Abstract
Cytochrome P450 enzymes are able to oxidize substrates that are more inert than their own surrounding protein framework. Now, a quantitative understanding has emerged as to how the enzymes accomplish this remarkable feat.
How does nature break strong C–H bonds? By making stronger O–H bonds. This formalism is the central paradigm in our understanding of enzymatic hydroxylation of saturated hydrocarbons. These metal-catalysed reactions allow organisms to grow on alkanes as a carbon source. Also, C–H hydroxylations mediated by enzymes of the cytochrome P450 family are central to phase I drug metabolism and steroid hormone biosynthesis1,2. The catalytic haem iron of cytochrome P450 enzymes — able to oxidize notoriously inert C–H bonds — have long been known to bear an unusual cysteine thiolate3 (Fig. 1a, left). Why sulfur? This chemically counterintuitive feature has been an enigma: thiolate is a strong electron donor to iron, and is easily oxidized, thus making for an unlikely candidate to generate an enzyme-bound oxidant strong enough to break the 100 kcal mol−1 C–H bond of a typical hydrocarbon (Fig. 1a).
Figure 1.
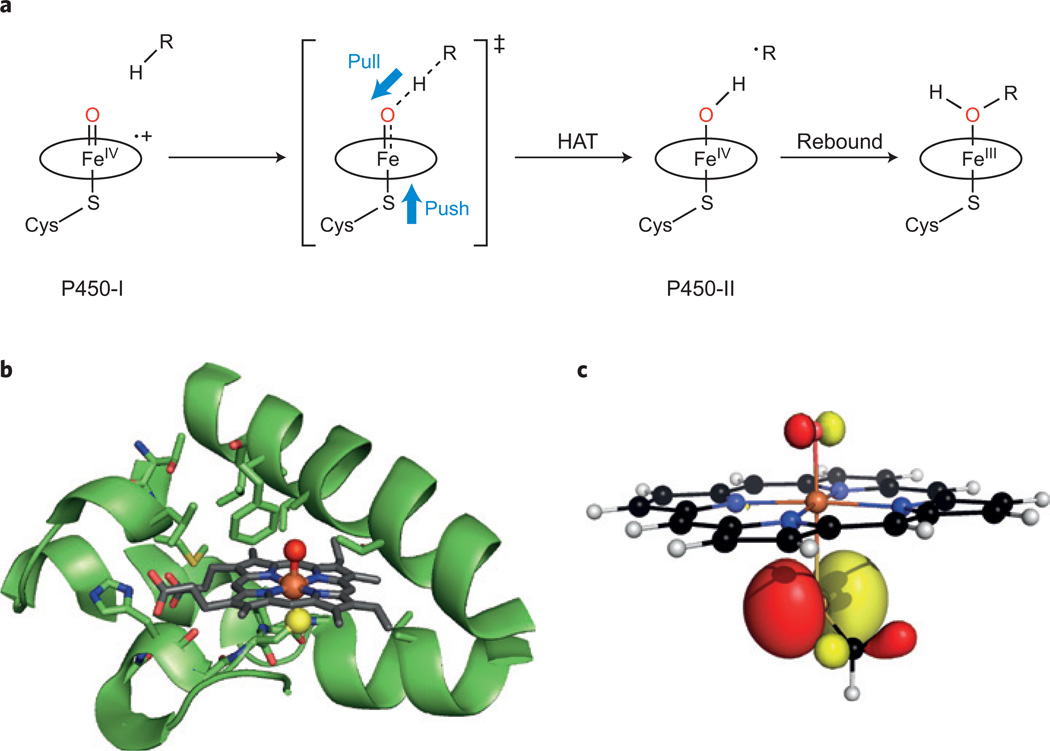
Oxidation of C–H bonds by P450 enzymes, emphasizing the role of the thiolate ligand. a, Oxygen rebound reaction scheme showing push–pull assistance of hydrogen atom transfer (HAT) mediated by the haem centre of a P450 enzyme, P+–(Cys–S)Fe(iv)=O (P450-I) to afford the Cys–S–Fe(iv)O–H species (P450-II) and a substrate radical, ∙R. The porphyrin macrocycle P coordinating the iron centre is represented by an oval for clarity purposes. b, Active-site structure and protein scaffold of CYP119 (PDB 1IO7). The haem iron(iii) (orange), cysteine thiolate sulfur (yellow) and axial water oxygen (red) are highlighted. c, DFT-calculated, doubly occupied HOMO orbital chosen to illustrate the sulfur electron push to the ferryl oxygen in a haem model H3C-S-Fe(iv)=O. We thank Dina Sharon for assistance with the DFT.
Finding an answer to this most intriguing question in oxidative catalysis has its origins in work by Green, Dawson and Gray on the haem–thiolate enzyme chloroperoxidase, with the recognition that its oxidized iron(iv) state compound II — reduced by one electron from the enzyme’s active intermediate (compound I) — was basic4. This meant that the Fe(iv) ferryl species was not the iron–oxo Cys–S–Fe(iv)=O, as had been presumed, but was instead a hydroxide, Cys–S–Fe(iv)–OH. The rationale for the basicity of the oxygen is that it derives from strong electron ‘push’ from the axial thiolate ligand. But how basic is the ferryl oxygen? And what does that have to do with the catalysis of C–H bond cleavage?
A more complete and quantitative answer has emerged from recent work5 by Michael Green and co-workers, as described in Science. The researchers used a bacterial cytochrome P450, CYP158, which harbours a large, solvent-accessible active site near a tyrosine residue. Several staple bioinorganic spectroscopic techniques were used in concert to probe the ferryl protonation, Cys–S–Fe(iv)=Oδ− → → Cys–S–Fe(iv)–OH.
Rapid-mixing pH-jump experiments allowed the exploration of compound II of CYP158 (CYP158-II) over a wide range of pH. Concurrent changes in the UV–visible absorption and 57Fe Mössbauer spectral signatures of the haem indicated that there were two forms of CYP158-II, interconnected by a remarkably basic pKa = 12. A similar exploration was also carried out with a second P450 enzyme, a CYP119-II variant (Fig. 1b). Although the two enzymes have different active-site environments and substrate affinities, they both showed similarly high values of pKa for their compound II, pointing to the generality of this feature for P450 enzymes’ thiolate–haem groups.
Both forms of CYP158-II displayed the ‘split’ Soret absorption band characteristic of thiolate binding in their UV–vis absorption spectra. Mössbauer spectra and the X-ray absorption edge data indicated that both forms were in the iron(iv) oxidation state. The protonated form, Cys–S–Fe(iv)–OH, showed an unusually large quadrupole splitting (2.0 mm s−1), similar to that of a well characterized dimethoxyiron(iv) porphyrin (MeO–Fe(iv)–OMe; ref. 2). This similarity indicates that the Fe=O bonding has decreased. Notably, there was a prominent pre-edge feature in the XAS data for the deprotonated Fe=O state that was much weaker in the protonated form at lower pH, consistent with a more centrosymmetric structure on protonation. Indeed, fitting of the X-ray absorption fine structure data showed that protonation of Cys–S–Fe(iv)=O lengthened the Fe–O bond and shortened the Fe–S bond.
Why is this basic Cys–S–Fe(iv)O–H pKa so informative of the mechanistic strategy for C–H bond cleavage by cytochrome P450? Because it is this FeO–H bond that is created during the scission of the substrate’s C–H bond, and it is this FeOH group that captures the resulting substrate radical in the product-forming oxygen rebound step (Fig. 1a). Finally, it is this FeO–H bond strength (D(OH)) that determines the thermodynamic driving force for hydrogen atom transfer from the substrate.
Strong C–H bonds have D(CH) ~100 kcal mol−1. Recently, another enzyme family — the haem–thiolate aromatic peroxygenases (APO) — has been discovered to rival P450 enzymes in their ability to cleave strong C–H bonds. Kinetic analysis of hydrogen atom transfer rates in our group6,7 has shown that the bond strength D(OH) for APO-II must also be ~100 kcal mol−1. A similar value for P450 enzymes is thus expected. Thermodynamic considerations allow us to dissect the bond strength D(OH) into two components8: the Fe(iv)O–H pKa, and the one-electron reduction potential of P+–Fe(iv)=O (where P is the porphyrin macrocycle) in APO-I or P450-I (Fig. 1a). Thus, knowing the pKa of FeO–H allows the calculation of the reduction potential of compound I, which for APO-I is ~1.4 V (ref. 6).
Returning to cytochrome P450-I, we can now understand how an electron ‘push’ effect from the trans-thiolate ligand can lead to greater ‘pull’ from the ferryl oxygen for C–H bond cleavage (Fig. 1a,c). The enzyme has engineered a clever strategy to increase the basicity of the ferryl oxygen of P450-II while sacrificing some of its redox potential. Insightful theoretical aspects of these effects have been discussed and reviewed by Shaik, Theil and colleagues9.
A higher pKa in Fe(iv)O–H (that is, a more basic Fe=O) allows for a lower redox potential for the P+–Fe(iv)=O oxidant that is responsible for C–H bond cleavage. This arrangement will maintain the facile hydrogen atom transfer pathway, which can be seen as a kind of inner-sphere process (Fig. 1a), while discriminating against longer range, outer-sphere electron-transfer processes from the protein matrix. Tyrosine and tryptophan residues both have redox potentials around 1 V, or higher if they are buried in the protein. This means that the rate of electron transfer from those sites will depend on the reduction potential of P450-I and the distance from those residues.
Green and co-workers estimate that this trade-off between redox potential and a very basic pKa allows the cysteine thiolate-ligated CYP158 to produce an oxidant that is wired for C–H bond cleavage, taking advantage of the basic ferryl species while suppressing long-range electron transfer rates — by as much as 10,000-fold compared with neutral histidine ligation that is typical of electron-transfer haem proteins such as peroxidases. Insights like this are important to understand the range of subtle strategies the enzyme has exploited to accomplish these very difficult C–H oxidations and to apply those strategies to the development of new catalysts.
References
- 1.Ortiz de Montellano PR. Chem. Rev. 2010;110:932–948. doi: 10.1021/cr9002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Groves JT. In: Cytochrome P450: Structure, Mechanism, and Biochemistry. 3rd edn. Ortiz de Montellano PR, editor. Klewer Academic/Plenum; 2005. pp. 1–44. [Google Scholar]
- 3.Poulos TL, Finzel BC, Howard A. J. J. Mol. Biol. 1987;195:687–700. doi: 10.1016/0022-2836(87)90190-2. [DOI] [PubMed] [Google Scholar]
- 4.Green MT, Dawson JH, Gray H. B. Science. 2004;304:1653–1656. doi: 10.1126/science.1096897. [DOI] [PubMed] [Google Scholar]
- 5.Yosca TH, et al. Science. 2013;342:825–829. doi: 10.1126/science.1244373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, Peter S, Ullrich R, Hofrichter M, Groves J. T. Angew. Chem. Int. Ed. 2013;52:9238–9241. doi: 10.1002/anie.201302137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, Peter S, Kinne M, Hofrichter M, Groves J. T. J. Am. Chem. Soc. 2012;134:12897–12900. doi: 10.1021/ja3049223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warren JJ, Tronic TA, Mayer J. M. Chem. Rev. 2010;110:6961–7001. doi: 10.1021/cr100085k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shaik S, et al. Chem. Rev. 2010;110:949–1017. doi: 10.1021/cr900121s. [DOI] [PubMed] [Google Scholar]