Abstract
High-throughput experimental technologies are generating increasingly massive and complex genomic data sets. The sheer enormity and heterogeneity of these data threaten to make the arising problems computationally infeasible. Fortunately, powerful algorithmic techniques lead to software that can answer important biomedical questions in practice. In this Review, we sample the algorithmic landscape, focusing on state-of-the-art techniques, the understanding of which will aid the bench biologist in analysing omics data. We spotlight specific examples that have facilitated and enriched analyses of sequence, transcriptomic and network data sets.
Biological data are exploding, both in size and complexity. High-throughput instruments are now routinely used in individual laboratories around the world in basic science applications as well as in efforts to understand and treat human disease. This trend towards the democratization of genome-scale technologies means that large data sets are being generated and used by individual bench biologists. Several software platforms and database systems have been developed for basic data analysis and integration1–3 (BOX 1). However, for anyone to extract biological insights from these data sets, familiarity with increasingly sophisticated computational techniques is required. Further complicating matters is that new genomic data are often best interpreted in the context of the heterogeneous large-scale data sets that have already been deposited in publicly available repositories. Finally, efficient means for storing, searching and retrieving data are of foremost concern as they are necessary for any analysis to proceed. Fortunately, an arsenal of algorithmic ideas — applicable in a wide variety of biological settings — can be deployed to address these challenges.
Box 1 | Platforms for biological data analysis.
Software platforms for biological data analysis
A long-standing challenge for practitioners is the proper use of software, including, for example, choosing suitable algorithms, installing software, setting correct parameters and assembling multiple programs into an integrative pipeline. These issues have become even more serious in the omics era, when high-throughput experiments have facilitated numerous large-scale analyses, thus requiring increasingly sophisticated computational tools. To address this need, multiple integrative software platforms have been developed with user-friendly interfaces. Some of these tools, such as Galaxy and Bioconductor, also aim to increase the ease of reproducibility of analyses. Computational scientists can then easily distribute their programs through such platforms. Some representative platforms are listed in the table.
Biological database systems
Similarly to the accessibility of computational tools, coordination of omics data sets can be difficult. Data sets are usually generated by different laboratories and can have different dimensionalities and organization. There have been substantial efforts towards formatting, storing and calibrating data sets, from the early Protein Data Bank, the US National Center for Biotechnology Information (NCBI) sequence data sets and the University of California, Santa Cruz (UCSC) Genome Browser164, to very recent consortia, such as the well-known ENCODE165 and modENCODE166 projects. To allow better sharing of data, several biological database systems have been developed to provide easy access to heterogeneous data sets for biologists. Many of these systems have also been integrated with software platforms, such as the ones mentioned above, so that researchers can build workflows for their analyses without writing extra code to integrate multiple programs.
| Name | Main function | Open source? | URL |
|---|---|---|---|
| Software platforms | |||
| Bioconductor | General purpose | Yes | http://www.bioconductor.org |
| Taverna | General purpose | Yes | http://www.taverna.org.uk |
| Galaxy | Sequence analysis | Yes | http://www.galaxyproject.org |
| GenePattern | General purpose | Yes | http://www.broadinstitute.org/cancer/software/genepattern |
| Cytoscape | Network analysis | Yes | http://www.cytoscape.org |
| BioDAS | Structural biology | Yes | http://www.biodas.org |
| Database systems | |||
| BioMart | General database | Yes | http://www.biomart.org |
| Addama | Heterogeneous database | Yes | http://www.systemsbiology.org/addama |
| SDCubes | Heterogeneous database | Yes | http://www.semanticbiology.com/software/sdcube |
Nowhere is the data deluge more apparent than in the area of high-throughput sequencing. In the past two decades, improvements in genomic sequencing capability have led to an exponential growth in the amount of publicly available sequence data that far outstrips the growth predicted by Moore’s law4–6. Moore’s law says that computing power and storage capacity doubles every 18 months, whereas the volume of new sequence data has grown tenfold every year since 2002 (REFS 7–11). The widening gap between data generation and computing power implies that many of our established ways of analysing smaller data sets simply cannot scale, not even with faster computers or with cloud computing or parallel computing. Further, the increasing diversity of experimental techniques, the high dimensionality of the resulting data, the noise in high-throughput measurements and the nature of the underlying biology result in substantial additional challenges in omics data analyses. The goal of this Review is to highlight a range of fundamental algorithmic ideas that have been successful in tackling omics data sets and that serve as a launching point for extracting biological insights from these data. We focus on applications in three diverse but important areas — sequencing, transcriptomics and networks — as each showcases a distinct aspect of what we believe are the main computational challenges facing us: algorithmic efficiency to handle large data sets, sensitive signal extraction from multidimensional data and contextualization of new data within existing data sets. In this Review, we primarily focus on algorithms for these three areas, whereas other important challenges, such as metagenomic12 and proteomic analysis13, will not be covered. Further, within our areas of focus, it is our hope that a survey of the underlying computational techniques will be helpful in guiding practitioners in the analysis of their data sets.
We begin with a survey of problems that arise in high-throughput sequencing. We consider problems that arise at multiple stages in the assembly, mapping, storage and retrieval pipeline. We show how algorithmic insights involving sophisticated data structures, graph algorithms and data compression can be deployed to attack some of the computational bottlenecks in sequencing. Next, we describe sensitive data mining and machine learning techniques for making sense of transcriptional data sets and look at their applications to translational biomedicine. Finally, we discuss algorithms for integrating heterogeneous omics data within the context of biological networks, for which a rich set of graph-theoretical formulations and algorithms can be leveraged. We close with a brief discussion of related areas of research that are in need of better computational tools as omics data accumulate, as well as challenges that remain in developing such tools and disseminating them to the biological community. Note that links to Web pages and references for all of the software packages described in this paper are listed in TABLE 1.
Table 1.
Representative software
| Software | Data sets | Techniques | Problem | Availability | Refs |
|---|---|---|---|---|---|
| Storage and retrieval of large data sets | |||||
| CaBLAST and CaBLAT |
Genomic sequences and database |
Edit script compression | Genome sequence search |
http://cast.csail.mit.edu | 45 |
| BWA | NGS reads, reference genomes |
Burrows–Wheeler transformation |
Read alignment | http://bio-bwa.sourceforge.net | |
| Bowtie and Botwie2 |
NGS reads, reference genomes |
Burrows–Wheeler transformation |
Read alignment | http://bowtie-bio.sourceforge.net | 31,32 |
| SOAP and SOAP2 | NGS reads, reference genomes |
Burrows–Wheeler transformation |
Read alignment | http://soap.genomics.org.cn | 35,168 |
| mrsFAST | NGS reads, reference genomes |
Cache-oblivious data structures and algorithm |
Short read alignment | http://mrsfast.sourceforge.net | 38 |
| ABySS | NGS reads or sequence fragments |
De Bruijin graph | De novo assembly | http://www.bcgsc.ca/platform/bioinfo/software/abyss | 24 |
| Velvet | NGS reads or sequence fragments |
De Bruijin graph | De novo assembly | http://www.ebi.ac.uk/~zerbino/velvet | 21 |
| SGA | NGS reads or sequence fragments |
FM-index | De novo assembly | https://github.com/jts/sga/wiki/SGA-Design | 26 |
| Cactus | Whole-genome sequences |
Cactus graph | Multiple sequence alignment |
http://hgwdev.cse.ucsc.edu | 169 |
| Data mining for transcriptomics | |||||
| IDA | Gene expression and perturbations |
Causal graphical models | Building regulatory network |
http://cran.r-project.org/web/packages/pcalg | 89 |
| Concordia | Multiple gene expression data sets |
Recursive PCA | Gene expression analysis |
http://concordia.csail.mit.edu | 106 |
| CONEXIC | Gene expression and CNV data sets |
Module analysis | Cancer driver gene identification |
http://www.c2b2.columbia.edu/danapeerlab/html/conexic.html | 88 |
| PEER | GWAS and gene expression data |
Sparse signal recovery | Interpretation analysis of gene expression |
http://www.sanger.ac.uk/resources/software/peer | 95,96 |
| PARADIM and PARADIGM-SHIFT |
Gene expression, CNVs and pathways |
Bayesian networks | Pathway analysis | http://sbenz.github.com/Paradigm | 97,98 |
| Integrative interactomics | |||||
| HotNet | Networks and mutation data |
Flow and diffusion | Cancer subgraphs | http://compbio.cs.brown.edu/projects/hotnet | 148 |
| ResponseNet | Networks, expression data, genetic screens |
Flow-based approach | Pathway reconstruction |
http://bioinfo.bgu.ac.il/respnet | 122 |
| IsoRank and IsoRankN |
Networks across organisms |
Spectral graph algorithm, graph matching |
Network alignment | http://isobase.csail.mit.edu | 134,135, 140 |
| MATISSE | Networks and expression data |
Integrative subnetwork detection |
Subnetwork detection | http://acgt.cs.tau.ac.il/matisse | 155 |
| SPICI | Weighted networks | Local network clustering | Subnetwork detection | http://compbio.cs.princeton.edu/spici | 118 |
| STEINERNET | Networks and seed set of proteins |
Steiner tree | Subnetwork detection | http://fraenkel.mit.edu/steinernet | 124 |
| NetGrep | Networks and numerous protein annotations |
Frequent pattern mining | Pattern search in networks |
http://genomics.princeton.edu/singhlab/netgrep | 133 |
| DAPPLE | Networks and disease loci or disease genes |
Permutation test | Disease subnetwork association |
http://www.broadinstitute.org/mpg/dapple/dapple.php | 143 |
| PrincePlugin | Networks and disease genes |
Network flow | Finding disease- associated genes |
http://www.cs.tau.ac.il/~bnet/software/PrincePlugin | 145 |
| DADA | Networks and disease genes |
Random walk | Finding disease- associated genes |
http://compbio.case.edu/dada | 147 |
| NetBox | Networks, mutations and CNVs |
Shortest path and clustering |
Cancer subnetworks | http://cbio.mskcc.org/downloads/index.html | 149 |
BWA, Burrows–Wheeler Aligner; CNV, copy number variant; GWAS, genome-wide association study; NGS, next-generation sequencing; PCA, principal component analysis.
Processing, storage and retrieval
Efficient processing, storage and retrieval of large-scale sequencing data sets are crucially important for modern ‘big-data-driven’ life science. In this section, we describe current solutions to these problems on the basis of state-of-the-art approaches for indexing large-scale genomic data (BOX 2; FIG. 1). The key underlying idea of these approaches is that by smart pre-processing, it is possible to store sequence data in a form that makes subsequent computations significantly faster.
Box 2 | Indexing techniques for sequencing data.
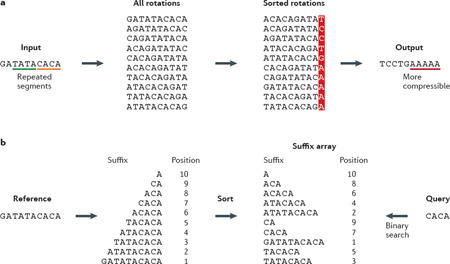
Burrows–Wheeler transformation (BWT) is a string transformation that converts highly redundant sequences (for example, the human genome) into a format that can easily be compressed (see below). It is carried out by: generating all rotations of the input such that each position becomes the starting position exactly once; sorting the rotated sequences in alphabetical order; and extracting the last column of all sorted rotations as the output sequence. If there are substrings that occur multiple times in the full sequence, then the transformation will cluster these substrings together, resulting in single-character repetitions in the final column; hence, the repetitive structure of the output sequence facilitates compression. Furthermore, BWT allows an efficient inverse transformation that can fully recover the original sequence from the transformed output. These features thus enable BWT to be a useful pre-processor for lossless data compression, such as bzip2 and Huffman encoding.
As opposed to rotations used in BWT, a suffix array is a sorted array that indexes all possible suffixes of a sequence (see below). The array is constructed by sorting all of the suffixes alphabetically. The order of each entry in the suffix array represents the ranking of that suffix in the sequence. By taking the similarity among suffixes, the suffix array can be constructed quite efficiently in practice. After a suffix array has been built, queries can be carried out by many different algorithms. For instance, binary search compares the query string and the middle element of the array and repeats the search on the left or right subarray according to the comparison; this requires only time O(m log(n)), where m is the length of the query, and n is the length of the original sequence167. As read length is typically very small (m < 100), and as the reference genome length n is substantial, the query time for the suffix array is significantly faster than naive sequence matching, which requires O(m n) time. Although further algorithmic advances have increased the speed of suffix array queries to O(m) time, the memory required for a suffix array for the whole human genome is very expensive; all suffixes need to be stored, and thus the size of the suffix array would be much larger than the size of the genome.
Advantageously, the FM-index is a hybrid of BWT and suffix arrays. The FM-index contains the information from all suffixes of the original sequence and allows fast subsequence mapping and counting in O(m) time36. See the original text in REF. 36 for technical details and illustrations on construction and query operations36 (also see Figure 1 of REF. 30). Compared with the space requirement of suffix arrays, only O(n) space is needed for the FM-index on a genome of length n36. These features make the FM-index ideal for short-read mapping, where the read length m is usually quite small, and the size of the reference genome is large. In addition to read mapping, this data structure has been applied for genome assembly26.
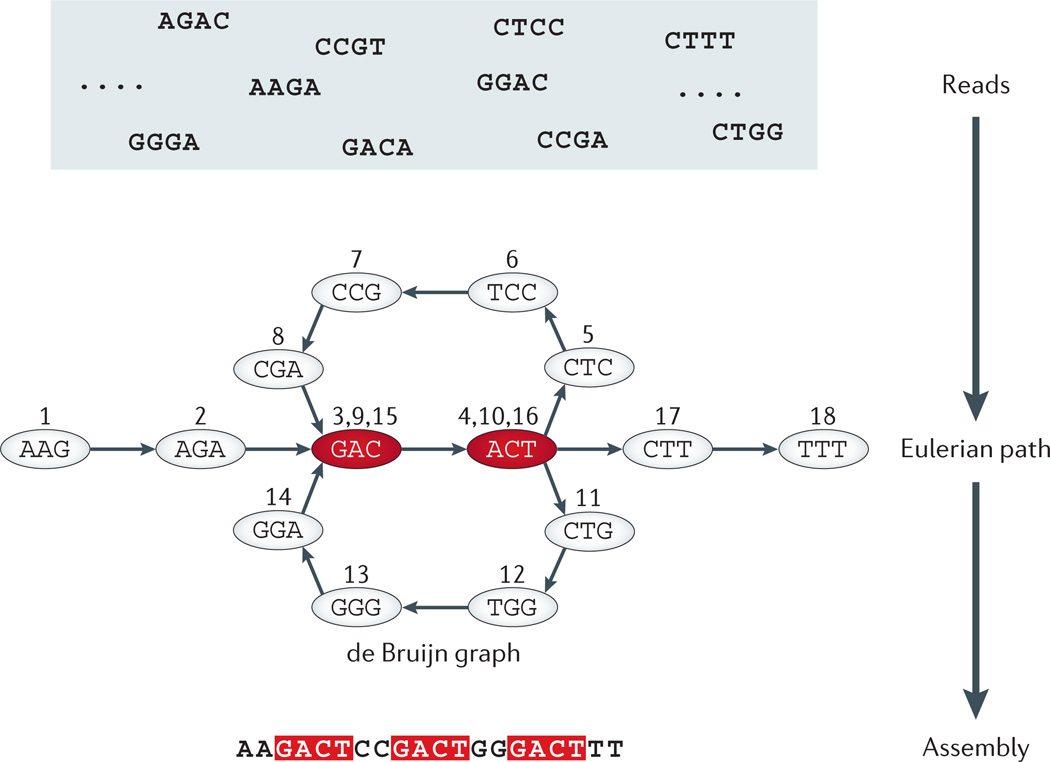
Figure 1. De Bruijn graph of DNA sequence assembly.
Each directed edge in a de Bruijn graph denotes a sequence read or a fragment of fixed length (4 bp in the figure); the source node of this edge is a prefix string of the read omitting the last nucleotide; the destination node of this edge is a suffix string of the same read (or sequence fragment) by omitting the first nucleotide. In the example shown in this figure, the top panel is a pool of representative short reads or fragments. In the middle panel, each node denotes a unique sequence prefix or suffix segment of length 3 bp found in the original reads of length 4 bp. The assembly of DNA sequences (segments) is thus represented as a de Bruijn graph. Assembling reads (or sequence fragments) in a de Bruijn graph reduces the problem to a fragment assembly problem that can be formulated as the goal to find a trail or Eularian path that visits each edge (read or fragment) in the (de Bruijn) graph exactly once. Nucleotides with a red background occur more than once in the sequence. Numbers on the edges represent an ordered Eulerian path through the de Bruijn graph, which can be followed to reconstruct the assembled sequence from the compact graph representation.
Genome assembly
Next-generation sequencing technologies, including 454, Illumina, SOLiD and ion semiconductor, can now yield hundreds of millions of short-read sequences (snippets of genomic sequences of typically less than several hundred base pairs)14,15 per human genome. With these evolving technologies, several important computational challenges have emerged. Among them, genome assembly is one of the most fundamental problems to address. Before any kind of genomic analysis can commence, it is important to generate a template sequence (that is, a reference genome) de novo, to which sequences from individuals and/or species can be compared against and with which variations can be analysed. Although the quality, depth and coverage of sequencing technologies have vastly advanced over the past several years, genome assembly from sequencing data remains a challenging task16,17. Accurate genome assembly requires sequencing at high depth, and assembling millions of these short reads into a full-length genome is computationally difficult as for each read, contiguous sequences need to be identified from a large unstructured pool of short reads. In addition to fast assembly algorithms, efficient storage techniques are especially important when carrying out assembly of large genomes, in which sequencing data can be in the terabytes.
Instead of comparing all possible pairs of sequence segments, most efficient assemblers, such as EULER18, ARACHNE19,20, Velvet21, SOAPdenovo22, ALLPATHS23 and ABySS24, have been developed using a graph-based data structure: the de Bruijn graph (FIG. 1). Assembling reads in a de Bruijn graph reduces fragment assembly to the classical graph-theoretical Eulerian path problem18,25. In this scenario, the goal of the Eulerian path problem is to find a trail (that is, a genome sequence or contig) that visits each edge (short read or sequence fragment) in the (de Bruijn) graph exactly once. There is a linear time algorithm for finding an Eulerian path in a de Bruijn graph that assembles contigs from sequence segments or reads. Finally, the assembled contigs are merged into a full-length genome sequence (see, for example, FIG. 1). Consequently, any such graph-based assembly algorithm will still take time at least linear in the number of reads and will require a substantial amount of memory.
Recently, several crucial evaluations of numerous popular genome assemblers, including de Bruijn-graph-based21–24 and FM-index-based (see BOX 2 and ‘Read mapping in next-generation sequencing’) assemblers26, have been carried out on four genomes with a wide phylogenetic range and varying degrees of difficulty27–29. Although these graph-theoretical methods have substantially advanced genome assembly, they still have great difficulty accurately assembling large genomes, such as the human genome; different algorithmic strategies lead to different trade-offs between correctness and contiguity (which is typically a measure of how much of the genome long contigs span). In large genomes, genomic repeats often introduce an exponential number of valid Eulerian paths in the assembly graph30. This complexity poses additional computational challenge for assemblers in choosing the best paths by using long-range genomic information, such as mate pairs (that is, two reads from the same clone) and other scaffolding information. Thus, the state of the art would seem to call for further improvements in sequencing technology, as well as algorithmic advances in assembly.
Read mapping in next-generation sequencing
Smart algorithms are also required to identify the genomic origin of sequencing reads (that is, to map reads to a reference genome). The naive string-matching approach, which compares reads with the whole-genome sequence for each nucleotide, would incur huge running times owing to the substantial number of reads to be aligned to a genome. To reduce the high computational cost, one solution is to pre-process the genome into a flexible and compact data format that allows fast indexing and alignment techniques, such as the FM-index (BOX 2).
The Burrows–Wheeler Aligner (BWA), Bowtie and SOAP31–35 are arguably the most widely used short-read alignment software programs. The core technique used by these programs is the FM-index, which is a compressed data structure for sequence data36 (BOX 2). By constructing a suffix-array-like data structure from a Burrows–Wheeler-transformed37 reference genome, the FM-index compactly represents its sequence more efficiently than standard suffix arrays and simultaneously indexes the reference genome for fast access and mapping (BOX 2). After indexing, the time required for read mapping is sublinear with respect to the size of the reference genome but at least linear with respect to the read data size31–35. The storage space requirement is linear with respect to the size of the reference genome, but it can be compressed to save space. In addition to read mapping, this data structure has also been applied for genome assembly26.
Hardware-accelerated algorithms are also used to speed up large-scale, but basic, arithmetic operations in read mapping. The recently developed Bowtie2 (REF. 31) implements parallel dynamic programming by fully exploiting the computational power of modern multicore central processing units (multicore CPUs), thereby accelerating gapped long-read alignment several-fold. Another read-mapping program, mrsFast38, uses a cache-oblivious algorithm. Together with efficient indexing data structures, these sophisticated computer algorithms make various large-scale whole-genome computational tasks — from read mapping to downstream analyses, such as structural variation detection and SNP base calling — possible even on personal computers.
Large-scale genome sequence compressed storage and search
As sequencing data rapidly accumulates, one challenge is to reduce the size of this data for storage and processing. The obvious place to turn is to algorithms that compress these genomic data sets, and in fact many such compression algorithms exist to reduce the space required for storage and transmission39–44. Reference-based compression methods39,45 align read sequences to a reference genome and then store only the differences between the new sequence and the reference genome. Such methods are ideal for the resequencing of well-studied genomes, and their compression factor increases almost linearly with the number of genome sequences. Non-reference-based methods, however, usually rely on string compression algorithms, which exploit repetitive DNA segments; most of them use well-known text compression algorithms, such as gzip, BWT and LZ77, in their implementations40,42,46,47. As an improvement tailored to sequencing data sets, SCALCE47 uses a locally consistent parsing technique that reorganizes the reads such that compression algorithms can achieve higher compression speed and rates. These string-based approaches can compress read data sets by factors of 2 to 15.
Compressive storage, however, addresses only a part of the problem, because these techniques generally require the data to be decompressed before computational analysis. In addition, any computational analysis, such as sequence search, that runs on the full genomic library — or even a constant fraction thereof — scales at least linearly in time with respect to the size of the library, and therefore these analyses effectively grow exponentially slower every year. Popular search algorithms such as BLAST48 are becoming too slow, and BLAT49 is soon to follow. To address this crucial problem, the paradigm of ‘compressive genomics’ was recently introduced45, whereby data are compressed in such a way that they can be efficiently and accurately searched without decompressing first (FIG. 2).
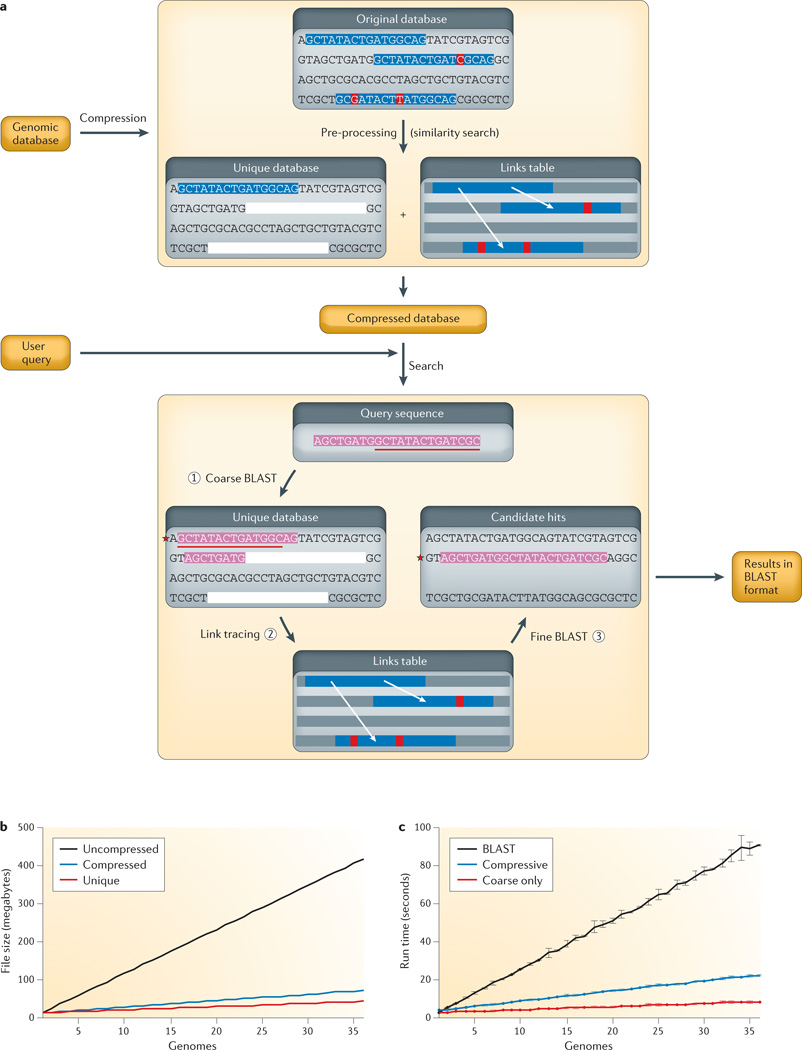
Figure 2. Application to sequence search.
a | Flow chart of CaBLAST. First, redundancy in the genomic database is identified and removed to create a unique database consisting of a smaller set of segmental exemplars. Only the first occurrence of repetitive sequence segments is retained, as represented by the blue background, and other similar regions are removed (blanked out in the pre-processing step) and encoded in a links table to their original locations. The red background indicates locations of unique bases within the repetitive element. An edit script compression technique45 is used to encode similar sequence fragments with reduced storage in a compressed database. After the compressed database is constructed, a coarse-to-fine strategy is adopted for sequence search. First, a BLAST search is run using the query sequence (shown with a pink background) against the unique database with a relaxed E value threshold to identify high-scoring hits. Second, the additional candidate hits are recovered by tracing through the links table. Third, BLAST is run against the final candidate hits. b,c | CaBLAST storage requirements and running time comparisons on uncompressed (black) and compressed (blue, full compressed database; red, only unique database or coarse search) genome sequence databases consisting of 1 to 36 yeast genomes45.
Compressive genomics exploits the redundancy of genomic sequences to allow parsimonious storage and fast access45. For example, as human genome sequences differ on average by only 0.5%50, 200 human genomes contain less than twice the unique data of one genome, as measured by the number of nucleotides of one full genome plus the nucleotides differing from it in the rest of the genomes (that is, the nucleotide substitutions, insertions, deletions and rearrangements that account for the change in each of the remaining genomes). Thus, although individual genomes are not very compressible, collections of related genomes are extremely compressible45. Intuitively, given highly similar genomes, any analysis carried out on one genome accounts for much of the computational work towards the same analysis on the others. These compressed large genomic data sets can be analysed with new algorithms that operate solely on the compressed non-redundant data without decompressing it. FIGURE 2 presents the CaBLAST and CaBLAT compressive genomics algorithms for BLAST and BLAT that have a similar accuracy to BLAST and BLAT yet have runtimes that scale sublinear to the total size of genomic data and almost linear to that of the non-redundant data.
High-throughput sequencers also provide a powerful approach for transcriptome quantification by RNA sequencing (RNA-seq)51–53. By sequencing the whole transcriptome, researchers are able to identify transcripts and to estimate gene expression levels. As with short-read DNA sequence analysis, short reads are either mapped to reference genomes and transcriptomes54 or assembled de novo55, before transcript counting and data normalization. The unique characteristics of RNA-seq data present substantial algorithmic challenges in their analysis. For instance, the existence of novel gene fusions and alternative splicing make mapping and assembly of RNA-seq data extremely difficult owing to ambiguity in read mapping54,55. Bioinformatic analysis of RNA-seq data sets is still in its infancy; in addition to quantification of expression levels, substantial work remains to be done on interpreting these data56.
Data mining for transcriptomics
RNA-seq51,52 and microarray57 experiments have produced large repositories of high-dimensional transcriptomic data58. Challenges include identifying cell-specific expression signals within tissue profiles, identifying regulatory and phenotypic genes and modules, and integrating multiple expression data sets for disease-related analysis. In this article, we focus on some of the most recent algorithmic developments in data modelling to decipher and to integrate multiple experiments over transcriptomic data sets. Also relevant are excellent review articles on more traditional expression analysis59,60, data normalization61 and RNA-seq data analysis62,56 (for example, integrated pipelines such as Tophat, Cufflinks and Cuffdiff54,63–65).
Identifying cell-specific expression signals
Heterogeneity of cell types may confound gene expression analysis. Transcriptomic expression methods, such as microarray techniques, require a large quantity of mRNA to obtain reliable expression levels. As a result, the tissue samples used for mRNA preparation often consist of several different cell types. Thus, expression levels based on tissue samples with varying cell type compositions are difficult to compare with or to interpret66,67. This issue is particularly problematic when carrying out differential analysis between complex disease and normal samples in clinical studies.
Similarly to deconvolution methods that are used in digital signal processing, and that were first used in electrical engineering, researchers have developed approaches to identify expression signals for each cell type from an expression study. Linear algebraic methods62 have proved to be effective for this task. These methods require measured cell type proportions. These proportions of cell types are used to weight a linear mixed model, which is a statistical model that is fit on the basis of the assumption that the overall observed expression signal can be constructed from a linearly weighted sum of the expression levels from each cell type (weighted according to the proportion of cells in the sample). After the model is fit, the expression profiles for each cell type are identified from overall expression signals. A similar method has also been designed to estimate the tissue components from surgical samples68. When the mixed cell type proportions are not available, methods based on matrix factorization69 or differential geometry70 have been developed to estimate simultaneously both mixture proportions and expression profiles for each component cell type.
Identifying regulatory genes and modules
Perhaps the most fundamental type of analysis is to detect the differential expression of gene sets in conditions of interest to infer key genes and pathways (for example, by identifying regulatory genes and linked pathways or to implicate genes or pathways in a disease-based analysis). To accomplish these tasks, numerous statistical methods, such as gene set enrichment analysis (GSEA)71, GenePattern72, joint clustering73,74 and DEseq75, have been devised and widely applied to analyse differentially expressed genes and gene sets56,57. A gene module consists of a group of genes that jointly carry out specific biological functions. Module discovery seeks to identify differentially expressed genes or dysregulated pathways in disease states, along with the regulatory relationships between them. To mine pathways and/or modules from transcriptome data sets, researchers have developed a number of mathematical models76–88. Among the most popular are probabilistic graphical models, which describe a distribution that can explain the observed transcriptome data. The nodes in the graph represent the genes (or modules), and the edges define the relationship between two genes (or modules). Graphical models can thus describe and uncover putative interactions between genes or modules and have been applied to these types of problems arising in gene expression analysis.
An important question is what the regulatory relationships or co-expression patterns are among genes. Over the past decade, graphical models have been extensively applied to this problem. IDA (for ‘interventional calculus when directed acyclic graph is absent’)89 and nested effects models90 are two recently developed graphical models that construct putative regulatory relationships between genes from transcriptomic data. Sparse learning is another recently introduced mathematical concept used to mine gene or module regulatory patterns. The key idea is that the gene regulatory network is sparsely structured; that is, the expression of any gene is directly regulated by only a few other genes, and this allows a concise representation of genes that explain the differential phenomena in gene expression. SPARCLE (for ‘sparse recovery of linear combinations of expression’) is a machine learning method that finds a gene set of minimum size such that its expression profile linearly fits the given genes of interest91. For example, genes that regulate specific pathways would correlate linearly with their targets. This idea is thus formulated as a compressed sensing problem and is solved through linear programming. This method is purely unsupervised (that is, no training data are required). In contrast to principle component analysis (PCA)92,93 or correlation-based methods93,94, SPARCLE is able to find robust gene sets of much smaller size from high-dimensional transcriptomes (that is, transcriptomes in which a large number of gene expression changes are observed), such that they can provide potential biological context for the given genes of interest. Experimental results also indicate that SPARCLE outperforms correlation-based approaches in predicting protein–protein interactions and genetic associations91. The sparse factor analysis method PEER95,96 has been developed to infer a small set of ‘hidden cellular phenotypes’ or expression patterns in the gene expression data that can explain the highest variability in gene expression across multiple samples. Similarly to SPARCLE, PEER enforces a sparsity constraint on the size of the hidden cellular phenotypes. By incorporating biological prior knowledge, the derived cellular phenotypes can be used to infer pathways or transcription factors.
Identifying gene expression alterations in disease
With the recent accumulation of cancer genomic data sets, another important problem is to identify genes and modules in two-way comparisons between tumour cells and normal cells76,88,97,98. For example, CONEXIC compares gene expression data in cancer tissue and normal tissue by extending the causal graphical model76, which connects gene sets with edges to represent their interactions, to elucidate dysregulated gene modules from cancer genomic data88. By leveraging gene expression data and the corresponding copy number variants (CNVs), CONEXIC builds such module networks to distinguish between the CNV-affected genes, which are believed to be main drivers of cancer, and abnormally regulated genes, which are affected by the dysregulation of gene expression and are difficult to identify.
Two other popular pieces of software, PARADIGM and PARADIGM-SHIFT, implement a Bayesian network to construct pathways from cancer transcriptomic profiling data sets97,98. By taking pathways into account, PARADIGM can identify weak but clinically relevant signals, which are often overlooked when only single genes are considered. PARADIGM-SHIFT has been extensively applied in recent cancer genomic research studies, including The Cancer Genome Atlas (TCGA)99–101. In the future, these ideas may find use in multiclass (that is, beyond simply case versus control studies) and cross-study analyses as well.
The lack of standardized nomenclature and annotation methods has made large-scale, multi-phenotype analyses of multiple tissues and disease states difficult. Large-scale gene expression investigations have had preliminary success at elucidating phenotypic gene expression signals102–104 and applying those signals to downstream analyses, such as drug repurposing104,105. However, such approaches still directly measure transcriptional differences between two phenotypes, inherently imposing subjective decisions about what constitutes an appropriate control population. This presupposition can limit the scope of such analyses to differentiate between biological processes that are unique to a particular phenotype or part of a larger process that is common to multiple phenotypes (for example, a generic ‘cancer pathway’). To address this limitation, one recently developed approach is Concordia106. Here, a ranked list of marker genes for a given complex phenotype is generated by querying a gene expression database sorted by gene expression intensity for each phenotype106,107. Concordia is able to classify tissue types with a high degree of accuracy, such as metastasized tumour samples, which strikingly resemble their tissue of origin106. Similar methods have also been developed with linear algebraic techniques to analyse existing disease signatures108 and to identify gene modules across multiple gene expression data sets109.
Integrative interactomics
Transcriptomic and other complex functional genomics data sets that are arising from high-throughput experimental biology benefit from analysis in the context of known cellular networks, which provide a holistic framework for interpretation. Although far from complete, large-scale networks have been determined for numerous organisms, including humans and other model organisms110–114. These networks, or interactomes, are commonly represented as graphs, in which nodes correspond to biological components (for example, genes, RNAs, proteins or metabolites), and edges correspond to known interactions among them (for example, physical, regulatory or genetic). Integrative interactomics analyses are typically premised on modularity, which is a key organizational property of cellular networks in which molecules that work together to carry out a specific biological process are enriched in interactions among themselves115.
Analysis of heterogeneous genomic data sets
Protein–protein and regulatory interaction networks provide a physical ‘scaffold’ with which to uncover modules that are specific to conditions of interest. Early pioneering work, now implemented as the jActive modules plugin for Cytoscape3, introduced the concept of active subnetworks, which consist of connected regions in physical interaction networks that manifest significant expression changes in specific contexts116. Numerous variations of this idea have since been introduced. For example, JACS allows for an arbitrary measure of similarity (for example, including, but not limited to, co-expression) between pairs of genes with the goal of uncovering connected subnetworks that exhibit high similarity117. Further, local clustering approaches, such as SPICi, that rapidly uncover densely interconnected sets of proteins corresponding to functional modules allow enumeration of context-specific modules when interactions are weighted by co-expression values that change depending on the condition of interest118. Better methods to determine the differences between modules uncovered across multiple conditions, and to reason about them, represent an important avenue for future work.
Network flow — a classic formulation in graph algorithms in which each edge has a capacity to carry flow that is pumped into the system from source nodes — has proved to be a powerful and general concept in integrative interactomics (FIG. 3b). An early application used flow to propagate biological process annotations over a protein–protein interaction network119. Flow can also be used to identify proteins that respond to a particular perturbation in high-throughput screens that are either noisy or incomplete and will therefore miss proteins of interest. For example, high-scoring hits from an RNA interference (RNAi)-based knockdown study in flies were mapped to the protein–protein interaction network, and to uncover the affected pathways the Influence Flow algorithm computed the simplest explanation of a signalling pathway perturbation that was consistent with both the network and RNAi data by constructing a set of constraints for which the solutions correspond to high-confidence estimates of the structure of the pathway120. Flow-based optimization is also used in ResponseNet to reconstruct pathways from protein–protein and protein–DNA interaction networks121,122. A minimum-cost flow approach connects the genetic interactors of a given gene with genes that have expression changes when this gene is knocked out. The prize-collecting Steiner tree problem provides an alternative theoretical formulation to the problem of interconnecting a seed set of proteins. In this case, each initial hit is associated with a prize, and each interaction is associated with a cost. The goal, as implemented in SteinerNet, is to identify a subset of the identified hits that are connected directly or through intermediate proteins in protein–protein and transcriptional regulatory networks, such that the sum of the cost of the chosen interactions and the prize of the hits not included is minimized123,124.
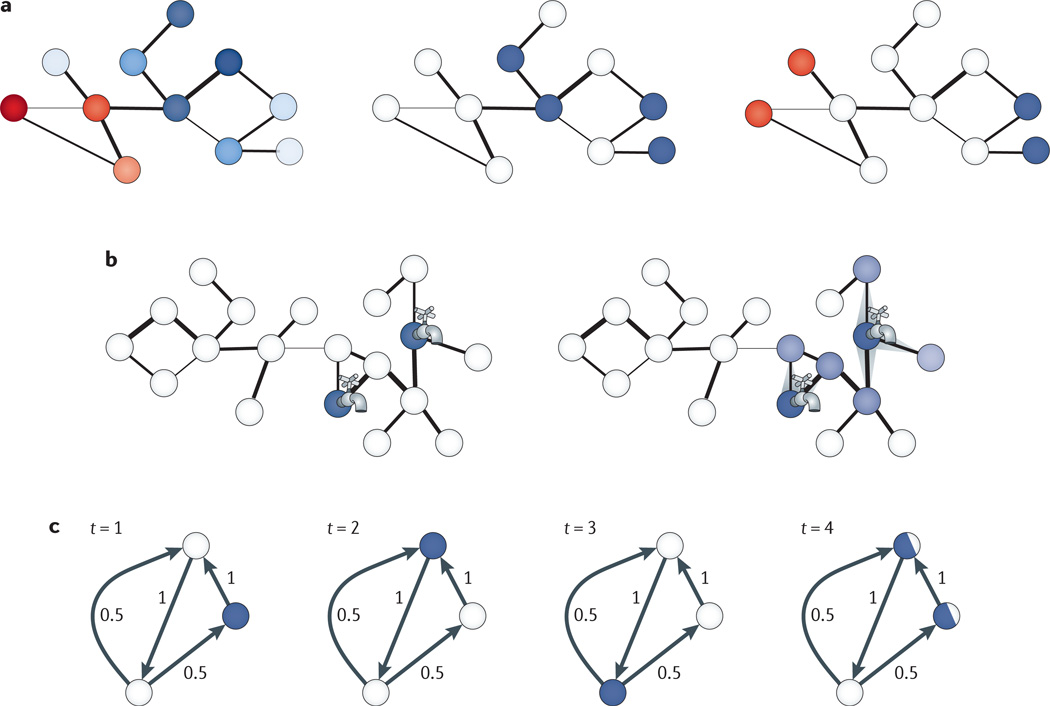
Figure 3. Integrative interactomics applications.
a | Schematics of computational formulations that arise when incorporating genomic data within a network context. Nodes correspond to biological components (for example, genes, proteins or other molecules), and edges correspond to known interactions among them (for example, physical, regulatory or genetic). In the left-hand panel, an attribute of interest has been measured for all molecules of the network (for example, differential gene expression values, shown in shades of blue and red). The goal is to uncover subnetworks that contain genes with similar values of attributes (for example, similarly differentially expressed, as shaded in similar colours in this panel). In the middle panel, a subset of genes has been identified as being of interest (for example, involved in some disease, shown in blue), and the goal is to uncover additional genes that take part in the same underlying pathway or functional module, as suggested by proximity in the network. In the right-hand panel, two subsets of genes, shown here in blue and red, have been identified (for example, corresponding to target genes, the expression values of which have changed and genes within loci that are associated with these targets), and the goal is to find paths in the network connecting these genes. b | Given an initial set of molecules (for example, genes that affect a phenotype of interest or that are involved in some disease), subnetworks containing additional genes of interest can be inferred using network flow approaches. The initial set of molecules comprises source nodes, from which fluid is pumped into the network, as represented by the taps. Each interaction between nodes can be weighted (for example, according to an estimate of the reliability of an interaction), and this weight can be used as a capacity to restrict the amount of flow that can go over the edge, as shown by the width of the edge. At each iteration of the algorithm, a node pumps flow to its neighbours while satisfying capacity constraints, and flow spreads through the network from the source nodes. Higher amounts of fluids through a node are shown with darker colours (right). In the classic network flow formulation, the amount of fluid in the network is maintained, whereas in other formulations fluid is pumped into the source nodes at a constant rate117,160. c | Random-walk-based approaches are also used to identify subnetworks from an initial set of molecules of interest. Starting from an initial node (for example, one of a set of known disease genes), a neighbour is repeatedly selected at random according to the distribution of transition probabilities between nodes, which can be set uniformly or based on estimates of the reliabilities of interactions or some other attribute of interest, such as co-expression between genes. In most applications, the walker also has some probability of staying at its current position at each step or jumping to any node chosen according to a pre-specified probability distribution (for example, to each disease gene with equal probability). In the shown example, at each time point, the distribution of the walker’s position is shown in blue. In the fourth time step (right), the walker is equally likely to be in one of two locations, and at each subsequent step, the probability of the walker being at each location can be estimated. After the probability estimates have converged, proteins are ranked (for example, as candidate disease genes) according to the probability that the walker is at the corresponding node.
Cellular networks also serve as a platform from which to infer causation in signalling and regulatory pathways. Probabilistic models have integrated network and expression data from gene knockout expression studies to predict cell-signalling cascades125. Random-walk-based approaches (FIG. 3c) have inferred causal genes driving expression variation within mapped expression quantitative trait loci (QTLs) by uncovering those genes that are visited more frequently in random walks initiated at target genes126. The flow of information from a locus to target genes has also been determined using electric current flow approaches127,128, in which each edge is associated with a conductance, and genes for which the nodes have the highest current through them are predicted to be causal; electric networks have previously been shown to be tightly linked to random walks on graphs129. By integrating protein annotations into network analysis, potential signal and regulatory pathways can be modelled as complex network schemas or patterns — with descriptions of proteins along with desired topologies and interactions among them — and matches rapidly uncovered in interactomes using NetGrep and other related tools130–133.
Comparative interactomics approaches are a powerful alternative approach for discovering modules and pathways across cellular networks. The central idea is that if a module is known in one organism, searching for homologues of its component proteins, along with conserved patterns of interactions, can yield information in other organisms134–139. The Isorank algorithm is one such network alignment approach; it combines sequence similarity and network similarity constraints to construct and to solve an eigenvalue problem. It has been used to align and to analyse networks based on protein–protein interaction134,135, genetic interaction140 and metabolic data141.
Interactome analysis of disease data sets
Some of the largest growing genomics data sets are arising in the context of human disease. These include genetic perturbations, such as mutations or copy number variations, observed in whole-genome or exome sequencing of afflicted individuals (for example, as observed in cancers) and variants identified as associated with disease through genome-wide association studies (GWASs), along with other high-throughput functional data (such as gene expression and DNA methylation data). The multifactorial nature of complex diseases suggests that although the genes underlying these diseases may differ among afflicted individuals, the pathways that are perturbed are likely to be shared, and thus proteins associated with the same disease have a tendency to interact142. Further, genes in loci identified by GWAS as being associated with complex diseases have been found to be ‘close’ within interaction networks135,143. Thus, network modularity can be leveraged to carry out disease gene prioritization and to uncover pathways associated with disease through a diverse set of methods144, including network flow145 and random walks146,147. In the context of analysing cancer genomes, approaches based on diffusion148 and shortest paths149 (Hotnet and Netbox) have been applied to identify subnetworks enriched in recurrently mutated proteins across patients. These network approaches complement approaches for disease gene prioritization based on assessing the impact of mutations on protein function150–152.
Permutation-based approaches, such as DAPPLE, are a useful means for testing the modularity of genes that are putatively associated with a specific disease143. Here, candidate disease genes are evaluated for proximity with respect to each other in the network. To assess significance, these values are compared with those computed on randomized networks that are obtained by shuffling protein names in an interaction-degree-preserving manner. Further, we note that network approaches may provide a powerful paradigm to recover potentially interesting associations in GWASs and other high-throughput experiments that are individually of marginal significance but are found in subnetworks enriched with other such genes and thus in aggregate suggest biological importance.
The molecular mechanisms that underlie disease can vary among affected individuals, and this can be evident at the mutational level or reflected in cellular measurements. Formulations based on extensions of the classic algorithmic problem of set cover have proved to be useful in considering this heterogeneity128,153,154. For example, in DEGAS, a set of dysregulated or mutated genes is determined for each individual, and the goal is to find a subnetwork in which each individual is ‘covered’ by some number of genes in the subnetwork, where a gene covers a disease individual if the gene is either dysregulated or mutated in that disease155. Such an approach naturally models both the modularity and the heterogeneity in disease. Nevertheless, consideration of patient heterogeneity represents a major challenge in further research in disease interactomics. A more complete discussion of the power of network biology approaches for understanding disease and disease heterogeneity can be found elsewhere156.
Conclusion and future prospects
It is clear that we are moving into an era in which diverse high-throughput data — genomes, transcriptomes, proteomes, interactomes and methylomes, among others — are routinely generated in individual laboratories. Understanding the algorithms underlying omics analyses will result in their correct application in answering biological questions. Here we outline some areas where further algorithms are needed to aid the bench biologist.
The recently developed compressive genomics approach45 represents an important milestone for designing compressive algorithmic frameworks that are adaptable to large-scale genomic data. Introducing compressive techniques for next-generation sequencing read data sets and their quality scores remains a major challenge. Such techniques would allow, for example, the type of meta-analyses across data sets that are routinely carried out using existing gene, protein and genome databases.
As transcriptomic data shifts from microarray to next-generation sequencing, we will also need to develop transcriptomic analysis methods to handle this new form of data. The experimental advantages of RNA-seq157 over microarrays — including, for example, the detection of transcript structure and alternative isoforms — add substantial complexity to these analyses. Further, high-throughput sequencing provides ‘read-out’ for a range of functional genomics experiments (for example, chromatin immunoprecipitation followed by high-throughput sequencing (ChIP–seq)158 to detect protein–DNA interactions, ultraviolet crosslinking and immunoprecipitation followed by sequencing (CLIP–seq) and related techniques159–162 to detect protein–RNA interactions and ribosome profiling163 to determine protein translation), and the tremendous amounts of data produced from these experiments, along with their specific attributes, will be a challenge to existing analysis paradigms.
As high-throughput technologies continue to improve, omics measurements will be made across organisms, individuals, cell types and conditions and eventually at the level of individual cells. Much future work in integrative interactomics will focus on characterizing the differences that distinguish individuals and cells from each other. These differences may reflect natural variation, differential functioning, disease pathology and other types of heterogeneity. Comprehensive, multimodal omics approaches are likely to be especially fruitful in uncovering the molecular landscapes underlying observed phenotypic variations.
Although several software platforms have been developed for basic data analysis and integration1–3, we close by highlighting the growing need for ‘translational’ computational biology. As new sophisticated algorithms continue to enter the toolbox of the most computationally expert biologists, wide adoption in the broader community will depend on user-friendly software tools and websites, with extensive documentation and easy-to-follow tutorials. Thus, although this Review has focused on the central role of algorithm development in high-throughput biology, it is clearly also essential to package effective methods into widely available software that can be easily and correctly used by scientists from a broad range of backgrounds.
Acknowledgements
The authors thank and L. Cowen for valuable feedback. B.B. thanks the US National Institutes of Health (NIH) for grant GM081871. M.S. thanks the NIH for grant GM076275 and US National Science Foundation (NSF) for grant ABI0850063.
Glossary
- Cloud computing
The use of computing resources distributed in the Internet to store, manage and analyse data, rather than doing so on a local server or personal computer.
- Parallel computing
A form of computation that allows numerous calculations to be carried out simultaneously, thereby accelerating computation. On the basis of this principle, many large-scale computational tasks can then be divided into smaller ones and solved on multiple machines concurrently.
- Machine learning techniques
Empirical data are taken as input, the relationship among the data is mathematically or statistically modelled, and patterns or predictions are generated. Supervised learning algorithms infer a function from labelled data features and predict labels on future input; unsupervised learning algorithms model the patterns or the distribution of a given unlabelled data set.
- Parallel dynamic programming
A technique that splits a large dynamic programming problem, usually by filling a table that can avoid redundant calculation, into a number of subproblems and computes all subproblems in parallel using multiple central processing units (CPUs). The computing speed-up scales almost linearly with the number of CPUs.
- Multicore computer processing units
(Multicore CPUs). Single computing processors with two or more independent computing units (called cores). Running multiple instructions on multiple cores at the same time can increase the overall speed of programs.
- Cache-oblivious algorithm
Takes advantage of the cache system of the central processing unit (that is, the local memory of frequently accessed data) to avoid expensive memory access operations and thus to improve efficiency; the intrinsic design of these algorithms does not require computer programs to be tuned for machines with different cache systems.
- Linear mixed model
A statistical model that models the observed effects from multiple different hidden factors; the effects are additively mixed according to the proportions of their corresponding factors.
- Matrix factorization
A method for decomposing a matrix into the product of two matrices. It can be applied to identify individual factors involved in a mixed observation.
- Differential geometry
A mathematical discipline for studying geometric objects, such as curves and surfaces, using the techniques of differential and integral calculus.
- Linear programming
A mathematical program for the optimization of a linear objective function, subject to linear constraints. Such functions capture the linear relationship between variables for the problem being optimized.
- Principle component analysis
A tool for transforming a set of observations with correlated variables into a set of linearly independent variables called principle components, making sure that the first principle component accounts for the largest variability of the data.
- Copy number variant
(CNV). Corresponds to abnormal number of copies of one or more segments in the genome. CNVs can be caused by structural rearrangements of the genome such as deletions, duplications, inversions and translocations.
- Bayesian network
A statistical model that describes the distribution of a set of random variables by a directed acyclic graph that represents the relationship among the random variables. For example, in a Bayesian network for a regulatory relationship for a set of genes, each variable represents a gene and each directed edge denotes either activating or repressing regulation between two genes.
- Steiner tree problem
Formulated on a network to find a minimum-length subnetwork that interconnects a set of seed nodes. Any two seed nodes may be connected by an edge or a path through other nodes.
- Random walk
A mathematical formulation of a number of successive random steps on a graph. It has been widely used to explain stochastic observations, such as diffusion in biological networks.
- Eigenvalue problem
The aim of this is to find a non-zero vector (that is, eigenvector), given a square matrix, such that the multiplication of the two is only different by a scalar factor.
- Set cover
Given a set of elements and subsets, the goal is to find the minimum number of subsets that cover all the elements.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
FURTHER INFORMATION
Bonnie Berger’s homepage: http://people.csail.mit.edu/bab
Mona Singh’s homepage: http://www.cs.princeton.edu/~mona
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goecks J, Nekrutenko A, Taylor J, Team G. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 5.Venter JC, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 6.Kircher M, Kelso J. High-throughput DNA sequencing — concepts and limitations. BioEssays. 2010;32:524–536. doi: 10.1002/bies.200900181. [DOI] [PubMed] [Google Scholar]
- 7.Kahn SD. On the future of genomic data. Science. 2011;331:728–729. doi: 10.1126/science.1197891. [DOI] [PubMed] [Google Scholar]
- 8.Gross M. Riding the wave of biological data. Curr. Biol. 2011;21:R204–R206. doi: 10.1016/j.cub.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 9.Huttenhower C, Hofmann O. A quick guide to large-scale genomic data mining. PLoS Comput. Biol. 2010;6:e1000779. doi: 10.1371/journal.pcbi.1000779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schatz M, Langmead B, Salzberg S. Cloud computing and the DNA data race. Nature Biotech. 2010;28:691–693. doi: 10.1038/nbt0710-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stein LD. The case for cloud computing in genome informatics. Genome Biol. 2010;11:207. doi: 10.1186/gb-2010-11-5-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tringe SG, Rubin EM. Metagenomics: DNA sequencing of environmental samples. Nature Rev. Genet. 2005;6:805–814. doi: 10.1038/nrg1709. [DOI] [PubMed] [Google Scholar]
- 13.Gstaiger M, Aebersold R. Applying mass spectrometry-based proteomics to genetics, genomics and network biology. Nature Rev. Genet. 2009;10:617–627. doi: 10.1038/nrg2633. [DOI] [PubMed] [Google Scholar]
- 14.Mardis ER. Next-generation DNA sequencing methods. Annu. Rev. Genom. Hum. Genet. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- 15.Metzker ML. Sequencing technologies — the next generation. Nature Rev. Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 16.Schatz MC, Delcher AL, Salzberg SL. Assembly of large genomes using second-generation sequencing. Genome Res. 2010;20:1165–1173. doi: 10.1101/gr.101360.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flicek P, Birney E. Sense from sequence reads: methods for alignment and assembly. Nature Methods. 2009;6:S6–S12. doi: 10.1038/nmeth.1376. [DOI] [PubMed] [Google Scholar]
- 18. Pevzner PA, Tang H, Waterman MS. An Eulerian path approach to DNA fragment assembly. Proc. Natl Acad. Sci. USA. 2001;98:9748–9753. doi: 10.1073/pnas.171285098. The EULER assembler introduces the de Bruijn graph and Eulerian path formulation for assembly, a paradigm used in the most popular assemblers.
- 19.Batzoglou S, et al. ARACHNE: a whole-genome shotgun assembler. Genome Res. 2002;12:177–189. doi: 10.1101/gr.208902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaffe DB, et al. Whole-genome sequence assembly for mammalian genomes: Arachne 2. Genome Res. 2003;13:91–96. doi: 10.1101/gr.828403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li R, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20:265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Butler J, et al. ALLPATHS: de novo assembly of whole-genome shotgun microreads. Genome Res. 2008;18:810–820. doi: 10.1101/gr.7337908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simpson JT, et al. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Compeau PE, Pevzner PA, Tesler G. How to apply de Bruijn graphs to genome assembly. Nature Biotech. 2011;29:987–991. doi: 10.1038/nbt.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simpson JT, Durbin R. Efficient de novo assembly of large genomes using compressed data structures. Genome Res. 2012;22:549–556. doi: 10.1101/gr.126953.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Earl D, et al. Assemblathon 1: a competitive assessment of de novo short read assembly methods. Genome Res. 2011;21:2224–2241. doi: 10.1101/gr.126599.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vezzi F, Narzisi G, Mishra B. Reevaluating assembly rvaluations with feature response curves: GAGE and Assemblathons. PLoS ONE. 2012;7:e52210. doi: 10.1371/journal.pone.0052210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salzberg SL, et al. GAGE: a critical evaluation of genome assemblies and assembly algorithms. Genome Res. 2012;22:557–567. doi: 10.1101/gr.131383.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kingsford C, Schatz MC, Pop M. Assembly complexity of prokaryotic genomes using short reads. BMC Bioinformatics. 2010;11:21. doi: 10.1186/1471-2105-11-21. This paper analyses complexity issues in genome assembly; the primary algorithmic challenge is that assembly can be complicated by short reads and genomic repeats.
- 31.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Langmead B, Trapnell C, Pop M, Salzberg S. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. Bowtie is probably the most widely used FM-index- or BWT-based short-read mapper. It demonstrates that the read-mapping problem can be done accurately even on a personal computer.
- 33.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H, Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li R, et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 36.Ferragina P, Manzini G. Indexing compressed text. JACM. 2005;52:552–581. [Google Scholar]
- 37.Burrows M, Wheeler DJ. A block-sorting lossless data compression algorithm. Digital Equipment Corporation; 1994. [Google Scholar]
- 38.Hach F, et al. mrsFAST: a cache-oblivious algorithm for short-read mapping. Nature Methods. 2010;7:576–577. doi: 10.1038/nmeth0810-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsi-Yang Fritz M, Leinonen R, Cochrane G, Birney E. Efficient storage of high throughput DNA sequencing data using reference-based compression. Genome Res. 2011;21:734–740. doi: 10.1101/gr.114819.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Christley S, Lu Y, Li C, Xie X. Human genomes as e-mail attachments. Bioinformatics. 2009;25:274–275. doi: 10.1093/bioinformatics/btn582. [DOI] [PubMed] [Google Scholar]
- 41.Pinho AJ, Pratas D, Garcia SP. GReEn: a tool for efficient compression of genome resequencing data. Nucleic Acids Res. 2012;40:e27. doi: 10.1093/nar/gkr1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tembe W, Lowey J, Suh E. G-SQZ: compact encoding of genomic sequence and quality data. Bioinformatics. 2010;26:2192–2194. doi: 10.1093/bioinformatics/btq346. [DOI] [PubMed] [Google Scholar]
- 43.Brandon MC, Wallace DC, Baldi P. Data structures and compression algorithms for genomic sequence data. Bioinformatics. 2009;25:1731–1738. doi: 10.1093/bioinformatics/btp319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang C, Zhang D. A novel compression tool for efficient storage of genome resequencing data. Nucleic Acids Res. 2011;39:e45. doi: 10.1093/nar/gkr009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Loh PR, Baym M, Berger B. Compressive genomics. Nature Biotech. 2012;30:627–630. doi: 10.1038/nbt.2241. This paper introduces ‘compressive genomics’, a general algorithmic paradigm that harnesses redundancy within data sets to speed up analyses by compressing data in such a way as to allow direct computation on the compressed data. Compressed versions of BLAST and BLAT demonstrate search times that scale linearly in the amount of non-redundant data without loss of accuracy.
- 46.Deorowicz S, Grabowski S. Compression of DNA sequence reads in FASTQ format. Bioinformatics. 2011;27:860–862. doi: 10.1093/bioinformatics/btr014. [DOI] [PubMed] [Google Scholar]
- 47.Hach F, Numanagic I, Alkan C, Sahinalp SC. SCALCE: boosting sequence compression algorithms using locally consistent encoding. Bioinformatics. 2012;28:3051–3057. doi: 10.1093/bioinformatics/bts593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 49.Kent WJ. BLAT—the BLAST-Like Alignment Tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Levy S, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nature Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 52.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ozsolak F, et al. Direct RNA sequencing. Nature. 2009;461:814–818. doi: 10.1038/nature08390. [DOI] [PubMed] [Google Scholar]
- 54.Trapnell C, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nature Biotech. 2012;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grabherr MG, et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nature Biotech. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garber M, Grabherr MG, Guttman M, Trapnell C. Computational methods for transcriptome annotation and quantification using RNA-seq. Nature Methods. 2011;8:469–477. doi: 10.1038/nmeth.1613. [DOI] [PubMed] [Google Scholar]
- 57.Brown PO, Botstein D. Exploring the new world of the genome with DNA microarrays. Nature Genet. 1999;21:33–37. doi: 10.1038/4462. [DOI] [PubMed] [Google Scholar]
- 58.Barrett T, et al. NCBI GEO: archive for functional genomics data sets-update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Butte A. The use and analysis of microarray data. Nature Rev. Drug Discov. 2002;1:951–960. doi: 10.1038/nrd961. [DOI] [PubMed] [Google Scholar]
- 60.Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis: from disarray to consolidation and consensus. Nature Rev. Genet. 2006;7:55–65. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- 61.Leek JT, et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nature Rev. Genet. 2010;11:733–739. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shen-Orr SS, et al. Cell type-specific gene expression differences in complex tissues. Nature Methods. 2010;7:287–289. doi: 10.1038/nmeth.1439. This work describes a linear algebraic approach to model the mixture of gene expression signals of multiple cell types from microarray experiments and to deconvolute the signals separately for each cell type.
- 63.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim D, Salzberg SL. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol. 2011;12:R72. doi: 10.1186/gb-2011-12-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Whitney AR, et al. Individuality and variation in gene expression patterns in human blood. Proc. Natl Acad. Sci. USA. 2003;100:1896–1901. doi: 10.1073/pnas.252784499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lu P, Nakorchevskiy A, Marcotte EM. Expression deconvolution: a reinterpretation of DNA microarray data reveals dynamic changes in cell populations. Proc. Natl Acad. Sci. USA. 2003;100:10370–10375. doi: 10.1073/pnas.1832361100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Y, et al. In silico estimates of tissue components in surgical samples based on expression profiling data. Cancer Res. 2010;70:6448–6455. doi: 10.1158/0008-5472.CAN-10-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gaujoux R, Seoighe C. Semi-supervised nonnegative matrix factorization for gene expression deconvolution: a case study. Infect. Genet. Evol. 2012;12:913–921. doi: 10.1016/j.meegid.2011.08.014. [DOI] [PubMed] [Google Scholar]
- 70.Clarke J, Seo P, Clarke B. Statistical expression deconvolution from mixed tissue samples. Bioinformatics. 2010;26:1043–1049. doi: 10.1093/bioinformatics/btq097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reich M, et al. GenePattern 2.0. Nature Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 73.Tanay A, Sharan R, Kupiec M, Shamir R. Revealing modularity and organization in the yeast molecular network by integrated analysis of highly heterogeneous genome-wide data. Proc. Natl Acad. Sci. USA. 2004;101:2981–2986. doi: 10.1073/pnas.0308661100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Narayanan M, Vetta A, Schadt EE, Zhu J. Simultaneous clustering of multiple gene expression and physical interaction datasets. PLoS Comput. Biol. 2010;6:e1000742. doi: 10.1371/journal.pcbi.1000742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Segal E, et al. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nature Genet. 2003;34:166–176. doi: 10.1038/ng1165. A probabilistic graphical model is constructed to identify regulatory modules, consisting of co-regulated or co-expressed genes, from gene expression data.
- 77.Kim D, Kim MS, Cho KH. The core regulation module of stress-responsive regulatory networks in yeast. Nucleic Acids Res. 2012;40:8793–8802. doi: 10.1093/nar/gks649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zinman GE, Zhong S, Bar-Joseph Z. Biological interaction networks are conserved at the module level. BMC Syst. Biol. 2011;5:134. doi: 10.1186/1752-0509-5-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rhrissorrakrai K, Gunsalus KC. MINE: Module Identification in Networks. BMC Bioinformatics. 2011;12:192. doi: 10.1186/1471-2105-12-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Colak R, et al. Module discovery by exhaustive search for densely connected, co-expressed regions in biomolecular interaction networks. PLoS ONE. 2010;5:e13348. doi: 10.1371/journal.pone.0013348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ali W, Deane CM. Functionally guided alignment of protein interaction networks for module detection. Bioinformatics. 2009;25:3166–3173. doi: 10.1093/bioinformatics/btp569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang Y, Xuan J, de los Reyes BG, Clarke R, Ressom HW. Reverse engineering module networks by PSO-RNN hybrid modeling. BMC Genomics. 2009;10(Suppl. 1):S15. doi: 10.1186/1471-2164-10-S1-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Michoel T, De Smet R, Joshi A, Van de Peer Y, Marchal K. Comparative analysis of module-based versus direct methods for reverse-engineering transcriptional regulatory networks. BMC Syst. Biol. 2009;3:49. doi: 10.1186/1752-0509-3-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Joshi A, De Smet R, Marchal K, Van de Peer Y, Michoel T. Module networks revisited: computational assessment and prioritization of model predictions. Bioinformatics. 2009;25:490–496. doi: 10.1093/bioinformatics/btn658. [DOI] [PubMed] [Google Scholar]
- 85.Wang X, Dalkic E, Wu M, Chan C. Gene module level analysis: identification to networks and dynamics. Curr. Opin. Biotechnol. 2008;19:482–491. doi: 10.1016/j.copbio.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hirose O, et al. Statistical inference of transcriptional module-based gene networks from time course gene expression profiles by using state space models. Bioinformatics. 2008;24:932–942. doi: 10.1093/bioinformatics/btm639. [DOI] [PubMed] [Google Scholar]
- 87.Litvin O, Causton HC, Chen BJ, Pe’er D. Modularity and interactions in the genetics of gene expression. Proc. Natl Acad. Sci. USA. 2009;106:6441–6446. doi: 10.1073/pnas.0810208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Akavia UD, et al. An integrated approach to uncover drivers of cancer. Cell. 2010;143:1005–1017. doi: 10.1016/j.cell.2010.11.013. The computational approach CONEXIC implements a module network to integrate different data sets, including CNVs and gene expression, from cancer studies and discover dysregulated genes.
- 89. Maathuis MH, Colombo D, Kalisch M, Buhlmann P. Predicting causal effects in large-scale systems from observational data. Nature Methods. 2010;7:247–248. doi: 10.1038/nmeth0410-247. This paper describes an algorithm to estimate the effects of perturbations from observational data in gene expression experiments in which the causal relationship is not known between genes.
- 90.Markowetz F, Kostka D, Troyanskaya OG, Spang R. Nested effects models for high-dimensional phenotyping screens. Bioinformatics. 2007;23:I305–I312. doi: 10.1093/bioinformatics/btm178. [DOI] [PubMed] [Google Scholar]
- 91.Prat Y, Fromer M, Linial N, Linial M. Recovering key biological constituents through sparse representation of gene expression. Bioinformatics. 2011;27:655–661. doi: 10.1093/bioinformatics/btr002. [DOI] [PubMed] [Google Scholar]
- 92.Yeung KY, Ruzzo WL. Principal component analysis for clustering gene expression data. Bioinformatics. 2001;17:763–774. doi: 10.1093/bioinformatics/17.9.763. [DOI] [PubMed] [Google Scholar]
- 93. Schmid M, et al. A gene expression map of Arabidopsis thaliana development. Nature Genet. 2005;37:501–506. doi: 10.1038/ng1543. Scalable methods are introduced here that associate expression patterns to phenotypes both to label new expression samples with and to identify marker genes for phenotypes.
- 94.Zhou X, Kao MC, Wong WH. Transitive functional annotation by shortest-path analysis of gene expression data. Proc. Natl Acad. Sci. USA. 2002;99:12783–12788. doi: 10.1073/pnas.192159399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Parts L, Stegle O, Winn J, Durbin R. Joint genetic analysis of gene expression data with inferred cellular phenotypes. PLoS Genet. 2011;7:e1001276. doi: 10.1371/journal.pgen.1001276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stegle O, Parts L, Piipari M, Winn J, Durbin R. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nature Protoc. 2012;7:500–507. doi: 10.1038/nprot.2011.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ng S, et al. PARADIGM-SHIFT predicts the function of mutations in multiple cancers using pathway impact analysis. Bioinformatics. 2012;28:i640–i646. doi: 10.1093/bioinformatics/bts402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vaske CJ, et al. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics. 2010;26:i237–i245. doi: 10.1093/bioinformatics/btq182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.The Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.The Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Heiser LM, et al. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc. Natl Acad. Sci. USA. 2012;109:2724–2729. doi: 10.1073/pnas.1018854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu X, Yu X, Zack DJ, Zhu H, Qian J. TiGER: a database for tissue-specific gene expression and regulation. BMC Bioinformatics. 2008;9:271. doi: 10.1186/1471-2105-9-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ogasawara O, et al. BodyMap-Xs: anatomical breakdown of 17 million animal ESTs for cross-species comparison of gene expression. Nucleic Acids Res. 2006;34:D628–D631. doi: 10.1093/nar/gkj137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sirota M, et al. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Sci. Transl. Med. 2011;3:96ra77. doi: 10.1126/scitranslmed.3001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lamb J. The Connectivity Map: a new tool for biomedical research. Nature Rev. Cancer. 2007;7:54–60. doi: 10.1038/nrc2044. [DOI] [PubMed] [Google Scholar]
- 106.Schmid PR, Palmer NP, Kohane IS, Berger B. Making sense out of massive data by going beyond differential expression. Proc. Natl Acad. Sci. USA. 2012;109:5594–5599. doi: 10.1073/pnas.1118792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Palmer NP, Schmid PR, Berger B, Kohane IS. A gene expression profile of stem cell pluripotentiality and differentiation is conserved across diverse solid and hematopoietic cancers. Genome Biol. 2012;13:R71. doi: 10.1186/gb-2012-13-8-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dudley JT, Tibshirani R, Deshpande T, Butte AJ. Disease signatures are robust across tissues and experiments. Mol. Syst. Biol. 2009;5:307. doi: 10.1038/msb.2009.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li W, et al. Integrative analysis of many weighted co-expression networks using tensor computation. PLoS Comput. Biol. 2011;7:e1001106. doi: 10.1371/journal.pcbi.1001106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Franceschini A, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Croft D, et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39:D691–D697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chatr-aryamontri A, et al. The BioGRID interaction database: 2013 update. Nucleic Acids Res. 2013;41:D816–D823. doi: 10.1093/nar/gks1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gerstein MB, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wong AK, et al. IMP: a multi-species functional genomics portal for integration, visualization and prediction of protein functions and networks. Nucleic Acids Res. 2012;40:W484–W490. doi: 10.1093/nar/gks458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402:C47–C52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 116.Ideker T, Ozier O, Schwikowski B, Siegel AF. Discovering regulatory and signalling circuits in molecular interaction networks. Bioinformatics. 2002;18(Suppl. 1):S233–S240. doi: 10.1093/bioinformatics/18.suppl_1.s233. [DOI] [PubMed] [Google Scholar]
- 117. Ulitsky I, Shamir R. Identification of functional modules using network topology and high-throughput data. BMC Syst. Biol. 2007;1:8. doi: 10.1186/1752-0509-1-8. This study uncovers modules in interaction networks such that the components within a module are also similar to each other with respect to expression or another attribute of interest.
- 118.Jiang P, Singh M. SPICi: a fast clustering algorithm for large biological networks. Bioinformatics. 2010;26:1105–1111. doi: 10.1093/bioinformatics/btq078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Nabieva E, Jim K, Agarwal A, Chazelle B, Singh M. Whole-proteome prediction of protein function via graph-theoretic analysis of interaction maps. Bioinformatics. 2005;21(Suppl. 1):i302–i310. doi: 10.1093/bioinformatics/bti1054. Network flow-based methods are introduced as a paradigm for propagating information within cellular networks.
- 120.Singh R, Berger B. Influence flow: integrating pathway-specific RNAi data and protein interaction data; International Society for Computational Biology; 2007. [online], http://www.iscb.org/cms_addon/conferences/uploaded/css/20070406163614_rsingh.pdf. [Google Scholar]
- 121.Yeger-Lotem E, et al. Bridging high-throughput genetic and transcriptional data reveals cellular responses to alpha-synuclein toxicity. Nature Genet. 2009;41:316–323. doi: 10.1038/ng.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lan A, et al. ResponseNet: revealing signaling and regulatory networks linking genetic and transcriptomic screening data. Nucleic Acids Res. 2011;39:W424–W429. doi: 10.1093/nar/gkr359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Huang SS, Fraenkel E. Integrating proteomic, transcriptional, and interactome data reveals hidden components of signaling and regulatory networks. Sci. Signal. 2009;2:ra40. doi: 10.1126/scisignal.2000350. This paper introduces a Steiner tree formulation to uncover subnetworks connecting a set of seed proteins.
- 124.Tuncbag N, McCallum S, Huang SS, Fraenkel E. SteinerNet: a web server for integrating ‘omic’ data to discover hidden components of response pathways. Nucleic Acids Res. 2012;40:W505–W509. doi: 10.1093/nar/gks445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yeang CH, Ideker T, Jaakkola T. Physical network models. J. Comput. Biol. 2004;11:243–262. doi: 10.1089/1066527041410382. [DOI] [PubMed] [Google Scholar]
- 126.Tu Z, Wang L, Arbeitman MN, Chen T, Sun F. An integrative approach for causal gene identification and gene regulatory pathway inference. Bioinformatics. 2006;22:e489–e496. doi: 10.1093/bioinformatics/btl234. [DOI] [PubMed] [Google Scholar]
- 127.Suthram S, Beyer A, Karp RM, Eldar Y, Ideker T. eQED: an efficient method for interpreting eQTL associations using protein networks. Mol. Syst. Biol. 2008;4:162. doi: 10.1038/msb.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kim YA, Wuchty S, Przytycka TM. Identifying causal genes and dysregulated pathways in complex diseases. PLoS Comput. Biol. 2011;7:e1001095. doi: 10.1371/journal.pcbi.1001095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Doyle PG, Snell JL. Random Walks and Electric Networks. Mathematical Association of America; 1984. [Google Scholar]
- 130.Steffen M, Petti A, Aach J, D’Haeseleer P, Church G. Automated modelling of signal transduction networks. BMC Bioinformatics. 2002;3:34. doi: 10.1186/1471-2105-3-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pandey J, et al. Functional annotation of regulatory pathways. Bioinformatics. 2007;23:i377–i386. doi: 10.1093/bioinformatics/btm203. [DOI] [PubMed] [Google Scholar]
- 132.Banks E, Nabieva E, Chazelle B, Singh M. Organization of physical interactomes as uncovered by network schemas. PLoS Comput. Biol. 2008;4:e1000203. doi: 10.1371/journal.pcbi.1000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Banks E, Nabieva E, Peterson R, Singh M. NetGrep: fast network schema searches in interactomes. Genome Biol. 2008;9:R138. doi: 10.1186/gb-2008-9-9-r138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Singh R, Xu J, Berger B. Global alignment of multiple protein interaction networks with application to functional orthology detection. Proc. Natl Acad. Sci. USA. 2008;105:12763–12768. doi: 10.1073/pnas.0806627105. This paper introduces global network alignment and pioneers the use of spectral methods to solve it. Led to IsoBase, a database of functionally related proteins across protein-protein, genetic interaction and metabolic networks, simultaneously incorporating both sequence and network data.
- 135.Liao CS, Lu K, Baym M, Singh R, Berger B. IsoRankN: spectral methods for global alignment of multiple protein networks. Bioinformatics. 2009;25:i253–i258. doi: 10.1093/bioinformatics/btp203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Flannick J, Novak A, Srinivasan BS, McAdams HH, Batzoglou S. Graemlin: general and robust alignment of multiple large interaction networks. Genome Res. 2006;16:1169–1181. doi: 10.1101/gr.5235706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Koyuturk M, et al. Pairwise alignment of protein interaction networks. J. Comput. Biol. 2006;13:182–199. doi: 10.1089/cmb.2006.13.182. [DOI] [PubMed] [Google Scholar]
- 138.Kelley BP, et al. Conserved pathways within bacteria and yeast as revealed by global protein network alignment. Proc. Natl Acad. Sci. USA. 2003;100:11394–11399. doi: 10.1073/pnas.1534710100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Atias N, Sharan R. Comparative analysis of protein networks: hard problems, practical solutions. Commun. Acm. 2012;55:88–97. [Google Scholar]
- 140.Park D, Singh R, Baym M, Liao CS, Berger B. IsoBase: a database of functionally related proteins across PPI networks. Nucleic Acids Res. 2011;39:D295–D300. doi: 10.1093/nar/gkq1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ma C-Y, et al. Reconstruction of phyletic trees by global alignment of multiple metabolic networks. BMC Bioinformatics. doi: 10.1186/1471-2105-14-S2-S12. (in the press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Goh KI, et al. The human disease network. Proc. Natl Acad. Sci. USA. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Rossin EJ, et al. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 2011;7:e1001273. doi: 10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Navlakha S, Kingsford C. The power of protein interaction networks for associating genes with diseases. Bioinformatics. 2010;26:1057–1063. doi: 10.1093/bioinformatics/btq076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Vanunu O, Magger O, Ruppin E, Shlomi T, Sharan R. Associating genes and protein complexes with disease via network propagation. PLoS Comput. Biol. 2010;6:e1000641. doi: 10.1371/journal.pcbi.1000641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Kohler S, Bauer S, Horn D, Robinson PN. Walking the interactome for prioritization of candidate disease genes. Am. J. Hum. Genet. 2008;82:949–958. doi: 10.1016/j.ajhg.2008.02.013. This paper introduces random-walk based approaches for prioritizing disease genes using interaction networks.
- 147.Erten S, Bebek G, Ewing RM, Koyuturk M. DADA: degree-aware algorithms for network-based disease gene prioritization. BioData Min. 2011;4:19. doi: 10.1186/1756-0381-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Vandin F, Upfal E, Raphael BJ. Algorithms for detecting significantly mutated pathways in cancer. J. Comput. Biol. 2011;18:507–522. doi: 10.1089/cmb.2010.0265. The authors develop a flow-based and statistical approach for analysing genes mutated in cancers within their network context in order to identify significantly mutated subnetworks.
- 149.Cerami E, Demir E, Schultz N, Taylor BS, Sander C. Automated network analysis identifies core pathways in glioblastoma. PLoS ONE. 2010;5:e8918. doi: 10.1371/journal.pone.0008918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 151.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nature Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yandell M, et al. A probabilistic disease-gene finder for personal genomes. Genome Res. 2011;21:1529–1542. doi: 10.1101/gr.123158.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Vandin F, Upfal E, Raphael B. De novo discovery of mutated driver pathways in cancer. Genome Res. 2012;22:375–385. doi: 10.1101/gr.120477.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chowdhury SA, Koyuturk M. Identification of coordinately dysregulated subnetworks in complex phenotypes. Pac. Symp. Biocomput. 2010;2010:133–144. doi: 10.1142/9789814295291_0016. [DOI] [PubMed] [Google Scholar]
- 155.Ulitsky I, Krishnamurthy A, Karp RM, Shamir R. DEGAS: de novo discovery of dysregulated pathways in human diseases. PLoS ONE. 2010;5:e13367. doi: 10.1371/journal.pone.0013367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Cho D-Y, Kim Y-A, Przytycka TM. Network biology approach to complex diseases. PLoS Comput. Biol. doi: 10.1371/journal.pcbi.1002820. (in the press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wang Z, Gerstein M, Snyder M. RNA-seq: a revolutionary tool for transcriptomics. Nature Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Furey TS. ChIP-seq and beyond: new and improved methodologies to detect and characterize protein– DNA interactions. Nature Rev. Genet. 2012;13:840–852. doi: 10.1038/nrg3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hafner M, Lianoglou S, Tuschl T, Betel D. Genome-wide identification of miRNA targets by PAR-CLIP. Methods. 2012;58:94–105. doi: 10.1016/j.ymeth.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wang ET, et al. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell. 2012;150:710–724. doi: 10.1016/j.cell.2012.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ascano M, Hafner M, Cekan P, Gerstberger S, Tuschl T. Identification of RNA-protein interaction networks using PAR-CLIP. Wiley Interdiscip. Rev. RNA. 2012;3:159–177. doi: 10.1002/wrna.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Jungkamp AC, et al. In vivo and transcriptome-wide identification of RNA binding protein target sites. Mol. Cell. 2011;44:828–840. doi: 10.1016/j.molcel.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Meyer LR, et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 2013;41:D64–D69. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.de Souza N. The ENCODE project. Nature Methods. 2012;9:1046–1046. doi: 10.1038/nmeth.2238. [DOI] [PubMed] [Google Scholar]
- 166.Gerstein MB, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE Project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Manber U, Myers G. Suffix Arrays — a new method for online string searches. Siam J. Comput. 1993;22:935–948. [Google Scholar]
- 168.Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 169.Paten B, et al. Cactus: algorithms for genome multiple sequence alignment. Genome Res. 2011;21:1512–1528. doi: 10.1101/gr.123356.111. [DOI] [PMC free article] [PubMed] [Google Scholar]