Abstract
Infant formula and breastfeeding are environmental factors that influence the incidence of Type 1 Diabetes (T1D) as well as the acidity of newborn diets. To determine if altering the intestinal microbiome is one mechanism through which an acidic liquid plays a role in T1D, we placed non-obese diabetic (NOD)/ShiLtJt mice on neutral (N) or acidified H2O and monitored the impact on microbial composition and diabetes incidence. NOD-N mice showed an increased development of diabetes, while exhibiting a decrease in Firmicutes and an increase in Bacteroidetes, Actinobacteria, and Proteobacteria from as early as 2 weeks of age. NOD-N mice had a decrease in the levels of Foxp3 expression in CD4+Foxp3+ cells, as well as decreased CD4+IL17+ cells, and a lower ratio of IL17/IFNγ CD4+ T-cells. Our data clearly indicates that a change in the acidity of liquids consumed dramatically alters the intestinal microbiome, the presence of protective Th17 and Treg cells, and the incidence of diabetes. This data suggests that early dietary manipulation of intestinal microbiota may be a novel mechanism to delay T1D onset in genetically pre-disposed individuals.
Keywords: type 1 diabetes, microbiota, mouse model, T-cells, diet
Introduction
The non-obese diabetic (NOD) mouse is currently one of the most commonly accepted models for Type 1 Diabetes (T1D) (Leiter 1997). This model shares both genetic and environmental pre-dispositions with human patients, and spontaneously develops diabetes by ~25 weeks of age. Current evidence has indicated that a shorter duration or lack of breastfeeding is a risk factor for the development of T1D later in life (Patelarou et al. 2012). Additionally, the incidence of diabetes in NOD mice appears to be sensitive to dietary alterations and these alterations have been reported to modify aerobic and microaerophilic culturable bacterial compositions, as well as immune mediators (Atkinson and Chervonsky 2012; Hara et al. 2012). However, whether it is actually a diet-induced dysbiosis that leads to the changes in diabetes incidence is not well defined. Consequently, understanding this potential dietary-associated dysbiosis may be a promising avenue to help prevent autoimmune diseases such as T1D (Brown et al. 2012; Mathis and Benoist 2012; Neu et al. 2010).
There are four dominant phyla that comprise the intestinal commensal microbiota of both humans and mice. These include Firmicutes and Bacteroidetes, which normally account for >90% of the bacterial populations in the colon, as well as Actinobacteria and Proteobacteria. The intestinal commensal populations in murine models of T1D, similar to human patients, are altered in comparison with non-diabetics (Giongo et al. 2011; Roesch et al. 2009). Individuals with T1D display lower populations of Firmicutes and increased Bacteroides in their GI tract when compared with healthy controls (Brown et al. 2011; Giongo et al. 2011). This dysbiosis may impact the presence of interleukin-17 (IL17)-producing T-helper cells (Th17) and regulatory T cells (Tregs), as the development of these cells is heavily reliant on the microbial composition (Atarashi et al. 2011; Ivanov et al. 2009; Kriegel et al. 2011).
Previous publications are in disagreement as to whether Th17s are protective or pathogenic in T1D; however, both animals and patients with T1D display imbalances between Th17 and Treg responses (Bradshaw et al. 2009; Ferraro et al. 2011; Lau et al. 2011; van den Brandt et al. 2010). The connection between the microbiota, Th17s, and Tregs in T1D patients is not well defined. Although T1D is a disease of the pancreas, mucosal immune responses to dietary and bacterial antigens may impact the pancreas, as areas of the transverse colon drain directly into the pancreatic lymph nodes (PLNs) (Carter and Collins 1974; Turley et al. 2005; Vaarala et al. 2008). In addition to this potential direct link, the mucosal immune system is also vital in maintaining balance and homeostasis throughout the rest of the systemic immune system (Dimmitt et al. 2010; McCracken and Lorenz 2001).
Breastfed infants have a lower gut pH (acidified environment) and a higher lactic acid concentration when compared with infants fed cow’s milk or formula (Ogawa et al. 1992). We hypothesized that this acidified environment shapes the intestinal microbiome, consequently modifying the mucosal and systemic immune responses, and ultimately impacting the incidence of T1D. NOD/ShiLtJt (NOD) mice from The Jackson Laboratory (Bar Harbor, ME) are routinely maintained on water acidified with hydrochloric acid (pH 2.8–3.1) to prevent the growth of microorganisms. Mice maintained on acidified H2O live longer lives and gain weight slower than mice maintained on neutral (pH ~7.0) H2O, but this has not been reported to have any significant effect on immune function (Hall et al. 1980; Harrison 2008; Hermann et al. 1982). Therefore, to test our hypothesis, we switched NOD breeding pairs from acidified to neutral water, and subsequently studied the microbiome of their offspring, their mucosal immune responses, and their incidence of diabetes. As breastfed babies have a decreased risk of development of T1D later in life (Mayer et al. 1988), this experiment offered the unique opportunity to determine if the intake of liquids that impact the acidity of the gut environment could alter not only mucosal microbiota and immune responses, but also a systemic autoimmune disease such as T1D.
Materials & Methods
Mice
NOD/ShiLtJt mice originally obtained from Jackson Laboratory (Bar Harbor, Maine) were bred and maintained under specific pathogen-free (SPF) conditions in Thoren Isolator racks (Hazelton, PA) under positive pressure, and were fed autoclaved NIH-31 rodent diet (Harlan Teklan, Madison, WI), and sterile water ad libitum. The original animals were acclimatized to our facility 2 weeks prior to mating. Water in the animal research building (RSB), is from the Birmingham city water supply and is chlorinated and autoclaved. Original breeding pairs were split between neutral (pH ~7, NOD-N) and acidified (pH ~3.2, NOD-A) H2O, and all pups born from these breeding pairs and thereafter were maintained on their specific water source. Acidified H2O is comprised of 1 mL of 1 N HCl per 500 mL of H2O (pH ~3.2). A minimum of two sets of founder mice, originally ordered from The Jackson Laboratory (Bar Harbor, Maine) at different times, were used to create each mouse population to ensure any changes we witness are not the result of a founder effect (Ubeda et al. 2012). The mice did not show any differences in weight gain on acidic vs. neutral water.
Incidence of Diabetes
NOD mice on either water source were evaluated from 9–10 weeks of age until 30 weeks of age for the onset of diabetes. Blood glucose from a tail bleed was taken weekly via OneTouch© Blood Glucose Meter (Greenwood Village, CO). Diabetes was defined if the mouse exhibited two weekly adjacent readings of over 200 mg/dl, or a single reading over 400 mg/dl. All experiments were approved by the UAB Institutional Care and Use Committee.
Pancreas Histology
Pancreatic tissue was removed from diabetic animals and placed in formalin for >24 hrs. Tissue was washed in 70% EtOH and embedded in HistoGel (Richard-Allan Scientific, Kalamazoo, MI). Tissue was cut into 5-μm sections and stained with standard H&E for histological examination. For immunofluorescence analysis, sections were stained with guinea pig anti-insulin to detect islet beta cells (Invitrogen, Carlsbad, CA; 4 g/L) at a 1/100 dilution, and with rabbit anti-glucagon to detect islet alpha cells (Cell Signaling, Beverly, MA; 100 µl) at a 1/200 dilution. For nuclear staining, Hoechst (Sigma-Aldrich, St. Louis, MO; 1 mg/ml) was used at a dilution of 1/2000. Secondary antibodies used were goat anti-guinea pig DyLightTM 594 (Jackson ImmunoResearch, West Grove, PA; 1.5 mg/ml) at a dilution of 1/100 and donkey anti-rabbit Alexa Fluor® 488 (Invitrogen; 2 mg/ml) used at a dilution of 1/100.
pH of Intestinal Compartments
NOD-N or NOD-A mice were fasted for 4 hrs. Mice were sacrificed and the stomach, duodenum, jejunum, ileum, cecum, and colon were each washed individually with 1 mL of filtered deionized water. Particulate matter was removed and the pH of the contents was measured via Corning Pinnacle 540 pH meter (Corning, NY). pH was converted to H+ concentration through the equation pH = −log10(H+).
Denaturing Gradient Gel Electrophoresis (DGGE)
Fecal contents, collected weekly from NOD mice, were stored at −20C until further use. Fecal pellets were weighed and the bacterial DNA extracted with phenol:chloroform, as previously described (Dimmitt et al. 2010; Schmitz et al. 2011). DNA was quantified using a NanoDrop 1000 (NanoDrop, Wilmington, DE). DNA was diluted to 150 ng/µl and subjected to polymerase chain reaction (PCR) using 16S universal bacterial primers: 341GC 5’- CGCCCGCCGCGCGCGGCGGGCGGGGGGGGCACGGGGGGCCTACGGGAGGCAGCAG-3’and 534R 5’-ATTACCGCGGCTGCTGG-3’ (Sigma-Aldrich). PCR was performed using TaKaRa ExTaq HotStart Taq Polymerase kit (Fisher, Pittsburg, PA). Thermal profile was set at 95C for 5 min; 95C for 1 min, 65C for 45 sec (decreasing 0.5C per cycle), 72C for 1 min, repeated for an additional 19 cycles; 95C for 1 min, 55C for 45 sec, 72C for 1 min, repeated for an additional 9 cycles; and a final extension of 72C for 5 min. Polyacrylamide gels were produced and samples ran as previously described (Schmitz et al. 2011). Briefly, PCR samples were diluted with gel loading dye and loaded onto the 60/35% gradient gels. Gels were loaded onto a Bio-Rad Dcode system (Bio-Rad Laboratories, Hercules, CA) and ran overnight at 58C and 58 V in 7 L 1× TAE solution. Gels were stained with ethidium bromide and imaged and analyzed via Bio-Rad ChemiDoc XRS and Image Lab Software (Bio-Rad Laboratories, Hercules, CA). Bands of interest were removed and the extracted DNA was subjected to another round of PCR with the same primers as described above that were tagged with M13 vector tails. DNA was sequenced by the UAB Heflin Genetics Center. Bacterial species were identified by sequence pairing through the NCBI BLAST database. Taxonomic specification was determined with a 75% homology to sequences within the BLAST database. Band similarity was analyzed and calculated using the GelCompar II program (Applied Maths Inc., Austin, TX).
454 Pyrosequencing
Pyrosequencing was performed on genomic DNA samples using the bacterial tag-encoded GS FLX-Titanium amplicon with primers 28f (5’-GAGTTTGATCNTGGCTCAG-3’) and 519r (5’-GTNTTACNGCGGCKGCTG-3’) (Dowd et al. 2008). Sequences were processed with the mothur software package (Schloss et al. 2009). Briefly, barcodes and primers were depleted and sequences with an average quality score of less than 30 were removed from the dataset. Sequences that were shorter than 250 base pairs, that contained ambiguous base-pair designations, or were greater than 8 homopolymers were also removed to maintain sequencing quality and aligned to the V1-V3 region of bacterial 16S RNA gene using the SILVA reference alignment as a template. Chimeric sequences were removed using the UCHIME algorithm (Edgar et al. 2011). Sequences were assigned taxonomically using the SILVA database. A distance matrix was created with a threshold of 0.15, which was used to cluster the remaining sequences into operational taxonomic units (OTU) using the average neighbor grouping method, with a cutoff of 95% sequence similarity. Finally, OTUs were classified into consensus taxonomies. Data quality was checked using α-diversity analysis. To estimate richness, Chao1 and abundance-based coverage estimation (ACE) indices were used. Diversity was estimated using both Shannon and Simpson indices . Rarefaction curves were also generated to estimate sequencing quality and coverage.
Bacterial Quantitative Real-Time PCR
Twenty-five ng of fecal extracted DNA was subject to quantitative real-time PCR. Briefly, 12.5 µl SYBR© Green (Clontech), 0.05 µl of both 20 µM forward and reverse primers, and 25 ng DNA were added per well, with sterile H2O used to bring the volume up to 25 µl. Samples were compared to a standard curve specific to the target bacteria starting at 1×108 copy numbers and serially diluted to 1×101 copy numbers. The thermal profile for the reaction was: 95C for 10 min followed by 95C for 15 sec, 56C for 18sec, 45C for 45 sec, repeated for an additional 44 cycles. The extension temperature was varied depending on the bacterial-specific primers of either total bacteria (Barman et al. 2008), Lactobacillus (Valladares et al. 2010), Clostridia (Valladares et al. 2010) or Bacteroides (Valladares et al. 2010). Bacterial-specific primers were purchased from Invitrogen.
Lamina Propria Preparation
The large and small bowels were removed from female 2-week-old and 8–10-week-old NOD-A or NOD-N mice and were digested in order to extract lamina propria lymphocytes (Dimmitt et al. 2010). Two NOD pups were pooled for each sample because of the small size and number of cells collected from 2-week-old pups. Briefly, GI tissue was opened longitudinally and cleared of fecal debris. Large and small intestines were handled separately, digested in HBSS media + 5 mM EDTA and filtered to remove epithelial cells. Tissue was minced and further digested with HBSS media + collagenase IV (Sigma-Aldrich), and the resulting solution was filtered through a 100-µm filter and collected. Cells were washed and re-suspended in 40% Percoll (Fisher, Pittsburg, PA) and layered onto 70% Percoll before centrifugation. The 40/70% Percoll interface containing the lymphocytes was collected and stored overnight at 4C to allow cells to recover their cell surface molecules.
Lymphocyte Activation and Flow Cytometry
For identification of IL17- and IFNγ-producing cells, lymphocytes were activated with 100 ng/ml phorbol myristate acetate (PMA) (Sigma-Aldrich), 1 µg/ml ionomycin, and 0.7 µl/ml Golgistop (BD Biosciences, San Jose, CA) in R-10P media (RPMI 1640 (Mediatech, Manassas, VA), 10% Fetal Calf Serum (Thermo Scientific, Rockford, IL), 1% penicillin/streptomycin, 0.1% β-mercaptoethanol, and 1% Glutamax (Fisher, Pittsburg, PA) for 5 hr at 37C. Staining was performed as previously described (Tanner et al. 2012). Briefly, the FcR were blocked via αCD16/32 (Biolegend, San Diego, CA) and CD4-APC was used as a cell-surface marker for Th1/Th17 lymphocytes. Permeabilization of cells allowed for the intracellular staining of IL-17-PE, IFNγ-FITC (Th17/Th1) and Foxp3-APC before fixation and FACS analysis on both Tregs and Teff cells (Biolegend). Cell-surface antibodies listed as CD4-FITC and CD25-PE were used as Treg markers.
Graphic and Statistical Analysis
Graphs were made using GraphPad Prism 5 (San Diego, CA). Significance was performed for quantitative bacterial copy number via Welch’s t-test. Significance for the incidence of diabetes was calculated using the Mantel-Cox test. Sequence data for each sample was converted into percentage data at the phylum and the genus levels, tested for normality using PROC Univariate, and analyzed using PROC Mixed in SAS (SAS Institute Inc., Cary, NC). Data that were not normally distributed were treated with PROC GLIMMIX through either Poisson or negative binomial distributions, with the Pearson chi-square/degrees of freedom ratio being applied to determine the goodness of fit for each non-normal distribution method. In order to evaluate further significant differences between the two treatments, partial least-squared discriminant analysis (PLS-DA) was used within the SIMCA P+ 13.0 software package (Umetrics, Umea, Sweden). Y variables were used to describe the two treatments, NOD-A and NOD-N, whereas X variables were used to represent the bacterial genera. The number of significant components was determined using R2 and Q2 values. Variable influence on projection value (VIP) was determined for each genus, and those with a VIP value below 0.3 was removed from the model. The score scatter plots and loading scatter plots were generated, and genera significantly associated with either treatment were determined by the PLS-regression coefficients and their plots (Fig. S3).
Results
NOD-N Mice have a Higher Incidence of Diabetes
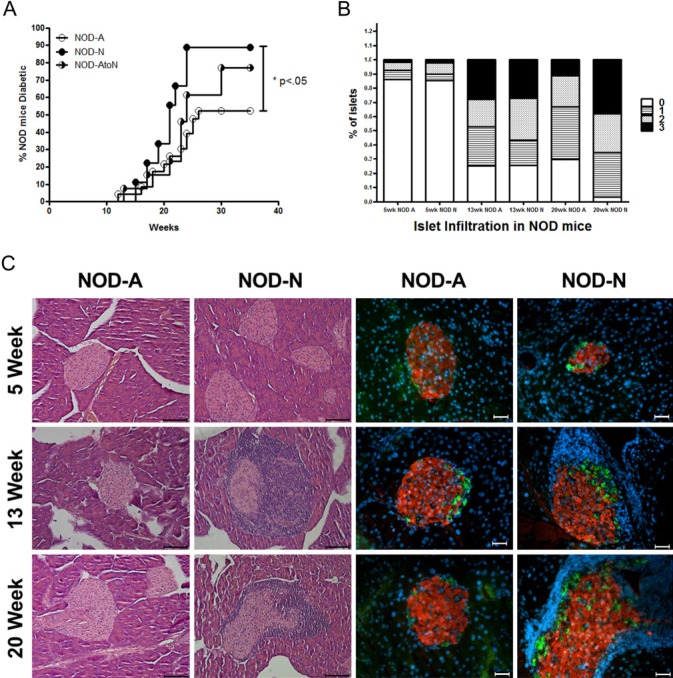
Female NOD mice on neutral (NOD-N) and acidified (NOD-A) H2O were followed for 30 weeks to ascertain the incidence of diabetes. NOD-N mice displayed a significantly higher incidence of diabetes. Only 11% of NOD-N mice remained diabetes-free at 30 weeks of age as compared with 46% of NOD-A mice (Fig. 1A). To evaluate the autoimmune activities leading to diabetes, pancreatic sections were evaluated for peri-insulitis and invasive immune infiltration into the pancreatic islets (insulitis) (Cardell 2006). NOD-N mice at 20 weeks displayed significantly increased insulitis compared with NOD-A mice (Fig. 1B). Histological and immunofluorescence analysis indicated that female NOD-N mice had increased islet infiltration starting at 13 weeks compared with female NOD-A mice, but that all mice had some remaining islets with functional alpha (glucagon-producing) and beta (insulin-producing) cells (Fig. 1C). Following a similar trend, NOD-N males also had a greater incidence of diabetes at 30 weeks (55% n=9) compared with NOD-A males (36% n=11). It has recently been reported that caging practices can influence microbial composition and diversity (Ubeda et al. 2012). Therefore, to confirm that changes in the incidence of diabetes seen in Figure 1 were not caused by a caging/founder effect, litters from multiple breeding pairs on acidified H2O were split at weaning, raising half of each litter on either acidified H2O, or switching them onto neutral H2O (NOD-AtoN). NOD-AtoN mice displayed a 77% incidence of diabetes, a result that is intermediate between mice raised on neutral H2O (89%) and those raised on acidified H2O (58%) (Fig. 1A). This demonstrates that the water pH is directly correlated with diabetes incidence. However, as the change in incidence was not complete, it also implies that the timing of the switch may also be critical, as these NOD-AtoN mice were exposed to the altered water source after weaning (instead of from birth).
Figure 1.
NOD mice on neutral drinking water have an increased incidence of diabetes and alterations in the diversity of their fecal microbiota. (A) Incidence of diabetes in NOD female mice on neutral water (NOD-N, open circles, n=9), acidified water (NOD-A, filled circles, n=23) or water switched from acidified to neutral at weaning (NOD-AtoN, half-filled circles, n=13). Data represent three individual experiments. Significance was determined by the Mantel-Cox Test. (B) Analysis of lymphocytic infiltrate and islet destruction in NOD mice on neutral or acidified water. 0 = no infiltration, 1 = peri-insulitis, 2 = <50% of islet infiltrated, 3 = >50% of islet infiltrated. At 20 weeks, the NOD-N mice had significantly more infiltrated islets (score of 2 or 3; p=0.03). An average of 80 islets per group was counted. (C) Representative H&E (left two panels; scale bar=100 µm) and immunofluorescence (right two panels; red = insulin, green = glucagon, blue = nucleus; scale bar =10 µm) images of pancreatic islets from NOD-A or NOD-N mice at the indicated ages.
NOD-N Mice have a Higher GI pH and Fewer Firmicutes
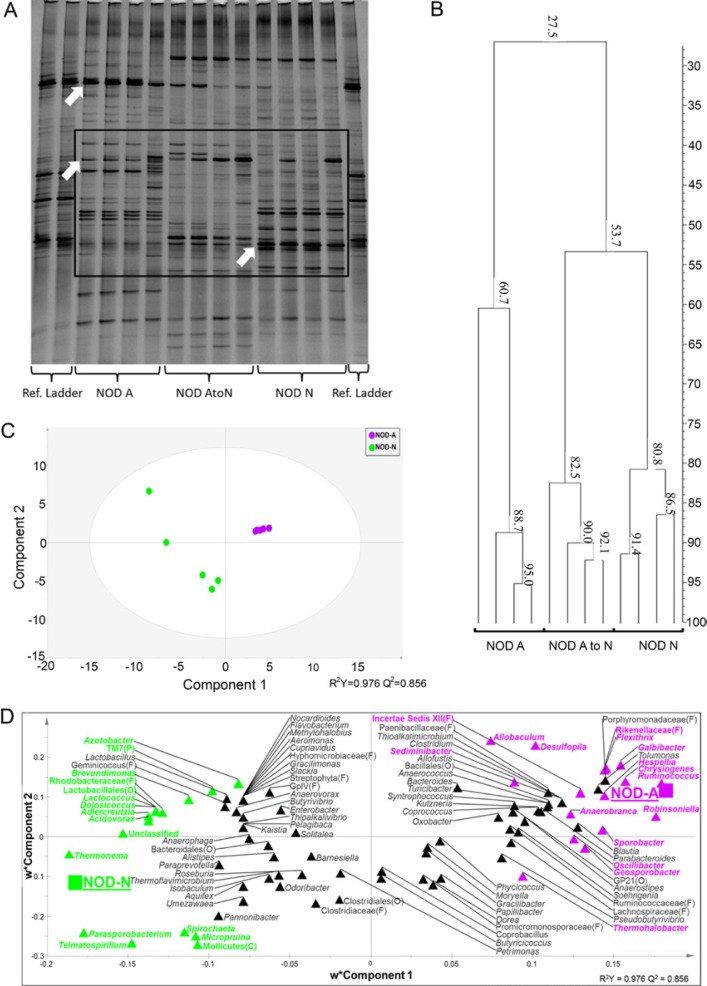
To determine if the acidity of the drinking water can actually alter the gastrointestinal (GI) luminal environment, the pH of the GI tract of 5-week-old female NOD-A and NOD-N mice was measured. NOD-N mice displayed a nearly 2-fold decreased concentration of H+ ions and thereby, a significantly higher pH in the duodenum, jejunum, cecum, and colon as compared with that from NOD-A mice (Fig. S1). The potential alterations in the microbial diversity associated with this altered pH were initially analyzed via DGGE. Ten-week-old NOD-N mice demonstrated dramatic shifts in the microbial communities compared with NOD-A mice (Fig. 2A). NOD-A and NOD-N mice only shared 27% band similarity compared with the 60.7% and 80.8% banding similarity (respectively) shared within each group (Fig. 2B). NOD-AtoN and NOD-N mice shared a higher degree of similarity (53.7%) as compared with NOD-AtoN and NOD-A mice. Unique bands (Fig. 2A) were removed and sequenced. Sequenced bands were discovered to belong to the groups of bacteria Lactobacillus, Bacteroides, and Clostridia cluster XIV. Although not quantitative, it is apparent that there are decreases in the populations of both Lactobacillus and Clostridia populations in NOD-N mice.
Figure 2.
Impact of neutral or acidified drinking water on the fecal microbiota composition of NOD-N and NOD-A mice. (A) A representative denaturing gradient gel electrophoresis (DGGE) analysis of banding patterns from 10-week-old female mice on neutral water (NOD-N, right), acidified water (NOD-A, left) or water switched from acidified to neutral at weaning (NOD-AtoN, middle). Top arrow points to a band identified by sequencing as Lactobacillus johnsonii. Middle arrow points to a band identified as Clostridia Cluster XIV species. Bottom arrow points to a band identified as Bacteroides sp. (B) Banding similarity analysis of the representative samples from 10-week-old NOD-A, NOD-AtoN and NOD-N mice indicates that their microbiota share a 27.5% similarity. NOD-AtoN and NOD-N mice share a higher degree of similarity (53.7%) comparedwith NOD-A mice. Banding analysis was conducted using the Pearson correlation analysis. Figure S1 demonstrates that NOD-A mice have a lower pH throughout their GI tract. Pyrosequencing data were subjected to partial least squares discriminant analysis (PLS-DA). (C) Score scatter plot representing individual animals from each treatment, grouped based on the composition of fecal microbiota. The R2 and Q2 of the model were 0.97 and 0.85, respectively. (D) Bacterial taxa plotted using weighted PLS component 1 and 2. Genera in the plot closer to either treatment are more strongly associated to it. Genera found to significantly contribute to the model prediction are shown in green (NOD-N) and magenta (NOD-A). When a sequence could not be classified to the genus level, the closest level of classification was given, followed by F (family), O (order), C (class), or P (phylum). Additional data showing differences between NOD-A and NOD-N mice at the genus level can be found in Figure S3 and Tables S2, S3 and S4.
In order to quantitate these results, fecal DNA from 10-week-old female NOD-A (5) and NOD-N (5) mice from two different litters/mothers per group was purified and analyzed via 454 pyrosequencing (Table S1). NOD-A mice, although non-significant, had numerically higher richness and diversity. Distinct grouping of the treatments based on sequencing information was shown via partial least-squared discriminant analysis (PLS-DA) in Figure 2c. The goodness of fit and predictive value of the model was tested using R2 (0.97) and Q2 (0.85), respectively. Genera most characteristic of NOD-A and NOD-N mice are shown in Figure 2D, with significantly associated genera highlighted per treatment. NOD-N mice displayed significantly decreased levels of Firmicutes, the phylum to which Lactobacillus and Clostridia genera belong. Only 50.6% of the reads in NOD-N fecal DNA belonged to Firmicutes, whereas Firmicutes in NOD-A samples were responsible for 68.5% of reads (Table 1). There was no significant change in the populations of the phylum Bacteroidetes which contains the genus Bacteroides. From NOD-N fecal DNA, 26.7% of the reads belonged to Bacteroidetes compared with 22.7% in the NOD-A fecal DNA. However, the phyla Actinobacteria and Proteobacteria were significantly higher in the NOD-N than NOD-A mice (6.5% vs 1.7% and 10.2% vs 4.0%, respectively). The shifts observed in the microbiome of pre-diabetic NOD-N mice were similar to previously described shifts in diabetic patients (Brown et al. 2011). Additional data showing differences between NOD-A and NOD-N mice at the genus level can be found in Figure S3 and Tables S2, S3 and S4.
Table 1.
Relative Abundance of Bacterial Phyla in the Fecal Microbiome of NOD-A and NOD-N Mice Generated from the Pyrosequencing Data.
| Percentages of Sequences in: |
||||
|---|---|---|---|---|
| Phyla2 | NOD-A | NOD-N | SEM | P-value2 |
| -------------------------------above 1% of population------------------------ | ||||
| Actinobacteria | 1.71 | 6.57 | 1.20 | 0.02G |
| Bacteroidetes | 22.78 | 26.73 | 4.58 | 0.56G |
| Firmicutes | 68.55 | 50.63 | 5.27 | 0.04G |
| Proteobacteria | 4.08 | 10.25 | 1.17 | 0.007P |
| ------------------------between 0.1 and 1% of population------------------ | ||||
| Acidobacteria | 0.53 | 0.21 | 0.20 | 0.29G |
| Aquificae | 1.13E-6 | 0.93 | 0.22 | 0.98P |
| Chrysiogenetes | 0.73 | 0.10 | 0.18 | 0.03G |
| Cyanobacteria | 0.08 | 0.86 | 0.27 | 0.19P |
| Nitrospira | 0.08 | 0.05 | 0.11 | 0.87N |
| Planctomycetes | 0.08 | 0.05 | 0.11 | 0.87N |
| Spirochaetes | 1.13E-6 | 0.26 | 0.11 | 0.98P |
| Tenericutes | 0.56 | 1.05 | 0.24 | 0.19G |
| TM7 | 0.09 | 0.61 | 0.29 | 0.24G |
| Verrucomicrobia | 0.47 | 0.58 | 0.23 | 0.74G |
Unclassified bacteria account for 0.26% of NOD-A and 1.21% of NOD-N sequences.
Method of analysis denoted by G (Gaussian), N (Negative Binomial), and P (Poisson).
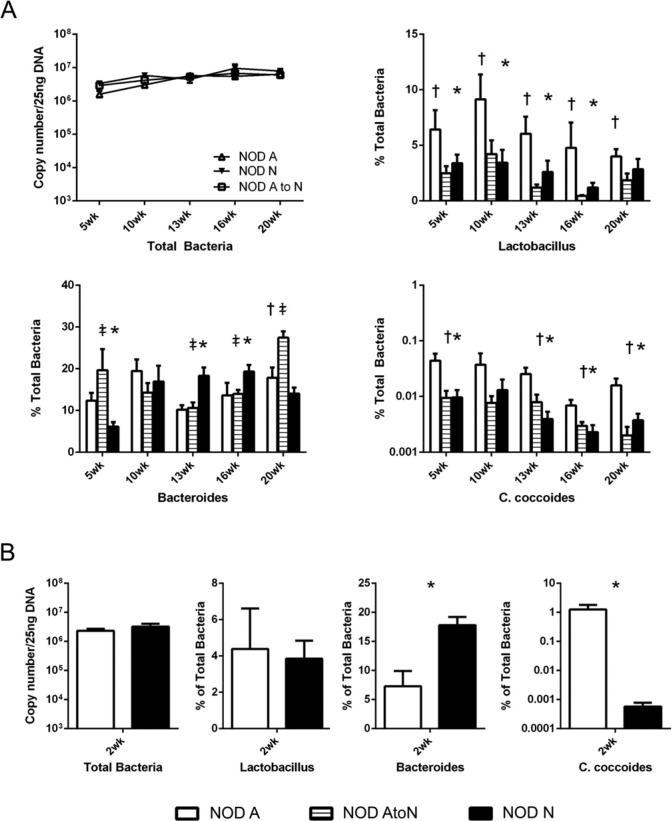
To evaluate if the differences in bacterial populations in 10-week-old NOD-N and NOD-A mice were maintained throughout the lifespan of the mouse, fecal DNA from NOD-N and NOD-A mice was subjected to qRT-PCR specific for individual groups of bacteria. The presence of Lactobacillus sp. and Clostridia coccoides (both Firmicutes), and Bacteroides sp. were investigated, as well a determination of total bacterial DNA to identify if there was a change in bacterial burden. Time points were chosen to represent pre-insulitis (5 weeks), post-insulitis, but before overt diabetes (10–13 weeks), and advanced progression of disease (16–20 weeks). There was little difference in the total bacterial populations between NOD-N and NOD-A mice over time (Fig. 3A). However, both Lactobacillus sp. and C. coccoides were significantly decreased at multiple ages in NOD-N mice compared with NOD-A mice (Fig. 3A). Conversely, there was no consistent change in the populations of Bacteroides sp. (Fig. 3A). In the NOD-AtoN mice, no significant differences were seen in the total bacterial numbers, but Lactobacillus sp. and C. coccoides from NOD-AtoN mice were significantly lower than their NOD-A counterparts and mirrored observations from NOD-N mice raised from birth on neutral water. Bacteroides populations were more similar to NOD-A mice and were significantly lower than that measured in NOD-N mice at 13 weeks and 16 weeks (Fig. 3A). Due to the observation that the NOD-AtoN mice did not completely mirror the diabetes susceptibility of the NOD-N mice, we studied 2-week-old neonatal mice to determine if there was a microbial dysbiosis early in life. At 2 weeks, there is a significant dysbiosis in NOD-N mice, with a >1000-fold decrease inC. coccoides and >50% increase in Bacteroides sp. as compared with NOD-A mice (Fig. 3B).
Figure 3.
qRT-PCR analysis of bacteria copy numbers in feces of NOD mice shows a significant dysbiosis in mice on neutral drinking water. (A) qRT-PCR analysis of the total copy numbers of NOD mice on acidified (NOD-A), acidified to neutral (NOD-AtoN), or neutral water (NOD-N) drinking water (upper left) at pre-insulitis (5 weeks), post-insulitis, but before overt diabetes (10–13 weeks), and advanced progression of disease (16–20 weeks). Bacterial populations of Lactobacillus sp. (upper right), Bacteroides sp. (lower left), and C. coccoides (lower right) in the feces of female NOD-A (white, n=8–10), NOD-AtoN (striped, n=5), or NOD-N, black, n=8–10) mice. (B) Analysis of fecal microbial populations between 2-week-old NOD-A and NOD-N mice comparing numbers of total bacteria, Lactobacillus sp., Bacteroides sp., and C. coccoides (NOD-A, n=4, NOD-N, n=5). Significance was determined using Welches’ t-test at p<0.05. Significance is indicated by *- NOD-A vs NOD-N, †- NOD-A vs NOD-AtoN, and ‡- NOD-N vs NOD-AtoN.
Level of Foxp3 Expression is Decreased in NOD-N Tregs
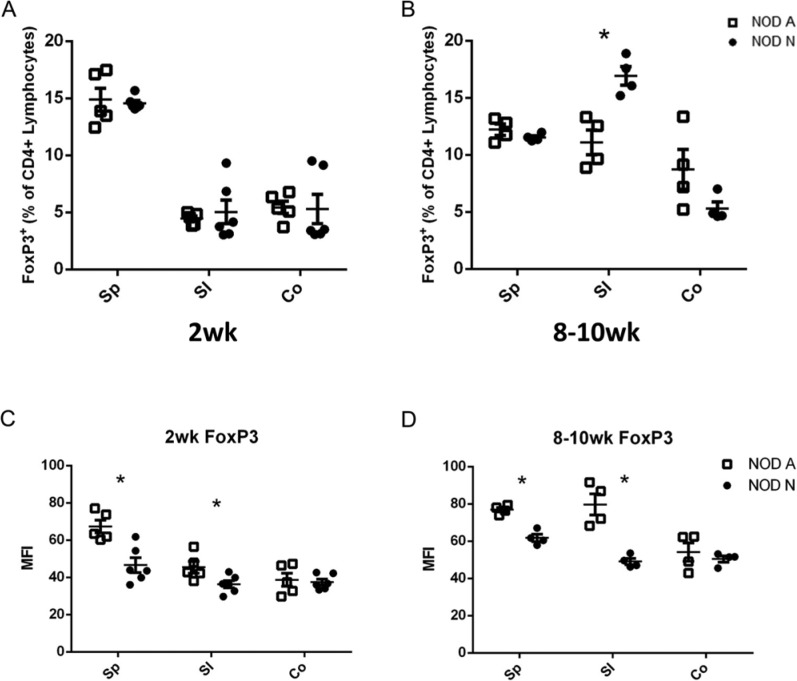
There is increasing evidence that autoimmune diseases, including T1D, are controlled by CD4+Foxp3+ Tregs (Sgouroudis and Piccirillo 2009). However, our results indicated that there was no discernible difference in the percentage or total number of CD4+Foxp3+ lymphocytes isolated from the spleen (Sp) or small intestinal (SI) and colonic (Co) lamina propria (LP) of 2-week-old NOD-A and NOD-N mice or in the Sp and Co LP of 8–10-week NOD-A and NOD-N mice (Fig. 4A, Fig. S2E). A significant increase in the percentage and total number of Foxp3+CD4+ Tregs in the SI LP of NOD-N mice was seen at 8–10 weeks (Fig. 4A; Fig. S2E). Recent data suggest that the level of Foxp3 expression in Tregs reflects their functional status, with increased expression being directly associated with increased regulatory function (Chauhan et al. 2009). Our data demonstrates that at both 2 weeks and 8–10 weeks, Foxp3 expression in CD4+ splenocytes and SI LP lymphocytes is significantly decreased (Fig. 4B). This indicates that in NOD-N mice, regardless of whether there is a similar (Sp) or higher (SI LP) number of Tregs, these cells should have decreased functional activity due to their lower Foxp3 expression. This conclusion is supported by the observation that NOD-A mice have an increase in colonic gene expression of the regulatory cytokine IL10 from 2–20 weeks, whereas there is no change in NOD-N mice (data not shown).
Figure 4.
NOD mice on neutral drinking water (NOD-N) have similar percentages of CD4+Foxp3+ Tregs, but decreased expression levels of Foxp3 as mice on acidic water (NOD-A). (A and B) The percentage of CD4+Foxp3+ Tregs in splenocytes (Sp) and small intestinal (SI) and colonic (Co) lamina propria (LP) lymphocytes from NOD-A and NOD-N mice at 2 weeks (NOD-A, n=5, NOD-N, n=6, left panel) and 8–10 weeks (NOD-A, n=4, NOD-N, n=4, right panel). (C and D) Mean fluorescence intensity (MFI) of Foxp3 expression in 2-week- (left panel) and 8–10-week-old (right panel) NOD-A and NOD-N mice. Significance was determined using Welches’ t-test at p<0.5. Absolute numbers of cells in each population can be seen in Figure S2.
NOD-N Mice have Decreased Expression of Intracellular IL17 in CD4+ T-cells
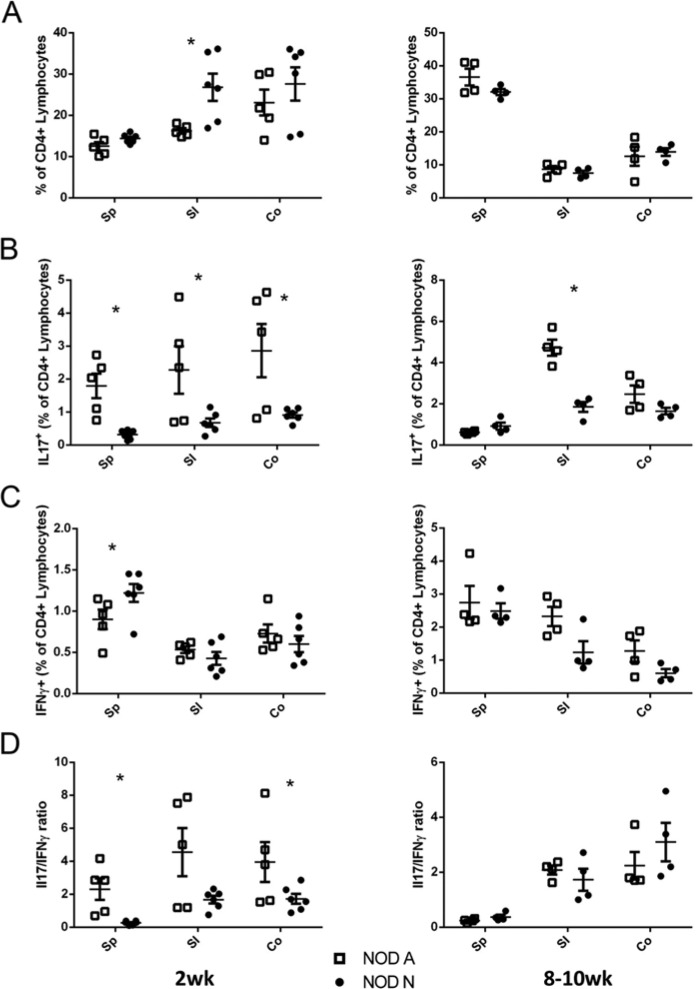
Several recent publications have demonstrated that components of the commensal microbiota, such as the common Firmicutes species, Lactobacillus and Clostridia, are able to induce IL17 expression and Treg cell expansion (Atarashi et al. 2011; Lau et al. 2011). However, the role of IL17 in autoimmunity and T1D is controversial, with reports showing both protective and pathogenic roles (Emamaullee et al. 2009; Kriegel et al. 2011; Lau et al. 2011; Nikoopour et al. 2010). To elucidate the effects of the dysbiosis in NOD-N mice, the SI and Co LP and Sp from 2-week- and 8–10-week-old NOD-A and NOD-N mice were analyzed via flow cytometry for the presence of CD4+ lymphocytes producing either IFNγ or IL17. The only significant difference in the percentage or total number of CD4+ cells was an increase in the NOD-N SI LP at 2 weeks of age (Fig. 5A and Fig. S2B). However, significant decreases were seen in the percentage of CD4+IL17+ cells within the Sp, SI and Co LP in 2 week NOD-N mice as compared with NOD-A mice (Fig. 5B). A decrease in the absolute numbers of CD4+IL17+ cells was also seen in the spleen and Co LP of NOD-N mice (Fig. S2B). By 8–10 weeks, decreased levels of CD4+IL17+ T cells were only seen in the SI LP of the NOD-N mice. There was no significant difference in the mean fluorescence intensity (MFI) of IL17 expression (data not shown). When the prototypical Th1 cytokine, IFNγ, was analyzed, the only significant change was an increase in CD4+IFNγ+ cells in the NOD-N Sp at 2 weeks (Fig. 5C). To determine the relationship between the Th17 and Th1 populations, we calculated the ratio of cells expressing IL17 to those expressing IFNγ. This analysis clearly demonstrated that NOD-N mice have dramatically fewer CD4+ T cells expressing IL17 than NOD-A mice (Fig. 5D). This altered ratio appears to be due to an increased percentage of CD4+ cells from NOD-A mice producing IL17 at 2 weeks. By 8–10 weeks, there was no difference in the ratio of IL17/IFNγ between the NOD-N and NOD-A mice. In addition, there was no histological inflammation seen in the colons of either the NOD-A or NOD-N mice (Figure S4).
Figure 5.
NOD mice on neutral drinking water (NOD-N) have decreased production of IL17 by CD4+ T cells as compared with mice on acidic water (NOD-A). (A) The percentage of CD4+ splenocytes and lymphocytes from the small intestine (SI) and colonic (Co) lamina propria (LP) of 2-week-old (left panel) and 8–10-week-old (right panel) NOD-A (open squares) and NOD-N (closed circles) mice. (B-C) Percentages of CD4+ lymphocytes that were also (B) IL17 or (C) IFNγ+. (D) Comparison of the ratio of CD4+IL17+ and CD4+IFNγ+ cells in 2-week-old (left panel) and 8–10-week-old (right panel) NOD-A and NOD-N mice. At 2 weeks: NOD-A, n=5; NOD-N, n=6. At 8–10 weeks: NOD-A, n=4; NOD-N, n=4). Significance was determined using Welches’ t-test at p<0.5. Absolute numbers of cells in each population can be seen in Figure S2.
Discussion
Microbial colonization of our gastrointestinal tract begins at birth, is primarily derived from maternal transmission, and profoundly impacts the development of the mucosal immune system (Dimmitt et al. 2010; McCracken and Lorenz 2001; Thum et al. 2012; Ubeda et al. 2012). The composition of this microbiota is shaped by the newborn diet, with breastfed babies having a more stable microbiome characterized by lower (more acidic) pH and higher lactic acid concentrations (Guaraldi and Salvatori 2012; Ogawa et al. 1992). As breastfed babies have a lower incidence of T1D, we designed a set of experiments to determine if this alteration in T1D risk could be correlated to changes in the microbiota and its subsequent impact on immunity. Our data demonstrates that the increased incidence of insulitis and diabetes in NOD-N mice is directly correlated with the changes in commensal microbiota caused by the shift from acidified to neutral H2O. This shift in microbiota is best characterized by a decrease in Firmicutes (including Lactobacillus sp. and Clostridia sp.) and increases in Bacteroides sp. prior to disease initiation.
There are thought to be two distinct phases of disease progression in NOD mice (Bettini and Vignali 2011; Eckenrode et al. 2004). Phase one occurs at 3–4 weeks of age and consists of peri-insulitis, whereas the second stage (8–12 weeks) involves progression to invasive insulitis (Cardell 2006). Insulin production becomes too low to regulate blood glucose levels after more than 90% of the pancreatic islet beta cells have been destroyed by this autoimmune attack, leading to the diagnosis of T1D. Our data indicates that the dysbiosis in 2-week-old mice caused by changing the gut pH (secondary to altered water sources for the dam) occurs prior to this first phase and implicates dysbiosis as an initial disease trigger. This idea is reinforced by the observation that the incidence of diabetes can be shifted by switching NOD mice from acidified to neutral water at weaning, but that the shift is not complete. Our data also support the conclusion that there are environmental exposures that occur early in life that play an important role in the risk of development of T1D, as it appears that the changes in the Lactobacillus sp., C. coccoides, and Bacteroides sp. early in life cause an imbalance in the mucosal immune system leading to increased susceptibility to diabetes. Other publications have demonstrated that expansion of Bacteroides sp. and a decrease in Clostridia sp. result in decreased colonic health and increased epithelial leakage, which lead to mucosal and systemic inflammation (Brinkworth et al. 2009; de Kort et al. 2011).
Others have shown that alterations to commensal microbial populations are strongly correlated with changes within the immune system (Ivanov et al. 2009). Our data indicate that 2-week-old NOD-N mice have a large decrease in IL17+CD4+ cells when compared with NOD-A mice. These findings suggest that, prior to the first phase of disease, Th17 populations confer protection against diabetes. This is consistent with multiple studies that have concluded that increased Th17 cells, induced by either various Firmicutes (including segmented filamentous bacteria (SFB) and Lactobacillus johnsonii) or by adjuvant immunotherapy, can delay the onset of T1D (Kriegel et al. 2011; Lau et al. 2011; Nikoopour et al. 2010). However, other studies in diabetic patients have shown that children with new onset T1D have higher numbers of Th17 cells (Ferraro et al. 2011; Marwaha et al. 2010). In addition, inhibition of IL17 after the initial phase of peri-insulitis (by either antibodies or diet change) has been shown to protect mice from T1D (Emamaullee et al. 2009). An explanation for these apparent contradictory reports may be that the reports on ‘protective’ Th17 cells were from models where these cells were present prior to the onset of T1D, whereas the ‘pathogenic’ Th17 cells were described in patients with diagnosed disease. This observation leads to the innovative paradigm that Th17 cells initially play a protective role, being induced by Firmicute colonization and contributing to the gastrointestinal epithelial barrier. However, after the stabilization of the microbiota and subsequent formation of the adult immune system, high levels of Th17 cells can then contribute to disease. It has been proposed that there are two different types of Th17 cells (classical and alternative) that are differentially developed in the presence of either TGFβ1 plus IL6 (classical/nonpathogenic) or TGFβ3 plus IL6 (alternative/pathogenic) (Lee et al. 2012). The colons of NOD-N mice show higher expression of the genes for both IL1β and IL6 at 2 weeks of age (compared with NOD-A colons from mice at 2 weeks; data not shown), which could indicate that even the small number of Th17 cells present may be of the alternative/pathogenic type instead of the classical/nonpathogenic type. This sub-classification of Th17 cells will need to be further explored in order to completely determine their role in disease.
The mechanism through which classical Th17 cells can protect NOD mice from T1D is unknown. Th17 cells can up-regulate intestinal epithelial barrier function, as well as help to promote effective contact-dependent suppression by Treg cells (Cao et al. 2012; Pappu et al. 2012). Although we did not witness any changes in frequency of Tregs between the NOD populations, we did find a significant decrease in the expression of Foxp3 in NOD-N Tregs. Lower levels of Foxp3 expression have been correlated with decreased function of Tregs in other models and this potential decrease in function may also contribute to disease in the NOD model, as functional defects in Tregs have also been described in patients with T1D (Ferraro et al. 2011).
We have noticed that NOD mice raised on acidified H2O in our facility have a lower incidence of diabetes compared with data for mice from Jackson Laboratories. This is probably secondary to microbiota differences and dietary changes between facilities (UAB diet = NIH-31; Jackson Laboratories diet = LabDiet 5K52/5K67). As there has been no previous complete sequencing of the microbiota of the NOD mouse, we are currently unable to compare the microbiota of our mice with other colonies. A recent publication demonstrated that natural colonization of NOD mice with segmented filamentous bacteria (SFB), caused attenuated disease progression/onset in mice; however, our NOD mice were tested and found to be SFB-negative (Kriegel et al. 2011).
The potent changes seen in the incidence of diabetes in NOD mice induced by something as simple as the pH of the water they drink clearly strengthens the hypothesis that subtle alterations within the GI microbiota early in development can have a significant impact on disease. It appears that the largest impacting factor in disease comes from the population shift between Firmicutes and Bacteroidetes within the GI tract and the subsequent induction of protective Th17 and Treg cells. Our research would suggest that whereas changes in microbiota initiated at weaning can alter the incidence of disease, the protective effect is truncated as compared with changes made at or before 2 weeks of age. Therefore, it may be necessary to expose children to protective bacteria prior to or immediately at the time of birth by giving pregnant mothers bacterotherapy. Infants whose mothers were taking probiotics before birth are colonized with the probiotic strains for at least 6 months after birth (Schultz et al. 2004; Thum et al. 2012). This could prove to be a novel and effective method in delivering protective bacteria to infants who may carry genetic susceptibility for diseases like T1D.
Acknowledgments
The authors would like to thank Wayne Duck and Dr. Charles O. Elson for assistance with DGGE and bacterial quantitation and Hubert Tse for advice on the NOD model. We would also like to thank Jamie McNaught for slide preparation and Mason Harris for animal husbandry, and members of the Lorenz lab for valuable advice. KJ.W. jointly conceived the study with R.G.L., performed all experiments, and prepared the initial draft of the manuscript; E.K. and R.H. did the 454 sequencing, analyzed the sequencing data, and helped prepare the manuscript; J.G.D. performed the histological and immunohistochemical analysis; S.M.T. completed lamina propria preparations on 2 week animals; R.G.L. analyzed the data and edited the manuscript.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported in part by the NIH grant P01 DK071176, the Juvenile Diabetes Research Foundation Grant #36-2008-930, the Crohn’s and Colitis Foundation of American Grant #26971, and the University of Alabama at Birmingham Digestive Diseases Research Development Center grant P30 DK064400. Aspects of this project were conducted in biomedical research space that was constructed with funds supported in part by NIH grant C06RR020136. Advice for statistical analysis was received from the UAB CCTS Biostatistics Clinic (UL1TR000165).
References
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, Taniguchi T, Takeda K, Hori S, Ivanov II, Umesaki Y, Itoh K, Honda K. (2011). Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331:337-341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson MA, Chervonsky A. (2012). Does the gut microbiota have a role in type 1 diabetes? Early evidence from humans and animal models of the disease. Diabetologia 55:2868-2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barman M, Unold D, Shifley K, Amir E, Hung K, Bos N, Salzman N. (2008). Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect Immun 76:907-915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettini M, Vignali DA. (2011). T cell-driven initiation and propagation of autoimmune diabetes. Curr Opin Immunol 23:754-760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw EM, Raddassi K, Elyaman W, Orban T, Gottlieb PA, Kent SC, Hafler DA. (2009). Monocytes from patients with type 1 diabetes spontaneously secrete proinflammatory cytokines inducing Th17 cells. J Immunol 183:4432-4439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkworth GD, Noakes M, Clifton PM, Bird AR. (2009). Comparative effects of very low-carbohydrate, high-fat and high-carbohydrate, low-fat weight-loss diets on bowel habit and faecal short-chain fatty acids and bacterial populations. Br J Nutr 101:1493-1502 [DOI] [PubMed] [Google Scholar]
- Brown CT, Davis-Richardson AG, Giongo A, Gano KA, Crabb DB, Mukherjee N, Casella G, Drew JC, Ilonen J, Knip M, Hyoty H, Veijola R, Simell T, Simell O, Neu J, Wasserfall CH, Schatz D, Atkinson MA, Triplett EW. (2011). Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PloS One 6:e25792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K, Decoffe D, Molcan E, Gibson DL. (2012). Diet-induced dysbiosis of the intestinal microbiota and the effects on immunity and disease. Nutrients 4:1095-1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao AT, Yao S, Gong B, Elson CO, Cong Y. (2012). Th17 cells upregulate polymeric Ig receptor and intestinal IgA and contribute to intestinal homeostasis. J Immunol 189:4666-4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 7(5):335-6. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardell SL. (2006). The natural killer T lymphocyte: a player in the complex regulation of autoimmune diabetes in non-obese diabetic mice. Clin Exp Immunol 143:194-202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter PB, Collins FM. (1974). The route of enteric infection in normal mice. J Exp Med 139:1189-1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan SK, Saban DR, Lee HK, Dana R. (2009). Levels of Foxp3 in regulatory T cells reflect their functional status in transplantation. J Immunol 182:148-153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kort S, Keszthelyi D, Masclee AA. (2011). Leaky gut and diabetes mellitus: what is the link? Obe Rev 12:449-458 [DOI] [PubMed] [Google Scholar]
- Dimmitt RA, Staley EM, Chuang G, Tanner SM, Soltau TD, Lorenz RG. (2010). Role of postnatal acquisition of the intestinal microbiome in the early development of immune function. J Pediatr Gastroenterol Nutr 51:262-273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd SE, Callaway TR, Wolcott RD, Sun Y, McKeehan T, Hagevoort RG, Edrington TS. (2008). Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol 8:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckenrode SE, Ruan Q, Yang P, Zheng W, McIndoe RA, She JX. (2004). Gene expression profiles define a key checkpoint for type 1 diabetes in NOD mice. Diabetes 53:366-375 [DOI] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194-2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emamaullee JA, Davis J, Merani S, Toso C, Elliott JF, Thiesen A, Shapiro AM. (2009). Inhibition of Th17 cells regulates autoimmune diabetes in NOD mice. Diabetes 58:1302-1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraro A, Socci C, Stabilini A, Valle A, Monti P, Piemonti L, Nano R, Olek S, Maffi P, Scavini M, Secchi A, Staudacher C, Bonifacio E, Battaglia M. (2011). Expansion of Th17 cells and functional defects in T regulatory cells are key features of the pancreatic lymph nodes in patients with type 1 diabetes. Diabetes 60:2903-2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giongo A, Gano KA, Crabb DB, Mukherjee N, Novelo LL, Casella G, Drew JC, Ilonen J, Knip M, Hyoty H, Veijola R, Simell T, Simell O, Neu J, Wasserfall CH, Schatz D, Atkinson MA, Triplett EW. (2011). Toward defining the autoimmune microbiome for type 1 diabetes. ISME J 5:82-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guaraldi F, Salvatori G. (2012). Effect of breast and formula feeding on gut microbiota shaping in newborns. Front Cell Infect Microbiol 2:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JE, White WJ, Lang CM. (1980). Acidification of drinking water: its effects on selected biologic phenomena in male mice. Lab Anim Sci 30:643-651 [PubMed] [Google Scholar]
- Hara N, Alkanani AK, Ir D, Robertson CE, Wagner BD, Frank DN, Zipris D. (2012). The role of the intestinal microbiota in type 1 diabetes. Clin Immunol 146:112-119 [DOI] [PubMed] [Google Scholar]
- Harrison D. (2008). Treatment data: distilled water. In http://research.jax.org/faculty/harrison/ger1vi_DistH20.html
- Hermann LM, White WJ, Lang CM. (1982). Prolonged exposure to acid, chlorine, or tetracycline in the drinking water: effects on delayed-type hypersensitivity, hemagglutination titers, and reticuloendothelial clearance rates in mice. Lab Anim Sci 32:603-608 [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. (2009). Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139:485-498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegel MA, Sefik E, Hill JA, Wu HJ, Benoist C, Mathis D. (2011). Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc Natl Acad Sci U S A 108:11548-11553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau K, Benitez P, Ardissone A, Wilson TD, Collins EL, Lorca G, Li N, Sankar D, Wasserfall C, Neu J, Atkinson MA, Shatz D, Triplett EW, Larkin J., 3rd (2011). Inhibition of type 1 diabetes correlated to a Lactobacillus johnsonii N6.2-mediated Th17 bias. J Immunol 186:3538-3546 [DOI] [PubMed] [Google Scholar]
- Lee Y, Awasthi A, Yosef N, Quintana FJ, Xiao S, Peters A, Wu C, Kleinewietfeld M, Kunder S, Hafler DA, Sobel RA, Regev A, Kuchroo VK. (2012). Induction and molecular signature of pathogenic TH17 cells. Nat Immunol 13:991-999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiter E. (1997). The NOD mouse: A model for insulin-dependent diabetes mellitus. In Current Protocols in Immunology. John Wiley & Sons, Hoboken, NJ: [DOI] [PubMed] [Google Scholar]
- Marwaha AK, Crome SQ, Panagiotopoulos C, Berg KB, Qin H, Ouyang Q, Xu L, Priatel JJ, Levings MK, Tan R. (2010). Cutting edge: Increased IL-17-secreting T cells in children with new-onset type 1 diabetes. J Immunol 185:3814-3818 [DOI] [PubMed] [Google Scholar]
- Mathis D, Benoist C. (2012). The influence of the microbiota on type-1 diabetes: on the threshold of a leap forward in our understanding. Immunol Rev 245:239-249 [DOI] [PubMed] [Google Scholar]
- Mayer EJ, Hamman RF, Gay EC, Lezotte DC, Savitz DA, Klingensmith GJ. (1988). Reduced risk of IDDM among breast-fed children. The Colorado IDDM Registry. Diabetes 37:1625-1632 [DOI] [PubMed] [Google Scholar]
- McCracken V, Lorenz R. (2001). The gastrointestinal ecosystem: a precarious alliance among epithelium, immunity and microbiota. Cell Microbiol 3:1-11 [DOI] [PubMed] [Google Scholar]
- Neu J, Lorca G, Kingma SD, Triplett EW. (2010). The intestinal microbiome: relationship to type 1 diabetes. Endocrinol Metab Clin North Am 39:563-571 [DOI] [PubMed] [Google Scholar]
- Nikoopour E, Schwartz JA, Huszarik K, Sandrock C, Krougly O, Lee-Chan E, Singh B. (2010). Th17 polarized cells from nonobese diabetic mice following mycobacterial adjuvant immunotherapy delay type 1 diabetes. J Immunol 184:4779-4788 [DOI] [PubMed] [Google Scholar]
- Ogawa K, Ben RA, Pons S, de Paolo MI, Bustos Fernandez L. (1992). Volatile fatty acids, lactic acid, and pH in the stools of breast-fed and bottle-fed infants. J Pediatr Gastroenterol Nutr 15:248-252 [DOI] [PubMed] [Google Scholar]
- Pappu R, Rutz S, Ouyang W. (2012). Regulation of epithelial immunity by IL-17 family cytokines. Trends Immunol 33:343-349 [DOI] [PubMed] [Google Scholar]
- Patelarou E, Girvalaki C, Brokalaki H, Patelarou A, Androulaki Z, Vardavas C. (2012). Current evidence on the associations of breastfeeding, infant formula, and cow’s milk introduction with type 1 diabetes mellitus: a systematic review. Nutr Rev 70:509-519 [DOI] [PubMed] [Google Scholar]
- Roesch LF, Lorca GL, Casella G, Giongo A, Naranjo A, Pionzio AM, Li N, Mai V, Wasserfall CH, Schatz D, Atkinson MA, Neu J, Triplett EW. (2009). Culture-independent identification of gut bacteria correlated with the onset of diabetes in a rat model. ISME J 3:536-548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537-7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz JM, Durham CG, Schoeb TR, Soltau TD, Wolf KJ, Tanner SM, McCracken VJ, Lorenz RG. (2011). Helicobacter felis-Associated Gastric Disease in Microbiota-Restricted Mice. J Histochem Cytochem 59:826-841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz M, Gottl C, Young RJ, Iwen P, Vanderhoof JA. (2004). Administration of oral probiotic bacteria to pregnant women causes temporary infantile colonization. J Pediatr Gastroenterol Nutr 38:293-297 [DOI] [PubMed] [Google Scholar]
- Sgouroudis E, Piccirillo CA. (2009). Control of type 1 diabetes by CD4+Foxp3+ regulatory T cells: lessons from mouse models and implications for human disease. Diabetes Metab Res Rev 25:208-218 [DOI] [PubMed] [Google Scholar]
- Tanner SM, Staley EM, Lorenz RG. (2012). Altered generation of induced regulatory T cells in the FVB.mdr1a-/- mouse model of colitis. Mucosal Immunol 10.1038/mi.2012.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thum C, Cookson AL, Otter DE, McNabb WC, Hodgkinson AJ, Dyer J, Roy NC. (2012). Can nutritional modulation of maternal intestinal microbiota influence the development of the infant gastrointestinal tract? J Nutr 142:1921-1928 [DOI] [PubMed] [Google Scholar]
- Turley SJ, Lee JW, Dutton-Swain N, Mathis D, Benoist C. (2005). Endocrine self and gut non-self intersect in the pancreatic lymph nodes. Proc Natl Acad Sci U S A 102:17729-17733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda C, Lipuma L, Gobourne A, Viale A, Leiner I, Equinda M, Khanin R, Pamer EG. (2012). Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J Exp Med 209:1445-1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaarala O, Atkinson MA, Neu J. (2008). The “perfect storm” for type 1 diabetes: the complex interplay between intestinal microbiota, gut permeability, and mucosal immunity. Diabetes 57:2555-2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valladares R, Sankar D, Li N, Williams E, Lai KK, Abdelgeliel AS, Gonzalez CF, Wasserfall CH, Larkin J, Schatz D, Atkinson MA, Triplett EW, Neu J, Lorca GL. (2010). Lactobacillus johnsonii N6.2 mitigates the development of type 1 diabetes in BB-DP rats. PloS One 5:e10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brandt J, Fischer HJ, Walter L, Hunig T, Kloting I, Reichardt HM. (2010). Type 1 diabetes in BioBreeding rats is critically linked to an imbalance between Th17 and regulatory T cells and an altered TCR repertoire. J Immunol 185:2285-2294 [DOI] [PubMed] [Google Scholar]