Abstract
BACKGROUND
Somatic mutations in the Janus kinase 2 gene (JAK2) occur in many myeloproliferative neoplasms, but the molecular pathogenesis of myeloproliferative neoplasms with nonmutated JAK2 is obscure, and the diagnosis of these neoplasms remains a challenge.
METHODS
We performed exome sequencing of samples obtained from 151 patients with myeloproliferative neoplasms. The mutation status of the gene encoding calreticulin (CALR) was assessed in an additional 1345 hematologic cancers, 1517 other cancers, and 550 controls. We established phylogenetic trees using hematopoietic colonies. We assessed calreticulin subcellular localization using immunofluorescence and flow cytometry.
RESULTS
Exome sequencing identified 1498 mutations in 151 patients, with medians of 6.5, 6.5, and 13.0 mutations per patient in samples of polycythemia vera, essential thrombocythemia, and myelofibrosis, respectively. Somatic CALR mutations were found in 70 to 84% of samples of myeloproliferative neoplasms with nonmutated JAK2, in 8% of myelodysplasia samples, in occasional samples of other myeloid cancers, and in none of the other cancers. A total of 148 CALR mutations were identified with 19 distinct variants. Mutations were located in exon 9 and generated a +1 base-pair frameshift, which would result in a mutant protein with a novel C-terminal. Mutant calreticulin was observed in the endoplasmic reticulum without increased cell-surface or Golgi accumulation. Patients with myeloproliferative neoplasms carrying CALR mutations presented with higher platelet counts and lower hemoglobin levels than patients with mutated JAK2. Mutation of CALR was detected in hematopoietic stem and progenitor cells. Clonal analyses showed CALR mutations in the earliest phylogenetic node, a finding consistent with its role as an initiating mutation in some patients.
CONCLUSIONS
Somatic mutations in the endoplasmic reticulum chaperone CALR were found in a majority of patients with myeloproliferative neoplasms with nonmutated JAK2. (Funded by the Kay Kendall Leukaemia Fund and others.)
The myeloproliferative neoplasms are chronic myeloid cancers that are characterized by the overproduction of mature blood cells, and that may evolve into acute myeloid leukemia.1,2 In addition to chronic myeloid leukemia with the BCR-ABL fusion gene, the three most common myeloproliferative neoplasms are essential thrombocythemia, polycythemia vera, and myelofibrosis.
Many patients with a BCR-ABL–negative myeloproliferative neoplasm carry a Janus kinase 2 (JAK2) V617F mutation.3-6 The JAK2 V617F mutation or JAK2 exon 12 mutations are found in most patients with polycythemia vera,7,8 whereas the JAK2 V617F mutation is found in only 50 to 60% of patients with essential thrombocythemia or myelofibrosis.9,10 Tests for JAK2 mutations have greatly simplified the diagnosis of myeloproliferative neoplasms and are now firmly embedded as front-line tests in national and international guidelines.11-14 However, distinguishing essential thrombocythemia with nonmutated JAK2 from the much more common reactive thrombocytosis remains a major diagnostic challenge.
Additional mutations have been identified in patients who have myeloproliferative neoplasms with or without JAK2 mutations. These mutations fall into two main classes: signaling mutations that activate the thrombopoietin receptor (MPL) or inactivate negative regulators such as LNK15,16 and mutations in epigenetic regulators of DNA methylation (TET2, DNMT3A, and IDH1/2)17-20 or chromatin structure (EZH2 and ASXL1).21,22 However, these additional mutations affect only a small minority of patients, genomewide data are lacking, and the pathogenesis of myeloproliferative neoplasms without JAK2 or MPL mutations remains obscure.
We have used massively parallel sequencing to characterize the mutational landscape of 151 patients with myeloproliferative neoplasms and have identified the gene encoding calreticulin (CALR) as a new cancer gene that is mutated in the majority of patients with myeloproliferative neoplasms and nonmutated JAK2.
METHODS
STUDY SAMPLES
We obtained samples from patients with myeloproliferative neoplasms and other cancers and from healthy controls after obtaining written informed consent. The study was approved by the ethics committee at each participating study center. For exome sequencing, we obtained tumor samples from blood granulocytes and constitutional samples from cultured T cells (in 34 patients), isolated T cells (in 42), or buccal swabs (in 75). (Additional details are provided in the Supplementary Appendix, available with the full text of this article at NEJM.org.) We classified all myeloproliferative neoplasms in accordance with the criteria of the British Committee for Standards in Haematology.11,12 The diagnoses of other hematologic neoplasms were made in accordance with 2008 World Health Organization categories.13
DNA SEQUENCING, CLONAL ANALYSIS, AND PROTEIN EXPRESSION
We performed exome sequencing of matched tumor and constitutional samples of DNA using standard protocols and conducted bioinformatic analyses to identify somatically acquired mutations, as described previously.23,24 We confirmed the presence of somatic sequence variants using custom target-capture methods and parallel sequencing combined with manual data curation. We used Sanger sequencing of CALR exon 9 combined with analyses of exome or genome sequencing data to determine the mutation status of CALR in an additional 1345 hematologic cancers, in 1517 other cancer samples, and in 550 control samples.
We cultured hematopoietic colonies, as described previously,25 and genotyped them using Sanger sequencing for all mutations identified on exome sequencing for that patient. Expression constructs, cell lines, and other methods are described in the Supplementary Appendix.
STATISTICAL ANALYSIS
We calculated the significance of recurrently mutated genes, taking into account mutation frequency, gene size, and mutation and sequence context. P values were adjusted for a false discovery rate and are expressed as a q value, with a value of less than 0.05 indicating statistical significance. Clonal heterogeneity was inferred from exome-sequencing data on the basis of a Bayesian Dirichlet process.26 We evaluated the statistical significance of differences between subtypes of myeloproliferative neoplasms using standard methods, which are detailed in the Supplementary Appendix.
RESULTS
MUTATIONAL LANDSCAPE IN THE PATIENTS
We analyzed the results of exome sequencing of DNA from granulocytes and constitutional DNA obtained from purified T cells or buccal cells in 168 patients with myeloproliferative neoplasms. The identification of appropriate constitutional DNA samples is a challenge among patients with myeloproliferative neoplasms, since circulating T cells and buccal cells may be contaminated by neoplastic cells (Fig. S1 in the Supplementary Appendix). For patients with the JAK2 V617F mutation, we measured the JAK2 V617F allele burden, using a quantitative polymerase-chain-reaction (PCR) assay to identify contaminated constitutional DNA samples, with in vitro T-cell cultures being necessary in some instances. In cases in which constitutional DNA was limited, the use of a combination of unamplified and whole-genome amplified DNA improved exome coverage (Fig. S2 in the Supplementary Appendix). We developed a bioinformatics workflow to enable the identification of high-confidence somatic variants (Fig. S3 in the Supplementary Appendix), and complemented this with orthogonal verification of mutations by targeted resequencing and manual curation.
Of the 168 samples of myeloproliferative neoplasms that were sequenced, 151 samples (48 samples of polycythemia vera, 62 of essential thrombocythemia, 39 of myelofibrosis, and 2 of unclassifiable myeloproliferative neoplasms) fulfilled the bioinformatic criteria and were taken forward for analysis (Fig. S3 and Table S1 in the Supplementary Appendix). The average exome sequencing coverage was 141×.
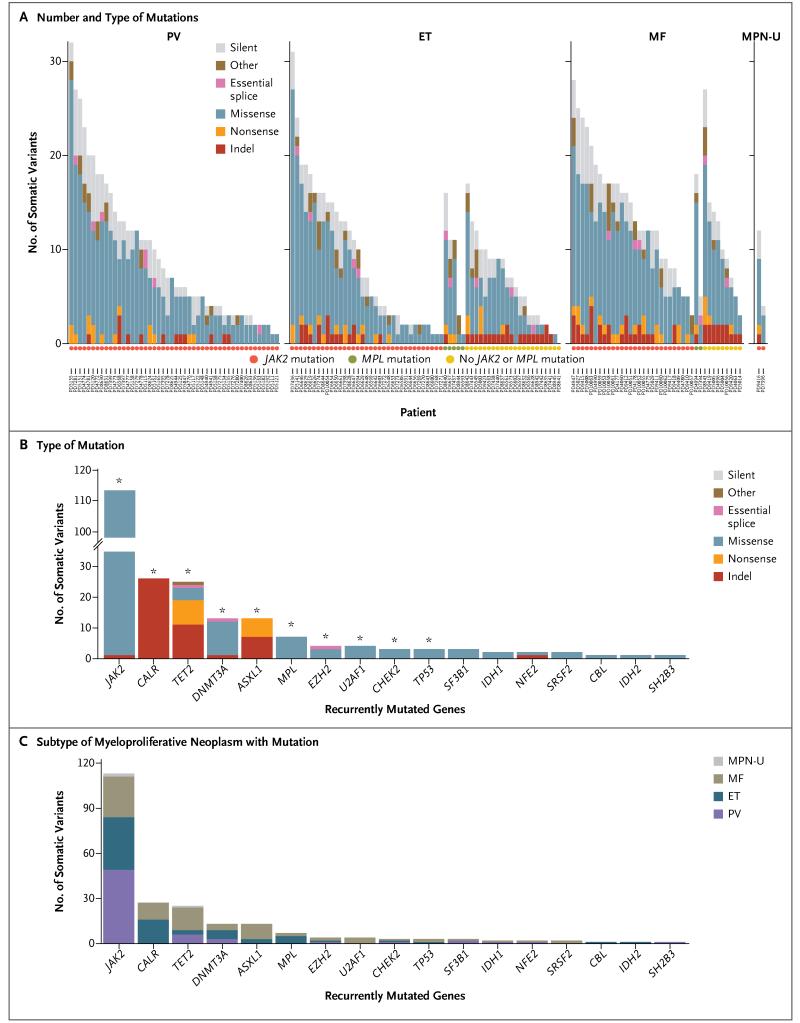
We identified and validated 1498 somatic mutations in 150 samples, with a range of 1 to 32 mutations per sample (Fig. 1A, and Table S2 in the Supplementary Appendix); no mutations were identified in the sample obtained from Patient PD8641, who had essential thrombocythemia. A total of 99 of the mutations we identified were insertions or deletions (indels) and 1399 were substitutions; 1405 mutations were found in coding sequences (1130 nonsynonymous changes), 22 in essential splice sites, 46 in introns within 10 base-pairs of splice junctions, and 25 in untranslated regions. The median number of mutations per patient was 6.5 in patients with polycythemia vera, 6.5 in those with essential thrombocythemia, and 13.0 in those with myelofibrosis (P = 0.008 by one-way analysis of variance). The significantly higher number of mutations in patients with myelofibrosis was consistent with the concept that this disorder is a more advanced stage of disease than either essential thrombocythemia or polycythemia vera (P<0.001 for the comparison of myelofibrosis with essential thrombocythemia and P = 0.008 for the comparison of myelofibrosis with polycythemia vera; pairwise t-test with Bonferroni correction for both comparisons). The mutational spectrum showed a predominance of C→T transitions (Fig. S4 in the Supplementary Appendix) and was similar to that observed in myelodysplasia and several epithelial cancers.23,27
Figure 1. Mutational Profile of 151 Myeloproliferative Neoplasms.
Panel A shows the number and type of mutations identified on exome sequencing in each sample obtained from 151 patients with myeloproliferative neoplasms. These patients included 48 with polycythemia vera (PV), 62 with essential thrombocythemia (ET), 39 with myelofibrosis (MF), and 2 with unclassifiable myeloproliferative neoplasms (MPN-U). The type of mutation is indicated in the key in each panel; the circles below the graphs indicate the patients’ mutational status: JAK2, MPL, or no JAK2 or MPL mutation. Also shown are the numbers of somatic mutations in recurrently mutated genes in this study as well as in genes previously reported to be mutated in myeloproliferative neoplasms, according to the type of mutation (Panel B) and subtype of myeloproliferative neoplasm (Panel C). In Panel B, the asterisks denote the significance of recurrently mutated genes (q<0.05). Indel denotes insertion or deletion mutation.
We identified mutations in several genes that have previously been implicated in myeloproliferative neoplasms or other myeloid cancers (Fig. 1B and 1C). JAK2 V617F was the most prevalent mutation and was found in all 48 patients with polycythemia vera, 35 of 62 patients with essential thrombocythemia (56%), and 27 of 39 patients with myelofibrosis (69%). Mutations in epigenetic regulators were as follows: TET2, 25 somatic variants in 22 patients; DNMT3A, 13 somatic variants in 12 patients; ASXL1, 13 somatic variants in 12 patients; EZH2 somatic variants in 4 patients; and IDH1/2 somatic variants in 3 patients (Fig. 1B). Mutations in genes encoding components of the splicing machinery were found in 9 patients (U2AF1 in 4 patients, SF3B1 in 3 patients, and SRSF2 in 2 patients), and mutations in the gene encoding the thrombopoietin receptor (MPL) were identified in 7 patients, all of whom had essential thrombocythemia or myelofibrosis with unmutated JAK2. Mutations in genes that are reported to be mutated in myeloproliferative neoplasms at low frequencies were found in 4 patients (CBL, in 1 patient; NFE2, in 2 patients; and SH2B3/LNK, in 1 patient) (Fig. 1B). Missense somatic mutations in CHEK2, which have not been reported previously in myeloproliferative neoplasms, were found in 1 patient each with polycythemia vera, essential thrombocythemia, and myelofibrosis (q = 0.008) (Table S3 in the Supplementary Appendix). We also assessed pairwise associations between genes mutated in myeloproliferative neoplasms (Fig. S5 in the Supplementary Appendix). Mutations in ASXL1 were found to be comutated with genes involved in RNA splicing (U2AF1 and SRSF2), and mutations in SRSF2 were comutated with genes that encode epigenetic modifiers (TET2, IDH1, and ASXL1) — a profile strikingly similar to the associations seen in myelodysplasia.28
RECURRENT SOMATIC CALR MUTATIONS
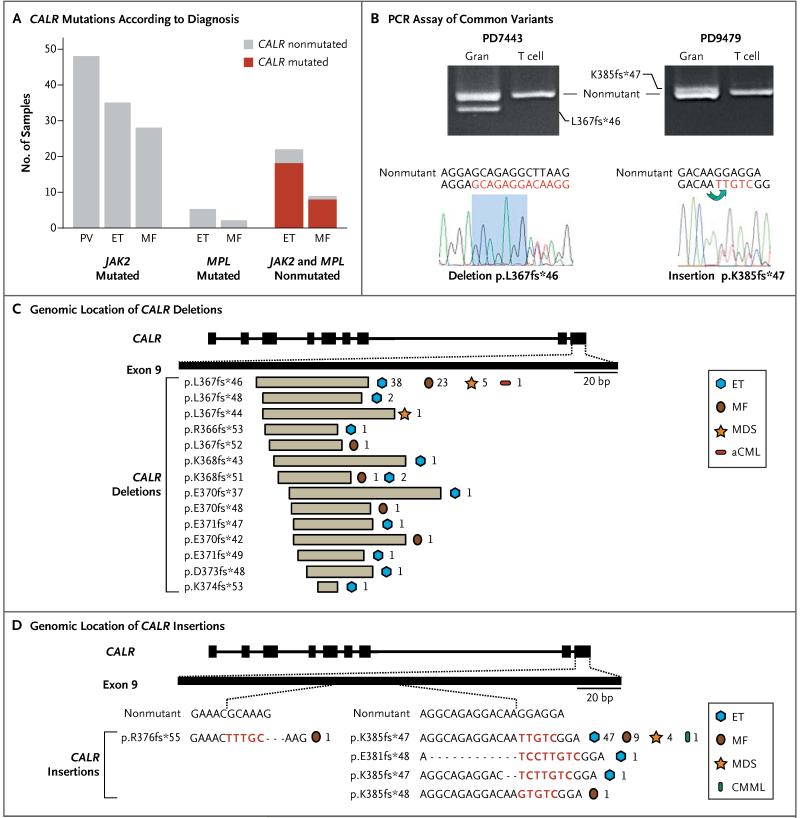
A striking pattern of somatic mutations was observed in CALR, a gene that had not previously been recognized as an oncogene mutated in any form of cancer. CALR mutations were identified in 26 patients (Fig. 1B and 1C). All the mutations were indels in exon 9 and had a remarkable association with disease, since they were found in 26 of 31 patients with essential thrombocythemia or myelofibrosis and nonmutated JAK2 or MPL (84%; 95% confidence interval [CI], 66 to 94) but in none of the 120 patients with JAK2 or MPL mutations (Fig. 2A, and Fig. S5 in the Supplementary Appendix). There were two common variants: L367fs*46, which resulted from a 52-bp deletion flanked by 7 base pairs of identical sequence; and K385fs*47, which resulted from a 5-bp insertion and represented an inverse duplication of the five nucleotides preceding the insertion (Fig. 2B).
Figure 2. CALR Mutations in Myeloproliferative Neoplasms.
Panel A shows the number of CALR mutations found in myeloproliferative neoplasms (polycythemia vera, PV; essential thrombocythemia, ET; or myelofibrosis, MF) that have mutated JAK2, mutated MPL, or nonmutated JAK2 or MPL. Panel B shows the results of validation on polymerase-chain-reaction (PCR) assay and Sanger sequencing of the two most common CALR variants — deletion (L367fs*46) and insertion (K385fs*47) — in patients with myeloproliferative neoplasms. The patients are indicated by their patient-identification numbers above the results of gel electrophoresis of PCR products of patients’ granulocytes (Gran) and T cells. The sequencing traces show the heterozygous mutation of CALR. The shaded region on the left highlights a homologous DNA sequence flanking the common CALR deletion, and the green arrow on the right highlights the inverse tandem duplication of five bases in the common CALR insertion. Panel C shows the genomic location of CALR deletions. The numbers indicate the number of patients with each disease. Panel D shows the genomic location of CALR insertions (shown in red). aCML denotes atypical chronic myeloid leukemia, CMML chronic myelomonocytic leukemia, and MDS myelodysplastic syndromes.
Exome sequencing revealed low coverage at CALR (median depth, 10 reads) (Fig. S6 in the Supplementary Appendix). We therefore also performed Sanger sequencing of exon 9, which detected CALR mutations in 10 to 15% of cells (Fig. S6 in the Supplementary Appendix). This confirmed the absence of CALR exon 9 mutations in all 120 patients who had JAK2 or MPL mutations. Of 6 patients initially lacking a mutation in CALR, JAK2, and MPL on exome sequencing, 1 patient was found to have low-level mutated CALR on Sanger sequencing.
Among patients with essential thrombocythemia, those with CALR mutations, as compared with those with JAK2 mutations, presented with significantly higher platelet counts (P<0.001 by the Wilcoxon rank-sum test) and lower hemoglobin levels (P = 0.02 by Student’s t-test) (Table S4 in the Supplementary Appendix). Patients with CALR mutations had a significantly higher incidence of transformation from essential thrombocythemia to myelofibrosis than did those with JAK2 mutations (P = 0.03 by Fisher’s exact test) (Table S4 in the Supplementary Appendix). There were no significant between-group differences in rates of survival, although the number of deaths was small.
Exome sequencing showed that 146 of 151 patients with myeloproliferative neoplasms (97%) had mutations in JAK2, MPL, or CALR in a mutually exclusive manner (q<0.01) (Fig. S5 in the Supplementary Appendix). The results of bone marrow histologic analyses were reviewed in 4 of the 5 patients lacking mutations in all three genes and were consistent with a myeloproliferative neoplasm. In addition, 4 of these patients (PD9420, PD8945, PD8635, and PD7441) had clonal somatic mutations, an indication that these patients had a clonal myeloid disorder (Table S2 in the Supplementary Appendix).
To further characterize CALR mutations in myeloid and other cancers, we performed Sanger sequencing of CALR exon 9 in 1397 samples (including 52 control samples) and evaluated wholeexome or whole-genome sequencing data for 502 solid tumors, 498 control samples, and 1015 cancer cell lines (Table 1). CALR mutations were present in 110 of 158 patients with myeloproliferative neoplasms lacking JAK2 or MPL mutations (70%; 95% CI, 62 to 77), including 80 of 112 patients with essential thrombocythemia (71%), 18 of 32 patients with primary myelofibrosis (56%), and 12 of 14 patients with progression of essential thrombocythemia to myelofibrosis (86%). No CALR mutations were found in 511 myeloproliferative neoplasms with JAK2 or MPL mutations. Secondary acute myeloid leukemia developed in 2 patients with CALR mutations (1 each with essential thrombocythemia and myelofibrosis, both with the K385fs*47 variant), with persistence of the CALR mutations after transformation. CALR mutations were identified in 10 of 120 patients with myelodysplastic syndromes (8%; 95% CI, 4 to 15) (Table 1). One patient each with chronic myelomonocytic leukemia and atypical chronic myeloid leukemia had CALR mutations. No mutations were found in control samples, lymphoid cancers, solid tumors, or cell lines (Table 1, and Table S5 in the Supplementary Appendix).
Table 1. Frequency of CALR Mutations in Samples Obtained from Patients with Myeloproliferative Neoplasms or Other Disorders and from Controls*.
| Tumor Type | No. of Samples | CALR Mutations | |
|---|---|---|---|
| Occurrence no./total no. | Frequency % (95% CI) | ||
| Exome screening | |||
| Myeloproliferative neoplasms | 151 | ||
| Polycythemia vera | 0/48 | 0 | |
| Essential thrombocythemia | |||
| With JAK2 or MPL mutation | 0/40 | 0 | |
| Without JAK2 or MPL mutation | 18/22 | 82 (59–94) | |
| Myelofibrosis | |||
| Primary myelofibrosis | |||
| With JAK2 or MPL mutation | 0/22 | 0 | |
| Without JAK2 or MPL mutation | 4/5 | 80 (30–99) | |
| After essential thrombocythemia | |||
| With JAK or MPL mutation | 0/2 | 0 | |
| Without JAK or MPL mutation | 4/4 | 100 (40–100) | |
| After polycythemia vera | 0/6 | 0 | |
| Unclassifiable myeloproliferative neoplasm | 0/2 | 0 | |
| Follow-up screening | |||
| Myeloproliferative neoplasm | 669 | ||
| Polycythemia vera | 0/217 | 0 | |
| Essential thrombocythemia | |||
| With JAK2 or MPL mutation | 0/138 | 0 | |
| Without JAK2 or MPL mutation | 80/112 | 71 (62–79) | |
| Myelofibrosis | |||
| Primary myelofibrosis | |||
| With JAK2 or MPL mutation | 0/97 | 0 | |
| Without JAK2 or MPL mutation | 18/32 | 56 (38–73) | |
| After essential thrombocythemia | |||
| With JAK2 or MPL mutation | 0/18 | 0 | |
| Without JAK2 or MPL mutation | 12/14 | 86 (56–98) | |
| After polycythemia vera | 0/41 | 0 | |
| Other myeloid disorder | 389 | ||
| Myelodysplastic syndromes | 10/120 | 8 (4–15) | |
| Refractory anemia | 5/53 | 9 (4–21) | |
| RARS | 3/27 | 11 (3–30) | |
| Refractory anemia with excess blasts | 2/17 | 12 (2–38) | |
| Refractory anemia (5q−) | 0/4 | 0 | |
| RARS and thrombocytosis | 0/6 | 0 | |
| RCMD | 0/10 | 0 | |
| RCMD and ringed sideroblasts | 0/3 | 0 | |
| Chronic myelomonocytic leukemia | 1/33 | 3 (0–18) | |
| Atypical chronic myeloid leukemia | 1/29 | 3 (0–20) | |
| Chronic myeloid leukemia | 0/28 | 0 | |
| Acute myeloid leukemia | 0/48 | 0 | |
| Eosinophilic disorder | 0/2 | 0 | |
| Systemic mastocytosis | 0/114 | 0 | |
| Idiopathic erythrocytosis | 0/5 | 0 | |
| Transient abnormal myelopoiesis | 0/10 | 0 | |
| Lymphoid cancer | 287 | ||
| Acute lymphoblastic leukemia | 0/62 | 0 | |
| Multiple myeloma | 0/73 | 0 | |
| Chronic lymphocytic leukemia | 0/77 | 0 | |
| Diffuse large B-cell lymphoma | 0/25 | 0 | |
| MALT lymphoma | 0/26 | 0 | |
| Splenic marginal-zone lymphoma | 0/24 | 0 | |
| Solid tumors | 502 | ||
| Breast | 0/190 | 0 | |
| Lung | 0/28 | 0 | |
| Prostate | 0/30 | 0 | |
| Colorectal | 0/110 | 0 | |
| Skin | 0/25 | 0 | |
| Bone | 0/80 | 0 | |
| Renal | 0/26 | 0 | |
| Meningioma | 0/13 | 0 | |
| Cell lines | 1015 | 0/1015 | 0 |
| Control samples | 550 | 0/550 | 0 |
Mutations in the gene encoding calreticulin (CALR) were identified from whole-exome sequencing and validated by targeted resequencing in the exome screening cohort. For the follow-up screening, CALR mutations were identified on Sanger sequencing of CALR exon 9 for myeloproliferative neoplasms, other myeloid disorders, lymphoid cancers, and a subset of 52 control samples. Solid tumors, cell lines, and the remaining 498 control samples were screened for CALR mutations from existing whole-exome or whole-genome sequencing data. MALT denotes mucosa-associated lymphoid tissue, RARS refractory anemia with ringed sideroblasts, and RCMD refractory cytopenias with multilineage dysplasia.
Overall, CALR exon 9 mutations were identified in 148 patients (Table S6 in the Supplementary Appendix). All mutations were indels with 19 distinct variants: 14 deletions, 2 insertions, and 3 complex indels (Fig. 2C and 2D). In these 148 patients, variants L367fs*46 and K385fs*47 were the most common CALR mutations (in 67 patients [45%] and 61 patients [41%], respectively). The remaining 20 patients were found to have 17 unique variants. L367fs*46 was found more frequently in myelofibrosis than was K385fs*47 (P = 0.009 by chi-square test).
MUTANT PROTEIN WITH A NOVEL C-TERMINAL
Calreticulin is a highly conserved protein with multiple reported functions. Within the endoplasmic reticulum, the protein ensures appropriate folding of newly synthesized glycoproteins and modulates calcium homeostasis.29,30 Outside the endoplasmic reticulum, calreticulin is also found in intracellular, cell-surface, and extracellular compartments, where it has been implicated in diverse biologic processes, including proliferation, apoptosis, and immunogenic cell death.31-34 Calreticulin has three main structural and functional domains: an N-terminal lectin-binding domain that is interrupted by a proline-rich P domain and an unstructured C-terminal acidic domain that contains multiple calcium-binding sites.30,35
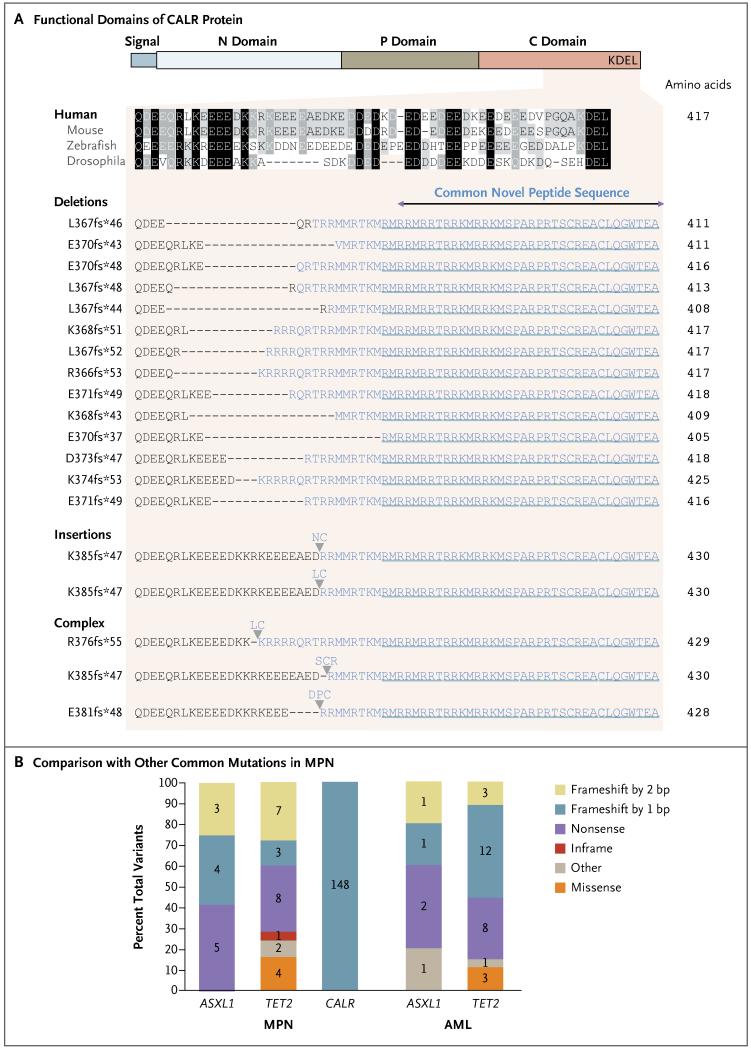
All the CALR indel mutations that were detected are predicted to generate mutant proteins with a novel C-terminal (Fig. 3A). The extent of the C-terminal alterations vary, but all 19 distinct variants share a loss of a sequence of 27 amino acids with a concomitant gain of a novel peptide consisting of 36 amino acids. These alterations result in the loss of most of the C-terminal acidic domain and the KDEL signal. (The KDEL amino acid sequence [Lys-Asp-Glu-Leu] is present on some resident endoplasmic reticulum proteins and enables retrieval of these proteins from the Golgi apparatus back to the endoplasmic reticulum.) This loss of function raises the possibility of compromised retention or retrieval in the endoplasmic reticulum. All CALR mutations shift the reading frame by one base pair, a pattern of mutation that is very different from that observed in known tumor-suppressor genes such as TET2 and ASXL1 (Fig. 3B).36,37 Such genes are affected by nonsense mutations and indels, with the latter generating +1 and +2 base-pair frame-shifts that frequently cause premature protein termination. The remarkably stereotypical pattern of CALR mutations implies a strong selective pressure to generate the mutant C-terminal.
Figure 3. Altered Protein Reading Frame with Novel C-Terminal Associated with CALR Mutations.
Panel A shows the functional domains of CALR protein, with (from left to right) signal sequence, N domain (N-terminal), P domain (proline-rich), C domain (C-terminal), and KDEL (endoplasmic reticulum retention signal). The conservation of the affected portion of the C domain across species is depicted by shading, with black indicating conserved regions and gray indicating partially conserved regions. The range of frameshift insertion and deletion mutations in CALR exon 9 are shown, all of which result in a common +1 base-pair–altered reading frame and predict a novel C-terminal peptide sequence lacking the KDEL motif. Panel B shows the mutational spectra of CALR in samples of myeloproliferative neoplasms (MPN) and of common loss-of-function genes such as ASXL1 and TET2 in myeloproliferative neoplasms and acute myeloid leukemia (AML). The numbers of each type of mutation are indicated for each gene. Data regarding myeloproliferative neoplasms are from the exome subgroup in this study, and AML data are from the Cancer Genome Atlas.36
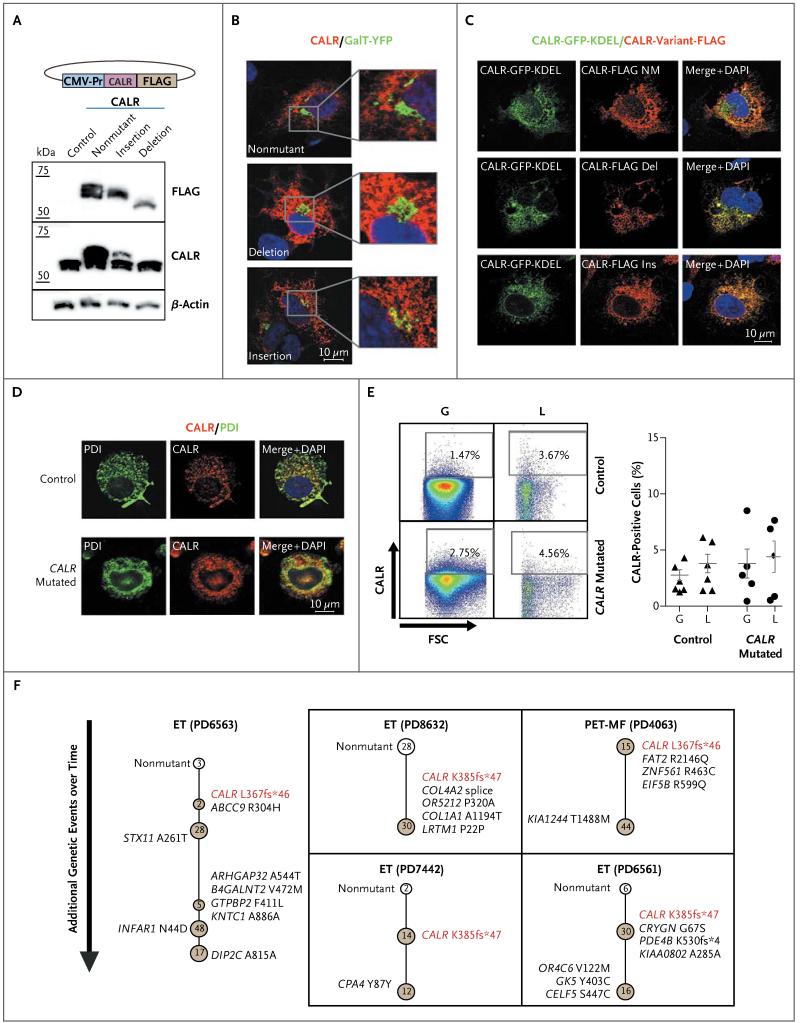
To confirm that CALR mutations result in a mutant protein product, CALR variant constructs (L367fs*46, deletion, and K385fs*47, insertion) were transiently expressed in human embryonic kidney (HEK) 293T cells. Immunoblotting confirmed the expression of mutant CALR proteins (Fig. 4A). Immunofluorescence studies of COS-7 cells showed that mutant CALR did not accumulate in the Golgi apparatus (Fig. 4B) and colocalized with exogenously expressed nonmutant CALR (Fig. 4C). Similar studies comparing mutated and nonmutated CALR myeloid cells from two patients with CALR-mutated essential thrombocythemia also showed no differences in the pattern of endogenous CALR (Fig. 4D).
Figure 4. Localization of Calreticulin and Clonal Heterogeneity in Patients with CALR Mutations.
Panel A shows immunoblotting of transiently transfected human embryonic kidney (HEK) 293T cells analyzed for FLAG (a polypeptide protein tag), CALR (detecting endogenous as well as transfected calreticulin), and beta-actin. The CALR deletion is the L367fs*46 variant, and the CALR insertion is the K385fs*47 variant. The CALR expression construct is shown above the immunoblots. CMV-Pr denotes CMV promoter. Panel B shows confocal photomicrographs of COS-7 cells transiently expressing FLAG-tagged CALR variants and a Golgi reporter (galactosyltransferase fused to yellow fluorescent protein [GalT-YFP]). Red indicates FLAG; green, Golgi; and blue, nucleus (4′,6-diamidino-2-phenylidole dihydrochloride [DAPI]). The images show that nonmutant and mutant CALR have an endoplasmic reticulum localization pattern with no increased accumulation in the Golgi. Panel C shows confocal photomicrographs of COS-7 cells transiently coexpressing nonmutant (NM) CALR tagged with green fluorescent protein (GFP) and FLAG-tagged CALR variants. Merge images show that FLAG-tagged CALR variants colocalize with nonmutant CALR to the endoplasmic reticulum. Panel D shows confocal photomicrographs of myeloid cells from CALR mutated and nonmutated granulocyte–macrophage colony-forming unit colonies derived from a patient with essential thrombocythemia with the CALR K385fs*47 mutation; the cells have been stained for protein disulfide isomerase (PDI, a resident protein of the endoplasmic reticulum) and endogenous CALR. Panel E shows flow cytometric analysis indicating the degree of CALR cell-surface expression in granulocytes (G) and lymphocytes (L) from a healthy control and from a patient with mutated CALR. The graph shows the percentage of viable cells expressing cell-surface CALR in peripheral blood from healthy controls (triangles) and patients with mutated CALR (circles). FSC denotes forward scatter. Panel F shows clonal structures in five patients (indicated by their patient-identification numbers) with myeloproliferative neoplasms with mutated CALR, as determined on genotyping of hematopoietic (erythroid) colonies. Each circle represents a clone, with nonmutant clones shown in white and mutant clones in brown. The earliest detectable clone is represented at the top of each diagram, with subsequent subclones shown below. Somatic mutations that were acquired in each subclone are indicated beside the respective nodes and represent those that were acquired in addition to mutations present in earlier subclones. Numbers of colonies that were identified for each node are shown inside the circles. ET denotes essential thrombo cythemia, and PET-MF post-ET myelofibrosis.
The up-regulation of cell-surface CALR is reported to mediate the phagocytosis of blasts38 in acute myeloid leukemia and might be a consequence of altered cellular localization of CALR due to loss of KDEL. We measured cell-surface CALR in hematopoietic 32D cells expressing mutant CALR variants (L367fs*46 and K385fs*47) and found no differences from nonmutant CALR expression (Fig. S7 in the Supplementary Appendix). Consistent with these results, there was no significant increase in cell-surface CALR in peripheral-blood leukocytes from five patients with mutated CALR (with estimated tumor burdens of 40 to 100% in granulocytes from exome sequencing), as compared with normal controls (Fig. 4E). Our data do not exclude the possibility of partial loss of mutant CALR from the endoplasmic reticulum through the secretory pathway.
CALR MUTATIONS IN THE HEMATOPOIETIC-STEM-CELL COMPARTMENT
CALR mutations were identified in patient-derived granulocyte–macrophage colonies and in erythroid colonies (Fig. S8 in the Supplementary Appendix), indicating that the mutations occur in a multipotent progenitor capable of generating both myeloid and erythroid progeny. Consistent with these results, genotyping studies of three patients showed that CALR mutations were present in flow-sorted, highly enriched hematopoietic stem cells (HSC; lin−CD34+CD38−CD45RA−CD90+), common myeloid progenitors (lin−CD34+CD38+ CD90−CD10−FLK2+CD45RA−), granulocyte-macrophage progenitors (lin−CD34+CD38+CD90−CD10− FLK2+CD45RA+), and megakaryocyte–erythroid progenitors (lin−CD34+CD38+CD90−CD10−FLK2− CD45RA−) (Fig. S8 in the Supplementary Appendix). These data are consistent with CALR mutations arising in the HSC compartment.
To ascertain whether mutation in CALR is an early event, we used exome-sequencing data to infer the fraction of cells bearing mutations and predict clonal relationships using a Bayesian Dirichlet process (Fig. S9 in the Supplementary Appendix). The results suggest that CALR mutation was an early event in most patients. However, such predictions should be interpreted cautiously because of the low sequencing coverage of CALR and because the approaches we used may not distinguish distinct tumor subclones with similar tumor burdens. Therefore, the order of mutation acquisition was determined in five patients with mutated CALR by genotyping 300 individual hematopoietic colonies for somatic mutations identified on exome sequencing. In all five patients, CALR mutations arose in the earliest phylogenetic node, consistent with mutation of CALR being an initiating event in these patients (Fig. 4F).
DISCUSSION
Understanding the molecular basis for essential thrombocythemia and myelofibrosis in patients without JAK2 mutations has been a major goal in the field of myeloproliferative neoplasms. In this study, we found that CALR, a previously unrecognized oncogene, is mutated in 70 to 84% of patients with essential thrombocythemia or myelofibrosis without JAK2 or MPL mutations. We also describe a comprehensive characterization of the mutational landscape of BCR-ABL–negative myeloproliferative neoplasms. We found that the median number of somatic mutations per patient was similar to that seen in other hematologic cancers (e.g., acute myeloid leukemia, acute lymphoblastic leukemia, and chronic lymphocytic leukemia, in which the median number of somatic mutations ranges from 8 to 13) but was substantially lower than the median number seen in most common epithelial cancers (range, 30 to 70).36,37 There was a long tail of cancer genes that have been implicated previously in myeloid cancers, with each gene mutated in a small fraction of patients with myeloproliferative neoplasms, as well as somatic mutations in CHEK2, which have not been described previously in myeloproliferative neoplasms.
CALR is present in the endoplasmic reticulum, where it forms a key component of the quality-control machinery that ensures proper glycoprotein folding and also contributes to calcium homeostasis.29,30 In addition, CALR functions outside the endoplasmic reticulum, in the cytoplasm, at the cell surface, and in the extracellular matrix, to influence a variety of processes, including proliferation, apoptosis, phagocytosis, and immune responses.31-33,37,39 Although calreticulin has been implicated in promoting the survival of malignant cells and in their ability to generate immune responses,32-34 to our knowledge, somatic mutations affecting endoplasmic reticulum chaperones have not been reported in any cancer. We describe 19 different CALR mutations (all deletions, insertions, or both) that generate a +1 base-pair frameshift predicted to replace the canonical C-terminal by a novel protein sequence. The normal CALR C-terminal domain is highly acidic and contains multiple calcium-binding sites together with a retrieval sequence (KDEL) that is typical of endoplasmic reticulum resident proteins. In contrast, the new predicted C-terminal domain of mutant CALR is basic, lacks multiple calcium-binding sites, and has no KDEL sequence. Mutant CALR did not have an altered Golgi or cell-surface accumulation of CALR, which was consistent with previously reported KDEL-independent retention mechanisms in the endoplasmic reticulum.40 However, our data indicate that mutant CALR probably has acquired novel functionality within or outside the endoplasmic reticulum.
Current diagnostic guidelines for patients with myeloproliferative neoplasms without JAK2 or MPL mutations rely on the ruling out of reactive causes of thrombocytosis combined with examination of bone marrow–biopsy specimens.11,13,14 In our exome subgroup, mutations in CALR, JAK2, or MPL were present in 146 of 151 patients with myeloproliferative neoplasms (97%). CALR mutations were present in 26 of 31 patients without JAK2 or MPL mutations in the exome subgroup (84%) and in 110 of 158 patients in the follow-up subgroup (70%), with the lower prevalence in the latter group possibly reflecting limitations of Sanger sequencing of whole-blood samples. Detection of CALR mutations in peripheral blood could potentially be used as a diagnostic tool in the same way that tests for JAK2 mutations have simplified and improved the accuracy of diagnosis of patients with myeloproliferative neoplasms worldwide.11-14 Further research is needed to explore these potential uses.
Supplementary Material
Acknowledgments
Supported by the Kay Kendall Leukaemia Fund (including a clinical research fellowship to Drs. Nangalia and Godfrey), the Wellcome Trust (including fellowships to Drs. Campbell and Ron), Leukemia and Lymphoma Research, Cancer Research UK, the National Institute for Health Research Cambridge Biomedical Research Centre, the Cambridge Experimental Cancer Medicine Centre, and the Leukemia and Lymphoma Society of America; a postdoctoral research grant from the Research Foundation–Flanders (to Dr. Van Loo); a grant from Associazione Italiana per la Ricerca sul Cancro (AIRC)–Gruppo Italiano Malattie Mieloproliferative (to Drs. Vannucchi and Guglielmelli); and a Canadian Institutes of Health Research Fellowship and the Lady Tata Memorial Trust grant (to Dr. Kent).
We thank members of the Cambridge Blood and Stem Cell Bank (Cambridge), Tayside Cancer Tissue Bank (Dundee), and Haemato-Oncology Diagnostics Laboratory, Addenbrooke’s Hospital (Cambridge) for technical assistance; Sarah Moody, Alex Clipson, Ming Wang, Giovanni Cazzaniga, Andrea Biondi, Jan Zuna, Helena Kempski, and Mike Groves for sample ascertainment; Stefan Marciniak, Steffen Preissler, and Bertie Gottgens for helpful advice and discussion; Janine Prick for technical assistance; and Hesham Eldaly and James Watkins for review of histologic analyses.
Appendix
The authors’ full names and degrees are as follows: Jyoti Nangalia, M.B.Chir., F.R.C.Path., Charles E. Massie, Ph.D., E. Joanna Baxter, Ph.D., Francesca L. Nice, M.Res., Gunes Gundem, Ph.D., David C. Wedge, Ph.D., Edward Avezov, Ph.D., Juan Li, Ph.D., Karoline Kollmann, Ph.D., David G. Kent, Ph.D., Athar Aziz, Ph.D., Anna L. Godfrey, B.M., B.Ch., F.R.C.Path., Jonathan Hinton, M.Sc., Inigo Martincorena, Ph.D., Peter Van Loo, Ph.D., Amy V. Jones, Ph.D., Paola Guglielmelli, M.D., Ph.D., Patrick Tarpey, Ph.D., Heather P. Harding, Ph.D., John D. Fitzpatrick, B.A., Calum T. Goudie, B.A., Christina A. Ortmann, M.D., Stephen J. Loughran, Ph.D., Keiran Raine, M.Sc., David R. Jones, M.Sc., Adam P. Butler, M.Sc., Jon W. Teague, M.Sc., Sarah O’Meara, B.Sc., Stuart McLaren, Dip.Mgmt., Michele Bianchi, D.N., Yvonne Silber, M.Sc., Danai Dimitropoulou, B.Sc., David Bloxham, M.Sc., F.R.C.Path., Laura Mudie, B.Sc., Mark Maddison, B.Sc., Ben Robinson, B.Sc., Clodagh Keohane, M.D., Cathy Maclean, M.Sc., Kate Hill, M.B., B.S., Kim Orchard, M.B., B.S., F.R.C.Path., Ph.D., Sudhir Tauro, F.R.C.Path., Ming-Qing Du, Ph.D., Mel Greaves, Ph.D., David Bowen, M.D., Brian J.P. Huntly, M.B., Ch.B., F.R.C.Path., Ph.D., Claire N. Harrison, D.M., F.R.C.Path., Nicholas C.P. Cross, F.R.C.Path., Ph.D., David Ron, M.D., Alessandro M. Vannucchi, M.D., Elli Papaemmanuil, Ph.D., Peter J. Campbell, M.B., Ch.B., Ph.D., and Anthony R. Green, F.R.C.Path., F.Med.Sci.
The authors’ affiliations are as follows: the Cambridge Institute for Medical Research and Wellcome Trust/MRC Stem Cell Institute (J.N., C.E.M., E.J.B., F.L.N., J.L., K.K., D.G.K., A.A., A.L.G., A.V.J., J.D.F., C.T.G., C.A.O., S.J.L., Y.S., D.D., B.J.P.H., A.R.G.), Department of Haematology (J.N., C.E.M., E.J.B., F.L.N., J.L., A.A., A.L.G., A.V.J., C.A.O., S.J.L., Y.S., D.D., C.M., B.J.P.H., P.J.C., A.R.G.), and the Division of Molecular Histopathology, Department of Pathology (M.-Q.D.), University of Cambridge, the Wellcome Trust Sanger Institute (J.N., C.E.M., G.G., D.C.W., J.H., I.M., P.V.L., P.T., K.R., D.R.J., A.P.B., J.W.T., S.O., S.M., L.M., M.M., B.R., E.P., P.J.C.), the Department of Haematology, Addenbrooke’s Hospital (J.N., A.L.G., M.B., D.B., C.M., B.J.P.H., P.J.C., A.R.G.), and the University of Cambridge Metabolic Research Laboratories and the National Institute for Health Research Cambridge Biomedical Research Centre (E.A., H.P.H., D.R.), Cambridge, the Faculty of Medicine, University of Southampton (A.V.J., N.C.P.C.), and the Department of Haematology, University Hospital Southampton (K.H., K.O.), Southampton, Guy’s and St. Thomas’ National Health Service Foundation Trust, Guy’s Hospital (C.K., C.N.H.), and the Haemato-Oncology Research Unit Division of Molecular Pathology, Institute of Cancer Research (M.G.), London, the Department of Hematology, Ninewells Hospital, Dundee (S.T.), and St. James Institute of Oncology, St. James Hospital, Leeds (D.B.) — all in the United Kingdom; the Department of Human Genetics, VIB and KU Leuven, Leuven, Belgium (P.V.L.); and the Dipartimento di Medicina Sperimentale e Clinica, University of Florence, Florence, Italy (P.G., A.M.V.).
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.Levine RL, Gilliland DG. Myeloproliferative disorders. Blood. 2008;112:2190–8. doi: 10.1182/blood-2008-03-077966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campbell PJ, Green AR. The myeloproliferative disorders. N Engl J Med. 2006;355:2452–66. doi: 10.1056/NEJMra063728. [DOI] [PubMed] [Google Scholar]
- 3.James C, Ugo V, Le Couédic JP, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144–8. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- 4.Baxter EJ, Scott LM, Campbell PJ, et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–61. doi: 10.1016/S0140-6736(05)71142-9. Erratum, Lancet 2005;366:122. [DOI] [PubMed] [Google Scholar]
- 5.Levine RL, Wadleigh M, Cools J, et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–97. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 6.Kralovics R, Passamonti F, Buser AS, et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352:1779–90. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- 7.Scott LM, Tong W, Levine RL, et al. JAK2 exon 12 mutations in polycythemia vera and idiopathic erythrocytosis. N Engl J Med. 2007;356:459–68. doi: 10.1056/NEJMoa065202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butcher CM, Hahn U, To LB, et al. Two novel JAK2 exon 12 mutations in JAK2V617F-negative polycythaemia vera patients. Leukemia. 2008;22:870–3. doi: 10.1038/sj.leu.2404971. [DOI] [PubMed] [Google Scholar]
- 9.Campbell PJ, Scott LM, Buck G, et al. Definition of subtypes of essential thrombocythaemia and relation to polycythaemia vera based on JAK2 V617F mutation status: a prospective study. Lancet. 2005;366:1945–53. doi: 10.1016/S0140-6736(05)67785-9. [DOI] [PubMed] [Google Scholar]
- 10.Campbell PJ, Griesshammer M, Döhner K, et al. V617F mutation in JAK2 is associated with poorer survival in idiopathic myelofibrosis. Blood. 2006;107:2098–100. doi: 10.1182/blood-2005-08-3395. [DOI] [PubMed] [Google Scholar]
- 11.Harrison CN, Bareford D, Butt N, et al. Guideline for investigation and management of adults and children presenting with a thrombocytosis. Br J Haematol. 2010;149:352–75. doi: 10.1111/j.1365-2141.2010.08122.x. [DOI] [PubMed] [Google Scholar]
- 12.McMullin MF, Reilly JT, Campbell P, et al. Amendment to the guideline for diagnosis and investigation of polycythaemia/erythrocytosis. Br J Haematol. 2007;138:821–2. doi: 10.1111/j.1365-2141.2007.06741.x. [DOI] [PubMed] [Google Scholar]
- 13.Swerdlow SH, Campo E, Harris NL, et al. WHO classification of tumours of haematopoietic and lymphoid tissues. IARC Press; Lyon, France: 2008. [Google Scholar]
- 14.Barbui T, Barosi G, Birgegard G, et al. Philadelphia-negative classical myeloproliferative neoplasms: critical concepts and management recommendations from European LeukemiaNet. J Clin Oncol. 2011;29:761–70. doi: 10.1200/JCO.2010.31.8436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pikman Y, Lee BH, Mercher T, et al. MPLW515L is a novel somatic activating mutation in myelofibrosis with myeloid metaplasia. PLoS Med. 2006;3(7):e270. doi: 10.1371/journal.pmed.0030270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lasho TL, Pardanani A, Tefferi A. LNK mutations in JAK2 mutation-negative erythrocytosis. N Engl J Med. 2010;363:1189–90. doi: 10.1056/NEJMc1006966. [DOI] [PubMed] [Google Scholar]
- 17.Delhommeau F, Dupont S, Della Valle V, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360:2289–301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 18.Pardanani A, Lasho TL, Finke CM, Mai M, McClure RF, Tefferi A. IDH1 and IDH2 mutation analysis in chronic- and blast-phase myeloproliferative neoplasms. Leukemia. 2010;24:1146–51. doi: 10.1038/leu.2010.77. [DOI] [PubMed] [Google Scholar]
- 19.Abdel-Wahab O, Pardanani A, Rampal R, Lasho TL, Levine RL, Tefferi A. DNMT3A mutational analysis in primary myelofibrosis, chronic myelomonocytic leukemia and advanced phases of myeloproliferative neoplasms. Leukemia. 2011;25:1219–20. doi: 10.1038/leu.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stegelmann F, Bullinger L, Schlenk RF, et al. DNMT3A mutations in myeloproliferative neoplasms. Leukemia. 2011;25:1217–9. doi: 10.1038/leu.2011.77. [DOI] [PubMed] [Google Scholar]
- 21.Ernst T, Chase AJ, Score J, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42:722–6. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 22.Carbuccia N, Murati A, Trouplin V, et al. Mutations of ASXL1 gene in myeloproliferative neoplasms. Leukemia. 2009;23:2183–6. doi: 10.1038/leu.2009.141. [DOI] [PubMed] [Google Scholar]
- 23.Papaemmanuil E, Cazzola M, Boultwood J, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365:1384–95. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stephens PJ, Tarpey PS, Davies H, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–4. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Godfrey AL, Chen E, Pagano F, et al. JAK2V617F homozygosity arises commonly and recurrently in PV and ET, but PV is characterized by expansion of a dominant homozygous subclone. Blood. 2012;120:2704–7. doi: 10.1182/blood-2012-05-431791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nik-Zainal S, Van Loo P, Wedge DC, et al. The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greenman C, Stephens P, Smith R, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–8. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papaemmanuil E, Gerstung M, Malcovati L, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013 Sep 12; doi: 10.1182/blood-2013-08-518886. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michalak M, Groenendyk J, Szabo E, Gold LI, Opas M. Calreticulin, a multiprocess calcium-buffering chaperone of the endoplasmic reticulum. Biochem J. 2009;417:651–66. doi: 10.1042/BJ20081847. [DOI] [PubMed] [Google Scholar]
- 30.Wang W-A, Groenendyk J, Michalak M. Calreticulin signaling in health and disease. Int J Biochem Cell Biol. 2012;44:842–6. doi: 10.1016/j.biocel.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 31.Gold LI, Eggleton P, Sweetwyne MT, et al. Calreticulin: non-endoplasmic reticulum functions in physiology and disease. FASEB J. 2010;24:665–83. doi: 10.1096/fj.09-145482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chao MP, Majeti R, Weissman IL. Programmed cell removal: a new obstacle in the road to developing cancer. Nat Rev Cancer. 2012;12:58–67. doi: 10.1038/nrc3171. [DOI] [PubMed] [Google Scholar]
- 33.Krysko DV, Garg AD, Kaczmarek A, Krysko O, Agostinis P, Vandenabeele P. Immunogenic cell death and DAMPs in cancer therapy. Nat Rev Cancer. 2012;12:860–75. doi: 10.1038/nrc3380. [DOI] [PubMed] [Google Scholar]
- 34.Luo B, Lee AS. The critical roles of endoplasmic reticulum chaperones and unfolded protein response in tumorigenesis and anticancer therapies. Oncogene. 2013;32:805–18. doi: 10.1038/onc.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pocanschi CL, Kozlov G, Brockmeier U, Brockmeier A, Williams DB, Gehring K. Structural and functional relationships between the lectin and arm domains of calreticulin. J Biol Chem. 2011;286:27266–77. doi: 10.1074/jbc.M111.258467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cancer Genome Atlas Research Network Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–74. doi: 10.1056/NEJMoa1301689. Erratum, N Engl J Med 2013;369:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chao MP, Jaiswal S, Weissman-Tsukamoto R, et al. Calreticulin is the dominant pro-phagocytic signal on multiple human cancers and is counterbalanced by CD47. Sci Transl Med. 2010;2(63):63ra94. doi: 10.1126/scitranslmed.3001375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pang WW, Pluvinage JV, Price EA, et al. Hematopoietic stem cell and progenitor cell mechanisms in myelodysplastic syndromes. Proc Natl Acad Sci U S A. 2013;110:3011–6. doi: 10.1073/pnas.1222861110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sönnichsen B, Füllekrug J, Nguyen Van P, Diekmann W, Robinson DG, Mieskes G. Retention and retrieval: both mechanisms cooperate to maintain calreticulin in the endoplasmic reticulum. J Cell Sci. 1994;107:2705–17. doi: 10.1242/jcs.107.10.2705. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.