Abstract
Pigmentation is a rapidly evolving trait that is under both natural and sexual selection in many organisms. In the quinaria group of Drosophila, nearly all of the 30 species have an abdomen that is light in color with distinct markings; D. tenebrosa is the exception in that it has a completely melanic abdomen with no visible markings. In this study, we use a combination of quantitative genetic and candidate gene approaches to investigate the genetic basis of abdominal pigmentation in D. tenebrosa. We find that abdominal pigmentation is invariant across wild-caught lines of D. tenebrosa and is not sexually dimorphic. Quantitative genetic mapping utilizing crosses between D. tenebrosa and the light-colored D. suboccidentalis indicates that two genomic regions together underlie abdominal pigmentation, including the X-chromosome and an autosome (Muller Element C/E). Further support for their central importance in pigmentation is that experimental introgression of one phenotype into the other species, in either direction, results in introgression of these two genomic regions. Finally, the expression of the X-linked gene yellow in the pupae exactly foreshadows the adult melanization pattern in the abdomen of both species, suggesting that changes in the regulation of yellow are important for the phenotypic divergence of D. tenebrosa from the rest of the quinaria group. These results contribute to a body of work that demonstrates how changes in expression of highly conserved genes can cause substantial phenotypic differences even between closely related species.
Keywords: adaptation, selection, development, yellow
Introduction
Pigmentation is a trait that shows tremendous variation among organisms and that can have substantial consequences for fitness (Protas and Patel, 2008). In insects, pigmentation has been associated with diverse traits such as crypsis, predator avoidance, thermoregulation, mate choice and parasite avoidance (Majerus, 1998). In Drosophila, pigmentation is one of the fastest evolving morphological traits, and both natural and sexual selection are thought to have a role in this diversification (reviewed by True (2003) and Wittkopp and Beldade (2009)). Because of this variation and the genetic tools that are available, Drosophila have become a model both for understanding the genetic mechanisms of pigmentation evolution and the consequences of this variation.
Here we focus on the genetic basis of abdominal pigmentation of the fly Drosophila tenebrosa, which is a member of the quinaria group in the subgenus Drosophila. The quinaria group is a young adaptive radiation that contains ∼30 species, most of which utilize mushrooms for a mating substrate and as a food source. The species in this group are highly diverse in terms of pigmentation and vary both in wing spots and abdominal pattern (for example, Dombeck and Jaenike, 2004; Werner et al., 2010). In nearly all quinaria group species, the abdomen is yellow with distinct dark spots or bands. The only exception to this is D. tenebrosa, whose abdomen is completely black, with no visible spots or bands (Figure 1). Both males and females have a black abdomen, and this dark abdominal pigmentation is likely a derived state, as D. tenebrosa is not a basal species in the quinaria group.
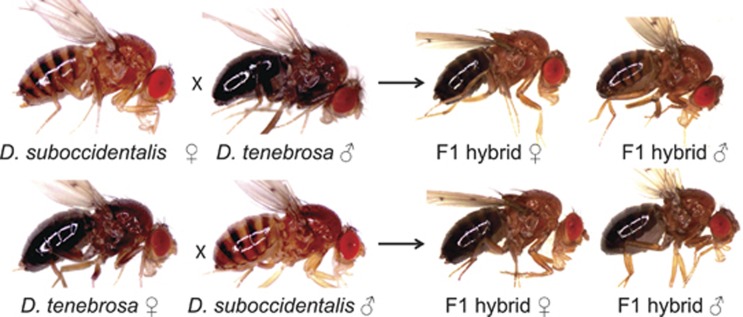
Figure 1.
Representative image of each species and the F1 hybrids from the reciprocal crosses.
D. tenebrosa has a unique geographical distribution relative to other quinaria group species: it is only found at the summits (>2500 m) of the sky island mountains of Arizona, New Mexico and Mexico. While several other species in the quinaria group are also found in these mountains, D. tenebrosa is the only species that is found primarily at high elevations. In other Drosophila species, dark pigmentation has been correlated with increased UV resistance and tolerance to desiccation and cold temperature, all of which are expected to be greater at higher elevation (reviewed by True (2003) and Wittkopp and Beldade (2009), but see Wittkopp et al. (2011)). Thus, it is possible that the dark pigmentation found in D. tenebrosa is a recently evolved adaptive trait.
In this study, we examine the genetic basis of the derived dark pigmentation in D. tenebrosa. We use both quantitative genetic and candidate gene approaches, including a combination of backcrosses, experimental introgression and expression analyses. Dark abdominal pigmentation does not vary across isofemale lines of D. tenebrosa, and so we use interspecific crosses to a closely related light-colored species, D. suboccidentalis, to elucidate the genetic basis of pigmentation. The backcrosses and introgressions indicate that two regions of the genome together underlie this trait, including the X-chromosome and an autosome. Expression analyses indicate that the yellow (y) gene is upregulated throughout the abdomen of D. tenebrosa relative to D. suboccidentalis, though the autosomal factor(s) remain unknown.
Materials and methods
Fly stocks and culturing
Our backcrossing, experimental introgression and expression studies used a single stock of D. tenebrosa collected in 2004 in the Chiricahua Mountains of Arizona and a single stock of D. suboccidentalis collected in 2004 near Bozeman Montana, each of which originated from a single wild-caught female. Seven additional isofemale lines of D. tenebrosa from two different geographic locations were used to survey variation in pigmentation within D. tenebrosa, which were collected more recently and are listed in Supplementary Table S1. Fly cultures and crosses were maintained on Instant Drosophila food (Carolina Biological, Burlington, NC, USA), supplemented with commercial mushroom (Agaricus bisporus). Flies were reared at 20 °C on a 14:10 light cycle and 60% relative humidity. Crosses were kept at a low density to prevent larval crowding.
Phenotypic characterization of pigmentation and its inheritance
For all experiments, abdominal pigmentation was scored on a scale of 1–8, with lower numbers representing lighter pigmentation and higher numbers representing darker pigmentation. All flies were 7 days old and scored by M Bray, using a dissecting microscope and a uniform level of light. A representative photo of each rank, with a description of the characteristics used to define that rank, is shown in Supplementary Figure S1. Images were taken under a dissecting microscope using a Zeiss ICc3 digital camera mounted on a Zeiss Stemi 2000 stereomicroscope (Carl Zeiss, Thornwood, NY, USA).
We first quantified the extent of natural variation in abdominal pigmentation within and among the lines of D. tenebrosa. Using the above ranking system, we scored at least 40 males and 40 females from each of the eight isofemale lines of D. tenebrosa. To determine the pattern of inheritance of the dark abdominal pigmentation, we completed reciprocal crosses between D. tenebrosa and D. suboccidentalis using the mapping lines described above. The F1 offspring were scored for abdominal pigmentation, as described above. The contribution of the X-chromosome to the pigmentation phenotype was estimated by comparing the difference in means of F1 males and females to the difference in the means of the F1 and parental females (Jones, 2001). As the hybrid F1 males were sterile and F1 females were fertile, we backcrossed the F1 hybrid females from the female D. suboccidentalis × male D. tenebrosa cross separately to male D. tenebrosa and D. suboccidentalis. Backcrossed progeny were collected on the day of eclosion and aged as virgins for 7 days before being scored for abdominal pigmentation, after which each fly was frozen for later genetic analysis.
Molecular methods and genotyping
To investigate the broad-scale genetic basis of dark pigmentation in D. tenebrosa, we used a total of 19 loci (Table 1), which included eight genes that control pigmentation or its regulation, two genes not involved in pigmentation and nine randomly chosen microsatellite loci. Primers for each locus are listed in Supplementary Table S2, and representative sequences for each species have been deposited to Genbank (accession KF751356-KF751381). We designed degenerate primers to conserved gene regions with GeneFisher2 (Giegerich et al., 1996), using the coding sequences for each gene from the D. melanogaster, D. pseudoobcura, D. virilis, D. grimshawii and D. mojavensis genomes, which were obtained from Flybase (www.flybase.org). In this way, we were able to amplify portions of black (b), bric-a-brac2 (bab2), bursicon (burs), ebony (e), engrailed (en), pale (ple), tan (t) and wingless (wg). Primers used to amplify the pigmentation gene yellow (y) and non-pigmentation genes mago nashi (mago) and plexin A (plexA) were previously available (Zilversmit et al., 2002; Dyer and Jaenike, 2004; Dyer et al., 2011). Based on the putative genomic locations of these 10 genes, we expected that every Muller Element including the dot chromosome would be included in our analysis.
Table 1. Map distances and single locus analysis for backcross to D. suboccidentalis.
| Locus | LG |
Putative |
Males |
Females |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Muller Element | Position (cM) | R2 | Effect | F | Pa | R2 | Effect | F | Pa | ||
| y | 1 | A | 0.00 | 0.616 | −3.51 | 311.45 | <0.0001 | 0.109 | −0.70 | 28.51 | <0.0001 |
| 2083 | 1 | A | 1.25 | 0.626 | −3.55 | 310.05 | <0.0001 | 0.088 | −0.61 | 21.68 | <0.0001 |
| t | 1 | A | 1.25 | 0.619 | −3.52 | 319.94 | <0.0001 | 0.084 | −0.61 | 21.67 | <0.0001 |
| 1082 | 1 | A | 1.50 | 0.607 | −3.55 | 290.73 | <0.0001 | 0.082 | −0.60 | 19.81 | <0.0001 |
| 3079 | 1 | A | 2.44 | 0.866 | −5.21 | 680.14 | <0.0001 | 0.072 | −0.55 | 18.20 | <0.0001 |
| wg | 2 | B | 0.00 | 0.026 | −0.63 | 4.04 | 0.046 | 0.033 | −0.39 | 7.14 | 0.008 |
| b | 2 | B | 5.30 | 0.036 | −0.86 | 7.39 | 0.007 | 0.011 | −0.22 | 2.65 | 0.105 |
| 1079 | 3 | C/E | 0.00 | 0.500 | −3.27 | 175.32 | <0.0001 | 0.583 | −1.62 | 293.21 | <0.0001 |
| 3038 | 3 | C/E | 5.11 | 0.533 | −3.32 | 164.11 | <0.0001 | 0.654 | −1.76 | 371.71 | <0.0001 |
| burs | 3 | C/E | 7.16 | 0.525 | −3.27 | 218.74 | <0.0001 | 0.637 | −1.68 | 420.96 | <0.0001 |
| mago | 3 | C/E | 8.29 | 0.505 | −3.18 | 206.79 | <0.0001 | 0.625 | −1.66 | 409.01 | <0.0001 |
| e | 3 | C/E | 10.32 | 0.505 | −3.18 | 207.09 | <0.0001 | 0.621 | −1.65 | 392.82 | <0.0001 |
| 2084 | 3 | C/E | 12.76 | 0.519 | −3.28 | 197.44 | <0.0001 | 0.628 | −1.65 | 391.09 | <0.0001 |
| en | 4 | C | 0.00 | 0.001 | −0.13 | 0.17 | 0.680 | 0.008 | −0.19 | 1.84 | 0.176 |
| 3090 | 5 | ? | 0.00 | 0.035 | 0.68 | 2.67 | 0.107 | 0.001 | −0.07 | 0.17 | 0.677 |
| 4073 | 5 | ? | 24.40 | 0.001 | 0.16 | 0.24 | 0.623 | 0.007 | −0.18 | 1.56 | 0.213 |
| bab2 | 6 | D | 0.00 | 0.001 | 0.13 | 0.17 | 0.684 | 0.002 | −0.10 | 0.55 | 0.459 |
| ple | 6 | D | 2.80 | 0.000 | 0.01 | 0.00 | 0.965 | 0.012 | −0.23 | 2.98 | 0.086 |
| plexA | 7 | F | 0.00 | 0.000 | −0.01 | 0.00 | 0.971 | 0.005 | 0.14 | 1.05 | 0.306 |
Abbreviations: LG, Linkage group; ?, unknown Muller Element.
Values in bold are significant after Bonferroni correction.
Bonferroni-corrected P-value of 0.05/19=0.00263.
DNA was extracted from single flies using the Qiagen Puregene kit (Qiagen, Valencia, CA, USA), standard PCR methods were used, and amplicons were sequenced in both directions. Base calls, including heterozygous sites, were confirmed using Sequencher (Gene Codes, Ann Arbor, MI, USA). BLAST algorithms were used to confirm that the amplified region was the intended gene. Genotyping of these loci for genetic mapping utilized either a length difference or a restriction site difference that was fixed between the mapping stocks, based on the sequences of four females from each stock (Supplementary Table S2).
We also used nine random microsatellite loci that were initially characterized from D. recens, but which our preliminary studies found also amplified in D. tenebrosa and D. suboccidentalis. These loci were each fixed for length difference between the two mapping stocks, based on genotyping four females from each stock. To genotype these loci for genetic mapping, one PCR primer was fluorescently labeled, amplicons were run in multiplex and concurrent with a size standard, and fragments were scored with GeneMarker (SoftGenetics, State College, PA, USA).
In total, we genotyped 736 F2 backcrossed offspring. From the backcross to D. suboccidentalis, this included 213 males and 251 backcrossed females, which were each genotyped at all 19 loci. From the backcross to D. tenebrosa, we genotyped 171 males and 101 backcrossed females at eight pigmentation-associated loci (all of the above except wg) as well as at mago and plexA.
Genetic association of loci with pigmentation
We constructed genetic maps independently for each backcrossed population, using all of the genotyped male and female F2 offspring. For each marker, segregation of alleles to the F2 generation was compared with 1:1 using a χ2 test, with the significance threshold determined by a Bonferroni correction. The linkage and order of markers was determined with the program MapDisto (Lorieux, 2012), using the Kosambi mapping function to assign linkage distances between markers (Kosambi, 1944). Based on earlier cytological analyses, we expect that D. suboccidentalis has the ancestral Drosophila karyotype of five rod chromosomes and a small dot chromosome, and that the karyotype of D. tenebrosa is identical except for a fusion of two autosomal elements (Patterson and Stone, 1952). Based on sequencing in males and females, the X-chromosome in D. tenebrosa and D. suboccidentalis is the same Muller Element (A) as the X-chromosome of D. melanogaster and most other species of Drosophila (Dyer, unpublished data). There is large-scale conservation of most genes to Muller Elements across Drosophila (Bhutkar et al., 2008).
We tested for single-locus effects on pigmentation with a one-way analysis of variance, as implemented in MapDisto, using a Bonferroni correction to determine the significance threshold. We conducted composite interval mapping with the program Windows QTL Cartographer v. 2.5 (Wang and Zeng, 2012). We used the linkage map determined from each relevant backcross and analyzed each sex of each backcross separately. Composite interval mapping used Model 6 in QTL Cartographer to test for the presence of a quantitative trait locus (QTL), computing the likelihood ratio every 0.5 cM, with five background markers chosen by forward regression using a window size of 10 cM. Significance thresholds were determined by 1000 permutations of the data (Churchill and Doerge, 1994).
Finally, we tested for epistatic effects between loci. As the linkage groups (LGs) were short, we used only one representative marker from each LG, which generally had the highest single locus effect for that group. This included y, b, burs, en, 3090, ple and plexA, with 3090 only included in the backcross to D. suboccidentalis. We tested for all two-way epistatic effects separately in each sex and backcross using PROC MIXED in SAS v. 9.3 (SAS Institute, Cary, NC, USA), fitting the model Y=M1+M2+M1*M2+ɛ. M1 and M2 are the genotypes of two different markers and were treated as fixed effects in the model, and ɛ is the random error term. The significance of the interaction term (M1*M2) was assessed using a Bonferroni correction.
Experimental introgression of pigmentation
We used repeated backcrossing to experimentally introgress the dark phenotype of D. tenebrosa into the D. suboccidentalis genetic background, and the light phenotype of D. suboccidentalis into the D. tenebrosa genetic background. We initiated each introgression with ∼100 flies of each sex. We collected the offspring of each generation as virgins, and then scored the flies for abdominal pigmentation when at least 7 days old. From the second generation on, depending on the introgression direction the 10–20 lightest or darkest females were chosen and backcrossed to pure species males. Males from generation 10 were scored for abdominal pigmentation phenotype and genotyped at the 19 loci used in the genetic mapping, as described above. This included 58 males from the introgression of the dark abdomen into D. suboccidentalis and 27 males from the introgression of the light abdomen into D. tenebrosa. We attempted to make the regions controlling pigmentation homozygous by full-sib inbreeding of introgression lines; however, this did not produce any offspring with the introgressed phenotype, which may be due to linked genetic incompatibilities. To test for an association of genotype with pigmentation, we used Fisher Exact Tests (FET) to determine whether the allele from the other species was over-represented in flies with the introgressed phenotype compared with flies that most closely resembled the species to which the line was backcrossed.
Expression analyses of candidate genes
Based on the results obtained from the above genetic analyses, we assayed three candidate genes—y, t and e—for expression in D. tenebrosa and D. suboccidentalis. To accomplish this, we used whole-mount in situs of P10–P15 pupal stages and freshly hatched adults (Bainbridge and Bownes, 1981). In other Drosophila species, these are the stages when the cuticle pigmentation is laid down (Walter et al., 1991; Kornezos and Chia, 1992). We cut the pupae and adults with a scalpel into left and right halves and fixed them as described by Wang and Yoder (2011). Using species-specific antisense RNA probes (Supplementary Table S3) we performed in situ hybridization of the mounted specimens, following the method of Werner et al. (2010). We imaged the abdomens at multiple focal planes with an Olympus DP72 camera attached to an Olympus SZX16 dissection microscope (Olympus, Center Valley, PA, USA) and z-stacked the images with Helicon Focus software.
Results
Phenotypic characterization of pigmentation inheritance
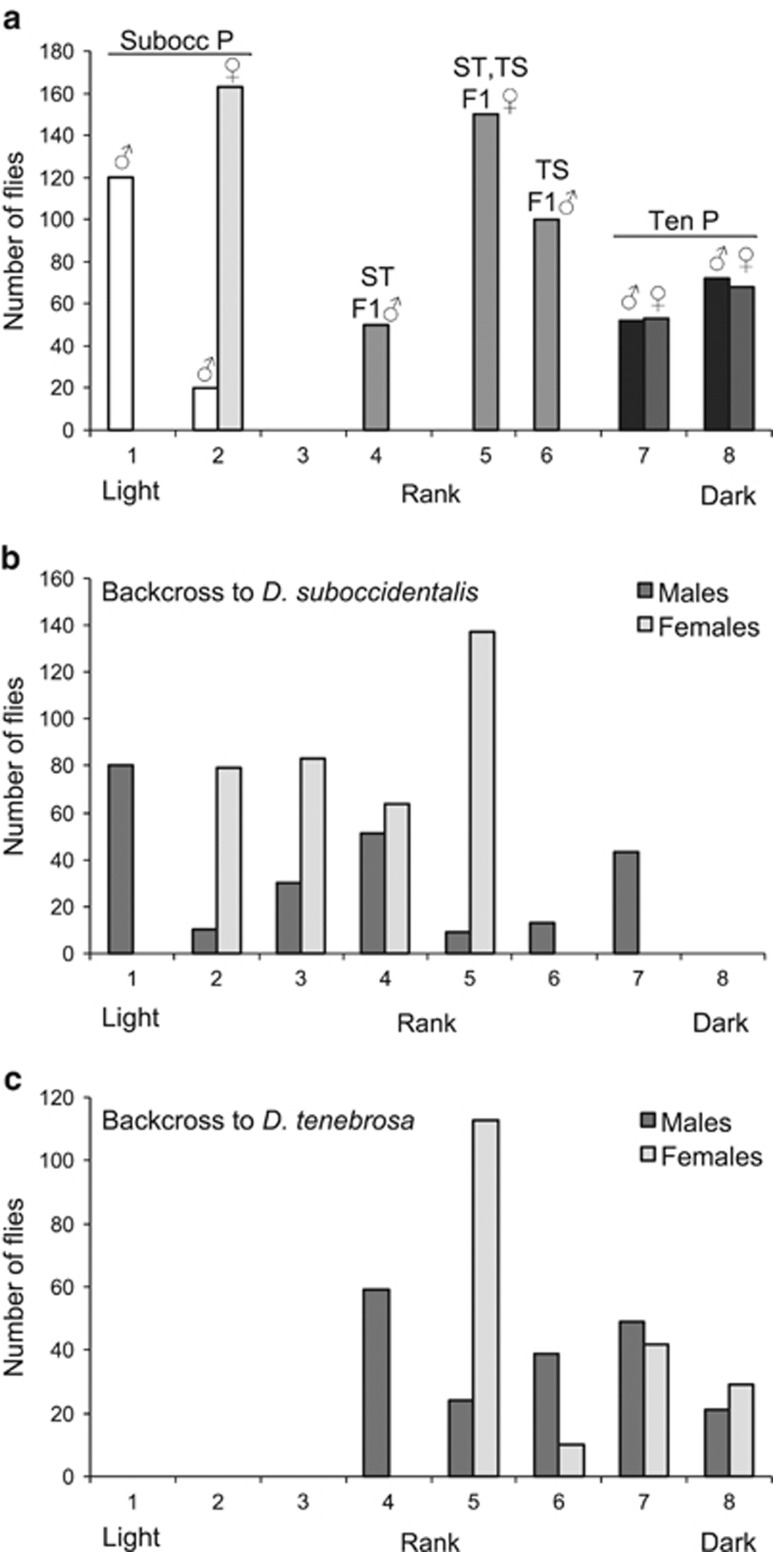
There was very little phenotypic variation within and among the eight isofemale lines of D. tenebrosa, which included the line used in mapping (Supplementary Table S1). Overall, the lines were consistently very dark in abdominal pigmentation, with no difference between males and females. Comparing the stocks of D. tenebrosa and D. suboccidentalis used in our genetic analyses, D. tenebrosa males averaged 7.56 and females 7.58 on the scale from 1 to 8, whereas D. suboccidentalis were very light, with males averaging 1.14 and females 2.00 (Supplementary Table S1, Figure 2a). In the reciprocal crosses of pure species D. tenebrosa and D. suboccidentalis, the pigmentation of the F1 hybrid females was the same regardless of the direction of the cross (rank 5, N=100 each cross; Figures 1 and 2a). This is close to the mid-parent values of 4.78 and 4.36 for the two reciprocal crosses, suggesting that pigmentation phenotype is mostly additive. In contrast, F1 males from each reciprocal cross appear more similar to their female parent: when the female parent was D. suboccidentalis, F1 hybrid males were all rank 4 (N=100), and when D. tenebrosa, F1 hybrid males were all rank 6 (N=50) (Figures 1 and 2a). As males only possess an X-chromosome from their mother, this pattern suggests that gene(s) on the X-chromosome contribute to the abdominal pigmentation phenotype. In line with this, based on the average pigmentation values of the parents and F1 males and females, the contribution of the X-chromosome to the abdominal phenotype is substantial, and is 40% when D. tenebrosa is the female parent and 33% when D. suboccidentalis is the female parent.
Figure 2.
Abdominal pigmentation phenotypes in pure species mapping lines, hybrids and backcrossed individuals. (a) Parents and F1 offspring from reciprocal crosses. Flies from the pure species mapping stocks are denoted ‘P.' For F1 hybrids, the female parent is listed first, where ST=D. suboccidentalis female × D. tenebrosa male, and TS=D. tenebrosa female × D. suboccidentalis male. (b) F2 offspring from backcross F1 hybrid female to D. suboccidentalis male and (c) F2 offspring from backcross of F1 hybrid female to D. tenebrosa male. See the text for details on the scoring of pigmentation.
As expected, the F2 backcross offspring showed substantial phenotypic variation (Figure 2). In the backcross to D. suboccidentalis, the females occurred in roughly equal numbers in ranks 2 through 5 (mean=3.71±0.06 (s.e.), N=363), and the males spanned from rank 1 through 7 (Figure 2b; mean=3.46±0.14, N=236). Among the offspring of the backcross to D. tenebrosa, females ranged between ranks 5 through 8 (mean=5.93±0.08, N=194), and males spanned from ranks 4 through 8 (Figure 2c; mean=5.77±0.10, N=192). The clearest phenotypic pattern among the backcross offspring is that the males from the backcross to D. suboccidentalis have a distinct trimodal distribution in phenotype (Figure 2b). This pattern is consistent with two loci of major effect where substituting a D. tenebrosa allele at either locus has a similar effect on pigmentation phenotype.
Genetic map and association of loci with pigmentation
The 19 loci formed seven LG in the backcross to D. suboccidentalis (Table 1). Results were consistent between the two different backcrosses, though the map from the backcross to D. tenebrosa contained fewer loci (Table 2). All loci that are on the same Muller Element in other Drosophila species were in linkage with each other in our analysis, with the exception of en, which was not linked to any other locus. Because of the low marker density, en may simply be far enough away from the other physically linked markers such that it segregates freely. Overall, each LG is very short, between 2 and 24 cM, which is likely due to inversion differences between our mapping stocks. Thus, to a large extent our genetic resolution is at the whole-chromosome level. In the backcross to D. suboccidentalis, two loci, b and wg on LG2, showed a significant excess of D. tenebrosa alleles (Supplementary Table S4). In the backcross to D. tenebrosa, four loci showed significant excess of D. suboccidentalis alleles, including b on LG2, and burs, e and mago on LG3. These may indicate the presence of genetic incompatibilities or segregation distorters.
Table 2. Map distances and single locus analysis for backcross to D. tenebrosa.
| Locus | LG |
Putative |
Males |
Females |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Muller Element | Position (cM) | R2 | Effect | F | Pa | R2 | Effect | F | Pa | ||
| y | 1 | A | 0.00 | 0.215 | 1.18 | 71.87 | <0.0001 | 0.003 | 0.11 | 0.29 | 0.590 |
| t | 1 | A | 0.39 | 0.188 | 1.09 | 59.12 | <0.0001 | 0.001 | −0.05 | 0.06 | 0.814 |
| b | 2 | B | 0.00 | 0.000 | 0.02 | 0.01 | 0.931 | 0.000 | −0.04 | 0.03 | 0.857 |
| mago | 3 | C/E | 0.00 | 0.429 | 1.75 | 197.07 | <0.0001 | 0.859 | 1.91 | 599.36 | <0.0001 |
| burs | 3 | C/E | 1.91 | 0.425 | 1.76 | 191.59 | <0.0001 | 0.858 | 1.90 | 584.85 | <0.0001 |
| e | 3 | C/E | 3.07 | 0.445 | 1.81 | 211.04 | <0.0001 | 0.859 | 1.90 | 596.54 | <0.0001 |
| en | 4 | C | 0.00 | 0.006 | −0.18 | 1.37 | 0.243 | 0.010 | −0.20 | 0.91 | 0.343 |
| bab2 | 6 | D | 0.00 | 0.001 | 0.07 | 0.18 | 0.672 | 0.035 | −0.38 | 3.56 | 0.062 |
| ple | 6 | D | 3.09 | 0.004 | −0.17 | 1.12 | 0.292 | 0.034 | −0.38 | 3.41 | 0.068 |
| plexA | 7 | F | 0.00 | 0.004 | −0.16 | 1.06 | 0.305 | 0.005 | −0.14 | 0.47 | 0.494 |
Abbreviation: LG, linkage group.
Values in bold are significant after Bonferroni correction.
Bonferroni-corrected P-value of 0.05/10=0.005.
Based on both single-marker analysis and composite interval mapping mapping, we identified two QTL with large effects on pigmentation (Tables 1, 2, 3). The first extends across the entire X-chromosome (LG1). The effect of this QTL is largest in males, where it accounts for ∼44% of the phenotypic variance in each backcross (Table 3). This QTL is also identified in females from the D. suboccidentalis backcross, where it accounts for 14% of the phenotypic variance in pigmentation, though it is not identified in females backcrossed to D. tenebrosa. The effect of this X-linked QTL is equal and opposite in the males from both backcrosses, and the effect in males is approximately twice that in females from the D. suboccidentalis backcross (Table 3); these patterns are both consistent with no dominance effect of this QTL.
Table 3. QTL identified with composite interval mapping.
| Backcross F2 Population | Sex | Linkage group | QTL | Peak LR | Effect (s.e.) | Effect/σP | R2 |
|---|---|---|---|---|---|---|---|
| To D. suboccidentalis | F | 1 | y—3079 | 123.75 | −0.81 (0.003) | −0.77 | 0.14 |
| 3 | 1079—e | 354.45 | −1.75 (0.001) | −1.67 | 0.69 | ||
| 6 | bab2—ple | 13.16 | −0.23 (0.007) | −0.22 | 0.01 | ||
| M | 1 | y—3079 | 431.95 | −3.03 (0.004) | −1.35 | 0.43 | |
| 3 | 1079—e | 288.47 | −2.42 (0.002) | −1.08 | 0.19 | ||
| To D. tenebrosa | F | 3 | mago—e | 210.44 | 1.95 (0.005) | 1.90 | 0.80 |
| M | 1 | t—y | 182.24 | 1.93 (na) | 1.39 | 0.44 | |
| 3 | mago—e | 135.74 | 1.93 (0.018) | 1.39 | 0.28 |
Abbreviations: LR, likelihood ratio; σP, standard deviation of the phenotype; R2, proportion of the variance accounted for by the QTL.
The second major QTL spans most of Muller Element C/E (LG3). This QTL is identified in both sexes and in both backcrosses. In males it accounts for 20–30% of the variance in phenotype, and in females it accounts for 70–80% of the variance in phenotype (Table 3). This QTL had larger effects in both males and females in the backcross to D. tenebrosa than to D. suboccidentalis, suggesting partial dominance of the D. suboccidentalis genotype. As dominance reduces the power to detect QTL, this may explain why we did not detect the X-linked QTL in females from the D. tenebrosa backcross. Our backcross analysis also identified one additional minor-effect QTL in females from the backcross to D. suboccidentalis on Muller Element D (LG6), though the effect is very small compared with the other chromosomes (R2=0.01). Within each sex and backcross, the QTLs we have identified account for a total of 60–85% of the variation in phenotype (Table 3).
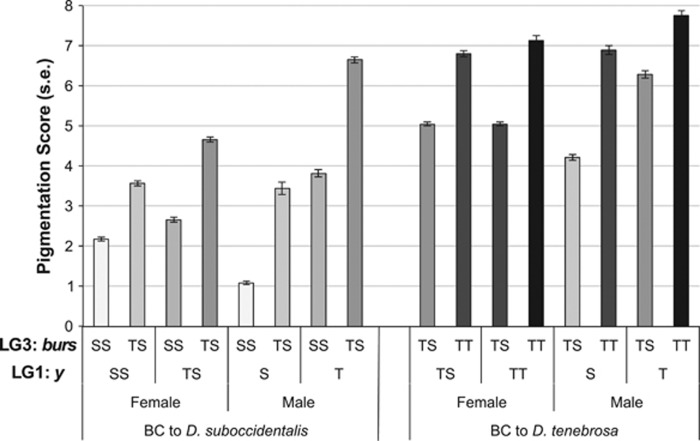
The phenotypic effects of the two major QTL are visualized in Figure 3, which shows the pigmentation phenotypes of the backcross flies given their genotype at y on the X-chromosome and burs on LG3. As we completed both backcrosses, we can compare the phenotypes of the genotypic classes from the two backcrosses that are identical in genotype at these markers but that may differ in the composition of the rest of their genome. This includes males that have the D. suboccidentalis y allele and are heterozygous at burs, males that have the D. tenebrosa y allele and are heterozygous at burs, and females that are heterozygous at both makers. For each of these comparisons, the pigmentation phenotypes of the two backcrosses are nearly identical, supporting the conclusion that these two QTL explain most of the variation in phenotype.
Figure 3.
Mean pigmentation scores of the F2 haplotypes derived from the two QTL with large effects on pigmentation. The letters denote the two alleles (S: D. suboccidentalis, T: D. tenebrosa), with S and T indicating hemizygous state of the X-chromosome in males, and SS, ST and TT indicating autosomes and the X-chromosome in females. A representative locus from each of the LGs was used, including y from the LG1 (the X-chromosome) and burs from LG3 (Muller Element C/E). Individuals are separated by backcross and by sex. The shading of the bars indicates an increasing number of D. tenebrosa alleles.
We also find evidence for some epistasis between these two QTL. After correcting for multiple tests, there are significant epistatic effects between y and burs in F2 females from the backcross to D. suboccidentalis as well as F2 males from the backcross to D. tenebrosa (Table 4). In both cases, the magnitude but not the direction of the reaction norm slopes differed. Among the females from the D. suboccidentalis backcross, flies that are heterozygous at both y and burs are darker than expected based on the individual allele affects. Likewise, among males from the D. tenebrosa backcross, flies that carry a D. suboccidentalis allele at y and are heterozygous at burs are lighter than expected based on the single-locus effects. We also find that in F2 females backcrossed to D. tenebrosa, there is a significant epistatic interaction between burs (LG3) and en (LG 4), though LG4 is not identified as a QTL in other analyses. In this interaction, the reaction norms cross, and carrying a D. suboccidentalis allele for en results in darker flies when the burs genotype is homozygous for the D. tenebrosa allele, whereas the opposite is true when the burs genotype is heterozygous. This may indicate the existence of a background-dependent modifier on pigmentation. Overall, our mapping results indicate that the X-chromosome and one autosome confer the differences in pigmentation, and that there is some epistasis between them. As our genetic mapping analysis is limited to the whole chromosome, further work is necessary to disentangle the epistatic effects because these interactions may change substantially if more than one gene within each QTL contributes to pigmentation.
Table 4. Significant epistatic effects that affect pigmentation between D. tenebrosa and D. suboccidentalis.
| Backcross F2 population | Sex | Marker 1 (LG) | Marker 2 (LG) | Effect (s.e.) | Effect/σP | P-valuea |
|---|---|---|---|---|---|---|
| To D. suboccidentalis | F | y (1) | burs (3) | 0.548 (0.16) | 0.521 | 0.0009 |
| y (1) | en (4) | 0.859 (0.32) | 0.818 | 0.0082 | ||
| To D. tenebrosa | F | burs (3) | en (4) | −0.516 (0.14) | −0.503 | 0.0004 |
| burs (3) | ple (6) | 0.463 (0.15) | 0.451 | 0.0034 | ||
| M | y (1) | burs (3) | −1.271 (0.33) | −0.918 | 0.0002 | |
| y (1) | b (2) | 0.876 (0.41) | 0.633 | 0.0327 | ||
| en (4) | b (2) | −1.233 (0.51) | 0.890 | 0.0174 |
Abbreviations: LG, linkage group; σP, standard deviation of the phenotype.
Effects that are significant after Bonferroni correction are in bold.
Bonferroni correction to D. suboccidentalis: 0.05/20=0.0025 and to D. tenebrosa: 0.05/15=0.0033.
Experimental introgression of pigmentation
We used experimental backcrossing to introgress the dark phenotype into the light background, and vice versa. Overall, there is a clear pattern that the X-chromosome and LG3 introgressed more often than the other LGs, regardless of the direction of introgression (Table 5 and Supplementary Table S5). In the introgression of dark pigmentation into D. suboccidentalis, there is an over-representation of the X-chromosome and LG3 from D. tenebrosa in the darkest and medium pigmentation males relative to the lightest individuals (all FET P<0.009). Interestingly, among the medium pigmentation-ranked flies, males had either the D. tenebrosa allele for the X-chromosome or LG3, but never both, consistent with the equal effects of substituting either of these chromosomes as seen in the QTL mapping analyses (Supplementary Table S5). In the introgression of light pigmentation into D. tenebrosa, the males with the lightest pigmentation all had the X-chromosome and LG3 from D. suboccidentalis (Table 5 and Supplementary Table S5), though this is only significant for the X-chromosome (X-chromosome: FET P=0.002; LG3: FET P=0.18). Of 16 medium-ranked pigmentation flies (rank 7), none carried the D. suboccidentalis X-chromosome (FET P=1.0), and all carried LG3 from D. suboccidentalis (FET P=0.048). Across both series of crosses, no other LG was over-represented in the introgressed phenotype class in both introgression directions, again suggesting that the X-chromosome and LG3 contribute the most to pigmentation.
Table 5. Summary of genotypes of males from introgressions, summarized by linkage group (LG).
| Introgression/phenotype | Rank | LG1 | LG2 | LG3 | LG4 | LG5 | LG6 | LG7 | Total flies |
|---|---|---|---|---|---|---|---|---|---|
| Dark into D. suboccidentalis | |||||||||
| Lightest | 1 | 0.00 | 0.40 | 0.07 | 0.00 | 0.10 | 0.37 | 0.00 | 30 |
| Medium | 3 | 0.24 | 0.57 | 0.76 | 0.00 | 0.05 | 0.29 | 0.00 | 21 |
| Darkest | 6 | 1.00 | 1.00 | 1.00 | 0.00 | 0.00 | 0.14 | 0.00 | 7 |
| Light into D. tenebrosa | |||||||||
| Darkest | 8 | 0.00 | 0.00 | 0.60 | 0.00 | 0.00 | 0.00 | 0.00 | 5 |
| Medium | 7 | 0.00 | 0.13 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 16 |
| Lightest | 4 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 6 |
Abbreviations: LG, linkage group; Rank, pigmentation rank as described in the text.
Shown is the fraction of males with the allele of the species with the introgressed phenotype. Numbers in bold indicate linkage groups with a significant difference (P<0.01) using a Fisher Exact Test of the allele representation versus the flies that are most similar to each pure species (that is, lightest in the introgression into D. suboccidentalis, and darkest in the introgression into D. tenebrosa).
Expression analyses of candidate genes
To test whether the y, t and e genes are expressed in the pupal abdomens of D. tenebrosa and D. suboccidentalis, we performed in situ hybridization experiments at the pupal stages P10–P15 and in newly hatched adults with species-specific RNA probes. We found that the X-linked y gene is strongly expressed in patterns precisely foreshadowing the future black abdominal pigment in both species at stage P10 (Figure 4), with a weaker pattern apparent at stage P11 and nearly undetectable at stage P12 and later. Specifically, in D. tenebrosa, y was expressed throughout the abdominal tergites, while in D. suboccidentalis, y expression appeared in the shape of the future black stripes on the posterior half of each tergite. The y expression signal was detectable as early as 10 min after we added the staining solution. In contrast, the e and t genes did not show any significant signal in either species from P10 to eclosion, indicating either that our probes did not work or that these genes are not expressed at the stages we assayed. In sum, our results suggest that y is an important gene that contributes to the adult pigmentation patterns in both species by being differentially expressed.
Figure 4.
y gene expression foreshadows the black pigment on adult abdomens. (a) D. tenebrosa adult, showing the uniformly black tergites on the abdomen. (b) D. tenebrosa, abdominal epidermis at pupal stage P10. The in situ hybridization, using a species-specific probe, shows that the y gene expression pattern correlates with the future black melanin pattern. (c) D. suboccidentalis adult, showing that tergites of the adult abdomen are yellow, each adorned with a black stripe along the posterior edge. (d) D. suboccidentalis, abdominal epidermis at pupal stage P10. In situ hybridization with a species-specific probe reveals that the y gene expression pattern foreshadows the black stripes in the adult cuticle.
Discussion
In this study, we investigated the dark abdominal pigmentation that is found only in D. tenebrosa and not in any other species in the quinaria group of Drosophila. While our study is limited by the presence of chromosomal inversions that are fixed between our mapping lines, we show that the derived dark pigmentation in D. tenebrosa can be attributed to one or more loci on the X-chromosome, and to one or more loci on an autosome, specifically Muller Element C/E (LG3). Thus, the dark abdominal pigmentation of D. tenebrosa may have a fairly simple genetic basis, with as few as two genes controlling the majority of the phenotype. Furthermore, our finding of some epistasis between the QTL is consistent with the underlying genes being in the same pathway(s). Our in situ expression analysis reveals that the X-linked gene yellow is highly expressed throughout the abdomen of D. tenebrosa, whereas in D. suboccidentalis yellow expression is limited to the locations where dark pigmentation will later be present. This pattern of yellow expression is consistent with dark pigmentation only occurring in the abdomen of D. tenebrosa. Thus, our results suggest that the yellow gene contributes to the dark pigmentation of D. tenebrosa. However, there may be additional factors on the X-chromosome that also contribute to the dark phenotype, possibly by regulating yellow expression.
The yellow gene is conserved across insects (Ferguson et al., 2011) and has been implicated in the synthesis of melanic pigment in Drosophila wings, abdomen, thorax and mouthparts, as well as in certain aspects of behavior (Biessmann, 1985; Wittkopp et al., 2002a; Wittkopp et al., 2002b; Drapeau et al., 2003; Gompel et al., 2005; Prud'homme et al., 2006). yellow has also been implicated in the pigmentation of other insects, for example the silkmoth Bombyx mori and Heliconius butterflies (Futahashi et al., 2008; Hines et al., 2012). Further analyses are necessary to characterize the sites responsible for the change in expression of yellow between D. tenebrosa and D. suboccidentalis. At the amino-acid level, the portion of yellow we have sequenced is identical in these two species and >95% identical to D. melanogaster; however, because we have not sequenced the entire coding region from both D. tenebrosa and D. suboccidentalis, we cannot infer with certainty whether the change in yellow expression is due to coding sequence changes or to cis-regulatory changes. Based on work in other species, we suggest that non-coding changes probably underlie this difference in expression, as all other instances where yellow has been implicated in pigmentation changes have been due to regulatory changes (Gompel et al., 2005; Jeong et al., 2006; Prud'homme et al., 2006; Jeong et al., 2008; Werner et al., 2010).
In this study, we were not able to identify the autosomal factor(s) that contribute to the dark pigmentation of D. tenebrosa. One obvious gene in LG3 to consider is ebony, though we did not detect any expression of this gene in the pupal stages we surveyed. More telling is that in our QTL mapping, the LOD values in the region of ebony were significant for both sexes and backcrosses, but they were not the highest LOD values for that QTL. For instance, in males from the D. suboccidentalis backcross, the average LOD near ebony was 44, but was 273 at the other loci on that LG. This suggests that other loci on that chromosome probably contribute more to variation in phenotype than ebony. Several other genes known to be involved in pigmentation are on Muller Elements C and E in other Drosophila species, for example, abd-A, Abd-B, yellow-e, burs and Dat.
Our analyses rule out the involvement of many other characterized genes known to be involved in pigmentation in Drosophila. For example, this includes wingless on Muller Element B, which was previously shown to induce yellow expression to cause the formation of melanin on the wing in other species of the quinaria group (for example, D. guttifera) (Werner et al., 2010). However, our genetic mapping did not indicate that wingless had a significant role in the dark pigmentation of D. tenebrosa. This suggests that even in closely related species there are multiple mechanisms by which dark pigmentation can evolve.
Based on our assays here and collections of wild D. tenebrosa, this species displays very little variation in abdominal pigmentation. This is in contrast to other quinaria group species, which harbor substantial genetic variation for abdominal pigmentation (Dombeck and Jaenike, 2004; K. Dyer, personal observation). This may be due to directional selection or, in contrast, it may indicate the fixation of dark alleles due to drift, for example, due to a low effective population size in D. tenebrosa. As both of the major QTL we identified have effects in the same direction (Table 3), this lends support that the phenotype may be adaptive. In addition, in crosses of D. tenebrosa to other quinaria group species, the pattern of abdominal spots that is recovered in the hybrids is always similar to that of the lighter species, suggesting that the dark pigmentation does not simply mask underlying variation (Dyer, unpublished results). Once the genetic basis is characterized at the fine scale, it will be interesting to determine the evolutionary history of dark pigmentation in D. tenebrosa, particularly whether the alleles show a signature of positive selection. It will also be interesting to compare the alleles that confer dark pigmentation in D. tenebrosa to the variation found in other closely related species to determine whether these changes occurred de novo in D. tenebrosa or whether these alleles also occur as standing genetic variation in other quinaria group species.
The physiological effects of the dark abdominal pigmentation in D. tenebrosa, if there are any, remain untested. In other Drosophila species, dark pigmentation has been associated with UV resistance, cold tolerance, desiccation resistance and resistance to parasites (reviewed by Wittkopp and Beldade (2009)). There may also be energetic tradeoffs associated with increased melanin, such as reduced reproductive investment or longer developmental time (Roff and Fairbairn, 2013). In no Drosophila species except where pigmentation is sex-specific is the ecological force that selected for the dark phenotype known. Dark pigmentation of some Drosophila species tends to associate with higher altitudes, where the coloration may help the flies regulate temperature or prevent desiccation through the control of cuticular water loss (Gibert et al., 1998; Brisson et al., 2005; Pool and Aquadro, 2007; Rajpurohit et al., 2008). However, based on climate data, the geographic range of D. tenebrosa is not generally colder or drier compared with the boreal forests of the northern North America, where D. suboccidentalis and several other lightly colored and closely related quinaria group species occur. Finally, Dombeck and Jaenike (2004) showed that in D. falleni, another quinaria group species, susceptibility to infection by Howardula nematodes was correlated with the intensity of abdominal pigmentation. It is not known whether D. tenebrosa is infected with Howardula nematodes in the wild, though in the lab this species can be infected with H. aoronymphium (J Jaenike, personal communication). Other species that are both sympatric and closely related to D. tenebrosa are naturally infected with Howardula nematodes, including D. macroptera and D. munda (Perlman et al., 2003). One mechanism by which dark pigmentation may be selected for is if the genes that underlie this phenotype also have pleiotropic effects on other traits, which alternatively may be the actual target of selection. If the same regions of the genome that confer dark pigmentation also influence other physiological traits, this could indicate that dark pigmentation was either directly or indirectly favored by selection (True, 2003). Thus, determining the adaptive value of the dark abdominal pigmentation of D. tenebrosa awaits further genetic analysis.
In conclusion, our result that yellow contributes to the unique abdominal pigmentation pattern of D. tenebrosa adds to the body of work that demonstrates that changes in expression of highly conserved genes can cause rapid evolution and substantial phenotypic differences, even between closely related species (reviewed by Prud'homme et al. (2007) and Carroll (2008)). Further genetic analysis is necessary to determine which other factors are also involved in the unique and potentially adaptive pigmentation in D. tenebrosa. There is substantial variation in pigmentation both within and between species in the quinaria group, particularly in the abdominal pattern, wing spots and the presence of sexually polymorphic pigmentation. As this is also a very young species radiation (10–15 million years old), many species can still be crossed to form fertile hybrid offspring (Patterson and Stone, 1952; K Dyer, unpublished data). Thus, it will be useful to compare the genetic basis of these morphological differences in the quinaria group with other groups of Drosophila and insects more broadly to determine the extent of parallel changes in the genetic pathway that underlies this phenotype. Furthermore, the species in this group are ecologically very tractable relative to many other species of Drosophila, which will enable the dissection of the ecological relevance of these traits to place them in the broader context of the organism's evolutionary history.
Data archiving
QTL mapping data available from the Dryad Digital Repository: Doi:10.5061/dryad.5jh27 and DNA sequences from Genbank: accession numbers KF751356-KF751381.
Acknowledgments
We are grateful to J Lopez, B White and T Womack for laboratory assistance; J Jaenike and R Unckless for sharing fly stocks, and D Hall and A Sweigart for feedback on the manuscript. Funding for this work was from the Center for Undergraduate Research Opportunities at UGA to MJB, Michigan Technological University to TW, and grants from the Ellison Medical Foundation and National Science Foundation to KAD.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Bainbridge SP, Bownes M. Staging the metamorphosis of Drosophila melanogaster. J Embryol Exp Morphol. 1981;66:57–80. [PubMed] [Google Scholar]
- Bhutkar A, Schaeffer SW, Russo SM, Xu M, Smith TF, Gelbart WM. Chromosomal rearrangement inferred from comparisons of 12 Drosophila genomes. Genetics. 2008;179:1657–1680. doi: 10.1534/genetics.107.086108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biessmann H. Molecular analysis of the yellow Gene (y) Region of Drosophila melanogaster. Proc Natl Acad Sci USA. 1985;82:7369–7373. doi: 10.1073/pnas.82.21.7369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisson JA, De Toni DC, Duncan I, Templeton AR. Abdominal pigmentation variation in drosophila polymorpha: geographic variation in the trait, and underlying phylogeography. Evolution. 2005;59:1046–1059. [PubMed] [Google Scholar]
- Carroll SB. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombeck I, Jaenike J. Ecological genetics of abdominal pigmentation in Drosophila falleni: A pleiotropic link to nematode parasitism. Evolution. 2004;58:587–596. [PubMed] [Google Scholar]
- Drapeau MD, Radovic A, Wittkopp PJ, Long AD. A gene necessary for normal male courtship, yellow, acts downstream of fruitless in the Drosophila melanogaster larval brain. J Neurobiol. 2003;55:53–72. doi: 10.1002/neu.10196. [DOI] [PubMed] [Google Scholar]
- Dyer KA, Jaenike J. Evolutionarily stable infection by a male-killing endosymbiont in Drosophila innubila: Molecular evidence from the host and parasite genomes. Genetics. 2004;168:1443–1455. doi: 10.1534/genetics.104.027854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer KA, White BE, Bray MJ, Pique DG, Betancourt AJ. Molecular evolution of a Y chromosome to autosome gene duplication in Drosophila. Mol Biol Evol. 2011;28:1293–1306. doi: 10.1093/molbev/msq334. [DOI] [PubMed] [Google Scholar]
- Ferguson LC, Green J, Surridge A, Jiggins CD. Evolution of the insect yellow gene family. Mol Biol Evol. 2011;28:257–272. doi: 10.1093/molbev/msq192. [DOI] [PubMed] [Google Scholar]
- Futahashi R, Sato J, Meng Y, Okamoto S, Daimon T, Yamamoto K, et al. yellow and ebony are the responsible genes for the larval color mutants of the silkworm Bombyx mori. Genetics. 2008;180:1995–2005. doi: 10.1534/genetics.108.096388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibert P, Moreteau B, Moreteau J-C, Parkash R, David JR. Light body pigmentation in Indian Drosophila melanogaster: a likely adaptation to a hot and arid climate. J Genet. 1998;77:13–20. [Google Scholar]
- Giegerich R, Meyer F, Schleiermacher C. GeneFisher—software support for the detection of postulated genes. Proc Int Conf Intell Syst Mol Biol. 1996;4:68–77. [PubMed] [Google Scholar]
- Gompel N, Prud'homme B, Wittkopp PJ, Kassner VA, Carroll SB. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature. 2005;433:481–487. doi: 10.1038/nature03235. [DOI] [PubMed] [Google Scholar]
- Hines HM, Papa R, Ruiz M, Papanicolaou A, Wang C, Nijhout HF, et al. Transcriptome analysis reveals novel patterning and pigmentation genes underlying Heliconius butterfly wing pattern variation. BMC Genomics. 2012;13:288. doi: 10.1186/1471-2164-13-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S, Rebeiz M, Andolfatto P, Werner T, True J, Carroll SB. The evolution of gene regulation underlies a morphological difference between two Drosophila sister species. Cell. 2008;132:783–793. doi: 10.1016/j.cell.2008.01.014. [DOI] [PubMed] [Google Scholar]
- Jeong S, Rokas A, Carroll SB. Regulation of body pigmentation by the Abdominal-B Hox protein and its gain and loss in Drosophila evolution. Cell. 2006;125:1387–1399. doi: 10.1016/j.cell.2006.04.043. [DOI] [PubMed] [Google Scholar]
- Jones CD. Extension of the Castle-Wright effective factor estimator to sex linkage and haplodiploidy. J Hered. 2001;92:274–276. doi: 10.1093/jhered/92.3.274. [DOI] [PubMed] [Google Scholar]
- Kornezos A, Chia W. Apical secretion and association of the Drosophila yellow gene product with developing larval cuticle structures during embryogenesis. Mol Gen Genet. 1992;235:397–405. doi: 10.1007/BF00279386. [DOI] [PubMed] [Google Scholar]
- Kosambi D. The estimation of map distances from recombination values. Ann Eugen. 1944;12:172–175. [Google Scholar]
- Lorieux M. MapDisto: fast and efficient computation of genetic linkage maps. Mol Breed. 2012;30:1231–1235. [Google Scholar]
- Majerus MEN. Melanism: Evolution in Action. Oxford University Press: Oxford, NY, USA; 1998. [Google Scholar]
- Patterson JT, Stone WS. Evolution in the Genus Drosophila. Macmillan: New York, NY, USA; 1952. [Google Scholar]
- Perlman SJ, Spicer GS, Shoemaker DD, Jaenike J. Associations between mycophagous Drosophila and their Howardula nematode parasites: a worldwide phylogenetic shuffle. Mol Ecol. 2003;12:237–249. doi: 10.1046/j.1365-294x.2003.01721.x. [DOI] [PubMed] [Google Scholar]
- Pool JE, Aquadro CF. The genetic basis of adaptive pigmentation variation in Drosophila melanogaster. Mol Ecol. 2007;16:2844–2851. doi: 10.1111/j.1365-294X.2007.03324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protas ME, Patel NH. Evolution of coloration patterns. Annu Rev Cell Dev Biol. 2008;24:425–446. doi: 10.1146/annurev.cellbio.24.110707.175302. [DOI] [PubMed] [Google Scholar]
- Prud'homme B, Gompel N, Carroll SB. Emerging principles of regulatory evolution. Proc Natl Acad Sci USA. 2007;104 (Suppl 1:8605–8612. doi: 10.1073/pnas.0700488104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud'homme B, Gompel N, Rokas A, Kassner VA, Williams TM, Yeh SD, et al. Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature. 2006;440:1050–1053. doi: 10.1038/nature04597. [DOI] [PubMed] [Google Scholar]
- Rajpurohit S, Parkash R, Ramniwas S. Body melanization and its adaptive role in the thermoregulation and tolerance against dessiccating conditions in drosophilids. Entomol Res. 2008;38:49–60. [Google Scholar]
- Roff DA, Fairbairn DJ. The costs of being dark: the genetic basis of melanism and its association with fitness-related traits in the sand cricket. J Evol Biol. 2013;26:1406–1416. doi: 10.1111/jeb.12150. [DOI] [PubMed] [Google Scholar]
- True JR. Insect melanism: the molecules matter. Trends Ecol Evol. 2003;18:640–647. [Google Scholar]
- Walter MF, Black BC, Afshar G, Kermabon AY, Wright TRF, Biessmann H. Temporal and spatial expression of the yellow gene in correlation with cuticle formation and dopa decarboxylase activity in Drosophila development. Dev Biol. 1991;147:32–45. doi: 10.1016/s0012-1606(05)80005-3. [DOI] [PubMed] [Google Scholar]
- Wang SCJB, Zeng Z-B.2012Windows QTL Cartographer 2.5 Department of Statistics, North Carolina State University, Raleigh, NC . http://statgen.ncsu.edu/qtlcart/WQTLCart.htm .
- Wang W, Yoder JH. Drosophila pupal abdomen immunohistochemistry. J Vis Exp. 2011;56:3139. doi: 10.3791/3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Koshikawa S, Williams TM, Carroll SB. Generation of a novel wing colour pattern by the Wingless morphogen. Nature. 2010;464:1143–1157. doi: 10.1038/nature08896. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Beldade P. Development and evolution of insect pigmentation: genetic mechanisms and the potential consequences of pleiotropy. Semin Cell Dev Biol. 2009;20:65–71. doi: 10.1016/j.semcdb.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Smith-Winberry G, Arnold LL, Thompson EM, Cooley AM, Yuan DC, et al. Local adaptation for body color in Drosophila americana. Heredity. 2011;106:592–602. doi: 10.1038/hdy.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, True JR, Carroll SB. Reciprocal functions of the Drosophila yellow and ebony proteins in the development and evolution of pigment patterns. Development. 2002;129:1849–1858. doi: 10.1242/dev.129.8.1849. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Vaccaro K, Carroll SB. Evolution of yellow gene regulation and pigmentation in Drosophila. Curr Biol. 2002;12:1547–1556. doi: 10.1016/s0960-9822(02)01113-2. [DOI] [PubMed] [Google Scholar]
- Zilversmit M, O'Grady P, Desalle R. Shallow genomics, phylogenetics, and evolution in the family Drosophilidae. Pac Symp Biocomput. 2002;7:512–523. doi: 10.1142/9789812799623_0048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.