SUMMARY
Fragment-based ligand design (FBLD) approaches have become more widely used in drug discovery projects from both academia and industry, and are even often preferred to traditional high-throughput screening (HTS) of large collection of compounds (>105). A key advantage of FBLD approaches is that these often rely on robust biophysical methods such as NMR spectroscopy for detection of ligand binding, hence are less prone to artifacts that too often plague the results from HTS campaigns. In this article, we introduce a screening strategy that takes advantage of both the robustness of protein NMR spectroscopy as the detection method, and the basic principles of combinatorial chemistry to enable the screening of large libraries of fragments (>105 compounds) preassembled on a common backbone. We used the method to identify compounds that target protein-protein interactions.
INTRODUCTION
In recent years, fragment-based ligand discovery (FBLD) approaches, also known as fragment-based drug discovery (FBDD), have become popular alternative strategies to conventional high-throughput screening (HTS) campaigns in both academic and industrial drug discovery projects (Congreve et al., 2008; Fischer and Hubbard, 2009; Hajduk and Greer, 2007; Murray and Rees, 2009). The basic idea behind FBDD approaches is to initially identify, usually by screening small focused libraries of low molecular weight compounds (fragments) via biophysical methods, key chemical substructures or pharmacophores sufficient to confer a minimal yet specific interaction with the given target. Subsequently, these fragment hits are matured into more potent binders by a variety of approaches, most often guided by structural studies using X-ray crystallography or nuclear magnetic resonance (NMR) spectroscopy (Congreve et al., 2008; Dalvit, 2009; Hubbard, 2008; Murray and Blundell, 2010; Pellecchia et al., 2002, 2004, 2008). Compared to HTS libraries, fragments libraries contain lower molecular weight compounds (MW < 300 Da), and the resulting hits are consequently of weak binding affinity (with dissociation constants in the micromolar to millimolar range). NMR spectroscopy has been the most widely applied method in FBDD given its unique advantages of (1) detecting fragment hits of weak binding affinity (Kd values up to mM level) with little ambiguity (when spectra of the target are obtained in the presence and absence of a test compound), and (2) providing crude but insightful information on the binding sites of hit compounds (Pellecchia et al., 2002, 2008). Binding information is usually obtained by using chemical shift mapping techniques with 15N and/or uniformly or selectively 13C labeled protein, provided that resonance assignments for the target and its three-dimensional structure are known.
Using protein-based NMR approaches, fragment libraries of up to 10,000 compounds are routinely screened in a relatively short time (from several hours to several days). Compounds are usually tested in mixtures of 10–20, but higher throughput is unlikely to be possible given the limitations of sample consumption and the relatively long measurement times required. Hence, HTS libraries, which usually contain more than 105 compounds, cannot be screened by using NMR or other biophysical approaches, as these methods have a limited throughput. Generally, plate-based spectrophotometric assays are used in HTS. Unfortunately, these methods often select for hundreds or even thousands of misleading compounds, including nonspecific hits, promiscuous aggregators, or other assay-related artifacts, that render follow-up optimizations time-consuming, tedious, and often unproductive and unsuccessful (Böcker et al., 2011; Feng et al., 2005, 2007; Shoichet, 2006a, 2006b).
Regardless of a large number of false hits, HTS has the advantage of testing large libraries rapidly. On the other hand, FBDD has the advantage of using a biophysical/analytical method, such as NMR spectroscopy, to detect binding. These methods are less prone to false hits, but can only be applied to test small libraries, leading to fairly weak binding hits as starting points. As a consequence, maturing the initial hits or linking multiple fragments together into a more potent hit is necessary to obtain a compound with sufficient potency to be used in subsequent hit-to-lead optimizations. Maturing the fragments or linking multiple fragments into a more potent binder is not a trivial task and presents several challenges.
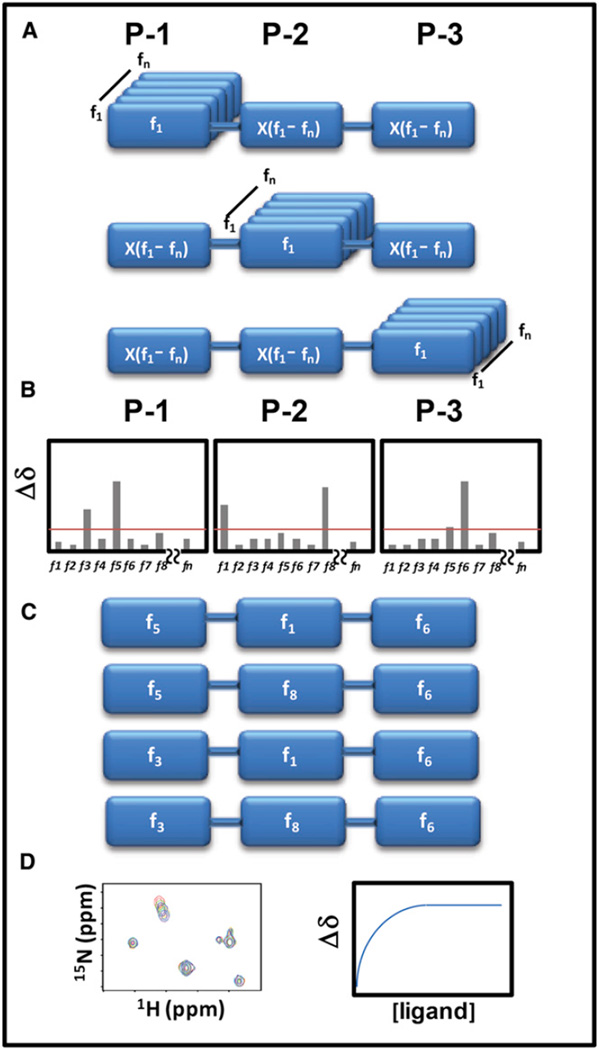
Here, we sought to combine the advantages of both approaches in a screening strategy that we named HTS by NMR. The approach combines basic combinatorial chemistry principles with the advantages of using NMR spectroscopy as the screening method, to screen larger libraries of compound fragments that are preassembled on a common backbone. In order to reduce the number of samples to be screened, hence making the method amenable to NMR-based screening techniques, the library is assembled in mixtures in which each position of a common backbone is systematically fixed while the other positions are populated by all possible functionalities, a technique termed “positional scanning” (Dooley and Houghten, 1993; Dooley et al., 1998; Houghten et al., 1991; Pinilla et al., 1992). For example, a given library with a common backbone that has three positions of diversity wherein each of those diversity positions could be 1 of 100 functionalities (i.e., fragments) and could include up to 106 molecules (100 × 100 × 100) to be synthesized and tested. However, if prepared in positional scanning mixtures, those fragments could be arranged and tested systematically using 100 + 100 + 100 mixtures. Hence, only 300 samples are needed to evaluate 106 compounds, and a sample set of 300 is highly amenable for screening by NMR in a relatively short time (Figure 1). Obviously, mixtures populated with the most effective fragment at a given position would produce the largest signal changes in the NMR spectra of the target, thus allowing the indirect identification of the preferred combinations of scaffolds (Figure 1). Subsequent synthesis and testing of individual compounds would result in the identification of the most active compounds among the possible 106 molecules (Figure 1). Because the final individual scaffolds identified by the approach are already arranged and linked in a specific order, these hits are immediately amenable to subsequent empirical medicinal chemistry strategies for hit-to-lead optimizations backed up by robust NMR-based binding data—without concern about artifact data and without the need to determine the structure of the complex to guide fragment linking or growing. Moreover, because NMR is an unbiased screening technique, ligands for different areas of the protein surface could be identified simultaneously, possibly delineating a protein’s hot spots or allosteric sites that were not known previously.
Figure 1. Schematic Representation of the HTS by NMR Approach.
(A) First, a positional scanning library of compounds needs to be assembled. In the example, a three-positions synthetic combinatorial library is prepared. With a library of n fragments, there will be 3 × n mixtures, each containing n × n compounds. Hence, rather than synthesizing and testing n × n × n individual compounds, the approach would result in testing 3 × n mixtures. For example, a library of 100 fragments assembled at three different positions could be sampled by screening 300 mixtures (100 + 100 + 100), rather than by synthesizing and testing 1,000,000 (100 × 100 × 100) individual compounds.
(B) Chemical shift perturbations induced by each mixture are measured and reported as function of the fixed fragment at each position.
(C) Based on a determined Δδ cutoff, preferential fragments for each position are selected and final individual compounds are synthesized.
(D) The binding affinity of the synthesized compounds is then determined via 2D heteronuclear NMR titration experiments.
See also Tables S1 and S2 reporting the composition of libraries used in the manuscript for the method.
As an initial proof-of-concept application, we explored experimental conditions for the HTS by NMR approach and examined its feasibility by targeting the baculovirus inhibitor of apoptosis protein (IAP) repeat 3 (BIR3) domain of the antiapoptotic protein XIAP, which is known to bind the high-affinity tetrapeptide ligand AVPF (Fesik and Shi, 2001; Sharma et al., 2006). In addition, we further applied the HTS by NMR in a de novo ligand discovery program against the ligand-binding domain (LBD) of the EphA4 receptor tyrosine kinase. Our data demonstrate the feasibility of the approach, and furthermore suggest that the method may be more successful than conventional HTS campaign in designing effective inhibitors of protein-protein interactions (PPI).
RESULTS AND DISCUSSION
HTS by NMR: Approach and Proof of Concept
As mentioned above, the main concept of the HTS by NMR is to identify fragment hits that are already preassembled on a common scaffold or backbone, so that the resulting hits would have the characteristics of HTS compounds but also the verified ability to specifically interact with the target provided by the NMR-based screening data. The general strategy is depicted schematically in Figure 1. In order to perform the HTS by NMR, a positional scanning synthetic combinatorial library is first designed in which each mixture is composed of thousands to millions of compounds, all of which have one position fixed by a common functionality (i.e., a given fragment), while other positions are diversified by all fragment components (Figure 1A). Binding of the mixtures to a protein target is subsequently detected via protein-based NMR chemical-shift perturbation experiments. The chemical shift of a given nucleus represents its local environment, which can be perturbed upon the binding of a compound in its proximity, causing the corresponding signals in the protein spectrum to change in position. If the binding falls into a fast exchange on the chemical-shift time scale, which is generally the case for binders with dissociation constants in the millimolar to nanomolar range, then the observed chemical shifts are the weight average of the signals of the protein in the free and bound states as shown in Equation 2 in Experimental Procedures (Pellecchia, 2005; Smet et al., 2005). Compared with the protein concentration in the NMR sample (usually between 5 and 100 µM), each compound in the mixture has a concentration that could practically reach only nanomolar or lower concentrations, depending on the number of fragments and positions scanned. Hence, one apparent challenge for the success of the HTS by NMR approach is whether NMR is sensitive enough to detect binding of compounds in such mixture-based samples under these experimental conditions. It is obvious that if the concentration of binding compounds is too low, compared to the protein concentration and the dissociation constant, then chemical shift differentials between the free and bound states are small and difficult to detect. However, if even low percentages of compounds in a mixture bind to the protein, then the large number of compounds containing a “hit” fragment at a given position collectively may contribute to produce observable and unambiguous perturbations in the NMR spectra of the target. Given the dominating concentration of the fixed fragment in a mixture, we hypothesize that the fixed fragment is the main contributor to the observed chemical shift perturbations. Therefore, each fixed fragment could be evaluated based on the resulting chemical shift perturbations in the various mixtures (Figure 1B). In theory, the most potent compound can be deduced from the combination of the mixtures producing the highest chemical shift perturbations at each position, assuming that a given scaffold adopts the same binding mode when present in different compounds. This is not necessarily true because compounds could, in principle, bind in different orientations. For example, if P1 is a binder for the target, either the mixture P1-X-X or the mixture X-X-P1 could position the P1 fragment at the same subpocket within the protein surface, and that would produce the erroneous suggestion to synthesize a P1-X-P1 series. Although this phenomenon could produce inactive combinations, the chance of this to occur diminishes with the complexity of the fragments. For example, if the X-X fragments in the P1-X-X or X-X-P1 examples are mere linkers or small fragments (let’s say a Gly-Gly linker, for example), the P1 could likely more freely occupy the same subpocket while embedded in either the P1-X-X or the X-X-P1 molecules. However, when, as in our case, the X-X are more complex and functionalized side chains, the probability of P1 being free to find the same subpocket regardless of its position, while still possible, it is diminished by steric contacts that other adjacent side chains can create. On balance, to find the most potent compounds, it is best to choose different fragment combinations (Figure 1C), synthesize them, and subsequently test the resulting individual compounds experimentally by means of two-dimensional (2D) heteronuclear NMR titrations (Figure 1D).
The scanning approach has been extensively used and validated in the identification of antigen-specific and protease-specific synthetic peptides from screening of large positional scanned libraries (Judkowski et al., 2011; Lim and Craik, 2009; Lustgarten et al., 2006; Pinilla et al., 1999; Reddy et al., 2011; Sospedra et al., 2010). However, unlike these previous positional scanning assay studies, which depend mostly on either fluorescence or absorbance readouts (Diamond, 2007; Lim and Craik, 2009), using protein-based NMR to perform positional scanning does not require specific knowledge of protein function for assay establishment. Moreover, the HTS by NMR approach not only identifies preferred scaffolds and initial hit compounds, but also determines their binding affinity and, in most cases, the site of binding, allowing for more direct follow-up hit-to-lead optimizations. In fact, protein-based NMR chemical shift perturbation can be used not only as a screening technique, but also to map compound binding sites on a protein structure when combined with resonance assignments, therefore providing initial structural information for future hit optimizations. Moreover, as mentioned, compound aggregators, redox, or otherwise reactive small molecules cause false hits that plague screening results from most spectrophotometric assay platforms (Baell and Holloway, 2010; Shoichet, 2006a, 2006b) and even other biophysical screening techniques such as surface plasmon resonance (Giannetti et al., 2008). However, because NMR is a powerful analytical technique to assess the integrity of the protein target, compounds (or mixtures in the primary screens) that cause aggregation or protein denaturation are readily identified and eliminated. Of note is that of the tested mixtures, only one seemed to cause aggregation of the LBD of EphA4 under the experimental conditions used. Again, these “aggregators” are readily identified, and hence not considered as hits.
Application of the HTS by NMR against the Bir3 Domain of XIAP as Proof of Concept
To assess the feasibility of the HTS by NMR approach, we applied it on a test case in which a target protein and its binders have been well studied. The target protein chosen in the training set is the BIR3 domain of the antiapoptotic protein XIAP, which binds directly to the N terminus of caspase-9 to inhibit programmed cell death (Fesik and Shi, 2001; Shiozaki et al., 2003; Wang et al., 2004). It has been shown that in the cell, this interaction can be displaced by the protein SMAC (second mitochondrial activator of caspases) and that the N-terminal tetrapeptide region (AVPF) of SMAC is responsible for the binding (Shiozaki et al., 2003; Srinivasula et al., 2001). Previous studies have also indicated that Ala and Pro are absolutely conserved at the positions P1 and P3 (from the N terminus) in the consensus AVPF motif, which is sufficient for binding to BIR3. Modifications of this AVPF motif have recently led to the generation of numerous XIAP antagonists as anticancer agents, some of which are currently under clinical investigation (Bank et al., 2008; Flygare and Fairbrother, 2010; Li et al., 2004). To evaluate whether the contribution of Ala and Pro to the binding could have been detected via HTS by NMR, we selected mixtures from combinatorial libraries of synthetic peptides of different lengths (Table S1 available online), including peptide mixtures in which either Ala was fixed at the P1 position or Pro was fixed at the P3 position. As controls, different mixtures with Gly fixed at P1 were also tested in the training set. Regardless of the diverse components and number of compounds included (Table S1), nine different mixtures were dissolved in DMSO as stock solutions with the overall concentration of a given mixture close to 150 mM relative to the fixed position.
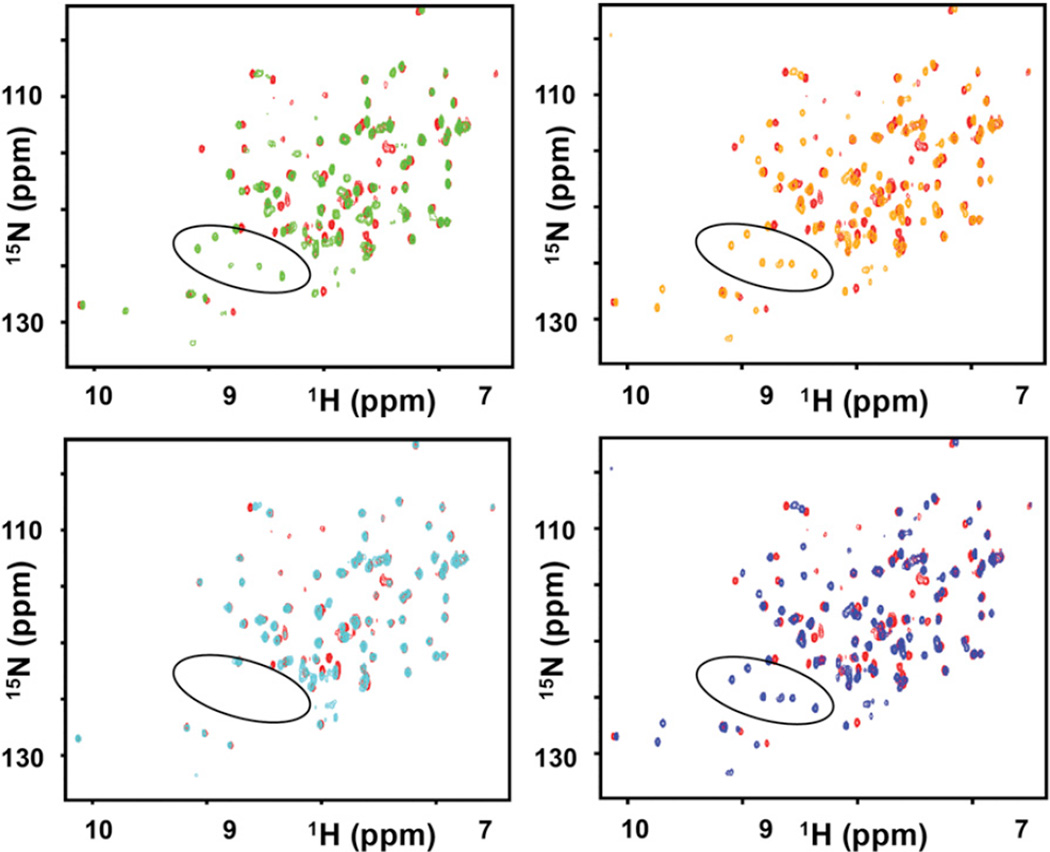
To detect the binding of the mixtures, we collected a series of 2D [15N, 1H] heteronuclear single quantum correlation (HSQC) spectra of 50 µM 15N-labeled BIR3 in the absence and presence of 1 mM mixtures. Assuming a hit rate as low as 1%, the overall concentration of a binding fragment at a fixed position would reach 10 µM, which is comparable to the 50 µM concentration of BIR3, thus possibly generating significant chemical shift perturbations. As shown in Figures 2A and 2B, the overall HSQC spectra of BIR3 experienced significant changes with the addition of the AXXXXX and XXPXXX mixtures, especially in the appearance of a few additional peaks. The same changes were also observed with the addition of the control peptide (AVPFGYSAYPDSVPMMSK), which contains a consensus AVPF motif at the N terminus (Figure 2D). It has been reported that these new occurring peaks around 122–127 ppm (15N) and 8–9 ppm (1HN) result from residues in the flexible loop nearby the BIR3 binding pocket once its conformation is stabilized by the bound peptide (de Souza et al., 2010; Moore et al., 2009). Therefore, the chemical shift perturbation data indicated that AXXXXX and XXPXXX mixtures contain ligands that interact with BIR3 in a way that is similar to the binding of the known AVPF motif. When the fixed residue Ala is replaced by Gly at P1, however, only minor chemical shift perturbations were observed and the new occurring peaks mentioned above did not appear (Figure 2C). Hence, significantly different binding behavior of AXXXXX and GXXXXX indicates that the Ala residue at position P1 is critical for binding to BIR3. This is in agreement with well-documented studies with AVPF and related small molecule inhibitors currently in the clinic, all containing an Ala or an Ala mimetic at the P1 position. Similarly, the tetrapeptide mixtures AXXX and XXPX caused significant chemical shift perturbations, while GXXX showed no significant binding (Figure S1), under the same experimental conditions. The shorter tripeptide mixtures AXX and XXP, however, only caused smaller shifts, presumably due to the lack of the fourth consensus amino acid (Figure S1). The above results are in agreement with the previous conclusion that Ala and Pro are important for binding to BIR3 when peptides are four residues or longer (Sharma et al., 2006). These proof-of-concept results clearly indicate that the HTS by NMR is an effective approach to identify critical “fragments” in complex mixtures of hundreds of thousands compounds even if the individual concentration of each compound is relatively low. Expanding on this approach to small-molecule-like libraries or peptide-mimetic libraries holds great promise as a novel method for hit generation, as next example will demonstrate.
Figure 2. Application of the HTS by NMR to the BIR3 Domain of XIAP.
Superposition of 2D [15N, 1H]-HSQC spectra of BIR3 domain if XIAP, measured in the absence and presence of the mixtures from a hexapeptide positional scanning library. The spectrum for apo-BIR3 is colored in red. The spectrum for BIR3 in the presence of AXXXXX is colored in green, in the presence of XXPXXX is colored in orange, in the presence of GXXXXX is colored in cyan and in the presence of the control peptide AVPFGYSA YPDSVPMMSK is colored in blue. X represents a possible fragment, whereas A, P, and G represent alanine, proline, and glycine, respectively. The new occurring peaks with the addition of control peptide, the AXXXXX mixture, and the XXPXXX mixture, are circled in black. See also Figure S1 and Table S1.
Application of the HTS by NMR Approach for De Novo Ligand Identification
To test the applicability of the HTS by NMR approach to de novo ligand identification, we used it with the EphA4 LBD. EphA4 belongs to the Eph family of receptor tyrosine kinases, which together with their membrane-bound ligands, the ephrins (Eph receptor-interacting proteins), generate bidirectional signals controlling a multitude of cellular processes during development and in the adult (Pasquale, 2005, 2008, 2010). The critical roles of EphA4 in various physiological and pathological processes have been reported in previous studies validating EphA4 as a promising target for the development of small molecule drugs to treat human diseases, such as abnormal blood clotting, spinal cord injury, amyotrophic lateral sclerosis, and certain types of cancer (Noberini et al., 2008, 2011a, 2011b; Qin et al., 2008, 2010).
Previous structural studies indicate that the EphA4 LBD contains a hydrophobic pocket surrounded by four flexible loops (BC, DE, GH, and JK; see later, Figure 5), which confer large structural plasticity to accommodate different binding partners (Bowden et al., 2009). Several 12-amino-acid-long peptide binders that selectively block ephrin ligands from binding to EphA4 have been reported (Murai et al., 2003). For instance, the APY, KYL, and VTM peptides (which were named based on the first three amino acids of their sequences) bind to EphA4 tightly with Kd values in the low micromolar range (Lamberto et al., 2012; Murai et al., 2003). In addition, a few small molecular weight compounds that inhibit ephrin binding to EphA4 at low micromolar concentration have also been reported from HTS campaigns (Giorgio et al., 2011; Noberini et al., 2008, 2011a, 2011b; Qin et al., 2008). However, their detailed mechanism of action remains unclear and likely complex, possibly involving compound oxidations or covalent binding, which are typical issues encountered in traditional HTS hits (Baell and Holloway, 2010; Noberini et al., 2008, 2011a, 2011b).
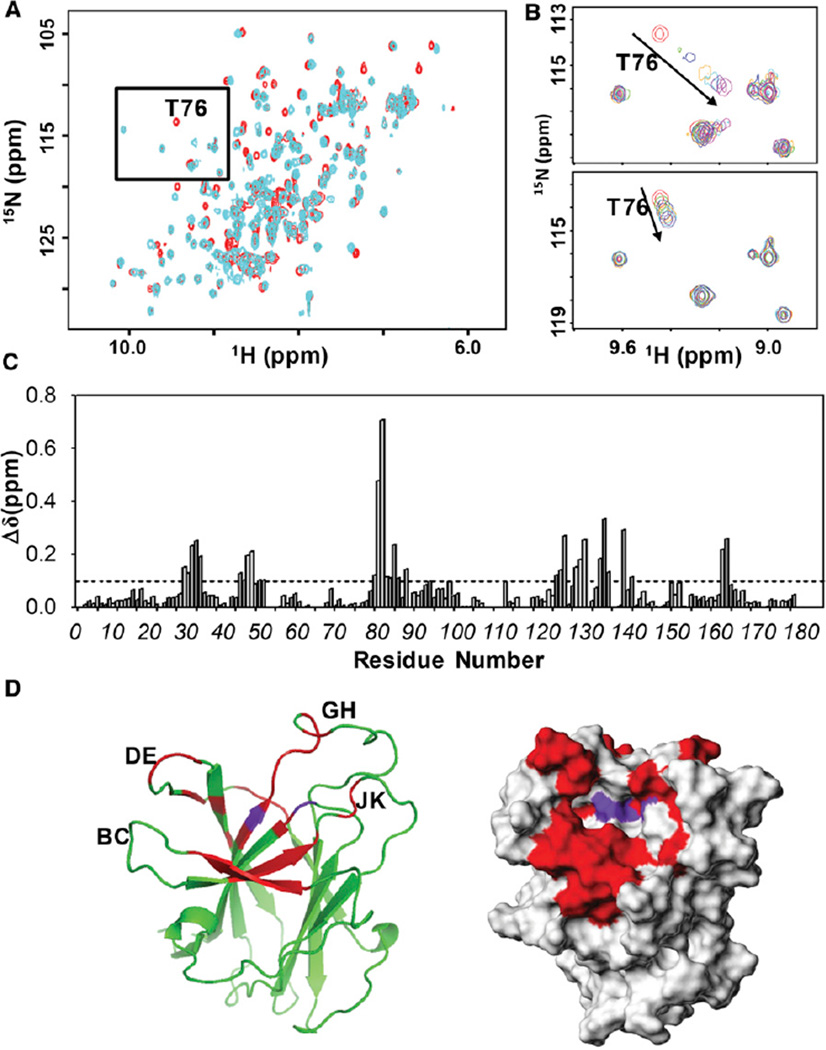
Figure 5. Chemical Shift Mapping Studies with Initial Hits against EphA4 LBD.
(A) Superposition of 2D [15N,1H]-HSQC spectra for 50 µM EphA4 LBD in the absence (red) and presence (cyan) of 300 µM compound 6.
(B) Close-up view of the region boxed in (A) showing an overlap of 2D [15N,1H]-HSQC spectra for 50 µM EphA4 LBD in presence of various concentration of compound 6 (top panel) or compound 1 (bottom panel).
(C) Summary of the chemical shift perturbation resulting from the binding of 300 µM compound 6 to the EphA4 LBD. The average chemical shift changes of each residue were calculated based on Equation 1 in Experimental Procedures.
(D) Chemical shift mapping for the binding of compound 6 on the ribbon representation (left panel) or surface representation (right panel) of the EphA4 LBD. Residues with averaged Δδ above 0.1 ppm are colored in red; residues experiencing intermediate exchange are colored in purple. The loops in the ribbon structure of the EphA4 LBD are also labeled. The surface structure of the EphA4 LBD is from PDB code 3CKH; Swiss-Model 3D was used to add the missing loops to the crystal structure.
See also Figure S8.
In this study, we screened a positional scanning library made up of the combinations of 58 amino acids at each position. In order to increase the drug likeness and the diversity of the compounds in the library, in addition to the natural L-amino-acids, we also included several nonnatural amino acids (Table S2). These 58 amino acids led to increased position diversity while producing average molecular weights of compounds in each mixture of about 500 Da or less (Table S2). Hence, a total of 174 mixtures (58 + 58 + 58) were obtained, each of which contained 3,364 compounds (1 × 58 × 58), with one fixed position and two positions where all combinations of the different 58 amino acids are incorporated.
Because screening compound libraries by NMR can be time and material intensive, we also sought to develop a simple strategy to prescreen a chosen library against a target in order to quickly evaluate the probability of finding hits using this approach. Hence, we prepared a single sample containing the entire collection, in which all three positions contain all amino acids (58 × 58 × 58 compounds). Therefore, the sample used in the prescreening procedure includes all the combinations of the fragments in the screening library. The idea behind the prescreening is that if multiple compounds binding to the target protein exist in the screening library, then detectable chemical shift perturbations should take place because various binders would cumulatively contribute to these shifts. Obviously, the pre-screening has critical requirements for protein and compound mixture concentrations. This type of pre-screening has been reported to be successful for other assays, including in vivo models, and it has been termed “scaffold ranking” (Houghten et al., 2008; Ranjit et al., 2010; Reilley et al., 2010; Rideout et al., 2011).
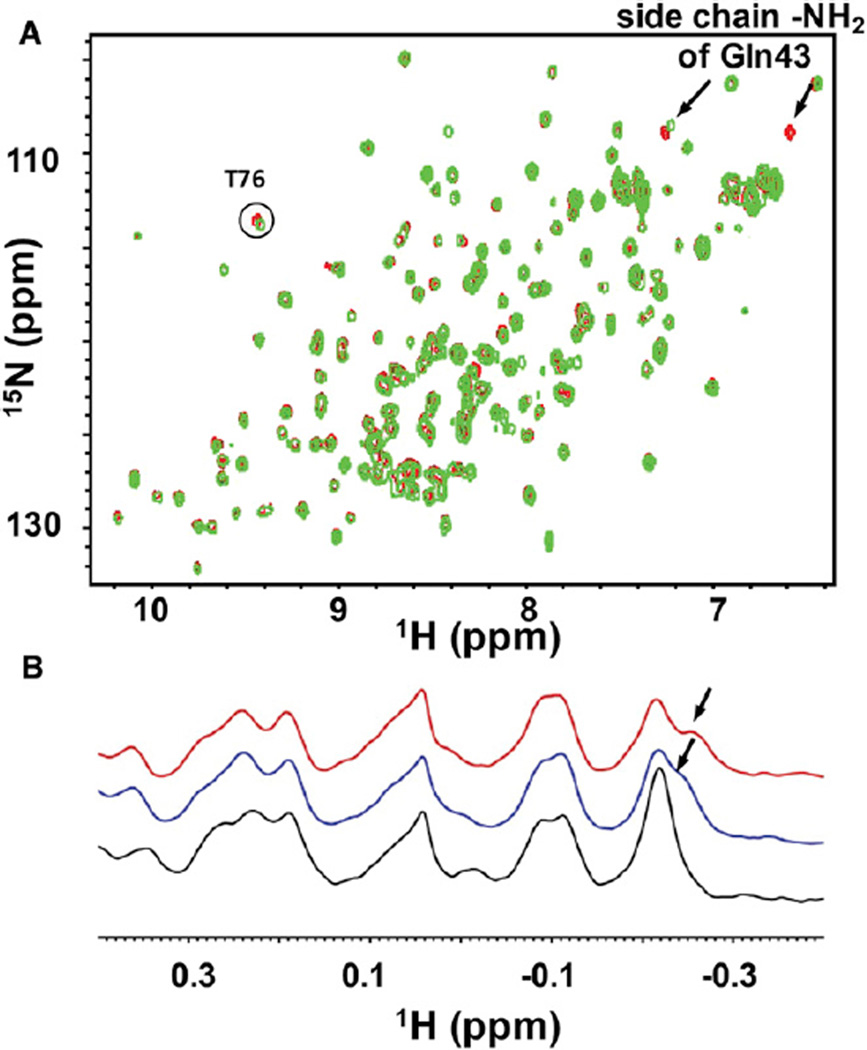
The mixture used in the prescreening contained in theory all 195,112 possible compounds in the screening library. The capability of such XXX mixture components to interact with the EphA4 LBD was determined by comparing the cross peak changes on the 2D [15N, 1H]-HSQC spectra of 15N-labeled EphA4 LBD at 5 µM concentration in the absence and presence of the mixture at 4 mM. In such conditions, and with a typical high-field NMR instrument (600–800 MHz with cryogenic probes), protein-based NMR spectra are collected within several hours, depending on the protein molecular weight, the experimental setup, and the observed nuclei (1H, 15N, 13C). As shown in Figure 3A, addition of the XXX mixture to a sample of 15N-labeled EphA4 LBD caused chemical shift perturbations that are similar to those caused by known inhibitors, inducing changes that are more noticeable in the side chain of residue Gln43 and other residues (Qin et al., 2008). Hence, the simple prescreening assay indicated the existence of binding compounds in the screening library, providing the motivation for performing the complete screen of the individual positional scanned mixtures to identify them. Because of the unbiased nature of the NMR-based screen, we propose that such a prescreening approach is suitable for assessing the druggability of novel targets and/or to identify novel binding surfaces on known targets.
Figure 3. Pretesting of the Mixture Library by NMR.
(A) Superposition of 2D 15N-HSQC spectra of EphA4 LBD in the absence (red) and presence (green) of the prescreening mixture XXX. The binding site resonances shifting upon exposing the protein to the mixture are labeled by arrows or a circle.
(B) 1D 1H aliphatic spectra of 5 µM EphA4 LBD in the apo form (black) and in the presence of 2 mM XXX (blue) and 4 mM XXX (red). The region of the spectrum affected by the presence of the mixture XXX is labeled by an arrow. See also Table S2.
To increase the throughput of the NMR-based screen and to reduce the amount of protein needed, we also monitored the aliphatic region (at 1 ppm and below) of the EphA4 LBD target in simple one-dimensional (1D) 1H-NMR spectra. This region contains resonances from the methyl groups of protein residues and it is rarely populated by signals from organic molecules, which conveniently allows it to be used as an effective primary screening tool to detect compound binding (Stebbins et al., 2007). As shown in Figure 3B, the 1D 1H-aliphatic-NMR spectrum of the EphA4 LBD presented a well separated region that was not affected by DMSO (up to 5%) or small changes in buffer conditions, making the identification of potential compounds straightforward and unambiguous. Hence, 1D 1H-aliphatic-NMR measurements were performed by using a 5 µM protein sample and the XXX prescreening mixture at two different concentrations (2 and 4 mM total concentration). Under these conditions, a new signal gradually shifted out of the overlapped peaks at −0.2 ppm, indicating binding events taking place upon titration of the mixture (Figure 3B). Because collecting 1D 1H spectra generally requires significantly less protein and relatively shorter measurement times (typically from a few minutes to a few hours, depending on the spectrometer used and the protein’s molecular weight) than typical 2D [15N,1H]-HSQC and/or [13C,1H]-HMQC spectra, we believe this approach can extend significantly the use of NMR for screening larger libraries of compounds. Hence, we named the overall method “HTS by NMR.”
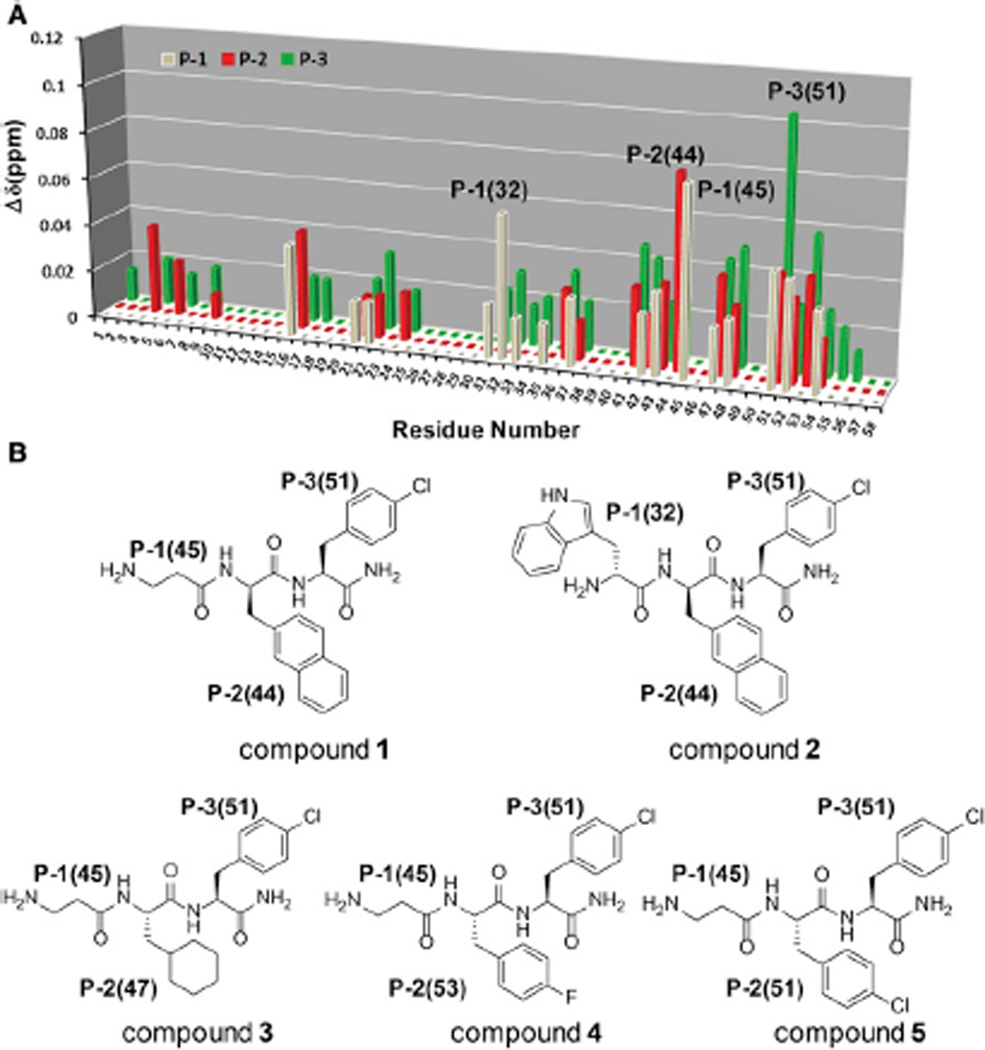
Based on these results, we next performed a screen of the full library by collecting a series of 1D 1H-aliphatic-NMR spectra of 5 µM EphA4 in the presence of each of the 174 mixtures comprising the tripeptide positional scanning library at an overall concentration of 1 mM each (0.3 µM for each compound in a mixture). To roughly evaluate and rank order the binding preference of fragments in the mixtures, chemical shift perturbations for each position were measured and summarized in a score matrix (Figure 4A). Each position was then analyzed separately, resulting in the selection of fragments 45 and 32 for the P-1 position, fragments 16, 47, 51, and 53 for the P-2 position, and fragment 51 for the P-3 position. The individual compounds with various fragment combinations (Figure 4) were subsequently synthesized and tested for binding by NMR and subsequently further characterized and validated by other biophysical and biochemical means (see below). To validate the binding of the synthesized compounds, a series of 2D [15N, 1H]-HSQC spectra of 50 µM EphA4 LBD in the absence and presence of 100 µM of the individual compounds were collected. The resulting chemical shift differences for the binding compounds were quantified as described in Experimental Procedures, and the shifts induced on the backbone amide of residue T76 located at the bottom of the binding pocket were used to roughly rank order the compounds and estimate the binding affinity of each compound for the receptor.
Figure 4. Application of the HTS by NMR in the Identification of Ligands against EphA4 LBD.
(A) Score matrix illustrating the results from the HTS by NMR with the EphA4 LBD. The chemical shift changes on the monitored peak are plotted versus the fixed fragment numbers on different positions in which P-1 is colored in gray, P-2 is colored in red, and P-3 is colored in green. The fragments with the highest induced chemical shift perturbation at each position as well as the second highest induced chemical shift perturbation at P-2 were labeled in bracket.
(B) Structures of compounds 1 to 5. The combination of the fragments with the highest score at each position resulted in compound 1. The combination of the fragments with the highest score at P-1 and P-3, and the second highest score at P-2, resulted in compound 2.
Compound 1 (Figure 4B) is the molecule carrying the fragments that induced the largest Δδ perturbations in the mixtures. Compound 1 caused small but significant chemical shift perturbations under these conditions (Figure S2), implying that it possesses moderate affinity for the EphA4 LBD compared to the other initial compounds tested. The dissociation constant (Kd) value of compound 1 binding to the EphA4 LBD was calculated to be 227 µM via NMR titration experiments, conducted by tracing the chemical shifts for the backbone 1HN/15N nuclei of residue T76 in the 2D [15N, 1H]-HSQC spectra of EphA4 LBD (data not shown). Mapping the chemical shift changes along the primary sequence and the three-dimensional structure of the EphA4 LBD indicated that compound 1 targets its ligandbinding pocket (Figure S2). When fragment 45 on P-1 in compound 1 is replaced by fragment 32 (compound 2; Figure 4B), however, no significant binding is observed. This supports the notion that a combination of fragments with high scores at each position does not always and necessarily yield a potent compound, as explained earlier, likely due to different binding orientations of the fragments when embedded in different molecules. Nonetheless, the method is significant as long as at the least one reasonably potent hit is found.
In view of the importance of fragment 45 at P-1 position and the larger chemical shift perturbations caused by fragment 51 at P-3, the effects of P-2 diversity on binding to the EphA4 LBD were evaluated in compounds 3–6, all of which are analogs of compound 1. Compound 6 possesses the strongest binding affinity for the EphA4 LBD (Table 1), inducing significant changes in its 2D [15N, 1H]-HSQC spectra (Figure 5A). Interestingly, different from compound 1, which exhibits a characteristic “peak walking” upon titration, typical of a fast exchange binding pattern (Figure 5B), upon titrating compound 6 into EphA4, the peaks corresponding to residues in the binding site residues loosed intensity at low ligand concentration and then gradually reappeared at different chemical shifts during the titration (Figure 5B). This is a typical intermediate exchange pattern on the chemical shift time scale for protein-ligand interactions, which usually suggests stronger binding affinities (dissociation constants approaching the low micromolar range). Mapping the most perturbed residues on the surface of EphA4 LBD indicated that two residues experiencing intermediate exchange upon binding are located at the bottom of the ephrin binding pocket while the other perturbed residues are located in a subpocket formed by the nearby DE and GH loops (Figures 5C and 5D). Given the plasticity of these loops as well as the flexibility of the backbone of compound 6, a conformational rearrangement around the pocket might occur during the binding of compound 6, which may also account for the observed slow exchange behavior in the titration.
Table 1.
Structure-Activity Relationship Studies for the Reported EphA4 LBD Ligands
| Hit structure at each modification stage |
Name | M.W. | R1 in defined position |
IC50 by FPA (µM)* |
Kd by ITC (µM) |
|---|---|---|---|---|---|
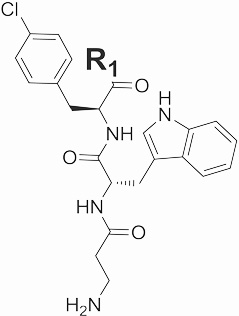 |
6 | 455 | -NH2 | 388 ± 30 | - |
| 7 | 584 | L-lysine-NH2 | 206 ± 27 | - | |
| 8 | 585 | L-glutamate-NH2 | 398 ± 75 | - | |
| 9 | 603 | L-phenylalanine-NH2 | 273 ± 117 | 41 | |
| 10 | 555 | L-valine-NH2 | 262 ± 75 | - | |
| 11 | 618 | L-4-methylphenylalanine-NH2 | - | - | |
| 12 | 605 | L-4-pyridylalanine-NH2 | 100 ± 32 | 20 | |
| 13 | 637 | L-4-chlorophenylalanine-NH2 | ND | ND | |
| 14 | 622 | L-4-fluorophenylalanine-NH2 | 99 ± 34 | - | |
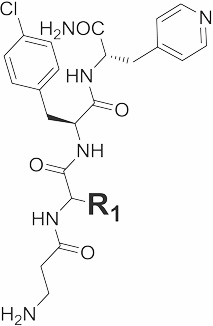 |
15 | 620 | 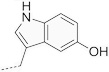 |
24 ± 6.2 | 12 |
| 16 | 635 | 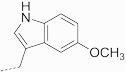 |
29 ± 6.4 | 14.9 | |
| 17 | 622 | 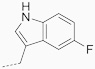 |
73 ± 36 | - | |
| 18 | 622 | 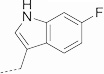 |
54 ± 23 | - | |
| 19 | 619 | 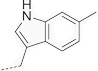 |
118 ± 67 | - | |
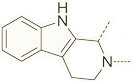 |
20 | 617 | 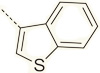 |
N.B. | - |
| 21 | 622 | 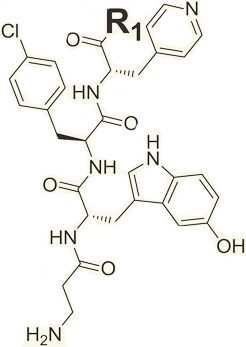 |
86 ± 13 | - | |
| 22 | 806 | D-tryptophan-NH2 | 3.77 ± 0.5 | 1.2 | |
| 23 | 806 | L-tryptophan-NH2 | 21 ± 4.4 | - | |
| 24 | 733 | D-isoleucine-NH2 | 26 ± 12 | - | |
| 25 | 733 | L-isoleucine-NH2 | 44 ± 16 | - | |
| 26 | 719 | D-valine-NH2 | 25 ± 9.7 | - | |
| 27 | 719 | L-valine-NH2 | 96 ± 32 | - | |
| 28 | 733 | D-leucine-NH2 | 60 ± 27 | - | |
| 29 | 733 | L-leucine-NH2 | 87 ± 35 | - | |
| KYLPYWPVLSSL | KYL | 1465 | 4.22 ± 0.5 | 1.3 |
See also Figure S4–S6 and Table S4.
When a protein-ligand interaction falls in slow or intermediate exchange on the NMR time scale, integrating cross peaks on 2D [15N, 1H]-HSQC spectra is required to calculate the molar fraction of nuclei in the free and bound states. This is generally difficult, which limits the application of NMR titration as a method for accurately measure dissociation constants. To circumvent this limitation, we applied an additional method based on fluorescence polarization to compare the binding potencies of the synthesized compounds. The fluorescence polarization assay (FPA) measures changes in the polarization of the light that results from a free tumbling fluoresceinated reference molecule and the same molecule in complex with a larger protein, which slows its rotational correlation times in solution (Jameson and Sawyer, 1995; Nasir and Jolley, 1999; Stewart et al., 2010). To study the interactions between the compounds and EphA4, 5 µM FITC-labeled control KYL peptide (KYLPYWPVLSSL) was incubated with 2 µM EphA4 LBD. After a 30 min incubation, the compounds were added, causing the release of the bound FITC-KYL molecules and a decrease in fluorescence polarization. By fitting the polarization changes at the various concentrations of the compounds, IC50 values can be estimated. The FPA measurements indicated that compound 6 possesses a stronger EphA4 binding potency than compound 1, consistent with what we observed in the NMR titration experiments. As a comparison, the IC50 value of unlabeled KYL peptide was also determined (Table 1). Taken together, these results indicate that compounds 1 and 6 target the ligand-binding pocket of EphA4.
Interestingly, we tested if individual side chains of compound 6, namely 3-methylindole and 4-chlorotoluene occupying positions P-2 and P-3, respectively, bind to the EphA4 LBD. Upon titration, these fragments caused small chemical shift perturbations in the 2D [15N, 1H]-HSQC spectra of EphA4 LBD, but these were observable at only higher ligand concentrations (3 mM), suggesting weak binding affinities against the protein (Figures S3 and S8). Chemical shift mapping indicated that the 3-methylindole mainly affected residues on the side of the pocket composed of loops BC and DE (Figure S8). On the contrary, the 4-chlorotoluene produced largest chemical shift perturbations (Δppm above 0.03 ppm) that localized in a nearby subpocket composed of loops GH and JK (Figure S8). These data suggest that the two fragments indeed occupy adjacent subpockets.
In addition of testing these side chains individually, we also tested the binding of compounds with the common scaffold carrying only one fragment at one position and two Gly residues at the two other positions (i.e., G-16-G and G-G-51). Again, binding of these test molecules can be observed only at millimolar ligand concentrations, with only minor shifts observed (Figure S3).
These binding studies revealed two points: first, the individual fragments exhibit weak binding affinities for the target protein, which is difficult to detect; second, incorporating a given fragment hit into the library, as in our approach, allows its unambiguous detection even at lower concentrations. These results support our hypothesis that preloading the fragments on a common backbone, arranged in positional scanning mixtures libraries, provides an effective mean for hit identification via NMR.
Similar to other fragment-based techniques, such as the SAR by NMR (Shuker et al., 1996), the approach is unbiased toward a particular pocket and does not require preconceived assays. However, unlike other fragment-based approaches, no structural characterization of the fragment’s target complex and/or systematic linker optimizations is necessary to obtain initial hit compounds.
Hit-to-Lead Optimizations
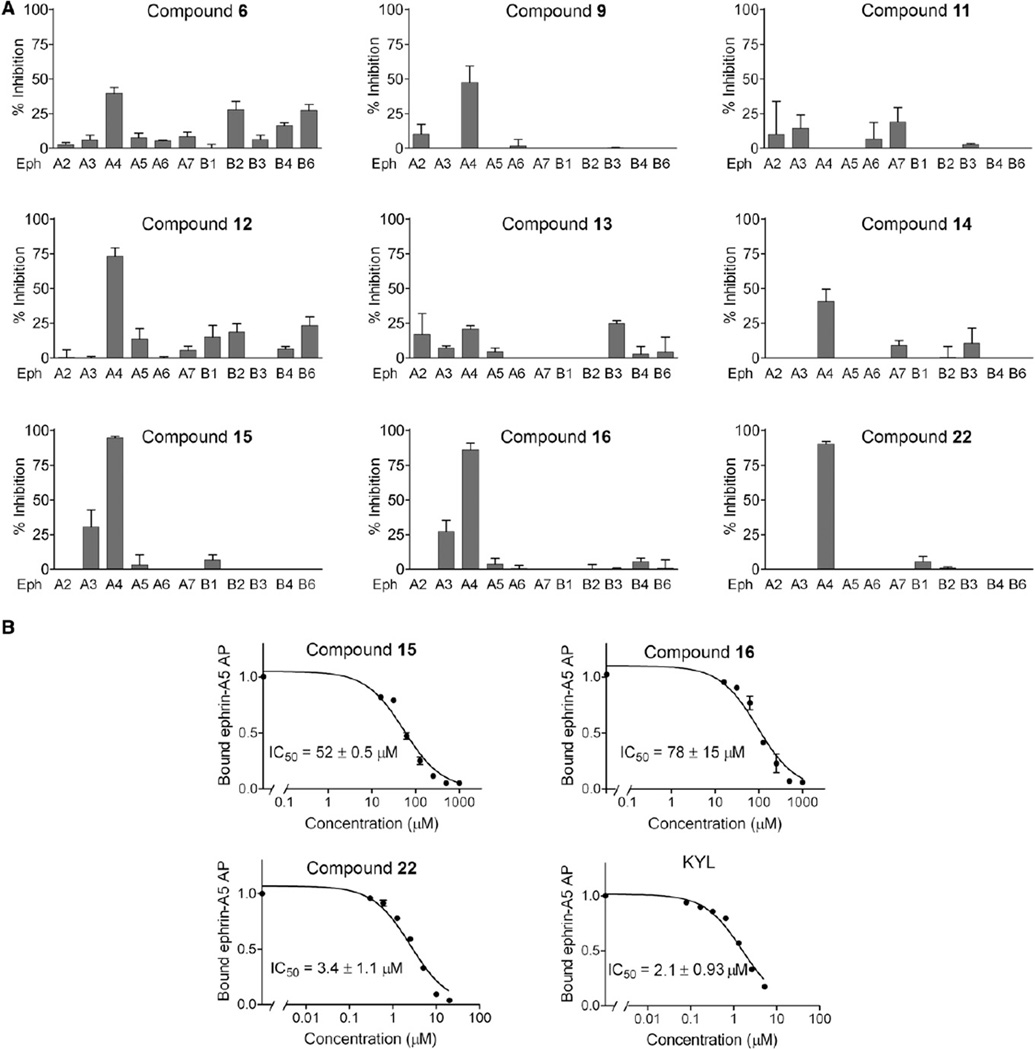
To further enhance the binding potency of the compounds, compound 6 was subsequently used as a starting point for iterative optimizations, mainly by incorporating additional heavy atoms and selecting fragment analogs to investigate the structure-activity relationships (SAR) of the compounds. First, four analogs of compound 6, namely compounds 7–10, were synthesized by elongating compound 6 at position P-4 with either a Lys, Glu, Phe, or Val amino acid, as representatives of four different residue types (positively charged, negatively charged, aromatic, and aliphatic, respectively). Because the compounds were first identified, and hence validated, by NMR, at this hit-to-lead optimization stage we can use the FPA and/or ELISA to more rapidly monitor progress, although NMR and/or isothermal titration calorimetry (ITC) validations could be conducted in parallel, and must be conducted at the least on key compounds. FPA studies revealed that, except for compound 8 with Glu on position P-4, compounds 7, 9, and 10 all exhibited significant improvements over compound 6, with IC50 values around 200–300 µM (Table 1). When using 1D 1H spectra of EphA4 to validate their binding, we noticed that only compound 9 with Phe on P-4 caused shifts in the intermediate exchange on the monitored peak at −0.2 ppm (data not shown), possibly suggesting a stronger binding affinity of compound 9 compared to compounds 7 and 10. Therefore, compound 9 was selected in this iteration for further evolutions.
In a second iteration, four derivatives of compound 9 (compounds 11–14) with analogs of Phe at position P-4 were synthesized and SAR data were obtained by FPA and ELISA. FPAs yielded IC50 values of about 100 µM for compounds 12 and 14, which are approximately 2-fold better than the initial hit compound 9 (Table 1). IC50 values could not be obtained for compounds 11 and 13 due to their poor solubility at concentrations above 200 µM; hence, these were no longer considered. Additional ELISA competition assays revealed that compounds 12 and 14 competed for the binding of ephrin-A5 alkaline phosphatase (AP) to EphA4, with 75% inhibition at 500 µM for compound 12 and 40% inhibition for compound 14 (Figure 6A). Consistent with the results from FPAs, ELISAs also indicated that compounds 11 and 13 exhibited no or weak inhibition (less than 25% inhibition) under these experimental conditions. All the above results suggested that compound 12 possesses higher affinity for the target compared to compounds 11, 13, and 14. Therefore, compound 12 was chosen as the hit compound for the next iteration.
Figure 6. Compounds Identified by the HTS by NMR Inhibit Ephrin Ligand Binding to EphA4 and Ephrin-Dependent EphA4 Tyrosine Phosphorylation in Cells.
(A) Inhibition of ephrin-A5 AP binding to immobilized EphA receptor Fc fusion proteins and ephrin-B2 AP binding to immobilized EphB receptor Fc fusion proteins in the presence of the peptides relative to no peptide. All the compounds were tested at 500 µM, with the exception of compound 22, which was tested at 15 µM. Error bars represent the standard errors from two to four measurements.
(B) Inhibition of ephrin-A5 AP binding to immobilized EphA4 Fc by compounds 15, 16, and 22 and by the KYL peptide. Error bars represent the standard errors from two to four measurements. R2 values indicating the goodness of fit for the inhibition curves were 0.97 for peptide 15, 0.95 for peptide 16, 0.93 for peptide 22 and 0.97 for KYL.
See also Figures S5–S7 and Table S4.
Because varying the fragments at position P-2 resulted in compounds with improved affinities, we further derivatized the indole ring at position P-2 of compound 12 with other analogs (compounds 15–21; Table 1). Among seven synthesized molecules, compounds 15 and 16 exhibited the most significant improvement in displacing the reference peptide, with IC50 values of 24 and 29 µM, respectively (by FPA), which corresponded to a 3- to 4-fold improvement compared to compound 12 (IC50 ~100 µM). The IC50 values for compounds 15 and 16 were also determined in dose-response ELISA measuring the ability of the compounds to displace the natural ligand, ephrin-A5, from EphA4. In this assay, we observed IC50 values of 170 µM for compound 12, 50 µM for compound 15, and 71 µM for compound 16 (Figure 6B and data not shown). Thus, the three compounds inhibited not only binding of the KYL peptide to EphA4, but also the binding of the natural ephrin ligand. Although the IC50 values obtained by two methods are slightly different, likely because of the higher EphA4 binding affinity of ephrin-A5 compared to the KYL peptide, the same ranking is clearly observed in that compounds 15 and 16 exhibit more pronounced inhibition than compound 12.
Compounds 15 and 16 share some structural features with the KYLPYWPVLSSL reference peptide, such as a positively charged group on the first position (side chain of Lys in KYLPYWPVLSSL versus N terminus of β-Ala in compound 15 and 16) and an aromatic ring on the second position (Tyr in KYL versus Trp analogs in compounds 15 and 16). However, in the KYLPYWPVLSSL peptide, there are several additional hydrophobic residues in the central part of the sequence. Hence, to further improve the binding potency of compound 15, we incorporated an extra hydrophobic group on the C terminus of compound 15 and synthesized compounds 22–30. Among these compounds, compound 22 displayed a Kd value of 3.77 µM (by ITC, see below) and an IC50 value for inhibition of EphA4-ephrin-A5 binding in ELISA of 3.4 µM (Figure 6B, see below). These values are 10- to 20-fold lower than those obtained with compounds 15 and 16 and comparable to the 12-mer KYLPYWPVLSSL peptide.
To further confirm the binding of compounds 12 and 15, 2D [15N, 1H]-HSQC spectra of 50 µM 15N-labeled EphA4 LBD recorded in the absence and presence of 100 µM of each compound were collected. After the addition of compounds 12, 15, or 22, significant chemical shift perturbations were observed and the perturbed residues were similar to those affected by compound 6, consistent with targeting of the ephrin-binding pocket of EphA4 (Figures S4 and S5). The dissociation constants of the compounds for the EphA4 LBD were then determined via ITC (see also Figure S6 and Table S3), which yielded Kd values of 20, 12, and 14.9 µM for compounds 12, 15, and 16, respectively. In addition, the control 12-mer peptide KYLPYWPVLSSL was also tested by ITC, which yielded a Kd of 1.3 µM under the same experimental conditions. The parameters derived from the ITC experiments indicated that the interaction between the binders (including KYLPYWPVLSSL and the synthesized compounds) and EphA4 LBD is enthalpy driven. Although the Kd values of compounds 15 and 16 are weaker than the control KYLPYWPVLSSL peptide, compounds 15 and 16 exhibit significantly better ligand efficiencies (0.151 for compound 15 versus 0.086 for KYL) because of their smaller molecular weight. Of note is that the tetrapeptide KYLP derived from the KYLPYWPVLSSL sequence resulted inactive by NMR and FPA under similar experimental conditions.
Moreover, together with the improved binding affinity, the compound selectivity is also markedly improved. As shown in Figure 6A, the first-generation compound 6 inhibited the binding of ephrin-A5 AP to EphA4 by 45% at 500 µM, but also with some-what lower potency other Eph receptors such as EphB2, EphB4, and EphB6 (inhibitions between15%and 30%). Compound 9 exhibited not only higher inhibition of EphA4-ephrin-A5 binding (more than 50% at 500 µM), but also higher selectivity, and only inhibited EphA2 by less than 15% besides EphA4. Compounds 15 and 16 exhibited even improved inhibition of ephrin binding to EphA4 (around 90%) among the EphA and EphB receptors examined, with some minor inhibition of EphA3 (25%) and no or weak inhibition of other Eph receptors (less than 5%). This is confirmed by NMR experiments showing that compound 15 caused significant chemical shift perturbations in EphA4 spectra and only minor changes in EphA3 LBD spectra, while no significant perturbations were detected in the EphA2 LBD spectra under the same experimental conditions (data not shown).
In addition to having improved potency, compound 22 showed high selectivity for EphA4 as at 15 µM it inhibited only EphA4 among the receptors tested (Figure 6). Compounds 15, 16, and 22 also appear to be remarkably resistant to proteases present in biological fluids, as assessed by measuring the ability of the compounds to inhibit EphA4-ephrin-A5 interaction in ELISAs after incubation in cell culture medium or mouse serum (Figure S7). In these stability assays, compound 15 had a half-life of ~12 hr in mouse serum, while compound 16 appeared to be even more stable and retained ~80% of its antagonistic activity after a 24 hr incubation in mouse serum. In addition, the three compounds retained ~60%–70% of their efficacy after a 72 hr incubation in medium conditioned by PC3 prostate cancer cells. This is in contrast with the lower stability observed for the KYLPYWPVLSSL peptide (Lamberto et al., 2012), which in our assay showed half-lives of ~0.6 and ~7 hr in mouse serum and in cell culture medium, respectively (Figure S7). Taken together, these results suggest that the compounds we have identified by using the HTS by NMR approach represent a worthy starting point for the development of EphA4 antagonists with markedly improved drug-like properties over existing peptides and small molecules.
Conclusions
To our knowledge, we reported on a new fragment screening strategy, HTS by NMR. We demonstrated the feasibility of this method by applying it first to a test case and subsequently to a de novo ligand-discovery program against the EphA4 LBD. An overall screening procedure was first established and tested against the EphA4 LBD, resulting in compound 22. This compound exhibits significant binding affinity and selectivity for the targeted ligand binding domain of EphA4, providing convincing proof-of-concept data that support the feasibility of the approach and the establishment of effective screening and optimization protocols. Our data clearly demonstrate the effectiveness and usefulness of our strategy for the rapid identification and optimization of compounds interacting with a target protein. Currently, we are evolving the approach using nonpeptide libraries arranged in the same positional scanning format (Houghten et al., 2008; Judkowski et al., 2011). Based on the data reported, we believe the approach may find its utility especially in the identification of inhibitors of PPIs, allosteric inhibitors, and their binding sites, and for establishing the overall druggability of targets.
SIGNIFICANCE
The challenge of FBLD approaches is that the evolution of initial weakly interacting fragments into more mature compounds with low micromolar affinity (usually the starting point for subsequent hit-to-lead optimizations) is not trivial and often involves attaining properly linked compound fragments. Approaches such as the SAR by NMR (Shuker et al., 1996) may also require structural studies and several iterations. Our idea is to combine the robustness of protein NMR spectroscopy as the detection method with the basic principles of combinatorial chemistry to enable the screening of large libraries of preassembled fragments (>105 compounds) on a common backbone. Hence, we term the approach HTS by NMR. The approach seems particularly suited to target larger protein-protein interaction surfaces. We indeed demonstrated the feasibility of HTS by NMR using a well-studied target, the baculovirus IAP repeat 3 (BIR3) domain, and further, we used the approach to identify novel (to our knowledge) and potent compounds that target the ligand binding domain of the EphA4 receptor.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The human EphA4 LBD (residues 29–209) was prepared as described previously (Qin et al., 2008). Briefly, the pET32á vector containing the EphA4 LBD cDNA fragment (kindly provided by Dr. Song) was transformed into Escherichia coli Rosetta-gami (DE3) cells (Novagen). The transformed cells were then transferred into L-Broth medium and were grown at 37°C. A total of 0.4 mM isopropyl 1-thio-D-galactopyranoside was added into the growing cells when optical density reached 0.7 and continued to grow at 20°C overnight. The overexpressed protein was purified using Ni2+ affinity chromatography. The generation of the isotope-labeled proteins for NMR studies followed a similar procedure except that the bacteria were grown in M9 medium with the addition of (15NH4)2SO4 for 15N labeling.
Library and Peptide Synthesis
The positional scanning libraries were prepared at the Torrey Pines Institute for Molecular Studies as described previously (Pinilla et al., 1992, 1994) using the simultaneous multipeptide synthesis method (Pinilla et al., 1994). All libraries used were peptide-like and arranged in an OXn format, where O represents one of the component in a defined position and X represents a mixture of all the components. The hexapeptide positional scanning library (TPI 1069) is made up of all the combinations of 19 natural amino acids with the exception of cysteine; the tetrapeptide positional scanning library (TPI 367/378) contains 52 components, at each of the four diversity positions, composed of L (16), D (14), and unnatural (22) amino acids; the tripeptide positional scanning library (TPI 1455) contains 58 components, at each of the three diversity positions, composed of L (17), D (16), and unnatural (25) amino acids (Table S2). Each mixture in the hexapeptide, tetrapeptide, and tripeptide libraries is composed of 2.5 million, 140,680, and 3,364 compounds, respectively. All mixtures of the libraries are N-terminal free and C-terminal amide.
Fluorescence Polarization Assays
An EphA4 KYL peptide (KYLPYWPVLSSL) (Murai et al., 2003) was labeled at the N terminus with fluorescein isothiocyanate (FITC) and purified by high-performance liquid chromatography. For competitive binding assays, 1 µl of 200 µM EphA4 LBD was preincubated with the tested compounds at various concentrations in 98 µl PBS (pH = 7.2) in 96-well black plates at room temperature for 10 min, and then 1 µl of 500 µM FITC labeled EphA4 peptide was added to produce a final volume of 100 µl. The KYL and DMSO were incubated in each assay as positive and negative controls, respectively. After 30 min of incubation at room temperature, the polarization values in millipolarization units were measured at excitation/emission wavelengths of 480/535 nm with a multilabel plate reader (PerkinElmer, Waltham, MA, USA). IC50 was determined by fitting the experimental data to a Sigmoidal dose-response (variable slope) nonlinear regression model (GraphPad Prism version 5.01 for Windows, GraphPad Software, San Diego, CA, USA).
NMR Spectroscopy
NMR spectra were acquired on 600 and 700 MHz Bruker Avance spectrometer equipped with either TCI probe and z-shielded gradient coils or a TCI cryoprobe. All NMR data were processed and analyzed using TOPSPIN2.1 (Bruker Biospin, Billerica, MA, USA) and SPARKY3.1 (University of California, San Francisco, CA, USA). 2D-[15N, 1H]-HSQC experiments were acquired using 32 scans with 2,048 and 128 complex data points in the 1H and 15N dimensions at 300 K. Compound binding was detected at 27°C by comparing the 2D-[15N, 1H]-HSQC spectra of 50 µM EphA4 LBD in the absence and presence of compounds at mole ratio 2:1, respectively. The chemical shift changes were calculated using the following Equation 1 (Farmer et al., 1996):
| (1) |
Dissociation equilibrium constants (Kd) of compounds against EphA4 were determined by monitoring the protein chemical shift perturbations as function of compound concentration. For instance, equivalent amounts of compounds were added to a 50 µM sample of EphA4 to yield 1:1, 2:1 stoichiometries of protein/ligand concentration. Titration analysis was done by fitting chemical shift data into a quadratic equation as described in the following Equation 2 (Pellecchia, 2005; Smet et al., 2005):
| (2) |
where Δobs is the observed chemical shift perturbation value at each titration point, Δmax is the maximum chemical shift perturbation value of the fully complexed protein, and [L]0 and [P]0 are the total concentrations of compound and protein.
Binding Constant Determination by ITC
Isothermal titration calorimetry was performed on a VP-ITC calorimeter from Microcal (Northampton, MA, USA). When indicated, measurements were performed in a reverse way—i.e., the protein was titrated into the compound solution. A total of 8 µl EphA4 solution (1.65 mM) was injected into the cell containing 165 µM compound per injection. In each experiment, 37 injections were made. All titrations were performed at 25°C in PBS buffer supplemented with 10% DMSO. Experimental data were analyzed using Microcal Origin software provided by the ITC manufacturer (Microcal).
ELISAs
Protein A-coated wells (Pierce Biotechnology, Rockford, IL, USA) were used to immobilize Eph receptor Fc fusion proteins (R&D Systems, Minneapolis, MN, USA) incubated at 1 µg/ml in TBST (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.01% Tween-20). The compounds were added to the plates for 2 hr before adding culture supernatants from transfected 293HEK cells containing ephrins fused to alkaline phosphatase (ephrin-A5 AP, 0.005 nM final concentration; ephrin-B2 AP, 0.01 nM final concentration) (Koolpe et al., 2002; Noberini et al., 2008). The culture supernatants were diluted in TBST and incubated for an additional 20 min in the presence or in the absence of compounds. The amount of bound AP fusion protein was quantified using pNPP as the substrate. Ephrin-AP concentrations were calculated from AP activity (Cullen, 2000; Flanagan et al., 2000). Unless otherwise specified, all the binding and washing steps were performed in TBST. IC50 values were calculated using nonlinear regression and the program GRAPHPAD (PRISM, La Jolla, CA, USA).
Measurement of Compound Stability in Mouse Serum and Cell Culture Medium
PC3 prostate cancer cells were grown in RPMI 1640 medium (Mediatech, Herndon, VA, USA) with 10% FBS (Hyclone, Logan, UT, USA), penicillin, and streptomycin. The compounds were added to mouse serum or culture medium conditioned by PC3 cells and incubated at 37°C for different times. Serum and culture medium were then diluted 1:20 (corresponding to a final concentration 150 µM) in ELISA wells and incubated for 2 hr in the presence of 0.005 nM ephrin-A5 AP. Inhibition of EphA4-ephrin-A5 binding was measured as described above. Absorbance from wells coated with Fc and incubated with ephrin-A5 AP and serum or culture medium was subtracted as the background. Absorbance obtained from wells incubated with mouse serum or conditioned medium not containing any compound was used to determine the 0% inhibition level (efficacy = 0), and absorbance obtained in the presence of the compounds mixed with serum or medium immediately before adding them to the ELISA wells was used for normalization (efficacy = 1).
Supplementary Material
ACKNOWLEDGMENTS
We thank the NIH for generous support (Grant NIDA 1R01DA031370-01 to R.A.H. and M.G.; Multiple Sclerosis National Research Institute to C.P.; and partially funded by the State of Florida, Executive Office of the Governor’s Office of Tourism, Trade, and Economic Development to R.A.H. and M.G.; Grant NCI P01CA138390 to E.B.P. and M.P.). We also thank Dr. Jianxing Song and Dr. Haina Qin at the University of Singapore for kindly sharing the expression plasmid for EphA4. Finally, we thank Dr. Andrey Bobkov of the SBMRI Protein Analysis Facility for precious support with ITC measurements. M.P. is a founder of AnCoreX Therapeutics, LLC and a member of its scientific advisory board.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes eight figures and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.chembiol.2012.10.015.
REFERENCES
- Baell JB, Holloway GA. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 2010;53:2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- Bank A, Wang P, Du C, Yu J, Zhang L. SMAC mimetics sensitize nonsteroidal anti-inflammatory drug-induced apoptosis by promoting caspase-3-mediated cytochrome c release. Cancer Res. 2008;68:276–284. doi: 10.1158/0008-5472.CAN-07-5242. [DOI] [PubMed] [Google Scholar]
- Böcker A, Bonneau PR, Edwards PJ. HTS promiscuity analyses for accelerating decision making. J. Biomol. Screen. 2011;16:765–774. doi: 10.1177/1087057111407763. [DOI] [PubMed] [Google Scholar]
- Bowden TA, Aricescu AR, Nettleship JE, Siebold C, Rahman-Huq N, Owens RJ, Stuart DI, Jones EY. Structural plasticity of eph receptor A4 facilitates cross-class ephrin signaling. Structure. 2009;17:1386–1397. doi: 10.1016/j.str.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Congreve M, Chessari G, Tisi D, Woodhead AJ. Recent developments in fragment-based drug discovery. J. Med. Chem. 2008;51:3661–3680. doi: 10.1021/jm8000373. [DOI] [PubMed] [Google Scholar]
- Cullen BR. Utility of the secreted placental alkaline phosphatase reporter enzyme. Methods Enzymol. 2000;326:159–164. doi: 10.1016/s0076-6879(00)26053-9. [DOI] [PubMed] [Google Scholar]
- Dalvit C. NMR methods in fragment screening: theory and a comparison with other biophysical techniques. Drug Discov. Today. 2009;14:1051–1057. doi: 10.1016/j.drudis.2009.07.013. [DOI] [PubMed] [Google Scholar]
- de Souza TL, Sanches D, Gonçalves RB, da RochaPita SS, Pascutti PG, Bianconi ML, de Almeida FC, Silva JL, de Oliveira AC. Conformational selection, dynamic restriction and the hydrophobic effect coupled to stabilization of the BIR3 domain of the human X-linked inhibitor of apoptosis protein by the tetrapeptide AVPI. Biophys. Chem. 2010;152:99–108. doi: 10.1016/j.bpc.2010.08.005. [DOI] [PubMed] [Google Scholar]
- Diamond SL. Methods for mapping protease specificity. Curr. Opin. Chem. Biol. 2007;11:46–51. doi: 10.1016/j.cbpa.2006.11.021. [DOI] [PubMed] [Google Scholar]
- Dooley CT, Houghten RA. The use of positional scanning synthetic peptide combinatorial libraries for the rapid determination of opioid receptor ligands. Life Sci. 1993;52:1509–1517. doi: 10.1016/0024-3205(93)90113-h. [DOI] [PubMed] [Google Scholar]
- Dooley CT, Ny P, Bidlack JM, Houghten RA. Selective ligands for the mu, delta, and kappa opioid receptors identified from a single mixture based tetrapeptide positional scanning combinatorial library. J. Biol. Chem. 1998;273:18848–18856. doi: 10.1074/jbc.273.30.18848. [DOI] [PubMed] [Google Scholar]
- Farmer BT, 2nd, Constantine KL, Goldfarb V, Friedrichs MS, Wittekind M, Yanchunas J, Jr, Robertson JG, Mueller L. Localizing the NADP+ binding site on the MurB enzyme by NMR. Nat. Struct. Biol. 1996;3:995–997. doi: 10.1038/nsb1296-995. [DOI] [PubMed] [Google Scholar]
- Feng BY, Shelat A, Doman TN, Guy RK, Shoichet BK. High-throughput assays for promiscuous inhibitors. Nat. Chem. Biol. 2005;1:146–148. doi: 10.1038/nchembio718. [DOI] [PubMed] [Google Scholar]
- Feng BY, Simeonov A, Jadhav A, Babaoglu K, Inglese J, Shoichet BK, Austin CP. A high-throughput screen for aggregation-based inhibition in a large compound library. J. Med. Chem. 2007;50:2385–2390. doi: 10.1021/jm061317y. [DOI] [PubMed] [Google Scholar]
- Fesik SW, Shi Y. Structural biology. Controlling the caspases. Science. 2001;294:1477–1478. doi: 10.1126/science.1062236. [DOI] [PubMed] [Google Scholar]
- Fischer M, Hubbard RE. Fragment-based ligand discovery. Mol. Interv. 2009;9:22–30. doi: 10.1124/mi.9.1.7. [DOI] [PubMed] [Google Scholar]
- Flanagan JG, Cheng HJ, Feldheim DA, Hattori M, Lu Q, Vanderhaeghen P. Alkaline phosphatase fusions of ligands or receptors as in situ probes for staining of cells, tissues, and embryos. Methods Enzymol. 2000;327:19–35. doi: 10.1016/s0076-6879(00)27264-9. [DOI] [PubMed] [Google Scholar]
- Flygare JA, Fairbrother WJ. Small-molecule pan-IAP antagonists: a patent review. Expert Opin. Ther. Pat. 2010;20:251–267. doi: 10.1517/13543770903567077. [DOI] [PubMed] [Google Scholar]
- Giannetti AM, Koch BD, Browner MF. Surface plasmon resonance based assay for the detection and characterization of promiscuous inhibitors. J. Med. Chem. 2008;51:574–580. doi: 10.1021/jm700952v. [DOI] [PubMed] [Google Scholar]
- Giorgio C, Hassan Mohamed I, Flammini L, Barocelli E, Incerti M, Lodola A, Tognolini M. Lithocholic acid is an Eph-ephrin ligand interfering with Eph-kinase activation. PLoS ONE. 2011;6:e18128. doi: 10.1371/journal.pone.0018128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajduk PJ, Greer J. A decade of fragment-based drug design: strategic advances and lessons learned. Nat. Rev. Drug Discov. 2007;6:211–219. doi: 10.1038/nrd2220. [DOI] [PubMed] [Google Scholar]
- Houghten RA, Pinilla C, Blondelle SE, Appel JR, Dooley CT, Cuervo JH. Generation and use of synthetic peptide combinatorial libraries for basic research and drug discovery. Nature. 1991;354:84–86. doi: 10.1038/354084a0. [DOI] [PubMed] [Google Scholar]
- Houghten RA, Pinilla C, Giulianotti MA, Appel JR, Dooley CT, Nefzi A, Ostresh JM, Yu Y, Maggiora GM, Medina-Franco JL, et al. Strategies for the use of mixture-based synthetic combinatorial libraries: scaffold ranking, direct testing in vivo, and enhanced deconvolution by computational methods. J. Comb. Chem. 2008;10:3–19. doi: 10.1021/cc7001205. [DOI] [PubMed] [Google Scholar]
- Hubbard RE. Fragment approaches in structure-based drug discovery. J. Synchrotron Radiat. 2008;15:227–230. doi: 10.1107/S090904950705666X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson DM, Sawyer WH. Fluorescence anisotropy applied to biomolecular interactions. Methods Enzymol. 1995;246:283–300. doi: 10.1016/0076-6879(95)46014-4. [DOI] [PubMed] [Google Scholar]
- Judkowski V, Bunying A, Ge F, Appel JR, Law K, Sharma A, Raja-Gabaglia C, Norori P, Santos RG, Giulianotti MA, et al. GM-CSF production allows the identification of immunoprevalent antigens recognized by human CD4+ T cells following smallpox vaccination. PLoS ONE. 2011;6:e24091. doi: 10.1371/journal.pone.0024091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koolpe M, Dail M, Pasquale EB. An ephrin mimetic peptide that selectively targets the EphA2 receptor. J. Biol. Chem. 2002;277:46974–46979. doi: 10.1074/jbc.M208495200. [DOI] [PubMed] [Google Scholar]
- Lamberto I, Qin H, Noberini R, Premkumar L, Bourgin C, Riedl SJ, Song J, Pasquale EB. Distinctive binding of three antagonistic peptides to the ephrin-binding pocket of the EphA4 receptor. Biochem. J. 2012;45:47–56. doi: 10.1042/BJ20120408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li Q, Xie C, Zhou H, Wang Y, Zhang N, Shao H, Chan SC, Peng X, Lin SC, Han J. Beta-actin is required for mitochondria clustering and ROS generation in TNF-induced, caspase-independent cell death. J. Cell Sci. 2004;117:4673–4680. doi: 10.1242/jcs.01339. [DOI] [PubMed] [Google Scholar]
- Lim MD, Craik CS. Using specificity to strategically target proteases. Bioorg. Med. Chem. 2009;17:1094–1100. doi: 10.1016/j.bmc.2008.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustgarten J, Dominguez AL, Pinilla C. Identification of cross-reactive peptides using combinatorial libraries circumvents tolerance against Her-2/neu-immunodominant epitope. J. Immunol. 2006;176:1796–1805. doi: 10.4049/jimmunol.176.3.1796. [DOI] [PubMed] [Google Scholar]
- Moore CD, Wu H, Bolaños B, Bergqvist S, Brooun A, Pauly T, Nowlin D. Structural and biophysical characterization of XIAP BIR3 G306E mutant: insights in protein dynamics and application for fragment-based drug design. Chem. Biol. Drug Des. 2009;74:212–223. doi: 10.1111/j.1747-0285.2009.00862.x. [DOI] [PubMed] [Google Scholar]
- Murai KK, Nguyen LN, Koolpe M, McLennan R, Krull CE, Pasquale EB. Targeting the EphA4 receptor in the nervous system with biologically active peptides. Mol. Cell. Neurosci. 2003;24:1000–1011. doi: 10.1016/j.mcn.2003.08.006. [DOI] [PubMed] [Google Scholar]
- Murray CW, Rees DC. The rise of fragment-based drug discovery. Nat. Chem. 2009;1:187–192. doi: 10.1038/nchem.217. [DOI] [PubMed] [Google Scholar]
- Murray CW, Blundell TL. Structural biology in fragment-based drug design. Curr. Opin. Struct. Biol. 2010;20:497–507. doi: 10.1016/j.sbi.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Nasir MS, Jolley ME. Fluorescence polarization: an analytical tool for immunoassay and drug discovery. Comb. Chem. High Throughput Screen. 1999;2:177–190. [PubMed] [Google Scholar]
- Noberini R, Koolpe M, Peddibhotla S, Dahl R, Su Y, Cosford ND, Roth GP, Pasquale EB. Small molecules can selectively inhibit ephrin binding to the EphA4 and EphA2 receptors. J. Biol. Chem. 2008;283:29461–29472. doi: 10.1074/jbc.M804103200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noberini R, De SK, Zhang Z, Wu B, Raveendra-Panickar D, Chen V, Vazquez J, Qin H, Song J, Cosford ND, et al. A disalicylic acid-furanyl derivative inhibits ephrin binding to a subset of Eph receptors. Chem. Biol. Drug Des. 2011a;78:667–678. doi: 10.1111/j.1747-0285.2011.01199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noberini R, Mitra S, Salvucci O, Valencia F, Duggineni S, Prigozhina N, Wei K, Tosato G, Huang Z, Pasquale EB. PEGylation potentiates the effectiveness of an antagonistic peptide that targets the EphB4 receptor with nanomolar affinity. PLoS ONE. 2011b;6:e28611. doi: 10.1371/journal.pone.0028611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquale EB. Eph receptor signalling casts a wide net on cell behaviour. Nat. Rev. Mol. Cell Biol. 2005;6:462–475. doi: 10.1038/nrm1662. [DOI] [PubMed] [Google Scholar]
- Pasquale EB. Eph-ephrin bidirectional signaling in physiology and disease. Cell. 2008;133:38–52. doi: 10.1016/j.cell.2008.03.011. [DOI] [PubMed] [Google Scholar]
- Pasquale EB. Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nat. Rev. Cancer. 2010;10:165–180. doi: 10.1038/nrc2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellecchia M. Solution nuclear magnetic resonance spectroscopy techniques for probing intermolecular interactions. Chem. Biol. 2005;12:961–971. doi: 10.1016/j.chembiol.2005.08.013. [DOI] [PubMed] [Google Scholar]
- Pellecchia M, Sem DS, Wüthrich K. NMR in drug discovery. Nat. Rev. Drug Discov. 2002;1:211–219. doi: 10.1038/nrd748. [DOI] [PubMed] [Google Scholar]
- Pellecchia M, Becattini B, Crowell KJ, Fattorusso R, Forino M, Fragai M, Jung D, Mustelin T, Tautz L. NMR-based techniques in the hit identification and optimisation processes. Expert Opin. Ther. Targets. 2004;8:597–611. doi: 10.1517/14728222.8.6.597. [DOI] [PubMed] [Google Scholar]
- Pellecchia M, Bertini I, Cowburn D, Dalvit C, Giralt E, Jahnke W, James TL, Homans SW, Kessler H, Luchinat C, et al. Perspectives on NMR in drug discovery: a technique comes of age. Nat. Rev. Drug Discov. 2008;7:738–745. doi: 10.1038/nrd2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinilla C, Appel JR, Blanc P, Houghten RA. Rapid identification of high affinity peptide ligands using positional scanning synthetic peptide combinatorial libraries. Biotechniques. 1992;13:901–905. [PubMed] [Google Scholar]
- Pinilla C, Appel JR, Houghten RA. Investigation of antigen-antibody interactions using a soluble, non-support-bound synthetic decapeptide library composed of four trillion (4 × 10(12) sequences. Biochem. J. 1994;301:847–853. doi: 10.1042/bj3010847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinilla C, Martin R, Gran B, Appel JR, Boggiano C, Wilson DB, Houghten RA. Exploring immunological specificity using synthetic peptide combinatorial libraries. Curr. Opin. Immunol. 1999;11:193–202. doi: 10.1016/s0952-7915(99)80033-8. [DOI] [PubMed] [Google Scholar]
- Qin H, Shi J, Noberini R, Pasquale EB, Song J. Crystal structure and NMR binding reveal that two small molecule antagonists target the high affinity ephrin-binding channel of the EphA4 receptor. J. Biol. Chem. 2008;283:29473–29484. doi: 10.1074/jbc.M804114200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Noberini R, Huan X, Shi J, Pasquale EB, Song J. Structural characterization of the EphA4-Ephrin-B2 complex reveals new features enabling Eph-ephrin binding promiscuity. J. Biol. Chem. 2010;285:644–654. doi: 10.1074/jbc.M109.064824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjit DK, Rideout MC, Nefzi A, Ostresh JM, Pinilla C, Segall AM. Small molecule functional analogs of peptides that inhibit lambda site-specific recombination and bind Holliday junctions. Bioorg. Med. Chem. Lett. 2010;20:4531–4534. doi: 10.1016/j.bmcl.2010.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy MM, Wilson R, Wilson J, Connell S, Gocke A, Hynan L, German D, Kodadek T. Identification of candidate IgG biomarkers for Alzheimer’s disease via combinatorial library screening. Cell. 2011;144:132–142. doi: 10.1016/j.cell.2010.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilley KJ, Giulianotti M, Dooley CT, Nefzi A, McLaughlin JP, Houghten RA. Identification of two novel, potent, low-liability antinociceptive compounds from the direct in vivo screening of a large mixture-based combinatorial library. AAPS J. 2010;12:318–329. doi: 10.1208/s12248-010-9191-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rideout MC, Boldt JL, Vahi-Ferguson G, Salamon P, Nefzi A, Ostresh JM, Giulianotti M, Pinilla C, Segall AM. Potent antimicrobial small molecules screened as inhibitors of tyrosine recombinases and Holliday junction-resolving enzymes. Mol. Divers. 2011;15:989–1005. doi: 10.1007/s11030-011-9333-2. [DOI] [PubMed] [Google Scholar]
- Sharma SK, Straub C, Zawel L. Development of Peptidomimetics Targeting IAPs. Int. J. Pept. Res. Ther. 2006;12:21–32. doi: 10.1007/s10989-005-9003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozaki EN, Chai J, Rigotti DJ, Riedl SJ, Li P, Srinivasula SM, Alnemri ES, Fairman R, Shi Y. Mechanism of XIAP-mediated inhibition of caspase-9. Mol. Cell. 2003;11:519–527. doi: 10.1016/s1097-2765(03)00054-6. [DOI] [PubMed] [Google Scholar]
- Shoichet BK. Interpreting steep dose-response curves in early inhibitor discovery. J. Med. Chem. 2006a;49:7274–7277. doi: 10.1021/jm061103g. [DOI] [PubMed] [Google Scholar]
- Shoichet BK. Screening in a spirit haunted world. Drug Discov. Today. 2006b;11:607–615. doi: 10.1016/j.drudis.2006.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuker SB, Hajduk PJ, Meadows RP, Fesik SW. Discovering high-affinity ligands for proteins: SAR by NMR. Science. 1996;274:1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- Smet C, Duckert JF, Wieruszeski JM, Landrieu I, Buée L, Lippens G, Déprez B. Control of protein-protein interactions: structure-based discovery of low molecular weight inhibitors of the interactions between Pin1 WW domain and phosphopeptides. J. Med. Chem. 2005;48:4815–4823. doi: 10.1021/jm0500119. [DOI] [PubMed] [Google Scholar]
- Sospedra M, Zhao Y, Giulianotti M, Simon R, Pinilla C, Martin R. Combining positional scanning peptide libraries, HLA-DR transfectants and bioinformatics to dissect the epitope spectrum of HLA class II cross-restricted CD4+ T cell clones. J. Immunol. Methods. 2010;353:93–101. doi: 10.1016/j.jim.2009.12.006. [DOI] [PubMed] [Google Scholar]
- Srinivasula SM, Hegde R, Saleh A, Datta P, Shiozaki E, Chai J, Lee RA, Robbins PD, Fernandes-Alnemri T, Shi Y, Alnemri ES. A conserved XIAP-interaction motif in caspase-9 and Smac/DIABLO regulates caspase activity and apoptosis. Nature. 2001;410:112–116. doi: 10.1038/35065125. [DOI] [PubMed] [Google Scholar]
- Stebbins JL, Zhang Z, Chen J, Wu B, Emdadi A, Williams ME, Cashman J, Pellecchia M. Nuclear magnetic resonance fragment-based identification of novel FKBP12 inhibitors. J. Med. Chem. 2007;50:6607–6617. doi: 10.1021/jm0707424. [DOI] [PubMed] [Google Scholar]
- Stewart ML, Fire E, Keating AE, Walensky LD. The MCL-1 BH3 helix is an exclusive MCL-1 inhibitor and apoptosis sensitizer. Nat. Chem. Biol. 2010;6:595–601. doi: 10.1038/nchembio.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Cuddy M, Samuel T, Welsh K, Schimmer A, Hanaii F, Houghten R, Pinilla C, Reed JC. Cellular, biochemical, and genetic analysis of mechanism of small molecule IAP inhibitors. J. Biol. Chem. 2004;279:48168–48176. doi: 10.1074/jbc.M405022200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.