing the mRNA (9, 46–54). In fact, the 2nd generation shRNA design is subtly different from the endogenous miR-30 gene in two aspects. First, it utilizes a perfectly double-stranded duplex that omits natural mismatches present in the natural miR-30 gene (4, 44, 45). Second, it places the guide strand, the sequence that is antisense to the target mRNA sequence, on the 3' arm of the shRNA. This design contrasts with the natural miR-30 gene that causes miR-30-5p to accumulate from 3 – 300 fold greater than miR-30-3p in multiple vertebrate cell types (55, 56). Although 2nd generation shRNAs do mature and incorporate into RISC, they may not fully exploit all the subtle features of the secondary structure of miR-30 that has been conserved from frogs to mammals (Fig. 1B).
In this study, we provide a method that we call the Organic shRNA platform (OshR): a Do-It-Yourself strategy for investigators to rationally design more natural shRNAs targeting any gene of interest. Drawing from the analogy of organically grown foods, which have not replaced conventional foods but instead provide a useful and desirable alternative, the OshR platform is a desirable alternative to current shRNA designs. OshRNAs perform well in always specifying the production of the desired Guide strand because the natural miR-30 features in our design more consistently suppress Passenger strand accumulation. Furthermore, we demonstrate in cell culture that Organic shRNAs are effective at targeting the Open Reading Frame (ORF) of vertebrate genes for knockdown studies, however a much higher probability of obtaining functional shRNAs can be achieved by targeting the 3'UnTranslated Region (3’UTR) of a target gene. Finally, we compare the OshR platform to a current 2nd generation shRNA platform and an emerging miR-451-backbone platform for versatility and gene silencing capacity.
MATERIALS AND METHODS
Design and cloning of shRNA constructs
Candidate shRNAs were designed by following the OshR workflow (Figure 2) and a custom Microsoft Excel Worksheet (Table S1). Oligonucleotides corresponding to the template and restriction enzyme sites were ordered from Integrated DNA Technologies. A 100 µl PCR reaction with 0.2 µM of template and oligonucleotides was subjected to 30 rounds of extension with a 60°C annealing temperature. A single amplicon was verified on agarose gel electrophoresis and purified on a spin column before restriction enzyme digestion. The digested PCR amplicon was then purified on spin columns and cloned into a CMV promoter-driven expression vector called pGSH0 that has an intron interrupting the coding sequence of GFP. This intron has a multiple cloning site for insertion of the shRNA PCR amplicon in a single direction. The parent shRNA expression plasmid described in this study as pGSH0 is available from Addgene. All plasmids were confirmed by sequencing and scaled up for transfection grade purification by a Midiprep column (Epoch Biosciences).
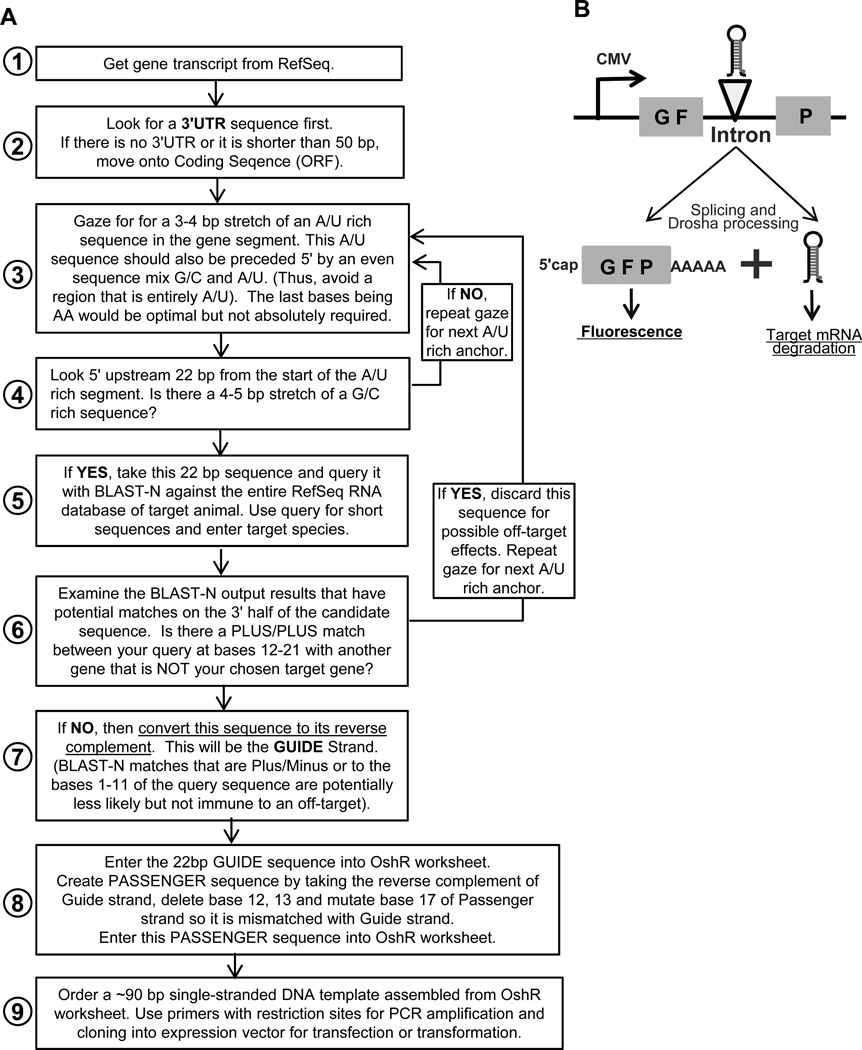
Figure 2. The design and decision workflow of the OshR platform.
A) Flowchart for how to pick a target sequence and design custom shRNAs based on the OshR platform. See also Table S1 for the worksheet. B) Diagram of the shRNA expression vector, pGSH0, used in this study. The shRNA is cloned into the intron of GFP and is generated after splicing and Drosha processing.
Cell culture and transfection
HEK293T cells were grown in DMEM medium (Invitrogen) supplemented with 10% heat-inactivated FBS (Lonza) and incubated in a humidified 37°C, 5% CO2 incubator. All shRNA constructs (1µg DNA per well of a 6-well dish) were co-transfected with a 200ng of a plasmid expressing myc-tagged Nek2 or 3xFLAG-tagged Xt_Tyrosinase into HEK293T cells according to the FuGENE®HD Transfection Reagent protocol (Promega) for the Xt_Tyr transgene or by the calcium phosphate method for the Nek2 transgene (57). Transfection efficiency was monitored by GFP fluorescence. The Nek2 cDNA was a kind gift from Erich Nigg.
Western blottling
Cells were washed twice with PBS and lysed in RIPA buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS) supplemented with protease inhibitor cocktail (Roche) or in 3X Sample Buffer (6% SDS, 0.1% bromophenol blue, 150 mM Tris pH 6.8, 30% glycerol, and 10% beta-mercaptoethanol). Protein was separated by SDS-PAGE, followed by transfer to PVDF or nitrocellulose membranes. Membranes were incubated in blocking buffer (5% nonfat milk in PBS- 0.05%Tween-20) for 1 hr and immunoblotted with primary antibody diluted in blocking buffer for 1 hr at room temperature. Primary antibodies include mouse anti-FLAG (M2, Sigma, 1:1000 dilution), mouse anti-α-tubulin (E-7a, Developmental Studies Hybridoma Bank, 1:10000 dilution), anti-beta-actin (Abcam, 1:5000), anti-myc (Sigma, 1:500), and rabbit anti-GFP (a gift from Michael Blower at Harvard Medical School, 1:1000). Membranes detected with ECL (GE Healthcare) and films were probed with HRP-conjugated goat-anti-mouse and goat-anti-rabbit IgG secondary antibodies (KPL, 1:5000 dilution). Membranes detected by the Odyssey Imaging system were probed with anti-mouse IR800CW (Licor; 1:10000) or anti-rabbit IR680LT (Licor; 1:10000) secondary antibodies.
Small RNA Northern blotting
Total RNA was isolated with TRI reagent RT (Molecular Research Center), followed by ethanol precipitation overnight at −20°C. RNA samples were separated on 15% urea polyacrylamide denaturing gels, transferred onto Hybond N+ membrane (GE Healthcare) for UV crosslinking or Hybond NX membrane (GE Healthcare) for chemical crosslinking with 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC, Sigma) , and probed with γ-32P-labeled DNA oligonucleotides antisense to guide strand or passenger strand of the individual shRNA overnight at 42°C. The membranes were washed with 3 × SSC/5% SDS/10xDenhardt’s solution/25mM NaH2PO4, pH7.5 at 42°C four times, 10 min for the first two washes and 30 min for another two washes, and were last washed with 1 × SSC/1% SDS for 5 minutes. The radioactive signals were then detected by phosphorimaging. For reprobing, the membrane was stripped using 1%SDS in water at 88°C for 40 minutes prior to hybridization.
RESULTS
The Organic shRNA (OshR): a Do-It-Yourself shRNA design
Although there are many web-based portals for shRNA design as well as multiple genome-wide repositories for purchasing sets of pre-designed shRNAs against a gene of interest, there is a lack of transparency of how each of these commercial shRNA designs are generated and whether the shRNAs are validated to produce the desired guide strand. Occasionally, an investigator may find that an entire panel of shRNAs purchased from these commercial sources fails to exhibit gene silencing even after controlling for transfection or transduction efficiency. In addition, investigators may want a less expensive alternative to commercial RNAi products, or require complete knowledge of the shRNA construct for the purposes of their experimental design. To address this need for an open-access alternative to shRNA design, we have devised the Organic shRNA (OshR) strategy that investigators can follow from the workflow in Figure 2.
This workflow begins by taking the complete mRNA sequence of the gene of interest and first determining if the mRNA has a stable and extensive 3'UTR that most metazoan genes possess. Our experience has been that targeting a gene’s 3'UTR increases the probability that an shRNA will not encounter refractory mRNA sequences, however ORFs are the next logical sequence to examine should a 3'UTR sequence be unavailable or too short (<50bp). The workflow then describes a sequence-gazing method that can be conducted manually to pick an optimal sequence base composition (Fig. 2A, Step 3 – 4), and a BLAST-N query approach to try to minimize off-target sequences (Step 5 – 7). An iterative procedure is followed to find optimal candidate GUIDE strand sequences of 22nt long. A PASSENGER strand sequence is then designed by taking the reverse complement of the Guide strand, deleting bases 12 and 13; and then mutating base 17 of Passenger strand so it is mismatched with Guide strand. These GUIDE and PASSENGER strand sequences are typed into a spreadsheet that concatenates the sequences into a ~90bp single-stranded DNA template that is ordered from an oligonucleotide vendor (See Table S1).
A set of primers that contain restriction enzyme (RE) sites are used to carry out a PCR reaction to generate an amplicon that can be cloned directionally into an animal expression vector. To express the OshRs in HEK293T cells, we cloned the OshR into a multiple cloning site of an intron that bisects GFP driven by a strong CMV promoter (Fig. 2B). This format allows the maturation of the shRNA to occur independently of GFP expression, and further mimics the natural occurrence of many mammalian miRNAs that reside in the introns of host genes (22). We believe that the OshR construct will also function in other expression vectors that insert shRNAs into the 3'UTR of a GFP transcript, as other groups have successfully demonstrated for other shRNA formats (4, 58–61). However, natural miR-30 appears to be transcribed as an RNA Pol II transcript (62, 63), thus we cannot predict how our OshR format would perform if inserted into 1st generation shRNA vectors that use a U6 snRNA promoter that drives RNA Pol III transcription (16, 17, 21). We verified by sequencing all plasmids and transfection-grade quality plasmids were prepared by ion exchange columns such as a commercial midiprep kit. Plasmids were then introduced by standard transfection protocols into cells along with epitope-tagged transgenes to monitor gene silencing.
The OshR platform enhances guide strand accumulation over the passenger strand
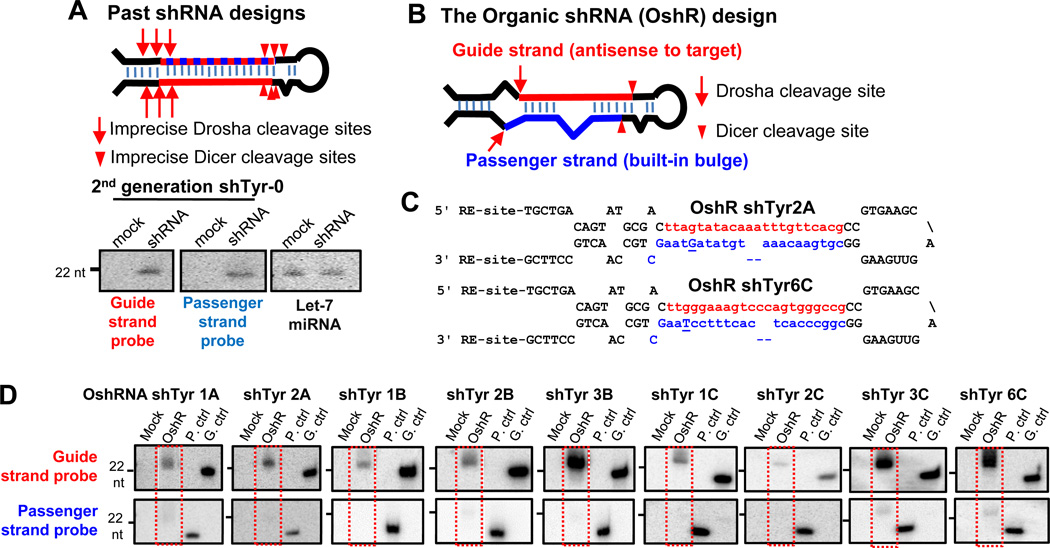
Although current shRNA designs contain the human miR-30a loop and RNA unwinding thermodynamics features that favor the guide sequence, these designs lack all internal bulges and instead utilize a complete duplex that places the guide strand on the 3' arm of the hairpin (4, 30, 31, 64). Generally, the 2nd generation shRNAs are effective in generating the proper guide strand for gene silencing, but occasionally undesired passenger strands can be produced. When we originally designed a 2nd generation shRNA to target the Xenopus tropicalis tyrosinase gene (Tyr) that synthesizes melanin and is implicated in albinism (65, 66), we also encountered issues of Guide strand specificity (Figure 3). The Passenger strand accumulated as abundantly as the Guide strand as detected by Northern blotting, and this is undesirable because off-target effects could result from the Passenger strand.
Figure 3. The OshR platform ensures proper Guide strand production.
A) Current shRNA designs can display variability in specifying the accumulation of both the Guide strand and Passenger strand. B) The Organic shRNA design builds in mismatches to direct specific Drosha and Dicer cleavage sites. C) Secondary structures of the OshR 2A and 6C against the X.tropicalis Tyrosinase (Tyr) mRNA. Guide strand is in red, Passenger strand is in blue, and intentional mismatched bases are underlined and uppercase. D) Northern blots indicate OshR designs yield predominant accumulation of the Guide Strand over Passenger strand. Mock is a mock transfection; OshR is the shRNA expression vector transfection; P.ctrl: synthetic Passenger strand mimic; G.ctrl: synthetic Guide strand mimic. Red dashed rectangles mark lanes of shRNA production.
We reasoned that a truly Organic shRNA design should completely mimic the base stem and the loop features of miR-30 to include all the features that naturally direct major accumulation of miR-30-5p from 3–50 fold over the mir-30-3p (55, 56) (Fig. 1B). Therefore, we devised our OshR scheme to place a programmable Guide strand on the 5' arm of the hairpin in place of mirR-30-5p, and we maintained key mismatched bases within the stem, and the loop (Fig. 3B). Next, we placed the Passenger strand on the 3' arm of the hairpin in place of miR-30- 3p, but instead of a perfect duplex, we engineered a mismatched base at the 3' end of the Passenger strand and deleted two bases in the middle, which are also natural features of miR-30-3p that ensure it is degraded simultaneously with the biased accumulation of miR-30-5p. Finally, we formed the rest of the miR-30a backbone using sequences that happened to derive from the X.tropicalis mirR-30a gene, but are equally tolerated in mouse and human cells.
To test whether the OshR platform would encourage Guide strand production while suppressing passenger accumulation, we constructed 9 additional shRNAs against the X.tropicalis Tyr gene and transfected each plasmid into HEK293T cells. We observed equivalent, highly efficient transfection for all plasmids as judged by >90% of the cells brightly fluorescing green (data not shown). By Northern blotting with probes that either detected the Guide strand or the Passenger strand, we consistently detected strong accumulation of the Guide strand sequence and very low or negligible production of the passenger strand (Fig. 3D). The size of the shRNA signal is consistent with mature small RNAs that would be incorporated into the RISC (21–23 nt long). These data support the conclusion that our rational Organic shRNA design enforces predominant Guide strand production while significantly preventing passenger strands from accumulating and causing off-target effects.
Targeting the 3'UTR of a transcript increases probability of shRNA efficacy
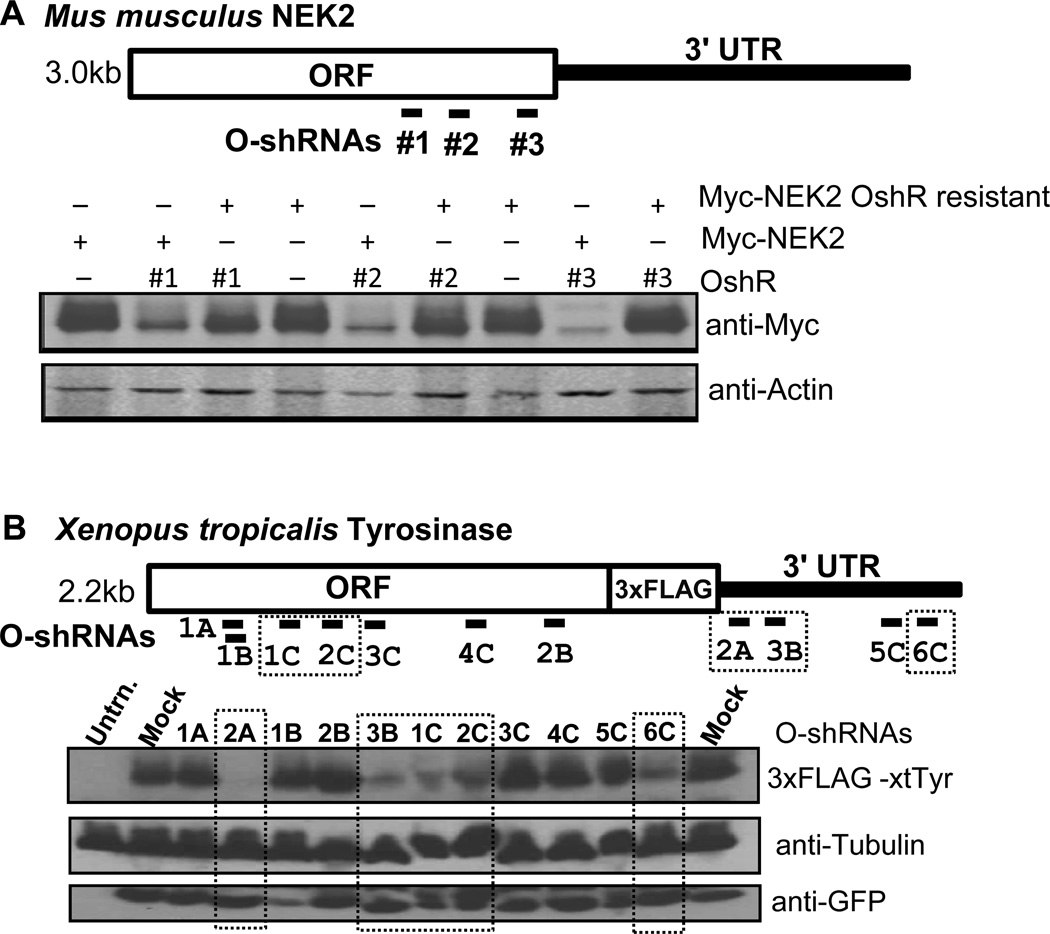
Theoretically, any Guide strand with perfect complementarity to a target transcript should direct RISC to degrade the target mRNA efficiently. In reality, certain mRNA sites can be refractory to RNAi in the cell because mRNA secondary structure, stalled ribosomes and other RNA binding proteins could conceivably block the RISC. These factors are still difficult to determine other than through empirical data, such as using an epitope-tagged transgene assay described here. To test the gene silencing efficacy of three organic shRNAs targeting the ORF of the mouse NIMA related protein kinase 2 (NEK2) gene, we co-transfected the OshR plasmids along with a myc-tagged NEK2 into HEK293T cells (Figure 4A). All three of the OshRs against NEK2 exhibited strong knockdown of the myc-tagged NEK2 transgene, and this knockdown could be circumvented with rescue constructs of NEK2 which had neutral mutations that altered the mRNA from base pairing perfectly with the shRNA. By validating these OshRs with epitope-tagged wild-type and rescue mutant transgenes (because antibodies are not always available for endogenous genes), we have established a bank of tools that will enable knocking down endogenous wild-type NEK2, testing for function, and then rescuing the function with shRNA-resistant transgenes. Thus, all the target sites we tested for NEK2 yielded effective gene knockdown by our organic shRNAs.
Figure 4. Gene silencing efficacy of various organic shRNAs.
A) Diagram of OshR target sites against mouse NEK-2 (top) and Western blots showing knockdown of Myc-tagged NEK2 (myc-NEK2) by all three OshRNAs (bottom). Actin is a loading control. Myc-NEK2 rescue constructs with mismatched bases to the shRNAs are resistant to knockdown by the OshRs. B) Diagram of OshR target sites against Xenopus Tyr (top) and Western blots showing knockdown of FLAG-tagged Tyr by OshRs marked by dashed rectangles (bottom). Tubulin is a loading control and GFP indicates proper expression of the OshR vector.
However, we observed a very different scenario of variable OshR silencing efficacy when targeting our FLAG epitope-tagged Xenopus Tyr transgene (Fig. 4B). Only 2 out of 7 shRNAs targeting the ORF of Tyr were able to elicit strong gene silencing despite our confirmation that sufficient guide strand was being produced (Fig. 3D), and these two ORF-targeting shRNAs targeted the same general locale of the Tyr mRNA. Strikingly, 3 out of 4 shRNAs targeting the 3’UTR significantly downregulated Tyr protein expression. In every transfection experiment, we confirmed nearly equal expression each of the shRNA constructs in the HEK293T cells as judged by GFP fluorescence (data not shown) and Western blot signal (Fig. 4B). Although we do not know if the region in the ORF targeted by Tyr shRNAs OshRs 1C and 2C is less likely to form secondary structures, the fact that the majority of OshRs targeting the 3'UTR were highly effective at gene silencing is consistent with the natural targeting proficiency of miRNAs against the 3'UTR of their targets. Therefore, our OshR design workflow recommends investigators to focus the sequence search first against a gene’s 3'UTR, and if the 3'UTR is too short or undefined then the ORF obviously follows. To fully determine the reliability of this process for shRNA selection will demand future studies of endogenous mRNA accessibility in the cell.
Comparison of the OshR platform to 2nd generation and miR-451-backbone shRNA designs
The best way to directly compare specific Guide strand production and targeting efficacy between our OshR platform and other shRNA formats was to take our two most effective OshRs 2A and 6C (Fig. 3C) and remake them as 2nd generation shRNAs. We did this by first placing the Guide strands of 2A and 6C onto the 3' arm and setting the duplex to be completely complementary (Figure 5). We cloned these 2nd generation shRNAs into the intron cloning site of our GFP expressing vector, and co-transfected shRNA vectors along with the 3xFLAG tagged Tyr transgene into HEK293T cells. In this case, both 2nd generation shRNAs generated the Guide strand as abundantly and predominantly over the Passenger strand as the OshRNAs (Fig. 5E); yet both Guide strands from the 2nd generation format where mainly 22nt long and about 1nt shorter than OshR Guide strands. In fact, nearly all Guide strands generated by the OshR platform that we tested by Northern blots accumulate as a ~23-24nt long (Fig. 3D), which is also the most abundant length of natural miR-30a (55).
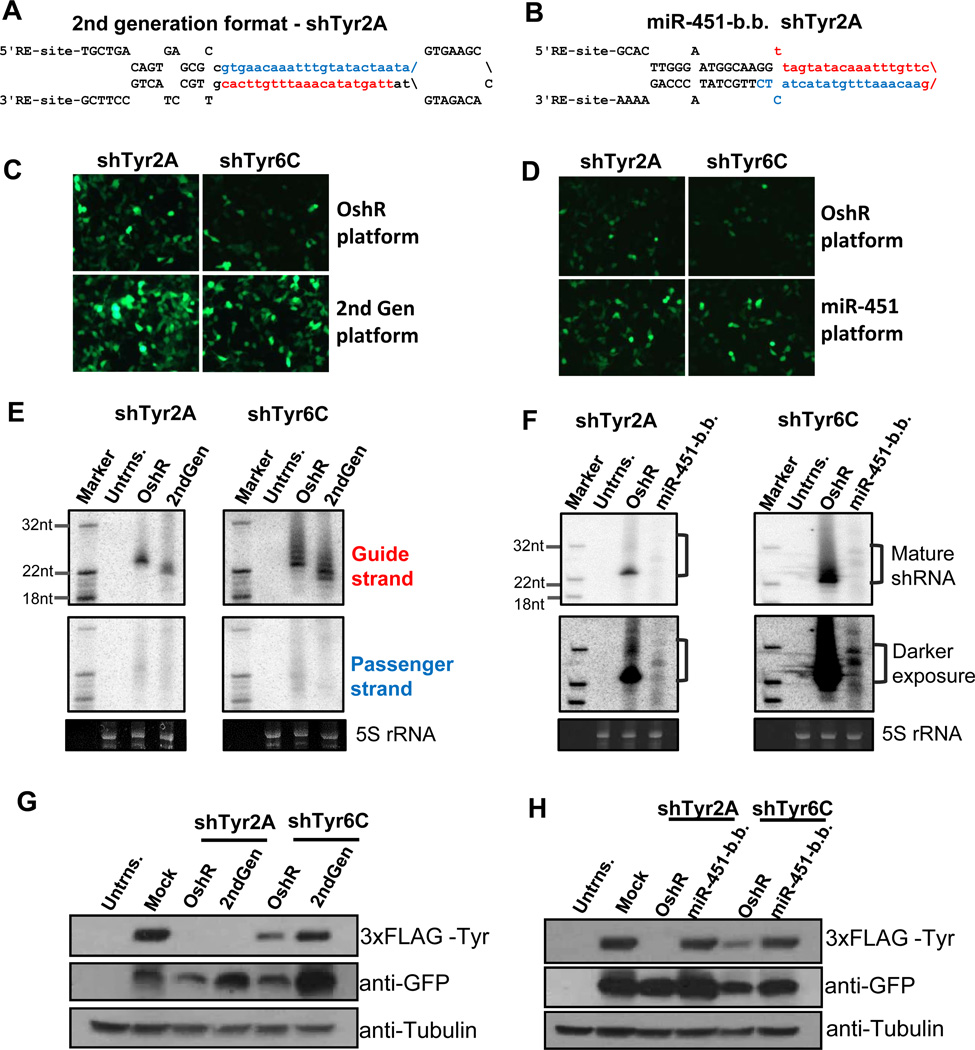
Figure 5. Comparing the OshR platform to the 2nd generation and miR-451 backbone platforms.
Secondary structures of shTyr-2A in the 2nd generation (A) and the miR-451 backbone (B). GFP expression of the shRNAs in the 2nd generation (C) and miR-451 backbone (D) directly compared with the analogous sequence in the OshR platform. Northern blots detecting shRNA production in the 2nd generation (E) and miR-451 backbone (F) directly compared with the analogous sequence in the OshR platform. Passenger strands were not assayed in (F) and 5S rRNA is a loading control. Western blots testing the knockdown of FLAG-tagged Tyr by shRNAs in the 2nd generation (G) and miR-451 backbone (H) directly compared with the analogous sequence in the OshR platform. Tubulin is a loading control and GFP indicates proper expression of the shRNA vector. Abbreviations: miR-451-b.b.: miR-451 backbone; Untrns.: untransfected cells; Mock: the empty vector.
Western blots indicated that the shTyr2A was equally robust at silencing the 3xFLAG-Tyr transgene regardless of whether it was in a 2nd generation or OshR format. However, the shTyr6C was more effective at gene silencing when expressed from the OshR format compared to the 2nd generation shRNA (Fig. 5G), despite the fact that there was nearly equal Northern blot signal from the OshR and 2nd generation shTyr6C Guide strands (Fig. 5E). Between sequences, shTyr2A was consistently more effective at silencing the transgene to nearly undetectable levels, which may be attributed to the sequence of shTyr2A being less G/C rich than shTyr6C. One theory is that underlying base compositions within siRNAs and miRNAs influence the multiple-turnover activity of Ago proteins (8, 67), and if a particular shRNA is so effective at silencing like shTyr2A, it does not matter whether it is expressed from a 2nd generation shRNA backbone or the more organic OshR backbone. With other shRNAs like shTyr6C, the OshR enhances sequences with perhaps lower targeting efficacy because the longer length of the major Guide strand contributes to stronger gene silencing.
Next, we compared our OshR platform to a newly emerging shRNA platform that uses the exemplary backbone of miR-451, a vertebrate-specific miRNA with a non-canonical stem loop that skips Dicer processing and instead matures through the Slicer activity of Ago2 (58, 61, 68, 69). The extraordinarily short stem of miR-451 directs only the 5' arm to be incorporated into RISC as the guide strand, and it has been shown that a variety of different sequences can reprogram a miR-451-based shRNA with virtually no passenger strand production. Furthermore, the Lai group has fused the miR-451 backbone with substrates of the RNaseZ and Integrator enzymes to create small RNA constructs that can be matured without requiring any RNaseIII activity (70, 71). Although the miR-451backbone theoretically only allows the Guide strand to enter Ago2 (58, 61, 68), and has a relatively simpler structure compared to the OshR backbone, we wondered whether the miR-451-backbone would truly be as widely versatile to sequence reprogramming and targeting efficacy.
We created miR-451-backbone shRNAs of our Tyr-targeting sequences 2A and 6C by maintaining necessary lower stem sequences and secondary structure features of miR-451 (i.e. Fig. 5E). Although we were worried that shTyr-2A’s A/T rich mid-section lacked a GC clamp region that is potentially required for Ago2 cleavage activity (61), we could temper this concern with shTyr-6C which retains the G/C pairs when inserted into the miR-451-backbone. As expected, we could much more readily detect the signal for the shTyr-6C mature shRNA compared to the shTyr-2A when inserted in the miR451 backbone constructs as well as the OshR backbone. However, the OshR backbone consistently yielded an order of magnitude greater accumulation of the mature guide strand than the miR-451 backbone (Fig. 5G), despite the fact that fluorescence and western blotting signals of GFP from miR-451 backbone vectors were greater than the OshR vectors (Fig. 5F). This suggested that the poor accumulation of Guide strands from the miR-451-backbone is more likely explained by issues with incorporation into RISC rather than the miR-451 backbone effects on the precursor RNA, which contributes effectively to GFP expression.
Finally, we compared the knockdown capacities of our organic shRNAs and the miR-451 backbone via co-transfection of the shRNA plasmid and the 3xFLAG tagged Tyr into HEK293T cells. Although some shRNAs like OshR-2C accumulate poorly but can efficiently trigger gene silencing (see Fig. 3D and 4B), we were surprised that we did not observe appreciable gene silencing by shTyr-2A and -6C in the miR-451 backbone, perhaps because of the relatively low accumulation of the guide strand (Fig. 5G and 5H). These results suggest that our OshR platform is more flexible for shRNA design compared to the miR-451-based platform because it does not constrain the shRNA to only enter Ago2, but rather may allow for the shRNA to accumulate to more significant levels in the other Argonaute proteins.
DISCUSSION
There is still a need for shRNA design improvements that enable investigators to create their own custom shRNAs when commercial shRNAs fail for some reason. We hypothesize these reasons could be when the passenger strand is produced instead of the guide strand, when exogenous shRNAs outcompete endogenous miRNAs for the RNAi machinery, or when mRNA sequences are refractory to targeting by shRNAs due to secondary structures or RNA binding proteins blocking access to the RISC. Although commercial 2nd generation shRNAs resources can be effective for gene silencing, investigators still need more options at hand for knocking down their gene of interest more completely and with fewer off-target effects. We still maintain caution and mindfulness that off-target effects could result from OshR format shRNAs since the BLAST-N workflow is currently manual, but using multiple shRNAs against a single gene should aid in differentiating an on-target phenotype from an off-target effect.
One new approach to circumvent sites refractory to shRNA targeting has been the development of a massively-parallel sensor assay that selects the sequences with the greatest potency from a random pool of 2nd generation shRNAs (32, 59). This sensor assay platform has contributed to the design and generation of transgenic mice with conditionally-expressed shRNAs for gene knockdown studies in tissues (60). However, this massively-parallel sensor assay requires a large up-front commitment of resources to generate the diverse random pools of shRNAs for a single gene and having fluorescence activated cell sorter to perform the selection. Therefore, we believe our rational design of organic shRNAs will be very attractive to many investigators looking for cost-effective procedures to construct a small pool of custom shRNAs to target their gene of interest.
Although our Organic shRNA design effectively specifies Guide strand accumulation and suppress the passenger strand, we could not pinpoint why so many of the OshRs designed against the Tyr ORF failed to exhibit strong gene silencing. Many studies have suggested that thermodynamic analyses of target regions of ORFs that exhibit less secondary structures are correlated with effective siRNAs (9, 46–54, 72). If secondary structure somehow blocks access to RISC, these structures do not seem to inhibit translation of the Tyr transgene (Fig. 4B). Perhaps it is the processivity of the translating ribosome that prevents RISC from accessing a transcript except at a region downstream of a translational pause. Therefore, the increased frequency of observing robust silencing when targeting a gene’s 3'UTR may reflect RISC not having to compete with translation. We postulate that the poor knockdown capacity of OshR shTyr5C against the Tyr 3'UTR might reflect RNA binding proteins or secondary structures that block the shRNA-guided RISC. Our pursuit of improving the shRNA platform offers an incremental and important improvement in the suite of tools for gene function analysis in animal cells. As the OshR platform exhibits a more ‘organic’ affinity with miRNAs, we foresee these and future studies with the OshR system will inform on miRNA processing biology and endogenous small RNA-mediated gene silencing mechanisms.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Robert Grainger for initial discussions and sharing of unpublished data that inspired this project. We also thank Stephen Elledge for suggesting we directly compare our shRNA platform to the original second generation platform. Finally, we thank members of the Lau lab for comments on this manuscript. This work was supported by a SPROUT grant from the Brandeis Virtual Incubator to M.Z. and from the National Institute of Health for grants R21DA026977 to S.P., EY018000 and EY017400 to T.N., and R00HD057298 to N.C. L.. N.C. L. is a Searle Scholar.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Lau NC. Int J Biochem Cell Biol. 2010 [Google Scholar]
- 2.Carthew RW, Sontheimer EJ. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perrimon N, Ni JQ, Perkins L. Cold Spring Harb Perspect Biol. 2. 2010 doi: 10.1101/cshperspect.a003640. a003640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang K, Elledge SJ, Hannon GJ. Nat Methods. 2006;3:707–714. doi: 10.1038/nmeth923. [DOI] [PubMed] [Google Scholar]
- 5.Bernards R, Brummelkamp TR, Beijersbergen RL. Nat Methods. 2006;3:701–706. doi: 10.1038/nmeth921. [DOI] [PubMed] [Google Scholar]
- 6.Khvorova A, Reynolds A, Jayasena SD. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 7.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Helwak A, Kudla G, Dudnakova T, Tollervey D. Cell. 2013;153:654–665. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leung AK, Young AG, Bhutkar A, Zheng GX, Bosson AD, Nielsen CB, Sharp PA. Nat Struct Mol Biol. 2011;18:237–244. doi: 10.1038/nsmb.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 13.Elbashir SM, Lendeckel W, Tuschl T. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cleary MA, Kilian K, Wang Y, Bradshaw J, Cavet G, Ge W, Kulkarni A, Paddison PJ, Chang K, Sheth N, Leproust E, Coffey EM, Burchard J, McCombie WR, Linsley P, Hannon GJ. Nat Methods. 2004;1:241–248. doi: 10.1038/nmeth724. [DOI] [PubMed] [Google Scholar]
- 15.Paddison PJ, Hannon GJ. Curr Opin Mol Ther. 2003;5:217–224. [PubMed] [Google Scholar]
- 16.Brummelkamp TR, Bernards R, Agami R. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 17.Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paddison PJ, Caudy AA, Hannon GJ. Proc Natl Acad Sci U S A. 2002;99:1443–1448. doi: 10.1073/pnas.032652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harborth J, Elbashir SM, Vandenburgh K, Manninga H, Scaringe SA, Weber K, Tuschl T. Antisense Nucleic Acid Drug Dev. 2003;13:83–105. doi: 10.1089/108729003321629638. [DOI] [PubMed] [Google Scholar]
- 20.Kim VN, Han J, Siomi MC. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 21.Gu S, Jin L, Zhang Y, Huang Y, Zhang F, Valdmanis PN, Kay MA. Cell. 2012;151:900–911. doi: 10.1016/j.cell.2012.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartel DP. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 23.Ghildiyal M, Zamore PD. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forstemann K, Tomari Y, Du T, Vagin VV, Denli AM, Bratu DP, Klattenhoff C, Theurkauf WE, Zamore PD. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang JS, Phillips MD, Betel D, Mu P, Ventura A, Siepel AC, Chen KC, Lai EC. RNA. 2010;17:312–326. doi: 10.1261/rna.2537911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okamura K, Liu N, Lai EC. Mol Cell. 2009;36:431–444. doi: 10.1016/j.molcel.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okamura K, Phillips MD, Tyler DM, Duan H, Chou YT, Lai EC. Nat Struct Mol Biol. 2008;15:354–363. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tyler DM, Okamura K, Chung WJ, Hagen JW, Berezikov E, Hannon GJ, Lai EC. Genes Dev. 2008;22:26–36. doi: 10.1101/gad.1615208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paddison PJ, Silva JM, Conklin DS, Schlabach M, Li M, Aruleba S, Balija V, O'Shaughnessy A, Gnoj L, Scobie K, Chang K, Westbrook T, Cleary M, Sachidanandam R, McCombie WR, Elledge SJ, Hannon GJ. Nature. 2004;428:427–431. doi: 10.1038/nature02370. [DOI] [PubMed] [Google Scholar]
- 30.Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, Sheth N, Bradshaw J, Burchard J, Kulkarni A, Cavet G, Sachidanandam R, McCombie WR, Cleary MA, Elledge SJ, Hannon GJ. Nat Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 31.Stegmeier F, Hu G, Rickles RJ, Hannon GJ, Elledge SJ. Proc Natl Acad Sci U S A. 2005;102:13212–13217. doi: 10.1073/pnas.0506306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan X, Lu ZJ, Gao G, Xu Q, Hu L, Fellmann C, Li MZ, Qu H, Lowe SW, Hannon GJ, Elledge SJ. Proc Natl Acad Sci U S A. 2012;109:869–874. doi: 10.1073/pnas.1119873109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adamson B, Smogorzewska A, Sigoillot FD, King RW, Elledge SJ. Nat Cell Biol. 2012;14:318–328. doi: 10.1038/ncb2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu G, Kim J, Xu Q, Leng Y, Orkin SH, Elledge SJ. Genes Dev. 2009;23:837–848. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Solimini NL, Xu Q, Mermel CH, Liang AC, Schlabach MR, Luo J, Burrows AE, Anselmo AN, Bredemeyer AL, Li MZ, Beroukhim R, Meyerson M, Elledge SJ. Science. 2012;337:104–109. doi: 10.1126/science.1219580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ni JQ, Zhou R, Czech B, Liu LP, Holderbaum L, Yang-Zhou D, Shim HS, Tao R, Handler D, Karpowicz P, Binari R, Booker M, Brennecke J, Perkins LA, Hannon GJ, Perrimon N. Nat Methods. 2011;8:405–407. doi: 10.1038/nmeth.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 38.Neumuller RA, Richter C, Fischer A, Novatchkova M, Neumuller KG, Knoblich JA. Cell Stem Cell. 2011;8:580–593. doi: 10.1016/j.stem.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Root DE, Hacohen N, Hahn WC, Lander ES, Sabatini DM. Nat Methods. 2006;3:715–719. doi: 10.1038/nmeth924. [DOI] [PubMed] [Google Scholar]
- 40.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, Carpenter AE, Foo SY, Stewart SA, Stockwell BR, Hacohen N, Hahn WC, Lander ES, Sabatini DM, Root DE. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 41.Fish RJ, Kruithof EK. BMC Mol Biol. 2004;5:9. doi: 10.1186/1471-2199-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 43.Lund E, Sheets MD, Imboden SB, Dahlberg JE. Genes Dev. 2011;25:1121–1131. doi: 10.1101/gad.2038811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McBride JL, Boudreau RL, Harper SQ, Staber PD, Monteys AM, Martins I, Gilmore BL, Burstein H, Peluso RW, Polisky B, Carter BJ, Davidson BL. Proc Natl Acad Sci U S A. 2008;105:5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gu S, Jin L, Zhang F, Huang Y, Grimm D, Rossi JJ, Kay MA. Proc Natl Acad Sci U S A. 2011;108:9208–9213. doi: 10.1073/pnas.1018023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li L, Lin X, Khvorova A, Fesik SW, Shen Y. RNA. 2007;13:1765–1774. doi: 10.1261/rna.599107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bergauer T, Krueger U, Lader E, Pilk S, Wolter I, Bielke W. Oligonucleotides. 2009;19:41–52. doi: 10.1089/oli.2008.0154. [DOI] [PubMed] [Google Scholar]
- 48.Obernosterer G, Tafer H, Martinez J. Biochem Soc Trans. 2008;36:1216–1219. doi: 10.1042/BST0361216. [DOI] [PubMed] [Google Scholar]
- 49.Tafer H, Ameres SL, Obernosterer G, Gebeshuber CA, Schroeder R, Martinez J, Hofacker IL. Nat Biotechnol. 2008;26:578–583. doi: 10.1038/nbt1404. [DOI] [PubMed] [Google Scholar]
- 50.Dickins RA, McJunkin K, Hernando E, Premsrirut PK, Krizhanovsky V, Burgess DJ, Kim SY, Cordon-Cardo C, Zender L, Hannon GJ, Lowe SW. Nat Genet. 2007;39:914–921. doi: 10.1038/ng2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nielsen CB, Shomron N, Sandberg R, Hornstein E, Kitzman J, Burge CB. RNA. 2007;13:1894–1910. doi: 10.1261/rna.768207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robins H, Li Y, Padgett RW. Proc Natl Acad Sci U S A. 2005;102:4006–4009. doi: 10.1073/pnas.0500775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, le Sage C, Nagel R, Voorhoeve PM, van Duijse J, Orom UA, Lund AH, Perrakis A, Raz E, Agami R. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 54.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 55.Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, Blelloch R, Schroth GP, Nusbaum C, Bartel DP. Genes Dev. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Faunes F, Almonacid LI, Melo F, Larrain J. Genesis. 2012;50:260–270. doi: 10.1002/dvg.22012. [DOI] [PubMed] [Google Scholar]
- 57.Xia Z, Dudek H, Miranti CK, Greenberg ME. J Neurosci. 1996;16:5425–5436. doi: 10.1523/JNEUROSCI.16-17-05425.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fellmann C, Zuber J, McJunkin K, Chang K, Malone CD, Dickins RA, Xu Q, Hengartner MO, Elledge SJ, Hannon GJ, Lowe SW. Mol Cell. 2011;41:733–746. doi: 10.1016/j.molcel.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Premsrirut PK, Dow LE, Kim SY, Camiolo M, Malone CD, Miething C, Scuoppo C, Zuber J, Dickins RA, Kogan SC, Shroyer KR, Sordella R, Hannon GJ, Lowe SW. Cell. 2011;145:145–158. doi: 10.1016/j.cell.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang JS, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, Papapetrou EP, Sadelain M, O'Carroll D, Lai EC. Proc Natl Acad Sci U S A. 2010;107:15163–15168. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cai X, Hagedorn CH, Cullen BR. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Siolas D, Lerner C, Burchard J, Ge W, Linsley PS, Paddison PJ, Hannon GJ, Cleary MA. Nat Biotechnol. 2005;23:227–231. doi: 10.1038/nbt1052. [DOI] [PubMed] [Google Scholar]
- 65.King RA, Pietsch J, Fryer JP, Savage S, Brott MJ, Russell-Eggitt I, Summers CG, Oetting WS. Hum Genet. 2003;113:502–513. doi: 10.1007/s00439-003-0998-1. [DOI] [PubMed] [Google Scholar]
- 66.Oetting WS. Pigment Cell Res. 2000;13:320–325. doi: 10.1034/j.1600-0749.2000.130503.x. [DOI] [PubMed] [Google Scholar]
- 67.Garcia DM, Baek D, Shin C, Bell GW, Grimson A, Bartel DP. Nat Struct Mol Biol. 2011;18:1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, Wolfe SA, Giraldez AJ. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang JS, Lai EC. Cell Cycle. 2010;9:4455–4460. doi: 10.4161/cc.9.22.13958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang JS, Lai EC. Mol Cell. 2011;43:892–903. doi: 10.1016/j.molcel.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang JS, Maurin T, Lai EC. RNA. 2012;18:945–957. doi: 10.1261/rna.032938.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gu S, Jin L, Zhang F, Sarnow P, Kay MA. Nat Struct Mol Biol. 2009;16:144–150. doi: 10.1038/nsmb.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.