Abstract
We analyzed the prevalence and characteristics of Vibrio parahaemolyticus among patients with acute infectious diarrhea in the southern coastal region of China. V. parahaemolyticus was the leading cause of bacterial infectious diarrhea in this region during 2007–2012. Serotype O3:K6 strains were most common, followed by serotypes O4:K8 and O3:K29.
Keywords: Vibrio parahaemolyticus, infectious diarrhea, epidemic, China, bacteria, Vibrio
Vibrio parahaemolyticus, a halophilic bacterium, is recognized as a major cause of acute gastroenteritis worldwide, often associated with the consumption of raw or undercooked shellfish. V. parahaemolyticus infections are caused by diverse serotypes; however, serotype O3:K6 has been reported to be dominant and has been a widespread serotype since 1997 (1).
V. parahaemolyticus has been the leading cause of foodborne outbreaks and bacterial infectious diarrhea in China since the 1990s, especially in coastal regions (2,3). Serotype O3:K6 was documented as the dominant serotype in Zhejiang Province, China, in 2002 and was proven to be a pandemic clone in 2008 (4). However, long-term fluctuation in the frequency of infections with the pandemic strains of V. parahaemolyticus remains unknown.
In 2007, laboratory-based surveillance for acute infectious diarrhea at 11 sentinel hospitals was established in Shenzhen City in the southern coastal region of China with V. parahaemolyticus as one of the target pathogens. To characterize V. parahaemolyticus infections and clarify its prevalence in this region, we analyzed all V. parahaemolyticus cases captured by this surveillance during 2007–2012.
The Study
Surveillance was conducted among outpatients who had >3 loose or liquid stools during a 24-hour period but lasting <14 days. A total of 1,488 V. parahaemolyticus infections were identified from 24,696 enrolled outpatients (6.0% of outpatients). More than half of the patients (835; 56.1%) were male. Patients ranged in age from 4 months to 84 years (median 27 years); 1,383 (92.9%) patients were 15–39 years of age. Most (914; 61.4%) patients were part of the large transient population living in Shenzhen. Of all patients with V. parahemoliticus infection, 1,150 (77.3%) had watery diarrhea, 1,176 (79.0%) had abdominal pain, 730 (49.1%) had vomiting, 206 (13.8%) had fever, and 4 (0.3%) had blood in stools.
Obvious monthly peaks of V. parahaemolyticus infections were found during the warmer months (June–October). Up to 30% of diarrhea cases covered by the surveillance could be attributed to V. paraheamolyticus infections during this period.
All 1,488 V. parahaemolyticusisolates were serotyped by slide agglutination by using a commercial serum (Denka-Seiken Ltd., Tokyo, Japan); 47 serotypes were detected. The O3:K6 serotype was dominant throughout the surveillance years (996 isolates; 66.9%), followed by O4:K8 (156 isolates; 10.5%) and O3:K29 (51 isolates; 3.4%). However, O3:K29 appeared more frequently during 2009 and 2010. Four other serotypes occurred as clusters in different years: O1:KUT in 2008, O1:K56 in 2010, O4:K68 in 2010, and O5:K68 in 2009 (Table 1).
Table 1. Serotype composition of 1,488 Vibrio parahaemolyticus isolates from patients with acute diarrhea, southern coastal region of China, 2007–2012.
| Serotype | No. isolates |
||||||
|---|---|---|---|---|---|---|---|
| 2007 | 2008 | 2009 | 2010 | 2011 | 2012 | Total | |
| O3:K6 | 153 | 94 | 180 | 210 | 198 | 161 | 996 |
| O4:K8 | 12 | 10 | 49 | 42 | 25 | 18 | 156 |
| O3:K29 | 2 | 1 | 24 | 18 | 5 | 1 | 51 |
| O1:KUT | 1 | 20 | 4 | 4 | 10 | 8 | 47 |
| O1:K56 | 0 | 0 | 0 | 28 | 0 | 1 | 29 |
| O1:K36 | 5 | 4 | 3 | 9 | 2 | 1 | 24 |
| O4:K9 | 1 | 5 | 6 | 1 | 3 | 7 | 23 |
| O4:K68 | 2 | 4 | 2 | 10 | 2 | 0 | 20 |
| O5:K68 | 2 | 0 | 10 | 3 | 1 | 0 | 16 |
| O1:K25 | 5 | 1 | 3 | 3 | 2 | 1 | 15 |
| O11:K36 | 0 | 0 | 5 | 7 | 1 | 1 | 14 |
| O2:K3 | 0 | 3 | 3 | 3 | 0 | 4 | 13 |
| O3:KUT | 0 | 4 | 3 | 1 | 0 | 1 | 9 |
| O4:K11 | 2 | 0 | 2 | 1 | 0 | 1 | 6 |
| O4:K55 | 0 | 1 | 0 | 2 | 2 | 1 | 6 |
| O1:K41 | 1 | 0 | 1 | 0 | 1 | 2 | 5 |
| O4:K13 | 0 | 1 | 4 | 0 | 0 | 0 | 5 |
| O4:KUT | 0 | 1 | 1 | 0 | 0 | 3 | 5 |
| O8:K41 | 1 | 1 | 0 | 0 | 0 | 3 | 5 |
| OUT:KUT | 1 | 1 | 1 | 0 | 0 | 1 | 4 |
| O6:K18 | 0 | 1 | 1 | 0 | 0 | 1 | 3 |
| O8:K21 | 0 | 0 | 2 | 0 | 0 | 1 | 3 |
| OUT:K68 | 3 | 0 | 0 | 0 | 0 | 0 | 3 |
| O1:K1 | 1 | 1 | 0 | 0 | 0 | 0 | 2 |
| O1:K68 | 0 | 0 | 0 | 2 | 0 | 0 | 2 |
| O1:K69 | 0 | 0 | 0 | 0 | 2 | 0 | 2 |
| O10:K52 | 0 | 0 | 0 | 0 | 0 | 2 | 2 |
| O3:K59 | 0 | 0 | 0 | 1 | 0 | 1 | 2 |
| O4:K12 | 0 | 0 | 0 | 0 | 0 | 2 | 2 |
| O1:K5 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| O10:KUT | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| O11:K19 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| O11:KUT | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| O2:KUT | 0 | 0 | 0 | 1 | 0 | 0 | 1 |
| O3:K1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| O3:K2 | 0 | 0 | 0 | 1 | 0 | 0 | 1 |
| O3:K28 | 0 | 0 | 0 | 1 | 0 | 0 | 1 |
| O3:K41 | 0 | 0 | 0 | 1 | 0 | 0 | 1 |
| O3:K45 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| O3:K54 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| O3:K56 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| O3:K7 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| O4:K63 | 0 | 0 | 0 | 1 | 0 | 0 | 1 |
| O8:KUT | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| OUT:K19 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| OUT:K44 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| OUT:K55 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
Representative strains were selected for PCR of the virulence genes thermostable direct hemolysin (tdh) and TDH-related hemolysin (trh) (5,6). A total of 833 isolates covering all 47 serotypes (1–560 isolates for each serotype) were screened. Most (788; 94.6%) strains were tdh+trh–; 28 isolates representing 5 serotypes were tdh+trh+ (O1:KUT, n = 18; O4:K12, n = 3; O1:K69, n = 4; O10:K52, n = 2; O4:K55, n = 1), and 17 isolates of 10 serotypes were tdh–trh– (O1:KUT, n = 5; O3:K6, n = 4; O1:K1, O1:KUT, O3:K1, O3:KUT, O4:K63, O10:KUT, OUT:K19, and OUT:K55, n = 1 each).
A total of 196 tdh+trh– isolates, representing the leading 10 serotypes (6–75 isolates for each serotype), were selected for group-specific PCR (GS-PCR) of toxRS/new sequence (7). Results demonstrated that pandemic genotype strains (110; 56.12%) prevailed among the leading 10 serotypes in Shenzhen. Most (68; 90.7%) O3:K6 isolates; all O1:K36, O4:K68, O5:K68, and O1:K25 isolates; and 6 (28.6%) O1:KUT isolates gave positive results by GS-PCR, whereas results were negative for the other 4 serotypes.
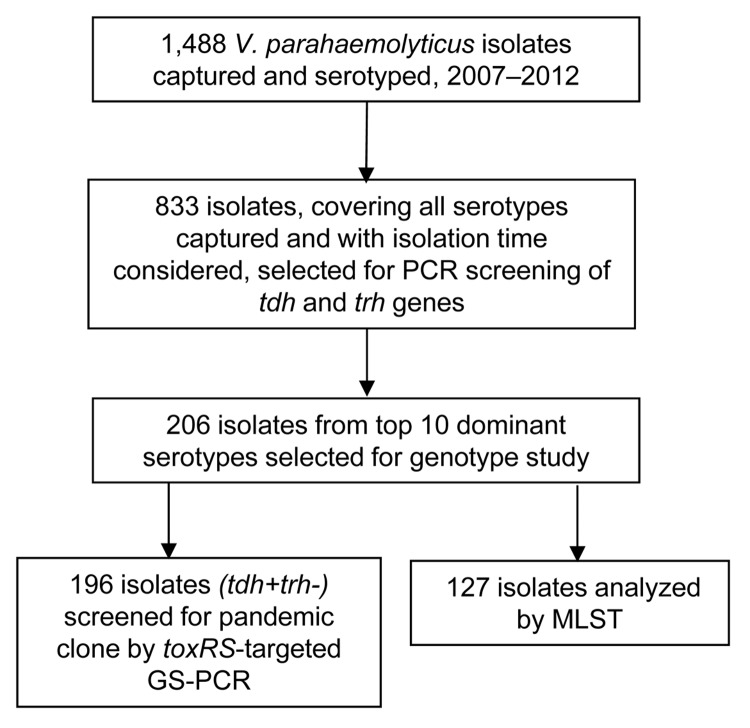
We analyzed 127 isolates by using the V. parahaemolyticusmultilocus sequence typing (MLST) scheme (http://pubmlst.org/vparahaemolyticus) (Figure). Clonal complex 3 (CC3) predominated (93 isolates; 73.2%). A new clonal complex, CC120, and sequence type (ST), 265, a presumed new ancestor of CC345, were identified (Table 2).
Figure.
Study design and eligibility for serotype and genotype analysis of Vibrio parahaemolyticus isolates from patients with acute diarrhea, southern coastal region of China, 2007–2012.
Table 2. Results of GS-PCR and MLST analysis of the 10 mostly commonly found serotypes of Vibrio parahaemolyticus isolates from patients with acute diarrhea, southern coastal region of China, 2007–2012*.
| Serotype | GS-PCR, n = 196 |
MLST, n = 127 |
||||
|---|---|---|---|---|---|---|
| No. positive, n = 110 | No. negative, n = 86 | No. isolates tested | ST (no. isolates) | CC | ||
| O3:K6 | 68 | 7 | 64 | ST3 (60), ST487 (1), ST489 (1), ST526 (1) | CC3 | |
| ST497 (1) | Singleton | |||||
| O4:K8 | 0 | 30 | 16 | ST265 (14), ST189 (1), ST438 (1) | CC345 | |
| O3:K29 | 0 | 14 | 13 | ST120 (11), ST480 (2) | CC120 | |
| O1:KUT | 6 | 15 | 3 | ST3 (3) | CC3 | |
| O1:K56 | 0 | 10 | 8 | ST8 (8) | CC8 | |
| O1:K36 | 9 | 0 | 7 | ST3 (7) | CC3 | |
| O4:K9 | 0 | 10 | 3 | ST332 (3) | Singleton | |
| O4:K68 | 10 | 0 | 5 | ST3 (5) | CC3 | |
| O5:K68 | 6 | 0 | 3 | ST3 (3) | CC3 | |
| O1:K25 | 11 | 0 | 5 | ST3 (4) | CC3 | |
| ST481 (1) | Singleton | |||||
*GS-PCR, group-specific PCR; MLST, multilocus sequence typing; ST, sequence type; CC, clonal complex.
Conclusions
During 2007–2012, V. parahaemolyticus was the dominant bacterial cause of acute diarrhea in the southern coastal region of China, surpassing Salmonella spp., diarrheagenic Escherichia coli, and Shigella spp. (data not shown). These findings differ from those for central and northern regions of China (8,9). Most case-patients in this surveillance were 15–39 years of age. The distribution of the 47 serotypes detected revealed the diversity of V. parahaemolyticus, which might explain the continuing epidemic of V. parahaemolyticus infections in this region.
V. parahaemolyticus serotype O3:K6, which emerged worldwide in 1997 as a pandemic clone and spread throughout Asia and to the Americas, Europe, and Africa (1), has been dominant in Shenzhen Province. Most (68; 90.7%) O3:K6 isolates tested were new clones, defined by toxRS-targeted GS-PCR, providing evidence that pandemic O3:K6 has spread to China. Most (63; 98.5%) serotype O3:K6 strains were identified as CC3 by MLST; serotype O1:KUT, O1:K36, O4:K68, O5:K68, and O1:K25 strains were positive by GS-PCR in our study and mostly documented as O3:K6 serovariants (1). These results indicate the long-term evolution of O3:K6 in Asia.
V. parahaemolyticus serotype O4:K8 has been an epidemic strain in Asia (10,11) and reported in Peru (12) but has been rarely seen in North America, Africa, and Europe. Serotype O4:K8 isolates from this study expressed a nonpandemic genotype. We presume that the evolution of O4:K8 was affected by local mutation and recombination rather than by a global pandemic, similar to a finding reported in Japan in 2007 (13). Notably, ST265 was predominant among strains with serotype O4:K8, whereas ST345, the previously considered founder of CC345, was not found (Table 2). Although ST265 might be a potential branch from ST345 in other regions, our findings strongly suggest that ST265 should be considered the epidemic clonal founder of CC345 in China. Overall, serotypes O3:K6 and O4:K8, stable subpopulations of the diverse V. parahaemolyticus population in our surveillance, have clearly been epidemic in China.
V. parahaemolyticus serotypes O3:K29 and O1:K56 were mainly reported in Asia, with nonpandemic groups identified in Japan (10,14). Our study showed that prevalence of serotype O3:K29 V. parahaemolyticus suddenly fluctuated during 2009 and 2010 and prevalence of O1:K56 fluctuated in 2010, but no focal outbreaks were confirmed; this finding indicates that sporadic outbreaks might have occurred. In addition, serotype O3:K29 isolates were identified as ST120 and the newly determined ST480, both belonging to CC120. Limited information could be obtained from the MLST database about the O1:K56 strains, and the isolates we tested were classified as ST8.
Further, our study found V. parahaemolyticus serotype O1:KUT might contain >1 character K antigens; however, 18 isolates harbored both tdh and trh genes, a combination that is not found frequently. Therefore, an O1:KUT epidemic clone might be prevalent in this region.
Whereas V. parahaemolyticus often is associated with the consumption of raw or undercooked shellfish, data from this surveillance program showed that most patients were transient residents who lived in rural areas and seldom ate seafood. However, epidemiologic data showed that V. parahaemolyticus infection was associated with eating outdoors and consumption of salad vegetables. Cross-contamination in food processing might be the source of infection; further epidemiologic investigation is under way.
In summary, V. parahaemolyticus has been prevalent for a long time in the southern coastal region of China, and diverse serotypes and multiple clones of the bacterium are circulating. On the basis of successful efforts to reduce prevalence of V. parahaemolyticus infections in Japan (15), we suggest holistic approaches involving regulations and guidance on fishery products and food hygiene to decrease the incidence of these infections in China.
Acknowledgments
We thank the personnel from 11 sentinel hospitals and participating district Centers for Disease Control and Prevention in Shenzhen for their participation in and contribution to our surveillance work.
This work was supported by China National Science and Technology Major Projects Foundation (no. 2012ZX10004215-003-005), National Natural Science Foundation of China (no. 81071433 to Q.H), and Shenzhen Public Service Platform of Pathogenic Microorganisms Repository.
Biography
Ms Li is a senior researcher in the Shenzhen Major Infectious Disease Control Key Laboratory, Shenzhen Center for Disease Control and Prevention. Her research interests include laboratory-based surveillance of foodborne pathogens and antimicrobial drug–resistant bacteria, particularly V. parahaemolyticus and diarrheagenic E. coli.
Footnotes
Suggested citation for this article: Li Y, Xie X, Shi X, Lin Y, Qiu X, Mou J, et al. Vibrio parahaemolyticus, southern coastal region of China, 2007–2012. Emerg Infect Dis [Internet]. 2014 Apr [date cited]. http://dx.doi.org/10.3201/eid2004.130744
References
- 1.Nair GB, Ramamurthy T, Bhattacharya SK, Dutta B, Takeda Y, Sack DA. Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin Microbiol Rev. 2007;20:39–48. 10.1128/CMR.00025-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin X, Ran L, Ma L, Wang Z, Feng Z. Analysis on the cases of infectious diarrhea (rather than cholera, dysentery, typhoid and paratyphoid) reported in China, 2010 [in Chinese]. Chinese Journal of Food Hygiene. 2011;23:385–9. [Google Scholar]
- 3.Liu X, Chen Y, Wang X, Ji R. Foodborne disease outbreaks in China from 1992 to 2001—national foodborne disease surveillance system [in Chinese]. Wei Sheng Yan Jiu. 2004;33:725–7. [PubMed]
- 4.Vongxay K, Pan Z, Zhang X, Wang S, Cheng S, Mei L, et al. Occurrence of pandemic clones of Vibrio parahaemolyticus isolates from seafood and clinical samples in a Chinese coastal province. Foodborne Pathog Dis. 2008;5:127–34. 10.1089/fpd.2007.0045 [DOI] [PubMed] [Google Scholar]
- 5.Blackstone GM, Nordstrom JL, Vickery MC, Bowen MD, Meyer RF, DePaola A. Detection of pathogenic Vibrio parahaemolyticus in oyster enrichments by real time PCR. J Microbiol Methods. 2003;53:149–55. 10.1016/S0167-7012(03)00020-4 [DOI] [PubMed] [Google Scholar]
- 6.Davis CR, Heller LC, Peak KK, Wingfield DL, Goldstein-Hart CL, Bodager DW, et al. Real-time PCR detection of the thermostable direct hemolysin and thermolabilehemolysin genes in a Vibrio parahaemolyticus cultured from mussels and mussel homogenate associated with a foodborne outbreak. J Food Prot. 2004;67:1005–8 . [DOI] [PubMed] [Google Scholar]
- 7.Matsumoto C, Okuda J, Ishibashi M, Iwanaga M, Garg P, Rammamurthy T, et al. Pandemic spread of an O3:K6 clone of Vibrio parahaemolyticus and emergence of related strains evidenced by arbitrarily primed PCR and toxRS sequence analyses. J Clin Microbiol. 2000;38:578–85 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qu M, Deng Y, Zhang X, Liu G, Huang Y, Lin C, et al. Etiology of acute diarrhea due to enteropathogenic bacteria in Beijing, China. J Infect. 2012;65:214–22. 10.1016/j.jinf.2012.04.010 [DOI] [PubMed] [Google Scholar]
- 9.Zhu M, Cui S, Lin L, Xu B, Zhao J, Xia S, et al. Analysis of the aetiology of diarrhoea in outpatients in 2007, Henan province, China. Epidemiol Infect. 2012;7:1–9 . 10.1097/01.ede.0000416615.21487.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chowdhury NR, Chakraborty S, Ramamurthy T, Nishibuchi M, Yamasaki S, Takeda Y, et al. Molecular evidence of clonal Vibrio parahaemolyticus pandemic strains. Emerg Infect Dis. 2000;6:631–6. 10.3201/eid0606.000612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiou CS, Hsu SY, Chiu SI, Wang TK, Chao CS. Vibrio parahaemolyticus serovar O3:K6 as cause of unusually high incidence of food-borne disease outbreaks in Taiwan from 1996 to 1999. J Clin Microbiol. 2000;38:4621–5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gavilan RG, Zamudio ML, Martinez-Urtaza J. Molecular epidemiology and genetic variation of pathogenic Vibrio parahaemolyticus in Peru. PLoS Negl Trop Dis. 2013;7:e2210. 10.1371/journal.pntd.0002210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhoopong P, Palittapongarnpim P, Pomwised R, Kiatkittipong A, Kamruzzaman M, Nakaguchi Y, et al. Variability of properties of Vibrio parahaemolyticus strains isolated from individual patients. J Clin Microbiol. 2007;45:1544–50. 10.1128/JCM.02371-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong HC, Liu SH, Wang TK, Lee CL, Chiou CS, Liu DP, et al. Characteristics of Vibrio parahaemolyticus O3:K6 from Asia. Appl Environ Microbiol. 2000;66:3981–6. 10.1128/AEM.66.9.3981-3986.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hara-Kudo Y, Saito S, Ohtsuka K, Yamasaki S, Yahiro S, Nishio T, et al. Characteristics of a sharp decrease in Vibrio parahaemolyticus infections and seafood contamination in Japan. Int J Food Microbiol. 2012;157:95–101. 10.1016/j.ijfoodmicro.2012.04.019 [DOI] [PubMed] [Google Scholar]