Abstract
Acellular vaccines against Bordetella pertussis were introduced in Australia in 1997. By 2000, these vaccines had replaced whole-cell vaccines. During 2008–2012, a large outbreak of pertussis occurred. During this period, 30% (96/320) of B. pertussis isolates did not express the vaccine antigen pertactin (prn). Multiple mechanisms of prn inactivation were documented, including IS481 and IS1002 disruptions, a variation within a homopolymeric tract, and deletion of the prn gene. The mechanism of lack of expression of prn in 16 (17%) isolates could not be determined at the sequence level. These findings suggest that B. pertussis not expressing prn arose independently multiple times since 2008, rather than by expansion of a single prn-negative clone. All but 1 isolate had ptxA1, prn2, and ptxP3, the alleles representative of currently circulating strains in Australia. This pattern is consistent with continuing evolution of B. pertussis in response to vaccine selection pressure.
Keywords: Bordetella pertussis, whooping cough, bacteria, outbreaks, pertactin, evolution, immunization, vaccination, vaccine, Australia
Bordetella pertussis is the gram-negative coccobacillus that causes the respiratory disease pertussis, also known as whooping cough. The incidence of pertussis infection and related deaths decreased dramatically after implementation of immunization with a whole-cell vaccine (WCV) during the 1950s. Because of side effects of WCV, such as high rates of fever and local reactions, and variable efficacy of WCVs, a less reactogenic acellular vaccine (ACV) was developed in the 1980s. ACVs have now replaced WCVs in many industrialized countries for primary and booster vaccinations against pertussis.
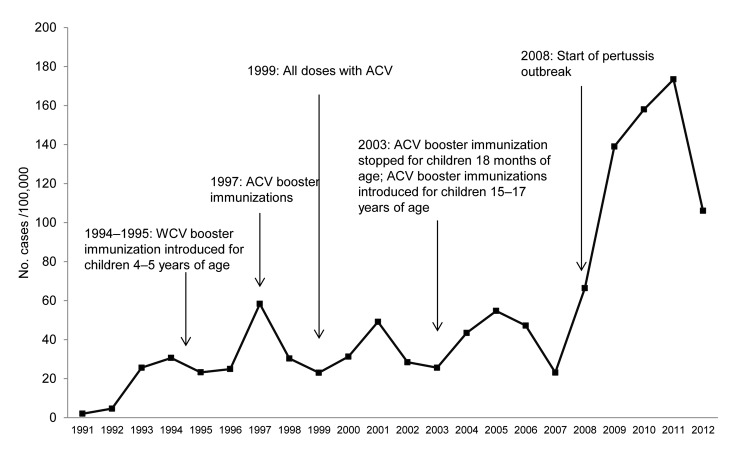
Although ACV formulations differ in the number of component pertussis antigens, the vaccine used in Australia contains pertussis toxin (ptx), pertactin (prn), and filamentous hemagglutinin (fha). A 5-component (ptx, prn, fha, fimbrial antigen [fim]2, and fim3) ACV is used for short periods in some regions (1). ACVs were introduced for the fourth and fifth doses in most states in Australia during 1997 and for all doses during 1999 (Figure 1). South Australia introduced ACVs for all doses in 1997. The current vaccination schedule for pertussis comprise 3 primary doses of ACV at 2, 4, and 6 months of age, and a booster vaccination at 4 years of age. A booster vaccination with ACV at 18 months of age, which was introduced in 1985, was removed from the National Immunization Program in Australia in 2003, and an adult-formulated ACV was introduced for children at 12–17 years of age in school-based programs in 2004 (2,3).
Figure 1.
Pertussis cases/100,000 population in Australia, 2008–20012, since mandatory reporting was instituted in 1991 and changes to pertussis vaccination schedule, including introduction of whole-cell vaccine (WCV) booster vaccinations for 4–5-year-old children in 1994–1995 and introduction of acellular vaccine (ACV) booster vaccinations in 1997. By 1999–2000, ACVs were used for all pertussis vaccinations. In 2003, the booster vaccinations for children 18 months of age was removed and replaced with a booster vaccination for children 15–17 years of age (3).
Since 1991, data on reported pertussis cases show that outbreaks occurred in Australia in 1996–1997, 2001, and 2004, and a series of outbreaks occurred in different regions starting in 2008 (Figure 1) (2,3). Multiple factors probably contributed to the resurgence of pertussis in high-income countries that had long-standing pertussis immunization programs. These factors include waning immunity (exacerbated by the change from WCVs to ACVs and, in Australia, cessation of the booster vaccination at 18 months of age) and increased use of more sensitive diagnostic tests, such PCR (4).
An additional possible contributing factor is evolution of B. pertussis through vaccine-driven adaptation (5). The most prominent recent changes in circulating B. pertussis strains are polymorphisms within genes encoding 2 of the 3 main virulence factors (ptx and prn) contained in the vaccine. Variations have also been reported in ptxP, the promoter of the ptx operon (6). In Australia, we have shown by single nucleotide polymorphism (SNP) typing that among B. pertussis isolates, ptxP3–containing strains predominate (7), and these strains belong to SNP cluster I (8,9).
Surveillance of recent B. pertussis isolates in several countries has identified prn deletions and gene disruptions, which lead to lack of expression of mature prn (10–13). This protein is a 69-kDa adhesin that aids B. pertussis attachment to epithelial cells and is one of the most polymorphic virulence genes within B. pertussis (it has 13 documented alleles) (5). SNPs and differences in the number of amino acid (GGFGP and PQP) repeats contribute to variation within the prn gene; variations are usually limited to 2 regions known as region 1 and region 2.
In this study, we identified B. pertussis isolates that do not express prn (prn negative) from a set of isolates collected in Australia during 1997–2012. We also characterized the causes of their lack of expression and evaluated trends in the proportion of prn-negative isolates over this period.
Methods
Bacterial Strains and Growth
A total of 453 B. pertussis isolates were available for this study; 133 isolates collected during 1997–2008 and 194 collected during 2008–2010 have been described (9). A total of 126 additional isolates collected from Westmead Hospital (Sydney, New South Wales, Australia) and Princess Margaret Hospital for Children (Perth, Western Australia, Australia) during 2011–2012 were also included this study. Although specific clinical information about the source of isolates was not available, isolates were obtained from patients who lived in large urban areas and who had PCR-confirmed pertussis infections. The number of available isolates in 2011–2012 was relatively small because several participating laboratories discontinued pertussis culture in favor of only direct PCR testing.
All B. pertussis isolates were grown on Bordet Gengou agar (Becton Dickinson, Sparks, MD, USA) supplemented with 10% defibrinated horse blood (Oxoid, Basingstoke, UK) at 37°C for 3–5 days before subculture and incubation at 37°C for 24 h. All cultures were examined for hemolytic activity indicating expression of the virulent (Bvg+) phase before being collected and resuspended in saline to an optical density at 650 nm = 1 for Western immunoblotting.
Western Immunoblotting
The ptx, prn, and fha proteins were detected by Western immunoblotting as described (10,11,14). Bacterial suspensions were mixed with Laemmli buffer containing 5% β-mercaptoethanol and boiled for 5 min. Proteins separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis were transferred to a polyvinylidene difluoride membrane at 100 V for 1 hr. Membranes were blocked with 5% (wt/vol) skim milk powder in wash buffer for 1 hr and incubated overnight with mouse polyclonal antibodies against ptx, fha, and prn diluted 1:1,000 with Tris-buffered saline (TBS) containing 1% Tween 20. After 3 washes with TBS containing 1% Tween 20, membranes were incubated for 1 h with sheep antimouse monoclonal antibodies in TBS plus 5% skim milk and 0.1% Tween 20. Antigen–antibody complexes were visualized by chemiluminescence on a LAS3000 imager (Fujifilm, Tokyo, Japan). The minimum detectable amount with this method was 1 ng of specific protein.
Genotyping and prn Gene Sequencing
Isolates were genotyped for fim3, prn, and ptxP alleles as described (7–9). Isolates that had not already been typed were characterized by SNP cluster and SNP profile as described by Octavia et al. (8), multilocus variable number tandem repeat analysis (MLVA) as described by Kurniawan et al. (1) typing of prn, fim3, and ptxP alleles (6,15). Relationships among SNP profiles and clusters were defined by Octavia et al. (8) and are shown in Technical Appendix Figure 1.
For isolates that did not express prn, overlapping primers reported by Fry et al. (16) were used to amplify a predicted 2,869-bp region that included the signal peptide region and the prn gene. The prn promoter region was also sequenced to detect any changes. Each PCR mixture contained ≈30 ng DNA, 10 mmol/L Tris-HCl (pH 8.3), 50 mmol/L KCl, 2.5 mmol/L MgCl2, 100 µmol/L of each deoxynucleotide, 10 pmol/L of each primer, 2.5 units of Taq polymerase, and milliQ water (Millipore, Billerica, MA, USA). Products were then sequenced on an Automated DNA Sequence Analyzer ABI3730 (Applied Biosystems, Foster City, CA, USA) to determine the complete prn gene, which included region 1 and region 2. All sequences were aligned against prn gene sequences identified by Mooi et al. (17).
Results
Identification and Distribution of B. pertussis Not Expressing prn
The 320 B. pertussis isolates obtained during 2008–2012 were from 5 states in Australia: New South Wales (116 isolates), Queensland (37), South Australia (47), Victoria (30), and Western Australia (90). All 96 (30) isolates identified by Western immunoblot as not expressing prn were obtained after 2008. Examples of Western immunoblots are shown in online Technical Appendix Figure 2. The other 133 isolates obtained before 2008 expressed prn and were from SNP clusters I and II or were unclustered. The distribution of prn-negative isolates in individual states is shown in the Table. Only isolates from Western Australia and New South Wales were available for all years during 2008–2012; no isolates were available from South Australia or Victoria during 2011–2012 or from Queensland during 2008–2009 and 2012. All isolates expressed ptx and fha.
Table. Distribution of pertactin-positive and protactin-negative Bordetella pertussis isolates in 5 states, Australia, 2008–2012*.
| Year | State |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| New South Wales |
Queensland |
South Australia |
Victoria |
Western Australia |
|||||||||||
| No. pos | No. neg | % Neg | No. pos | No. neg | % Neg | No. pos | No. neg | % Neg | No. pos | No. neg | % Neg | No. pos | No. neg | % Neg | |
| 2008 | 18 | 0 | 0 | – | – | – | 13 | 2 | 13 | 1 | 0 | 0 | 5 | 0 | 0 |
| 2009 | 52 | 0 | 0 | – | – | – | 17 | 9 | 35 | 10 | 1 | 9 | 18 | 2 | 10 |
| 2010 | 8 | 6 | 43 | 3 | 6 | 67 | 5 | 1 | 17 | 17 | 1 | 6 | 14 | 1 | 7 |
| 2011 | 6 | 17 | 74 | 21 | 7 | 25 | – | – | – | – | – | – | 8 | 15 | 65 |
| 2012 | 2 | 7 | 78 | – | – | – | – | – | – | – | – | – | 6 | 21 | 78 |
*Pos, positive; Neg, negative; –, no isolates were obtained.
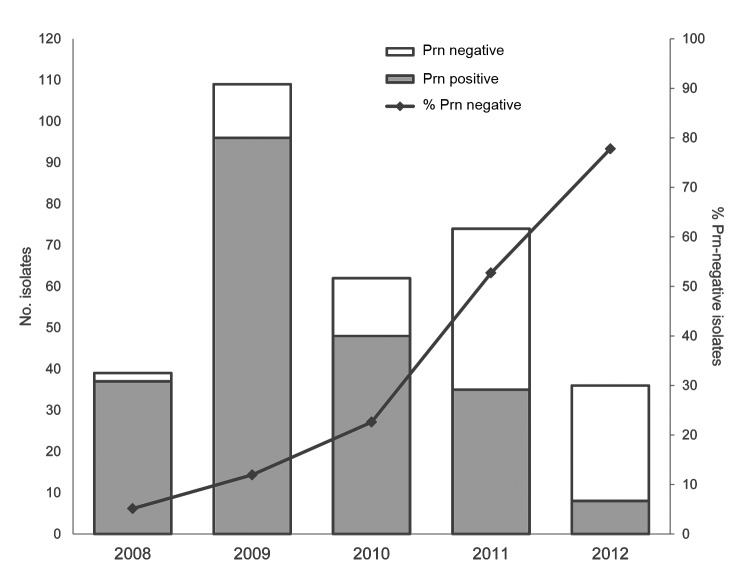
The prn-negative strains were first identified in isolates collected in 2008, when they made up 5% (2/39) of the isolates. By 2012, the proportion of prn-negative isolates had increased to 78% (28/36) (Figure 2). In Western Australia and New South Wales, where isolates were available for all years, there was a progressive increase from 3% in 2009 to 78% in 2012 (Technical Appendix Figure 3). Lack of isolates from Queensland, Victoria, and South Australia in various years was related to changes in laboratory practice (cultures no longer obtained) or decreased numbers in a post-epidemic period, rather than any systematic differences in collection. It is unlikely that different patterns of circulating B. pertussis differed in these regions.
Figure 2.
Number and percentage of pertactin (Prn)–negative Bordetella pertussis isolates in Australia, 2008–2012. During this period, 320 B. pertussis isolates obtained in New South Wales, Queensland, South Australia, Victoria, and Western Australia were identified as expressing prn or not expressing prn by using Western immunoblotting. The increasing percentage of prn-negative isolates each year during 2008–2012 was 5%, 12%, 23%, 53%, and 78% respectively. Data for individual states and years can be found in the Table. Gray bars indicate number of isolates expressing prn, and white bars indicate number of isolates not expressing prn. Error bars indicate 95% CIs.
The increase in prn-negative isolates during 2011–2012, in comparison with 2008, was significant (p<0.05, by Fisher exact test with multiple test correction). All but 1 prn-negative isolate had the ptxA1, prn2, and ptxP3 alleles and belonged to SNP cluster I; the exception, L1378, had ptxA1 and prn1 but not ptxP3, and was not assigned to any SNP cluster. In addition, the prn-negative isolates had new MLVA types that were closely related to MT27 and MT114, both of which are currently circulating in Australia (9), although MT27 still predominates.
Sequence Analysis of prn Gene of prn-deficient Isolates
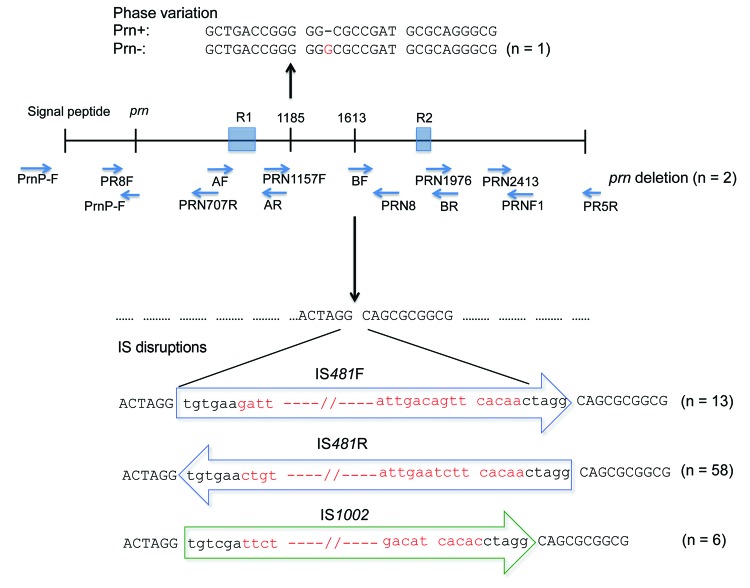
Mechanisms of disruption, identified by sequencing the prn region, including the signal peptide, of 80/96 prn-negative isolates, are shown in Figure 3. Seventy-seven (82%) isolates had IS elements located between region 1 and region 2; in prn, a 1049-bp IS481 was inserted in the forward direction in 13 isolates and in the reverse direction in 58 isolates. A 1,037-bp IS1002 was inserted in the forward direction in 6 isolates, which has not been described in the prn region. All IS element disruptions were at position 1613 and were flanked by a 6-bp repeat (ACTAGG) at the 5′ end and AGGCAG at the 3′ end (Figure 3).
Figure 3.
Variations in protactin (prn) gene of prn-negative Bordetella pertussis isolates, Australia, 2008–2012, Ninety-six B. pertussis isolates were identified as prn negative. Eighty of these isolates had 1 of 4 mechanisms of prn disruption: IS481 (in forward and reverse directions) and IS1002, which were inserted at the ACTAGG motif within prn, or an extended homopolymeric tract of G residues (n = 1). Lower case letters indicate residues that are conserved in all IS disruptions, and red letters indicate differences in IS disruptions. Positions of nucleotides have been numbered relative to the first start codon of sequence AJ011092 (17). The prn gene of 2 isolates was not amplified by PCR with a combination of primers from published studies (15–19), which indicated a deletion of the entire gene. Sixteen isolates that had no gene disruptions were also observed.
One isolate had no IS within prn but had an additional guanine residue at position 1185 between region 1 and region 2, which resulted in a stop codon at amino acid position 749. Two isolates from South Australia that had SNP profile SP13 were nontypeable. For prn; multiple pairs of PCR primers specific for the prn gene (15–19) failed to amplify a product, which indicated deletion of the entire gene. IS disruptions, deletions, or other variations were not detected in prn or the prn promoter region of 16 prn-negative isolates. Details of the 96 prn-negative isolates, including individual prn, fim, and ptxP alleles, SP, MLVA type, and mechanism of prn disruption, are shown in the online Technical Appendix Table.
Discussion
In the 2 regions of Australia where isolates were available for all years during 2008–2012, prn-negative B. pertussis isolates increased from >10% to ≈80% of B. pertussis isolates over this period. Prn-negative strains have been isolated in several countries that have high coverage for vaccination but have not been shown to constitute such a high proportion of circulating B. pertussis (12,13,20,21). Japan was the first country to implement ACVs against pertussis in 1981, and the proportion of prn-negative isolates reported from countrywide surveillance during 2005–2009 was 32% (18/57). In France, where ACVs have been used since 1998, originally as booster vaccinations, and then for all doses since 2002 (11), ptx-negative and fha-negative isolates were first obtained in 2003, although only prn-negative isolates have increased and were reported to make up 13.3% of 120 isolates analyzed in 2011 (10).
The prn-negative B. pertussis isolates have also been identified in Finland and the United States (13,21,22). The United States introduced ACVs as booster vaccinations in 1991, but not until 1997 were all 5 primary doses replaced with ACVs (23). Although Finland replaced WCVs with ACVs at a later time (booster vaccinations in 2003 and primary vaccinations in 2005), both countries detected prn-negative isolates during 2011–2012. Long-term temporal analysis has not been performed to determine whether such isolates are increasing over time.
In comparison, until 2001 and 2009, respectively, Russia (24) and Senegal (25), which currently use only WCVs, have not reported prn-negative isolates. However, it is difficult to draw a definitive conclusion on the correlation of timing of emergence of prn-negative strains with timing of introduction of ACVs. Extensive analysis of isolates from earlier years from different countries would be required.
Multiple mechanisms of lack of expression of prn have been reported (11,12). Insertion of IS481 into the prn gene in either the forward or reverse direction was still the main mechanism of disruption (73.9%). This disruption occurred at the same conserved site identified in 3 isolates from the United States (nt position 1613) (21) and 9 isolates from Japan (nt position 1598) (12). The 15-bp difference in position is caused by an additional GGFGP repeat in prn2 in the isolates in our study and those from the United States, compared with those from Japan, which have prn1.
Six isolates in our study had an additional IS1002 disruption at nucleotide position 1613 (Figure 2), which confirmed that the 6-bp repeat site flanking IS elements is conserved (26). The lower number of isolates with IS1002 disruptions could be caused by fewer copies of IS1002 than IS481 in the genome (6 for IS1002 in Tohama I compared with 238 for IS481). Disruption of virulence genes by IS1002 has been reported; it can result in B. pertussis isolates not expressing O antigen (27). All isolates in this study that had the IS1002 disruption were collected in 1 state in Australia (New South Wales) and might have arisen from a single outbreak. More isolates are needed to determine whether this finding is indicative of an expanding clone.
Another mechanism of disruption is an 84-bp deletion of the signal peptide (nucleotide position 26). This deletion was observed in 24 isolates from Japan (12) and 2 isolates from Finland (13) but was not observed in any of the isolates in our study. However, 2 of the isolates apparently had the entire prn gene deleted.
In 1 isolate (L1502), an additional G residue in a homopolymeric tract of G residues resulted in a downstream stop codon. Truncations caused by stop codons in the prn gene were reported in 7 isolates from the United States (21), but they were at nucleotide position 1273 and the actual base change was not specified. Phase variation has been associated with variation in other B. pertussis genes, including fim2, fim3, fimX, and bapC (28), and is a common mechanism of phase variation in other pathogenic microorganisms (29,30).
A large proportion (17%, 16/96) of prn-negative isolates had no sequence change detected in the prn gene or its promoter upstream, which indicated that other mechanisms must have been responsible for inactivating prn expression. These 16 isolates belong to 3 SNP profiles; 8, 7, and 1 isolates belonging to SP13, SP14, and SP18, respectively, which suggests 3 independent inactivating events. Inactivation of expression could have occurred at the transcriptional or translational level. Our preliminary investigations showed that 3 of these prn-negative isolates produced prn gene transcripts. A consequence of prn inactivation without sequence variation of the prn gene is that it can be detected only at the protein level. Until mechanisms are identified, culturing of isolates will still be needed to monitor B. pertussis.
The increase in isolates that do not express a specific antigen has been documented only recently in Australia and other countries that use ACVs. The predominant isolates we identified are from SP13, SP14, or SP16, and all but 1 had the ptxA1, prn2, ptxP3 genotype. We have also shown that isolates with different SNP profiles can be affected by the same IS disruptions, and conversely, different IS disruptions can affect isolates with the same SNP profiles.
Most of the recently isolated prn-negative strains from the United States have the prn2 allele, which has been the predominant type since the 1990s (21,22). However, mutations causing inactivation of expression of the prn gene differ from those reported in this study and elsewhere. Prn-negative isolates characterized by Otsuka et al. (12) had the prn1, ptxA2, ptxP1 genotype and were from MT186 or related MT194 or MT226. Our previous analysis showed that MT186 belongs to SNP cluster V; this type is unrelated to isolates examined in the current study, which belong to SNP cluster I (Technical Appendix Figure 1), but was affected by the same IS disruption mechanisms. Two isolates from Finland that had prn1 were also reported to be prn negative because of deletions, although prn2 is the current predominant allele (13). Thus, the combination of SNP typing, antigen gene typing, and prn gene disruption mechanisms clearly demonstrates that isolates that do not express the prn gene from Australia and other countries do not belong to the same clone and that the recent almost simultaneous appearance and expansion of prn-negative isolates in several countries were independent events rather than global spread of a single clone.
The multiple origins of prn-negative isolates also point strongly to selective pressure on the bacterium. Therefore, it is conceivable that these prn-negative isolates are more likely to evade a vaccine-induced immune response. However, the relative contribution of prn to pertussis disease has not been clearly established. Various studies using prn mutants have shown that mutants that do not express prn do not colonize mouse lungs as well as isolates that express prn (31) but were more invasive in epithelial cells and persist for a longer period (32). The prn-negative strains have a greater growth advantage in vitro than their Prn-positive counterparts (12). This growth advantage can be beneficial in maintaining a high level of transmissibility between hosts, which is consistent with increasing numbers of infections with prn-negative isolates identified in Australia and elsewhere.
Whether these isolates have greater or lesser virulence than prn-positive strains is unclear. In contrast to lack of production of ptx, loss of prn does not seem to affect B. pertussis lethality in mice, possibly because of the range of autotransporters within B. pertussis that can compensate for the role of prn (10). In a retrospective study, no differences were found in severity of symptoms or duration of hospitalization between infants infected with prn-positive and prn-negative strains in France (20); the only major difference observed was the longer period from onset of pertussis symptoms to time of hospitalization among infants whose B. pertussis isolate was prn negative. Regardless of prn expression, vaccination reduced the severity of disease and the likelihood of being admitted to intensive care, which suggests that even an incomplete course of primary vaccination provides some protection against severe pertussis (20).
The results in this study highlight the emerging trend of prn-deficient B. pertussis isolates circulating in Australia. In addition to changes observed in prn, ptxA, ptxP, and fim genes of currently circulating strains, this study and other studies have reported the increasing prevalence of isolates not expressing prn in many countries that have a high uptake of ACV. The overall effect of lack of expression of an antigen on herd immunity is unknown. Emergence of prn-negative isolates is a relatively recent phenomenon that has affected currently circulating B. pertussis isolates. Whether strains not expressing prn continue to increase locally or globally and affect vaccine effectiveness and bacterial pathogenicity is unknown. Continued monitoring of genotypic and phenotypic properties of B. pertussis is required to better understand the effects of vaccination on the evolution of the organism.
Characteristics of Pertactin-deficient Bordetella pertussis Isolates, Australia, 2008–2012.
Acknowledgments
We thank the research staff of the Vaccinology and Immunology Research Trials Unit at the Women’s and Children’s Hospital, Adelaide, Verity Hill, and Mary Walker for providing assistance; Narelle Raven for providing technical assistance; and N. Guiso for providing polyclonal antibodies against ptx, fha, and prn.
National serosurveys used pertussis toxin provided by GlaxoSmithKline. N.W. and H.M. have been investigators in studies supported by pharmaceutical companies, including GlaxoSmithKline or Sanofi-Pasteur. H.M. has been given travel support by GlaxoSmithKine to present scientific data at international meetings. N.W. has been given travel support by GlaxoSmithKine to attend an advisory board meeting. Institutional support for serologic testing has been provided by GlaxoSmithKIine for investigator-led vaccine safety and immunogenicity studies conducted by H.M., P.M., and N.W.
This study was supported by the National Health and Medical Research Council (grant 1011942). H.M. was supported by Career Development Fellowship no. 1016272.
Biography
Ms Lam is a doctoral candidate at the University of New South Wales, Sydney, Australia. Her research interests include the epidemiology and evolution of human pathogens.
Footnotes
Suggested citation for this article: Lam C, Octavia S, Ricafort L, Sintchenko V, Gilbert GL, Wood N, et al. Rapid increase in pertactin-deficient Bordetella pertussis isolates, Australia. Emerg Infect Dis [Internet]. 2014 Apr [date cited]. http://dx.doi.org/10.3201/eid2004.131478
References
- 1.Kurniawan J, Maharjan RP, Chan WF, Reeves PR, Sintchenko V, Gilbert GL, et al. Bordetella pertussis clones identified by multilocus variable-number tandem-repeat analysis. Emerg Infect Dis. 2010;16:297–300. 10.3201/eid1602.081707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quinn HE, Mahajan D, Hueston L, Campbell P, Menzies RI, Gilbert GL, et al. The seroepidemiology of pertussis in NSW: fluctuating immunity profiles related to changes in vaccination schedules. N S W Public Health Bull. 2011;22:224–9. 10.1071/NB11023 [DOI] [PubMed] [Google Scholar]
- 3.Campbell P, McIntyre P, Quinn H, Hueston L, Gilbert GL, McVernon J. Increased population prevalence of low pertussis toxin antibody levels in young children preceding a record pertussis epidemic in Australia. PLoS ONE. 2012;7:e35874. 10.1371/journal.pone.0035874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spokes PJ, Quinn HE, McAnulty JM. Review of the 2008–2009 pertussis epidemic in NSW: notifications and hospitalisations. N S W Public Health Bull. 2010;21:167–73. 10.1071/NB10031 [DOI] [PubMed] [Google Scholar]
- 5.Mooi FR, van der Maas NA, de Melker HE. Pertussis resurgence: waning immunity and pathogen adaptation: two sides of the same coin. Epidemiol Infect. 2013;13:1–10. 10.1017/S0950268813000071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mooi FR, van Loo IH, van Gent M, He Q, Bart MJ, Heuvelman KJ, et al. Bordetella pertussis strains with increased toxin production associated with pertussis resurgence. Emerg Infect Dis. 2009;15:1206–13. 10.3201/eid1508.081511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lam C, Octavia S, Bahrame Z, Sintchenko V, Gilbert GL, Lan R. Selection and emergence of pertussis toxin promoter ptxP3 allele in the evolution of Bordetella pertussis. Infect Genet Evol. 2012;12:492–5. 10.1016/j.meegid.2012.01.001 [DOI] [PubMed] [Google Scholar]
- 8.Octavia S, Maharjan RP, Sintchenko V, Stevenson G, Reeves PR, Gilbert GL, et al. Insight into evolution of Bordetella pertussis from comparative genomic analysis: evidence of vaccine-driven selection. Mol Biol Evol. 2011;28:707–15. 10.1093/molbev/msq245 [DOI] [PubMed] [Google Scholar]
- 9.Octavia S, Sintchenko V, Gilbert GL, Lawrence A, Keil AD, Hogg G, et al. Newly emerging clones of Bordetella pertussis carrying prn2 and ptxP3 alleles implicated in Australian pertussis epidemic in 2008–2010. J Infect Dis. 2012;205:1220–4. 10.1093/infdis/jis178 [DOI] [PubMed] [Google Scholar]
- 10.Hegerle N, Paris AS, Brun D, Dore G, Njamkepo E, Guillot S, et al. Evolution of French Bordetella pertussis and Bordetella parapertussis isolates: increase of Bordetellae not expressing pertactin. Clin Microbiol Infect. 2012;•••:E340–6 PubMed. 10.1111/j.1469-0691.2012.03925.x [DOI] [PubMed] [Google Scholar]
- 11.Bouchez V, Brun D, Cantinelli T, Dore G, Njamkepo E, Guiso N. First report and detailed characterization of B. pertussis isolates not expressing pertussis toxin or pertactin. Vaccine. 2009;27:6034–41. 10.1016/j.vaccine.2009.07.074 [DOI] [PubMed] [Google Scholar]
- 12.Otsuka N, Han HJ, Toyoizumi-Ajisaka H, Nakamura Y, Arakawa Y, Shibayama K, et al. Prevalence and genetic characterization of pertactin-deficient Bordetella pertussis in Japan. PLoS ONE. 2012;7:e31985. 10.1371/journal.pone.0031985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barkoff AM, Mertsola J, Guillot S, Guiso N, Berbers G, He Q. Appearance of Bordetella pertussis strains not expressing the vaccine antigen pertactin in Finland. Clin Vaccine Immunol. 2012;19:1703–4. 10.1128/CVI.00367-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weber C, Boursaux-Eude C, Coralie G, Caro V, Guiso N. Polymorphism of Bordetella pertussis isolates circulating for the last 10 years in France, where a single effective whole-cell vaccine has been used for more than 30 years. J Clin Microbiol. 2001;39:4396–403. 10.1128/JCM.39.12.4396-4403.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mooi FR, Hallander H, Wirsing von Konig CH, Hoet B, Guiso N. Epidemiological typing of Bordetella pertussis isolates: recommendations for a standard methodology. Eur J Clin Microbiol Infect Dis. 2000;19:174–81. 10.1007/s100960050455 [DOI] [PubMed] [Google Scholar]
- 16.Fry NK, Neal S, Harrison TG, Miller E, Matthews R, George RC. Genotypic variation in the Bordetella pertussis virulence factors pertactin and pertussis toxin in historical and recent clinical isolates in the United Kingdom. Infect Immun. 2001;69:5520–8. 10.1128/IAI.69.9.5520-5528.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mooi FR, van Oirschot H, Heuvelman K, van der Heide HG, Gaastra W, Willems RJ. Polymorphism in the Bordetella pertussis virulence factors P.69/pertactin and pertussis toxin in the Netherlands: temporal trends and evidence for vaccine-driven evolution. Infect Immun. 1998;66:670–5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boursaux-Eude C, Thiberge S, Carletti G, Guiso N. Intranasal murine model of Bordetella pertussis infection: II. Sequence variation and protection induced by a tricomponent acellular vaccine. Vaccine. 1999;17:2651–60. 10.1016/S0264-410X(99)00038-9 [DOI] [PubMed] [Google Scholar]
- 19.Kinnear SM, Boucher PE, Stibitz S, Carbonetti NH. Analysis of BvgA activation of the pertactin gene promoter in Bordetella pertussis. J Bacteriol. 1999;181:5234–41 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bodilis H, Guiso N. Virulence of pertactin-negative Bordetella pertussis isolates from infants, France. Emerg Infect Dis. 2013;19:471–4 . 10.3201/eid1903.121475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Queenan AM, Cassiday PK, Evangelista A. Pertactin-negative variants of Bordetella pertussis in the United States. N Engl J Med. 2013;368:583–4. 10.1056/NEJMc1209369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pawloski LC, Queenan AM, Cassiday PK, Lynch AS, Harrison M, Shang W, et al. Prevalence and molecular characterization of pertactin-deficient Bordetella pertussis in the US. Clin Vaccine Immunol. 2013. [Epub ahead of print].http:// [DOI] [PMC free article] [PubMed]
- 23.Schmidtke AJ, Boney KO, Martin SW, Skoff TH, Tondella ML, Tatti KM. Population diversity among Bordetella pertussis isolates, United States, 1935–2009. Emerg Infect Dis. 2012;18:1248–55. 10.3201/eid1808.120082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurova N, Njamkepo E, Brun D, Tseneva G, Guiso N. Monitoring of Bordetella isolates circulating in Saint Petersburg, Russia between 2001 and 2009. Res Microbiol. 2010;161:810–5. 10.1016/j.resmic.2010.09.013 [DOI] [PubMed] [Google Scholar]
- 25.Njamkepo E, Cantinelli T, Guigon G, Guiso N. Genomic analysis and comparison of Bordetella pertussis isolates circulating in low and high vaccine coverage areas. Microbes Infect. 2008;10:1582–6. 10.1016/j.micinf.2008.09.012 [DOI] [PubMed] [Google Scholar]
- 26.Stibitz S. IS481 and IS1002 of Bordetella pertussis create a 6-base-pair duplication upon insertion at a consensus target site. J Bacteriol. 1998;180:4963–6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parkhill J, Sebaihia M, Preston A, Murphy LD, Thomson N, Harris DE, et al. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat Genet. 2003;35:32–40. 10.1038/ng1227 [DOI] [PubMed] [Google Scholar]
- 28.Gogol EB, Cummings CA, Burns RC, Relman DA. Phase variation and microevolution at homopolymeric tracts in Bordetella pertussis. BMC Genomics. 2007;8:122. 10.1186/1471-2164-8-122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willems R, Paul A, van der Heide HG, ter Avest AR, Mooi FR. Fimbrial phase variation in Bordetella pertussis: a novel mechanism for transcriptional regulation. EMBO J. 1990;9:2803–9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salaün L, Snyder LA, Saunders NJ. Adaptation by phase variation in pathogenic bacteria. Adv Appl Microbiol. 2003;52:263–301. 10.1016/S0065-2164(03)01011-6 [DOI] [PubMed] [Google Scholar]
- 31.van Gent M, van Loo IH, Heuvelman KJ, de Neeling AJ, Teunis P, Mooi FR. Studies on prn variation in the mouse model and comparison with epidemiological data. PLoS ONE. 2011;6:e18014. 10.1371/journal.pone.0018014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bassinet L, Gueirard P, Maitre B, Housset B, Gounon P, Guiso N. Role of adhesins and toxins in invasion of human tracheal epithelial cells by Bordetella pertussis. Infect Immun. 2000;68:1934–41 . 10.1128/IAI.68.4.1934-1941.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characteristics of Pertactin-deficient Bordetella pertussis Isolates, Australia, 2008–2012.