Abstract
A number of well-known type II inhibitors (ATP non-competitive) that bind kinases in their DFG-out conformation were tested against wild-type LRRK2 and the most common Parkinson’s disease-linked mutation G2019S. We found that traditional type II inhibitors exhibit surprising variability in their inhibition mechanism between wild type (WT) and the G2019S mutant of LRRK2. The type II kinase inhibitors were found to work by an ATP-competitive fashion against the G2019S mutant, whereas they appear to follow the expected non-competitive mechanism against WT. Since the G2019S mutation lies in the DXG-motif (DYG in LRRK2 but DFG in most other kinases) of the activation loop, we explored the structural consequence of the mutation on loop dynamics using an enhanced sampling method called metadynamics. The simulations suggest that the G2019S mutation stabilizes the DYG-in state of LRRK2 through a series of hydrogen bonds, leading to an increase in the conformational barrier between the active and inactive forms of the enzyme and a relative stabilization of the active form. The conformational bias toward the active form of LRRK2 mutants has two primary consequences: 1) the mutant enzyme becomes hyperactive, a known contributor to the Parkinsonian phenotype, as a consequence of being “locked” into the activated state and 2) the mutation creates an unusual allosteric pocket that can bind type II inhibitors but in an ATP competitive fashion. Our results suggest that developing type II inhibitors, which are generally considered superior to type I inhibitors due to desirable selectivity profiles, might be especially challenging for the G2019S LRRK2 mutant.
Parkinson’s disease (PD) is a neurodegenerative disorder that affects over 1 million Americans and more than 60,000 patients are newly diagnosed each year. Loss of dopaminergic neurons in a part of the brain called the substantia nigra leads to lowered production of dopamine and the brain’s ability to control movement is compromised (1-4). Mutations in several genes have been genetically linked to PD in recent years. Among them, leucine-rich repeat kinase 2 (LRRK2) has emerged as a highly relevant gene to PD pathogenesis (5-7). At least 40 mutations in LRRK2 have been identified in the most common familial forms of PD, some sporadic forms of PD, and have been associated with typical idiopathic, late-onset PD (8-12).
LRRK2 is a large, multi-domain protein that encodes two distinct enzymes: a protein kinase and a GTPase (13-16). The most prevalent mutation is G2019S, which demonstrates increased kinase activity, is correlated with increased neurotoxicity. In recent studies, LRRK2 inhibitors have been shown to protect dopaminergic neuron loss in PD animal models (17-25), suggesting that kinase activity of LRRK2 plays a critical role in the pathogenesis of PD. Several type I kinase inhibitors that are capable of targeting the ATP binding hinge of the LRRK2 kinase in its active form (DYG-in) have been described but few mechanistic studies have been carried on type II (DYG-out) inhibitors that target an inactive conformation of the kinase.
The structural rearrangement needed for binding type II inhibitors involves movement of the activation loop bearing a conserved DXG motif (DFG in most kinases but DYG in LRRK2), where Asp and Phe/Tyr exchange positions (called as DXG-flip) that inactivates the kinase (26-31). G2019S is immediately adjacent to this bipositional switch, suggesting that it may directly affect the activation status of LRRK2. In this study, we test several type II kinase inhibitors against wild-type LRRK2 and the PD-linked mutant G2019S. While most of these molecules are shown to inhibit the WT enzyme in an ATP noncompetitive manner, suggesting binding to a DYG-out state of the enzyme, the same inhibitors appear to block the G2019S mutant by an ATP competitive mechanism.
In order to understand this unexpected and counterintuitive observation, we carried out temperature dependent kinetic studies, metadynamics simulations (32-34), and induced-fit docking. Metadynamics simulations support these experimental findings, suggesting that the mutation not only leads to a high-energy barrier for the activation loop transition but also preferentially stabilization the DYG-in state. The free energy surfaces and modeled structures from the metadynamics simulations rationalize the observations and provide mechanistic insights. Induced fit docking of type II inhibitors against mutant LRRK2 using the DYG-in state explains the atypical ATP competitive inhibition observed in the experimental studies.
Materials and Methods
Kinase assay
Truncated wild-type LRRK2 (residues 970-2527) and mutant G2019S (Invitrogen, Carlsbad, CA) expressed in baculovirus system were used in this study. The kinase assay for LRRKtide (RLGRDKYKTLRQIRQ) (American Peptide, Sunnyvale, CA) phosphorylation was conducted in buffer containing 20 mM HEPES (pH 7.4), 50 mM NaCl, 10 mM MgCl2, 1 mM DTT, BSA 0.5 mg/ml, 1 mM beta-Gly-PO4, LRRKtide, ATP and [γ-33P]-ATP (Perkin Elmer, Boston, MA). Detailed methodology of the assay and the analysis of data were published previous by Liu et al (35). The reactions were conducted in duplicate, initiated by the addition of 6 nM truncated LRRK2, and incubated at room temperature for 120 min. The reactions were stopped by the addition of 20 mM EDTA and the mixture was transferred to a multiscreen PH filtration plate (Millipore, Billerica, MA) and washed six times with 75 mM H3PO4. The plate was dried, filters were removed, and the samples were analyzed with a scintillation counter. Background reactions were conducted in the absence of LRRK2. In all cases, reaction progress curves for production of phospho-LRRKtide were linear over at last 60 minutes and allowed calculation of initial velocities.
Temperature dependent kinetics
We monitored kcat at various temperatures during LRRK2 catalyzed phosphorylation both by WT and G2019S. The experiment yields enthalpy, entropy, and free energy values for these enzyme forms. The experiments were carried out using a 1570 amino acid fragment of LRRK2 (residues 970-2527, which retained full catalytic activity. According to transition state theory, kcat (kc)depends on temperature as equation 1:
| (1) |
where ΔG‡, kB, h, and R are the Gibbs free energy of activation, the Boltzmann, Planck, and gas constants, respectively. The rearranged equation gives:
| (2) |
where ΔH‡ and ΔS‡ are the enthalpy and entropy of activation, respectively. A linear plot of ln(kch/kBT) vs 1/T has a slope of − ΔH‡/R and a y-intercept of ΔS‡/R, respectively.
Modeling of LRRK2 kinase domain and docking
The LRRK2 kinase domain, between residues 1859-2138, was modeled using Modeller v9.11 (36-39). Briefly, the main variables in homology modeling are template selection and sequence alignment between the target and the template. Modeling details have been published previously and described in details in reference 35 but briefly, B-raf kinase with 33% sequence identity to the LRRK2 kinase domain, was used as a template for homology modeling since this enzyme had the highest sequence similarity with LRRK2 around the active site region and the ATP binding hinge compared to the other kinases. In addition, B-raf and LRRK2 inhibitors could cross-inhibit each other supporting our choice of template selection for modeling (details to be published elsewhere). Recently, kinase domain of Roco4 from Dictyostelium discoidium (PDB code: 4F0G), a closely related member of the LRRK2 family, was published and we constructed a model of LRRK2 with this as a template and compared with our previous model. The comparison of the two LRRK2 models revealed that the overall C-alpha atoms RMSD was less than < 1.3Å.
Docking of the ATP molecule in the binding site was carried out using Glide v2.2 (40), which treats the receptor rigidly. Docking of type II inhibitors were carried out using the Induced Fit Docking (IFD) (41) protocol implemented in the Schrödinger suite (41). Briefly, in the first stage of IFD 20 initial poses are generated using Glide with a softened potential to allow for clashes with the receptor. For each of the top 20 poses from the initial softened-potential docking step, a cycle of protein side chain prediction and full residue minimization is performed using Prime to generate 20 induced-fit receptor structures (42). All residues having at least one atom within 5 Å of any atom from the 20 ligand poses are refined. Finally, the ligand is re-docked using Glide with default settings into each induced-fit receptor structure and a composite score that accounts for the protein/ligand interaction energy (GlideScore) and the total energy of the system (Prime energy) is used to rank the induced-fit structures.
Modeling of the active and inactive conformations of LRRK2
Starting with the homology model of the active form of WT LRRK2 described above, the DYG-out state of the activation loop between residues 2015-2027 was modeled using Prime v2.0 (details in Supporting Information). The lowest energy structure with a DYG-out conformation was chosen for further modeling experiments. The protein domain motion server HingeProt (43) was used to generate the “open” conformation of the kinase using the Prime generated DYG-out structure as the input model, where the N-terminal β-sheet and C-helix were repositioned resulting in disruption of the salt-bridge between K1096 and E1920.
The PD linked mutation G2019S was modeled both in the DYG-in (closed) and DYG-out (open) conformation using the mutagenesis script of PyMOL (44) and the resulting structure was subjected to 2000 cycles energy minimization using Desmond v3.0 (45). In order to determine if the modeled structures were thermally stable, short MD simulations (5 ns) of the two conformational states of the kinase (WT and G2019S) described above were carried out using the Desmond v3.0 and the OPLS_2005 force field (46) (details in Supporting Information). The model structures for LRRK2 WT and G2019S were found to be thermally stable over the course of the short simulation with RMSD of C-alpha atoms less than 1.2 Å from the starting structure.
Metadynamics simulations
Metadynamics (47-57) simulations were carried out with Desmond v3.0 using two collective variables (CVs). In order to test for robustness of the simulations and choice of CVs, we chose two independent pairs of CVs (one pair with distances and the other pair with torsions) and ran separate simulations. We chose the distance between the center of mass of a collection of atoms in the β-sheet (residues 1878-1906) and the N-terminal C-helix (residues 1915-1928) as the first CV denoted [s]. For the second CV denoted [z], we chose and the center of mass of a subset of residues in the activation loop (Y2018, A2021, C2024, R2026, and M2027) and the center of mass for the ATP binding hinge comprised of residues (M1947, L1949, and K1952) . The distance between the N-terminal β-sheet domain and C-helix in the N-terminal domain describes the opening and closing motion of LRRK2 around the hinge region. The center of mass of the activation loop is the primary degree of freedom of interest for this study because it directly involves the transition from active to inactive form.
For the torsion CVs, we chose Ramachandran ϕ/ψ dihedrals of residue 2019 (Gly in WT and Ser in the mutant) in the activation loop. These torsions are known to undergo substantial changes in the transition form DFG-in to DFG-out in other kinases. See Figure S1 (Supporting Information) and Table S1 (Supporting Information) for an illustration of the location of these CVs on the LRRK2 modeled structure.
The default equilibration protocol in Desmond was run before the metadynamics simulations, which relaxes the system in a gradual way with progressively weaker harmonic restraints and heating of the system from 0 K to 300 K. The total length of each production simulation was 250 ns at a temperature of 300 K and pressure of 1 atm. A Gaussian biasing potential with a height of 0.03 kcal/mol was injected at 0.09 ps intervals throughout the simulation and the free-energy surface (FES) was reconstructed from these Gaussians. Multiple simulations were carried out using different initial activation loop conformations. At the end of 250ns, all simulations converged and the FES constructed using these simulations were virtually identical. Clusters of conformations corresponding to the various energy minima observed in FES were extracted and were analyzed for structural details.
Results
Inhibition of LRRK2 by type II Inhibitors
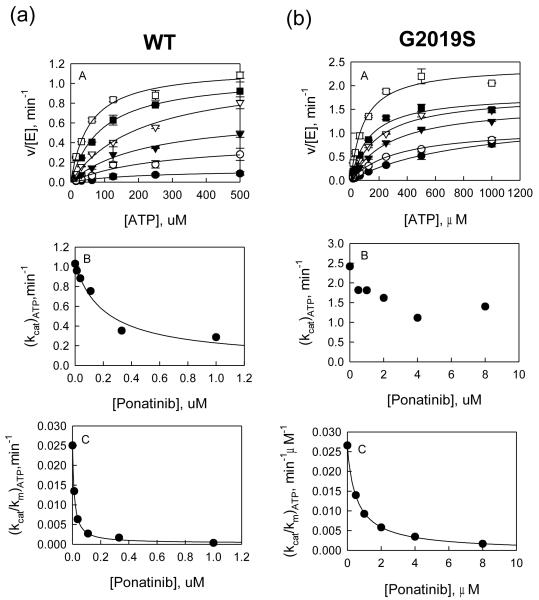
Type II (or DFG-out) inhibitors are known to bind kinases at an allosteric site that exists when the kinase switches to an inactive DFG-out form. The advantage of type II inhibitors is that they bind noncompetitively with ATP and therefore have the potential advantage of avoiding selectivity problems arising from binding to the highly conserved ATP pocket in all kinases. Four known type II inhibitors of other kinases (sorafenib, ponatinib, bosutinib, and imatinib) were tested against LRRK2 for affinity and inhibition mechanism (Table 1, Figure 1) and (Figure S2, supporting information). In addition, a type I inhibitor (GSK3-XIII) was tested as a control (Table 1). Inhibition was measured in a dose-response manner with WT and G2019S for each of the five compounds at concentrations of LRRKtide and ATP around their Km values. The dependence of initial velocity on inhibitor concentration followed the simple inhibition expression in the general form: vinhib = vcontrol/(1 + [I]/Ki,app). All compounds except sorafenib behaved as classic inhibitors with no sign of partial inhibition and no need of including higher-order terms to attain good data fitting (Figure 1) and (Figure S2, Supporting Information). Partial inhibition was observed for sorafenib and it is more evident with the mutant G2019S at higher [ATP]. The determined IC50 values are summarized in Table 1.
Table 1.
Docking score and Ki,app values of DYG-out inhibitors of LRRK2
| Compounds | Dominant Binding mode to WT LRRK2 |
Ki,app, μM | IFD Scores | ||
|---|---|---|---|---|---|
| WT | G2019S | WT | G2019S | ||
| Ponatinib | DFG-out + hinge | 0.031 ± 0.004 NC | 0.2 ± 0.03 C | −528.6 | −525.2 |
| Sorafenib | DFG-out | 0.7 ± 0.3 NC | 9.7 ± 3.2 C | −523.2 | −517.35 |
| Bosutinib | Hinge | 0.3 ± 0.02 C | 0.2 ± 0.02 C | −525.8 | −525.1 |
| Imatinib | DFG-out + Hinge | 5.1 ± 1.4 NC | 5.6 ± 0.4 C | −515.2 | −513.5 |
| GSK3-XIII | Hinge | 0.1 ± 0.01 | 0.1 ± 0.01 | −520.4 | −520.3 |
C → competitive inhibition, NC → Non-competitive inhibition, IFD → Induced fit docking
Figure 1.
(a) Inhibition study of wt LRRK2 catalyzed phosphorylation of LRRKtide by Ponatinib. A: Plot of initial velocities vs [ATP] at [Ponatinib] = 1 (●), 0.3 (엯), 0.1 (▾), 0.04 (▿), 0.1 (∎), and 0 μM (□) all at a fixed LRRKtide concentration of 50 μM. B & C: Ponatinib concentration dependencies of (kcat)ATP and (kcat/Km)ATP apparent values derived from analysis of the data of panel A. (b) Inhibition study of the G2019S mutant catalyzed phosphorylation of LRRKtide by Ponatinib. A: Plot of initial velocities vs [ATP] at [Ponatinib] = 8 (●), 4 (엯), 2 (▾), 1 (▿), 0.5 (∎), and 0 μM (□) all at a fixed LRRKtide concentration of 50 μM. B & C: Ponatinib concentration dependencies of (kcat)ATP and (kcat/Km)ATP apparent values derived from analysis of the data of panel A.
The mechanism of inhibition of the four DYG-out inhibitors was studied and data were analyzed using the methods of replots as described in detail previously (58). Briefly, initial velocities of LRRK2-catalyzed LRRKtide phosphorylation were measured as a function of inhibitor concentration [I] at varied ATP concentrations and at a fixed LRRKtide concentration. The shape of replots of (kcat)ATP vs. [I] and (kcat/Km)ATP vs. [I] were used to determine the inhibition mechanism. As expected based on their mechanism of action with their targeted kinase (see Table 1), ponatinib, sorafenib, and imatinib show ATP non-competitive inhibition toward WT (Table 1). Surprisingly however, ponatinib, sorafenib, and imatinib inhibit the G2019S mutant via an ATP competitive mechanism (Table 1, Figure 1 and Figure S2, Supporting Information). The atypical switching of the mode of action for these three compounds motivated us to pursue kinetic and structural studies to elucidate the origin of this behavior. Bosutinib was the only exception that showed ATP competitive binding toward both WT and G2019S (Table 1 and Figure S2, Supporting Information). This is not surprising since there is precedence for bosutinib being able to bind both DFG-in and DFG-out states of cAbl (59).
Temperature dependent kinetics
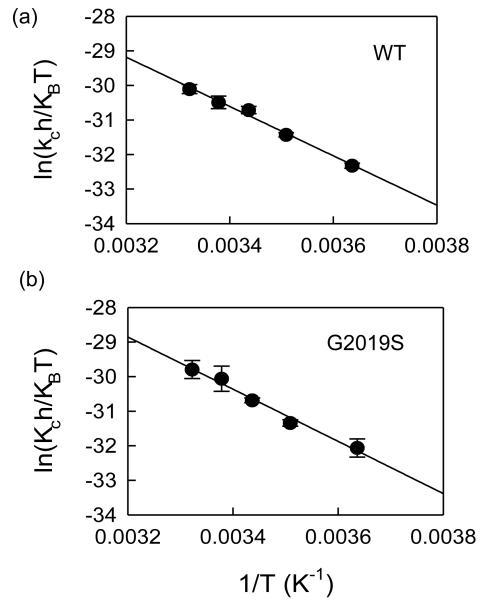
We monitored kcat at various temperatures during LRRK2-catalyzed phosphorylation both by WT and G2019S to determine enthalpy, entropy, and free energy values for these enzyme forms (see Methods). Here we chose two peptides, LRRKtide and LRRKtideS to evaluate activation parameters associated with both chemical transfer step and product release step. Previously, we reported that G2019S has increased activity in catalyzing both peptide substrates compared to WT and the rate-limiting steps of the process governed by kcat are different for these two peptides— product release step is slow for LRRKtide and chemical transfer is slow for LRRKtideS. The temperature dependencies of kcat for LRRKtide phosphorylation catalyzed by both wild type and the mutant are shown in Figure 2 and activation energy parameters for phosphorylation of both LRRKtide and LRRKtideS are summarized in Table 2. These results show that G2019S has a lower energy barrier to activation relative to WT in both reactions, consistent with its increased activity. The relatively small difference in free energy between WT and the mutant G2019S are associated with unexpected significant changes in enthalpy and entropy, suggesting formation of new hydrogen bonds for the mutant G2019S during the steps of chemical transfer and product release.
Figure 2.
Temperature dependencies for WT LRRK2 and mutant-catalyzed LRRKtide and LRRKtideS phosphorylation. The Eyring plots show kcat dependence of temperature for WT LRRK2 (a), the mutant G2019S (b). The kcat was the average of four independent measurements. The data from these plots are summarized in table 1.
Table 2.
Activation energy parameters of kc for LRRK2-catalyzed LRRKtide phosphorylation
| LRRK2 | ΔG‡ , kJ | ΔH‡, kJ | aTΔS‡, kJ |
|---|---|---|---|
| WT | −75.0 ± 0.1 | −63.4 ± 4 | 11.6 ± 4.1 |
| G2019S | −73.8 ± 0.1 | −69.8 ± 6.7 | 4.0 ± 1 |
aT = 298 K
Structural models of WT and the G2019S mutation
The observation that the same inhibitor targets WT LRRK2 and the G2019S mutant by different mechanisms suggests that the mutation in the DYG-motif of the activation loop could have an effect on the ability of this loop to switch between active and inactive conformations. A homology model (Figure S3a, Supporting Information) of the LRRK2 kinase domain between residues 1859-2138 was constructed using B-raf (33% sequence identity) as a template and compared with the x-ray structure of the LRRK2 homolog Roco 4 kinase. Our model based on B-raf and the model based on Roco 4 kinase were found to be virtually identical with less than 1.3Å root mean squared deviation (RMSD) Cα atoms. In order to investigate the robustness of the homology model, we compared modeled structures predicted from four homology modeling programs (Modeller, Prime, SwissModel, and Rosetta) (41, 60-62) and found that the root mean squared deviation (RMSD) of Cα atoms was less than 1.2 Å for all pairwise comparisons. The model of LRRK2 built with Modeller v9.11 (Figure S3, Supporting Information) shows a typical kinase domain composed of an N-terminal N lobe and a C-terminal C lobe forming an ATP and substrate-binding active site at the interlobe cleft (Figure S3b, Supporting Information). The activation loop spans residues 2016-2036 and is modeled in the DYG-in conformation. Spatial comparison of key residues in the LRRK2 model with high-resolution x-ray structures (previously published in ref 35) of other kinases in the catalytic site residues, hinge region and the conserved hydrophobic spines described by Taylor and co-workers indicate that the model has no large errors associated with it.
Using the WT DYG-in model generated by Modeller as a starting point, we introduced the G2019S mutation using PyMOL and energy minimized the structure using Desmond (45). The energy-minimized structure of the G2019S mutant when subjected to molecular dynamics (MD) thermal equilibration was found to be stable in water during the course of 5 ns simulation with a maximum backbone RMSD < 1.3 Å over the simulation. In addition, the mutation was well accommodated at this site. In the initial model, S2019 was found to make hydrogen bonds with R1918 and E1920. Similar observations were made by Wittinghoffer and colleagues based on their model of LRRK2 constructed using the Roco4 (63). In the Roco4 homolog, the G1179S mutation in the activation loop (the side chain of the structurally equivalent serine) was found to make a hydrogen bond with R1077 (63) . However, upon thermal relaxation of the model using short MD simulations (5 ns, see Materials and Methods for details), the relaxed structure retained only the hydrogen bond between S2019 and E1920 after equilibration.
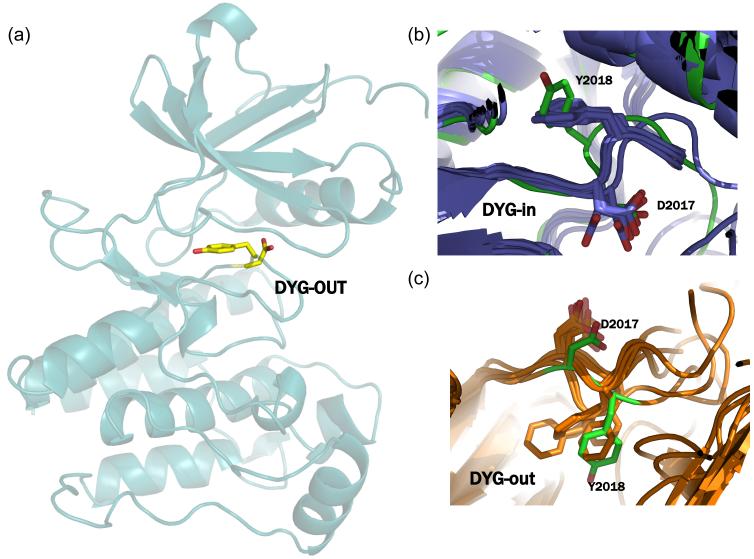
In order to generate the LRRK2 structure in the inactive conformation we carried out loop sampling between residues 2016-2036 using Prime v2.0 (64). We saved 50 structures from the loop sampling and chose the lowest energy structure with the activation loop in the DYG-out conformation (Figure 3a). Structural overlay and comparison of the ϕ/ψ angles of the DYG motif of the modeled LRRK2 (DYG-in and out) with x-ray structures of other known kinases (Table S1, Supporting Information) indicate that the model torsion angle values are within acceptable range for this motif (Figures 3b and 3c).
Figure 3.
(a) Homology of model of LRRK2 with the activation loop in the DYG-out conformation generated using Prime. (b) superposition of the DYG-in model of LRRK2 with x-ray structures of cABL, cKIT, Aurora, B-raf, EPHA3, SRC, LCK and MK14 in their DFG-in conformation. The overall RMSD between any two pairs of structures is < 1.9Å and the DYG-in of LRRK2 (shown in green) model is conformationally very similar to these x-ray structures. (c) superposition of the DYG-out model of LRRK2 (shown in green) generated using Prime with x-ray structures of cABL, cKIT, Aurora, B-raf, EPHA3, SRC, LCK and MK14 in their DFG-out conformation. The overall RMSD between any two pairs of structures is < 2.0Å and the DYG-out of LRRK2 model is conformationally very similar to these x-ray structures.
Free energy surface for activation loop transition
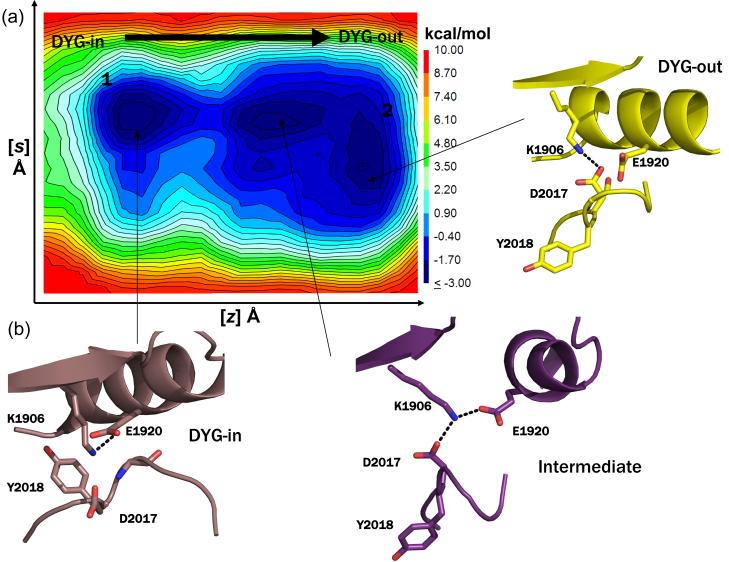
In order to determine the energy barrier between the active and inactive states of the activation loop, we used an enhanced sampling molecular dynamics method called metadynamics with two collective variables (CV) to describe the transition of the loop between DYG-in and DYG-out states (see Materials and Methods for details on the collective variables). Multiple metadynamics simulations starting with slightly different activation loop conformations were carried out for a total of 250 ns for both WT LRRK2 and the mutant G2019S under identical simulation conditions. Free energy surfaces (FES) for these simulations are shown in Figure 4. For WT, the FES shows two low energy wells (blue) corresponding to the active and inactive conformations with a small barrier between the DYG-in and DYG-out states (Figure 4). However, the FES for G2019S shows a much larger energetic barrier between the active and inactive forms, with an energetic preference for the active form (Figure 5). The increased stabilization of the active state in G2019S relative to WT appears to come from an enhanced hydrogen bonding network observed with the serine and the reduced conformational flexibility of the activation loop. As a robustness check and to determine the sensitivity to the choice of CVs, we carried out a second set of metadynamics simulations using the ϕ/ψ Ramachandran angles of G2019 (WT) and S2019 (G2019S) as CVs and find similar results to those observed with the distance CVs described above. The FES for the torsional CVs is shown in Figure S4 (Supporting Information).
Figure 4.
(a) Representative free energy surface (FES) generated from 250 ns metadynamics simulation of WT LRRK2 generated by following the opening and closing of the enzyme involving the ATP binding β-sheet domain and C-helix as the first collective variable (z) and the motion of the center of mass of the activation loop as the second collective variable (s) as the kinase switches between active and inactive form. The FES shows that opening and closing motion of the enzyme occurs unhindered and the activation loop can switch between the active and inactive form easily (a contagious low energy path connects the two conformations forms). (b) Snapshots of structures extracted from local clustering of conformations within the simulation corresponding to the three local minima observed during the course of the simulation are shown here. The states 1 and 2 correspond to the active (DYG-in) and inactive (DYG-out) states of the enzyme. An intermediate metastable transition is observed, where K1906 makes a shared hydrogen bond with both E1920 (C-helix) and D2017 (DYG-motif).
Figure 5.
(a) Representative free energy surface (FES) generated from a 250 ns metadynamic simulation run of G2019S mutant generated by following the opening and closing of the enzyme involving the ATP binding β-sheet domain and C-helix as the first collective variable (z) and the motion of the center of mass of the activation loop as the second collective variable (s) as the kinase switches between active and inactive form. The simulations were carried out under identical conditions as that of the WT LRRK2 described above. The FES plot shows that there is a high energy barrier that separates the active and inactive form of the kinases indicating that higher amount of energy is required for the mutant to switch between active and inactive conformations relative to WT. However, independently both the active and inactive form of the enzyme can switch between open and closed forms. (b) Snapshots of structures extracted from local clustering of conformations within the simulation corresponding to the three local minima are shown here. In state 1, the G2019S mutation results in a side-chain of E1920 and S2019. This offers additional stability to this state. In state 2, the side-chain of S2019 makes a hydrogen bond with the backbone of D2017 and appears to be a metastable state. A transient stable intermediate state 3 is observed in the simulation where the side chain of S2019 makes a hydrogen bond with backbone of Y2018.
Induced fit docking of type II inhibitors to DYG-in and DYG-out conformations of LRRK2
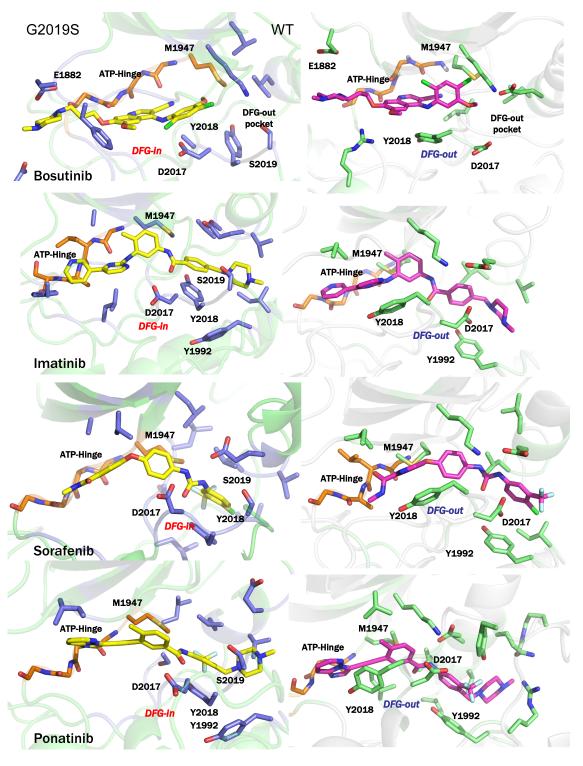
Finally, to understand the structural details of a type II inhibitor binding to an active form of mutant LRRK2, we carried out Induced Fit Docking (IFD) calculations on the inhibitors using both DYG-in and DYG-out structures of LRRK2 (details in Materials and Methods and Supporting Information). We docked the four type II inhibitors in this study (ponatinib, sorafenib, imatinib, and bosutinib) to the DYG-out structure of WT LRRK2 and the DYG-in structure of the G2019S mutant. As seen in Figure 6 (right panel), the ligands docked to the WT DYG-out conformation all adopt a traditional binding mode for type II kinase inhibitors. However, for the G2019S mutant, ponatinib, sorafenib, and imatinib all make a hinge interaction (Figure 6, left panel) . The IFD scores are reported in Table 1 and the key interactions made by the inhibitors are shown along with a ligand-interaction diagrams (Figure S5, Supporting Information).
Figure 6.
(left panels) Structural representations of induced fit docked conformations for the known DFG-out inhibitors bosutinib, imatinib, sorafenib and ponatinib on the DFG-in conformation of G2019S mutant of LRRK2. (right panels) Structural representations of induced fit docked conformations for the known DFG-out inhibitors bosutinib, imatinib, sorafenib and ponatinib on the DFG-out conformation of WT LRRK2. In this figure, only few residues are labeled to preserve clarity. Figure S5 in Supporting Information accompanies this figure showing all protein inhibitor interactions.
Discussion
Mutations in the leucine-rich repeat kinase 2 (LRRK2) gene are the most common known genetic cause of PD, with the most prevalent LRRK2 mutation (G2019S) estimated to be associated with 5%–6% of familial PD and 1%–2% of idiopathic cases in populations of European descent (11, 65-85). Multiple reports have indicated that the mutation leads to a hyperactive kinase, which might be directly linked to PD pathology; therefore, inhibition of mutant LRRK2 represents an attractive therapeutic strategy in the PD field.
A large number of ATP competitive type I inhibitors of LRRK2 have been reported from screening studies and recently reviewed (21, 86, 87). In addition, homology models of LRRK2 built by other research groups have been used successfully to drive SAR studies for ATP-competitive inhibitors (88). However, there is little discussion about modeling of ATP non-competitive type II inhibitors of LRRK2 that bind to the DYG-out state. In this study, we used a number of well-known type II (DFG-out) inhibitors of other kinases and test them against LRRK2 (both WT and the G2019S mutant). Experimental findings of the inhibition mechanism revealed an unexpected observation that type II inhibitors work by an ATP competitive mechanism against G2019S whereas they behave as expected (noncompetitive inhibitors) against WT. In addition, temperature-dependent kinetic studies conducted under the condition of kcat revealed that the G2019S mutant is more entropically favorable than WT (smaller negative ΔS for the mutant than WT) suggesting a greater population of the enzyme maybe existing in its active form for the mutant compared to the WT.
In order to explain the inhibition mechanism and observed kinetics, we built homology models of LRRK2 in the DYG-in (active) and DYG-out (inactive) forms for both WT and G2019S. The models revealed the ability of the S2019 side chain in the mutant to make hydrogen bonding interactions with E1920 that could potentially explaining the increased stability for the DYG-in state and the observed kinetic behavior. Similar observations have been made by Wittinghofer and co-workers with the recent x-ray structure of LRRK2 homolog roco4 (63).
To gain further insights into the consequence of this mutation on the energetics of the activation loop, metadynamics simulations were run to determine the free energy surface (FES) associated with the activation loop transition in WT and G2019S. The simulations revealed significant differences in the FES of the activation loop between WT and G2019S. The FES for WT shows two nearly equienergetic basins for the DYG-in and DYG-out states (numbered 1, 2 in Figure 4) along with a relatively low-energy transition path between the two states. Similar analysis of the FES for G2019S shows that the DYG-out form is less prominent (shallower well) when compared to the DYG-in state (Figure 5) and there is a high-energy barrier that separates the two states. Analysis of the structure in the DYG-in state of G2019S suggests that the side chain of S2019 participates in a stable hydrogen bond with E1920 (Figure 5), which is not present in WT and contributes to the observed increased barrier for transition (Figure 4 ).
In addition, G2019 in WT has more backbone conformational flexibility as compared with S2019 in the mutant. The loss in side chain and backbone entropy of the mutant is accompanied by unfavorable changes in the enthalpy, suggesting newly formed specific interactions upon the mutation. In order to measure the change in conformational torsional freedom upon mutation (entropic changes), we carried out metadynamics simulations using the torsion angles (ϕ/ψ) of G2019 (WT) and S2019 (G2019S) as collective variables (Figure S4, Supporting Information). Comparison of the FES of WT and G2019S mutant indicates that the mutant shows a large reduction in torsional conformational space accessible to the loop indicating that this mutation would lower the entropy of the loop and is in agreement with the temperature-dependent studies (Figure 2 and Table 2).
The combination of enzyme mechanism studies, temperature dependent kinetics, and metadynamics simulations suggests G2019S may not easily switch to a DYG-out form and yet is able to bind traditional type II DFG-out inhibitors. To predict the binding mode for these compounds, we carried out Induced Fit Docking calculations with four type II inhibitors (Table 1) against WT (DYG-out state) and G2019S (DYG-in state). The docking calculations reveal that the common feature between the two forms is the similar orientation of the ATP-binding hinge region and major differences lie in the allosteric pocket. It is therefore reasonable to expect that the number of interactions made by the inhibitor with the hinge as opposed to the allosteric pocket would have a major impact on the inhibition mechanism. The type II inhibitor sorafenib, which makes no H-bond interactions with the hinge region (Figure 6 and Supporting Information Figure S5) was effective against WT but was poorly effective against the G2019S mutant (Table 1). In addition, in the DYG-out state, sorafenib makes an aromatic stack with Y2018 and an additional hydrogen bond with the backbone L1885, which are both absent in the DYG-in form of the mutant. This contributes to the potency of the inhibitor towards WT enzyme but not the mutant. The type II inhibitor bosutinib, on the other hand, makes three hydrogen bond in the hinge region and very few interactions in the DYG-out pocket (Figure S5, Supporting Information). This also explains why bosutinib is equally potent against WT and the G2019S mutant and inhibits both enzyme forms by an ATP competitive mechanism (Table 1 and Figure S5, Supporting Information). Ponatinib is capable of making one hydrogen bond in the hinge region and a number of hydrophobic interactions in the DFG-out pocket of WT. Structural comparison of docked structures of ponatinib with WT and G2019S reveal that in both cases ponatinib in the allosteric pocket makes strong aromatic stacking interactions with tyrosine residues. Y1992 is involved in this aromatic stacking in WT but in case of the mutant, Y2018 occupies the same virtual position as Y1992 making nearly identical set of interactions. This makes ponatinib not only exceptionally potent against WT but also retain fair amount potency against the mutants due to hinge interactions (Table 1 and Figure S5, Supporting Information). A control type I kinase inhibitor GSK-3-XIII showed nearly identical potency against WT and the G2019S mutant, consistent with expectations that it binds to the DYG-in conformation of LRRK2. Similar observation were made by Gray and others that have described ATP-competitive inhibitors of LRRK2 that target the hinge region of the kinase (21).
This model emerging from the combination of experimental and computational studies suggests that the G2019S mutant stabilizes the DYG-in conformation, which has an allosteric pocket that cannot fully accommodate inhibitors that are DFG-out pocket dominant binders (sorafenib for example). As such, type II inhibitors that cannot make sufficient hinge interactions with the kinase lose considerable potency when binding to G2019S. Interestingly, type II kinase inhibitors with significant hinge-binding characteristics, such as bosutinib, are able to bind G2019S in an ATP competitive fashion and show nearly identical potency against WT and G2019S. This dual characteristic of the unique allosteric pocket of G2019S opens up the possibility of a new design strategy to generate G2019S specific inhibitors, since traditional type I kinase inhibitors tend to lack selectivity and type II inhibitors were found in this work to either not bind to G2019S or to adopt an ATP-competitive binding mode.
While the x-ray structure for LRKK2 is yet to be determined (a closely related roco kinase domain is now available), enzyme mechanism studies and molecular modeling have provided significant insights into the nature of LRRK2 and the PD-linked mutation G2019S. With further studies, possibly including x-ray crystallography or long timescale molecular dynamics simulations, it may eventually be possible to understand the nature of the allosteric pocket in the G2019S mutant enzyme and develop highly selective inhibitors. However, given the unique insights gained in this work with traditional type II kinase inhibitors binding to the G2019S mutant in an ATP-competitive way, it is conceivable that a new class of inhibitors (e.g. type III) will be needed to gain the desired affinity and selectivity to target G2019S in the pursuit to treat Parkinson’s disease. The presence of two exposed cysteine residues (C2024 and C2025) close to the ATP binding site (see supporting information) opens up one such possibility of developing covalent inhibitors for this enzyme. Recent ab initio binding simulation of kinase inhibitor complexes by D.E Shaw and co-workers have revealed previously ligand unprecedented binding sites within the C-terminal of kinases (45). It is conceivable that similar sites can be discovered on LRRK2 as well that can form hot-spots for type III inhibitor design.
Supplementary Material
Acknowledgements
We wish to thank Dmitry Lupyan (Schrodinger LLC.) for help with setting up metadynamics simulations. We wish to thank Karl Ruping (Proteus Discovery) and Patrick Kleyn (Proteus Discovery) for useful discussions at various stages of manuscript preparation. We also wish to acknowledge help from Ms. Kristine Vernon in preparation of the manuscript.
Footnotes
Supporting information paragraph: Additional details of experimental methods, figures and tables are provided in the supporting information section. The supporting information associated with this manuscript is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Bosco DA, LaVoie MJ, Petsko GA, Ringe D. Proteostasis and movement disorders: Parkinson’s disease and amyotrophic lateral sclerosis. Cold Spring Harb Perspect Biol. 2011;3:a007500. doi: 10.1101/cshperspect.a007500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martin I, Dawson VL, Dawson TM. Recent Advances in the Genetics of Parkinson’s Disease. Annu Rev Genomics Hum Genet. 2010 doi: 10.1146/annurev-genom-082410-101440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hardy J, Cookson MR, Singleton A. Genes and parkinsonism. Lancet Neurol. 2003;2:221–228. doi: 10.1016/s1474-4422(03)00350-8. [DOI] [PubMed] [Google Scholar]
- 4.Cookson MR. Pathways to Parkinsonism. Neuron. 2003;37:7–10. doi: 10.1016/s0896-6273(02)01166-2. [DOI] [PubMed] [Google Scholar]
- 5.Benamer HT. The ancestry of LRRK2 Gly2019Ser parkinsonism. Lancet Neurol. 2008;7:769–770. doi: 10.1016/S1474-4422(08)70178-9. author reply 770-761. [DOI] [PubMed] [Google Scholar]
- 6.Cookson MR. The role of leucine-rich repeat kinase 2 (LRRK2) in Parkinson’s disease. Nat Rev Neurosci. 2010;11:791–797. doi: 10.1038/nrn2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cookson MR, Dauer W, Dawson T, Fon EA, Guo M, Shen J. The roles of kinases in familial Parkinson’s disease. J Neurosci. 2007;27:11865–11868. doi: 10.1523/JNEUROSCI.3695-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark LN, Wang Y, Karlins E, Saito L, Mejia-Santana H, Harris J, Louis ED, Cote LJ, Andrews H, Fahn S, Waters C, Ford B, Frucht S, Ottman R, Marder K. Frequency of LRRK2 mutations in early- and late-onset Parkinson disease. Neurology. 2006;67:1786–1791. doi: 10.1212/01.wnl.0000244345.49809.36. [DOI] [PubMed] [Google Scholar]
- 9.Cookson MR, Bandmann O. Parkinson’s disease: insights from pathways. Hum Mol Genet. 2010;19:R21–27. doi: 10.1093/hmg/ddq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cookson MR, Xiromerisiou G, Singleton A. How genetics research in Parkinson’s disease is enhancing understanding of the common idiopathic forms of the disease. Curr Opin Neurol. 2005;18:706–711. doi: 10.1097/01.wco.0000186841.43505.e6. [DOI] [PubMed] [Google Scholar]
- 11.Correia Guedes L, Ferreira JJ, Rosa MM, Coelho M, Bonifati V, Sampaio C. Worldwide frequency of G2019S LRRK2 mutation in Parkinson’s disease: a systematic review. Parkinsonism Relat Disord. 2010;16:237–242. doi: 10.1016/j.parkreldis.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 12.Dachsel JC, Farrer MJ. LRRK2 and Parkinson disease. Arch Neurol. 2010;67:542–547. doi: 10.1001/archneurol.2010.79. [DOI] [PubMed] [Google Scholar]
- 13.Mata IF, Wedemeyer WJ, Farrer MJ, Taylor JP, Gallo KA. LRRK2 in Parkinson’s disease: protein domains and functional insights. Trends Neurosci. 2006;29:286–293. doi: 10.1016/j.tins.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 14.Daniels V, Vancraenenbroeck R, Law BM, Greggio E, Lobbestael E, Gao F, De Maeyer M, Cookson MR, Harvey K, Baekelandt V, Taymans JM. Insight into the mode of action of the LRRK2 Y1699C pathogenic mutant. J Neurochem. 2011;116:304–315. doi: 10.1111/j.1471-4159.2010.07105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greggio E, Taymans JM, Zhen EY, Ryder J, Vancraenenbroeck R, Beilina A, Sun P, Deng J, Jaffe H, Baekelandt V, Merchant K, Cookson MR. The Parkinson’s disease kinase LRRK2 autophosphorylates its GTPase domain at multiple sites. Biochem Biophys Res Commun. 2009;389:449–454. doi: 10.1016/j.bbrc.2009.08.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis PA, Greggio E, Beilina A, Jain S, Baker A, Cookson MR. The R1441C mutation of LRRK2 disrupts GTP hydrolysis. Biochem Biophys Res Commun. 2007;357:668–671. doi: 10.1016/j.bbrc.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luzon-Toro B, Rubio de la Torre E, Delgado A, Perez-Tur J, Hilfiker S. Mechanistic insight into the dominant mode of the Parkinson’s disease-associated G2019S LRRK2 mutation. Hum Mol Genet. 2007;16:2031–2039. doi: 10.1093/hmg/ddm151. [DOI] [PubMed] [Google Scholar]
- 18.West AB, Moore DJ, Biskup S, Bugayenko A, Smith WW, Ross CA, Dawson VL, Dawson TM. Parkinson’s disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc Natl Acad Sci U S A. 2005;102:16842–16847. doi: 10.1073/pnas.0507360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith WW, Pei Z, Jiang H, Dawson VL, Dawson TM, Ross CA. Kinase activity of mutant LRRK2 mediates neuronal toxicity. Nat Neurosci. 2006;9:1231–1233. doi: 10.1038/nn1776. [DOI] [PubMed] [Google Scholar]
- 20.Greggio E, Jain S, Kingsbury A, Bandopadhyay R, Lewis P, Kaganovich A, van der Brug MP, Beilina A, Blackinton J, Thomas KJ, Ahmad R, Miller DW, Kesavapany S, Singleton A, Lees A, Harvey RJ, Harvey K, Cookson MR. Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol Dis. 2006;23:329–341. doi: 10.1016/j.nbd.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 21.Deng X, Dzamko N, Prescott A, Davies P, Liu Q, Yang Q, Lee JD, Patricelli MP, Nomanbhoy TK, Alessi DR, Gray NS. Characterization of a selective inhibitor of the Parkinson’s disease kinase LRRK2. Nat Chem Biol. 2011;7:203–205. doi: 10.1038/nchembio.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maekawa T, Mori S, Sasaki Y, Miyajima T, Azuma S, Ohta E, Obata F. The I2020T Leucine-rich repeat kinase 2 transgenic mouse exhibits impaired locomotive ability accompanied by dopaminergic neuron abnormalities. Mol Neurodegener. 2012;7:15. doi: 10.1186/1750-1326-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ujiie S, Hatano T, Kubo SI, Imai S, Sato S, Uchihara T, Yagishita S, Hasegawa K, Kowa H, Sakai F, Hattori N. LRRK2 I2020T mutation is associated with tau pathology. Parkinsonism Relat Disord. 2012 doi: 10.1016/j.parkreldis.2012.03.024. [DOI] [PubMed] [Google Scholar]
- 24.Deng H, Le W, Davidson AL, Xie W, Jankovic J. The LRRK2 I2012T, G2019S and I2020T mutations are not common in patients with essential tremor. Neurosci Lett. 2006;407:97–100. doi: 10.1016/j.neulet.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 25.Lu CS, Simons EJ, Wu-Chou YH, Fonzo AD, Chang HC, Chen RS, Weng YH, Rohe CF, Breedveld GJ, Hattori N, Gasser T, Oostra BA, Bonifati V. The LRRK2 I2012T, G2019S, and I2020T mutations are rare in Taiwanese patients with sporadic Parkinson’s disease. Parkinsonism Relat Disord. 2005;11:521–522. doi: 10.1016/j.parkreldis.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 26.Taylor SS, Kornev AP. Protein kinases: evolution of dynamic regulatory proteins. Trends Biochem Sci. 2011;36:65–77. doi: 10.1016/j.tibs.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor SS, Kim C, Vigil D, Haste NM, Yang J, Wu J, Anand GS. Dynamics of signaling by PKA. Biochim Biophys Acta. 2005;1754:25–37. doi: 10.1016/j.bbapap.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 28.Johnson DA, Akamine P, Radzio-Andzelm E, Madhusudan M, Taylor SS. Dynamics of cAMP-dependent protein kinase. Chem Rev. 2001;101:2243–2270. doi: 10.1021/cr000226k. [DOI] [PubMed] [Google Scholar]
- 29.Taylor SS, Radzio-Andzelm E, Madhusudan, Cheng X, Ten Eyck L, Narayana N. Catalytic subunit of cyclic AMP-dependent protein kinase: structure and dynamics of the active site cleft. Pharmacol Ther. 1999;82:133–141. doi: 10.1016/s0163-7258(99)00007-8. [DOI] [PubMed] [Google Scholar]
- 30.Taylor SS, Radzio-Andzelm E. Protein kinase inhibition: natural and synthetic variations on a theme. Curr Opin Chem Biol. 1997;1:219–226. doi: 10.1016/s1367-5931(97)80013-0. [DOI] [PubMed] [Google Scholar]
- 31.Cox S, Radzio-Andzelm E, Taylor SS. Domain movements in protein kinases. Curr Opin Struct Biol. 1994;4:893–901. doi: 10.1016/0959-440x(94)90272-0. [DOI] [PubMed] [Google Scholar]
- 32.Besker N, Gervasio FL. Using metadynamics and path collective variables to study ligand binding and induced conformational transitions. Methods Mol Biol. 2012;819:501–513. doi: 10.1007/978-1-61779-465-0_29. [DOI] [PubMed] [Google Scholar]
- 33.Fiorin G, Pastore A, Carloni P, Parrinello M. Using metadynamics to understand the mechanism of calmodulin/target recognition at atomic detail. Biophys J. 2006;91:2768–2777. doi: 10.1529/biophysj.106.086611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masetti M, Cavalli A, Recanatini M, Gervasio FL. Exploring complex protein-ligand recognition mechanisms with coarse metadynamics. J Phys Chem B. 2009;113:4807–4816. doi: 10.1021/jp803936q. [DOI] [PubMed] [Google Scholar]
- 35.Liu M, Kang S, Ray S, Jackson J, Zaitsev AD, Gerber SA, Cuny GD, Glicksman MA. Kinetic, mechanistic, and structural modeling studies of truncated wild-type leucine-rich repeat kinase 2 and the G2019S mutant. Biochemistry. 2011;50:9399–9408. doi: 10.1021/bi201173d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fiser A, Sali A. ModLoop: automated modeling of loops in protein structures. Bioinformatics. 2003;19:2500–2501. doi: 10.1093/bioinformatics/btg362. [DOI] [PubMed] [Google Scholar]
- 37.John B, Sali A. Comparative protein structure modeling by iterative alignment, model building and model assessment. Nucleic Acids Res. 2003;31:3982–3992. doi: 10.1093/nar/gkg460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eswar N, John B, Mirkovic N, Fiser A, Ilyin VA, Pieper U, Stuart AC, Marti-Renom MA, Madhusudhan MS, Yerkovich B, Sali A. Tools for comparative protein structure modeling and analysis. Nucleic Acids Res. 2003;31:3375–3380. doi: 10.1093/nar/gkg543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanchez R, Sali A. Evaluation of comparative protein structure modeling by MODELLER-3. Proteins Suppl. 1997;1:50–58. doi: 10.1002/(sici)1097-0134(1997)1+<50::aid-prot8>3.3.co;2-w. [DOI] [PubMed] [Google Scholar]
- 40.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 41.Sherman W, Day T, Jacobson MP, Friesner RA, Farid R. Novel procedure for modeling ligand/receptor induced fit effects. J Med Chem. 2006;49:534–553. doi: 10.1021/jm050540c. [DOI] [PubMed] [Google Scholar]
- 42.Jacobson MP, Pincus DL, Rapp CS, Day TJ, Honig B, Shaw DE, Friesner RA. A hierarchical approach to all-atom protein loop prediction. Proteins. 2004;55:351–367. doi: 10.1002/prot.10613. [DOI] [PubMed] [Google Scholar]
- 43.Emekli U, Schneidman-Duhovny D, Wolfson HJ, Nussinov R, Haliloglu T. HingeProt: automated prediction of hinges in protein structures. Proteins. 2008;70:1219–1227. doi: 10.1002/prot.21613. [DOI] [PubMed] [Google Scholar]
- 44.Lill MA, Danielson ML. Computer-aided drug design platform using PyMOL. J Comput Aided Mol Des. 2011;25:13–19. doi: 10.1007/s10822-010-9395-8. [DOI] [PubMed] [Google Scholar]
- 45.Shaw DE, Maragakis P, Lindorff-Larsen K, Piana S, Dror RO, Eastwood MP, Bank JA, Jumper JM, Salmon JK, Shan Y, Wriggers W. Atomic-level characterization of the structural dynamics of proteins. Science. 2010;330:341–346. doi: 10.1126/science.1187409. [DOI] [PubMed] [Google Scholar]
- 46.Guvench O, MacKerell AD., Jr. Comparison of protein force fields for molecular dynamics simulations. Methods Mol Biol. 2008;443:63–88. doi: 10.1007/978-1-59745-177-2_4. [DOI] [PubMed] [Google Scholar]
- 47.Barducci A, Bonomi M, Parrinello M. Linking well-tempered metadynamics simulations with experiments. Biophys J. 2010;98:L44–46. doi: 10.1016/j.bpj.2010.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barducci A, Bussi G, Parrinello M. Well-tempered metadynamics: a smoothly converging and tunable free-energy method. Phys Rev Lett. 2008;100:020603. doi: 10.1103/PhysRevLett.100.020603. [DOI] [PubMed] [Google Scholar]
- 49.Barducci A, Chelli R, Procacci P, Schettino V, Gervasio FL, Parrinello M. Metadynamics simulation of prion protein: beta-structure stability and the early stages of misfolding. J Am Chem Soc. 2006;128:2705–2710. doi: 10.1021/ja057076l. [DOI] [PubMed] [Google Scholar]
- 50.Berteotti A, Barducci A, Parrinello M. Effect of urea on the beta-hairpin conformational ensemble and protein denaturation mechanism. J Am Chem Soc. 2011;133:17200–17206. doi: 10.1021/ja202849a. [DOI] [PubMed] [Google Scholar]
- 51.Bonomi M, Barducci A, Gervasio FL, Parrinello M. Multiple routes and milestones in the folding of HIV-1 protease monomer. PLoS One. 2010;5:e13208. doi: 10.1371/journal.pone.0013208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bonomi M, Parrinello M. Enhanced sampling in the well-tempered ensemble. Phys Rev Lett. 2010;104:190601. doi: 10.1103/PhysRevLett.104.190601. [DOI] [PubMed] [Google Scholar]
- 53.Branduardi D, Gervasio FL, Parrinello M. From A to B in free energy space. J Chem Phys. 2007;126:054103. doi: 10.1063/1.2432340. [DOI] [PubMed] [Google Scholar]
- 54.Bussi G, Laio A, Parrinello M. Equilibrium free energies from nonequilibrium metadynamics. Phys Rev Lett. 2006;96:090601. doi: 10.1103/PhysRevLett.96.090601. [DOI] [PubMed] [Google Scholar]
- 55.Leone V, Marinelli F, Carloni P, Parrinello M. Targeting biomolecular flexibility with metadynamics. Curr Opin Struct Biol. 2010;20:148–154. doi: 10.1016/j.sbi.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 56.Limongelli V, Bonomi M, Marinelli L, Gervasio FL, Cavalli A, Novellino E, Parrinello M. Molecular basis of cyclooxygenase enzymes (COXs) selective inhibition. Proc Natl Acad Sci U S A. 2010;107:5411–5416. doi: 10.1073/pnas.0913377107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Limongelli V, Marinelli L, Cosconati S, La Motta C, Sartini S, Mugnaini L, Da Settimo F, Novellino E, Parrinello M. Sampling protein motion and solvent effect during ligand binding. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1112181108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu M, Choi S, Cuny GD, Ding K, Dobson BC, Glicksman MA, Auerbach K, Stein RL. Kinetic studies of Cdk5/p25 kinase: phosphorylation of tau and complex inhibition by two prototype inhibitors. Biochemistry. 2008;47:8367–8377. doi: 10.1021/bi800732v. [DOI] [PubMed] [Google Scholar]
- 59.Levinson NM, Boxer SG. Structural and spectroscopic analysis of the kinase inhibitor bosutinib and an isomer of bosutinib binding to the Abl tyrosine kinase domain. PLoS One. 2012;7:e29828. doi: 10.1371/journal.pone.0029828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman K, Renfrew PD, Smith CA, Sheffler W, Davis IW, Cooper S, Treuille A, Mandell DJ, Richter F, Ban YE, Fleishman SJ, Corn JE, Kim DE, Lyskov S, Berrondo M, Mentzer S, Popovic Z, Havranek JJ, Karanicolas J, Das R, Meiler J, Kortemme T, Gray JJ, Kuhlman B, Baker D, Bradley P. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009;37:D387–392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eswar N, Eramian D, Webb B, Shen MY, Sali A. Protein structure modeling with MODELLER. Methods Mol Biol. 2008;426:145–159. doi: 10.1007/978-1-60327-058-8_8. [DOI] [PubMed] [Google Scholar]
- 63.Gilsbach BK, Ho FY, Vetter IR, van Haastert PJ, Wittinghofer A, Kortholt A. Roco kinase structures give insights into the mechanism of Parkinson disease-related leucine-rich-repeat kinase 2 mutations. Proc Natl Acad Sci U S A. 2012;109:10322–10327. doi: 10.1073/pnas.1203223109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhu K, Pincus DL, Zhao S, Friesner RA. Long loop prediction using the protein local optimization program. Proteins. 2006;65:438–452. doi: 10.1002/prot.21040. [DOI] [PubMed] [Google Scholar]
- 65.Aasly JO, Toft M, Fernandez-Mata I, Kachergus J, Hulihan M, White LR, Farrer M. Clinical features of LRRK2-associated Parkinson’s disease in central Norway. Ann Neurol. 2005;57:762–765. doi: 10.1002/ana.20456. [DOI] [PubMed] [Google Scholar]
- 66.Abdalla-Carvalho CB, Santos-Reboucas CB, Guimaraes BC, Campos M, Pereira JS, de Rosso AL, Nicaretta DH, Marinho e Silva M, dos Santos MJ, Pimentel MM. Genetic analysis of LRRK2 functional domains in Brazilian patients with Parkinson’s disease. Eur J Neurol. 2010;17:1479–1481. doi: 10.1111/j.1468-1331.2010.03039.x. [DOI] [PubMed] [Google Scholar]
- 67.Alcalay RN, Mejia-Santana H, Tang MX, Rosado L, Verbitsky M, Kisselev S, Ross BM, Louis ED, Comella CL, Colcher A, Jennings D, Nance MA, Bressman S, Scott WK, Tanner C, Mickel SF, Andrews HF, Waters CH, Fahn S, Cote LJ, Frucht SJ, Ford B, Rezak M, Novak K, Friedman JH, Pfeiffer R, Marsh L, Hiner B, Siderowf A, Caccappolo E, Ottman R, Clark LN, Marder KS. Motor phenotype of LRRK2 G2019S carriers in early-onset Parkinson disease. Arch Neurol. 2009;66:1517–1522. doi: 10.1001/archneurol.2009.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bardien S, Marsberg A, Keyser R, Lombard D, Lesage S, Brice A, Carr J. LRRK2 G2019S mutation: frequency and haplotype data in South African Parkinson’s disease patients. J Neural Transm. 2010;117:847–853. doi: 10.1007/s00702-010-0423-6. [DOI] [PubMed] [Google Scholar]
- 69.Bar-Shira A, Hutter CM, Giladi N, Zabetian CP, Orr-Urtreger A. Ashkenazi Parkinson’s disease patients with the LRRK2 G2019S mutation share a common founder dating from the second to fifth centuries. Neurogenetics. 2009;10:355–358. doi: 10.1007/s10048-009-0186-0. [DOI] [PubMed] [Google Scholar]
- 70.Belarbi S, Hecham N, Lesage S, Kediha MI, Smail N, Benhassine T, Ysmail-Dahlouk F, Lohman E, Benhabyles B, Hamadouche T, Assami S, Brice A, Tazir M. LRRK2 G2019S mutation in Parkinson’s disease: a neuropsychological and neuropsychiatric study in a large Algerian cohort. Parkinsonism Relat Disord. 2010;16:676–679. doi: 10.1016/j.parkreldis.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 71.Benamer HT, de Silva R. LRRK2 G2019S in the North African population: a review. Eur Neurol. 2010;63:321–325. doi: 10.1159/000279653. [DOI] [PubMed] [Google Scholar]
- 72.Bonifati V. Parkinson’s disease: the LRRK2-G2019S mutation: opening a novel era in Parkinson’s disease genetics. Eur J Hum Genet. 2006;14:1061–1062. doi: 10.1038/sj.ejhg.5201695. [DOI] [PubMed] [Google Scholar]
- 73.Bras J, Guerreiro R, Ribeiro M, Morgadinho A, Januario C, Dias M, Calado A, Semedo C, Oliveira C, Hardy J, Singleton A. Analysis of Parkinson disease patients from Portugal for mutations in SNCA, PRKN, PINK1 and LRRK2. BMC Neurol. 2008;8:1. doi: 10.1186/1471-2377-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Carmine Belin A, Westerlund M, Sydow O, Lundstromer K, Hakansson A, Nissbrandt H, Olson L, Galter D. Leucine-rich repeat kinase 2 (LRRK2) mutations in a Swedish Parkinson cohort and a healthy nonagenarian. Mov Disord. 2006;21:1731–1734. doi: 10.1002/mds.21016. [DOI] [PubMed] [Google Scholar]
- 75.Change N, Mercier G, Lucotte G. Genetic screening of the G2019S mutation of the LRRK2 gene in Southwest European, North African, and Sephardic Jewish subjects. Genet Test. 2008;12:333–339. doi: 10.1089/gte.2007.0098. [DOI] [PubMed] [Google Scholar]
- 76.Cho JW, Kim SY, Park SS, Jeon BS. The G2019S LRRK2 Mutation is Rare in Korean Patients with Parkinson’s Disease and Multiple System Atrophy. J Clin Neurol. 2009;5:29–32. doi: 10.3988/jcn.2009.5.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Criscuolo C, De Rosa A, Guacci A, Simons EJ, Breedveld GJ, Peluso S, Volpe G, Filla A, Oostra BA, Bonifati V, De Michele G. The LRRK2 R1441C mutation is more frequent than G2019S in Parkinson’s disease patients from Southern Italy. Mov Disord. 2011;26:1732–1736. doi: 10.1002/mds.23735. [DOI] [PubMed] [Google Scholar]
- 78.Deng H, Le W, Guo Y, Hunter CB, Xie W, Jankovic J. Genetic and clinical identification of Parkinson’s disease patients with LRRK2 G2019S mutation. Ann Neurol. 2005;57:933–934. doi: 10.1002/ana.20510. [DOI] [PubMed] [Google Scholar]
- 79.Di Fonzo A, Tassorelli C, De Mari M, Chien HF, Ferreira J, Rohe CF, Riboldazzi G, Antonini A, Albani G, Mauro A, Marconi R, Abbruzzese G, Lopiano L, Fincati E, Guidi M, Marini P, Stocchi F, Onofrj M, Toni V, Tinazzi M, Fabbrini G, Lamberti P, Vanacore N, Meco G, Leitner P, Uitti RJ, Wszolek ZK, Gasser T, Simons EJ, Breedveld GJ, Goldwurm S, Pezzoli G, Sampaio C, Barbosa E, Martignoni E, Oostra BA, Bonifati V. Comprehensive analysis of the LRRK2 gene in sixty families with Parkinson’s disease. Eur J Hum Genet. 2006;14:322–331. doi: 10.1038/sj.ejhg.5201539. [DOI] [PubMed] [Google Scholar]
- 80.Funalot B, Nichols WC, Perez-Tur J, Mercier G, Lucotte G. Genetic screening for two LRRK2 mutations in French patients with idiopathic Parkinson’s disease. Genet Test. 2006;10:290–293. doi: 10.1089/gte.2006.10.290. [DOI] [PubMed] [Google Scholar]
- 81.Gandhi PN, Chen SG, Wilson-Delfosse AL. Leucine-rich repeat kinase 2 (LRRK2): a key player in the pathogenesis of Parkinson’s disease. J Neurosci Res. 2009;87:1283–1295. doi: 10.1002/jnr.21949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gao L, Gomez-Garre P, Diaz-Corrales FJ, Carrillo F, Carballo M, Palomino A, Diaz-Martin J, Mejias R, Vime PJ, Lopez-Barneo J, Mir P. Prevalence and clinical features of LRRK2 mutations in patients with Parkinson’s disease in southern Spain. Eur J Neurol. 2009;16:957–960. doi: 10.1111/j.1468-1331.2009.02620.x. [DOI] [PubMed] [Google Scholar]
- 83.Goldwurm S, Di Fonzo A, Simons EJ, Rohe CF, Zini M, Canesi M, Tesei S, Zecchinelli A, Antonini A, Mariani C, Meucci N, Sacilotto G, Sironi F, Salani G, Ferreira J, Chien HF, Fabrizio E, Vanacore N, Dalla Libera A, Stocchi F, Diroma C, Lamberti P, Sampaio C, Meco G, Barbosa E, Bertoli-Avella AM, Breedveld GJ, Oostra BA, Pezzoli G, Bonifati V. The G6055A (G2019S) mutation in LRRK2 is frequent in both early and late onset Parkinson’s disease and originates from a common ancestor. J Med Genet. 2005;42:e65. doi: 10.1136/jmg.2005.035568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Goldwurm S, Tunesi S, Tesei S, Zini M, Sironi F, Primignani P, Magnani C, Pezzoli G. Kin-cohort analysis of LRRK2-G2019S penetrance in Parkinson’s disease. Mov Disord. 2011 doi: 10.1002/mds.23807. [DOI] [PubMed] [Google Scholar]
- 85.Ishihara L, Gibson RA, Warren L, Amouri R, Lyons K, Wielinski C, Hunter C, Swartz JE, Elango R, Akkari PA, Leppert D, Surh L, Reeves KH, Thomas S, Ragone L, Hattori N, Pahwa R, Jankovic J, Nance M, Freeman A, Gouider-Khouja N, Kefi M, Zouari M, Ben Sassi S, Ben Yahmed S, El Euch-Fayeche G, Middleton L, Burn DJ, Watts RL, Hentati F. Screening for Lrrk2 G2019S and clinical comparison of Tunisian and North American Caucasian Parkinson’s disease families. Mov Disord. 2007;22:55–61. doi: 10.1002/mds.21180. [DOI] [PubMed] [Google Scholar]
- 86.Ramsden N, Perrin J, Ren Z, Lee BD, Zinn N, Dawson VL, Tam D, Bova M, Lang M, Drewes G, Bantscheff M, Bard F, Dawson TM, Hopf C. Chemoproteomics-based design of potent LRRK2-selective lead compounds that attenuate Parkinson’s disease-related toxicity in human neurons. ACS Chem Biol. 2011;6:1021–1028. doi: 10.1021/cb2002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kramer T, Lomonte F, Goring S, Amombo GMO, Schmidt B. Small molecule kinase inhibitors for LRRK2 and their application to parkinson’s disease models. ACS Chem. Neurosci. 2012;3:151–160. doi: 10.1021/cn200117j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen H, Chan BK, Drummond J, Estrada AA, Gunzner-Toste J, Liu X, Liu Y, Moffat J, Shore D, Sweeney ZK, Tran T, Wang S, Zhao G, Zhu H, Burdick DJ. Discovery of Selective LRRK2 Inhibitors Guided by Computational Analysis and Molecular Modeling. J Med Chem. 2012;55:5536–5545. doi: 10.1021/jm300452p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.