Abstract
The SaPIs and their relatives are a family of genomic islands that exploit helper phages for high frequency horizontal transfer. One of the mechanisms used by SaPIs to accomplish this molecular piracy is the redirection of the helper phage DNA packaging machinery. SaPIs encode a small terminase subunit that can be substituted for that of the phage. In this study we have determined the initial packaging cleavage sites for helper phage 80α which uses the phage-encoded small terminase subunit, and for SaPI1, which uses the SaPI-encoded small terminase subunit. We have identified a 19 nt SaPI1 sequence that is necessary and sufficient to allow high frequency 80α transduction of a plasmid by a terminase carrying the SaPI1-encoded small subunit. We also show that the hybrid enzyme with the SaPI1 small terminase subunit is capable of generalized transduction.
Keywords: Pathogenicity island, SaPI, Terminase, DNA Packaging, Helper phage, Pac site
1. Introduction
SaPIs are a family of mobile Staphylococcus aureus pathogenicity islands that represent a large group of elements, widely distributed among Gram-positive bacteria, which collectively are designated as phage-related chromosomal islands. SaPI mobility occurs via an unusual high efficiency transduction mechanism that involves specific exploitation of helper bacteriophages (Christie and Dokland, 2012; Novick et al., 2010). The SaPI excision-replication-packaging cycle is induced either by phage infection or by induction of a helper prophage in a SaPI-containing strain. SaPI induction involves derepression by specific phage-encoded antirepressors (Tormo-Más et al., 2010), which leads to expression of SaPI excision and replication functions (Mir-Sanchis et al., 2012; Ubeda et al., 2007, 2012). SaPI DNA is then encapsidated in virions comprised of phage-encoded structural proteins (Tallent et al., 2007; Tormo et al., 2008). SaPI1 hijacks the phage capsid assembly process to direct the formation of smaller capsids that are too small to accommodate complete helper phage genomes (Ruzin et al., 2001). This capsid size redirection involves two SaPI1-encoded proteins that are associated with procapsids and form an alternative internal scaffold (Damle et al., 2012; Dearborn et al., 2011; Poliakov et al., 2008). Finally, SaPIs manipulate the DNA packaging specificity of the helper phage. SaPIs encode their own small subunit of terminase (TerS) which redirects packaging specificity to SaPI DNA. The phage-encoded small terminase subunit is completely dispensable for SaPI packaging (Ubeda et al. 2009). The SaPI TerS and helper phage-encoded large terminase subunit (TerL) are believed to form a hybrid terminase complex that recognizes a specific packaging initiation signal (pac) on the SaPI genome. Some SaPIs further interfere with helper phage packaging by directly inhibiting the phage small terminase subunit (Ram et al., 2012).
The virion DNAs of SaPI1 (Ruzin et al., 2001) and the phages that mobilize it - 80, 80α and ϕ11 (Bachi, 1980; Lofdahl et al., 1981; Stewart et al., 1985) - are all partially circularly permuted. This is characteristic of phages that package their DNA by a headful mechanism in which there is an initial cleavage at a specific pac site sequence on a linear concatemer, followed by processive packaging of a limited number of slightly larger than unit length genome fragments into virus particles. The specificity for pac site recognition generally resides in the small subunit of the terminase complex, while the large subunit has ATP-binding, prohead binding, and DNA cleavage activities (reviewed in Feiss and Rao, 2012). Since high frequency SaPI1 transduction is dependent upon redirection of packaging specificity by a terminase complex carrying a SaPI1-encoded small subunit (Ubeda et al., 2009), we predicted that the helper phages and SaPIs each contain unique pac site sequences that are specifically recognized by terminase complexes containing their cognate small subunits. In this study we have localized the sites of initial cleavage in SaPI1 and helper phage 80α. The critical determinants for SaPI1-specific packaging were further localized by deletion analysis to a small region upstream of the promoter for SaPI1 operon 1. This is strikingly different from the pac cleavage site used by the helper phage terminase, which maps to within the small terminase gene itself. The SaPI1 pac site sequence is necessary and sufficient for high frequency transduction that depends upon SaPI1 TerS.
2. Materials and Methods
2.1 Bacterial strains and growth conditions
With the exception of the clinical isolate carrying wild-type SaPI1, S. aureus strains used in this study are all derivatives of the restriction-defective S. aureus strain RN4220 (Kreiswirth et al., 1983) and all strains are listed in Table S1. Routine growth of S. aureus strains was at 32°C on tryptic soy agar. E. coli DH5α™ (Invitrogen) and Stellar™ (Clontech) cells were used as the intermediate bacterial hosts for plasmid construction. All E. coli strains were cultured in LB medium either in liquid with shaking (200 rpm) or on agar plates at 37ºC. Whenever required, antibiotics were added to the media as follows: 10 μg/ml of erythromycin for S. aureus (plasmid selection), 5 μg/ml of tetracycline for SaPI1, and 100 μg/ml of ampicillin for E. coli. Transduction and transformation of S. aureus were performed as described previously (Novick, 1991). For transduction analysis, plasmids were introduced by electroporation into S. aureus RN4220 and a derivative lysogenic for 80α ΔterS (ST24). Cells were infected with 80α or the prophage was induced by treatment with 2μg/ml mitomycin C. The resulting lysates were titered on RN4220 for plaque-forming units and for erythromycin resistant colonies. Transduction of SaPI1 tst::tetM and SaPI1 deletion mutants was performed after prophage induction by UV, with selection for tetracycline resistant colonies.
2.2 DNA methods
General DNA manipulations were performed using standard procedures (Sambrook and Russell, 2006). Enzymes were used according to manufacturer’s instructions. Qiagen kits were used for the isolation of plasmid DNA, purification of PCR products and digested fragments, and extraction of DNA fragments from agarose gels. Oligonucleotide primers used for PCR amplification are listed in Table S2, and were obtained from Integrated DNA Technologies Inc. PCR amplification for subcloning was performed using high fidelity Phusion polymerase (NEB), and all cloned fragments were verified by DNA sequencing (UMBC Biopolymer Laboratory, Baltimore MD or MWG Biotech). The SaPI1 deletions used in the experiment shown in Table 1 were introduced into the SaPI1-containing 80α lysogen RN10628 by allelic exchange using derivatives of pMAD (Arnaud et al., 2004), as described previously (Poliakov et al., 2008).
Table 1.
Effect of SaPI1 deletions on SaPI1 transduction.
|
|
||
|---|---|---|
| Transduction Titera | ||
|
| ||
| SaPI genotypeb | 80α | 80α ΔterS |
| SaPI1 | 3.7 × 107 | 2.3 × 107 |
| SaPI1 Δpac | 4.4 × 105 | 1.4 × 105 |
| SaPI1 ΔterS | 1.2 × 105 | NDc |
Lysates were titered on RN4220 following UV induction of the 80α prophage. Results shown in this table were reproducible.
Number of TetR transductants per ml of lysate
All SaPI1 strains carry tst::tetM
Not detectable. No colonies were detected using 0.1 ml of undiluted lysate.
2.3 Plasmid construction
Plasmids used in this study are listed in Table S3. Inserts were generated by amplification with specific primers and digestion with appropriate restriction enzymes or were generated by annealing complementary oligonucleotides that included appropriate overhangs for subsequent ligation. The digested (or annealed) products were purified from agarose gels and ligated with the correspondingly digested vectors. Alternatively, PCR fragments were incorporated into the vector using a Gibson Assembly reaction (Gibson et al., 2009). In that case, fragments and linearized vector DNA containing 15 bp overlaps were assembled using the In-Fusion Cloning Kit (Clontech).
2.4 Preparation of virion DNA
Large scale lysates of 80α grown on RN4220 and SaPI1-containing strain RN4282 were prepared as described previously (Novick, 1991). Particles were purified by PEG precipitation followed by centrifugation in a CsCl step gradient (1.3 – 1.6). DNA-containing virions were collected from the 1.4–1.5 interface, disrupted by treatment with formamide and EDTA (Sambrook and Russell, 2006) and the DNA was precipitated with ethanol, redissolved in 10 mM Tris pH 8, 1 mM EDTA, and extracted with phenol. Following a second ethanol precipitation, the virion DNA was again redissolved in TE for subsequent restriction analysis and pac cleavage site mapping.
2.5 Cleavage site mapping
The location of the first headful 5′ cut site was first estimated from the lengths of the submolar fragments generated from restriction of virion DNA. A pair of nested primers, annealing downstream of and directed towards the 5′ end, were designed for use with the Universal GenomeWalker™ kit (Clontech Laboratories, Inc.). Purified genomic DNA was ligated with GenomeWalker adapters which then served as a template for amplification with adapter primer AP1 (provided with the kit) and either 80αPAC1A (for 80α) or SaPAC1a (for SaPI1). The unpurified products from this PCR reaction were then used as a template for amplification with nested primers AP2 and either 80αPAC1B or SaPAC1b. The 80α amplicons were gel purified and ligated with the TOPO-TA cloning vector pCR® 2.1 (InVitrogen) for sequence analysis of individual clones, and also subjected to direct sequencing using primer 80αPAC1B. The final SaPI1 products from the second PCR reaction were used directly in the TOPO cloning reaction for subsequent sequence analysis of the linker-virion DNA junctions.
3. Results
3.1 Mapping the phage 80α packaging cleavage site
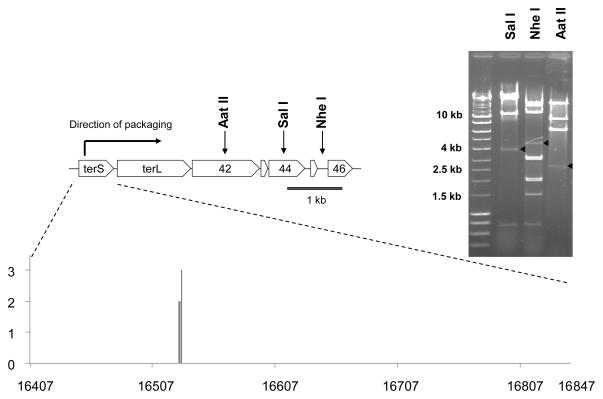
Early restriction mapping of 80α revealed a circularly permuted genome as well as the presence of minor submolar bands (Stewart et al., 1985). This is characteristic of phages that utilize a pac site for recognition and subsequent cleavage of DNA followed by processive headful packaging from a linear concatemer. The submolar bands correspond to those fragments that have one end generated by the packaging initiation cut. The availability of the 80α genome sequence (Christie et al., 2010) permitted us to revisit this strategy and led to the identification of submolar fragments consistent with an initial cleavage in or near the gene encoding the small terminase subunit (Fig 1). In order to locate precisely the site of this cleavage, we ligated a GenomeWalker adapter onto the 5′ ends of linear 80α DNA isolated from virions and then used nested PCR with adapter primers and phage-specific primers to amplify the adapter-phage junction fragment. The adapters will ligate onto all ends generated by the initial and subsequent cleavages. However, use of a phage-specific primer that anneals near the cleavage site that was mapped by restriction analysis allows preferential amplification from those adapters attached to the ends generated by the initial cleavage. DNA sequence analysis of the pooled amplicons, using primer 80αPAC2, revealed a sequence matching that of the terS gene until genome position 16529, where the signal became degenerate. This was indicative of at least some heterogeneity at the phage-adapter junction. The amplified junction fragments were then introduced into the TOPO vector pCR® 2.1 and the sequence of the adapter-phage junction was determined for seven independent clones. The junction mapped to four adjacent nucleotides (16527–16530) within the 80α terS coding sequence (Fig 1), revealing a highly specific initial cleavage event.
Figure 1. Mapping the 80α packaging cleavage site.
At the top is a region of the 80α map showing the start of the morphogenetic operon and the locations of the restriction sites corresponding to the submolar fragments identified by the arrows in the gel to the right. We previously confirmed the direction of packaging suggested by the restriction analysis (Ubeda et al., 2009). Below is an expanded map of the terS coding region, illustrating the results from the Genomewalker™ experiment. The x-axis represents the terS gene (genome coordinates 16407 to 16847; Genbank accession number DQ517338). The ends determined by the adaptor-phage junctions of seven independent clones mapped to three neighboring nucleotides in terS, indicative of cleavage on the 3′ side of bases 16526, 16527 and 16529; the height of the line corresponds to the number of sequences obtained at each site (2 or 3).
3.1 Mapping the SaPI1 packaging cleavage site
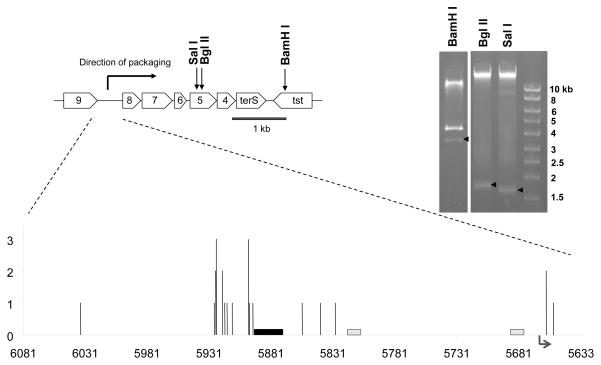
The same approach used for 80α was used to determine the initial cleavage site on SaPI1 virion DNA. Earlier restriction analysis established that SaPI1 DNA was also circularly permuted, and provided an initial rough estimate of the quadrant of the genome containing the apparent molecule ends (Ruzin et al., 2001). We repeated this analysis, focusing on enzymes that cut SaPI1 only once or twice. As shown in Figure 2, this restriction analysis localized the site of initial cleavage to a region upstream of the operon in which the SaPI1 small terminase subunit is encoded, and far from the terminase gene itself. GenomeWalker adapters were ligated onto the ends of wild type SaPI1 virion DNA, and nested primers within the ORF8 coding sequence were used to amplify the adapter-SaPI1 junction. Initial sequence analysis of a small number of independent clones indicated that these ends were much more heterogeneous than what we had observed for 80α, so additional cloned fragments were sequenced. Twenty-three ends were scattered across 401 nucleotides in the intergenic region between ORF9 and ORF8. This region also contains an SOS-inducible promoter for operon 1 (ORF8-terS), which encodes functions involved in SaPI1 encapsidation including the capsid size redirection proteins and the SaPI1 small terminase subunit (Ubeda et al., 2007; 2009; Poliakov et al., 2008; Dearborn et al., 2011; Damle et al., 2012). The adaptor junction sequences indicate that most of the cleavage occurs within a region of 35 nucleotides just upstream of the minimal pac site mapped in this study; 16/23 or 70% of the ends fell in this region (Fig 2), upstream of the SOS-inducible operon 1 promoter and the LexA binding site consensus sequences. The difference in initial cleavage of 80α DNA and SaPI1 DNA is consistent with the utilization of terminase complexes carrying different TerS subunits, and suggests fundamental differences in the way initial cleavage is directed.
Figure 2. Mapping the SaPI1 packaging cleavage site.
At the top is a region of the SaPI1 map showing operon 1 and the upstream intergenic region, with the locations of the restriction sites corresponding to the submolar fragments identified by the arrows in the gel to the right. Bgl II and Sal I each have a unique cleavage site in the SaPI1 genome while BamHI cuts once more outside of the region shown. Below is an expanded view of the intergenic region between ORF9 and ORF8, summarizing the Genomewalker™ results. The x-axis represents the intergenic region from 6081 to 5633 (Genebank accession number U93688; numbering is backwards from the conventional SaPI1 map orientation). The ends determined by adapter-phage junctions of 23 sequenced clones are shown. The bars indicate the location and their height reflects the relative number of ends mapped to each cleavage site. The black box indicates the size and location of the pac site identified in this study. Grey boxes indicate the two LexA binding sites in the operon 1 promoter region, and the arrow indicates the start and direction of operon 1 transcription.
3.2 Sequence determinants for SaPI1 – specific packaging
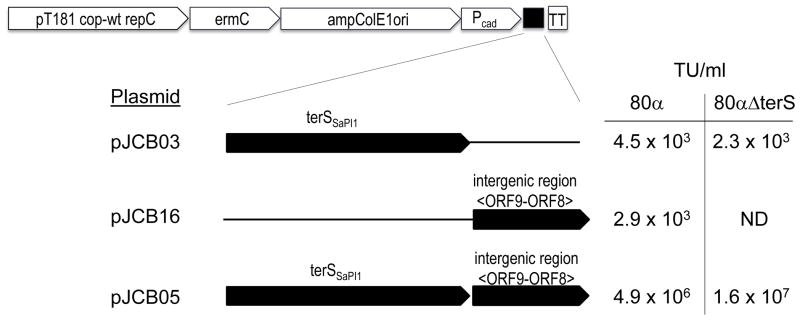
To characterize further the requirements for SaPI1-specific packaging, we developed a plasmid assay for high frequency SaPI1 transduction. S. aureus phages have been reported to transduce plasmids as linear concatemers of plasmid DNA (Novick et al., 1986). This same study reported that plasmid-initiated replication is required for their generation but additive interplasmid recombination is not. Concatemers are probably generated by the interaction of one or more phage functions with replicating plasmid DNA. The effect of the SaPI1 small terminase and the putative SaPI1 pac determinants were assayed using the expression plasmid pCN51 (Charpentier et al., 2004). We introduced the 449 bp ORF 8 – ORF 9 intergenic region, which contains the SaPI1 cleavage site, with and without the SaPI1 terS gene. Expression of the cloned terS gene was driven by the basal activity of the Pcad promoter in pCN51; cadmium induction did not appreciably increase plasmid transduction frequency (data not shown). The resulting plasmids were introduced into SaPI-negative S. aureus strain RN4220 and infected with helper phage 80α. As shown in Figure 3, the plasmids carrying either terS alone or the intergenic region alone showed characteristic levels of generalized transduction by 80α while the plasmid carrying both terS and the intergenic region was transduced at a frequency three orders of magnitude higher. Data are presented as transducing units per ml of culture, rather than the more traditional transducing units per plaque forming unit (pfu), to allow direct comparison with the results using 80α ΔterS, since this mutant is not capable of plaque formation. However, wild type phage titers in these experiments were all approximately 1010/ml, yielding transducing titers in the 10−7 per pfu range which is consistent with earlier reported frequencies for generalized plasmid transduction (Novick et al., 1986). Deletion of the 80α terS gene did not affect transduction frequency if SaPI1 terS was present on the plasmid. This demonstrates that only SaPI1 terS is required for high frequency transduction using the SaPI1 packaging site present in the intergenic region. In the absence of the intergenic region, the plasmid expressing SaPI1 terS was transduced at similar frequencies by 80α wt and 80α ΔterS, indicating that the hybrid terminase carrying the SaPI1 small subunit is also capable of generalized transduction. As expected, no plasmid transduction was observed in the absence of both small terminase subunits.
Figure 3. Transduction analysis of plasmids carrying the SaPI1 packaging determinants.
Derivatives of plasmid pCN51 carrying the small terminase gene from SaPI1 and/or the SaPI1 intergenic region between ORF 9 and 8, as indicated, were introduced into S. aureus strain RN4220 and analyzed for transduction after infection with 80α or introduced into strain ST24 and assayed after induction of the 80α ΔterS prophage. Representative data are shown; assays were performed in triplicate and were reproducible. The transducing titer (TU) is expressed as the number of TetR transductants per ml of lysate rather than the more conventional ratio of the transducing titer to phage titer, since the 80α ΔterS mutant does not produce plaques. ND indicates no plaques were detected in 0.1 ml of undiluted lysate.
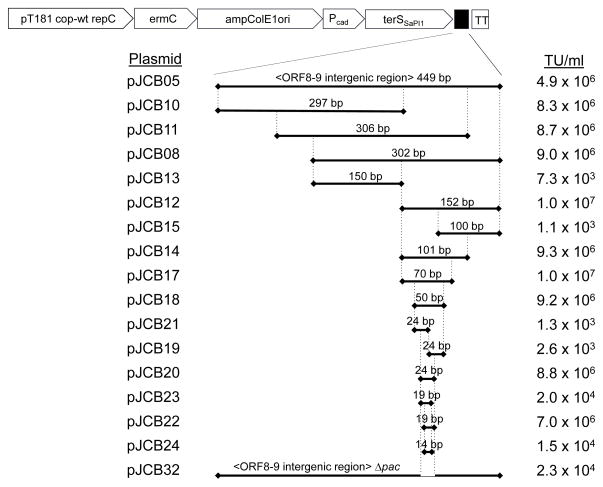
A series of overlapping smaller fragments from the intergenic region were introduced into the plasmid carrying SaPI1 terS in order to narrow down the sequence determinants for recognition by the SaPI1-specific packaging machinery. A 19 bp fragment carrying SaPI1 nucleotides 5894–5876 (coordinates from GenBank U93688) was sufficient to confer high frequency transduction (Fig 4). A fragment carrying the entire intergenic region except for 24 bp between 5899–5876 exhibited only generalized transduction. Thus, we have identified a small sequence that is both necessary and sufficient for SaPI1-directed packaging in the context of plasmid transduction. Comparison of the sequence in the plasmid carrying the 24 bp region that supports high frequency transduction with the equivalent region from those carrying the 19 and 14 bp sequences reveals that the common sequence shared by all four constructs is a 16 bp sequence that has an inverted repeat (Fig 5). We postulate that it is this inverted repeat that constitutes the actual SaPI1 TerS recognition sequence.
Figure 4. Deletion analysis of the SaPI1 pac region.
Fragments from the intergenic region between ORF 9 and 8 were cloned into plasmid pJCB03, as indicated, and introduced into S. aureus strain RN4220. Strains were infected with helper phage 80α and the resulting lysate was titered for plasmid transduction (TU), as shown in the right column.
Figure 5. DNA sequence of the SaPI1 pac site.
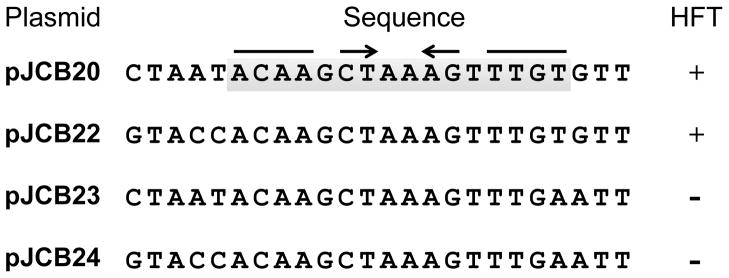
Arrows indicate the inverted repeat in the 16 bp common sequence (boxed in gray) shared by the constructs that retain high frequency transduction (HFT) by 80α in the presence of SaPI1 TerS.
To determine the relevance of the pac site described above on the transduction of SaPI1 in its native context, we introduced the 24 bp deletion into strain RN10628 (RN4220 SaPI1 tst::tetM, lysogenic for 80α) by allelic exchange using a derivative of the plasmid pMAD (Arnaud et al., 2004). Deletion of the SaPI1 pac site reduced transduction of SaPI1 by 2 orders of magnitude (Table 1). This is similar to the reduction seen in a SaPI1 ΔterS mutant, consistent with this small region containing the determinants required for terminase recognition by the SaPI1-encoded subunit. Residual generalized SaPI1 transduction by 80α is higher than was seen for the plasmids, but consistent with previous observations (Ubeda et al., 2009). Again, as was seen for plasmid transduction, in the absence of 80αterS the SaPI1 terS supports generalized transduction while deletion of both terminases eliminates all SaPI1 transfer.
DISCUSSION
A critical step in the assembly of double-stranded DNA bacteriophages is the packaging of newly synthesized genomes into procapsids. For most dsDNA phages characterized thus far, the packaging substrate is head-to-tail concatemers that are cleaved during packaging by the terminase. Cleavage and packaging of phage genomes from such concatemers is initiated by the binding of the small terminase subunit to the DNA. For those phages in which the initial cleavage site and the packaging recognition determinants have been established, there is variability in both the initial cleavage and the arrangement of the packaging recognition region. In phage λ, which uses a specific sequence for both initiation and termination of packaging, a staggered nick generates 12 bp cohesive ends at both the beginning and end of the packaged DNA and the recognition sequence, termed cos, consists of three discrete subsites (Catalano et al, 1995). Most phages that carry out headful packaging also recognize a specific site, termed pac, to direct the initial cleavage while the cleavage at the termination of packaging occurs after slightly more than one unit length genome has been packaged, and is controlled by the capacity of the prohead. In B. subtilis phage SPP1 the initial cleavage is highly specific, generating 1 bp staggered nicks at a site flanked by upstream and downstream recognition regions encompassing about 270 bp (Chai et al., 1995). The primary determinants for P22 packaging, on the other hand, are contained within a 16 bp asymmetric sequence that leads to heterogeneous initial cleavage acoss a region of about 120-bp flanking the pac site (Wu et al., 2002). Similar diversity in both size and location of pac site determinants and in the heterogeneity of the cleavage event itself has been described for several other phages, including T3, P1, and Mu (Groenen and van de Putte, 1985; Hashimoto and Fujisawa, 1992; Sternberg and Coulby, 1987). In most cases the packaging determinants are near or within the genes for the packaging machinery. This was also the case for phage 80α; the packaging cleavage site mapped in this study is within the gene for the small terminase subunit itself. Furthermore, this cleavage appears to be highly specific; all of the ends detected were clustered over just 3 nucleotides. SaPI1 cleavage, on the other hand, was less precise. The ends were mapped over a range of several hundred nucleotides, although most were clustered within a region of 35 nucleotides just upstream of the pac site, and both the recognition site and the cleavages were located far from the SaPI1 terS gene. A small sequence containing an inverted repeat appears to contain all of the specific packaging determinants for recognition by the SaPI1 small terminase. This is actually somewhat surprising, since packaging is directional and this is generally reflected in asymmetry of the pac sites. However, the existing sample size of well-studied packaging systems is still fairly small, leaving ample room for as yet unexplored variations in this process. Furthermore, the SaPI1 sequence is not perfectly symmetric. We also have not examined polarity in the context of the plasmid system; it is formally possible that with these small inserts packaging is not actually directional. Finally, we cannot rule out a contribution of flanking sequences in determining directionality even if not essential for the initial recognition.
While the site of initial cleavage for 80α has been mapped, the packaging determinants remain to be defined. We did not attempt to use the same approach employed here for SaPI1 because of potential complications introduced by the homology between cloned 80α fragments and the phage itself. As reported previously (Novick et al., 1986), such fragments, even as small as 300 bp, would be expected to result in high frequency transduction by 80α due to recombination between the phage and the homologous fragment on the plasmid. Inspection of the region of the 80α genome flanking the cleavage site did not reveal any obvious sequence features (direct or inverted repeats or a difference in G+C content) that would suggest locations for binding or bending of the DNA. We expect the phage recognition determinants to be different from those used by SaPI1, but further studies will be required to define these sequence elements.
In most dsDNA viruses, the terminase enzymes are hetero-oligomers composed of small and large subunits, assembled in a complex with stoichiometry that varies among different viruses. The large terminase subunit contains the DNA cleavage activity, the ATPase responsible for DNA translocation, and binds to the portal protein complex on the procapsid. The small subunit confers DNA recognition specificity and regulates the large subunit ATPase activity (Feiss and Rao, 2012). Based on the relatively small number of terminase structures known thus far, it appears that the small terminase subunits are more structurally diverse than the large subunits (Teschke, 2012). The 80α and SaPI1 small terminase subunits both belong to the SPP1-like small terminase family and share 31% amino acid identity; they are no more similar to each other than they are to the SPP1 small terminase (32% identity for 80α; 35% identity for SaPI1). The SaPI1 small terminase has a C-terminal extension not present in these other family members (see Fig S1A). The recent structure of another small terminase closely related to that of SPP1, from B. subtilis phage SF6 (Büttner et al., 2012), provides a scaffold for modeling of these proteins by I-TASSER (Roy et al., 2010). While the overall folds in the conserved regions are similar, there are several striking differences in the predicted structures (Fig S1B). These include an extra helix in the SaPI1 small terminase at the end of the C-terminal β-barrel domain and additional sequence in the β1/β2 region of the central oligomerization domain, suggesting that these different small terminases may oligomerize differently.
Our understanding of the powerful molecular motors that are responsible for viral DNA packaging is still in its infancy. The SaPI-helper system provides a unique and attractive model for the study of terminase structure/function. The SaPI1 and 80α small terminases bind to the same large terminase but recognize different DNA sequences, and direct different patterns of DNA cleavage. In addition, the phage small terminase subunit is a target for packaging interference by the SaPI Ppi function (Ram et al., 2012). Teasing apart the residues responsible for DNA binding in the phage terminases has been complicated by the fact that mutations affecting the terminase coding sequence also potentially alter the recognition determinants due to the location of these determinants within the terS gene itself. The location of the SaPI1 pac site away from its terS gene will facilitate genetic analysis of each component separately. Identifying how each of these small terminases interacts with the same TerL and stimulates a precise cleavage in one case and heterogeneous cuts in the other will also provide valuable insight into terminase function.
Supplementary Material
We mapped packaging cleavage sites for phage 80α and pathogenicity island SaPI1
Packaging directed by 80α TerS initiates within the 80α terS coding sequence
SaPI1 TerS-specific packaging initiates in an intergenic region
The SaPI1 pac site determinants were mapped using a plasmid-based assay
Deletion of the pac site from SaPI1 eliminates high frequency transduction
Acknowledgments
This work was performed in part under FDA's Medical Countermeasures Initiative, Contract #HHSF2232010000521 and NIH 1R56 AI081837 (to GEC). JCB was supported by the Portuguese Institute Fundação para a Ciência e Tecnologia (FCT), PhD Fellowship SFRH/BD/66250/2009. EKR was supported by the National Academies National Research Council; his contribution to this study was carried out at NICHHD, NIH, Bethesda MD, in the laboratory of the late Robert A Weisberg. Plasmid pCN51 (in strain NRS613) and strain RN4282 (NRS145) were obtained through the Network of Antimicrobial Resistance in Staphylococcus aureus (NARSA) program supported under NIAID/NIH contract #HHSN272200700055C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Joana C Bento, Email: jcbento@vcu.edu.
Kristin D Lane, Email: lanekd@vcu.edu.
Erik K Read, Email: Erik.Read@fda.hhs.gov.
Nuno Cerca, Email: nunocerca@ceb.uminho.pt.
References
- Arnaud M, Chastanet A, Débarbouillé M. New Vector for Efficient Allelic Replacement in Naturally Gram-Positive Bacteria. Applied and Environmental Microbiology. 2004;70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachi B. Physical mapping of the BglI, BglII, PstI and EcoRI restriction fragments of staphylococcal phage phi 11 DNA. Molecular & General Genetics. 1980;180:391–398. doi: 10.1007/BF00425853. [DOI] [PubMed] [Google Scholar]
- Bowdens DW, Modrich P. In Vitro Maturation of Circular Bacteriophage P2 DNA. The Journal of Biological Chemistry. 1985;260:6999–7007. [PubMed] [Google Scholar]
- Büttner CR, Chechik M, Ortiz-Lombardía M, Smits C, Ebong IO, Chechik V, Jeschke G, Dykeman E, Benini S, Robinson CV, Alonso Juan C, Antson A. Structural basis for DNA recognition and loading into a viral packaging motor. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:811–816. doi: 10.1073/pnas.1110270109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalano CE, Cue D. MicroReview Virus DNA packaging : the strategy used by phage. Molecular Microbiology. 1995;16:1075–1086. doi: 10.1111/j.1365-2958.1995.tb02333.x. [DOI] [PubMed] [Google Scholar]
- Chai S, Lurz R, Alonso JC. The small subunit of the terminase enzyme of Bacillus subtilis bacteriophage SPP1 forms a specialized nucleoprotein complex with the packaging initiation region. Journal of Molecular Biology. 1995;252:386–398. doi: 10.1006/jmbi.1995.0505. [DOI] [PubMed] [Google Scholar]
- Charpentier E, Anton AI, Barry P, Alfonso B, Fang Y, Novick RP. Novel Cassette-Based Shuttle Vector System for Gram-Positive Bacteria. Applied and Environmental Microbiology. 2004;70:6076–6085. doi: 10.1128/AEM.70.10.6076-6085.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie GE, Matthews AM, King DG, Lane KD, Olivarez NP, Tallent SM, Gill SR, Novick RP. The complete genomes of Staphylococcus aureus bacteriophages 80 and 80alpha--implications for the specificity of SaPI mobilization. Virology. 2010;407:381–390. doi: 10.1016/j.virol.2010.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie GE, Dokland T. Pirates of the Caudovirales. Virology. 2012;434:210–21. doi: 10.1016/j.virol.2012.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damle PK, Wall EA, Spilman MS, Dearborn AD, Ram G, Novick RP, Dokland T, Christie GE. The roles of SaPI1 proteins gp7 (CpmA) and gp6 (CpmB) in capsid size determination and helper phage interference. Virology. 2012;432:277–82. doi: 10.1016/j.virol.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dearborn AD, Spilman MS, Damle PK, Chang JR, Monroe EB, Saad JS, Christie GE, Dokland T. The Staphylococcus aureus Pathogenicity Island 1 Protein gp6 Functions as an Internal Scaffold during Capsid Size Determination. Journal of Molecular Biology. 2011;412:710–722. doi: 10.1016/j.jmb.2011.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiss M, Rao VB. The bacteriophage DNA packaging machine. Advances in Experimental Medicine and Biology. 2012;726:489–509. doi: 10.1007/978-1-4614-0980-9_22. [DOI] [PubMed] [Google Scholar]
- Groenen MA, Van de Putte P. Mapping of a site for packaging of bacteriophage Mu DNA. Virology. 1985;144:520–522. doi: 10.1016/0042-6822(85)90292-2. [DOI] [PubMed] [Google Scholar]
- Hashimoto C, Fujisawa H. DNA sequences necessary for packaging bacteriophage T3 DNA. Virology. 1992;187:788–795. doi: 10.1016/0042-6822(92)90480-d. [DOI] [PubMed] [Google Scholar]
- Kreiswirth BN, Lofdahl S, Betley MJ, O’Neill M, Schlievert PM, Bergdoll MS, Novick RP. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305:709–12. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- Lofdahl S, Zabielski J, Philipson L. Structure and restriction enzyme maps of the circularly permuted DNA of staphylococcal bacteriophage phi 11. Journal of Virology. 1981;37:784–794. doi: 10.1128/jvi.37.2.784-794.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir-Sanchis I, Martínez-Rubio R, Martí M, Chen J, Lasa Í, Novick RP, Tormo-Más MÁ, Penadés JR. Control of Staphylococcus aureus pathogenicity island excision. Molecular Microbiology. 2012;85:833–845. doi: 10.1111/j.1365-2958.2012.08145.x. [DOI] [PubMed] [Google Scholar]
- Novick RP, Edelman I, Lofdahl S. Small Staphylococcus aureus plasmids are transduced as linear multimers that are formed and resolved by replicative processes. Journal of Molecular Biology. 1986;192:209–220. doi: 10.1016/0022-2836(86)90360-8. [DOI] [PubMed] [Google Scholar]
- Novick RP, Christie GE, Penadés JR. The phage-related chromosomal islands of Gram-positive bacteria. Nature Reviews Microbiology. 2010;8:541–551. doi: 10.1038/nrmicro2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick RP. Genetic Systems in Staphylococci. Methods of Enzymology. 1991;204:587–636. doi: 10.1016/0076-6879(91)04029-n. [DOI] [PubMed] [Google Scholar]
- Poliakov A, Chang JR, Spilman MS, Damle PK, Christie GE, Mobley JA, Dokland T. Capsid size determination by Staphylococcus aureus pathogenicity island SaPI1 involves specific incorporation of SaPI1 proteins into procapsids. Journal of Molecular Biology. 2008;380:465–475. doi: 10.1016/j.jmb.2008.04.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram G, Chen J, Kumar K, Ross HF, Ubeda Carles, Damle PK, Lane Kristin D, Penadés JR, Christie GE, Novick RP. Staphylococcal pathogenicity island interference with helper phage reproduction is a paradigm of molecular parasitism. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:16300–16305. doi: 10.1073/pnas.1204615109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nature Protocols. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzin A, Lindsay J, Novick RP. Molecular genetics of SaPI1--a mobile pathogenicity island in Staphylococcus aureus. Molecular Microbiology. 2001;41:365–377. doi: 10.1046/j.1365-2958.2001.02488.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning. 3 2006. [Google Scholar]
- Sternberg N, Coulby J. Recognition and cleavage of the bacteriophage P1 packaging site (pac). II. Functional limits of pac and location of pac cleavage termini. Journal of Molecular Biology. 1987;194:469–479. doi: 10.1016/0022-2836(87)90675-9. [DOI] [PubMed] [Google Scholar]
- Stewart PR, Waldron HG, Lee JS, Matthews PR. Molecular relationships among serogroup B bacteriophages of Staphylococcus aureus. Journal of Virology. 1985;55:111–116. doi: 10.1128/jvi.55.1.111-116.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallent SM, Langston TB, Moran RG, Christie GE. Transducing Particles of Staphylococcus aureus Pathogenicity Island SaPI1 Are Comprised of Helper Phage-Encoded Proteins. Journal of Bacteriology. 2007;189:7520–7524. doi: 10.1128/JB.00738-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschke CM. Themes and Variations of Viral Small Terminase Proteins. Structure. 2012;20:1291–1292. doi: 10.1016/j.str.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tormo MA, Ferrer MD, Maiques E, Ubeda C, Selva L, Lasa I, Calvete JJ, Novick RP, Penades JR. Staphylococcus aureus pathogenicity island DNA is packaged in particles composed of phage proteins. Journal of Bacteriology. 2008;190:2434–2440. doi: 10.1128/JB.01349-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tormo-Más MÁ, Mir I, Shrestha A, Tallent SM, Campoy S, Lasa Í, Barbé J, Novick RP, Christie GE, Penadés JR. Moonlighting bacteriophage proteins derepress staphylococcal pathogenicity islands. Nature. 2010;465:779–782. doi: 10.1038/nature09065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda C, Barry P, Penadés JR, Novick RP. A pathogenicity island replicon in Staphylococcus aureus replicates as an unstable plasmid. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:14182–14188. doi: 10.1073/pnas.0705994104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda C, Maiques E, Barry P, Matthews A, Tormo MA, Lasa I, Novick RP, Penadés JR. SaPI mutations affecting replication and transfer and enabling autonomous replication in the absence of helper phage. Molecular Microbiology. 2008;67:493–503. doi: 10.1111/j.1365-2958.2007.06027.x. [DOI] [PubMed] [Google Scholar]
- Ubeda C, Olivarez NP, Barry P, Wang H, Kong X, Matthews A, Tallent SM, Christie GE, Novick RP. Specificity of staphylococcal phage and SaPI DNA packaging as revealed by integrase and terminase mutations. Molecular Microbiology. 2009;72:98–108. doi: 10.1111/j.1365-2958.2009.06634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda C, Tormo-Más MÁ, Penadés JR, Novick RP. Structure-function analysis of the SaPIbov1 replication origin in Staphylococcus aureus. Plasmid. 2012;67:183–190. doi: 10.1016/j.plasmid.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Sampson L, Parr R, Casjens S. The DNA site utilized by bacteriophage P22 for initiation of DNA packaging. Molecular Microbiology. 2002;45:1631–1646. doi: 10.1046/j.1365-2958.2002.03114.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.