Abstract
Microbes and viruses provoke immune responses because certain of their molecular determinants engage and activate dendritic cells (DC). However, evidence is growing for lymphocyte activation by tissue dysregulation. Thus, murine γδ T cells and NK cells can respond rapidly in vivo to Major Histocompatibility Complex (MHC) class I–related “stress-antigens” displayed by cells experiencing DNA damage and/or other physico-chemical stress. Such “lymphoid stress-surveillance” (LSS) can limit tumor formation, but may also promote immunopathology. MICA is a highly polymorphic human stress-antigen implicated in tumor-surveillance, inflammation, and transplant rejection. However, neither the generality of LSS in humans, nor a functional context for MICA polymorphism has been established. Here we show that MICA coding-sequence polymorphisms substantially affect RNA and protein expression. All donors tested showed LSS responses of γδ T and NK cells, but unexpectedly each was individually “tuned”. Hence, some responded optimally to high MICA expression, while others responded better to poorly-expressed MICA alleles, challenging the orthodoxy that higher stress-antigen levels promote greater responsiveness. The routine clinical monitoring of individual tuning should provide practical insight into individual variation in tumor immune-surveillance, transplant rejection and inflammation, and introduce new perspectives on immuno-evasion and immune-suppression in these scenarios.
Introduction
There is increasing acknowledgement of the overlap between the recognition of foreign moieties, as would characterize infection, and the recognition of self that has been dysregulated by non-microbial challenges such as irradiation or oxidative stress. Thus, MICA and other MHC-I-like ligands for the activating NKG2D receptor, expressed by NK cells, γδ T cells and some cytolytic CD8(+) αβ T cells, are upregulated by viruses, by some bacteria, and by sterile stresses such as cell transformation (1). Such lymphoid stress-surveillance (LSS) may complement the capacity of myeloid cells to recognize microbes via Toll-like receptors, provoking the rapid eradication of infected and transformed stromal cells (2, 3). In support of this, the experimental epidermis-specific upregulation of a single transgenic murine NKG2D ligand, Rae-1, was sufficient in vivo to rapidly activate local γδ T cells that conferred resistance to cutaneous chemical carcinogenesis (3). Further attesting to their importance, NKG2D ligands are frequently targeted by immune-evasion mechanisms of diverse viruses and tumors (4-6).
There is attractive clinical potential in harnessing a system that may protect against non-microbial damage; that is non-MHC-restricted; that may regulate tissue inflammation; and that may be efficacious against agents, such as HIV-1 that do not readily activate conventional antigen-presenting DC. Hence, this aspect of immunology is attracting intense translational interest (7). Nonetheless, the generality of LSS beyond the mouse has not been established, with several significant unresolved issues limiting the capacity to accept the NKG2D axis as a major mediator of LSS in humans. For example, in a highly reductionist system, NKG2D engagement alone provoked increased NK cell adherence to targets, but failed to promote cytotoxicity, instead requiring co-engagement of the activating receptors 2B4 and NKp46 (8, 9). There are likewise conflicting reports as to whether NKG2D expressed by human γδ cells functions as a primary activator or a co-stimulator for T cell receptor (TCR)-mediated responses (10-12). However, while such studies investigated hierarchies of receptor-ligand interactions for NK and γδ T cell activation, they neither established not refuted the potential for NKG2D-mediated LSS in humans.
Added to this MICA is highly polymorphic, with over 75 documented alleles. While this has been associated with the evolution of the host response to viral variation and immuno-evasion (13), there has been scant functional assessment of its impact on LSS: for example, are some alleles better than others at evoking NK or γδ T cell responses? Indeed, some reports argue that MICA*008, the most abundant Caucasian MICA allele, is non-functional by virtue of premature termination in the transmembrane anchor (14, 15). Likewise, are all individuals more or less responsive to their own versus allotypic MICA alleles, and are they comparably responsive to non-polymorphic NKG2D ligands, such as ULBP2? Collectively, these issues demand a re-examination of human cellular responses to NKG2D ligands.
Results
MICA polymorphisms determine RNA and protein expression levels
To examine the impact of MICA polymorphism on recognition by NKG2D(+) cells, we adopted a functional genomics system whereby MICA cDNAs encoded by different alleles were stably integrated (via an FRT recombinase) as single copies into the identical genomic site of Chinese Hamster Ovary (CHO) epithelial cells. By eliminating variation among transfectants caused by random integration and copy number, and by using CHO cells that do not express HLA-ligands for human inhibitory NK receptors, the biology of the different MICA alleles could be directly compared (Fig. S1; Fig. 1A). We examined the following alleles: MICA*008, the most common Caucasian allele, which carries a premature stop-codon in the transmembrane anchor that is predicted to limit its cell surface expression and to possibly abrogate function (14-16); MICA*027 which carries an identical ectodomain to MICA*008 (thus controlling for structural interactions with NKG2D) but with a full trans-membrane anchor; MICA*009, which couples a full-length transmembrane anchor to an ectodomain that differs from MICA*027 in four residues, and which has been genetically associated with Behçet’s Disease; and MICA*009v, which is our term for an unreported allele derived from a tumor cell line, MOU that differs from MICA*009 by two amino acids in the ectodomain (Fig. S2), and by just one amino acid from MICA*006. The cDNA fragments used to transfect cells contained negligible 5′ and 3′ untranslated sequences that were identical in each case.
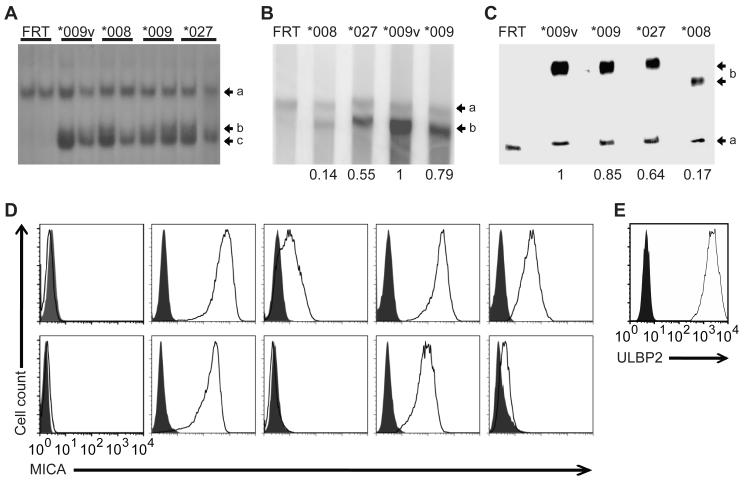
Figure 1. Generation and analysis of CHO transfectants.
(A) Southern Blot of duplicate BlpI-digested genomic DNA from CHO-FRT cells stably transfected with MICA*009, *009v, *008 and *027 cDNAs (see Fig. S1) probed with full-length MICA cDNA. Arrows: (a) a probe-homologous sequence in the CHO genome; (b) the 5′ flanking region of the integration site detected by those sequences in the MICA cDNA probe 5′ to the BlpI site; (c) the 4,745bp fragment diagnostic for MICA cDNA integration into the FRT site. (B) Northern blot of 10μg of RNA run on a formaldehyde gel, probed simultaenously for β-actin mRNA [(a) 1892 bp] and MICA mRNA [(b) 1201 bp]. Numbers reflect quantitation normalized to β-actin and relative to MICA*009v, determined by image scanning. (C) Western blot analysis of 10μg of cell lysates loaded on a 4-20% polyacrylamide gradient gel; transferred to nitorcellulose and probed simultaneously with anti-antibodies for β-actin and MICA, for which detected bands (a) (b) migrated at the sizes generally observed (β-actin, 47kD; MICA, 76kD; MICA*008, 71kD). Quantification was performed as above. (D) MICA expression of denoted stable CHO-FRT transfectants as assessed by flow cytometry on intact (upper panels) or permeabilized cells (lower panels). Shaded histograms correspond to isotype control stainings. (E) Cell surface expression of CHO cells stably transfected with ULBP2.
Upon integration of cDNA into the FRT-site, recombinants lose zeocin resistance but acquire hygromycin resistance. Stable transfectants derived in this way were assessed by Southern blot (Fig. 1A). A 4,745bp BlpI fragment (c) was diagnostic for the introduction of the MICA cDNA into the lacZ-zeo cassette present in the parental CHO cell line (see Fig. S1). The faint band of ~5kb (b) is a single flanking fragment confirming that the integration site is into the same site in each transfectant, while the absence of any other band [apart from a cross-reacting sequence present in the CHO genome, (a)] confirms that all transfectants carry a single-copy of the cDNA with no extraneous, random integrations. Analogous transfectants were generated using cDNA for the non-polymorphic NKG2D ligand, ULBP2.
The FRT system directs expression of each allele from identical transcriptional promoter and termination elements, such that variation in expression owes directly to intragenic coding-sequence variation. In this regard, RNA expression quantitated by Northern blot with a conserved probe showed a clear hierarchy in several independent transfectants, with MICA*009v RNA expressed at 1.3x the level of MICA*009 RNA, at 1.8x the level of MICA*027 RNA, and 7.1x the level of MICA*008 RNA (Fig. 1B). Thus, perhaps surprisingly, intragenic MICA polymorphisms substantively affect RNA expression levels, presumably via transcript elongation and/or stability (see Discussion). This quantitative variation in mRNA expression was observed in independent stable transfectants, and extrapolated almost precisely to steady-state protein expression, as quantitated by Western blot (Fig. 1C (b): note that MICA*008 protein migrates faster because of its truncated sequence). The hierarchy was conserved but exaggerated in cell surface expression (Fig. 1D): for example, most transfectants expressed ~20-fold less MICA*008 at the cell surface than MICA*009. Clearly this may reflect variable stability in the plasma membrane. Note, it did not reflect variable shedding or cleavage of MICA proteins into the medium (data not shown). Moreover, whereas poor surface expression of MICA*008 has been attributed to poor translocation to the plasma membrane, almost all MICA*008 protein was found at the cell surface rather than intracellularly (Fig 1D). By contrast, for highly expressed MICA*009 and *009v, ~10%-20% of protein was detected intracellularly at steady state. ULBP2 was very highly expressed at the cell surface of FRT-CHO transfectants (Fig. 1E). The range of surface expression of MICA displayed by the CHO-transfectants reflected that observed on primary human cells and on tumor cell lines (17-19) (Fig. S3).
MICA provokes specific killing
The capacity of different MICA alleles or ULBP to render stably-transfected CHO cells targets for killing was assessed by incubating effector cells for 12 hours with a 50:50 mixture of MICA or ULBP FRT-CHO transfectants labeled with a high concentration of the membrane intercalating dye, CFSE, and control FRT-vector-transfected CHO cells labeled with a low concentration of CFSE: after incubation, the ratio of CFSEhi to CFSElow cells was taken as a measure of specific target killing. The NKG2D(+) human cell line, NKL, showed dose-dependent specific cytolysis of all MICA and ULBP2 transfectants (Fig. 2A). Although MICA*009 targets were killed better than MICA*008 targets, the difference was marginal relative to the substantial differences in surface expression.
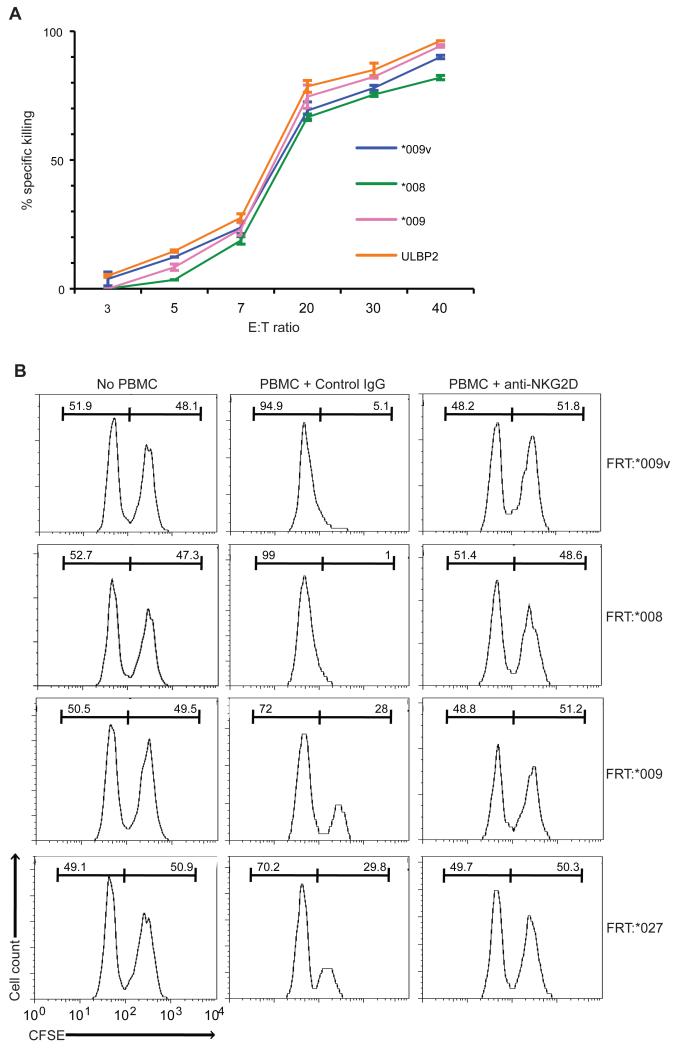
Figure 2. Efficient NKG2D-mediated killing of MICA and ULBP2 transfectants.
(A) Killing over a 12h period of CHO-MICA and -ULBP2 cells by NKL cells at indicated E:T ratios for 12 hours, assessed by CFSE. Specific killing determined by flow cytometry of the resultant control : transfectant ratio, relative to input. Data are means of a triplicate experiment +/− SD. (B) CFSE assay performed as described except that PBMC from a healthy donor were used (20:1 E:T ratio). Percentages of CFSE-stained control and transfected cells (listed above each histogram) were recorded before adding PBMC (left panels) and 12h after co-culture in the presence of isotype control (middle panels) or blocking mouse anti-human NKG2D antibody (10μg/mL, right panels).
CHO-FRT-MICA transfectants were also targets for primary peripheral blood mononuclear cells (PBMC) from individual donors, with all specific killing completely inhibited by a blocking anti-NKG2D antibody (Fig. 2B). However, while PBMC of more than forty donors examined in this and other ongoing studies all showed some capacity to kill CHO-FRT-MICA cells, there was unexpectedly high inter-individual variation in the hierarchy of specific cytolysis, that for any one donor was largely stable longitudinally, for at least four months (Fig. 3A,B). Thus, whereas Donor AD consistently targeted MICA*008(+) cells better than MICA*009(+) cells, this was not the case for donor MS. The extreme variation is evident from the analysis of 22 donors shown in Fig. 3B. Of note, ~30% of donors specifically targeted MICA*008(+) cells more efficiently than MICA*009v(+) cells, despite the former expressing the lowest and the latter expressing the highest levels of surface MICA among all transfectants. There was likewise no correlation of MICA*009 and MICA*027 expression levels with the capacity of MICA*009(+) and MICA*027(+) cells to be targeted.
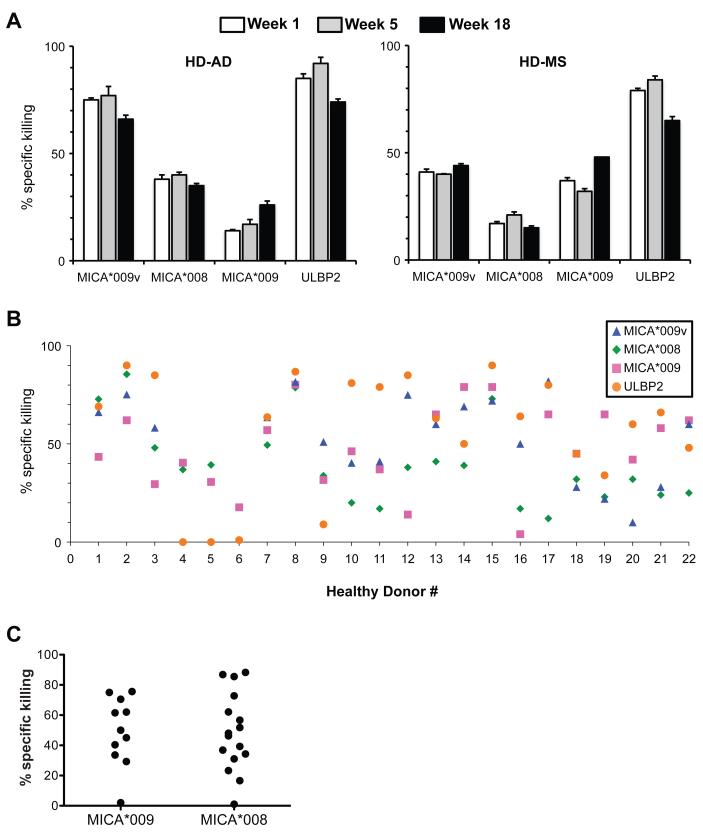
Figure 3. Stable donor-dependent variation in killing target cells.
(A) Killing of transfectants by PBMC of two healthy donors at three time points, as measured by CFSE assay, as in Fig. 2B. (B) Killing assay data from 22 healthy donors (not all donors were tested against all targets). (C) Killing of MICA*009 or MICA*008 targets was further analyzed for MICA*009 or MICA*008 heterozygous or homozygous donors. All data are means of triplicate experiments. SD was consistently <5%, but is not shown so as to preserve the clarity of the figures.
Additionally, PBMC from some individuals (e.g. 2, 8, 15) targeted transfectants very effectively, whereas PBMC from other donors (e.g. 4, 5, 6, 9) were less effective. This had no obvious relationship to hierarchies: thus, donor 8 killed all targets at 80-90% effectiveness; donor 2 killed MICA*008 targets at ~90% effectiveness; but donor 17, while killing MICA*009v(+) targets at ~85% effectiveness, showed only ~10% killing of MICA*008(+) targets. Nonetheless, it did appear that the least effective PBMC were particularly poor at targeting cells expressing ULBP2, a highly expressed non-polymorphic NKG2D ligand.
Killing hierarchies also did not obviously relate to donor ligand or receptor haplotype. For example, MICA*008(+) and MICA*009(+) individuals showed as broad a range of targeting MICA*008(+) cells and MICA*009(+) cells, respectively, as did the unstratified population (Fig. 3C), and there was considerable functional variation within groups of individuals with identical NKG2D haplotypes (Table S1). The lack of correlation with MICA haplotype was consistent with the inter-individual variation in targeting ULPB2 (see Discussion). Furthermore, hierarchies were not obviously explained by either the frequencies of NKG2D-expressing NK cells or γδ T cells or the level of NKG2D expression, for which there was very little variation (Table S2; Fig. S4; see below).
NK cells and γδ T cells mount Stress-Surveillance
A qualification of the CFSE assay is that outcomes may be affected by variation in the non-specific lysis of the FRT-control cells. We therefore assayed PBMC responses by a complementary assay measuring the direct response of PBMC to target cells via cell surface display of CD107a induced by cytolytic granule exocytosis (20). In this assay, measure can be made of the percentage of cells that degranulate (commonly from ~5% to ~20%); the degree to which they degranulate (by MFI); and the phenotype of cells that degranulate by co-staining for markers on CD107a(+) cells. The assay invariably shows a background response of PBMC even in the absence of target cells. Nonetheless, NK cells from each of two donors (A and E) clearly degranulated above background in response to NKG2D-ligand expressing CHO-FRT cells with targeting hierarchies consistent with the CFSE assay (Fig. 4A, B): that is, donor A targeted MICA*008(+) and MICA*027(+) cells poorly as these cells displayed very little specific CD107a; whereas donor E effectively targeted all transfectants, and each promoted comparable degranulation. This was the case for all other donors examined [e.g. Fig. S5]. Although the percentages of cells that degranulated may seem low, these cells were clearly efficacious in killing large numbers of cells, perhaps consistent with the “rapid-hit” model of killing provoked by NK receptors (21).
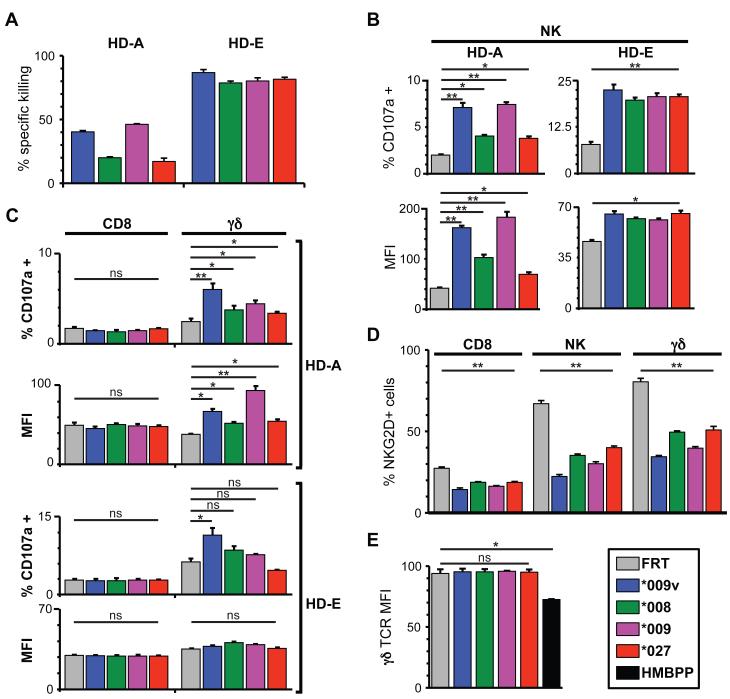
Figure 4. Identification of effector subsets that target transfectants.
(A) CFSE assay data for two donors (HD-A; HD-E), as described in Fig. 3. (B) CD107a data for the same two donors (HD-A; HD-E); target cells were incubated with PBMC (1:1) for 5h in the presence of a PE anti-CD107a antibody. Cells were then stained with APC-anti-CD3 to exclude T cells and FITC-anti-CD56 to identify NK cells and the percentage of activated cells (CD107a(+), top panel) and their mean fluorescence intensity (MFI, lower panel) measured by flow cytometry. (C) Cells from the assay performed in (B) were also stained with APC-anti-CD3 and FITC-anti-CD8 or APC-antiCD3 and FITC-anti-pan γδ TCR. (D) NKG2D downregulation assessed by staining PBMC with PE anti-NKG2D after 5h incubation with target cells and staining for specific subpopulations, as described in (B) and (C). (E) Surface TCR-staining of γδ T cells that had been incubated with the indicated transfectants or with HMBPP. All results are means of triplicate stimnulations +/− SD, representative of six donors. *: p<0.05, **: p<0.01, ns: not significant (Student’s t-test).
Importantly, every donor showed NK and γδ T cell degranulation in response to at least one NKG2D-ligand-expressing target. Commonly, a lower percentage of γδ T cells than NK cells degranulated, but MFIs were often comparable (Fig. 4C). By contrast, there was no example of NKG2D(+) CD8(+) αβ T cell degranulation (Fig. 4C), despite NKG2D down-regulation on the CD8(+) T cells providing evidence that target engagement had occurred (Fig. 4D). Conversely, responses of γδ T cells in the absence of overt TCR engagement were validated by the lack of TCR down-regulation, compared to a positive control of cells exposed to the Vγ9Vδ2 TCR agonist HMBPP (Fig. 4E). These data illustrate a key distinction between unconventional T cells and conventional adaptive T cells, with the former able to mount functional responses to NKG2D ligands, while the latter presumably require TCR engagement.
NKG2D-ligand responses show evidence of tuning
In seeking to explain inter-individual variation in functional responsiveness, we noted that for many donors the hierarchy of targeting was related to the level of MICA expression, albeit set at different optima. Thus, some donors (e.g. HD-MS; HD-17, Fig. 3) targeted cells in direct proportion to MICA expression levels (MICA*009v highest; MICA*008 lowest); rare donors (e.g. HD-5, Fig. 3B; GD011, Fig. 5C [below]) targeted cells inversely proportional to expression levels; while several donors (e.g. HD-14, Fig 3B) optimally targeted cells expressing intermediate levels of MICA, e.g. MICA*027, while targeting very high and very low expressers less well. This provoked the hypothesis that individual responses might be “tuned” to different “dose bandwidths” as schematized in Fig. S6 (see Discussion).
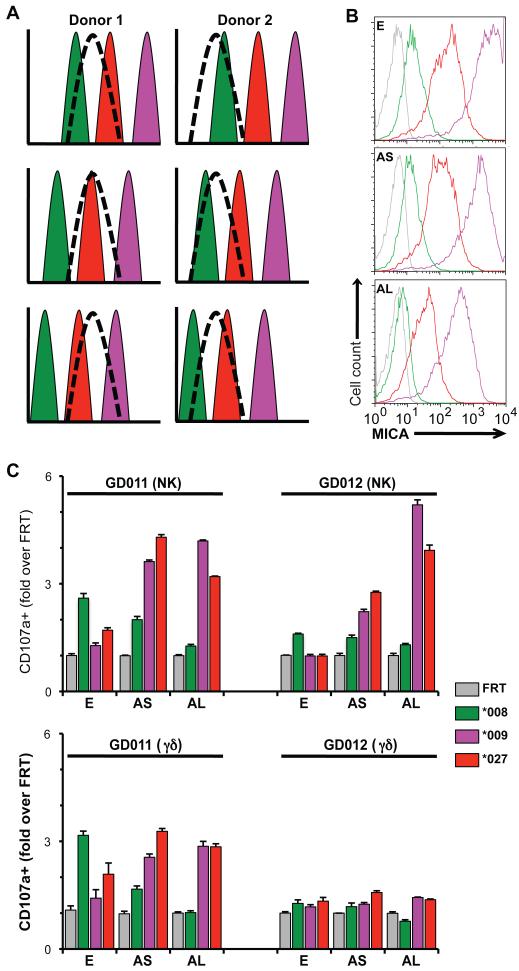
Figure 5. Contribution of tuning to the preferential recognition of MICA alleles.
(A) Model for a bell-shaped response of effector cells from 2 donors tuned to different ranges (dashed curves) of MICA expression levels, set against the levels of MICA expressed by CHO-MICA*008 (green), CHO-MICA*027 (red), and CHO-MICA*009 (purple) cells (top panels). As the expression of these allelic forms is gradually reduced (middle and bottom panels), the capacity to be targeted by donor 1 and donor 2 effector cells is increased for MICA*027 and MICA*009 cells while targeting of MICA*008 cells is decreased. (B) CHO cells were collected with PBS-10mM EDTA (E) or following a short (5min, AS) or long (20min, AL) treatment with Accutase and stained for cell surface MICA expression. Colors coded according to key box shown. (C) Target cells from panel B were utilized in a CD107a assay with NK (top panel) and γδ T cells (bottom panels) from two healthy donors (GD011 and GD012). Responses were normalized to the percentage of CD107a(+) cells in the presence of control vector-transfected FRT cells. Data are means of triplicate stimulations +/− SD.
The tuning hypothesis predicts bell-shaped response curves for many donors that will ignore cells expressing MICA levels outside the bandwidth. Thus, counter-intuitively, recognition may increase if MICA levels are reduced (Fig 5A). To test this, the density of MICA*008, MICA*009, and MICA*027 on FRT transfectants was gradually reduced by increasing exposure to accutase, which partially strips proteins from cell surfaces (Fig. 5B). By this means, poor NK and γδ T cell responses of donors GD011 and GD012 to MICA*009 cells were each increased as the expression of this molecule was decreased (Fig 5C). The same was true for MICA*027, although for GD011, the response peaked as surface expression was initially reduced, but declined again as more protein was stripped from the surface. Thus, there was an optimum level of surface MICA*027 expression to which donor GD011 responded: importantly, this was not the highest level of expression, and it differed from the optimum for donor GD012. Decreasing MICA*008 levels, which were low to begin with, invariably reduced responses (Fig. 5C), as if it were dropping to the left of the optimum bandwidth (Fig. 5A). Because for each donor the NKG2D-mediated response to some ligands increased while others simultaneously decreased, it was improbable that targeting efficiency was responding to changing surface expression of other unknown moieties, e.g. adhesion molecules. In sum, the data are most consistent with a model response of the kind shown in Fig. S5. Nonetheless, there is still some qualitative effect of allele polymorphism on targeting since, for example, MICA*009 on accutase-treated cells (Fig. 5B-AL) is expressed at levels similar to MICA*027 in untreated cells (Fig. 5B-E), but is targeted more efficiently (Fig. 5C). Such qualitative effects may reflect affinity for NKG2D; microdomain localization in the plasma membrane; or other MICA-dependent properties affecting the capacity to engage effector cells.
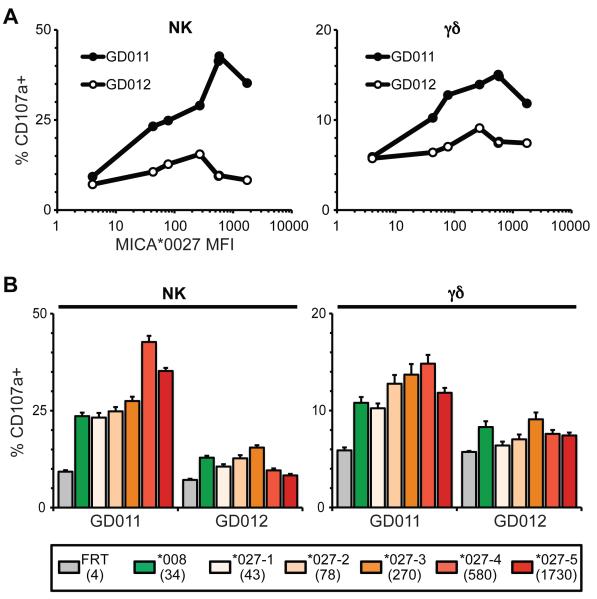
To further investigate the potential contribution of tuning, a set of CHO-transfectants was established in which a single MICA allele, MICA*027, was stably expressed at various levels, as determined by flow cytometry (Fig. 6A). Donors GD011 and GD012 both showed bell-shaped dose-response curves: that is, optimal responses were to cells expressing neither the highest nor lowest levels of MICA*027. The peak responses were closely equivalent in the NK and γβ T cell compartments, but the peak for each donor was different. At the same time, qualitative effects of MICA alleles were again apparent, in that for both GD011 and GD012, the decline in response to MICA*027 as surface expression decreased successively from 270 MFI units to 78 MFI units to 43 MFI units, contrasted with a higher responsiveness to MICA*008 cells with MFI of only 34 units (Fig. 6B). Because MICA*027 and MICA*008 have identical ectodomains, the qualitative differences may reflect different intra-membrane organization dictated by the respective trans-membrane domains, as in the report that MICA*008 preferentially localizes to exosomes (22).
Figure 6. Quantitative and Qualitative aspects of responses to CHO-MICA cells.
(A) MICA*027 was subcloned into the pcDNA3.1 vector to generate stable cloned cell lines with a spectrum of MICA expression. Such clones were assessed for their capacity to stimulate CD107a upregulation by NK cells (left panel) and γδ T cells (right panel) as a function of MICA expression levels (x-axis): cells from the same donors as in Fig. 5. SD is not shown to improve figure clarity but was consistently <5%. (B) Comparison of responses of NK cells (left panel) and γδ T cells (right panel) to the different MICA*027 clones as well as to the MICA*008 cell line. Numbers in parentheses indicate the MICA MFI measured by flow cytometry. Data are means of triplicate stimulations +/− SD.
Discussion
NKG2D-mediated recognition of “stress-ligands” such as MICA or members of the ULBP family is strongly implicated in infection, inflammatory and autoimmune disease, and tumor surveillance, and is a common target of immune-evasion. It may also be germane to transplant rejection (7). Thus, LSS has attracted much biological and translational interest. This study further justifies that interest by showing that γδ T and NK cells from all individuals tested could mount LSS, emphasizing the potential of tissue dysregulation and stress-antigens to initiate and/or sustain immune activity. Moreover, LSS can be very sensitive with individuals responding to low levels of MICA as presented by CHO-MICA*008(+) cells. Hence, MICA*008, the most common allele among Caucasians, is unlikely to be without function, as has been considered (14-16). LSS responses were not contributed by NKG2D(+) CD8(+) αβ T cells despite their engaging MICA(+) targets. This distinction between rapid, stress-antigen driven responses and slower clonal activation of the antigen-receptor dependent adaptive response is a key facet of LSS. The differential expression by unconventional versus conventional T cells of signaling regulators (23) may in part account for the qualitatively distinct activation requirements of these different lymphocyte subsets.
LSS showed very high inter-individual variation. Different killing efficiencies will reflect multiple factors, including polymorphisms in the cytoloytic machinery (24); and variable expression by PBMC of receptors (e.g. integrins) that may promote target engagement, versus those that may inhibit it. Additionally, independent studies have shown that NK cell responsiveness to NKG2D engagement is established by the types of activating and inhibitory receptor-ligand engagements that “educate” the cells during development (25). The multiplicity of such engagements leads to a continuum of responsiveness from low to high: the so-called “rheostat model”. Importantly, because the determinants for some such engagements are both variable and polymorphic, e.g. those between inhibitory receptors (expressed by NK and γδ T cells) and MHC Class I, the percentage of cells that will respond well to NKG2D engagement would be predicted to show individual variation, as demonstrated here. Hence, persons will differ in the efficacy of LSS-mediated immune activation. Indeed, were some donors’ PBMC to be co-incubated with only one or few MICA(+) targets, there might be no apparent targeting, potentially provoking false negative results that may account for some apparent conflicts in experimental outcomes.
While such considerations predict observed differences in killing efficiency, they do not predict the overt inter-individual variation in the hierarchy of allelic targeting, by both NK and γδ T cells. There may be qualitative contributions to this. For example, NKG2D of one individual may interact better with particular allelic forms of MICA based on affinity or on the context (e.g. membrane microdomain) in which that allelic form is presented (15, 22). However, because phenotypes do not segregate with NKG2D haplotypes, this cannot fully explain the data.
Instead, the preference of some individuals’ cells for lower levels of MICA expression, as typified by CHO- MICA*008(+) cells, is consistent with “Reversible Tuning”, an extension of the rheostat model that is based on independent studies of NK cells and T cells (25). This considers that cell responses are not to absolute levels of ligand, but to the change above background – the “delta value”. This will be set by the MICA haplotype, and by multiple factors regulating gene and protein expression that will vary among individuals. For example, three MICA*008 homozygotes may upregulate MICA to different degrees in response to environmental stress. As a result their cells, which will have been variably educated in the context of different levels and types of receptor-ligand engagement, will become dynamically tuned to low, medium, and high levels of MICA expression, respectively. This can explain this study’s findings that functional responses do not segregate directly with MICA haplotype, and that some individuals cannot even target cells expressing high levels of the non-polymorphic ligand, ULBP2.
The molecular basis of reversible tuning has yet to be clearly elucidated. Possibly, cells that respond to high MICA delta values rarely seen in a particular person die by neglect. Furthermore, excessive activating signals may evoke cell death or anergy, as in the detection of high levels of activation-induced RAP1-GTP in anergic T cells (26). The important predictions of reversible tuning are that individuals will mount distinct bell-shaped responses corresponding to different bandwidths of MICA expression, and that, counter-intuitively, reducing MICA expression may evoke higher responses. Both predictions were validated in this study. Tuning likely affects other functions of T and NK cells (e.g. cytokine secretion) that were not examined here. Moreover, it may be a common immunological process. For example, the cytotoxic response of macrophages to CD47 expression is governed by their prior exposure to CD47 during development (27).
Tuned NK and γδ T cell compartments would provide an individual with optimum responsiveness to the levels of MICA dysregulation that they most often encounter, while maintaining tolerance of unperturbed self. However, they offer new possibilities for immuno-evasion. For example, very high MICA levels are found on many human tumors. While some such tumors may have blocked immune detection by MICA shedding or cleavage, this study’s findings suggest that immuno-evasion may also occur when cells express NK or γδ T cell ligands outside an individual’s response bandwidth. This confounds the orthodoxy that NKG2D-mediated responses are simply proportional to the amount of ligand expression induced. Moreover, were MICA expression levels to change, for example because of epigenetic effects, an inappropriate LSS response could become a contributory cause of inflammation. Possibly this underpins putative linkages to inflammatory pathologies, such as that of MICA*009 to Behçet’s disease (28).
Inter-individual variation in LSS typifies processes that may go unnoticed in experimental model systems. In practical terms, LSS may be routinely and longitudinally monitored, affording insight into a subject’s changing responsiveness to the dynamics of MICA expression on tumors, grafts, or infected tissues. Importantly, seeming failures of immune surveillance may not be attributable to NK and/or γδ T cells, but to target tissues sitting outside the bandwidth for detection. Such a scenario may pertain to the report by Dranoff and colleagues of improved tumor immunotherapy in patients who generated their own antibodies to MICA following GM-CSF treatment (29). Such antibodies may be efficacious not just by absorbing shed MICA, but by reducing perceived MICA expression levels on tumor cells into alignment with the patients’ functional bandwidth for LSS. Conversely, the monitoring of MICA levels on a graft vis-à-vis a recipient’s LSS bandwidth may guide the decision as to whether immunosuppressants are appropriate to limit LSS.
Whereas polymorphic MHC molecules qualitatively vary the cargo they present to conventional T cells, our analysis of MICA coding-sequence variation has re-emphasised the centrality of expression levels as a key means by which NKG2D ligands are discerned by the immune system. The different expression levels dictated by different alleles may have been selected by the collective benefit of spanning a wide range of MICA expression levels induced by different viruses, microbes, and/or non-microbial stresses. The manner by which small coding-sequence variations affect RNA stability of RNA is under investigation. Moreover, these marked effects may be supplemented by 5′ and/or 3′ non-coding tracts that were not examined here.
Nonetheless, a full understanding of human LSS will also require the puzzling multiplicity of NKG2D ligands to be addressed. While MICA and, to a lesser degree, MICB are polymorphic, ULBPs are not, evoking αβTCR ligands among which peptide-presenting MHC molecules are highly polymorphic, while lipid-presenting CD1 molecules are not. While such distinctions are currently unexplained, we would note that tuning may both be affected by non-polymorphic NKG2D ligand expression and affect the responses to such ligands. This is particularly important given recent evidence that individual ULBPs evoke distinct immunoprotective responses not evoked by other ULBPs or MICA/MICB (30).
Materials and Methods
Antibodies and reagents
All antibodies were from Biolegend except for unlabeled mouse anti-human MICA, HRP-goat anti-mouse IgG and -rabbit anti-goat IgG (R&D systems), unlabeled goat anti-mouse beta actin (Abcam), and PE mouse anti-human CD107a (BD Biosciences). All reagents were from Sigma-Aldrich except for HMBPP (kindly provided by Dr. H. Jomaa, Justus-Liebig-Universität, Giessen, Germany).
Cell culture and PBMC isolation
All cell lines used were maintained in RPMI 1640 + 10% FCS (complete medium). NKL cell line (kindly provided by Pr. S. Khakoo, Imperial College London, London, UK) was maintained in complete medium with 100 U/ml IL-2. All cell culture reagents were from Invitrogen except for Accutase (PAA laboratories). Peripheral blood mononuclear cells (PBMC) were freshly isolated by Ficoll gradient, and were used immediately. Blood samples from healthy volunteers were collected after informed consent was obtained in agreement with the Declaration of Helsinki.
Cloning of MICA and ULBP2
Total RNA was isolated using Trizol (Invitrogen, UK) from C1R (MICA*009, Sigma-Aldrich, UK), MOU (MICA*009v, Sigma-Aldrich, UK), Int407 (MICA*008, ATCC), SWEIG007 (MICA*027, ATCC) and HaCat cells (ULBP2, ATCC) which were UV treated for 24 hours (Human keratinocyte cell line). cDNA was synthesized using SuperScript II Reverse Transcriptase (Invitrogen Life-Technologies, UK). Amplification of full length MICA and ULBP2 was performed by RT-PCR using the following primers: MICA Forward: 5′ CGGGGCCATGGGGCTGGGCCCGGT 3′ MICA Reverse: 5′ AGCCGCCTGGCTGTAGAGTCTAGG 3′ ULBP2 Forward: 5′ TACCGTCGTCGGCGGCGATG 3′ ULBP2 Reverse: 5′ TCAGATGCCAGGGAGGATGAA 3′ cDNAs were directly cloned after PCR into the pCR2.1 Topo vector using the TA cloning system (Invitrogen, UK) according to the manufacturer’s instructions. Sequences were checked and the cDNAs were then subcloned into the pcDNA5/FRT vector using the BstXI restriction site.
CHO cell transfection
CHO cells (2.106/mL) were electroporated in 4mm cuvettes in a Bio-Rad Gene Pulser II (Molecular BioProducts, UK) according to protocols optimized for Flp-In recombination (Invitrogen, UK). Electroporated cells were transferred to 6 well-plates containing 3mL of pre-warmed complete medium. Hygromycin (600μg/mL) was added 48h later.
Southern Blot
Genomic DNA was extracted from each cell line using phenol:chloroform; 20μg was digested with BlpI; the digests separated on a 1% agarose gel; blotted onto a Hybond N+ membrane (Amersham, UK); and probed with full length MICA cDNA (1158bp), 32P-labelled with the Rediprime II DNA labelling system (Amersham). The membrane was exposed to Kodak Biomax MS film (Sigma-Aldrich). Quantification was performed using ImageQuant system (GE Healthcare).
Northern blot
Northern blot analysis was performed by loading 10μg of RNA on a formaldehyde gel; transfer to a Hybond N+ membrane (Amersham, UK); and probing with MICA cDNA and full length beta actin (1892bp) 32P-labelled (as above). The membrane was exposed and quantified as above.
Western Blot
Cells were lysed in 1% NP40, 50mM Tris-HCl pH 7.2, 150mM NaCl, 10mM EDTA, with pan-protease inhibitors (Roche Diagnostics). After centrifugation (20000g, 10min, 4°C) supernatants were collected and protein concentration determined by micro BCA protein assay (Thermoscientific, UK). Equal amounts were separated on a 4-20% gradient polyacrylamide gel; transferred onto a nitrocellulose membrane; and proteins detected with mouse anti-human MICA monoclonal and goat polyclonal anti-mouse beta-actin antibodies, using secondary horseradish peroxidase (HRP)-conjugated anti-mouse IgG and HRP-conjugated anti-goat IgG, respectively. Antibody binding to membranes was visualized by ECL chemiluminescence (Amersham) and quantification was performed as above.
Flow cytometry
Phenotypes of lymphocyte sub-populations (NK and γδ) were determined by staining 200000 cells with the indicated monoclonal antibodies or isotype controls in FACS buffer (PBS 5% FCS). Samples were washed once in FACS buffer, re-suspended in 300μl FACS buffer, and analyzed on a FACScalibur flow cytometer (BD).
CFSE assay
Determination of specific killing of target cells was carried out using the CFSE method as previously described (31). Briefly, control cells (CHO-FRT) and target cells (CHO-MICA/ULBP2) were mixed at a 1:1 ratio after labeling with a low (1μM) or high (10μM) concentration of CFSE, respectively. Effector cells (PBMCs) were labeled with 2μM DDAO-SE and added to target cells and incubated for 12h at 37°C, 5%CO2. Specific lysis of transfectants was determined after measuring the resulting control:target cell ratios by flow cytometry on a FACScalibur (BD) and using the following calculation: % Specific lysis = 100 × (1-[(control ratio)/(experimental ratio)]) whereby the control and experimental ratios refer to the CFSElow: CFSEhi fractions in the absence or presence of effector cells, respectively.
CD107a assay
Activation of effector cells by measuring the cell-surface translocation of the CD107a marker was performed as described previously (20) except that Brefeldin A was omitted. Briefly, target cells and PBMC were mixed at a 1:1 ratio in the presence of the PE anti-CD107a antibody (1/30 final dilution) and incubated for 5h at 37°C, 5% CO2. Cells were then washed with FACS buffer and stained with combinations of APC anti-CD3 and FITC anti-CD56 (NK cells), APC anti-CD3 and FITC anti-CD8 (CD8 T cells), or APC-anti-CD3 and FITC anti-pan γδTCR (γδ T cells). Data were acquired on a FACScalibur (BD) and are means of triplicate cultures +/− SD.
MICA haplotyping
All healthy donors were MICA typed in the Clinical Transplantation Department, Guy’s Hospital, London, UK, using Labtype SSO luminex kits (One lambda, USA).
Supplementary Material
Acknowledgements
We thank Drs. S. Khakoo, H. Jomaa, and Y. Bryceson for kind gifts of reagents and advice, and all donors for their collaboration.
Funding: This work was supported by a Wellcome Trust Programme Grant (AH); the European Community’s Seventh Framework Programme FP7/2007-2013 under grant agreement #PIEF-GA-2009-255285 (PV); Cancer Research UK; the British Eye Research Fund and Fight for Sight (SS, GW, MS); together with infrastructure support from the NIHR Biomedical Research Centre, and the Medical Research Council Centre for Transplantation Biology.
Footnotes
This manuscript has been accepted for publication in Science Translational Medicine. This version has not undergone final editing. Please refer to the complete version of record at http://stm.sciencemag.org/content/3/113/113ra124. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
Competing interests: The authors declare no competing interests.
Supplementary materials and methods.
Figure S1: Schematic representation of the Flp-in system (Invitrogen).
Figure S2: Alignment of the protein sequences of the MICA alleles used in this study.
Figure S3: MICA expression levels in various contexts.
Figure S4: Representative NKG2D staining on NK cells from 2 donors.
Figure S5: Killing of CHO-MICA cells and respective activation of NKG2D(+) subsets from an additional donor.
Figure S6: A schematic of tuning.
Table S1: NKG2D genotyping from 8 donors used in this study.
Table S2: Phenotyping of NKG2D(+) γδ T and NK cells in PBMC from healthy donors.
Supplementary references.
References
- 1.Gleimer M, Parham P. Stress management: MHC class I and class I-like molecules as reporters of cellular stress. Immunity. 2003;19:469–477. doi: 10.1016/s1074-7613(03)00272-3. [DOI] [PubMed] [Google Scholar]
- 2.Hayday AC. Gammadelta T cells and the lymphoid stress-surveillance response. Immunity. 2009;31:184–196. doi: 10.1016/j.immuni.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Strid J, et al. Acute upregulation of an NKG2D ligand promotes rapid reorganization of a local immune compartment with pleiotropic effects on carcinogenesis. Nat Immunol. 2008;9:146–154. doi: 10.1038/ni1556. [DOI] [PubMed] [Google Scholar]
- 4.Fernandez-Messina L, et al. Differential mechanisms of shedding of the glycosylphosphatidylinositol (GPI)-anchored NKG2D ligands. J Biol Chem. 2010;285:8543–8551. doi: 10.1074/jbc.M109.045906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waldhauer I, et al. Tumor-associated MICA is shed by ADAM proteases. Cancer Res. 2008;68:6368–6376. doi: 10.1158/0008-5472.CAN-07-6768. [DOI] [PubMed] [Google Scholar]
- 6.Wilkinson GW, et al. Modulation of natural killer cells by human cytomegalovirus. J Clin Virol. 2008;41:206–212. doi: 10.1016/j.jcv.2007.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flegel WA. Will MICA glitter for recipients of kidney transplants? N Engl J Med. 2007;357:1337–1339. doi: 10.1056/NEJMe078165. [DOI] [PubMed] [Google Scholar]
- 8.Bryceson YT, Ljunggren HG, Long EO. Minimal requirement for induction of natural cytotoxicity and intersection of activation signals by inhibitory receptors. Blood. 2009;114:2657–2666. doi: 10.1182/blood-2009-01-201632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bryceson YT, March ME, Ljunggren HG, Long EO. Synergy among receptors on resting NK cells for the activation of natural cytotoxicity and cytokine secretion. Blood. 2006;107:159–166. doi: 10.1182/blood-2005-04-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Das H, et al. MICA engagement by human Vgamma2Vdelta2 T cells enhances their antigen-dependent effector function. Immunity. 2001;15:83–93. doi: 10.1016/s1074-7613(01)00168-6. [DOI] [PubMed] [Google Scholar]
- 11.Nedellec S, Sabourin C, Bonneville M, Scotet E. NKG2D costimulates human V gamma 9V delta 2 T cell antitumor cytotoxicity through protein kinase C theta-dependent modulation of early TCR-induced calcium and transduction signals. J Immunol. 2010;185:55–63. doi: 10.4049/jimmunol.1000373. [DOI] [PubMed] [Google Scholar]
- 12.Rincon-Orozco B, et al. Activation of V gamma 9V delta 2 T cells by NKG2D. J Immunol. 2005;175:2144–2151. doi: 10.4049/jimmunol.175.4.2144. [DOI] [PubMed] [Google Scholar]
- 13.Bahram S. MIC genes: from genetics to biology. Adv Immunol. 2000;76:1–60. doi: 10.1016/s0065-2776(01)76018-x. [DOI] [PubMed] [Google Scholar]
- 14.Eleme K, et al. Cell surface organization of stress-inducible proteins ULBP and MICA that stimulate human NK cells and T cells via NKG2D. J Exp Med. 2004;199:1005–1010. doi: 10.1084/jem.20032194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suemizu H, et al. A basolateral sorting motif in the MICA cytoplasmic tail. Proc Natl Acad Sci U S A. 2002;99:2971–2976. doi: 10.1073/pnas.052701099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fodil N, et al. Allelic repertoire of the human MHC class I MICA gene. Immunogenetics. 1996;44:351–357. doi: 10.1007/BF02602779. [DOI] [PubMed] [Google Scholar]
- 17.Borchers MT, et al. NKG2D ligands are expressed on stressed human airway epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2006;291:L222–231. doi: 10.1152/ajplung.00327.2005. [DOI] [PubMed] [Google Scholar]
- 18.Eagle RA, et al. Cellular expression, trafficking, and function of two isoforms of human ULBP5/RAET1G. PLoS One. 2009;4:e4503. doi: 10.1371/journal.pone.0004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molinero LL, et al. Activation-induced expression of MICA on T lymphocytes involves engagement of CD3 and CD28. J Leukoc Biol. 2002;71:791–797. [PubMed] [Google Scholar]
- 20.Betts MR, et al. Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. J Immunol Methods. 2003;281:65–78. doi: 10.1016/s0022-1759(03)00265-5. [DOI] [PubMed] [Google Scholar]
- 21.Deguine J, et al. Intravital imaging reveals distinct dynamics for natural killer and CD8(+) T cells during tumor regression. Immunity. 2010;33:632–644. doi: 10.1016/j.immuni.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 22.Ashiru O, et al. Natural killer cell cytotoxicity is suppressed by exposure to the human NKG2D ligand MICA*008 that is shed by tumor cells in exosomes. Cancer Res. 2010;70:481–489. doi: 10.1158/0008-5472.CAN-09-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pennington DJ, et al. The inter-relatedness and interdependence of mouse T cell receptor gammadelta+ and alphabeta+ cells. Nat Immunol. 2003;4:991–998. doi: 10.1038/ni979. [DOI] [PubMed] [Google Scholar]
- 24.de Saint Basile G, Menasche G, Fischer A. Molecular mechanisms of biogenesis and exocytosis of cytotoxic granules. Nat Rev Immunol. 2010;10:568–579. doi: 10.1038/nri2803. [DOI] [PubMed] [Google Scholar]
- 25.Brodin P, Karre K, Hoglund P. NK cell education: not an on-off switch but a tunable rheostat. Trends Immunol. 2009;30:143–149. doi: 10.1016/j.it.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 26.Boussiotis VA, et al. Maintenance of human T cell anergy: blocking of IL-2 gene transcription by activated Rap1. Science. 1997;278:124–128. doi: 10.1126/science.278.5335.124. [DOI] [PubMed] [Google Scholar]
- 27.Wang H, et al. Lack of CD47 on nonhematopoietic cells induces split macrophage tolerance to CD47null cells. Proc Natl Acad Sci U S A. 2007;104:13744–13749. doi: 10.1073/pnas.0702881104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wallace GR, et al. MIC-A allele profiles and HLA class I associations in Behcet’s disease. Immunogenetics. 1999;49:613–617. doi: 10.1007/s002510050656. [DOI] [PubMed] [Google Scholar]
- 29.Jinushi M, Hodi FS, Dranoff G. Therapy-induced antibodies to MHC class I chain-related protein A antagonize immune suppression and stimulate antitumor cytotoxicity. Proc Natl Acad Sci U S A. 2006;103:9190–9195. doi: 10.1073/pnas.0603503103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lanca T, et al. The MHC class Ib protein ULBP1 is a nonredundant determinant of leukemia/lymphoma susceptibility to gammadelta T-cell cytotoxicity. Blood. 2010;115:2407–2411. doi: 10.1182/blood-2009-08-237123. [DOI] [PubMed] [Google Scholar]
- 31.Oppenheim DE, et al. Sustained localized expression of ligand for the activating NKG2D receptor impairs natural cytotoxicity in vivo and reduces tumor immunosurveillance. Nat Immunol. 2005;6:928–937. doi: 10.1038/ni1239. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.