Abstract
Marginal Zone (MZ) B cells play an important role in the clearance of blood-borne bacterial infections via rapid T-independent IgM responses. We have previously demonstrated that MZ B cells respond rapidly and robustly to bacterial particulates. To determine the MZ-specific genes that are expressed to allow for this response, MZ and Follicular (FO) B cells were sort-purified and analyzed via DNA microarray analysis. We identified 181 genes that were significantly different between the two B cell populations. 99 genes were more highly expressed in MZ B cells while 82 genes were more highly expressed in FO B cells. To further understand the molecular mechanisms by which MZ B cells respond so rapidly to bacterial challenge, idiotype positive and negative MZ B cells were sort-purified before (0 hour) or after (1 hour) i.v. immunization with heat killed Streptococcus pneumoniae, R36A, and analyzed via DNA microarray analysis. We identified genes specifically up regulated or down regulated at 1 hour following immunization in the idiotype positive MZ B cells. These results give insight into the gene expression pattern in resting MZ vs. FO B cells and the specific regulation of gene expression in antigen-specific MZ B cells following interaction with antigen.
Keywords: MZ B cell, FO B cell, microarray, cytokine, idiotype
Introduction
Mature B lymphocytes play an integral role in the adaptive immune response via antigen presentation and antibody secretion. The mature splenic B cell population is divided into the marginal zone (MZ) and follicular (FO) B cell subsets based on anatomical location, cellular surface molecules, and functional immune responses [reviewed in (1)]. MZ B cells respond primarily to T-independent antigens and are proposed to bridge the gap between the rapid antigen non-specific response and the delayed antigen-specific response. FO B cells respond primarily to T-dependent antigens and are responsible for the generation of long-term memory. However, the exact molecular mechanism by which each subset of B cells function is not fully understood.
MZ B cells are primarily non-recirculating, located at the outer limit of the white pulp region, and characterized by the expression of IgMhiIgDloCD1d+CD21hiCD23lo. The MZ B cell repertoire is enriched with B cells expressing germline-encoded B cell receptors (2-4), some of which have a low level of self-reactivity. Following activation, MZ B cells increase B7-1 and B7-2 expression, develop into plasmablasts more readily, and are more sensitive to LPS stimulation than their FO counterparts (5, 6). In addition to rapid production of IgM antibody, MZ B cells also possess the ability to capture and shuttle antigen to follicular dendritic cells (7) as well as efficiently activate naive T cells directly (8), suggesting a potential role for MZ B cells in T cell-dependent antibody responses as well. In addition to anatomical location and cellular functions, MZ and FO B cells differentially express a number of cell surface molecules. We have previously shown that CD9, a member of the tetraspanin family, is expressed by MZ and B1 B cell populations but not by FO B cells (9). Additionally, we identified Fc Receptor Homolog 3 (FcRH3) as a potentially immunoregulatory molecule expressed by MZ and B1 cells, but not by FO B cells (10). Recently the scavenger receptor, CD36, was identified as a marker predominantly expressed by MZ B cells (11). Taken together, it is clear that MZ B cells fill a specific niche in the splenic environment through unique expression and regulation of specific genes.
The development of DNA microarray technology has allowed for the rapid analysis of genome wide gene expression profiles. Using this technology, we set out to identify differentially regulated genes between FO and MZ B cells as well as the genes specifically up regulated or down regulated following activation. DNA microarray analysis of FACS-sorted resting MZ and FO B cells from MD4 mice revealed 181 genes that are differentially expressed in the resting B cell populations. 99 genes were more highly expressed in MZ B cells while 82 genes were more highly expressed in FO B cells. In addition, a comparative DNA microarray analysis of FACS-sorted MZ idiotype positive and negative B cells at 0 and 1 hr following i.v. immunization with heat killed Streptococcus pneumoniae, R36A, revealed genes specifically up regulated or down regulated following activation. These results give new insight into the differences between MZ and FO B cells and reveal new candidate genes and pathways to study.
Materials and Methods
Animals
SWR/J and C3H/HeJ samples were kindly provided by T. Waldschmidt (University of Iowa, Iowa City, Iowa) and were from mice housed at the University of Iowa in specific pathogen-free conditions. MD4 anti-HEL conventional transgenic mice were originally obtained from Dr. C. Goodnow (Australian National University, Canberra, Austrailia) (12). MD4 transgenic mice are on a C57BL/6 (B6) background. M167 Tg mice have been described previously (13). The IL-10/Thy1.1 reporter mice were generously provided by Casey Weaver (University of Alabama at Birmingham, Birmingham, Alabama), as described previously (14). IL-10/Thy1.1 mice were crossed with M167 Tg mice. All mice were bred and housed within the pathogen-free facility at The University of Alabama at Birmingham and used at 6 to 8 weeks of age according to approved animal protocols.
DNA Microarray Analysis
Microarray analysis was performed as described previously (15). Briefly, total RNA was isolated from sort-purified cell populations using an Rneasy Mini Kit with on-column Dnase digestion (Qiagen Inc., Valencia, CA), and, in accordance with the Expression Analysis Technical Manual (Affymetrix, Santa Clara, CA), cDNA was synthesized. CRNA was synthesized with BioArray High-Yield Transcript Labeling kit (Enzo, New York, NY). Labeled cRNA (~15 μg) was chemically fragmented for 35 min. at 94°C. Affymetrix MG U74Av2 oligonucleotide GeneChips (Affymetrix, Santa Clara, CA) were probed, hybridized, stained, washed, and scanned according to the manufacturer’s protocol at the University of Minnesota Biomedical Genomics Center facility. Each sort-purified cell population was processed independently as true biological replicates.
Flow Cytometry and Cell Sorting
FACS analysis was performed as described previously (16). Briefly, total splenocytes were collected, red blood cells lysed with ammonium chloride, and stained with different combinations of the following antibodies: fluorescein (FITC), phycoerythrin (PE), or allophycocyanin (APC) conjugated anti-mouse CD21, CD23, Thy1.1, CD19 (eBiosciences, San Diego, CA), goat anti-human RGS10 (Santa Cruz Biotechnology, Inc.), goat anti-mouse D6 beta chemokine receptor, and rabbit anti-human Sharp2/Stra13 (abcam Cambridge, MA). All anti-human antibodies cross react with mouse targets. For intracellular FACS analysis, cells were then washed, fixed, and permeablized using the Cytofix/Cytoperm (BD Biosciences) kit according to manufacturer’s directions. All samples were analyzed using a FACSCalibur flow cytometer or FACSAria cell sorter (BD Biosciences, San Jose, CA). The data were analyzed using FLOWJO software (Tree Star, Inc.).
Western Blot Analysis
Western blot analysis was performed as described previously (17). Briefly, following B cell isolation, cells were lysed, total protein quantitated using a protein quantitation assay (BioRad, Inc.), and protein samples (5-20 μg) were resolved by electrophoresis on 10% polyacrylamide gels (BioRad, Inc.), transferred to Immobilon-P PVDF membranes (Millipore), probed with either goat anti-human RGS10, anti-actin (Santa Cruz Biotechnology, Inc.), goat anti-mouse D6 beta chemokine receptor (abcam Cambridge, MA), and detected with horseradish peroxidase (HRP)-labeled anti-mouse, goat, and rabbit antibodies (Santa Cruz Biotechnology, Inc.), and developed with the LumiGlo Detection Kit (Cell Signaling).
RNA isolation and PCR
Total RNA was isolated from approximately 5 × 105 sort-purified MZ B cells using TRIZOL Reagent (Invitrogen, Carlsbad, CA) following manufacturer’s directions. RT-PCR was performed using Omniscript RT Kit (Qiagen, Valencia, CA) following manufacturers directions. The following gene-specific primers were used to amplify the cDNA obtained from the RT Kit using Fisher Taq and PCR products were resolved using a 1% agarose gel and visualized using Ethidium Bromide. Primers: β-actin - 5′-TACAGCTTCACCACCACAGC-3′ and 5′-AAGGAAGGCTGGAAAAGAGC-3′; D6 - 5′-CTTCCAGCTGAACCTTCTGG-3′ and 5′-CGAGTGCAGAAACAAGGTGA-3′; RGS10 - 5′-GCCTTAAGAGCACAGCCAAG-3′ and 5′-CTTTTCCTGCATCTGCTTCC-3′; Thy1.1 - 5′-ACCAAAACCTTCGCCTGGACTG-3′ and 5′-TCCTTGGGGTCTTCTACCTTTCTC-3′; IL-10 - F-CATGGGTCTTGGGAAGAGAA, R-CATTCCCAGAGGAATTGCAT; Stra13 - 5′-GGATTTGCCCACATGTACC-3′ and 5′-TCAATGCTTTCACGTGCTTC-3′ (60°C annealing temperature for all primers).
Data Processing
GeneData Expressionist Pro 1.0 (GeneData Inc., Waltham, MA) was used to generate relative expression values for each transcript using the MAS 5.0 algorithm, default settings, and a scaling factor of 1500 to control for minor cross-chip differences in hybridization intensities. GeneData Expressionist and Microsoft Excel (Microsoft Corp., Seattle, WA) were used for statistical analysis. Hierarchical clustering analysis was performed using CLUSTER and visualized in TREEVIEW, as described previously (18).
Statistics
Data with three or more groups were analyzed by a one-way ANOVA and statistical significance was determined by a p value of <0.05. Data with two groups were analyzed by a two-tailed paired t test and statistical significance was determined by a p value of <0.02.
Results
DNA Microarray Analysis of resting MZ and FO B cells
The mature splenic B cell population is divided into MZ and FO B cells based on anatomical location, cellular surface molecule expression, and functional immune responses [reviewed in (1)]. DNA microarray analysis was employed to determine differences in gene expression profiles between MZ and FO B cell populations. Splenocytes from B6 MD4 transgenic mice were sort-purified to obtain paired MZ (B220+, CD21hi, CD23low) and FO (B220+, CD21int, CD23pos) B cell samples. Post-sort analysis revealed greater than 95% purity of each B cell population (data not shown). MD4 mice carry a heavy and light chain transgene specific for hen egg lysozyme antigen (12) and were used because greater than 90% of their B cells express the transgenic B cell receptor, thereby potentially reducing the variability due to a polyclonal repertoire. Gene expression was assessed in three replicates of each B cell population using Affymetrix U74A mouse GeneChip microarray, representing approximately 11,000 transcripts. Expression levels were quantified using GeneData Expressionist Pro 1.0 software and the data from each array was analyzed to identify the genes that were differentially expressed between the MZ and FO B cell populations. Differential expression was defined as a mean fold change > 2 and p < 0.02 by Student’s T test.
Based on this definition, we identified 181 transcripts differentially expressed between the two populations. 99 transcripts (approximately 55% of total) were more highly expressed in MZ B cells relative to FO B cells while 82 transcripts (approximately 45% of total) were more highly expressed in FO B cells relative to MZ B cells. To better visualize the data, each expression value was divided by the mean expression of all six samples of that transcript and converted into log2 space. The data was then analyzed by unsupervised hierarchical clustering, as described previously (18). The data showed tight clustering of the three replicates of each cell type with a coefficient of correlation between any two replicate samples greater than 0.98. The 181 gene transcripts identified were grouped into the following broad functional classifications: Figure 1 (A) motility/adhesion, (B) immune response, (C) apoptosis, (D) proliferation, Figure 2 (A) transcription factors, (B) signal transduction, metabolism (data not shown), or miscellaneous (data not shown). All 181 genes are listed in Table 1.
Figure 1. Expression profile of differentially expressed genes between FO and MZ B cells.
DNA microarray analysis identified 181 genes that were significantly different in sort-purified follicular (FO) vs. marginal zone (MZ) B cells from MD4 transgenic mice (B6 background). The identified transcripts have a fold change > 2 and a p value < 0.02 by T-test. The differentially expressed genes were grouped into various functional categories (A) Motility/Adhesion, (B) Immune Response, (C) Apoptosis, and (D) Proliferation. Shown are normalized expression values greater than (yellow), near (black), or less than (blue) the mean of that gene. Each column represents one independent sample of sort-purified FO or MZ B cells. Genes or transcripts are represented in rows. Clustering of the genes is unsupervised.
Figure 2. Expression profile of differentially expressed genes between FO and MZ B cells.
DNA microarray analysis identified 181 genes that were significantly different in sort-purified follicular (FO) vs. marginal zone (MZ) B cells from MD4 transgenic mice (B6 background). The identified transcripts have a fold change > 2 and a p value < 0.02 by T-test. The differentially expressed genes were grouped into various functional categories (A) Transcription Factors and (B) Signal Transduction. Shown are normalized expression values greater than (yellow), near (black), or less than (blue) the mean of that gene. Each column represents one independent sample of sorted FO or MZ B cells. Genes or transcripts are represented in rows. Clustering of the genes is unsupervised.
Table 1.
Genes differentially expressed between FO and MZ B cells in B6, SWR,and C3H mouse strains.
| Relative Expression | Relative Expression | Relative Expression | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Affy ID | Gene Symbol | Gene Title | B6 FO | B6 MZ | Fold Difference (MZ/FO) | SWR FO | SWR MZ | Fold Difference(MZ/FO) | C3H FO | C3H MZ | Fold Difference (MZ/FO) |
| 93430 at | Cmkor1 | chemokine orphan receptor 1 | 78 | 4369 | 56.0 | 12 | 2081 | 178.5 | 107 | 3147 | 29.3 |
| 97967 at | Plxnd1 | plexin D1 | 70 | 3800 | 54.4 | 70 | 107 | 1.5 | 44 | 50 | 1.1 |
| 102910 at | Abcb1a | ATP-binding cassette, sub-family B (MDR/TAP) | 48 | 953 | 19.8 | 28 | 716 | 25.3 | 52 | 596 | 11.6 |
| 92217 s at | Gp49 | glycoprotein 49 A/B | 161 | 2762 | 17.2 | 559 | 2146 | 3.8 | 582 | 2470 | 4.2 |
| 101587 at | Ephx1 | epoxide hydrolase 1, microsomal | 235 | 3410 | 14.5 | 3682 | 4366 | 1.2 | 4074 | 4099 | 1.0 |
| 100325 at | Gp49 | glycoprotein 49 A/B | 394 | 5124 | 13.0 | 716 | 4134 | 5.8 | 1009 | 4883 | 4.8 |
| 96865 at | Marcks | myristoylated alanine rich protein kinase C substrate | 1183 | 14871 | 12.6 | 952 | 3840 | 4.0 | 1085 | 5057 | 4.7 |
| 97105 at | C230027N18Rik | RIKEN cDNA C230027N18 gene | 269 | 3142 | 11.7 | 564 | 2822 | 5.0 | 417 | 2519 | 6.0 |
| 101923 at | P1a2g7 | phospholipase A2, group VII | 284 | 2408 | 8.5 | 24 | 669 | 28.2 | 89 | 1230 | 13.9 |
| 160495 at | Ahr | aryl-hydrocarbon receptor | 31 | 249 | 8.0 | 461 | 1477 | 3.2 | 737 | 2594 | 3.5 |
| 93411 at | Sema7a | semaphorin 7A | 788 | 6214 | 7.9 | 1038 | 4241 | 4.1 | 826 | 2926 | 3.5 |
| 102722 g at | IgG3 | Ig gamma-3 heavy chain precursor | 310 | 2436 | 7.9 | 829 | 1541 | 1.9 | 1281 | 2699 | 2.1 |
| 100912 at | Dph5 | DPH5 homolog (S. cerevisiae) | 1229 | 9553 | 7.8 | 687 | 3390 | 4.9 | 954 | 4768 | 5.0 |
| 98309 at | Ccbp2 | Chemokine binding protein 2 | 481 | 3549 | 7.4 | 85 | 895 | 10.5 | 343 | 526 | 1.5 |
| 97487 at | Serpine2 | serine (or cysteine) peptidase inhibitor | 219 | 1377 | 6.3 | 148 | 747 | 5.1 | 203 | 1708 | 8.4 |
| 103422 at | CD1d | CD1d antigen | 1999 | 12449 | 6.2 | 1339 | 8025 | 6.0 | 2393 | 5418 | 2.3 |
| 161058 f at | R74862 | expressed sequence R74862 | 52 | 313 | 6.1 | 17 | 131 | 7.8 | 79 | 184 | 2.3 |
| 92356 at | Ptpn22 | protein tyrosine phosphatase, non-receptor type 22 | 1775 | 10160 | 5.7 | 4668 | 13103 | 2.8 | 7670 | 20357 | 2.7 |
| 95462 at | Bzw2 | basic leucine zipper and W2 domains 2 | 1482 | 8442 | 5.7 | 1678 | 4786 | 2.9 | 3611 | 17670 | 4.9 |
| 95661 at | CD9 | CD9 antigen | 480 | 2677 | 5.6 | 127 | 2212 | 17.4 | 550 | 2737 | 5.0 |
| 104701 at | Bh1hb2 | basic helix-loop-helix domain containing | 272 | 1490 | 5.5 | 5828 | 8002 | 1.4 | 5729 | 6426 | 1.1 |
| 160629 at | Rgs10 | regulator of G-protein signalling 10 | 450 | 2368 | 5.3 | 135 | 1234 | 9.2 | 419 | 838 | 2.0 |
| 97740 at | Dusp16 | dual specificity phosphatase 16 | 787 | 4075 | 5.2 | 1214 | 3889 | 3.2 | 1173 | 4992 | 4.3 |
| 102924 at | Dtx1 | deltex 1 homolog (Drosophila) | 7962 | 39206 | 4.9 | 9975 | 29520 | 3.0 | 7015 | 31023 | 4.4 |
| 93101 s at | Nedd4 | neural precursor cell expressed | 660 | 3176 | 4.8 | 480 | 1244 | 2.6 | 645 | 1626 | 2.5 |
| 93195 at | Mfhas1 | malignant fibrous histiocytoma amplified sequence | 1143 | 5462 | 4.8 | 2857 | 2424 | 0.8 | 3446 | 3592 | 1.0 |
| 101584 at | Rsu1 | Ras suppressor protein 1 | 2028 | 9591 | 4.7 | 2007 | 4411 | 2.2 | 1809 | 72 86 | 4.0 |
| 102721 at | IgG3 | Ig gamma-3 heavy chain precursor | 812 | 3415 | 4.2 | 1344 | 1948 | 1.4 | 1766 | 2882 | 1.6 |
| 98433 at | Bid | BH3 interacting domain death agonist | 1622 | 6473 | 4.0 | 689 | 1926 | 2.8 | 495 | 1277 | 2.6 |
| 101516 at | CD59a | CD59a antigen | 527 | 2091 | 4.0 | 410 | 1617 | 3.9 | 533 | 1817 | 3.4 |
| 102644 at | Mdfic | MyoD family inhibitor domain containing | 784 | 3083 | 3.9 | 533 | 1751 | 3.3 | 1426 | 2847 | 2.0 |
| 99071 at | Mpeg1 | macrophage expressed gene 1 | 2402 | 8938 | 3.7 | 491 | 1982 | 4.0 | 1743 | 5743 | 3.3 |
| 160487 at | My14 | myosin, light polypeptide 4 | 942 | 3407 | 3.6 | 2627 | 6593 | 2.5 | 2335 | 6783 | 2.9 |
| 102223 at | Pp1 | periplakin | 1294 | 4673 | 3.6 | 1482 | 2177 | 1.5 | 1404 | 2906 | 2.1 |
| 96283 at | Itm2c | integral membrane protein 2C | 1458 | 5254 | 3.6 | 881 | 1943 | 2.2 | 1073 | 3249 | 3.0 |
| 161765 f at | Rgs10 | regulator of G-protein signalling 10 | 535 | 1792 | 3.4 | 365 | 963 | 2.6 | 290 | 733 | 2.5 |
| 94958 at | 1110013L07Rik | RIKEN cDNA 1110013LC7 gene | 486 | 1575 | 3.2 | 220 | 548 | 2.5 | 129 | 336 | 2.6 |
| 101897 g at | CD1d | CD1d antigen | 5039 | 16158 | 3.2 | 2039 | 6961 | 3.4 | 3129 | 7065 | 2.3 |
| 102289 r at | CD21 | complement receptor 2 | 931 | 2964 | 3.2 | 2116 | 4053 | 1.9 | 1348 | 3034 | 2.3 |
| 97460 at | Ube2r2 | ubiquitin-coniugating enzyme E2R 2 | 9117 | 28949 | 3.2 | 5367 | 13029 | 2.4 | 10403 | 17285 | 1.7 |
| 102914 s at | Bc12a1 | B-cell leukemia/lymphoma 2 related protein A1 | 3284 | 10193 | 3.1 | 15571 | 28095 | 1.8 | 22037 | 32334 | 1.5 |
| 95084 f at | Grhpr | glyoxylate reductase/hydroxypyruvate reductase | 2362 | 7175 | 3.0 | 1612 | 3071 | 1.9 | 1565 | 2941 | 1.9 |
| 160711 at | Decr1 | 2,4-dienoyl CoA reductase 1, mitochondrial | 187 | 553 | 3.0 | 267 | 270 | 1.0 | 201 | 437 | 2.2 |
| 100397 at | DAP12 | TYRO protein tyrosine kinase binding protein | 4658 | 13714 | 2.9 | 751 | 2767 | 3.7 | 2109 | 3985 | 1.9 |
| 96735 at | Stard10 | START domain containing 10 | 2538 | 7449 | 2.9 | 2026 | 2932 | 1.4 | 1349 | 1734 | 1.3 |
| 92587 at | Fdx1 | ferredoxin 1 | 1631 | 4714 | 2.9 | 1510 | 2957 | 2.0 | 2305 | 3744 | 1.6 |
| 104298 at | 2310044G17Rik | RIKEN cDNA 2310044G17 gene | 1290 | 3689 | 2.9 | 1797 | 2292 | 1.3 | 1320 | 4032 | 3.1 |
| 104299 at | Zdhhc14 | zinc finger, DHHC domain containing 14 | 1176 | 3359 | 2.9 | 328 | 651 | 2.0 | 794 | 1865 | 2.4 |
| 160941 at | Pde8a | phosphodiesterase 8A | 383 | 1085 | 2.8 | 690 | 1016 | 1.5 | 414 | 1013 | 2.4 |
| 98822 at | G1p2 | interferon, alpha-inducible protein | 1381 | 3885 | 2.8 | 1335 | 2609 | 2.0 | 1244 | 3258 | 2.6 |
| 98033 at | 1100001H23Rik | RIKEN cDNA 1100001H23 gene | 4159 | 11695 | 2.8 | 4474 | 7701 | 1.7 | 6414 | 9391 | 1.5 |
| 94186 at | Traf1 | Tnf receptor-associated factor 1 | 1612 | 4530 | 2.8 | 1740 | 4258 | 2.4 | 1538 | 4368 | 2.8 |
| 160069 at | Gmnn | geminin | 422 | 1174 | 2.8 | 376 | 323 | 0.9 | 229 | 445 | 1.9 |
| 95758 at | Scd2 | stearoyl-Coenzyme A desaturase 2 | 4483 | 12233 | 2.7 | 1094 | 2501 | 2.3 | 703 | 1147 | 1.6 |
| 100880 at | 9830147J24Rik | RIKEN cDNA 9830147J24 gene | 1271 | 3463 | 2.7 | 733 | 951 | 1.3 | 676 | 1657 | 2.5 |
| 92850 at | Rrbp1 | ribosome binding protein 1 | 2821 | 7681 | 2.7 | 3262 | 6051 | 1.9 | 2147 | 5191 | 2.4 |
| 93013 at | Id2 | inhibitor of DNA binding 2 | 2417 | 6577 | 2.7 | 2786 | 11282 | 4.0 | 6513 | 15250 | 2.3 |
| 93261 at | Lgmn | legumain | 2555 | 6932 | 2.7 | 1582 | 3143 | 2.0 | 1838 | 2832 | 1.5 |
| 93833 s at | Hist1h2bc | histone 1, H2bc | 773 | 2093 | 2.7 | 617 | 697 | 1.1 | 1189 | 536 | 0.5 |
| 96688 at | Tmem77 | transmembrane protein 77 | 728 | 1935 | 2.7 | 320 | 860 | 2.7 | 572 | 1211 | 2.1 |
| 160762 at | Abr | active BCR-related gene | 706 | 1845 | 2.6 | 736 | 2020 | 2.7 | 620 | 1084 | 1.7 |
| 161788 f at | S1P1 | sphingolipid G-protein-coupled receptor 1 | 565 | 1476 | 2.6 | 1332 | 747 | 0.6 | 936 | 562 | 0.6 |
| 93483 at | Hck | hemopoietic cell kinase | 5919 | 15436 | 2.6 | 2869 | 9495 | 3.3 | 2498 | 6070 | 2.4 |
| 94995 at | A030007L17Rik | RIKEN cDNA A030007L17 gene | 848 | 2186 | 2.6 | 926 | 734 | 0.8 | 1184 | 817 | 0.7 |
| 92925 at | Cebpb | CCAAT/enhancer binding protein (C/EBP), beta | 1911 | 4882 | 2.6 | 12638 | 12321 | 1.0 | 20455 | 13386 | 0.7 |
| 100516 at | Chka | choline kinase alpha | 874 | 2195 | 2.5 | 1728 | 1646 | 1.0 | 880 | 728 | 0.8 |
| 104712 at | Myc | myelocytomatosis oncogene | 906 | 2242 | 2.5 | 4293 | 11096 | 2.6 | 4967 | 12611 | 2.5 |
| 92352 at | S1P3 | sphingolipid G-protein-coupled receptor 3 | 1522 | 3765 | 2.5 | 1223 | 1791 | 1.5 | 1370 | 2343 | 1.7 |
| 98931 at | Gns | glucosamine (N-acetyl)-6-sulfatase | 2595 | 6366 | 2.5 | 3560 | 4612 | 1.3 | 2685 | 4575 | 1.7 |
| 102410 at | Hs3st1 | heparan sulfate (glucosamine) 3-O-sulfotransferase | 575 | 1392 | 2.4 | 510 | 2503 | 4.9 | 9768 | 14854 | 1.5 |
| 97949 at | Fg12 | fibrinogen-like protein 2 | 314 | 752 | 2.4 | 126 | 541 | 4.3 | 234 | 1144 | 4.9 |
| 101495 at | CD81 | CD81 antigen | 8639 | 20584 | 2.4 | 5250 | 7579 | 1.4 | 5062 | 8355 | 1.7 |
| 98092 at | P1ac8 | placenta-specific 8 | 56686 | 134293 | 2.4 | 37938 | 75591 | 2.0 | 29999 | 77352 | 2.6 |
| 98417 at | Mx1 | myxovirus (influenza virus) resistance 1 | 260 | 608 | 2.3 | 190 | 309 | 1.6 | 104 | 206 | 2.0 |
| 103459 at | S1c39a6 | solute carrier family 39 (metal ion transporter) | 862 | 2007 | 2.3 | 1337 | 2067 | 1.5 | 971 | 1825 | 1.9 |
| 95358 at | Pip5k2a | phosphatidylinositol-4-phc sphate 5-kinase | 4119 | 9541 | 2.3 | 2763 | 5125 | 1.9 | 3479 | 6084 | 1.7 |
| 93084 at | S1c25a4 | solute carrier family 25 (adenine translocator) | 4789 | 11081 | 2.3 | 3596 | 5997 | 1.7 | 6120 | 7659 | 1.3 |
| 102217 at | Gprk5 | G protein-coupled receptor kinase 5 | 772 | 1784 | 2.3 | 450 | 1351 | 3.0 | 601 | 668 | 1.1 |
Identification of strain-specific differences in gene expression between resting FO and MZ B cells
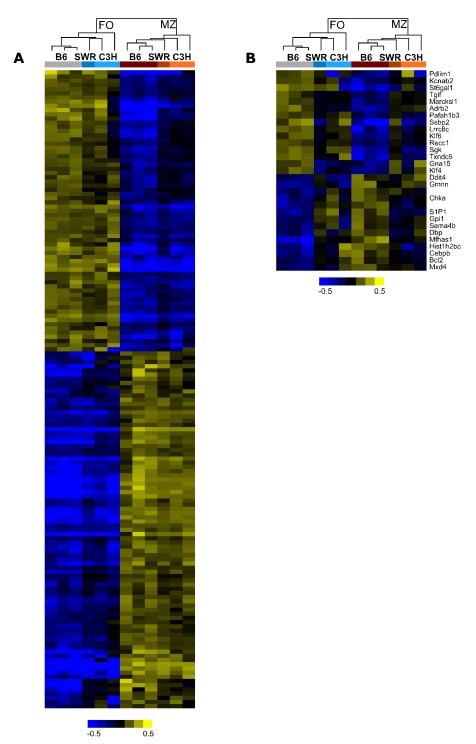
To determine if any strain-specific differences exist between MZ and FO B cell gene expression profiles, we expanded our gene expression analysis to include two additional mouse strains, C3H/HeJ (C3H) and SWR/J (SWR). C3H mice have an enlarged MZ B cell population relative to B6 mice while SWR mice have a smaller MZ B cell population relative to B6 mice (data not shown). The 181 transcripts found to be significantly different between FO and MZ B cells were analyzed for their expression levels in C3H and SWR mice, respectively. While the absolute signal intensities varied across strains (Table 1), the fold changes between MZ and FO B cell gene expression were comparable (Fig. 3A). We identified 29 genes (approximately 16% of total) that appeared to have a different expression profile between FO and MZ B cells in the C3H and SWR strains relative to the B6 strain (Fig. 3B and Table 2). These strain-specific differences might reflect changes in genes regulating MZ B cell size, strain-specific functional differences, or polymorphisms that influence probe hybridization but have no functional consequences.
Figure 3. Identification of strain-specific differences in gene expression profiles between FO and MZ B cells.
Gene expression profile of splenic FO and MZ B cells from B6, SWR, and C3H mice. The profile includes 181 gene transcripts with a fold change > 2 and a p value < 0.02 by T-test. (A) Hierarchical analysis of 152 genes with consistent regulation across the three mouse strains. (B) Hierarchical analysis of 29 genes with strain-specific differences in MZ vs. FO gene expression profiles. Shown are normalized expression values greater than (yellow), near (black), or less than (blue) the mean of that strain. Each column represents one sample of sorted FO or MZ B cells. Genes or transcripts are represented in rows. Clustering of genes is unsupervised.
Table 2.
Genes differentially expressed between FO and MZ B cells from B6, SWR, and C3H strains of mice.
| Relative Expression | Relative Expression | Relative Expression | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Affy_ID | Gene Symbol | Gene Title | B6FO | B6MZ | Fold Difference (MZ/FO) | SWR FO | SWR MZ | Fold Difference (MZ/FO) | C3H FO | C3H MZ | Fold Difference (MZ/FO) |
| 93195_at | Mfhas1 | malignant fibrous histiocytoma amplified sequence | 1143 | 5462 | 4.8 | 2857 | 2424 | 0.85 | 3446 | 3592 | 1.04 |
| 160069 at | Gmnn | geminin | 422 | 1174 | 2.8 | 376 | 323 | 0.86 | 229 | 445 | 1.94 |
| 93833_s_at | Hist1h2bc | histone 1, H2bc | 773 | 2093 | 2.7 | 617 | 697 | 1.13 | 1189 | 536 | 0.45 |
| 161788_f_at | S1P1 | sphingolipid G-protein-coupled receptor 1 | 565 | 1476 | 2.6 | 1332 | 747 | 0.56 | 936 | 562 | 0.60 |
| 94995 at | RIKEN cDNA A030007L17 gene | 848 | 2186 | 2.6 | 926 | 734 | 0.79 | 1184 | 817 | 0.69 | |
| 92925_at | Cebpb | CCAAT/enhancer binding protein (C/EBP), beta | 1911 | 4882 | 2.6 | 12638 | 12321 | 0.97 | 20455 | 13386 | 0.65 |
| 100516_at | Chka | choline kinase alpha | 874 | 2195 | 2.5 | 1728 | 1646 | 0.95 | 880 | 728 | 0.83 |
| 160841_at | Dbp | D site albumin promoter binding protein | 239 | 540 | 2.3 | 226 | 129 | 0.57 | 293 | 336 | 1.15 |
| 102104_f_at | est | 2147 | 4844 | 2.3 | 1411 | 2614 | 1.85 | 1284 | 1133 | 0.88 | |
| 99024 at | Mxd4 | Max dimerization protein 4 | 5977 | 12958 | 2.2 | 6053 | 6769 | 1.12 | 6873 | 6672 | 0.97 |
| 95387_f_at | Sema4b | semaphorin 4B | 8465 | 17775 | 2.1 | 4483 | 3837 | 0.86 | 4046 | 4411 | 1.09 |
| 103460_at | Ddit4 | DNA-damage-inducible transcript 4 | 3274 | 6711 | 2.0 | 1631 | 1280 | 0.78 | 1275 | 3062 | 2.40 |
| 100573_f_at | Gpi1 | glucose phosphate isomerase 1 | 1410 | 2889 | 2.0 | 3126 | 1986 | 0.64 | 1911 | 2109 | 1.10 |
| 98868_at | Bcl2 | B-cell leukemia/lymphoma 2 | 1203 | 2437 | 2.0 | 1411 | 1261 | 0.89 | 1030 | 833 | 0.81 |
| 9443l_at | St6gal1 | beta galactoside alpha 2,6 sialyltransferase 1 | 1562 | 331 | 4.7 | 922 | 410 | 2.25 | 850 | 872 | 0.98 |
| 103504_at | Ssbp2 | single-stranded DNA binding protein 2 | 1538 | 335 | 4.6 | 85 | 224 | 0.38 | 496 | 182 | 2.72 |
| 98918 at | Txndc5 | thioredoxin domain containing 5 | 5318 | 1447 | 3.7 | 745 | 893 | 0.83 | 3928 | 1955 | 2.01 |
| 104523_at | Lrrc8c | leucine rich repeat containing 8 family, member C | 1533 | 483 | 3.2 | 905 | 1098 | 0.82 | 970 | 693 | 1.40 |
| 97890_at | Sgk | serum/glucocorticoid regulated kinase | 1107 | 351 | 3.2 | 1857 | 1923 | 0.97 | 1674 | 717 | 2.33 |
| 93193_at | Adrb2 | adrenergic receptor, beta 2 | 5831 | 1961 | 3.0 | 8025 | 9318 | 0.86 | 3784 | 5394 | 0.70 |
| 98083_at | Klf6 | Kruppel like factor 6 | 4002 | 1522 | 2.6 | 14189 | 19034 | 0.75 | 18319 | 14977 | 1.22 |
| 99622_at | Klf4 | Kruppel-like factor 4 | 697 | 278 | 2.5 | 10472 | 25698 | 0.41 | 18855 | 23655 | 0.80 |
| 102892_at | Kcnab2 | potassium voltage-gated channel | 2707 | 1133 | 2.4 | 2525 | 2350 | 1.07 | 1751 | 1970 | 0.89 |
| 100554_at | Pdlim1 | PDZ and LIM domain 1 (elfin) | 2352 | 1009 | 2.3 | 443 | 336 | 1.32 | 147 | 243 | 0.61 |
| 97203 at | Marcksl1 | MARCKS-like 1 | 2027 | 900 | 2.3 | 5719 | 7047 | 0.81 | 5684 | 6025 | 0.94 |
| 94753_at | Gna15 | guanine nucleotide binding protein, alpha 15 | 689 | 315 | 2.2 | 51 | 136 | 0.37 | 84 | 136 | 0.61 |
| 98335_at | Recc1 | replication factor C 1 | 2339 | 1104 | 2.1 | 1421 | 1647 | 0.86 | 1758 | 1339 | 1.31 |
| 101502_at | Tgif | TG interacting factor | 2690 | 1292 | 2.1 | 27576 | 36312 | 0.76 | 23520 | 24132 | 0.97 |
| 100576_at | Pafah1b3 | platelet-activating factor acetylhydrolase | 3106 | 1545 | 2.0 | 1069 | 1280 | 0.84 | 1627 | 988 | 1.65 |
D6 Beta Chemokine Receptor and RGS10 Are More Highly Expressed in MZ than FO B cells
MZ B cells provide a rapid response to blood-borne bacterial particulates, in part because of their localization in the spleen. For example, blood-borne antigens accumulate within the splenic marginal zone as early as 30 min. following i.v. immunization (8), giving an opportunity for MZ B cells to sample blood and respond rapidly to an antigen. A number of factors have been shown to play a role in MZ B cell localization within the splenic microenvironment including S1P1 (19), the presence of marginal zone macrophages (20), and integrins (21). In addition, in vivo injection of pertussis toxin disrupts MZ localization, suggesting involvement of G protein-coupled receptor(s) (22). The current microarray data identified a number of molecules that are potentially involved in the migration, localization, and/or retention of MZ B cells in the splenic marginal zone. Two proteins more highly expressed in MZ B cells relative to FO B cells were the D6 beta chemokine receptor (D6) and the regulator of G-protein signaling (RGS10) protein. To confirm that these two proteins are indeed more highly expressed in MZ B cells, resting splenic MZ and FO B cells were sort-purified and analyzed for the level of D6 and RGS10 mRNA (Fig. 4A) and protein (Fig. 4B-D) by RT-PCR, Western blot, and FACS, respectively. Thus, resting MZ B cells express D6 and RGS10 at higher levels than FO B cells, with the potential to be involved in MZ B cell localization.
Figure 4. MZ B cells express higher levels of D6 and RGS10 relative to FO B cells.
Resting splenic MZ and FO B cells were sort-purified and total RNA and protein isolated. Resting MZ B cells express higher (A) mRNA and (B) protein levels of D6 and RGS10, as determined by RT-PCR and Western blot, respectively. Total splenocytes were isolated and analyzed via FLOW cytometry. The expression level of (C) D6 Isotype 0.3%, MZ B cell 88.8%, and FO B cell 11.8% and (D) RGS10 Isotype 0.2%, MZ B cell 82.1%, and FO B cell 1.1% are displayed as a histogram plot.
Gene Expression Profile of MZ Id+ B cells before and after stimulation
In addition to the differential phenotype of resting FO and MZ B cells, MZ B cells respond very differently to antigen than FO B cells. Following activation with antigen, MZ B cells increase B7-1 and B7-2 expression, develop into plasmablasts more readily, and are more sensitive to LPS stimulation than their FO counterparts (5, 6). In addition to rapid production of IgM antibody, MZ B cells also possess the ability to efficiently activate naive T cells (8). However, the genes that are rapidly up regulated and down regulated in MZ B cells following activation with antigen have not been fully characterized. To determine the gene expression profile of antigen (idiotype) positive MZ B cells before and after activation, M167 Tg mice were immunized i.v. with heat-killed Streptococcus pneumoniae, R36A, and Id+ and Id− MZ B cells were sort-purified at 0 and 1 hour following immunization. The samples were analyzed via DNA microarray analysis as described for the resting MZ vs. FO B cell microarray above. The gene transcripts identified to significantly increase or decrease were grouped into the following broad functional classifications: Figure 5 (A) chemokines, (B) chemokine receptors, (C) cytokines, (D) cytokine receptors, Figure 6 (A) apoptosis (B) immune cell markers.
Figure 5. Regulated genes in Idiotype Positive MZ B cells after Activation.
MZ Id+ (Ag+) and Id− (Ag−) B cells were isolated from M167 Tg mice at 0 and 1 hour after i.v. immunization with heat killed S. pneumoniae, R36A. DNA microarray analysis identified genes that were significantly up regulated and down regulated in the Id+ MZ B cells 1 hr after activation. The genes specifically regulated in the Id+ MZ B cells were grouped into various functional categories (A) Chemokines, (B) Chemokine Receptors, (C) Cytokines, and (D) Cytokine Receptors. Shown are normalized expression values greater than (yellow), near (black), or less than (blue) the mean of that gene. Each column represents one independent sample. Genes or transcripts are represented in rows. Clustering of the genes is unsupervised.
Figure 6. Regulated genes in Idiotype Positive MZ B cells after Activation.
MZ Id+ (Ag+) and Id− (Ag−) B cells were isolated from M167 Tg mice at 0 and 1 hour after i.v. immunization with heat killed S. pneumoniae, R36A. DNA microarray analysis identified genes that were significantly up regulated and down regulated in the Id+ MZ B cells 1 hr after activation. The genes specifically regulated in the Id+ MZ B cells were grouped into various functional categories (A) Apoptosis and (B) Immune Cell Markers. Shown are normalized expression values greater than (yellow), near (black), or less than (blue) the mean of that gene. Each column represents one independent sample. Genes or transcripts are represented in rows. Clustering of the genes is unsupervised.
We focused on the antigen responsive Id+ MZ B cells and the genes that were regulated following i.v. immunization with R36A. The Id+ MZ B cell gene expression profile exhibited an activated phenotype at 1 hr post immunization, as would be predicted. The Id+ MZ B cells rapidly down regulated pro-apoptotic genes such as multiple caspase proteins, annexin A4, and programmed cell death proteins while concurrently up regulating anti-apoptotic genes such as Bcl-like proteins. MZ B cells also up regulated a number of cytokine genes including IL-10, IL-6, TGF-β, and IL-1β while down regulating many cytokine receptors. Chemokine ligands such as CXCL10, CXCL2, CCL3, CXCL5, and CCL4 were up regulated while chemokine receptors were either up regulated (CCR7) or down regulated (D6, CCR5, and RDC-1). In addition, we cross-referenced the 99 transcripts that were more highly expressed in the MZ B cells relative to FO B cells with the expression profile in the Id+ MZ B cells 1 hour after activation to determine if any significant changes occurred (Table 3). 6 of 99 genes (6 %) more highly expressed in MZ B cells were up regulated after activation in the MZ Id+ B cells while 17 of 99 genes (17 %) were down regulated. Taken together, these results suggest that Id+ MZ B cells have a unique gene expression profile following i.v. immunization with R36A.
Table 3.
Genes more highly expressed in MZ B cells relative to FO B cells and specifically regulated in MZ Id+ B cells following activation.
| Relative Expression | Relative Expression | |||||||
|---|---|---|---|---|---|---|---|---|
| Affy_ID | Gene Symbol | Gene Title | B6 FO | B6 MZ | Fold Difference (MZ/FO) | MZ Id+ 0hr | MZ Id+ 1hr | Fold Difference (1hr/0hr) |
| 95462_at | Bzw2 | basic leucine zipper and W2 domains 2 | 1482 | 8442 | 5.7 | 3855 | 13947 | 3.6 |
| 92217_s_at | Gp49 | glycoprotein 49 A/B | 161 | 2762 | 17.2 | 2530 | 13563 | 5.4 |
| 100325_at | Gp49 | glycoprotein 49 A/B | 394 | 5124 | 13.0 | 2530 | 13563 | 5.4 |
| 93013_at | Id2 | inhibitor ol DNA binding 2 | 2417 | 6577 | 2.7 | 9220 | 46793 | 5.1 |
| 95661_at | CD9 | CD9 antigen | 480 | 2677 | 5.6 | 8265 | 16347 | 2.0 |
| 104701_at | Bhlhb2 | basic helix-loop-helix domain containing | 272 | 1490 | 5.5 | 6307 | 22934 | 3.6 |
| 102914_s_at | Bcl2a1 | B-cell leukemia/lymphoma 2 related protein A1 | 3284 | 10193 | 3.1 | 30696 | 152568 | 5.0 |
| 161788_f_at | S1P1 | sphingolipid G-protein-coupled receptor 1 | 565 | 1476 | 2.6 | 129 | 18 | 0.14 |
| 97740_at | Dusp16 | dual specilicity phosphatase 16 | 787 | 4075 | 5.2 | 312 | 9 | 0.03 |
| 93101_s_at | Nedd4 | neural precursor cell expressed | 660 | 3176 | 4.8 | 3668 | 565 | 0.15 |
| 99071_at | Mpeg1 | macrophage expressed gene 1 | 2402 | 8938 | 3.7 | 55 | 13 | 0.24 |
| 95758_at | Scd2 | stearoyl-Coenzyme A desaturase 2 | 4483 | 12233 | 2.7 | 6858 | 821 | 0.12 |
| 160711_at | Decr1 | 2,4-dienoyl CoA reductase 1, mitochondrial | 187 | 553 | 3.0 | 670 | 364 | 0.54 |
| 160069_at | Gmnn | geminin | 422 | 1174 | 2.8 | 905 | 216 | 0.24 |
| 102410_at | Hs3st1 | heparan sullate (glucosamine) 3-O-sullotranslerase | 575 | 1392 | 2.4 | 2749 | 254 | 0.09 |
| 97949_at | Fgl2 | fibrinogen-like protein 2 | 314 | 752 | 2.4 | 570 | 242 | 0.42 |
| 98417_at | Mx1 | myxovirus (influenza virus) resistance 1 | 260 | 608 | 2.3 | 428 | 233 | 0.54 |
| 160841_at | Dbp | D site albumin promoter binding protein | 239 | 540 | 2.3 | 516 | 121 | 0.23 |
| 95387_f_at | Sema4b | semaphorin 4B | 8465 | 17775 | 2.1 | 1495 | 271 | 0.18 |
| 98026_g_at | Evi2a | ecotropic viral integration site 2a | 3528 | 7178 | 2.0 | 618 | 271 | 0.44 |
| 93430_at | Cmkor1 | chemokine orphan receptor 1 | 78 | 4369 | 56.0 | 8291 | 1092 | 0.13 |
| 160495_at | Ahr | aryl-hydrocarbon receptor | 31 | 249 | 8.0 | 735 | 404 | 0.55 |
| 98309_at | Ccbp2 | Chemokine binding protein 2 | 481 | 3549 | 7.4 | 3011 | 310 | 0.10 |
| 103422_at | CD1d | CD1d antigen | 1999 | 12449 | 6.2 | 22540 | 4484 | 0.20 |
Id+ MZ B cells upregulate IL-10 and Stra13 in response to R36A immunization
A number of interesting genes were identified by DNA microarray analysis on sort-purified MZ Id+ and Id− B cells at 0 and 1 hour following i.v. immunization with R36A. Two of these genes that warranted further investigation were IL-10 and Stra13. IL-10 is an immunoregulatory cytokine that plays a role in negatively regulating inflammatory immune responses and B cells have been shown to secrete IL-10 (23). To confirm whether Id+ MZ B cells are activated to secrete IL-10 in response to R36A, we crossed the M167 heavy chain immunoglobulin tg mouse with an IL-10/Thy1.1 reporter mouse in which all IL-10+ cells are Thy1.1+ (14), immunized with R36A, and analyzed isolated Id+ MZ B cells for the presence of IL-10 and Thy1.1 mRNA (Fig. 7A) and the Thy1.1 reporter protein (Fig. 7B). As expected, IL-10 and Thy1.1 mRNA increased only in the Id+ MZ B cells following immunization with R36A. The difference in degree of induction between IL-10 and Thy1.1 is most likely due to the copy number of the Thy1.1 transgene, which is estimated to be at least 12 copies (14). Stra13 is a basic helix-loop-helix domain containing class B2 protein that is thought to be a negative regulator of B cells (24). To confirm that Stra13 is up regulated following activation of MZ B cells, MZ Id+ B cells were isolated before and after immunization with R36A and analyzed for the level of Stra13 mRNA (Fig. 7A). As expected, Stra13 mRNA increased only in the Id+ MZ B cells following immunization with R36A. Thus, the DNA microarray analysis of Id+ and Id− MZ B cells at 0 and 1 hours following immunization with R36A identified multiple genes of interest that were rapidly up regulated or down regulated after activation including IL-10 and Stra13.
Figure 7. IL-10 and Stra13 are increased following R36A immunization.
M167 tg mice were crossed with an IL-10/Thy1.1 reporter mouse and immunized i.v. with R36A. MZ Id+ B cells were sort-purified at 0, 1, and 4 hours after immunization and total RNA was isolated. (A) RT-PCR was performed using gene-specific primers for IL-10, Thy1.1, Stra13, and actin. The expression level of (B) Thy1.1 was determined via FACS analysis on gated MZ Id+ B cells at 24 hours following R36A immunization. MZ Id+ B cells were approximately 5% (PBS) and 20% (R36A) positive for Thy1.1 respectively.
Discussion
Previous data studying FO and MZ B cells have shown that these B cell subsets differ based on their anatomical location in the spleen, cellular surface molecule expression, and effector functions [reviewed in (1)]. We set out to identify new genes and pathways that are differentially expressed between FO and MZ B cells and those that were specifically up regulated or down regulated within each subset following activation. DNA microarray allows for a high throughput analysis of genomic expression differences between two sample populations. This approach identified 181 genes differentially expressed between resting MZ and FO B cells. 99 genes were more highly expressed in MZ B cells while 82 genes were more highly expressed in FO B cells. In addition, DNA microarray analysis of MZ Id+ and Id− B cells before (0hr) and after (1hr) R36A immunization revealed many new genes and pathways specifically regulated in the MZ Id+ B cells. These findings further our understanding of MZ and FO B cell biology while at the same time identifying new candidate genes and pathways to study.
The MZ vs. FO B cell microarray used cells that were isolated by gating on B220 and then sorted based on surface expression patterns of CD21hiCD23lo for MZ B cells and CD21loCD23hi for FO B cells. As expected, our DNA microarray results showed higher mRNA expression of CD21 and lower expression of CD23 in MZ B cells relative to FO B cells. MZ B cells are known to express surface CD9 and CD1d, while FO B cells express little to no CD9 and CD1d (9, 25). Similarly, our microarray results showed a higher expression of CD9 and CD1d on MZ B cells relative to FO B cells. Furthermore, S1P1 and S1P3 were previously shown to be expressed at higher levels on MZ B cells relative to FO B cells, while S1P4 being expressed higher on FO B cells (19). Our data was again consistent with what has been shown in the literature, showing higher expression of S1P1 and S1P3 on MZ B cells and higher expression of S1P4 on FO B cells. Taken together, it appears that our DNA microarray data agrees with what has been shown in the literature with respect to known phenotypic differences between MZ and FO B cells, suggesting that our sorted B cell populations were pure and our DNA microarray method of analysis is valid.
One interesting gene more highly expressed in MZ B cells relative to FO B cells was RGS10. RGS10 has been previously confirmed to be specifically expressed in MZ B cells (mRNA) as well as plasma cells (26). RGS10 attenuates signaling pathways via increased GTPase activity to specific G-alpha subunits (27). Phosphorylation by PKA induces its localization to the nucleus (28). Recently, RGS10−/− mice were reported and exhibited severe osteopetrosis and impaired osteoclast differentiation resulting from the loss of [Ca2+]i oscillation regulation (29), though no immune characterization was reported. While RGS10 was more highly expressed in resting MZ B cells relative to FO B cells, RGS10 mRNA was not found to be regulated following activation in MZ Id+ B cells. However, since chemokine receptors are G-protein coupled, a protein that regulates their signaling capacity might play an important role in localization, maintenance, or migration of MZ B cells. For example, RGS1, RGS3, and RGS4 introduction into B cell lines dramatically alters chemokine-induced cell migration (30-32). Taken together, MZ B cell-specific expression of RGS10 potentially plays a role in regulating the ability to respond to chemokine signals and might play a role in MZ B cell localization.
An additional gene more highly expressed in MZ B cells was D6. D6 is proposed to be a decoy chemokine receptor that has the ability to bind, internalize, and degrade chemokine ligands through a β-arrestin-dependent mechanism, a function termed chemokine scavenging (33-36). Interestingly, our results show that D6 is more highly expressed in resting MZ B cells relative to FO B cells and D6 is rapidly downregulated (10-fold) following activation. Given its proposed property of a chemokine sink, and the fact that D6 has not been shown to signal intracellularly, the potential exists that D6 expression on the surface of MZ B cells is involved in keeping them properly localized within the splenic microenvironment. Rapid down regulation of D6 after activation potentially enhances the migration of MZ B cells to the T:B cell border. D6 expression has been reported in B cells previously (37), though the differential expression in B cell subsets was not investigated. Thus, D6 appears to be an interesting candidate gene potentially involved in MZ B cell localization, maintenance, and/or migration.
Our second DNA microarray experiment was aimed at identifying genes that were specifically up regulated or down regulated following activation. Using the M167 tg mouse, we sort-purified MZ Id+ and Id− B cells at 0 and 1 hr post i.v. immunizaton with R36A. S1P1 transcripts were rapidly down regulated (7.0-fold) following activation only in the Id+ MZ B cells, which is in agreement with Cyster et. al. (19). S1P1 has been shown to play a role in the migration of MZ B cells to the T:B border following activation. In addition, the MZ B cell-specific marker, CD9, was increased 2.0-fold following activation, consistent with our previous findings (9). Of additional interest, only the MZ Id+ and not MZ Id− B cells rapidly increased anti-apoptotic genes and decreased pro-apoptotic genes, a phenotype consistent with cellular activation. This shows a remarkable degree of antigen specificity in that virtually no concurrent increases were detected in the MZ Id+ and Id− B cell populations at 1 h. Our microarray results appear to agree with a number of well studied genes already published in the literature with respect to MZ B cells, indicating that both our cell sort and microarray analyses are accurate. Consequently, further analysis and weight can be given to the other genes found to be regulated following immunization.
One interesting gene identified by microarray analysis to be specifically increased only in the Id+ MZ B cells was IL-10. Interestingly, MZ B cells have been suggested to play an immunoregulatory role through secretion of IL-10 (38). IL-10 is an immunoregulatory cytokine that plays an important role in negatively regulating inflammatory immune responses. A variety of cells are capable of producing IL-10 including Th2, Treg, B-1 and MZ B cells (39). The effects of IL-10 are mainly immunosuppressive, but also depend on what cell type is being affected by IL-10. In EAE, an experimental model of MS, one study suggested that B cells regulate Treg cells via B7 and IL-10 to suppress autoimmune inflammation (40). Besides the immunosuppressive role of IL-10, it has been suggested to play a role in B cell antibody production. Addition of IL-10 to human B cell cultures is reported to increase class switch recombination and production of IgA and IgG (41-43). However, the specific B cell subset, its location in the spleen before and after stimulation, and the signals required to produce IL-10 are not fully understood.
Stra13 was another interesting gene that was rapidly up regulated following activation. Stra13 is a basic helix-loop-helix domain containing class B2 protein that is thought to be a negative regulator of B cells (24). Stra13−/− mice develop autoimmune disease characterized by accumulation of spontaneously activated T and B cells, circulating autoantibodies, infiltration of T and B cells into several organs and immune complex deposition in glomeruli (44). Stra13 transgenic mice show impaired development of T and B cells, with the expansion of progenitor B and T cells most strongly affected (45). Of interest, Stra13 is developmentally regulated in B cells and decreases after activation in germinal center B cells (45). Our results in Id+ MZ B cells show that Stra13 increases after activation, although the functional relevance of this regulation is currently unknown.
The goal of this study was two fold; to identify genes that were differentially expressed between resting FO and MZ B cells, and to identify genes that were specifically regulated in MZ Id+ B cells following activation. The results generated give a genome wide look at the genes differentially expressed in FO and MZ B cells that potentially account for their differences in localization and function. Furthermore, the second microarray gave a comparative snapshot at one hour of the gene expression profiles between antigen-specific versus non-specific MZ B cells. One major problem with DNA microarray analysis is that many of the genes reported have not been studied, making conclusions difficult. However, a multitude of data is present here with respect to FO and MZ B cell biology, which will facilitate identification of new genes and pathways to explore.
Acknowledgements
The authors gratefully acknowledge Dr. Casey Weaver (University of Alabama at Birmingham) for generously sharing the Thy1.1/IL-10 reporter mice.
Footnotes
This work was supported by research funds from the National Institutes of Health (NIH) Grant AI14782. N.W.K. is a recipient of a Training Grant Postdoctoral Fellowship Award from NIH Grant T32 AI7051.
References
- 1.Martin F, Kearney JF. Marginal-zone B cells. Nat Rev Immunol. 2002;2:323–335. doi: 10.1038/nri799. [DOI] [PubMed] [Google Scholar]
- 2.Dammers PM, Visser A, Popa ER, Nieuwenhuis P, Kroese FG. Most marginal zone B cells in rat express germline encoded Ig VH genes and are ligand selected. J Immunol. 2000;165:6156–6169. doi: 10.4049/jimmunol.165.11.6156. [DOI] [PubMed] [Google Scholar]
- 3.Bendelac A, Bonneville M, Kearney JF. Autoreactivity by design: innate B and T lymphocytes. Nat Rev Immunol. 2001;1:177–186. doi: 10.1038/35105052. [DOI] [PubMed] [Google Scholar]
- 4.Kearney JF. Innate-like B cells. Springer Semin Immunopathol. 2005;26:377–383. doi: 10.1007/s00281-004-0184-0. [DOI] [PubMed] [Google Scholar]
- 5.Oliver AM, Martin F, Gartland GL, Carter RH, Kearney JF. Marginal zone B cells exhibit unique activation, proliferative and immunoglobulin secretory responses. Eur J Immunol. 1997;27:2366–2374. doi: 10.1002/eji.1830270935. [DOI] [PubMed] [Google Scholar]
- 6.Oliver AM, Martin F, Kearney JF. IgMhighCD21high lymphocytes enriched in the splenic marginal zone generate effector cells more rapidly than the bulk of follicular B cells. J Immunol. 1999;162:7198–7207. [PubMed] [Google Scholar]
- 7.Cinamon G, Zachariah MA, Lam OM, Foss FW, Jr., Cyster JG. Follicular shuttling of marginal zone B cells facilitates antigen transport. Nat Immunol. 2008;9:54–62. doi: 10.1038/ni1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Attanavanich K, Kearney JF. Marginal zone, but not follicular B cells, are potent activators of naive CD4 T cells. J Immunol. 2004;172:803–811. doi: 10.4049/jimmunol.172.2.803. [DOI] [PubMed] [Google Scholar]
- 9.Won WJ, Kearney JF. CD9 is a unique marker for marginal zone B cells, B1 cells, and plasma cells in mice. J Immunol. 2002;168:5605–5611. doi: 10.4049/jimmunol.168.11.5605. [DOI] [PubMed] [Google Scholar]
- 10.Won WJ, Foote JB, Odom MR, Pan J, Kearney JF, Davis RS. Fc receptor homolog 3 is a novel immunoregulatory marker of marginal zone and B1 B cells. J Immunol. 2006;177:6815–6823. doi: 10.4049/jimmunol.177.10.6815. [DOI] [PubMed] [Google Scholar]
- 11.Won WJ, Bachmann MF, Kearney JF. CD36 Is Differentially Expressed on B cell Subsets during Development and In Responses to Antigen. J Immunol. 2007;179 doi: 10.4049/jimmunol.180.1.230. [DOI] [PubMed] [Google Scholar]
- 12.Goodnow CC, Crosbie J, Adelstein S, Lavoie TB, Smith-Gill SJ, Brink RA, Pritchard-Briscoe H, Wotherspoon JS, Loblay RH, Raphael K, et al. Altered immunoglobulin expression and functional silencing of self-reactive B lymphocytes in transgenic mice. Nature. 1988;334:676–682. doi: 10.1038/334676a0. [DOI] [PubMed] [Google Scholar]
- 13.Storb U, Pinkert C, Arp B, Engler P, Gollahon K, Manz J, Brady W, Brinster RL. Transgenic mice with mu and kappa genes encoding antiphosphorylcholine antibodies. J Exp Med. 1986;164:627–641. doi: 10.1084/jem.164.2.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maynard CL, Harrington LE, Janowski KM, Oliver JR, Zindl CL, Rudensky AY, Weaver CT. Regulatory T cells expressing interleukin 10 develop from Foxp3+ and Foxp3− precursor cells in the absence of interleukin 10. Nat Immunol. 2007;8:931–941. doi: 10.1038/ni1504. [DOI] [PubMed] [Google Scholar]
- 15.Liu J, Karypis G, Hippen KL, Vegoe AL, Ruiz P, Gilkeson GS, Behrens TW. Genomic view of systemic autoimmunity in MRLlpr mice. Genes Immun. 2006;7:156–168. doi: 10.1038/sj.gene.6364286. [DOI] [PubMed] [Google Scholar]
- 16.Kin NW, Sanders VM. CD86 Regulates IgG1 Production via a CD19-Dependent Mechanism. J Immunol. 2007;179:1516–1523. doi: 10.4049/jimmunol.179.3.1516. [DOI] [PubMed] [Google Scholar]
- 17.Kin NW, Sanders VM. CD86 stimulation on a B cell activates the phosphatidylinositol 3-kinase/Akt and phospholipase C gamma 2/protein kinase C alpha beta signaling pathways. J Immunol. 2006;176:6727–6735. doi: 10.4049/jimmunol.176.11.6727. [DOI] [PubMed] [Google Scholar]
- 18.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cinamon G, Matloubian M, Lesneski MJ, Xu Y, Low C, Lu T, Proia RL, Cyster JG. Sphingosine 1-phosphate receptor 1 promotes B cell localization in the splenic marginal zone. Nat Immunol. 2004;5:713–720. doi: 10.1038/ni1083. [DOI] [PubMed] [Google Scholar]
- 20.Karlsson MC, Guinamard R, Bolland S, Sankala M, Steinman RM, Ravetch JV. Macrophages control the retention and trafficking of B lymphocytes in the splenic marginal zone. J Exp Med. 2003;198:333–340. doi: 10.1084/jem.20030684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu TT, Cyster JG. Integrin-mediated long-term B cell retention in the splenic marginal zone. Science. 2002;297:409–412. doi: 10.1126/science.1071632. [DOI] [PubMed] [Google Scholar]
- 22.Guinamard R, Okigaki M, Schlessinger J, Ravetch JV. Absence of marginal zone B cells in Pyk-2-deficient mice defines their role in the humoral response. Nat Immunol. 2000;1:31–36. doi: 10.1038/76882. [DOI] [PubMed] [Google Scholar]
- 23.Fillatreau S, Sweenie CH, McGeachy MJ, Gray D, Anderton SM. B cells regulate autoimmunity by provision of IL-10. Nat Immunol. 2002;3:944–950. doi: 10.1038/ni833. [DOI] [PubMed] [Google Scholar]
- 24.Seimiya M, Bahar R, Wang Y, Kawamura K, Tada Y, Okada S, Hatano M, Tokuhisa T, Saisho H, Watanabe T, Tagawa M, J. OW. Clast5/Stra13 is a negative regulator of B lymphocyte activation. Biochem Biophys Res Commun. 2002;292:121–127. doi: 10.1006/bbrc.2002.6605. [DOI] [PubMed] [Google Scholar]
- 25.Roark JH, Park SH, Jayawardena J, Kavita U, Shannon M, Bendelac A. CD1.1 expression by mouse antigen-presenting cells and marginal zone B cells. J Immunol. 1998;160:3121–3127. [PubMed] [Google Scholar]
- 26.Haller C, Fillatreau S, Hoffmann R, Agenes F. Structure, chromosomal localization and expression of the mouse regulator of G-protein signaling10 gene (mRGS10) Gene. 2002;297:39–49. doi: 10.1016/s0378-1119(02)00883-1. [DOI] [PubMed] [Google Scholar]
- 27.Hunt TW, Fields TA, Casey PJ, Peralta EG. RGS10 is a selective activator of G alpha i GTPase activity. Nature. 1996;383:175–177. doi: 10.1038/383175a0. [DOI] [PubMed] [Google Scholar]
- 28.Burgon PG, Lee WL, Nixon AB, Peralta EG, Casey PJ. Phosphorylation and nuclear translocation of a regulator of G protein signaling (RGS10) J Biol Chem. 2001;276:32828–32834. doi: 10.1074/jbc.M100960200. [DOI] [PubMed] [Google Scholar]
- 29.Yang S, Li YP. RGS10-null mutation impairs osteoclast differentiation resulting from the loss of [Ca2+]i oscillation regulation. Genes Dev. 2007;21:1803–1816. doi: 10.1101/gad.1544107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bowman EP, Campbell JJ, Druey KM, Scheschonka A, Kehrl JH, Butcher EC. Regulation of chemotactic and proadhesive responses to chemoattractant receptors by RGS (regulator of G-protein signaling) family members. J Biol Chem. 1998;273:28040–28048. doi: 10.1074/jbc.273.43.28040. [DOI] [PubMed] [Google Scholar]
- 31.Moratz C, Kang VH, Druey KM, Shi CS, Scheschonka A, Murphy PM, Kozasa T, Kehrl JH. Regulator of G protein signaling 1 (RGS1) markedly impairs Gi alpha signaling responses of B lymphocytes. J Immunol. 2000;164:1829–1838. doi: 10.4049/jimmunol.164.4.1829. [DOI] [PubMed] [Google Scholar]
- 32.Reif K, Cyster JG. RGS molecule expression in murine B lymphocytes and ability to down-regulate chemotaxis to lymphoid chemokines. J Immunol. 2000;164:4720–4729. doi: 10.4049/jimmunol.164.9.4720. [DOI] [PubMed] [Google Scholar]
- 33.Nibbs RJ, Wylie SM, Pragnell IB, Graham GJ. Cloning and characterization of a novel murine beta chemokine receptor, D6. Comparison to three other related macrophage inflammatory protein-1alpha receptors, CCR-1, CCR-3, and CCR-5. J Biol Chem. 1997;272:12495–12504. doi: 10.1074/jbc.272.19.12495. [DOI] [PubMed] [Google Scholar]
- 34.Fra AM, Locati M, Otero K, Sironi M, Signorelli P, Massardi ML, Gobbi M, Vecchi A, Sozzani S, Mantovani A. Cutting edge: scavenging of inflammatory CC chemokines by the promiscuous putatively silent chemokine receptor D6. J Immunol. 2003;170:2279–2282. doi: 10.4049/jimmunol.170.5.2279. [DOI] [PubMed] [Google Scholar]
- 35.Galliera E, Jala VR, Trent JO, Bonecchi R, Signorelli P, Lefkowitz RJ, Mantovani A, Locati M, Haribabu B. beta-Arrestin-dependent constitutive internalization of the human chemokine decoy receptor D6. J Biol Chem. 2004;279:25590–25597. doi: 10.1074/jbc.M400363200. [DOI] [PubMed] [Google Scholar]
- 36.Weber M, Blair E, Simpson CV, O’Hara M, Blackburn PE, Rot A, Graham GJ, Nibbs RJ. The chemokine receptor D6 constitutively traffics to and from the cell surface to internalize and degrade chemokines. Mol Biol Cell. 2004;15:2492–2508. doi: 10.1091/mbc.E03-09-0634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKimmie CS, Graham GJ. Leucocyte expression of the chemokine scavenger D6. Biochem Soc Trans. 2006;34:1002–1004. doi: 10.1042/BST0341002. [DOI] [PubMed] [Google Scholar]
- 38.Lenert P, Brummel R, Field EH, Ashman RF. TLR-9 activation of marginal zone B cells in lupus mice regulates immunity through increased IL-10 production. J Clin Immunol. 2005;25:29–40. doi: 10.1007/s10875-005-0355-6. [DOI] [PubMed] [Google Scholar]
- 39.Lund FE, Garvy BA, Randall TD, Harris DP. Regulatory roles for cytokine-producing B cells in infection and autoimmune disease. Curr Dir Autoimmun. 2005;8:25–54. doi: 10.1159/000082086. [DOI] [PubMed] [Google Scholar]
- 40.Mann MK, Maresz K, Shriver LP, Tan Y, Dittel BN. B cell regulation of CD4+CD25+ T regulatory cells and IL-10 via B7 is essential for recovery from experimental autoimmune encephalomyelitis. J Immunol. 2007;178:3447–3456. doi: 10.4049/jimmunol.178.6.3447. [DOI] [PubMed] [Google Scholar]
- 41.Malisan F, Briere F, Bridon JM, Harindranath N, Mills FC, Max EE, Banchereau J, Martinez-Valdez H. Interleukin-10 induces immunoglobulin G isotype switch recombination in human CD40-activated naive B lymphocytes. J Exp Med. 1996;183:937–947. doi: 10.1084/jem.183.3.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Briere F, Servet-Delprat C, Bridon JM, Saint-Remy JM, Banchereau J. Human interleukin 10 induces naive surface immunoglobulin D+ (sIgD+) B cells to secrete IgG1 and IgG3. J Exp Med. 1994;179:757–762. doi: 10.1084/jem.179.2.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fujieda S, Saxon A, Zhang K. Direct evidence that gamma 1 and gamma 3 switching in human B cells is interleukin-10 dependent. Mol Immunol. 1996;33:1335–1343. doi: 10.1016/s0161-5890(96)00092-2. [DOI] [PubMed] [Google Scholar]
- 44.Sun H, Lu B, Li RQ, Flavell RA, Taneja R. Defective T cell activation and autoimmune disorder in Stra13-deficient mice. Nat Immunol. 2001;2:1040–1047. doi: 10.1038/ni721. [DOI] [PubMed] [Google Scholar]
- 45.Seimiya M, Wada A, Kawamura K, Sakamoto A, Ohkubo Y, Okada S, Hatano M, Tokuhisa T, Watanabe T, Saisho H, Tagawa M, J OW. Impaired lymphocyte development and function in Clast5/Stra13/DEC1-transgenic mice. Eur J Immunol. 2004;34:1322–1332. doi: 10.1002/eji.200324700. [DOI] [PubMed] [Google Scholar]