Abstract
Inhibiting expression of eukaryotic translation initiation factor 4G (eIF4G) arrests normal development but extends lifespan when suppressed during adulthood. In addition to reducing overall translation, inhibition alters the stoichiometry of mRNA translation in favor of genes important for responding to stress and against those associated with growth and reproduction in C. elegans. In humans, aberrant expression of eIF4G is associated with certain forms of cancer and neurodegeneration. Here we review what is known about the roles of eIF4G in molecular, cellular, and organismal contexts. Also discussed are the gaps in understanding of this factor, particularly with regard to the roles of specific forms of expression in individual tissues and the importance of understanding eIF4G for development of potential therapeutic applications.
Keywords: ageing, protein synthesis, eIF4G, IFG-1, translation initiation, longevity
1. Introduction
While the functional importance of many translation factors is known in a general sense, more appreciation is being given to their role in determining which mRNAs are given preference for translation, both spatially and temporally, under different conditions. Regulating gene expression at the level of translation acts as an important point of control for diverse processes including growth, cellular differentiation, programmed cell death, and for responding to environmental changes. Most of this control is exerted during the steps immediately preceding peptide synthesis, called translation initiation, which is usually the rate-limiting step of mRNA translation (Hershey et al., 2012).
Some of the interest in the specificity of mRNAs selected for translation has undoubtedly been generated by studies carried out in animal models showing that altering expression of translation factors and machinery can increase lifespan (Chen et al., 2007; Chiocchetti et al., 2007; Curran and Ruvkun, 2007; Hansen et al., 2007; Henderson et al., 2006; Pan et al., 2007; Steffen et al., 2008; Syntichaki et al., 2007). Reducing eukaryotic translation initiation factor 4G (eIF4G) in yeast (Smith et al., 2008) and during adulthood in nematodes (Curran and Ruvkun, 2007; Hansen et al., 2007; Henderson et al., 2006; Pan et al., 2007) leads to a robust increase in lifespan. In mammals, elevated expression of eIF4G is associated with cancer. However, regular eIF4G expression is essential early in life for a normal rate of growth. In addition, diminished levels under certain conditions may lead to neurodegeneration. Here, we briefly review the general role of eIF4G in translation initiation and then discuss what is known about the functional importance of the timing, tissue-specificity, and form (homolog, splice variant, or cleavage product) of this factor, especially with regard to lifespan determination and age-related disease.
1.1. eIF4G and Translation Initiation
Regulation of protein synthesis is essential during organismal development and for responding to environmental input. Translation involves an intricate three stage process of initiation, elongation, and termination. Although translation is subject to regulation at each stage, the rate-limiting step is usually initiation, which centers on recruitment of ribosomal subunits to mature mRNA (Hershey et al., 2012). Translation initiation involves protein-RNA and protein-protein interactions synchronized by multiple translation initiation factors. eIF4G acts as a major hub in initiation and mediates recruitment of additional initiation factors, providing a scaffold for ribosome/mRNA-bridging (Sonenberg et al., 1978).
mRNA is co-transcriptionally modified by the addition of a methyl-guanosine (m7GpppX) on the 5’ end, which protects the mRNA from exonuclease activity (Izaurralde et al., 1992) and serves to recruit factors important for initiating translation (Muthukrishnan et al., 1975). The methyl-guanosine addition to the transcript is called the “5’ cap” and translation predominately occurs as a cap-dependent process. Although capped-mRNA is favored for protein synthesis, translation can occur in a cap-independent process that relies on alternate mRNA features (Svitkin et al., 2005). eIF4G plays a crucial role in both processes.
In cap-dependent translation, the cap-binding protein eIF4E binds to eIF4G, which further enhances the affinity of eIF4E for the cap (Haghighat and Sonenberg, 1997). eIF4G also helps recruit the mRNA helicase eIF4A. Together, these three factors make up eIF4F, also known as the cap-binding complex. Through this complex, eIF4G bridges the 5’ untranslated region (UTR) with the polyadenylated 3’ UTR via polyA binding protein (PABP). Association of PABP with eIF4G induces mRNA circularization so that eIF4E at the 5’ cap links with the PABP and the 3’ tail (Tarun Jr and Sachs, 1996). Circularization of mRNA enhances initiation and mRNA stability (Gallie, 1991).
Once the cap-binding complex has bound and circularized the mRNA, eIF4G then helps mediate recruitment of the 40S ribosomal subunit to the mRNA through its association with ribosomal binding protein eIF3 (Lamphear et al., 1995). Prior to this recruitment, the 40S ribosomal subunit associates with eIF3, mRNA scanning proteins eIF1A and eIF1, and the ternary complex, composed of eIF2, GTP, and the methionyl-tRNA initiator (met-tRNAi), together forming the 43S pre-initiation complex (Marchione et al., 2013). eIF3 affixes the 43S pre-initiation complex onto the mRNA, assisted through eIF4G and eIF4E interactions (Etchison et al., 1982; Magnuson et al., 2012). The joined mRNA, pre-initiation complex, and eIF4F complex comprise the 48S pre-initiation complex. The 48S pre-initiation complex scans the 5’ UTR and migrates in a 5’-3’ fashion assisted by the selectivity of eIF1A and eIF1 for the start codon. Upon identification of the start codon, eIF5 triggers initiation by stimulating eIF2 hydrolysis of the ternary complex GTP and then detachment of all 40S-bound initiation factors (Unbehaun et al., 2004). Hydrolysis of GTP from the ternary complex triggers dissociation of initiation factors in the 48S pre-initiation complex and positions met-tRNAi into the P-site on the 40S ribosomal subunit associated with the start codon (Unbehaun et al., 2004). Ribosomal protein S6 within the 40S subunit is phosphorylated by S6 kinase, promoting the joining of both the 40S and 60S subunits, and formation of the 80S ribosome (Jefferies et al., 1994; Magnuson et al., 2012). Successful assembly of the ribosome completes initiation and protein synthesis begins, leaving eIF4G and other initiation factors available to begin a new round of translation initiation.
Cap-independent translation may take place if an internal ribosomal entry site (IRES) is present within the 5’ UTR of the mRNA (Pelletier and Sonenberg, 1988). However, capped mRNAs compete for translation against IRES-containing mRNAs (Svitkin et al., 2005). In the presence of an IRES, eIF4G is capable of initiating translation in the absence of a functional 5’ cap-binding complex (Ali et al., 2001), as is the case when eIF4E is sequestered by eIF4E Binding Protein 1 (4EBP1). When available eIF4E becomes limited, the sub-complex eIF4G/4A is able to bind to IRES-containing mRNAs to mediate translation (Svitkin et al., 2005). If eIF4G is unable to bind eIF4A, then IRES directed translation is abrogated (Lomakin et al., 2000). Therefore eIF4G plays an important role in both cap-dependent and cap-independent mediated translation.
2. Biological Effects of eIF4G at the Level of Tissues and the Whole Organism
eIF4G was first characterized through a mammalian in vitro lysate experiment testing 5’ cap-binding during mRNA translation (Sonenberg et al., 1978). Further testing determined that removing eIF4G from this in vitro translation system effectively hinders initiation complex formation, thereby preventing protein synthesis (Ali et al., 2001). In vivo, eIF4G is essential, as studies in yeast and nematodes demonstrate that absence of expression results in developmental arrest and lethality (Contreras et al., 2008; Goyer et al., 1993). However, reducing its expression after development can increase lifespan, while overexpression is associated with malignant transformation of cells. In addition to modulation by transcription, there are homologs, isoforms, and cleavage products of eIF4G that alter the rate of translation and type of mRNA species that are translated. The following subsections address the timing, form, and tissue-specific effects of eIF4G expression in the context of development, longevity, response to stress, and age-associated pathology.
2.1. Importance of eIF4G during and after development and in response to stress
Expression of eIF4G is crucial during organismal development. Complete knockout of eIF4G in the yeast Saccharomyces cerevisiae is lethal (Goyer et al., 1993). A strain bearing a null mutation in the C. elegans eIF4G gene, ifg-1(ok1211), must be maintained as a heterozygote, as animals homozygous for the knockout mutation cannot develop past the second larval stage of development (Contreras et al., 2008). Subsequently, knock-down of eIF4G via RNAi feeding in C. elegans late in larval development diminishes fecundity and arrests growth in the subsequent generation (Long et al., 2002; Pan et al., 2007). RNAi knock-down is also associated with enlarged vesicle formation in the intestinal cells of offspring which is accompanied with severe intestinal atrophy (Long et al., 2002). Loss of function mutants exhibit increased apoptosis in germ cells concomitant with a shift in mRNA translation that reinforces the apoptotic cascade (Contreras et al., 2011). Thus, eIF4G removal or suppression is associated with negative development and growth of offspring.
In contrast to the deleterious effects of inhibiting eIF4G during development in C. elegans, multiple studies have concluded that reduced expression during adulthood leads to a significant lifespan extension (Curran and Ruvkun, 2007; Hansen et al., 2007; Henderson et al., 2006; Pan et al., 2007). Congruently, in yeast a deletion to the eIF4G gene, TIF4631, also produced a long lived mutant (Smith et al., 2008). At the organismal level, reducing overall eIF4G expression in adult nematodes, in addition to globally reducing translation, results in a modest shift in translation efficiency from transcripts associated with growth to those associated with homeostasis and increased longevity (Rogers et al., 2011). This broad level change in translation efficiency is correlated with mRNA length, connecting gene function with transcript size. In yeast, two eIF4G closed-loop structures during translation initiation have been identified that are biased to mRNA length in a manner dependent on the level of eIF4G present (Amrani et al., 2008). Depletion of eIF4G in yeast, while greatly diminishing overall protein synthesis, results in a small but widespread effect on translational efficiency (Park et al., 2011). Thus, eIF4G appears to control differential mRNA translation, and differentially regulated mRNAs are functionally connected at the level of the biological processes they modulate.
The ontological connection between eIF4G level and differentially translated mRNA may help explain the observation that nematodes have increased survival in the absence of food when eIF4G is inhibited (Pan et al., 2007; Rogers et al., 2011). Taken together with the fact that wild-type animals exhibit diminished eIF4G protein levels when removed from food (Rogers et al., 2011), these results suggests that controlling expression of this highly conserved translation factor may be important for survival during periods of nutrient deprivation. Additional support for this idea comes from the observation that the same genes essential for longevity and increased survival in the absence of food when eIF4G is inhibited are also important for lifespan extension in a genetic model of dietary restriction (Rogers et al., 2011). eIF4G protein levels are diminished during glucose starvation in yeast (Berset et al., 1998) and upon inhibition of the nutrient-responsive Target of Rapamycin (TOR) pathway in mammalian cells (Ramirez-Valle et al., 2008). TOR controls a number of cellular processes, including mRNA translation (Kapahi et al., 2010). TOR has also been shown to regulate differential translation in yeast in a manner that is dependent on eIF4G (Thoreen et al., 2012). The connection between eIF4G and the nutrient sensing TOR pathway, together with results showing that expression of eIF4G is related to nutrient availability, suggest that changing the amount of eIF4G may be a conserved adaptive response to conditions of limited food resources.
eIF4G overexpression prevents autophagy, apoptosis and senescence induced by ionizing radiation in breast cancer epithelial cells, while reducing eIF4G decreases viability (Badura et al., 2012). Under heat stress, eIF4G in mammalian cells dissociates from eIF4E, indicating a halt to cap-dependent translation (Vries, 1997). Other studies have documented sequestration of eIF4G into cytoplasmic stress granules of cultured cells during heat or oxidative stress (Brown et al., 2011; Cuesta et al., 2000), suggesting a cellular need to halt protein synthesis under stress conditions. Resistance to heat and oxidative stress in C. elegans was not significantly increased beyond control levels when eIF4G was knocked down via RNAi for 48 hours in young adult worms (Pan et al., 2007; Rogers et al., 2011). However, another study showed that four days of RNAi feeding was able to increase thermotolerance (Hansen et al., 2007), possibly indicating that several days of inhibition are required to sufficiently reduce eIF4G protein to a level that increases resistance to stress. A looming question is whether effects on lifespan and stress tolerance depend on the specific form of eIF4G and/or its expression in specific tissues.
2.2. Form dependent function of eIF4G
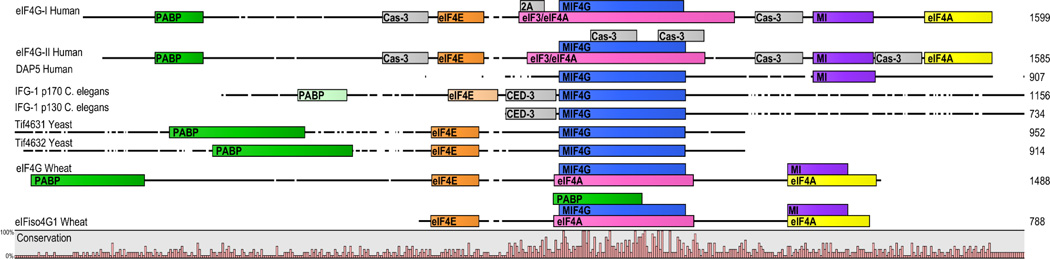
Two distinct eIF4G proteins were first identified in wheat, referred to as eIF4G and eIFiso4G (Browning et al., 1992, 1987). The two forms differ in size with eIF4G being considerably larger (165 kDa) than eIFiso4G (86 kDa) (Browning et al., 1987; Gallie and Browning, 2001) (Figure 1). Although both proteins are capable of initiating cap-dependent and cap-independent translation in vitro, evidence suggests differences for their roles in translation. The complex formed by inclusion of the 165 kDa eIF4G homolog more efficiently initiates translation when the 5’ UTR contains a high degree of secondary structure or even when the 5’ cap is missing compared with that formed by the 86 kDa eIFiso4G homolog, which exhibits preferential binding to mRNA containing a less structured 5’ UTR (Gallie and Browning, 2001). The importance of understanding such differences is made particularly interesting by the fact that the complexes formed between the standard m7GpppX cap and eIFiso4F more closely resemble the complexes formed between the cap analogue and mammalian eIF4F (Carberry et al., 1991). Total knockout of eIFiso4G forms in Arabidopsis thaliana leads to reduced germination rates, slower growth, diminished fertility and reduced long term seed viability, indicating eIFiso4G has a role in translation of proteins involved in plant growth and development (Lellis et al., 2010). These findings are in agreement with eIF4G deletions in other organismal models, namely yeast and C. elegans.
Figure 1. Comparisons of eIF4G across species.
Protein structures of eIF4G for different species aligned using the most highly conserved middle domain of eukaryotic initiation factor 4G (MIF4G; blue). Percent sequence conservation across the forms is indicated at the bottom of the alignment. DAP5 was included based on homology with both human eIF4G-I and –II. Conserved binding domains for eIF4E (orange), eIF3 and/or eIF4A (pink; yellow for second eIF4A binding site), and PABP (green) are shown. Putative PABP (light green) and eIF4E (light orange) binding sites are also indicated in the C. elegans IFG-1 isoform A and D (p170 and p130 respectively). The conservation of putative binding domains for eIF4A and eIF3, although presumably present, were not definitively identified in C. elegans and yeast. Cleavage sites for protease 2A, caspase-3, and caspase CED-3 (C. elegans) are shown in grey. The MA-3 and eIF4G domain (MI; purple) represents a homologous sequence between human and wheat eIF4G isoforms which has not been identified in C. elegans or yeast. See Supplemental Table 1 for amino acid binding sites. CLC Workbench 6.8.2 alignment.
Mammals also encode two separate homologs of eIF4G called eIF4G-I and eIF4G-II, both of which contain binding sites for PABP, eIF3, eIF4A, and eIF4E (Figure 1, Supplemental Table 1; Gradi et al., 1998) The concentration of eIF4G homologs varies between cell types, but eIF4G-I is consistently expressed at a higher concentration than eIF4G-II (Coldwell et al., 2012). Both eIF4G-I and -II initiate cap-dependent translation and are susceptible to cleavage by virally encoded proteases, albeit at different locations (Marissen et al., 2000). However, as with wheat, evidence suggests that each form displays differences in the efficiency with which translation is carried out that is dependent on the mRNA. eIF4G-I is important for proliferation, bioenergetics, and mitochondrial activity and is important for translating mRNAs related to these processes (Ramirez-Valle et al., 2008). This form, which is associated with cancer in mammals (covered in greater depth in section 2.3), was found to promote the translation of transcripts with low abundance and those containing upstream open reading frames in the aforementioned study. Translation of mRNA subsets specifically regulated by the TOR pathway are greatly diminished only when eIF4G-I is depleted, but not after removal of eIF4G-II (Thoreen et al., 2012). However, eIF4G-I depletion does not significantly alter global translation rate (Ramirez-Valle et al., 2008), indicative of a role for other homologs, including eIF4G-II and DAP5, in contributing to a significant portion of translation.
One report distinguishing the function of mammalian eIF4G-I and -II, found that eIF4G-II is selectively recruited to capped mRNA at the initiation of differentiation in cells (Caron et al., 2004). The researchers also provided evidence that cytokines can differentially regulate the activity of both homologs. One year earlier, a phosphorylation screen aimed at identifying targets of Ca2+/calmodulin-dependent protein kinase I found that eIF4G-II was a target (Qin, 2003). These studies suggest a role for eIF4G-II in development and translation. Further evidence for such a role came from a more recent study showing that this homolog is required for male fertility in mice, specifically for meiotic exit during spermatogenesis (Sun et al., 2010; referred to by Ensembl designation eIF4G3 in that study). More studies elucidating the distinct roles of the two homologs are needed to identify an impact on growth, development, and ageing.
In yeast, two eIF4G paralogs exist called TIF4631 and TIF4632 (Figure 1, Table 1). Individual manipulations lead to varied effects on growth and development (Goyer et al., 1993; Winstall et al., 2000). Yeast containing gene disruptions to TIF4631 display slow growth and cold sensitivity which is not exhibited for disruptions in TIF4632 (Goyer et al., 1993). However, altering the highly conserved leucine region to alanine residues within either Tif4631 or Tif4632 results in a temperature sensitive growth phenotype (Tarun and Sachs, 1997). Another study removed both eIF4G paralogs and replaced them with either a single Tif4631 or Tif4632 bearing the leucine to alanine change as the sole source of eIF4G and found temperature sensitivity or lethality depending on the location of the mutation (Winstall et al., 2000). Additionally, TIF4631 has been identified as an ageing regulator via the deletion mutant tif4631δ, representing a long lived yeast model (Smith et al., 2008). All together, these findings implicate eIF4G as a yeast factor influencing growth and ageing in a paralog dependent manner.
Table 1.
eIF4G homologs across species with MIF4G region identified by amino acid location.
| Sequence Name (Amino Acid Length) |
Binding Domain |
Region | Reference |
|---|---|---|---|
| eIF4G-I/eIF4G1 Human (1599) | MIF4G | 565–792 | Uniprot: Q04637 |
| Isoform 1 (1600) | MIF4G | 762–987 | NCBI: NP_886553.3 |
| Isoform 2 (1512) | MIF4G | 674–899 | NCBI: NP_937887.1 |
| Isoform 3 (1435) | MIF4G | 597–822 | NCBI: NP_937885.1 |
| Isoform 4 (1404) | MIF4G | 566–791 | NCBI: NP_004944.3 |
| Isoform 5 (1599) | MIF4G | 761–986 | NCBI: NP_937884.1 |
| Isoform 6 (1606) | MIF4G | 768–993 | NCBI: NP_001181875.1 |
| Predicted Isoform X3 (1600) | MIF4G | 762–987 | NCBI: XP_005247244.1 |
| Predicted Isoform X4 (1600) | MIF4G | 762–987 | NCBI: XP_005247245.1 |
| Predicted Isoform X12 (1404) | MIF4G | 566–791 | NCBI: XP_005247253.1 |
| Predicted Isoform X13 (1404) | MIF4G | 566–791 | NCBI: XP_005247254.1 |
| eIF4G-II/eIF4G3 Human (1585) | MIF4G | 755–983 | Uniprot: O43432 |
| Isoform 1 (1621) | MIF4G | 791–1016 | NCBI: NP_001185730.1 |
| Isoform 2 (1591) | MIF4G | 761–986 | NCBI: NP_001185731.1 |
| Isoform 3 (1585) | MIF4G | 755–980 | NCBI: NP_003751.2 |
| Isoform 4 (515) | NCBI: NP_001185732.1 | ||
| eIF4G-I/eIF4G1 Mouse (1600) | MIF4G | 765–993 | Uniprot: Q6NZJ6 |
| Isoform A (1600) | MIF4G | 765–993 | NCBI: NP_666053.2 |
| Isoform B (1593) | MIF4G | 758–986 | NCBI: NP_001005331.1 |
| eIF4G-II/eIF4G3 Mouse (1579) | MIF4G | 750–978 | Uniprot: Q80XI3 |
| Isoform 1 (1578) | MIF4G | 768–993 | NCBI: NP_766291.2 |
| Isoform 2 (1567) | MIF4G | 757–982 | NCBI: NP_001243124.1 |
| Isoform 3 (1464) | MIF4G | 757–982 | NCBI: NP_001243127.1 |
| eIF4G-I/eIF4G1 Rabbit (1402) | MIF4G | 567–793 | NCBI: P41110.1 |
| eIF4G-II/eIF4G3 Rabbit (1579) | Uncharacterized | Uniprot: G1SHL6 | |
| IFG-1 p170 C. elegans (1156) | MIF4G | 518–748 | Uniprot: Q21531 |
| Isoform A (1156) | MIF4G | 518–748 | NCBI: CAA90261.1 |
| Isoform B (1155) | MIF4G | 517–747 | NCBI: CAE47470.1 |
| Isoform C (1042) | MIF4G | 404–634 | NCBI: CAR31495.1 |
| Isoform D (734) | MIF4G | 96–326 | NCBI: NP_001129820 |
| Isoform E (688) | MIF4G | 50–280 | NCBI: CAR31497.1 |
| IFG-1 p130 C. elegans (765) | MIF4G | 127–357 | (Contreras et al., 2011) |
| eIF4G-I/eIF4G1 Fruit Flya (1666) | MIF4G | 751–999 | NCBI: AAC38985.1 |
| Isoform A (1666) | MIF4G | 751–999 | NCBI: AAF59403.3 |
| Isoform B (1919) | MIF4G | 1004–1252 | NCBI: ABV53593.1 |
| Isoform C (1666) | MIF4G | 751–999 | NCBI: NP_001137857.1 |
| Tif4631 Yeastb (952) | MIF4G | 607–850 | Uniprot: P39935 |
| Tif4632 Yeastb (914) | MIF4G | 567–810 | Uniprot: P39936 |
| eIF4G1 Wheatc (1488) | MIF4G | 883–1106 | Uniprot: G5CEW6 |
| eIFiso4G1 Wheatc (788) | MIF4G | 211–436 | Uniprot: Q03387 |
| eIFiso4G2 Wheatc (787) | MIF4G | 210–435 | Uniprot: Q41583 |
| eIF4G Arabidopsisd (1727) | MIF4G | 1096–1319 | NCBI: AEE80028.1 |
| eIFiso4G1 Arabidopsisd (780) | MIF4G | 216–440 | NCBI: Q93ZT6.1 |
| eIFiso4G2 Arabidopsisd (747) | MIF4G | 176–400 | NCBI: O82233.1 |
Drosophila melanogaster,
Saccharomyces cerevisiae,
Triticum aestivum,
Arabidopsis thaliana
A single gene called ifg-1 in C. elegans encodes two characterized isoforms of eIF4G, referred to as p170 and p130 (Contreras et al., 2008; Long et al., 2002), although current Ensembl projections include 6 splice variants (Ensembl Acc: NP_001022259). However, of the two isoforms investigated, both are present and equally distributed throughout the germline and somatic tissues (Contreras et al., 2008). p170 is similar to full length versions of eIF4G in other species, while p130 lacks the N-terminal domain containing the eIF4E and PABP-binding sites (Contreras et al., 2011, 2008), which is suggestive of a role in initiating cap-independent translation. Although both isoforms are susceptible to cleavage during apoptosis and viral infection, the p130 isoform contains only 65 more amino acids on the N-terminal domain than the apoptotically cleaved forms of p130 and p170 (Figure 1; Contreras et al., 2011). Whether the additional stretch of amino acids in p130 functionally separates it from the cleaved form is unknown.
The short isoform and cleaved version of eIF4G in C. elegans are similar to cleavage products that exist in other species, which can appear during viral infection and apoptosis. During picornaviral infection, cleaved eIF4G initiates translation of viral RNA, which does not contain a 5’ cap but relies on IRES-mediated translation (Pelletier and Sonenberg, 1988). These viruses hijack much of the host’s translation machinery by cleaving eIF4G and separating eIF4E and PABP binding domains from the domains that bind other initiation factors important for ribosomal recruitment (Ali et al., 2001). The overall effect is that cap-dependent translation decreases while IRES-mediated translation of non-capped mRNAs increases (Ohlmann et al., 1995), allowing viral mRNA an advantage over host mRNA for translation (Ali et al., 2001). Interestingly, in an in vitro system, picornaviral RNA remains untranslated in the presence of intact eIF4G (Borman et al., 1997), suggesting that viral infection relies on the cleaved form of eIF4G for translation.
Hypoxia also induces a switch from cap-dependent to cap-independent translation. Such is the case in breast cancer epithelial cells and HeLa cells in which cap-binding of eIF4E is inhibited by 4EBP (Braunstein et al., 2007; Koritzinsky et al., 2006). Although restricting its access to the 5’ cap decreases overall protein synthesis, eIF4G-I is able to stimulate cap-independent IRES-mediated translation (Braunstein et al., 2007). During hypoxia, eIF4G is not noticeably cleaved in HeLa cells (Koritzinsky et al., 2006), indicating that IRES-mediated translation of host mRNA might not rely on truncated eIF4G for initiation. Under hypoxic conditions, eIF4G-II readily dissociates from the cap while eIF4G-I is slower to disassociate, and small levels are still detected at the 5’ cap after 16 hours of hypoxia (Koritzinsky et al., 2006). IRES-containing mRNAs involved in promoting tumor angiogenesis and survival such as vascular endothelial growth factor (VEGF), hypoxia inducible factor 1α, and B-cell lymphoma 2 protein are preferentially translated by eIF4G-I in a cap-independent manner (Braunstein et al., 2007). Silencing eIF4G-I expression in tumor cells prevents hypoxia induced expression of VEGF (Braunstein et al., 2007). Together, these results suggest that the eIF4G-I homolog preferentially mediates changes in preferred mRNA translation in response to hypoxia.
During development and under certain environmental conditions, the protease caspase-3 cleaves eIF4G-I and eIF4G-II in mammalian cells, shutting down cap-dependent translation and inducing apoptosis (Marissen and Lloyd, 1998; Marissen et al., 2000). Despite cleavage kinetics being almost identical for both forms of eIF4G in a human cell line, caspase-3 cleavage occurs at more sites in eIF4G-II than eIF4G-I (Bushell et al., 1999; Marissen et al., 2000). Similarly, the caspase CED-3 in C. elegans cleaves both IFG-1 p170 and p130 to induce programmed removal of cells during development (Figure 1; Contreras et al., 2011). In tetracycline-inducible cell lines expressing an eIF4G-I C-terminal fragment unable to participate in cap-mediated binding, eIF4G potently stimulated expression of the IRES-containing angiogenesis factor VEGF and tumor promoting protein c-myc (Kaiser et al., 2008). Under nutrient stress in yeast, several invasive yeast growth messengers are induced by cap-independent translation utilizing potent IRES elements and eIF4G binding (Gilbert et al., 2007). Thus, the switch from cap-dependent to cap-independent translation has a powerful influence on cellular fate.
The mammalian EIF4G-I gene contains three active promoters which produce six eIF4G-I mRNA transcripts that give rise to five documented isoforms (Byrd, 2005) each with a different role in regulating translation (Coldwell and Morley, 2006). Similarly, the mammalian EIF4G-II gene produces multiple isoforms (Table 1). However, due to six documented promoters, numerous splicing events and the use of an unusual CUG initiation codon the exact number of eIF4G-II isoforms is not yet known (Coldwell et al., 2012). Multiple isoforms of eIF4G expression from one gene is also apparent in non-mammalian models such as C. elegans. Although only the p170 and p130 IFG-1 isoforms have been documented in vivo (Contreras et al., 2008), Ensembl lists the existence of 6 transcript variants (Flicek et al., 2012; Ensembl Acc: 3298).
Other intracellular proteins also share a similar domain with eIF4G, and a small eIF4G family has come to be recognized, defined by the presence of the highly homologous middle domain of eIF4G (MIF4G in Figure 1, and Table 2). This family includes death associated protein (DAP5; previously known as p97 and NAT1 and referred to as eIF4G2 in Ensembl Acc: 3297), PABP interacting protein-1 (PAIP-1), cap-binding protein 80 (CBP80)/20-dependent translation initiation factor (CTIF), CWC22, nucleolar MIF4G domain-containing protein (NOM1), up-frame shift suppressor 2 homolog (UPF2), and SLBP-interacting protein 1 (SLIP1; Table 2).
Table 2.
Blast search of the middle domain of eukaryotic initiation factor 4G region across species, with MIF4G identified by amino acid location.
| Sequence Namea (Amino Acid Length) |
Binding Domain |
Region | Reference |
|---|---|---|---|
| Human MIF4G Family | |||
| CWC22 (908) | MIF4G | 163–346 | Uniprot: Q9HCG8 |
| DAP5/eIF4G2 (907) | MIF4G | 78–308 | Uniprot: P78344 |
| NCBP-1/CBP80 (790) | MIF4G | 28–240 | NCBI: Q09161.1 |
| NOM1 (860) | MIF4G | 362–559 | Uniprot: Q5C9Z4 |
| PAIP-1 (479) | MIF4G | 159–376 | Uniprot: Q9H074 |
| SLIP1 (222) | MIF4G | 3–205 | Uniprot: A9UHW6 |
| UPF2 (1272) | MIF4G1 | 168–431 | Uniprot: Q9HAU5 |
| MIF4G2 | 569–758 | Uniprot: Q9HAU5 | |
| MIF4G3 | 773–986 | Uniprot: Q9HAU5 | |
| Mouse MIF4G Family | |||
| CWC22 (908) | MIF4G | 161–344 | Uniprot: Q8C5N3 |
| DAP5/eIF4G2 (906) | MIF4G | 78–308 | Uniprot: Q62448 |
| NOM1 (854) | MIF4G | 356–553 | Uniprot: Q3UFM5 |
| PAIP-1 (400) | MIF4G | 80–297 | Uniprot: Q8VE62 |
| SLIP1 (222) | MIF4G | 3–205 | Uniprot: Q3UBZ5 |
| UPF2 (1269) | MIF4G1 | 167–362 | NCBI: NP_001074601.1 |
| MIF4G2 | 569–756 | NCBI: NP_001074601.1 | |
| MIF4G3 | 771–984 | NCBI: NP_001074601.1 | |
| Rabbit MIF4G Family | |||
| DAP5/eIF4G2 | MIF4G | 78–308 | NCBI: NP_001075848.1 |
| Predicted UPF2 (1269) | MIF4G1 | 160–355 | NCBI: 291401994 |
| MIF4G2 | 563–750 | NCBI: 291401994 | |
| MIF4G3 | 765–978 | NCBI: 291401994 | |
| C. elegans MIF4G Family | |||
| F44A2.5 Isoform B (390) | MIF4G | 173–372 | NCBI: CCD71191.1 |
| NCBP-1/CBP80 (798) | MIF4G | 28–241 | NCBI: NP_491850.2 |
| LET-858/CWC22 ortholog (897) | MIF4G | 195–382 | NCBI: CAB04256.1 |
| SMG-3/UPF2 ortholog (1142) | MIF4G1 | 461–601 | NCBI: ABQ96385.1 |
| MIF4G2 | 650–827 | NCBI: ABQ96385.1 | |
| Y52B11A.10/NOM1 (819) | MIF4G | 332–529 | NCBI: CAB63391.1 |
| Fruit Flyb MIF4G Family | |||
| CBP80 (800) | MIF4G | 31–243 | NCBI: Q7K4N3.1 |
| CG7907 Isoform B (753) | MIF4G | 63–243 | NCBI: ABW08721.1 |
| CG7907 Isoform C (753) | MIF4G | 63–243 | NCBI: ABW08722.1 |
| CWC22 (1330) | MIF4G | 421–602 | NCBI: 74869383 |
| DAP5/eIF4G2 (2072) | MIF4G | 1274–1514 | NCBI: AAF56194.2 |
| NOM1 (854) | MIF4G | 352–553 | NCBI: 74871964 |
| Upf2 (1241) | MIF4G1 | 104–300 | NCBI: AAF46314.1 |
| MIF4G2 | 518–701 | NCBI: AAF46314.1 | |
| MIF4G3 | 716–934 | NCBI: AAF46314.1 | |
| Yeastc MIF4G Family | |||
| Cwc22p (577) | MIF4G | 24–182 | NCBI: NP_011794.3 |
| Upf2p | MIF4G1 | 34–240 | NCBI: AAA66521.1 |
| MIF4G2 | 374–561 | NCBI: AAA66521.1 | |
| MIF4G3 | 578–780 | NCBI: AAA66521.1 | |
| Sgd1p/NOM1 ortholog (899) | MIF4G | 350–540 | NCBI: NP_013440.1 |
| Sto1/NCBP-1 ortholog (861) | MIFG4 | 36–264 | Uniprot: P34160 |
| Arabidopsisd MIF4G Family | |||
| At1g80930/F23A5_23 (900) | MIF4G | 363–546 | NCBI: AEE36468.1 |
| MIF4G Domain Protein (223) | MIF4G | 9–198 | NCBI: AEE33963.1 |
| MIF4G/MA3 Domain Protein (784) | MIF4G | 304–499 | NCBI: AED92488.1 |
| NCBP-1/CBP80 (848) | MIF4G | 20–201 | NCBI: AEC06238.1 |
| UPF2 (1182) | MIF4G1 | 57–208 | NCBI: AEC09651.1 |
| MIF4G2 | 468–656 | NCBI: AEC09651.1 | |
| MIF4G3 | 671–872 | NCBI: AEC09651.1 | |
| MIF4G Protein/PUB57 (371) | MIF4G | 4–166 | NCBI: AEE33333.1 |
| MIF4G Protein/PUB58 (485) | MIF4G | 25–200 | NCBI: AEE33334.1 |
Wheat was excluded from the table due to uncharacterized MIF4G family members.
Drosophila melanogaster,
Saccharomyces cerevisiae,
Arabidopsis thaliana
DAP5 is homologous to the C-terminal two thirds of eIF4G but does not contain eIF4E and PABP-binding sites (Figure 1, and Supplemental Table 1). Although originally shown to be an inhibitor of cap-dependent and independent translation (Imataka and Sonenberg, 1997; Yamanaka et al., 1997), subsequent studies have shown instances in which DAP5 promotes cap-independent/IRES-mediated translation. In addition to being implicated in driving IRES-mediated translation under various cellular stresses, it has been demonstrated to be important for initiating synthesis of cell cycle factors (Lee and McCormick, 2006) and in mediating non-stressed cell survival during mitosis (Liberman et al., 2009). DAP5 can be proteolytically cleaved to form a P86 form and both the full length and cleaved forms are able to initiate translation (Nousch et al., 2007). Furthermore, DAP5 can promote translation of DAP5 mRNA via its own IRES under ER stress in a caspase independent manner while its cleaved P86 form is responsible for selective translation of human inhibitor of apoptosis protein-2 under the same condition (Lewis et al., 2007).
PAIP-1 is thought to be a translational co-activator due to the lack of an eIF4E binding site, but maintains the ability to bind eIF4A and shows 39% similarity to human MIF4G (Craig et al., 1998). More recently, evidence suggested that eIF3-PAIP-1 stabilizes the interaction between PABP and eIF4G (Martineau et al., 2008). This study established the presence of three isoforms of PAIP-1 named according to their size (P45, P51, and P65). Although P65 is the dominant form expressed in HeLa cells, the P45 form is able to induce translation in a reporter system that is two times that of either the P51 or P65 isoforms (Martineau et al., 2008). Little is known about whether these forms have preferred activity for mRNAs based on characteristics of transcripts such as size, UTR structure, or the presence of cis-regulatory elements.
The other eIF4G family members CTIF, NOM1, CWC22, UPF2 and SLIP1 have not been as heavily studied as DAP5 and PAIP-1. The CTIF protein displays similar characteristics of eIF4G in addition to containing a MIF4G. CTIF is highly enriched in the perinuclear region, interacts with the exon junction complex (EJC), and initiates cap-binding dependent translation of mRNAs after nuclear export (Kim et al., 2009). Identified in humans, NOM1 is nuclear, integral to rRNA biogenesis, and is considered an eIF4G-like nucleolar translation partner (Alexandrov et al., 2011). The protein CWC22, responsible for EJC formation, is involved in pre-mRNA splicing, and shares sequence similarity to NOM1 (Steckelberg et al., 2012). Unlike the protein synthesis favoring properties of eIF4G, UPF2 possesses three MIF4G regions and is involved in mRNA degradation when translation initiation is inhibited or premature nonsense codons are encountered (Cui et al., 1995). Mice with UPF2 allele knockout exhibit early embryonic lethality (Weischenfeldt et al., 2008). Mammalian SLIP1 activates translation of mRNAs with a histone stem-loop in the nucleus, and also interacts with both eIF4G-I and –II, supporting the role of SLIP1 in translation (Cakmakci et al., 2007).
2.3. Disease and aberrant expression of eIF4G in specific tissues
Despite controlling growth, development, and lifespan regulation, eIF4G is also linked to several processes and development of disease. For a number of years, aberrant or overexpression of eIF4G has been associated with cancer in humans and in initiating tumorigenesis in tissue culture and in studies with mice. Overexpressing human eIF4G-I in mouse fibroblasts is sufficient to induce cellular transformation (Fukuchi-Shimogori et al., 1997). The resulting cells lack contact inhibition and continue to grow, exhibiting colony formation akin to malignant phenotypes. Injection of the eIF4G-I overexpressing cells into mice is also sufficient to induce tumor formation (Fukuchi-Shimogori et al., 1997).
Aberrant increased expression of eIF4G-I is found in hypopharyngeal and nasopharyngeal carcinoma (Cromer et al., 2003) (Fang et al., 2008). A later study found that nasopharyngeal carcinoma patients with the highest levels of eIF4G-I have the lowest survival rates (Tu et al., 2010). In fact, Tu and colleagues showed that eIF4G-I expression could be used as a biomarker for the prognosis of patients with this type of cancer. The same study also showed that knocking down eIF4G-I expression reduces cell migration and matrigel invasion in culture and dramatically decreases tumor size in xenografted mice. In the absence of eIF4G-I, cells exhibit an increase in levels of programmed cell death 4, a key tumor suppressing protein (Tu et al., 2010).
eIF4G is also overexpressed in squamous cell lung carcinoma (Bauer et al., 2002; Comtesse et al., 2007). In particular, biochemical analysis indicates overexpression of eIF4G-I in a majority of surgically removed lung cancer tumors (Bauer et al., 2002). eIF4G-I mRNA expression is elevated in human squamous cell lung carcinomas, consistent with the previous findings of eIF4G protein overexpression (Comtesse et al., 2007). Interestingly, gene expression of EIF2B, EIF4A1 and EIF4B were also elevated in squamous cell carcinomas, large cell carcinomas, adenocarcinomas and small cell carcinomas, but no increase in the eIF4G binding partner eIF4E was found (Comtesse et al., 2007).
Biopsies from breast cancer patients exhibit drastically increased eIF4G expression in large advanced carcinomas (Braunstein et al., 2007). Overexpression of eIF4G-I increases IRES-mediated translation of mRNAs encoding the cell surface anchoring protein catenin and promotes inflammatory breast cancer tumor formation (Silvera et al., 2009), a primary breast cancer with the highest rate of lethality. Regression analysis suggests that eIF4G-I is the only significant predictor of inflammatory breast cancer of those tested. Using shRNA targeted to eIF4G-I reduced its expression by 90% in SUM149 cells, which are used as a model of inflammatory breast cells. Knock-down of eIF4G-I is sufficient to impair inflammatory breast cancer tumor growth in a nude mouse xenograft model and to inhibit characteristics important for invasiveness despite only a 15% reduction in overall protein synthesis. Further analysis shows that selective translation inhibition of mRNA-IRES elements in the 5’ UTR is responsible for the impaired tumor growth (Silvera et al., 2009).
The relatively small decrease in overall protein synthesis when eIF4G-I is inhibited is also observed for other cell types, including an immortalized breast epithelial cell line, primary human neonatal dermal fibroblasts, and BT-474 breast cancer cells (Ramirez-Valle et al., 2008). Inhibiting eIF4G-I induces autophagy and diminishes mitochondrial function similar to TOR pathway inhibition (Ramirez-Valle et al., 2008). It also reduces ATP levels, activating AMPK. Silencing other translation factors, including eIF4G family members, does not show the same effect on energy status. Interestingly, although silencing eIF4G-I has also been shown to increase expression of eIF4G-II in breast cancer epithelial cell lines (Badura et al., 2012), simultaneous eIF4G-I and eIF4G-II silencing does not decrease protein synthesis beyond the results of silencing eIF4G-I alone, while silencing both eIF4G-I and the eIF4G family member DAP5 together results in a 60% reduction in translation (Ramirez-Valle et al., 2008). Silencing eIF4G-I leads to a 50-fold decrease in epithelial cell viability after ionizing radiation, while silencing eIF4G-II or eIF4E resulted in only a partial reduction in viability (Badura et al., 2012). The same study showed that apoptosis in the absence of ionizing radiation is also significantly increased with eIF4G-I silencing.
After ischemic brain injury, protein synthesis decreases in neurons, correlated with degradation of eIF4G in vivo (Neumar et al., 1998; Vosler et al., 2011). Overexpression of eIF4G-I in cultured neurons results in increased protein synthesis, and provides protection from ischemia induced neuronal death (Vosler et al., 2011). Silencing neuronal expression of eIF4G-I results in increased cell death after ischemia (Vosler et al., 2011). Ultimately, preserving eIF4G-I levels in vivo prevents tissue loss after brain injury (Neumar et al., 1998; Vosler et al., 2011). Thus, effects of expression levels of eIF4G, even in a post-developmental context, are tissue- and condition-specific.
A mutation in the eIF4G-I gene is linked to familial Parkinson’s disease in a French family (Chartier-Harlin et al., 2011). The mutant allele affects the primary structure in the regions interacting with eIF3 and eIF4A, yet only eIF3 binding appears diminished. eIF4G-I encoded by the mutant allele is unable to associate with eIF4E, possibly due to conformational changes (Chartier-Harlin et al., 2011). A separate study confirmed that the EIF4G-I variation R1205H found in this study seems to be a strong risk factor for Parkinson’s disease (Nuytemans et al., 2013). However, a study of 425 Parkinson’s disease cases in the Chinese Han population did not detect this mutation (Yuan et al., 2013), neither is it found to be a genetic determinant of this disease in a larger screen of Parkinson’s patients in another European population (Lesage et al., 2012). While the latter study did document several other mutations in the eIF4G-I allele, some of which are present in normal controls and others only in Parkinson’s patients, the findings are insufficient to support a direct link between eIF4G-I mutations and pathogenesis (Lesage et al., 2012). Further research is required to elucidate the role of specific eIF4G alleles in neurodegenerative disorders like, and including, Parkinson’s.
3. Conclusion
eIF4G is an essential mediator of protein synthesis, required for growth and development that also contributes to age-related decline and disease susceptibility. Reducing overall eIF4G expression in adult nematodes, in addition to increasing lifespan and globally reducing translation, differentially alters translation efficiency away from transcripts associated with growth toward those associated with homeostasis and increased longevity (Rogers et al., 2011). A deletion in yeast also results in increased lifespan (Smith et al., 2008), with depletion leading to a widespread effect on translation efficiency (Park et al., 2011). However, this factor comes in different forms, which evidence suggests function differently. These differences alter translation efficiency, further testing of which will reveal how altered expression via these homologs and isoforms of eIF4G affect ageing and the progression of age-related disease.
Mechanisms controlling translation have been shown to play an important role in age-related disease, such as cancer. It has been demonstrated that the rate limiting factor for tumor growth and development is angiogenesis triggered by hypoxia (Bergers and Benjamin, 2003). Interestingly, eIF4G functions as a hypoxia-activated switch to facilitate cap-independent mRNA translation in mammalian tumors (Braunstein et al., 2007) and mild hypoxia is associated with an increase in lifespan in C. elegans (Lee et al., 2010). Indeed effectors of the hypoxic response have also been shown to influence lifespan in nematodes (Chen et al., 2009; Mehta et al., 2009). Whether an increase in cap-independent translation in C. elegans, possibly mediated by the short isoform or proteolytically cleaved long isoform of eIF4G, might promote longevity in the absence of an overall reduction in eIF4G levels is unknown. At the very least, further testing on hypoxia and eIF4G in C. elegans may yield insight into whether the mechanisms controlling mammalian tumor growth are the same or divergent from ageing models.
Evidence suggests that the form of eIF4G appears to be detrimental or beneficial according to the context in which it is regulated. In mammals, pathogenesis has come to be associated with the eIF4G-I homolog, with suppression of this form able to slow tumor growth. On the other hand, expression of eIF4G-I in damaged neural tissue promotes cell survival and potentially ameliorates effects of certain neurodegenerative disorders. Exactly how (or whether) these observations in mammals are connected to lifespan modulation in invertebrate models is unclear. However, multiple models of longevity regulation exist that are centered on translational control, which are now beginning to be established in mammals (e.g., Selman et al., 2009). Whole animal model screens allowing analysis of the role of specific forms of eIF4G in certain tissues and developmental phases would allow researchers to more fully characterize the contribution of eIF4G to the regulation of lifespan and may help elucidate its role in the initiation and progression of cancer, Parkinson’s disease, and other age-related diseases.
As this initiation factor remains conserved across species, fully understanding the mechanisms behind eIF4G regulation, specifically with regards to tissue location, is paramount. We speculate that eIF4G has a similar if not identical role in mammalian ageing and age-related disease susceptibility as exhibited in simpler models. Due to the experimental tractability, brief lifespan, and analogy to mammalian models, invertebrate models such including yeast, worms, and flies will likely pave the way for advanced understanding of eIF4G regulation in the health and vitality of the whole organism. Additional research into the importance of specific forms of eIF4G, their tissue-specific roles and the timing/circumstances of expression in organismal health hold the possibility of identifying therapeutic applications of eIF4G modulation to increase lifespan and slow age-related decline in humans.
Supplementary Material
Highlights.
We review the role of eIF4G in translation initiation.
We examine eIF4G with regards to expression timing, tissue-specificity, and form.
The role of eIF4G in promoting longevity and impact on disease is explored.
Specific forms of eIF4G across species are compared.
Acknowledgements
We thank Jarod Rollins and Matt Newsom for useful discussions and comments on the manuscript. Amber Howard and Aric N. Rogers are supported by NIH funding (5R00AG037621-04). Aric N. Rogers is also supported by funding from The Ellison Medical Foundation and the MDIBL Scientific Program Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexandrov A, Colognori D, Steitz JA. Human eIF4AIII interacts with an eIF4G-like partner, NOM1, revealing an evolutionarily conserved function outside the exon junction complex. Genes & Development. 2011;25:1078–1090. doi: 10.1101/gad.2045411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali IK, McKendrick L, Morley SJ, Jackson RJ. Truncated initiation factor eIF4G lacking an eIF4E binding site can support capped mRNA translation. EMBO J. 2001;20:4233–4242. doi: 10.1093/emboj/20.15.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrani N, Ghosh S, Mangus DA, Jacobson A. Translation factors promote the formation of two states of the closed-loop mRNP. Nature. 2008;453:1276–1280. doi: 10.1038/nature06974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badura M, Braunstein S, Zavadil J, Schneider RJ. DNA damage and eIF4G1 in breast cancer cells reprogram translation for survival and DNA repair mRNAs. Proceedings of the National Academy of Sciences. 2012;109:18767–18772. doi: 10.1073/pnas.1203853109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer C, Brass N, Diesinger I, Kayser K, Grasser FA, Meese E. Overexpression of the eukaryotic translation initiation factor 4G (eIF4G-1) in squamous cell lung carcinoma. International Journal of Cancer. 2002;98:181–185. doi: 10.1002/ijc.10180. [DOI] [PubMed] [Google Scholar]
- Bergers G, Benjamin LE. Angiogenesis: Tumorigenesis and the angiogenic switch. Nature Reviews Cancer. 2003;3:401–410. doi: 10.1038/nrc1093. [DOI] [PubMed] [Google Scholar]
- Berset C, Trachsel H, Altmann M. The TOR (target of rapamycin) signal transduction pathway regulates the stability of translation initiation factor eIF4G in the yeast Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences. 1998;95:4264–4269. doi: 10.1073/pnas.95.8.4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borman AM, Kirchweger R, Ziegler E, Rhoads RE, Skern T, Kean KM. elF4G and its proteolytic cleavage products: effect on initiation of protein synthesis from capped, uncapped, and IRES-containing mRNAs. RNA. 1997;3:186–196. [PMC free article] [PubMed] [Google Scholar]
- Braunstein S, Karpisheva K, Pola C, Goldberg J, Hochman T, Yee H, Cangiarella J, Arju R, Formenti SC, Schneider RJ. A Hypoxia-Controlled Cap-Dependent to Cap-Independent Translation Switch in Breast Cancer. Molecular Cell. 2007;28:501–512. doi: 10.1016/j.molcel.2007.10.019. [DOI] [PubMed] [Google Scholar]
- Brown JAL, Roberts TL, Richards R, Woods R, Birrell G, Lim YC, Ohno S, Yamashita A, Abraham RT, Gueven N, Lavin MF. A Novel Role for hSMG-1 in Stress Granule Formation. Molecular and Cellular Biology. 2011;31:4417–4429. doi: 10.1128/MCB.05987-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning KS, Lax SR, Ravel JM. Identification of two messenger RNA cap binding proteins in wheat germ. Evidence that the 28-kDa subunit of eIF-4B and the 26-kDa subunit of eIF-4F are antigenically distinct polypeptides. J. Biol. Chem. 1987;262:11228–11232. [PubMed] [Google Scholar]
- Browning KS, Webster C, Roberts JK, Ravel JM. Identification of an isozyme form of protein synthesis initiation factor 4F in plants. Journal of Biological Chemistry. 1992;267:10096–10100. [PubMed] [Google Scholar]
- Bushell M, McKendrick L, Jänicke RU, Clemens MJ, Morley SJ. Caspase-3 is necessary and sufficient for cleavage of protein synthesis eukaryotic initiation factor 4G during apoptosis. FEBS Lett. 1999;451:332–336. doi: 10.1016/s0014-5793(99)00614-6. [DOI] [PubMed] [Google Scholar]
- Byrd MP. Translation of Eukaryotic Translation Initiation Factor 4GI (eIF4GI) Proceeds from Multiple mRNAs Containing a Novel Cap-dependent Internal Ribosome Entry Site (IRES) That Is Active during Poliovirus Infection. Journal of Biological Chemistry. 2005;280:18610–18622. doi: 10.1074/jbc.M414014200. [DOI] [PubMed] [Google Scholar]
- Cakmakci NG, Lerner RS, Wagner EJ, Zheng L, Marzluff WF. SLIP1, a Factor Required for Activation of Histone mRNA Translation by the Stem-Loop Binding Protein. Molecular and Cellular Biology. 2007;28:1182–1194. doi: 10.1128/MCB.01500-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carberry SE, Goss DJ, Darzynkiewicz E. A comparison of the binding of methylated cap analogs to wheat germ protein synthesis initiation factors 4F and (iso) 4F. Biochemistry. 1991;30:1624–1627. doi: 10.1021/bi00220a026. [DOI] [PubMed] [Google Scholar]
- Caron S, Charon M, Cramer E, Sonenberg N, Dusanter-Fourt I. Selective Modification of Eukaryotic Initiation Factor 4F (eIF4F) at the Onset of Cell Differentiation: Recruitment of eIF4GII and Long-Lasting Phosphorylation of eIF4E. Molecular and Cellular Biology. 2004;24:4920–4928. doi: 10.1128/MCB.24.11.4920-4928.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartier-Harlin M-C, Dachsel JC, Vilariño-Güell C, Lincoln SJ, Leprêtre F, Hulihan MM, Kachergus J, Milnerwood AJ, Tapia L, Song M-S, Le Rhun E, Mutez E, Larvor L, Duflot A, Vanbesien-Mailliot C, Kreisler A, Ross OA, Nishioka K, Soto-Ortolaza AI, Cobb SA, Melrose HL, Behrouz B, Keeling BH, Bacon JA, Hentati E, Williams L, Yanagiya A, Sonenberg N, Lockhart PJ, Zubair AC, Uitti RJ, Aasly JO, Krygowska-Wajs A, Opala G, Wszolek ZK, Frigerio R, Maraganore DM, Gosal D, Lynch T, Hutchinson M, Bentivoglio AR, Valente EM, Nichols WC, Pankratz N, Foroud T, Gibson RA, Hentati F, Dickson DW, Destée A, Farrer MJ. Translation Initiator EIF4G1 Mutations in Familial Parkinson Disease. The American Journal of Human Genetics. 2011;89:398–406. doi: 10.1016/j.ajhg.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Pan KZ, Palter JE, Kapahi P. Longevity determined by developmental arrest genes in Caenorhabditis elegans. Aging Cell. 2007;6:525–533. doi: 10.1111/j.1474-9726.2007.00305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Thomas EL, Kapahi P. HIF-1 Modulates Dietary Restriction-Mediated Lifespan Extension via IRE-1 in Caenorhabditis elegans. PLoS Genetics. 2009;5:e1000486. doi: 10.1371/journal.pgen.1000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiocchetti A, Zhou J, Zhu H, Karl T, Haubenreisser O, Rinnerthaler M, Heeren G, Oender K, Bauer J, Hintner H, Breitenbach M, Breitenbach-Koller L. Ribosomal proteins Rpl10 and Rps6 are potent regulators of yeast replicative life span. Experimental Gerontology. 2007;42:275–286. doi: 10.1016/j.exger.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Coldwell MJ, Morley SJ. Specific Isoforms of Translation Initiation Factor 4GI Show Differences in Translational Activity. Molecular and Cellular Biology. 2006;26:8448–8460. doi: 10.1128/MCB.01248-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coldwell MJ, Sack U, Cowan JL, Barrett RM, Vlasak M, Sivakumaran K, Morley SJ. Multiple isoforms of the translation initiation factor eIF4GII are generated via use of alternative promoters, splice sites and a non-canonical initiation codon. Biochemical Journal. 2012;448:1–11. doi: 10.1042/BJ20111765. [DOI] [PubMed] [Google Scholar]
- Comtesse N, Keller A, Diesinger I, Bauer C, Kayser K, Huwer H, Lenhof H-P, Meese E. Frequent overexpression of the genes FXR1, CLAPM1 and EIF4G located on amplicon 3q26-27 in squamous cell carcinoma of the lung. International Journal of Cancer. 2007;120:2538–2544. doi: 10.1002/ijc.22585. [DOI] [PubMed] [Google Scholar]
- Contreras V, Friday AJ, Morrison JK, Hao E, Keiper BD. Cap-Independent Translation Promotes C. elegans Germ Cell Apoptosis through Apaf-1/CED-4 in a Caspase-Dependent Mechanism. PLoS ONE. 2011;6:e24444. doi: 10.1371/journal.pone.0024444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras V, Richardson MA, Hao E, Keiper BD. Depletion of the cap-associated isoform of translation factor eIF4G induces germline apoptosis in C. elegans. Cell Death & Differentiation. 2008;15:1232–1242. doi: 10.1038/cdd.2008.46. [DOI] [PubMed] [Google Scholar]
- Craig AW, Haghighat A, Yu AT, Sonenberg N. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature. 1998;392:520–523. doi: 10.1038/33198. [DOI] [PubMed] [Google Scholar]
- Cromer A, Carles A, Millon R, Ganguli G, Chalmel F, Lemaire F, Young J, Dembélé D, Thibault C, Muller D, Poch O, Abecassis J, Wasylyk B. Identification of genes associated with tumorigenesis and metastatic potential of hypopharyngeal cancer by microarray analysis. Oncogene. 2003;23:2484–2498. doi: 10.1038/sj.onc.1207345. [DOI] [PubMed] [Google Scholar]
- Cuesta R, Laroia G, Schneider RJ. Chaperone hsp27 inhibits translation during heat shock by binding eIF4G and facilitating dissociation of cap-initiation complexes. Genes & development. 2000;14:1460–1470. [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Hagan KW, Zhang S, Peltz SW. Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes Dev. 1995;9:423–436. doi: 10.1101/gad.9.4.423. [DOI] [PubMed] [Google Scholar]
- Curran SP, Ruvkun G. Lifespan Regulation by Evolutionarily Conserved Genes Essential for Viability. PLoS Genetics. 2007;3:e56. doi: 10.1371/journal.pgen.0030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchison D, Milburn SC, Edery I, Sonenberg N, Hershey JW. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J. Biol. Chem. 1982;257:14806–14810. [PubMed] [Google Scholar]
- Fang W, Li X, Jiang Q, Liu Z, Yang H, Wang S, Xie S, Liu Q, Liu T, Huang J, Xie W, Li Z, Zhao Y, Wang E, Marincola FM, Yao K. Transcriptional patterns, biomarkers and pathways characterizing nasopharyngeal carcinoma of Southern China. Journal of Translational Medicine. 2008;6:32. doi: 10.1186/1479-5876-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, Gil L, Garcia-Giron C, Gordon L, Hourlier T, Hunt S, Juettemann T, Kahari AK, Keenan S, Komorowska M, Kulesha E, Longden I, Maurel T, McLaren WM, Muffato M, Nag R, Overduin B, Pignatelli M, Pritchard B, Pritchard E, Riat HS, Ritchie GRS, Ruffier M, Schuster M, Sheppard D, Sobral D, Taylor K, Thormann A, Trevanion S, White S, Wilder SP, Aken BL, Birney E, Cunningham F, Dunham I, Harrow J, Herrero J, Hubbard TJP, Johnson N, Kinsella R, Parker A, Spudich G, Yates A, Zadissa A, Searle SMJ. Ensembl 2013. Nucleic Acids Research. 2012;41:D48–D55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuchi-Shimogori T, Ishii I, Kashiwagi K, Mashiba H, Ekimoto H, Igarashi K. Malignant transformation by overproduction of translation initiation factor eIF4G. Cancer research. 1997;57:5041–5044. [PubMed] [Google Scholar]
- Gallie DR. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes & Development. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- Gallie DR, Browning KS. eIF4G Functionally Differs from eIFiso4G in Promoting Internal Initiation, Cap-independent Translation, and Translation of Structured mRNAs. Journal of Biological Chemistry. 2001;276:36951–36960. doi: 10.1074/jbc.M103869200. [DOI] [PubMed] [Google Scholar]
- Gilbert WV, Zhou K, Butler TK, Doudna JA. Cap-Independent Translation Is Required for Starvation-Induced Differentiation in Yeast. Science. 2007;317:1224–1227. doi: 10.1126/science.1144467. [DOI] [PubMed] [Google Scholar]
- Goyer C, Altmann M, Lee HS, Blanc A, Deshmukh M, Woolford JL, Trachsel H, Sonenberg N. TIF4631 and TIF4632: two yeast genes encoding the high-molecular-weight subunits of the cap-binding protein complex (eukaryotic initiation factor 4F) contain an RNA recognition motif-like sequence and carry out an essential function. Molecular and cellular biology. 1993;13:4860–4874. doi: 10.1128/mcb.13.8.4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradi A, Imataka H, Svitkin YV, Rom E, Raught B, Morino S, Sonenberg N. A novel functional human eukaryotic translation initiation factor 4G. Molecular and cellular biology. 1998;18:334–342. doi: 10.1128/mcb.18.1.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghighat A, Sonenberg N. eIF4G dramatically enhances the binding of eIF4E to the mRNA 5′-cap structure. Journal of Biological Chemistry. 1997;272:21677–21680. doi: 10.1074/jbc.272.35.21677. [DOI] [PubMed] [Google Scholar]
- Hansen M, Taubert S, Crawford D, Libina N, Lee S-J, Kenyon C. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- Henderson ST, Bonafè M, Johnson TE. daf-16 protects the nematode Caenorhabditis elegans during food deprivation. The Journals of Gerontology Series A: Biological Sciences and Medical Sciences. 2006;61:444–460. doi: 10.1093/gerona/61.5.444. [DOI] [PubMed] [Google Scholar]
- Hershey JWB, Sonenberg N, Mathews MB. Principles of translational control: an overview. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a011528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imataka H, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Molecular and cellular biology. 1997;17:6940–6947. doi: 10.1128/mcb.17.12.6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E, Stepinski J, Darzynkiewicz E, Mattaj IW. A cap binding protein that may mediate nuclear export of RNA polymerase II-transcribed RNAs. The Journal of cell biology. 1992;118:1287–1295. doi: 10.1083/jcb.118.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferies HB, Reinhard C, Kozma SC, Thomas G. Rapamycin selectively represses translation of the “polypyrimidine tract” mRNA family. Proceedings of the National Academy of Sciences. 1994;91:4441–4445. doi: 10.1073/pnas.91.10.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C, Dobrikova EY, Bradrick SS, Shveygert M, Herbert JT, Gromeier M. Activation of cap-independent translation by variant eukaryotic initiation factor 4G in vivo. RNA. 2008;14:2170–2182. doi: 10.1261/rna.1171808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapahi P, Chen D, Rogers AN, Katewa SD, Li PW-L, Thomas EL, Kockel L. With TOR, Less Is More: A Key Role for the Conserved Nutrient-Sensing TOR Pathway in Aging. Cell Metabolism. 2010;11:453–465. doi: 10.1016/j.cmet.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KM, Cho H, Choi K, Kim J, Kim B-W, Ko Y-G, Jang SK, Kim YK. A new MIF4G domain-containing protein, CTIF, directs nuclear cap-binding protein CBP80/20-dependent translation. Genes & Development. 2009;23:2033–2045. doi: 10.1101/gad.1823409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koritzinsky M, Magagnin MG, van den Beucken T, Seigneuric R, Savelkouls K, Dostie J, Pyronnet S, Kaufman RJ, Weppler SA, Voncken JW, Lambin P, Koumenis C, Sonenberg N, Wouters BG. Gene expression during acute and prolonged hypoxia is regulated by distinct mechanisms of translational control. EMBO J. 2006;25:1114–1125. doi: 10.1038/sj.emboj.7600998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamphear BJ, Kirchweger R, Skern T, Rhoads RE. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J. Biol. Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- Lee SH, McCormick F. p97/DAP5 is a ribosome-associated factor that facilitates protein synthesis and cell proliferation by modulating the synthesis of cell cycle proteins. The EMBO journal. 2006;25:4008–4019. doi: 10.1038/sj.emboj.7601268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-J, Hwang AB, Kenyon C. Inhibition of Respiration Extends C. elegans Life Span via Reactive Oxygen Species that Increase HIF-1 Activity. Current Biology. 2010;20:2131–2136. doi: 10.1016/j.cub.2010.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lellis AD, Allen ML, Aertker AW, Tran JK, Hillis DM, Harbin CR, Caldwell C, Gallie DR, Browning KS. Deletion of the eIFiso4G subunit of the Arabidopsis eIFiso4F translation initiation complex impairs health and viability. Plant Molecular Biology. 2010;74:249–263. doi: 10.1007/s11103-010-9670-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage S, Condroyer C, Klebe S, Lohmann E, Durif F, Damier P, Tison F, Anheim M, Honoré A, Viallet F, Bonnet A-M, Ouvrard-Hernandez A-M, Vidailhet M, Durr A, Brice A. EIF4G1 in familial Parkinson’s disease: pathogenic mutations or rare benign variants? Neurobiology of Aging. 2012;33:2233.e1–2233.e5. doi: 10.1016/j.neurobiolaging.2012.05.006. [DOI] [PubMed] [Google Scholar]
- Lewis SM, Cerquozzi S, Graber TE, Ungureanu NH, Andrews M, Holcik M. The eIF4G homolog DAP5/p97 supports the translation of select mRNAs during endoplasmic reticulum stress. Nucleic Acids Research. 2007;36:168–178. doi: 10.1093/nar/gkm1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman N, Marash L, Kimchi A. The translation initiation factor DAP5 is a regulator of cell survival during mitosis. Cell Cycle. 2009;8:204–209. doi: 10.4161/cc.8.2.7384. [DOI] [PubMed] [Google Scholar]
- Lomakin IB, Hellen CU, Pestova TV. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Molecular and cellular biology. 2000;20:6019–6029. doi: 10.1128/mcb.20.16.6019-6029.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long X, Spycher C, Han ZS, Rose AM, Müller F, Avruch J. TOR Deficiency in <i>C. elegans</i> Causes Developmental Arrest and Intestinal Atrophy by Inhibition of mRNA Translation. Current biology. 2002;12:1448–1461. doi: 10.1016/s0960-9822(02)01091-6. [DOI] [PubMed] [Google Scholar]
- Magnuson B, Ekim B, Fingar DC. Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochemical Journal. 2012;441:1–21. doi: 10.1042/BJ20110892. [DOI] [PubMed] [Google Scholar]
- Marchione R, Leibovitch SA, Lenormand J-L. The translational factor eIF3f: the ambivalent eIF3 subunit. Cellular and Molecular Life Sciences. 2013 doi: 10.1007/s00018-013-1263-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marissen WE, Gradi A, Sonenberg N, Lloyd RE. Cleavage of eukaryotic translation initiation factor 4GII correlates with translation inhibition during apoptosis. Cell Death Differ. 2000;7:1234–1243. doi: 10.1038/sj.cdd.4400750. [DOI] [PubMed] [Google Scholar]
- Marissen WE, Lloyd RE. Eukaryotic translation initiation factor 4G is targeted for proteolytic cleavage by caspase 3 during inhibition of translation in apoptotic cells. Molecular and cellular biology. 1998;18:7565–7574. doi: 10.1128/mcb.18.12.7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martineau Y, Derry MC, Wang X, Yanagiya A, Berlanga JJ, Shyu A-B, Imataka H, Gehring K, Sonenberg N. Poly(A)-Binding Protein-Interacting Protein 1 Binds to Eukaryotic Translation Initiation Factor 3 To Stimulate Translation. Molecular and Cellular Biology. 2008;28:6658–6667. doi: 10.1128/MCB.00738-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta R, Steinkraus KA, Sutphin GL, Ramos FJ, Shamieh LS, Huh A, Davis C, Chandler-Brown D, Kaeberlein M. Proteasomal Regulation of the Hypoxic Response Modulates Aging in C. elegans. Science. 2009;324:1196–1198. doi: 10.1126/science.1173507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthukrishnan S, Both GW, Furuichi Y, Shatkin AJ. 5’-Terminal 7-methylguanosine in eukaryotic mRNA is required for translation. Nature. 1975;255:33–37. doi: 10.1038/255033a0. [DOI] [PubMed] [Google Scholar]
- Neumar RW, DeGracia DJ, Konkoly LL, Khoury JI, White BC, Krause GS. Calpain Mediates Eukaryotic Initiation Factor 4G Degradation During Global Brain Ischemia. Journal of Cerebral Blood Flow & Metabolism. 1998:876–881. doi: 10.1097/00004647-199808000-00007. [DOI] [PubMed] [Google Scholar]
- Nousch M, Reed V, Bryson-Richardson RJ, Currie PD, Preiss T. The eIF4G-homolog p97 can activate translation independent of caspase cleavage. RNA. 2007;13:374–384. doi: 10.1261/rna.372307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuytemans K, Bademci G, Inchausti V, Dressen A, Kinnamon DD, Mehta A, Wang L, Züchner S, Beecham GW, Martin ER. Whole exome sequencing of rare variants in EIF4G1 and VPS35 in Parkinson disease. Neurology. 2013;80:982–989. doi: 10.1212/WNL.0b013e31828727d4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlmann T, Rau M, Morley SJ, Pain VM. Proteolytic cleavage of initiation factor eIF-4 gamma in the reticulocyte lysate inhibits translation of capped mRNAs but enhances that of uncapped mRNAs. Nucleic Acids Res. 1995;23:334–340. doi: 10.1093/nar/23.3.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan KZ, Palter JE, Rogers AN, Olsen A, Chen D, Lithgow GJ, Kapahi P. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell. 2007;6:111–119. doi: 10.1111/j.1474-9726.2006.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E-H, Zhang F, Warringer J, Sunnerhagen P, Hinnebusch AG. Depletion of eIF4G from yeast cells narrows the range of translational efficiencies genome-wide. BMC genomics. 2011;12:68. doi: 10.1186/1471-2164-12-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- Qin H. Phosphorylation Screening Identifies Translational Initiation Factor 4GII as an Intracellular Target of Ca2+/Calmodulin-dependent Protein Kinase I. Journal of Biological Chemistry. 2003;278:48570–48579. doi: 10.1074/jbc.M308781200. [DOI] [PubMed] [Google Scholar]
- Ramirez-Valle F, Braunstein S, Zavadil J, Formenti SC, Schneider RJ. eIF4GI links nutrient sensing by mTOR to cell proliferation and inhibition of autophagy. The Journal of Cell Biology. 2008;181:293–307. doi: 10.1083/jcb.200710215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers AN, Chen D, McColl G, Czerwieniec G, Felkey K, Gibson BW, Hubbard A, Melov S, Lithgow GJ, Kapahi P. Life Span Extension via eIF4G Inhibition Is Mediated by Posttranscriptional Remodeling of Stress Response Gene Expression in C. elegans. Cell Metabolism. 2011;14:55–66. doi: 10.1016/j.cmet.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selman C, Tullet JMA, Wieser D, Irvine E, Lingard SJ, Choudhury AI, Claret M, Al-Qassab H, Carmignac D, Ramadani F, Woods A, Robinson ICA, Schuster E, Batterham RL, Kozma SC, Thomas G, Carling D, Okkenhaug K, Thornton JM, Partridge L, Gems D, Withers DJ. Ribosomal Protein S6 Kinase 1 Signaling Regulates Mammalian Life Span. Science. 2009;326:140–144. doi: 10.1126/science.1177221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvera D, Arju R, Darvishian F, Levine PH, Zolfaghari L, Goldberg J, Hochman T, Formenti SC, Schneider RJ. Essential role for eIF4GI overexpression in the pathogenesis of inflammatory breast cancer. Nature Cell Biology. 2009;11:903–908. doi: 10.1038/ncb1900. [DOI] [PubMed] [Google Scholar]
- Smith ED, Tsuchiya M, Fox LA, Dang N, Hu D, Kerr EO, Johnston ED, Tchao BN, Pak DN, Welton KL, Promislow DEL, Thomas JH, Kaeberlein M, Kennedy BK. Quantitative evidence for conserved longevity pathways between divergent eukaryotic species. Genome Research. 2008;18:564–570. doi: 10.1101/gr.074724.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Morgan MA, Merrick WC, Shatkin AJ. A polypeptide in eukaryotic initiation factors that crosslinks specifically to the 5’-terminal cap in mRNA. Proceedings of the National Academy of Sciences. 1978;75:4843–4847. doi: 10.1073/pnas.75.10.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steckelberg A-L, Boehm V, Gromadzka AM, Gehring NH. CWC22 Connects Pre-mRNA Splicing and Exon Junction Complex Assembly. Cell Reports. 2012;2:454–461. doi: 10.1016/j.celrep.2012.08.017. [DOI] [PubMed] [Google Scholar]
- Steffen KK, MacKay VL, Kerr EO, Tsuchiya M, Hu D, Fox LA, Dang N, Johnston ED, Oakes JA, Tchao BN, Pak DN, Fields S, Kennedy BK, Kaeberlein M. Yeast Life Span Extension by Depletion of 60S Ribosomal Subunits Is Mediated by Gcn4. Cell. 2008;133:292–302. doi: 10.1016/j.cell.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Palmer K, Handel MA. Mutation of Eif4g3, encoding a eukaryotic translation initiation factor, causes male infertility and meiotic arrest of mouse spermatocytes. Development. 2010;137:1699–1707. doi: 10.1242/dev.043125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin YV, Herdy B, Costa-Mattioli M, Gingras A-C, Raught B, Sonenberg N. Eukaryotic Translation Initiation Factor 4E Availability Controls the Switch between Cap-Dependent and Internal Ribosomal Entry Site-Mediated Translation. Molecular and Cellular Biology. 2005;25:10556–10565. doi: 10.1128/MCB.25.23.10556-10565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syntichaki P, Troulinaki K, Tavernarakis N. Protein Synthesis Is a Novel Determinant of Aging in Caenorhabditis elegans. Annals of the New York Academy of Sciences. 2007;1119:289–295. doi: 10.1196/annals.1404.001. [DOI] [PubMed] [Google Scholar]
- Tarun SZ, Jr, Sachs AB. Association of the yeast poly (A) tail binding protein with translation initiation factor eIF-4G. The EMBO Journal. 1996;15:7168. [PMC free article] [PubMed] [Google Scholar]
- Tarun SZ, Sachs AB. Binding of eukaryotic translation initiation factor 4E (eIF4E) to eIF4G represses translation of uncapped mRNA. Molecular and cellular biology. 1997;17:6876–6886. doi: 10.1128/mcb.17.12.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu L, Liu Z, He X, He Y, Yang H, Jiang Q, Xie S, Xiao G, Li X, Yao K. Research Over-expression of eukaryotic translation initiation factor 4 gamma 1 correlates with tumor progression and poor prognosis in nasopharyngeal carcinoma. 2010 doi: 10.1186/1476-4598-9-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unbehaun A, Borukhov SI, Hellen CU, Pestova TV. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes & development. 2004;18:3078–3093. doi: 10.1101/gad.1255704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosler PS, Gao Y, Brennan CS, Yanagiya A, Gan Y, Cao G, Zhang F, Morley SJ, Sonenberg N, Bennett MV. Ischemia-induced calpain activation causes eukaryotic (translation) initiation factor 4G1 (eIF4GI) degradation, protein synthesis inhibition, and neuronal death. Proceedings of the National Academy of Sciences. 2011;108:18102–18107. doi: 10.1073/pnas.1112635108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vries RGJ. Heat Shock Increases the Association of Binding Protein-1 with Initiation Factor 4E. Journal of Biological Chemistry. 1997;272:32779–32784. doi: 10.1074/jbc.272.52.32779. [DOI] [PubMed] [Google Scholar]
- Weischenfeldt J, Damgaard I, Bryder D, Theilgaard-Monch K, Thoren LA, Nielsen FC, Jacobsen SEW, Nerlov C, Porse BT. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes & Development. 2008;22:1381–1396. doi: 10.1101/gad.468808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winstall E, Sadowski M, Kuhn U, Wahle E, Sachs AB. The Saccharomyces cerevisiae RNA-binding protein Rbp29 functions in cytoplasmic mRNA metabolism. J. Biol. Chem. 2000;275:21817–21826. doi: 10.1074/jbc.M002412200. [DOI] [PubMed] [Google Scholar]
- Yamanaka S, Poksay KS, Arnold KS, Innerarity TL. A novel translational repressor mRNA is edited extensively in livers containing tumors caused by the transgene expression of the apoB mRNA-editing enzyme. Genes & Development. 1997;11:321–333. doi: 10.1101/gad.11.3.321. [DOI] [PubMed] [Google Scholar]
- Yuan L, Song Z, Xu H, Gu S, Zhu A, Gong L, Zhao Y, Deng H. EIF4G1 Ala502Val and Arg1205His variants in Chinese patients with Parkinson disease. Neuroscience Letters. 2013;543:69–71. doi: 10.1016/j.neulet.2013.02.056. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.