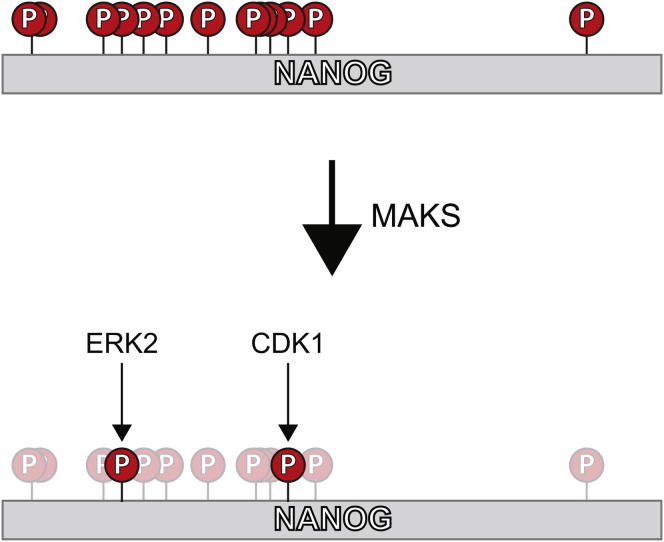
Summary
NANOG is a divergent homeobox protein and a core component of the transcriptional circuitry that sustains pluripotency and self-renewal. Although NANOG has been extensively studied on the transcriptional level, little is known regarding its posttranslational regulation, likely due to its low abundance and challenging physical properties. Here, we identify eleven phosphorylation sites on endogenous human NANOG, nine of which mapped to single amino acids. To screen for the signaling molecules that impart these modifications, we developed the multiplexed assay for kinase specificity (MAKS). MAKS simultaneously tests activity for up to ten kinases while directly identifying the substrate and exact site of phosphorylation. Using MAKS, we discovered site-specific phosphorylation by ERK2 and CDK1/CyclinA2, providing a putative link between key signaling pathways and NANOG.
Graphical Abstract
Highlights
-
•
We report 11 phosphorylation sites (9 localized) on endogenous, human NANOG
-
•
We introduce a multiplexed assay to identify kinases that modify a given substrate
-
•
We demonstrate that NANOG is directly phosphorylated by ERK2 and CDK1 in vitro
-
•
We connect cell-cycle- and growth-factor-mediated signaling to NANOG
NANOG is a core pluripotency factor. Here, Brumbaugh et al. show that NANOG is heavily phosphorylated on the N terminus and many of these sites are proline directed. To place these modifications in the context of signaling pathways, they developed the multiplexed assay for kinase specificity (MAKS) and show that NANOG is directly phosphorylated by ERK2 and CDK1 in vitro.
Introduction
Phosphorylation is a pervasive form of cell signaling that orchestrates numerous processes, including metabolism, cell mobility, cell cycle, and differentiation (Brumbaugh et al., 2011, Van Hoof et al., 2009, Xu and Fisher, 2012). Mass spectrometry has revealed the complexity of the phosphoproteome in pluripotent cells with great detail (Muñoz and Heck, 2011, Phanstiel et al., 2011, Rigbolt et al., 2011, Swaney et al., 2009, Van Hoof et al., 2009); however, determining the biological relevance of such data remains a major challenge. Mapping the kinases responsible for a specific phosphorylation event is instructive because it places that information in the context of signaling molecules that direct biological function. Traditionally, kinase assays are performed by detecting the transfer of a radioactive phosphoryl group to a given substrate following an in vitro reaction. This method provides a direct measure of phosphorylation but necessitates the use of hazardous materials, cannot directly localize phosphorylation to a single amino acid when more than one potential site is present, and cannot multiplex kinases and substrates. Recently, a mass-spectrometric method was introduced to profile phosphorylation on synthetic peptides treated with cell lysates (Yu et al., 2009). This method is ideal for profiling cell-type-specific phosphorylation, but does not directly determine the kinase responsible for phosphorylation. Another method assesses kinase activity but relies upon heavy isotope-labeled amino acids and is limited to testing one or two kinases at a time (Singh et al., 2012b). Several other methods have been developed to identify kinase consensus motifs or test a single kinase, but are not capable of multiplexed analysis (Hennrich et al., 2013, Kettenbach et al., 2012, Songyang et al., 1994, Xue et al., 2012). Thus, there remains a pressing need for a high-throughput method to screen for kinase(s) that phosphorylate a protein of interest.
In pluripotent cells, phosphorylation has a central role in directing cell identity by relaying growth-factor signaling through key pathways (i.e., fibroblast growth factor [FGF] and transforming growth factor β [TGF-β]) (Chen et al., 2011, Singh et al., 2012a, Vallier et al., 2005, Yu et al., 2011). The ultimate targets of these phosphorylation cascades are largely unknown, although recent works have provided some direction by mapping phosphorylation on the pluripotency factors OCT4 and SOX2 (Brumbaugh et al., 2012, Jeong et al., 2010, Phanstiel et al., 2011). Conspicuously absent in these studies is NANOG, a divergent homeobox transcription factor that promotes pluripotency by binding to DNA and regulating the expression of genes related to cell fate (Boyer et al., 2005, Chambers et al., 2003, Mitsui et al., 2003, Pan and Thomson, 2007). In mice, overexpression of NANOG permits extended culture of undifferentiated embryonic stem cells (ESCs) in the absence of otherwise obligatory extrinsic signaling factors such as LIF and BMP4 (Chambers et al., 2003, Pan and Thomson, 2007). Correspondingly, NANOG overexpression in human ESCs obviates the requirement for exogenous signaling through feeder cells in basal media or FGF in defined culture systems (Darr et al., 2006, Xu et al., 2008). Hence, NANOG has a conserved role in mediating growth-factor signals that are critical for pluripotency, and, intriguingly, its overexpression is sufficient to bypass these signaling pathways to maintain the ESC state. Still, a direct link between NANOG and the signaling molecules that determine cell state remains elusive.
In mouse, NANOG protein levels are dynamic (Chambers et al., 2007), and it was recently proposed that NANOG stability is tied to phosphorylation (Moretto-Zita et al., 2010). Several studies suggested that mouse NANOG exists as a phosphoprotein (Li et al., 2011, Moretto-Zita et al., 2010, Yates and Chambers, 2005); however, its unique primary sequence and relatively low abundance make it difficult to purify and detect in a physiologically relevant context (i.e., without overexpression and in pluripotent cell types). As a result, there are currently no known phosphorylation sites for NANOG from human pluripotent stem cells and only a single site for endogenous mouse NANOG (Li et al., 2011). To address this gap, we applied high-resolution mass spectrometry to show that NANOG is heavily phosphorylated at proline-directed sites on its N terminus. To place these modifications in the context of signaling molecules, we developed the multiplexed assay for kinase specificity (MAKS), and found that ERK2 and CDK1 differentially phosphorylate NANOG in vitro.
Results
Identification of NANOG Phosphorylation Sites
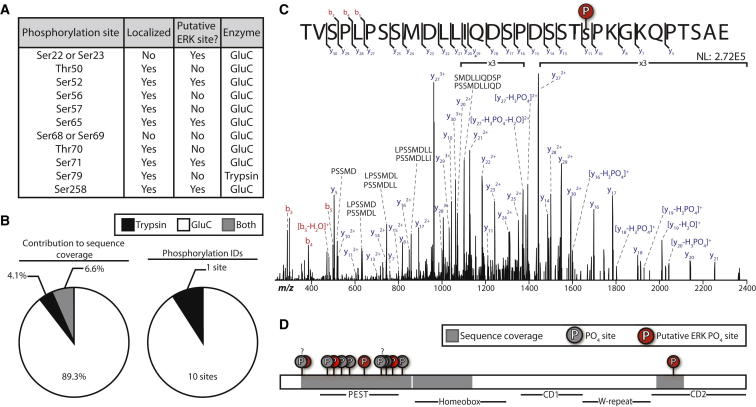
Due to the underrepresentation of NANOG in proteomic data sets, we adapted a highly efficient purification strategy that combines high-detergent lysis with multiple forms of affinity chromatography (see Supplemental Experimental Procedures available online; Brumbaugh et al., 2012). Consistent with previous results, traditional digestion methods that utilize trypsin in preparation for mass spectrometry yielded only one phosphopeptide identification (Figures 1A and 1B). Inspection of the primary sequence for NANOG revealed that tryptic cleavage sites were tightly clustered at the center of the protein, with few sites near either termini, resulting in a limited number of peptides that were amenable to mass spectrometry (Figure S1). We therefore tested several alternate proteases and found that GluC provided superior sequence coverage (Figures 1C and 1D; Swaney et al., 2010). In total, we sequenced over 40% of the endogenous NANOG protein and identified 11 phosphorylation sites (<1% false discovery rate), nine of which localized to a single residue (Figure 1D; Table S1). Note that the exact site of phosphorylation could not be distinguished between Ser22/23 and Ser68/69 by our stringent criteria, but were localized to adjacent residues in each case. Figure 1C shows a representative spectrum with a fully annotated, localized modification; all spectra were manually validated to confirm each phosphorylation site (Figure S2). Many of the peptides identified from this region were multiply phosphorylated, indicating that endogenous NANOG is hyperphosphorylated in the pluripotent state.
Figure 1.
Mass Spectrometry Identifies Phosphorylation on Human NANOG
(A) NANOG phosphorylation sites.
(B) The contribution to either sequence coverage or phosphorylation site identification for samples digested with trypsin or GluC.
(C) Representative spectrum showing unambiguous localization of phosphorylation on Ser71. Neutral loss (asterisk) and internal fragments are annotated.
(D) Schematic of NANOG with functional domains. Question marks denote phosphorylation sites that were indistinguishable between adjacent residues.
See also Figures S1 and S2, and Table S1.
To place NANOG phosphorylation sites in the context of cell signaling, we used the Motif-X program to search for kinase motifs in our data set (Schwartz and Gygi, 2005). Intriguingly, a preponderance of NANOG’s phosphorylation sites were proline directed (xP) and matched the minimal ERK motif (5/11 sites; Figure 1B) (Songyang et al., 1996). This finding is compelling from a biological standpoint because NANOG overexpression sustains pluripotency upon ERK inhibition (Xu et al., 2008) and activation of ERK2 maintains NANOG, even in differentiating cells (Yu et al., 2011). Further computational analysis using the NetPhorest platform substantiated the hypothesis that ERK2 phosphorylates these sites, but also pointed to CDK1 as a possible modifying enzyme (Miller et al., 2008). Note that these kinases share similar motifs, and the importance of applying both experimental and computational methods to determine kinase-substrate relationships has been documented (Linding et al., 2007). Thus, it is important to directly screen for the kinases that modify NANOG.
MAKS
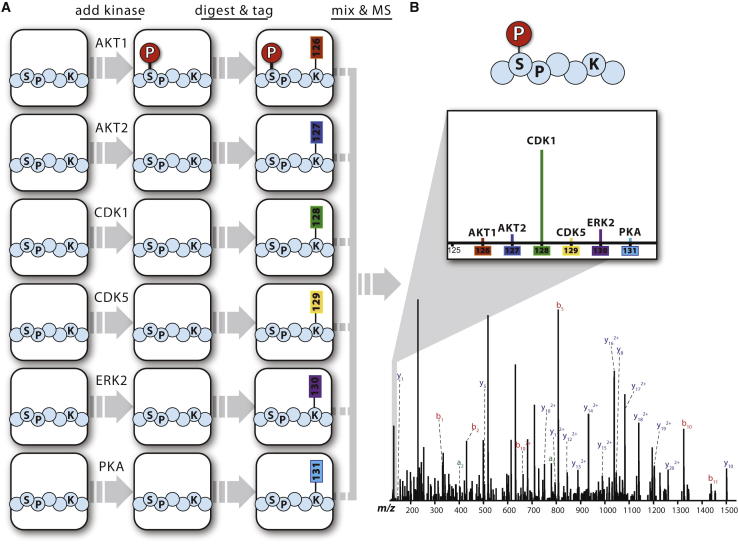
To directly identify kinases that target specific residues on NANOG, we developed a mass-spectrometric approach called MAKS. This in vitro assay leverages isobaric tagging technology to monitor the activity of up to ten kinases in a single experiment. In the first step, candidate kinases, ATP, and reaction buffer are added to purified protein(s) in separate but otherwise identical reactions (Figure 2A). Following incubation, each sample is digested in preparation for mass spectrometry and labeled with a different isobaric tag. This tag imparts a unique chemical signature to every peptide, linking it to the kinase-substrate reaction from which it originated. Samples are combined and subjected to quantitative tandem mass spectrometry to directly identify peptide sequences and sites of phosphorylation. During peptide fragmentation, the isobaric tags are cleaved, generating reporter ions that are detected in the low-mass region of tandem mass spectra (Figure 2B). The intensity of these reporter ions functions as a readout for the abundance of phosphorylated peptides from the corresponding kinase reaction.
Figure 2.
A Simplified Schematic for MAKS
(A) Identical protein samples are treated separately with kinases, labeled, and mixed prior to analysis. A six-plex experiment is shown, but ten samples can be simultaneously assayed.
(B) Peptide identity and phosphorylation site localization are determined using tandem mass spectrometry. Reporter tags are cleaved during tandem mass spectrometry and appear in the low-mass range (liftout). The relative intensity for each reporter tag is directly proportional to the amount of phosphorylation contributed by the corresponding kinase.
MAKS Reveals Site-Specific Phosphorylation by ERK2 and CDK1 on NANOG
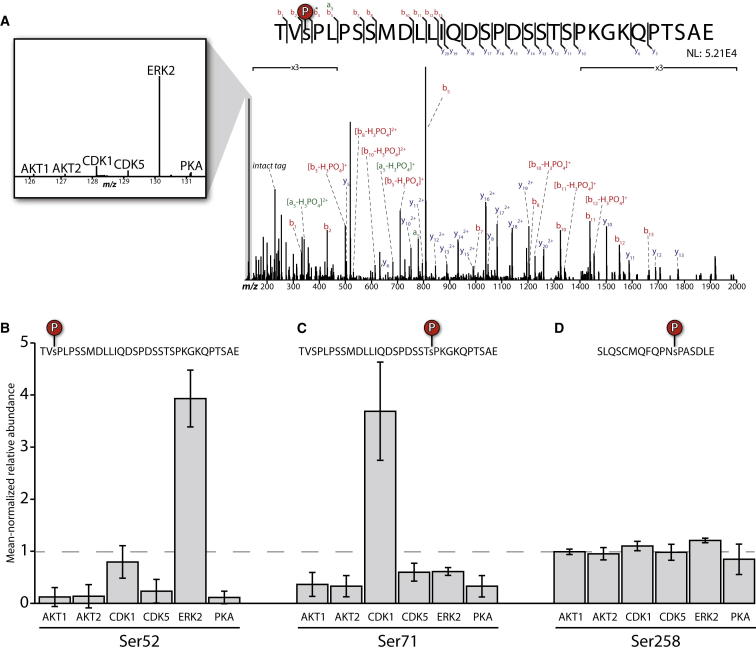
We selected AKT1, AKT2, CDK1/CyclinA2, CDK5/p35, ERK2, and PKA as candidate kinases for NANOG modification. We chose these kinases based primarily on motif analysis for phosphorylation sites on NANOG (ERK2 and CDK1/5), but also considered kinases involved in pluripotency and differentiation (AKT1/2 and PKA). This selection of kinases also covered candidates from both the AGC and CMCG kinase families. All kinases were active and capable of phosphorylating numerous control substrates (Table S2). We therefore performed our in vitro assay by incubating each kinase with full-length, recombinant NANOG. Following mass spectrometry, reporter ion intensities were mean normalized and adjusted using nonphosphorylated peptides to correct for pipetting error and sample loss (see Experimental Procedures; Figure S3A). In total, we quantified five phosphorylation sites, all of which corresponded to modifications on the endogenous protein (Figures 3 and S3B). As shown in Figure 3, ERK2 demonstrated a clear preference for Ser52, a putative ERK site that was also identified on the endogenous protein (Figure 3B). The abundance of phosphorylation for Ser52 was 5-fold higher for the ERK2 channel than for the next-closest channel. Interestingly, our data also showed that Ser71, another putative ERK site identified on endogenous NANOG, was instead preferentially phosphorylated by CDK1/CyclinA2 (Figure 3C), while Ser65 was modified by both ERK2 and CDK1/CyclinA2 (Figure S3B). This demonstrates the remarkable specificity achieved through this simple assay and also illustrates the importance of determining phosphorylation for individual sites. Phosphorylation at Ser23 and Ser258 was similar across all kinases (Figures 3D and S3B), suggesting that (1) a different kinase is responsible for phosphorylation at these sites, (2) cofactors not included in this assay are required for specificity, or (3) these sites can be phosphorylated by a variety of kinases. Importantly, this shows that the specificity observed at Ser52 and Ser71 is not the result of higher activity for the respective kinases. Phosphorylation sites that were identified on the endogenous protein but not quantified in the MAKS assay may be subject to regulation by kinases that were not included in our panel, or may require additional cofactors and/or subcellular localization.
Figure 3.
ERK2 and CDK1 Specifically Phosphorylate Different Sites on NANOG
(A) Mass spectrum from a typical kinase assay with Ser52 phosphorylation. Neutral loss (asterisk) is annotated. The liftout shows a zoomed view of the reporter ion region.
(B) ERK2 preferentially phosphorylates Ser52.
(C) CDK1 specifically phosphorylates Ser71.
(D) No kinase tested shows a preference for Ser258. Bar graphs represent the normalized abundance of at least three PSMs. Error bars represent the SD calculated from at least three PSMs across three independent experiments. The dashed line indicates the level that would be expected if the contribution for all channels were equal.
Discussion
Together with OCT4 and SOX2, NANOG functions as a key transcriptional regulator in pluripotency. To date, almost no information is available regarding posttranslational regulation of NANOG. Our data show that NANOG is multiply phosphorylated in human ESCs. Nine of 11 sites were observed in a serine/threonine-rich patch from residues 50–79, suggesting that this region may serve as a regulatory cluster. In support of this notion, recent computational work identified a putative PEST sequence (residues 47–72) on NANOG that overlaps greatly with the hyperphosphorylated region (Figure 1D; Ramakrishna et al., 2011). Deletion of the PEST sequence altered NANOG ubiquitination and degradation (Ramakrishna et al., 2011). Further, mutational analysis in mouse for sites homologous to Ser52, Ser65, and Ser71 suggested that phosphorylation of these residues stabilizes NANOG (Moretto-Zita et al., 2010). All of these results point to a role for this region in regulating NANOG protein levels. Our data provide direct evidence that these regulatory sites are phosphorylated on human NANOG and in a physiological context. The function of phosphorylation at Ser22/23 and Ser258 is currently unclear; however, Ser258 is located in the CD2 activation domain and may modulate NANOG’s ability to activate transcription (Do et al., 2009). Further work will be needed to clarify the exact role of each modification identified here.
For years, FGF has been a requisite component in human pluripotent stem cell media (Chen et al., 2011, Ludwig et al., 2006), and yet the mechanism by which FGF supports pluripotency was unknown. Our data show that ERK2, a downstream effector in the FGF pathway, directly phosphorylates NANOG at Ser52. A recent report demonstrated that phosphorylation of a homologous site on mouse NANOG facilitated interaction with Pin1, a prolyl isomerase that inhibits ubiquitination and subsequent proteosomal degradation by inducing conformational changes (Moretto-Zita et al., 2010). Together, these data suggest that ERK2 phosphorylation positively regulates NANOG by increasing its stability. The interplay between ERK and CDK signaling is also interesting in the context of pluripotency. Our data show that these kinases phosphorylate similar sequences on NANOG that are separated by only 19 amino acids. This result raises the possibility that CDK1 functions analogously to ERK, perhaps to sustain NANOG during S/M phase; however, this is only speculative at this point.
MAKS provides a multiplexed strategy for simultaneously screening multiple kinases and protein substrates. A major advantage of this assay is its capacity to directly assess kinase activity for individual phosphorylation sites. In the case of NANOG, this feature enabled us to dissect the specificity of ERK2 and CDK1/CyclinA2, even on nearby sites. This specificity may result from three-dimensional protein structure or cooperation with CyclinA2, although it is also possible that the primary sequence alone is sufficient to direct kinase activity. In this regard, a further benefit of MAKS is that it allows one to carry out kinase assays using whole proteins as a substrate. This preserves a potential phosphorylation site in the context of adjacent residues or protein structure. Other cofactors, subcellular localization, or additional modifications may be necessary to confer specificity. MAKS provides a means to test the role of many of these additional components, which can be added or removed from the kinase reaction to determine their effect. The multiplexing capacity is only limited by the number of isobaric tags available from the manufacturer (currently ten tags are commercially available), and because substrates are directly identified via mass spectrometry, multiple proteins of interest can be analyzed in a single reaction.
We note that the MAKS assay applies relative quantitation and therefore requires careful selection of candidate kinases. In some instances, for example, a physiological kinase for a given substrate may be absent from the panel tested. In such cases, MAKS will identify the kinase that exhibits the highest activity for a given substrate, if indeed there is a preference among the kinases tested. Computational methods will be critical for selecting the most likely candidate kinases, and testing a wide variety of kinases should help in identifying the most relevant kinases. To this end, we are currently working to couple kinase panels to automated mass spectrometry to increase the throughput of the MAKS assay and expand the number of kinases that can be tested to hundreds of kinases. The MAKS approach should also be applicable to any protein-modifying enzyme (i.e., acetylase, methyltransferase, etc.). Together, the simplicity and multiplexing capabilities of this assay make MAKS widely applicable in screening for enzymes that modify a substrate of interest.
Experimental Procedures
Kinase Assays
Kinase assays were carried out in 25 mM MOPS, pH 7.2, 12.5 mM beta-glycerol-phosphate, 25 mM MgCl2, 5 mM EGTA, 2 mM EDTA, 1 mM dithiothreitol (DTT), and 0.1 mM ATP. Approximately 1 μg of recombinant NANOG and 0.25 μg of AKT1, AKT2, CDK1, CDK5, or ERK2 (all from Promega), or 2.5 U of PKA (Sigma) were added for 30 min at 23°C. For control reactions, 0.5 μg of maltose-binding protein (MBP) and histone H1 (SignalChem) were added in place of NANOG. The reactions were then treated with 5 mM DTT at 37°C for 30 min, 10 mM iodoacetamide in the dark at 23°C for 30 min, and 10 mM DTT at 23°C for 30 min. Proteins were digested with GluC or chymotrypsin (both from Roche Diagnostics) at an enzyme-to-protein ratio of 1:200, purified by solid-phase extraction, and labeled with TMT isobaric tags (Thermo-Fisher) according to the manufacturer’s recommendations.
Data analysis
Phosphosite localization was evaluated using Phosphinator software (Swaney et al., 2009). This algorithm processes the theoretical fragment ion m/z ratios for all possible permutations of phosphopeptide isoforms based on sequence and the number of phosphorylation sites. It then compares each experimental spectrum to the theoretical product ions using a product mass tolerance of ± 0.02 Th. Phosphinator determines the significance of observed product ions that designate phosphorylation at a given amino acid. A localized site must have a p value of <0.05 for consideration. Further details about this algorithm were previously described (Swaney et al., 2009). Each localized phosphosite reported by Phosphinator was manually validated. Other high-scoring spectra with unlocalized phosphosites (as reported by Phosphinator) were manually interrogated to determine whether localization could be achieved. Theoretical product ions from other candidate phospho-isoforms were generated using the MS Product program in Protein Prospector (http://prospector.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msproduct), and localization and spectral chimerism were evaluated (Clauser et al., 1999). In all cases, product ions were required to be within 15 ppm of the theoretical m/z with a signal-to-noise ratio ≥ 3 to be used for sequence assignment. Both neutral loss product ions and internal fragments were considered during manual validation in addition to standard b- and y-type ions (HCD) and c- and z-type product ions (ETD).
TMT isotope purity corrections were applied using the TagQuant program in COMPASS (Wenger et al., 2011). Purity-corrected tag intensities were mean normalized for all peptide spectral matches (PSMs). We then took advantage of E. coli proteins that copurified with recombinant NANOG to further correct for pipetting error and sample loss. Briefly, E. coli PSMs were filtered to eliminate any peptide hit that contained a potential phosphorylation site (STY) or exhibited greater than 25% interference. The values for each channel were then averaged to establish the mean bias for that channel. Dividing one by each mean bias generated correction factors, which were used to normalize the quantitative values for each channel.
Acknowledgments
We are grateful to Krista Eastman for editing and Zhonggang Hou for discussion of the manuscript. This work was supported by the University of Wisconsin, the Beckman Foundation, and National Institutes of Health grants R01GM080148 (to J.J.C.) and P01GM081629 (to J.A.T. and J.J.C.). J.A.T. is a founder, stockowner, consultant, and board member of Cellular Dynamics International (CDI), and serves as scientific advisor to and has financial interests in Tactics II Stem Cell Ventures.
Published: January 9, 2014
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplemental Information includes Supplemental Experimental Procedures, three figures, and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2013.12.005.
Supplemental Information
References
- Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumbaugh J., Rose C.M., Phanstiel D.H., Thomson J.A., Coon J.J. Proteomics and pluripotency. Crit. Rev. Biochem. Mol. Biol. 2011;46:493–506. doi: 10.3109/10409238.2011.624491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumbaugh J., Hou Z., Russell J.D., Howden S.E., Yu P., Ledvina A.R., Coon J.J., Thomson J.A. Phosphorylation regulates human OCT4. Proc. Natl. Acad. Sci. USA. 2012;109:7162–7168. doi: 10.1073/pnas.1203874109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I., Colby D., Robertson M., Nichols J., Lee S., Tweedie S., Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., Robertson M., Vrana J., Jones K., Grotewold L., Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Chen G., Gulbranson D.R., Hou Z., Bolin J.M., Ruotti V., Probasco M.D., Smuga-Otto K., Howden S.E., Diol N.R., Propson N.E. Chemically defined conditions for human iPSC derivation and culture. Nat. Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauser K.R., Baker P., Burlingame A.L. Role of accurate mass measurement (+/- 10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal. Chem. 1999;71:2871–2882. doi: 10.1021/ac9810516. [DOI] [PubMed] [Google Scholar]
- Darr H., Mayshar Y., Benvenisty N. Overexpression of NANOG in human ES cells enables feeder-free growth while inducing primitive ectoderm features. Development. 2006;133:1193–1201. doi: 10.1242/dev.02286. [DOI] [PubMed] [Google Scholar]
- Do H.J., Lee W.Y., Lim H.Y., Oh J.H., Kim D.K., Kim J.H., Kim T., Kim J.H. Two potent transactivation domains in the C-terminal region of human NANOG mediate transcriptional activation in human embryonic carcinoma cells. J. Cell. Biochem. 2009;106:1079–1089. doi: 10.1002/jcb.22089. [DOI] [PubMed] [Google Scholar]
- Hennrich M.L., Marino F., Groenewold V., Kops G.J., Mohammed S., Heck A.J. Universal quantitative kinase assay based on diagonal SCX chromatography and stable isotope dimethyl labeling provides high-definition kinase consensus motifs for PKA and human Mps1. J. Proteome Res. 2013;12:2214–2224. doi: 10.1021/pr400074f. [DOI] [PubMed] [Google Scholar]
- Jeong C.H., Cho Y.Y., Kim M.O., Kim S.H., Cho E.J., Lee S.Y., Jeon Y.J., Lee K.Y., Yao K., Keum Y.S. Phosphorylation of Sox2 cooperates in reprogramming to pluripotent stem cells. Stem Cells. 2010;28:2141–2150. doi: 10.1002/stem.540. [DOI] [PubMed] [Google Scholar]
- Kettenbach A.N., Wang T., Faherty B.K., Madden D.R., Knapp S., Bailey-Kellogg C., Gerber S.A. Rapid determination of multiple linear kinase substrate motifs by mass spectrometry. Chem. Biol. 2012;19:608–618. doi: 10.1016/j.chembiol.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.R., Xing X.B., Chen T.T., Li R.X., Dai J., Sheng Q.H., Xin S.M., Zhu L.L., Jin Y., Pei G. Large scale phosphoproteome profiles comprehensive features of mouse embryonic stem cells. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.001750. M110 001750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linding R., Jensen L.J., Ostheimer G.J., van Vugt M.A., Jørgensen C., Miron I.M., Diella F., Colwill K., Taylor L., Elder K. Systematic discovery of in vivo phosphorylation networks. Cell. 2007;129:1415–1426. doi: 10.1016/j.cell.2007.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig T.E., Levenstein M.E., Jones J.M., Berggren W.T., Mitchen E.R., Frane J.L., Crandall L.J., Daigh C.A., Conard K.R., Piekarczyk M.S. Derivation of human embryonic stem cells in defined conditions. Nat. Biotechnol. 2006;24:185–187. doi: 10.1038/nbt1177. [DOI] [PubMed] [Google Scholar]
- Miller M.L., Jensen L.J., Diella F., Jørgensen C., Tinti M., Li L., Hsiung M., Parker S.A., Bordeaux J., Sicheritz-Ponten T. Linear motif atlas for phosphorylation-dependent signaling. Sci. Signal. 2008;1:ra2. doi: 10.1126/scisignal.1159433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K., Tokuzawa Y., Itoh H., Segawa K., Murakami M., Takahashi K., Maruyama M., Maeda M., Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- Moretto-Zita M., Jin H., Shen Z., Zhao T., Briggs S.P., Xu Y. Phosphorylation stabilizes Nanog by promoting its interaction with Pin1. Proc. Natl. Acad. Sci. USA. 2010;107:13312–13317. doi: 10.1073/pnas.1005847107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz J., Heck A.J. Quantitative proteome and phosphoproteome analysis of human pluripotent stem cells. Methods Mol. Biol. 2011;767:297–312. doi: 10.1007/978-1-61779-201-4_22. [DOI] [PubMed] [Google Scholar]
- Pan G., Thomson J.A. Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res. 2007;17:42–49. doi: 10.1038/sj.cr.7310125. [DOI] [PubMed] [Google Scholar]
- Phanstiel D.H., Brumbaugh J., Wenger C.D., Tian S., Probasco M.D., Bailey D.J., Swaney D.L., Tervo M.A., Bolin J.M., Ruotti V. Proteomic and phosphoproteomic comparison of human ES and iPS cells. Nat. Methods. 2011;8:821–827. doi: 10.1038/nmeth.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna S., Suresh B., Lim K.H., Cha B.H., Lee S.H., Kim K.S., Baek K.H. PEST motif sequence regulating human NANOG for proteasomal degradation. Stem Cells Dev. 2011;20:1511–1519. doi: 10.1089/scd.2010.0410. [DOI] [PubMed] [Google Scholar]
- Rigbolt K.T., Prokhorova T.A., Akimov V., Henningsen J., Johansen P.T., Kratchmarova I., Kassem M., Mann M., Olsen J.V., Blagoev B. System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci. Signal. 2011;4:rs3. doi: 10.1126/scisignal.2001570. [DOI] [PubMed] [Google Scholar]
- Schwartz D., Gygi S.P. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 2005;23:1391–1398. doi: 10.1038/nbt1146. [DOI] [PubMed] [Google Scholar]
- Singh A.M., Reynolds D., Cliff T., Ohtsuka S., Mattheyses A.L., Sun Y., Menendez L., Kulik M., Dalton S. Signaling network crosstalk in human pluripotent cells: a Smad2/3-regulated switch that controls the balance between self-renewal and differentiation. Cell Stem Cell. 2012;10:312–326. doi: 10.1016/j.stem.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S.A., Winter D., Bilimoria P.M., Bonni A., Steen H., Steen J.A. FLEXIQinase, a mass spectrometry-based assay, to unveil multikinase mechanisms. Nat. Methods. 2012;9:504–508. doi: 10.1038/nmeth.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songyang Z., Blechner S., Hoagland N., Hoekstra M.F., Piwnica-Worms H., Cantley L.C. Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr. Biol. 1994;4:973–982. doi: 10.1016/s0960-9822(00)00221-9. [DOI] [PubMed] [Google Scholar]
- Songyang Z., Lu K.P., Kwon Y.T., Tsai L.H., Filhol O., Cochet C., Brickey D.A., Soderling T.R., Bartleson C., Graves D.J. A structural basis for substrate specificities of protein Ser/Thr kinases: primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol. Cell. Biol. 1996;16:6486–6493. doi: 10.1128/mcb.16.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaney D.L., Wenger C.D., Thomson J.A., Coon J.J. Human embryonic stem cell phosphoproteome revealed by electron transfer dissociation tandem mass spectrometry. Proc. Natl. Acad. Sci. USA. 2009;106:995–1000. doi: 10.1073/pnas.0811964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaney D.L., Wenger C.D., Coon J.J. Value of using multiple proteases for large-scale mass spectrometry-based proteomics. J. Proteome Res. 2010;9:1323–1329. doi: 10.1021/pr900863u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallier L., Alexander M., Pedersen R.A. Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J. Cell Sci. 2005;118:4495–4509. doi: 10.1242/jcs.02553. [DOI] [PubMed] [Google Scholar]
- Van Hoof D., Muñoz J., Braam S.R., Pinkse M.W., Linding R., Heck A.J., Mummery C.L., Krijgsveld J. Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell. 2009;5:214–226. doi: 10.1016/j.stem.2009.05.021. [DOI] [PubMed] [Google Scholar]
- Wenger C.D., Phanstiel D.H., Lee M.V., Bailey D.J., Coon J.J. COMPASS: a suite of pre- and post-search proteomics software tools for OMSSA. Proteomics. 2011;11:1064–1074. doi: 10.1002/pmic.201000616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Fisher G.J. Receptor type protein tyrosine phosphatases (RPTPs)—roles in signal transduction and human disease. J. Cell Commun. Signal. 2012;6:125–138. doi: 10.1007/s12079-012-0171-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R.H., Sampsell-Barron T.L., Gu F., Root S., Peck R.M., Pan G., Yu J., Antosiewicz-Bourget J., Tian S., Stewart R., Thomson J.A. NANOG is a direct target of TGFbeta/activin-mediated SMAD signaling in human ESCs. Cell Stem Cell. 2008;3:196–206. doi: 10.1016/j.stem.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L., Wang W.H., Iliuk A., Hu L., Galan J.A., Yu S., Hans M., Geahlen R.L., Tao W.A. Sensitive kinase assay linked with phosphoproteomics for identifying direct kinase substrates. Proc. Natl. Acad. Sci. USA. 2012;109:5615–5620. doi: 10.1073/pnas.1119418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates A., Chambers I. The homeodomain protein Nanog and pluripotency in mouse embryonic stem cells. Biochem. Soc. Trans. 2005;33:1518–1521. doi: 10.1042/BST0331518. [DOI] [PubMed] [Google Scholar]
- Yu Y., Anjum R., Kubota K., Rush J., Villen J., Gygi S.P. A site-specific, multiplexed kinase activity assay using stable-isotope dilution and high-resolution mass spectrometry. Proc. Natl. Acad. Sci. USA. 2009;106:11606–11611. doi: 10.1073/pnas.0905165106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P., Pan G., Yu J., Thomson J.A. FGF2 sustains NANOG and switches the outcome of BMP4-induced human embryonic stem cell differentiation. Cell Stem Cell. 2011;8:326–334. doi: 10.1016/j.stem.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.