Abstract
Successful vaccine development for infectious diseases has largely been achieved in settings where natural immunity to the pathogen results in clearance in at least some individuals. HIV presents an additional challenge in that natural clearance of infection does not occur, and the correlates of immune protection are still uncertain. However, partial control of viremia and markedly different outcomes of disease are observed in HIV infected persons. Here we examine the antiviral mechanisms implicated by one variable that has been consistently associated with extremes of outcome, namely HLA class I alleles, and in particular HLA-B, and examine the mechanisms by which this modulation is likely to occur, and the impact of these interactions on evolution of the virus and the host. Studies to date provide evidence for both HLA-dependent and epitope-dependent influences on viral control and viral evolution, and have important implications for the continued quest for an effective HIV vaccine.
INTRODUCTION
HIV infection is typically associated with an acute viral syndrome, with subsequent resolution of symptoms and then an 8-10 year asymptomatic period until the development of AIDS, defined in adults by a decline in CD4+ T cell number to less than 200 cells/mm3 or the presence of certain AIDS-defining illnesses that reflect underlying immune deficiency. Remarkably, however, the kinetics of progression to AIDS differ from a course as short as one year or less, to a lack of disease progression after more than 35 years and counting in some rare individuals. The one parameter that has been repeatedly predictive of the course of disease is human leukocyte antigen (HLA) class I, clearly indicating a role for host genetics in outcome of HIV infection.
The major histocompatibility complex (MHC) coding region, known as HLA in humans and situated on the short arm of chromosome 6, is the most polymorphic of the entire human genome. The single most polymorphic gene locus within this region is the HLA class I locus, for which 4,269 different HLA-A, HLA-B and HLA-C molecules have been described (Robinson et al., 2011). The explanation for this extreme polymorphism is that differences between the HLA class I alleles expressed can have a major impact on the incidence of, and survival from, a wide variety of infectious diseases, inflammatory conditions, autoimmune diseases and malignancies (genome.gov/26525384).
The reason for these associations between HLA and disease outcome becomes clear when one examines the structure and function of HLA class I. In 1974, Zinkernagel and Doherty demonstrated that MHC class I molecules are directly involved in the ability of CD8+ T cells to recognize and kill virus infected cells (Zinkernagel and Doherty, 1974), a phenomenon termed MHC class I restriction, and subsequently proposed that infectious diseases drive MHC class I polymorphism (Doherty and Zinkernagel, 1975).
HLA molecules bind fragments of host or pathogen-derived proteins that have been processed intracellularly and transported into the endoplasmic reticulum by TAP (the transporter associated with antigen processing), where they associate with developing class I molecules and are then transported to the cell surface. CD8+ T cells that recognise self- peptides bound to MHC class I molecules and expressed on the cell surface are deleted by negative selection in the thymus, providing T cells a means to recognize non-self proteins and thus defend against intracellular pathogens. Thus HLA class I is integral to the immune system being able to differentiate infected from non-infected cells.
The advent of structural data on the HLA class I-peptide complex led to a clearer understanding of the specificity of immune recognition. In 1987, Bjorkman et al revealed the three-dimensional structure of the surface expressed HLA class I molecule, consisting of a highly variable heavy chain comprising three alpha domains in complex with a soluble invariant molecule, β2 microglobulin (Bjorkman et al., 1987a, b). The alpha 1 and alpha 2 domains combine to make a four-stranded β-sheet lined by two antiparallel helices, which together form a deep groove that binds self or foreign peptide. This structural analysis identified 57 accessible HLA residues within the alpha 1 and alpha 2 domains that form the peptide binding groove, and that potentially interact with bound peptide or T cell receptor. Calculation of numbers of synonymous (dS) and nonsynonymous (dN) nucleotide substitutions per site, a measure of selection pressure, demonstrated that, for these 57 codons encoding the peptide binding region, dN/dS ratios were strikingly high (more than 3 in each of HLA-A, -B and –C), whereas for the codons encoding the remainder of the α1 and α2 domains, dN/dS ratios in each case were <0.4 (Hughes and Nei, 1988). Thus, purifying selection operates to maintain HLA amino acid sequences outside the peptide-binding region, whereas positive selection resulting from dramatically divergent HLA-associated disease outcomes is focused on the residues that constitute the peptide-binding region.
In this Review, we will first evaluate the consistency of associations between expression of HLA class I molecules and HIV disease outcome, focusing in particular on HLA-B alleles, for which the strongest data exist. We will then consider the potential mechanisms underlying these associations, including the nature of protective HLA class I alleles themselves and their interactions with CD8+ T cells and other binding partners and the specific epitopes presented within the binding groove. Deciphering these, and how they might be manipulated to augment immune control, is central to efforts to contain HIV through vaccine development.
HLA-HIV disease outcome studies
HLA class I alleles have consistently been the strongest independent associations, genetic or otherwise, with differential rates of HIV disease outcome. The first large population study showing a modulating role for HLA class I alleles in HIV infection revealed a relationship between HLA profile and time to onset of AIDS, with HLA-B*27 and HLA-B*57 having the highest impact on slowing the course of disease (Kaslow et al., 1996). This was followed by numerous additional studies in both acute and chronic infection, ultimately providing multiple independent documentations of this HLA impact. The most consistent of these observations for HIV disease association in relation to particular HLA-A and HLA-B alleles are shown in Table 1.
Table 1.
Consistent HLA Associations with Variation in HIV Disease Progression
| HLA-B | References | Cohorts |
|---|---|---|
|
Protective HLA I Alleles | ||
| B*13:02 | Honeyborne et al., 2007; Fellay et al., 2009; Pereyra et al., 2010 | C clade South Africa; B clade Caucasian |
| B*14/14:02 | Pereyra et al., 2010; Lazaryan et al., 2011 | B clade Caucasian; African American |
| B*27/27:05 | Kaslow et al., 1996; O'Brien et al., 2001, Fellay et al., 2009; Pereyra et al., 2010 | B clade Caucasian |
| B*42:01 | Carlson et al., 2012 | C clade South Africa/Botswana/Zimbabwe |
| B*44:03 | Leslie et al., 2010; Carlson et al., 2012 | C clade, South Africa; South Africa/Botswana/Zimbabwe |
| B*51 | Kaslow et al.,1996; O'Brien et al., 2001 | B clade Caucasian |
| B*52:01 | Pereyra et al., 2010 | B clade Caucasian |
| B*57/B*57:01 | Kaslow et al., 1996; Migueles et al., 2000; O'Brien et al., 2001; Pereyra et al., 2010 | B clade Caucasian; Caucasian/Hispanic |
| B*57:02 | Leslie et al., 2010 | C clade South Africa |
| B*57:03 | Costello et al., 1999; Leslie et al., 2010 | A clade Rwanda; C clade South Africa |
| Tang et al., 2010; Carlson et al., 2012 | C clade Zambia; South Africa/Botswana/Zimbabwe | |
| B*57:03 | Pereyra et al., 2010; Lazaryan et al., 2011 | B clade African American |
| B*58:01 | Kiepiela et al., 2004; Lazaryan, 2006[ml1] | C clade South Africa; Zambia |
| Carlson et al., 2012 | C clade South Africa/Botswana/Zimbabwe | |
| B*81:01 | Kiepiela et al., 2004; Tang et al., 2010 | C clade, South Africa; Zambia; South Africa/Botswana/Zimbabwe |
| Carlson et al., 2012; Pereyra et al., 2010 | B clade African American; C clade | |
| A*25/25:01 | Kaslow et al., 1996; Pereyra et al., 2010 | B clade Caucasian |
| A*32/32:01 | Kaslow et al., 1996; Fellay et al., 2009 | B clade Caucasian; |
| Lazaryan et al., 2011 | B clade North American African American | |
| A*74/74:01 | Tang et al., 2010; Matthews et al., 2011; Lazaryan et al., 2011; Koehler et al., 2011[ml2] | C clade Zambia; South Africa/Botswana/Zimbabwe; B clade African American; clades A, C, D, Tanzania |
| Disease-Susceptible HLA I Alleles | ||
|---|---|---|
| B*07:02 | Pereyra et al., 2010 | B clade Caucasian |
| B*08/08:01 | Steel et al., 1988; Pereyra et al., 2010; | B clade Caucasian |
| Carlson et al., 2012 | C clade South Africa/Botswana/Zimbabwe | |
| B*18/18:01 | Leslie et al., 2010; Carlson et al., 2012 | C clade South Africa/Botswana/Zimbabwe |
| B*35 | Carrington et al., 1999; O'Brien et al., 2001 | B clade Caucasian |
| B*35:01 | Pereyra et al., 2010 | B clade North American European |
| B*35:02/35:03 | Gao et al., 2001; Fellay et al., 2009 | B clade Caucasian |
| B*45/45:01 | Tang et al., 2010; Carlson et al., 2012 | C clade Zambia; South Africa/Botswana/Zimbabwe |
| B*51:01 | Carlson et al., 2012 | C clade South Africa/Botswana/Zimbabwe |
| B*53:01 | Gao et al., 2001; Pereyra et al., 2010 | B clade Caucasian; African American |
| B*54/55/56 | Hendel et al., 1999; Dorak et al., 2003 | B clade Caucasian |
| B*58:02 | Kiepiela et al., 2004; Tang et al., 2010; | C clade South Africa; Zambia |
| Carlson et al., 2012 | South Africa/Botswana/Zimbabwe | |
| A*36:01 | Tang et al., 2010; Carlson et al., 2012 | C clade Zambia; South Africa/Botswana/Zimbabwe |
| B*07:02 | Pereyra et al., 2010 | B clade Caucasian |
| B*08/08:01 | Steel et al., 1988; Pereyra et al., 2010; | B clade Caucasian |
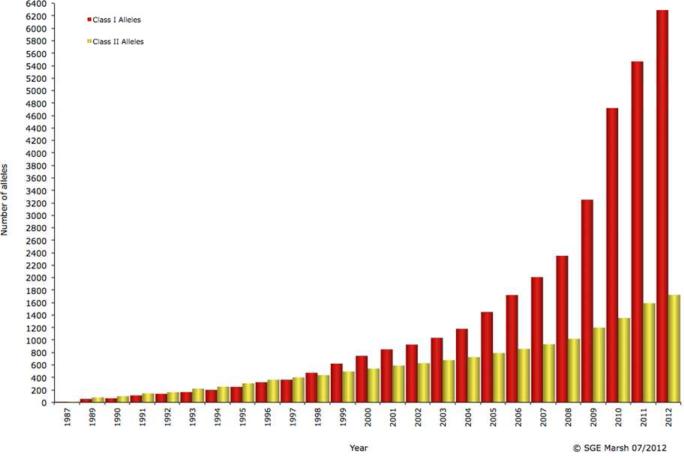
Improvements in HLA typing techniques have enhanced the ability to identify HLA associations with HIV disease outcome. Since the early 1990s, due to the advent of molecular rather than antibody-based typing, the numbers of potentially assignable class I alleles have increased 100-fold (Figure 1). This increase comes not just from detection of new alleles but also from the ability to molecularly differentiate alleles that were previously indistinguishable by antibody-based techniques. Current typing via direct sequencing of exons 2 and 3, which encode the α1 and α2 extracellular domains, is a substantial advance, with the first two digits specifying a group of alleles (for example, HLA-B*15), and the third through fourth or fifth specifying distinct HLA molecules (for example, HLA-B*15:01 through HLA-B*15:254, since there are 254 distinct HLA-B*15 molecules).
Figure 1.
Defined HLA class I and class II alleles by year from 1968-2012. Graph taken from http://www.ebi.ac.uk/imgt/hla/intro.html, copyright SGE Marsh 07/2012, with permission].
With improved typing technology to better define the extent of HLA diversity, and the use of large study cohorts, the ability to detect substantial HLA associations with HIV disease outcome, even for rare alleles, has improved. For most populations, where perhaps 60-70 different HLA class molecules are expressed at a phenotypic frequency of 1% or greater, a minimum of 500-1000 subjects are needed to have a realistic chance of detecting a significant effect for an individual HLA allele, depending on the frequency of the allele, and the size of the effect. However, an important challenge in determining the HLA allele responsible for a particular observed effect on HIV disease outcome is the issue of linkage disequilibrium (LD). For this reason the impact of individual HLA-C alleles is not included in Table 1 as it remains unclear the extent to which their appearance in HLA-HIV disease association studies is simply due to LD with certain HLA-B alleles. The HLA-C alleles that have been associated with protection against HIV disease progression tend to be in LD with protective HLA-B alleles (for example, the HLA-Cw*18:01 in LD with HLA-B*57:03 and with HLA-B*81:01), and similarly for disease-susceptible HLA-C alleles (such as HLA-Cw*07:02 and HLA-B*07:02). In Caucasian populations, where the disease-susceptible allele HLA-B*58:02 is largely absent, and the protective allele HLA-B*57:01 is in tight LD with HLA-Cw*06:02, HLA-Cw*06:02 may appear protective. Conversely, in African populations where HLA-B*57:01 is largely absent, HLA-B*58:02 is highly prevalent and in LD with HLA-Cw*06:02, so HLA-Cw*06:02 may appear disease-susceptible (Kiepiela et al., 2004; Leslie et al., 2010).
The dominant role of HLA-B
Some important observations may be made from the list of MHC I molecules shown in Table 1. First, HLA-B, which constitute 1,898 of the 4,269 distinct HLA class I A, B and C molecules described thus far, form the majority of the HLA class I alleles for which associations with either slow or rapid progression to HIV disease that have been identified. The most dramatic of these is HLA-B*57:01, which is the most highly enriched of all HLA alleles in persons who control HIV spontaneously (Emu et al., 2008; Migueles et al., 2000; Pereyra et al., 2008). Second, the protective HLA-B alleles are not broadly similar to each other, nor are the disease-susceptible HLA-B alleles similar to each other. Rather these arise from several different supertypes, which are defined by similarities in peptide binding motif (Sidney et al., 2008). Among these supertypes are B07 (HLA-B*07:02, HLA-B*35, HLA-B*42:01, HLA-B*51:01, HLA-B*53, HLA-B*81:01), B27 (HLA-B*14, HLA-B*27:05), B44 (HLA-B*18, HLA-B44:03, HLA-B*45:01), B58 (HLA-B*57, HLA-B*58:01, HLA-B*58:02), B62 (HLA-B*52:01) and unclassified (HLA-B*13:02). Of note, dramatic differences in outcome in HIV clade C virus infection are associated with HLA HLA-B*58:01 and HLA-B*58:02, both from the same supertype and differing by only 3 amino acids (Kiepiela et al., 2004; Ngumbela et al., 2008) . Third, HLA-B*51:01 appears both protective and disease-susceptible, depending on the geographic and evolutionary context. This reflects both the dynamic nature of HLA-HIV disease progression associations and also their HIV clade dependence. There is evidence that HLA-B*51:01 was protective early on in the B clade epidemic, but that the rapid accumulation of escape mutants at the population level within the dominant HLA-B*51:01-restricted CD8+ T cell epitope (TAFTIPSI, RT aa128-135) has reduced its protective effect (Kawashima et al., 2010; Kawashima et al., 2009). It appears also that this protective effect may be clade-specific, HLA-B*51:01 being linked with disease progression in C clade infection (Carlson et al., 2012). Similarly, HLA-B*35:01 is not disease susceptible in C clade infection, where the dominant Gag epitope NPPIPVGDIY (Gag 253-262) is highly targeted; however, in B clade infection, where HLA-B*35:01 is associated with rapid disease progression (Gao et al., 2001; Lazaryan et al., 2011; Pereyra et al., 2010) this Gag epitope is not targeted. The reason for this is that the B clade version NPPIPVGEIY is non-immunogenic, owing to the reduced peptide-MHC binding stability brought about by the D260E substitution at P8 in the epitope (our unpublished data).
HLA class I alleles are further divided into so-called private and public allotypes based on the particular amino acids expressed at residues 77 and 80-83 in the alpha 1 helix that determine the public allotypes Bw4 and Bw6. HLA residues 77, 80 and 81 make contributions to the F pocket of the peptide binding groove, which forms the C-terminal anchor, and therefore influence the epitopes that can be presented to CD8+ T cells; however this region also makes important interactions with NK cells. Although most protective HLA class I molecules carry the Bw4 motif, and most disease-susceptible molecules the Bw6 motif (Flores-Villanueva et al., 2001), there are several exceptions to this pattern: HLA-B*14, HLA-B*42:01 and HLA-B*81:01 are Bw6 alleles associated with delayed disease progression, and HLAB*53:01 and HLA-B*58:02 are Bw4 alleles associated with rapid progression. Also, although the few HLA-A alleles that have been linked with slow disease progression include HLA-A*25:01 and HLA-A*32:01, both expressing the Bw4 motif, other HLA-A molecules such as HLA-A*23 and HLA-A*24 that carry the Bw4 motif are not protective, and HLA-A*74:01, which differs from HLA-A*32:01 almost exclusively in not carrying the residues that constitute the Bw4 motif, is nonetheless consistently protective against HIV disease progression (Carlson et al., 2012; Koehler et al., 2010; Lazaryan et al., 2011; Matthews et al., 2011; Tang et al., 2010) .
Potential structural mechanisms of HLA class I-mediated control of HIV
The consistent observation that certain class I alleles are associated with relative protection from disease progression and others with risk of progression suggests that specific structural properties of this highly polymorphic complex might influence outcome.
Insights from host gene sequencing:
A number of genome wide association studies (GWAS) have shown that single nucleotide polymorphisms (SNPs) within the HLA class I region have the strongest genetic association with durable control of HIV (Dalmasso et al., 2008; Fellay et al., 2007; Limou et al., 2009; Pereyra et al., 2010), in particular a SNP that is in strong LD with HLA-B*57 and another associated with HLA-C expression . Although it had been already established that there was a strong HLA-B*57 effect on HIV disease progression (Kaslow et al., 1996; Migueles et al., 2000), these initial GWAS studies clearly showed that the major genetic signal associated with viral setpoint is exclusively within the HLA, and that no other genetic associations reached statistical significance, giving a sense of magnitude of the HLA effect. A major limitation of GWAS studies, however, is the issue of LD, since measurements are only made of specific tagging SNPs, each of which is in LD with multiple other SNPs that may be causal.
A higher power view of the genetic impact of HLA on viral control arose from coupling GWAS data with sequencing within the HLA locus (Pereyra et al., 2010). A novel imputation method to assign sequence within the HLA (de Bakker et al., 2006) allowed comparison of persons classified as HIV controllers (VL <2000 copies) with typical progressors (VL > 10,000 copies). That study, which extended previous GWAS studies by allowing assessment of the effect of individual amino acid positions on control, showed that the most significant independent genetic associations with viral control relate to specific amino acids within the HLA peptide binding groove that are involved in binding to the viral peptide. The strongest amino acid association with both control and lack of control is the allelic variant at position 97 within the HLA-B peptide binding cleft, which forms part of the C pocket and helps to anchor the viral peptide and thus likely influences the three dimensional structure of this complex. Interestingly, HLA-B*58:01 and HLA-B*58:02, associated with extremes of outcome in clade C virus infection, differ only at positions 94, 95 and 97 (Marsh et al., 2000). An important point is that position 97, which has 6 allelic variants, remains statistically significant even after controlling for HLA-B*57 and HLA-B*27, alleles known to be associated with host control (Pereyra et al., 2010). Amino acid variability at position 97 affects structure and flexibility of the peptide binding groove, being implicated both in HLA protein folding and also in cell surface expression (Blanco-Gelaz et al., 2006; Fagerberg et al., 2006).
Other amino acids markedly impacting viral load were found to line the HLA-B peptide binding groove, likewise interacting with bound peptide. An effect of HLA-C was also identified in these studies, but this contribution is likely due to variable expression levels modulated by a specific microRNA rather than the actual physicochemical properties of the structure itself (Kulkarni et al., 2011). The impact of HLA-C expression may relate to the fact that, unlike HLA-A and HLA-B, HLA-C is not downregulated by Nef, leaving infected cells still potentially susceptible to HLA-C restricted immune responses (Cohen et al., 1999). The exact mechanism whereby alterations in the HLA-B binding groove influence control remain unclear, but variability at the same amino acid positions in the HLA binding groove reconcile both protective and risk HLA-B alleles.
The potential importance of HLA binding motifs
The above studies suggest that specific structural features within the peptide binding groove of the HLA class I are important for immune control of immunodeficiency virus infection, and review of the peptide binding motifs of HLA class I molecules (Table 2) provides additional support for this hypothesis. These relate in particular to the nature of the B pocket, which binds position 2 (P2) of the peptide, and the F pocket, which binds the carboxy-terminal amino acid of the peptide (PC), with characteristics that are consistent across species. For example, the peptide-binding motif of the protective MHC class I allele Mamu-B*08 in simian immunodeficiency virus (SIV) infection is remarkably similar to that of the protective allele HLA-B*27 in HIV infection in humans, with a strict requirement for the self or viral peptide to contain Arg at P2, a strong preference for Arg (or Lys) at P1, and for medium-size hydrophobic residues at PC. This observation provides strong evidence for convergent evolution in these species, both evolving a similar binding motif . It has been proposed that the preference of Mamu-B*08, and HLA-B*27, for peptides carrying Arg both at P1 and P2, may mean that the epitopes restricted by these alleles are located in protein regions of particular structural significance, that is, containing adjacent Arg residues, and as a result that are highly resistant to escape (Mudd et al., 2011). There is also similarity in the peptide-binding groove of the protective macaque allele Mamu-B*17 and HLA-B*57/58:01, HLA-A*25 and HLA-A*32, all of which have a strong preference for Trp at PC. It is noteworthy that of the 50 HLA-B molecules for which peptide binding motifs are listed in The HLA Facts Book (Marsh et al., 2000), with one exception, only HLA-B*57 and HLA-B*58:01, both protective alleles, have Trp as a primary anchor residue, and accommodate no other amino acids at that position. The exception is HLA-B*15:13 which also contains Trp as a sole anchor at PC, but it is sufficiently rare that its capacity as a protective allele or otherwise is unknown. HLA-B*27 alleles are the only HLA-B alleles with Arg exclusively as a primary anchor residue.
Table 2.
Dominant Epitopes Restricted by MHC Class I Alleles Associated with Protection in HIV/SIV Infection
| Peptide-Binding Motif | Dominant HIV/SIV-Specific Epitope(s) | References | ||||||
|---|---|---|---|---|---|---|---|---|
| Position | P1 | P2 | P3 | P5-7 | PC | |||
| Binding Pocket | A | B | D | C | F | |||
| B*13:02 | QML | LIV | VQNLQGQMV | p24 Gag 135-143 | Honeyborne et al., 2007 | |||
| RQANFLGKI | p15 Gag 429-437 | Honeyborne et al., 2007 | ||||||
| B*14:02 | DE | RK | YFL | RH | L | DRFFKTLRA | p24 Gag 298-306 | Harrer et al., 1996 |
| B*27:05 | RK | R | YFWI | ILPV | RKHYFMLI | KRWIILGLNK | p24 Gag 262-270 | Nixon et al., 1988 |
| B*42:01 | PI | LMIF | TPQDLNTML | p24 Gag 180-189 | Goulder et al., 2000 | |||
| B*44:03 | DA | E | M | V | YF | AEQATQDVKNW | p24 Gag 306-316 | Kiepiela et al., 2007 |
| B*52:01 | Q | FYW | LIV | IV | RMYSPTSI | p24 Gag 275-282 | Brander et al., 1999 | |
| B*57:01 | K | ATS | KRLFY | FW | TSTLQEQIAW | p24 Gag 240-249 | Goulder et al., 1996 | |
| KAFSPEVIPMF | p24 Gag 162-172 | Goulder et al., 1996 | ||||||
| B*57:02 | K | ATS | DVPIM | FW | TSTLQEQIAW | p24 Gag 240-249 | ||
| ISPRTLNAW | p24 Gag 147-155 | Goulder et al., 1996 | ||||||
| B*57:03 | K | ATS | FY | FW | TSTLQEQIAW | p24 Gag 240-249 | ||
| KAFSPEVIPMF | p24 Gag 162-172 | |||||||
| B*58:01 | ATS | NLFY | FW | TSTLQEQIAW | p24 Gag 240-249 | Goulder et al., 1996 | ||
| B*81:01 | P | LMIF | TPQDLNTML | p24 Gag 180-189 | Leslie et al., 1996 | |||
| A*25:01 | DE | TVILF | W | ETINEEAAEW | p24 Gag 203-212 | Klenerman et al., 1996 | ||
| A*32:01 | IQ | W | PIQKETWETW | RT 559-568 | Harrer et al., 1996 | |||
| A*74:01 | IQ | R | GQMVHQAISPR | p24 Gag 140-150 | Matthews et al., 2011 | |||
| Mamu-A*01 | ST | P | FLIVM | CTPYDINQM | p24 Gag 181 -189 | Allen et al., 2001 | ||
| Mamu-B*08 | R | R | L | RRHRILDIYL | Nef 137-146 | Maness et al., 2008 | ||
| RRDNRRGL | Vif 172-179 | Maness et al., 2008 | ||||||
| RRAIRGEQL | Vif 123-131 | Maness et al., 2008 | ||||||
| Mamu-B*17 | RHL | W | IRYPKTFGW | Nef 165-173 | Mothe et al., 2002 | |||
| MHPAQTSQW | Nef 195-203 | Mothe et al., 2002 | ||||||
| HLEVQGYW | Vif 66-73 | Mothe et al., 2002 | ||||||
| Mane-A*10 | N/A | KKFGAEVVP | Gag 164-172 | Smith et al., 2005 | ||||
| Mamu-A1*043:01 (90-120-Ia) | N/A | IINEEAADWDL | Gag 206-216 | Nomura and Matano, 2012 | ||||
| Mamu-A1*065:01 (90-120-Ia) | N/A | SSVDEQIQW | Gag 241 -249 | Nomura and Matano, 2012 | ||||
This table contains the following references (exclusive of the main text): Allen et al. (2001), Brander et al. (1999), Goulder et al. (2000), Goulder et al. (1996), Harrer et al. (1996), Klenerman et al. (1996), Nixon et al., (1988), Nomura and Matano (2012), and Smith et al. (2005). Peptide-binding motifs are reviewed in Marsh et al., 2000. N/A, peptide-binding motifs not determined.
These observations prompt the hypothesis that there is a particular significance to the peptide-MHC interactions that involve either Arg or Trp and the complementary binding pocket into which these residues precisely fit. Both amino acids are unique in their respective ways. Arg is the most positively charged of all the amino acids (the pKa of the side chain is 12.48) and also has a 3-carbon aliphatic straight chain, providing a long finger-like projection with the positively charged guanidium group at its tip, fitting perfectly into the B pocket of HLA-B*27 (Madden et al., 1993). The fact that Arg appears to be irreplaceable in this position is evidence of the quality of fit. Studies of the stability of HLA-B*27 influenza, EBV and HIV-specific peptide-MHC complexes confirm that, in each case, the Arg at P2 contributes the most to peptide-MHC stability, even when the peptide carries Arg at PC, as it does in the EBV epitope (Stewart-Jones et al, personal communication. It is possible that the preference for Arg at both P1 and P2 for Mamu-B*08 and B*27 may critically increase the stability of such peptide-MHC complexes. Thus the complementarity of the anchor binding pocket to Arg appears critical, but what specific advantage such peptide stability might confer, for example on the avidity of TCR binding or on TCR signalling, is not clear.
Similarly, Trp has unique features, being the largest of the amino acids (molecular weight 203), and precise shape complementarity with the appropriate F pocket, such as provided by HLA-B*57, allows greater stability due to the higher number of interatomic van der Waal's interactions than would be generated by smaller anchor residues in smaller pockets. The HLA-B*57:03-KAFSPEVIPMF complex has 46 interatomic van der Waal's contacts between Phe at PC and the F pocket, the HLA-B*57:03-KAFSPEVI complex has 30, and the HLA-B*57:03-ISPRTLNAW complex has 86 van der Waal's contacts between Trp at the C terminus and the F-pocket (Stewart-Jones et al., 2005). These considerations prompt the speculation that those MHC class I molecules that have anchor-binding pockets that are precisely complementary to Arg (such as the HLA-B*27 and Mamu-B*08 B pockets, and perhaps also the HLA-A*74:01 F pocket) or to Trp (such as HLA-B*57/5801, HLA-A*25, HLA-A*32, Mamu-B*17 F pockets) may have the capacity to bind peptides more stably, and contribute to the immunodomination of CD8+ T cell responses restricted by these alleles (Altfeld et al., 2006).
One additional particular feature of Trp, distinct from all the other amino acids, is that it is encoded by only one codon, UGG. Single nucleotide changes from this result either in a Stop codon (UGA, UAG) or a codon encoding dramatically different amino acids such as Gly, Cys, Ser, or Arg, any of which would likely have very significant structural consequences were they to replace Trp. Thus mutational escape might be more problematic in this instance than is the case for Tyr-Phe or Ile-Leu substitutions, for example, which might be sufficient to reduce epitope recognition without significantly altering protein function.
The nature of the HLA peptide binding pockets may also modulate immune responses through an impact on thymic selection, which is governed by the strength of interaction of self-peptides presented within the peptide binding groove and the TCR. The signal delivered to the T cell has to be of sufficient magnitude to promote proliferation (positive selection), and yet not too strong or the clonal response will be deleted (negative selection). One computational study has shown that HLA alleles having the strongest association with protection from HIV disease progression, namely B*57 and B*27, are predicted to bind the fewest self-peptides of those HLA alleles evaluated, whereas alleles most associated with risk bind greater numbers of self peptides (Kosmrlj et al., 2010). T cells restricted by HLA-B*57 and B*27 would have undergone less negative selection since they are selected against fewer self peptides, and so a greater number would likely survive thymic selection and these would be predicted to be more likely to be cross reactive. Experimental data will be required to validate this hypothesis.
Kinetics and magnitude of HLA class I-peptide expression
Most data on HIV-specific CD8+ T cell responses in humans and in monkey models have been generated in the context of peptide-sensitized target cells rather than infected cells. Few studies have actually examined infected cells in which CTL recognition is dependent upon antigen processing (examples include (Chen et al., 2012a; Chen et al., 2011; Chen et al., 2009; Draenert et al., 2004; Migueles et al., 2002; Migueles et al., 2008; Sáez-Cirión et al., 2007; Yang et al., 1996; Yang et al., 2003)). Where antigen processing has been evaluated, variation in the kinetics of presentation of viral epitope are observed due to differences in processing that depend on the flanking sequences (Draenert et al., 2004; Le Gall et al., 2007), or are due to cell type-specific differences in processing (Lazaro et al., 2009; Steers et al., 2011). Moreover, it appears that there is a kinetic advantage to targeting Gag and Pol, since preformed Gag and Pol protein that enter the cell upon infection are rapidly degraded and processed through the class I pathway, whereas proteins such as Env must be endogenously processed(Chen et al., 2012a; Payne et al., 2010; Sacha et al., 2007a; Sacha et al., 2007b). The kinetics of epitope expression may be particularly important due to Nef-mediated downregulation of surface HLA class I expression. The selective downregulation of HLA-A and HLA-B by Nef protects infected cells from immune recognition by CTL (Collins et al., 1998; Yang et al., 2002), but because Nef does not downreglate HLA-C they are not susceptible to NK cell-mediated attack (Cohen et al., 1999). Interestingly, one study showed Nef downregulation of HLA-A was observed to be consistently more efficient than downregulation of HLA-B (Cohen et al., 1999), perhaps contributing to the dominance of HLA-B restricted responses associated with lowering of viremia. However, HLA-associated differences in susceptibility to Nef mediated downregulation was not observed in a careful comparative study of HLA-A*02 and HLA-B*57:01 (ref (Chen et al., 2012a; Chen et al., 2011)).
HLA effects on CD8+ T cell function
The proximate ligand for the peptide-HLA complex is the T cell receptor (TCR) on cytotoxic T lymphocytes, suggesting that HLA-dependent differences in CD8+ T cell function might account for the observed HLA-mediated effects. The functional profile of CD8+ T cells (effector cytokine secretion and degranulation) has been shown to be enhanced for HLA-B*27:05 HIV infected persons with both high and low viral loads, suggesting an HLA-specific effect on effector function (Almeida et al., 2007). Similar HLA-associated effects on HIV-specific CD8 T cell proliferation have also been suggested (Horton et al., 2006), and may also regulate the immunomodulatory effects of T regulatory cells (Elahi et al., 2011). The strongest correlation with in vivo control has been the in vitro proliferative capacity of CD8+ T cells following antigen-specific stimulation, as originally shown using autologous HIV-infected CD4+ T cells (Migueles et al., 2002). In these same experiments no differences were observed based on quantitative assessments by IFN gamma production or by tetramer staining. Notably, the strongest levels of proliferation have been associated with protective HLA alleles (Horton et al., 2006), and at least some studies have shown proliferative responses restricted by the most protective alleles, HLA-B*57 and HLA- B*27, to persist to some extent even with progressive infection (Horton et al., 2006; Migueles et al., 2002), suggesting HLA-dependent effects on CD8+ T cell proliferation. Indeed, in situations in which epitope-specific responses have been compared, proliferation associated with HLA-B*27 and HLA-B*57 restricted epitopes has been much greater than proliferation of other epitope-specific responses in the same individuals (Horton et al., 2006; Migueles et al., 2002). However, comparison of controllers and progressors expressing protective HLA alleles indicate that the effect is not only due to the HLA allele itself, since clear differences in proliferation have been shown comparing controllers to progressors (Migueles et al., 2002; Migueles et al., 2008). However, whether this difference is CD8+ T cell intrinsic or related to differences in CD4+ T helper cell function is unclear (Chen et al., 2012b; Lichterfeld et al., 2004).
A number of studies have begun to shed light on the mechanisms that may be responsible for the link between HLA, T cell function and viral control. Lytic granule loading by CD8+ T cells and delivery of granzyme B to infected target cells either after 6 days of stimulation in vitro or within as short a time as 30 minutes has been associated with enhanced killing of infected cells, and enhanced inhibition of HIV replication in vitro (Chen et al., 2012b; Hersperger et al., 2011; Migueles et al., 2002; Migueles et al., 2008). The potential role of specific TCR usage has also been examined, suggesting that public TCR usage, meaning TCRs specific for the same response that are identical or near identical in different individuals, may be more common in persons who control HIV. However, clonal composition does not appear to differentiate controllers compared to progressors (Chen et al., 2012b; Mendoza et al., 2012), and the role of avidity remains controversial (Bennett et al., 2008; Chen et al., 2012b; Iglesias et al., 2011).
In a study of HLA-B*27:05 positive persons infected persons, all of whom targeted the same epitope in Gag (designated KK10) and had no detectable escape mutations with that epitope, a comparison of HIV controllers compared to progressors revealed that the antiviral efficacy of the CTL differed significantly between these groups (Chen et al., 2012b). TCR in controllers not only had a greater ability to recognize infected cells and inhibit virus replication in an in vitro culture system, but were also more cross reactive with variants within the epitope that are known to arise in vivo. These studies show a significant impact of TCR usage in modulating the protective effect of class I alleles. Similar results were also found for the protective allele HLA-B*57 in that study. Interestingly, other parameters including magnitude, polyfunctionality and functional avidity did not correlate with control in the small cohort studied, indicating that those effects may be minor compared to the impact of TCR.
CTL engagement of target cells is comprised not just of a TCR-peptide-MHC interaction, but is also influenced by binding of the CD8 receptor to a region of the α3 domain of the HLA. The likelihood that this interaction has an influence on CTL efficacy is suggested by a recently completed GWAS and imputed amino acid sequence analysis of African Americans, which demonstrated not just residues in the HLA-B binding groove as being important, but also the allelic form of aa position 245, which is directly involved in CD8 binding to the peptide-MHC complex (McLaren et al., 2012). This position is monoallelic in Caucasians but biallelic in African-Americans, and suggests that residues outside the HLA binding groove are also important. It is noteworthy, however, that the protective HLA-B*81 is one of the HLA molecules, together with HLA-B*48 and HLA-A*68:01, carrying a variant at HLA residue 245 that reduces CD8 binding affinity. The relationship, if any, of this polymorphism to HIV disease progression thus remains unclear.
Other HLA binding partners
The HLA molecule interacts not just with the TCR on CTL through the α2 and α3 domains in the context of a presented viral peptide, but also with other binding partners through other cells. Of particular potential importance is their involvement in innate immunity as ligands for the killer cell immunoglobulin-like receptors (KIR), which modulate natural killer cell activity, the first line of defense against virally infected and cancer cells (Carrington et al., 2008). NK cells are actively inhibited by cells that express HLA Class I, but go on the attack against cells that are missing class I or have downregulated it, as occurs with certain cancers and virus infections. There are many different NK cell receptor families, but KIR genes are the most polymorphic, consistent with an active role in modulating HIV disease outcome. One of these interactions occurs between HLA class I and products of the KIR3DL1, KIR3DS1 genes. KIR3DL1, an inhibitory receptor, interacts with its binding partner Bw4 expressed on approximately 40% of HLA-B alleles. It is through this KIR-HLA interaction that Nef-mediated downregulation of HLA-B may sensitize infected cells for recognition by NK cells. Certain KIR alleles are associated with improved outcome, in particular KIR3DL1 in the presence of HLA-B molecules expressing Bw4 that contain an Ile at position (Bw4-80I) (Martin et al, 2007). KIR3DS1, which encodes an activating receptor, is also protective against disease progression in the setting of Bw4-80I (Martin et al., 2002). The mechanisms that are operating are not entirely clear (Altfeld and Goulder, 2007) but may involve direct lysis or antibody-dependent cell-mediated cytotoxicity (ADCC) (Parsons et al., 2012). Evidence suggests that the synergistic impact of these loci begins soon after infection, since the strongest effect was on CD4+ T cell decline . A link between innate immune function and disease outcome is further supported by the recent demonstration of viral evolution under NK-mediated immune selection pressure, demonstrated indirectly by sequence evolution that maps to NK cell immune pressure (Alter et al., 2011). These sequence polymorphisms in HIV have been shown to enhance binding of inhibitory KIRs to ligands on infected CD4+ T cells, thus reducing the antiviral effect NK cells, and further suggesting the importance of the KIR-HLA interaction in modulating of disease.
Another binding partner for HLA class I is a group of immunomodulatory molecules called leukocyte immunoglobulin-like receptors (LILRs) expressed on dendritic cells (DCs), which regulate the functional properties of DCs and thereby influence overall immune activation (Lichterfeld and Yu, 2012). It has been suggested that immune function may be influenced by binding of certain HLA alleles to ILT4, one of the LILRs on dendritic cells (Huang et al., 2010; Huang et al., 2009), but further studies are needed to define the relative contribution of this interaction with HLA to HIV control.
Epitope dependent mechanisms of MHC class I-mediated control of HIV
Recognition of infected cells depends on CD8+ T cell recognition of a complex consisting of viral peptide and HLA class I, thus the HLA-peptide complex clearly plays a role, as outlined above. However, additional variables including the kinetics of HLA-peptide expression and the impact of immune selection events on viral evolution that occur through this process, suggest that the actual epitopes presented by HLA class I alleles can be expected to play a role as well.
HIV epitope targeting and viral load
The dominant epitopes restricted by the MHC class I that have been linked with immune control of immunodeficiency virus infection are included in Table 2. In HIV infection, the approach typically adopted to evaluate the contribution of a particular epitope to immune control has been to compare viral loads in HLA-matched study subjects who do or do not make particular CD8+ T-cell responses (Kiepiela et al., 2004), although this requires large populations in order to adequately correct for multiple comparisons. The HLA-B*57 responses towards the Gag epitopes TSTLQEQIAW (‘TW10’, Gag 240-249) and KAFSPEVIPMF (‘KF11’, Gag 162-172) have been linked with substantially lower viral loads in HLA-B*57-positive subjects who show responses to these epitopes, whereas a response to the B*57-restricted Vif epitope LGHGVSIEW is associated with high viral loads (Kiepiela et al., 2007). Longitudinal studies in HLA-B*57:01 positive subjects with variable rates of disease progression, in this case defined by set point CD4 counts, also show epitope-specific targeting is associated with outcome: CD8+ T cell responses to TW10 and IW9 (ISPRTLNAW, Gag 147-155) are more polyfunctional and robust in those with delayed progression (Norstrom et al., 2012). Similarly, the HLA-B*27-restricted response towards the Gag epitope KRWIILGLNK (‘KK10’, Gag 263-272) is linked with lower viral loads in HLA-B*27-positive subjects (Ammaranond et al., 2011; Feeney et al., 2004; Goulder et al., 1997). Although transmission studies remain somewhat anecdotal, there is evidence that transmission to HLA-B*57 or HLA-B*27-positive persons of viruses carrying escape mutants in these particular epitopes results in more rapid progression (Crawford et al., 2009; Goulder et al., 2001).
The role of HIV-1 Gag
Numerous studies have linked viral load or disease outcome to CD8+ T cell targeting of HIV-1 Gag (Dinges et al., 2010; Edwards et al., 2002; Geldmacher et al., 2007; Kiepiela et al., 2007; Klein et al., 1995; Masemola et al., 2004; Novitsky et al., 2003; Ogg et al., 1998; Riviere et al., 1995; Streeck et al., 2007; Zuniga et al., 2006) . The mechanism of this effect may involve at least two different aspects related to Gag specificity. The first is that Gag is so highly abundant within the virus entering CD4+ T cell target cells that Gag epitopes can be processed, bind with developing class I molecules, and be expressed on the cell surface and recognised by Gag-specific CD8+ effector T cells within a few hours of infection (Chen et al., 2012a; Payne et al., 2010; Sacha et al., 2007a), comfortably prior to de novo synthesis of viral proteins including Nef, whose effects include down-regulation of HLA class I that impairs CD8+ T cell recognition (Collins et al., 1998). This same property appears to also apply to Pol which is present on incoming virions, though in smaller amounts (Chen et al., 2012a; Sacha et al., 2007b). The second aspect of a Gag-specific response, particularly if one considers the capsid protein, p24 Gag, is that the amino acid sequence is highly conserved, implying that a significant fitness cost would likely result from any sequence changes (Martinez-Picado et al., 2006; Miura et al., 2009a). Thus, although escape mutations may be selected by the virus, enabling it to evade potent CD8+ T-cell responses, the consequence of these mutations in p24 Gag may be a reduction in viral replicative capacity (RC) and therefore an overall benefit to the host.
HLA associated viral mutations and viral fitness
In the case of the HLA-B*57 and HLA-B*27 p24 Gag epitopes mentioned above, in each instance evaluated, the escape mutations that are selected do indeed reduce viral RC (Crawford et al., 2009; Martinez-Picado et al., 2006; Miura et al., 2009b; Schneidewind et al., 2008; Schneidewind et al., 2007). The most commonly occurring escape mutations within the HLA-B*27 epitope, KK10, involving the substitution at P2 of Arg by Lys, causes such a reduction in RC that this escape mutation evidently cannot be accommodated by the virus without a compensatory mutation (S173A) arising simultaneously 91 amino acids upstream in a location closely adjacent in the 3-D structure of the capsid protein to the R264K escape mutation (Schneidewind et al., 2007). This requirement for two mutations simultaneously may in part explain the many years it typically takes for someone who has HLA-B*27 to lose immune control of HIV infection. In the case of the HLA-B*57 p24 Gag epitopes, there are 3 of these, and accumulating mutations in these epitopes each contributes to progressive loss of viral RC such that, once all the mutations are in place, a substantially attenuated virus remains (Crawford et al., 2009). Even in elite controllers there is evidence of escape from CD8+ T cell responses (Bailey et al., 2009; Miura et al., 2009c), with some data indicating that the rarely observed escape mutants, which inflict greatest damage to viral RC, are those that tend accompany maintained control of viraemia (Miura et al., 2009a; Miura et al., 2009b). This may be related to an enhanced ability of the particular TCR to recognise the commonly arising variants that inflict only modest degree of damage to viral RC (Chen et al., 2012b).
It is noteworthy that, with the exception of HLA-A*32, the dominant response restricted by all the protective HLA alleles is in p24 Gag (Table 2). In contrast, the dominant responses restricted by the HLA alleles associated with rapid progression are more often in Nef or Env (Kiepiela et al., 2007; Ngumbela et al., 2008). Numerous studies have demonstrated that a Gag-specific response, and especially the breadth of the Gag-specific CD8+ T-cell response, is inversely correlated with viral load (Dinges et al., 2010; Edwards et al., 2002; Geldmacher et al., 2007; Kiepiela et al., 2007; Masemola et al., 2004; Novitsky et al., 2003; Ogg et al., 1998; Riviere et al., 1995; Streeck et al., 2007; Zuniga et al., 2006) while the reverse is the case for Nef- or Env-specific responses (Dinges et al., 2010; Kiepiela et al., 2007; Masemola et al., 2004; Riviere et al., 1995). There is also evidence that escape mutants in Env epitopes tend to have little impact on viral RC, in comparison to those that arise within Gag (Troyer et al., 2009), such that HLA alleles presenting Env epitopes may have less antiviral efficacy.
Micropolymorphism studies, in which HLA class I molecules are compared that differ by one, or a small number of, amino acids, have been illuminating here. Two closely-related HLA-B alleles, HLA-B*42:01 and HLA-B*42:02 differ by a single amino acid, with Tyr and His at HLA position 9, respectively, which lies at the base of the peptide binding groove (Marsh et al., 2000). Both express the Bw6 motif and therefore do not act as KIR ligands. Both are in LD with the same HLA-A and HLA-C alleles (HLA-A*30:01 and HLA-Cw*17:01) and therefore the impact via LD of other HLA alleles on the differential effects observed for HLA-B*42:01 and HLA-B*42:02 is minimised. Although the peptide binding motif of these two molecules appears indistinguishable, in practice the epitopes targeted by individuals expressing HLA-B*42:01 and HLA-B*42:02 differ dramatically. Most notably, HLA-B*42:01 presents multiple Gag epitopes, including TPQDLNTML (‘TL9’, Gag 180-189), whereas HLA-B*42:02 presents no Gag epitopes. These differences are also associated with a 0.52 log10 lower viral load set point in subjects with HLA-B*42:01, consistent with the notion of Gag responses generally affording some protection against disease progression (Kloverpris et al., 2012). For comparison, the average reduction in viral set point resulting from expressing of HLA-B*57:03 is approximately 0.75 log10 (Carlson et al., 2012).
Monkey studies also support a role for Gag targeting in the context of MHC class I alleles and immune control. Burmese rhesus macaque animals carrying the 90-120-Ia haplotype vaccinated with a DNA-prime, Gag-expressing Sendai virus vector boost and then challenged with SIVmac239 were able to control viraemia to undetectability, compared to controls whose viral set points were in the 104−106 range (Matano et al, 2004). The epitopes identified as responsible were IINEEAADWDL (‘IL11’, Gag 206-216) and SSVDEQIQW (‘SW9’, Gag 241-249). Escape mutations arise early in the course of infection in IW11 (L216S) and later in SW9 (D244E) that both reduce viral RC (Kawada et al., 2007; Matano et al., 2004). Moreover, infection of vaccinated 90-120-Ia-positive macaques with a virus carrying these mutants resulted in failure of these animals to control viraemia (Kawada et al., 2008). Animals expressing the 90-120-Ia haplotype who were vaccinated with a construct expressing the single epitope SW9 were able to control post-challenge viraemia (Tsukamoto et al., 2009), as were animals vaccinated with a construct expressing the single epitope IL11 (Ishii et al., 2012), indicating that induction of a single epitope-specific response, properly targeted in the correct MHC context, may be sufficient for a vaccine to be effective. These experiments closely resemble what is observed in HLA-B*57-positive humans who control HIV infection, in whom the dominant response in acute infection is towards an epitope TSTLQEQIAW (‘TW10’) that is precisely analogous to the protective SIV epitope SSVDEQIQW (‘SW9’) (Table 2).
Data such as these suggest a role for protein-specificity and also for the particular epitope being targeted, independent of the MHC effect. The epitopes identified as playing a part in immune control of SIV infection and of HIV infection are, as illustrated above, in many cases virtually identical, in spite of the vast phylogenetic distance between the MHC class molecules restricting them, and the sizeable distance between HIV and SIV. In addition to Mamu-A1*065:01-SW9 and HLA-B*57/5801-TW10, these include Mamu-A1*043:01-IW11 and HLA-A*25-EW10, Mane-A*10-KP9 and HLA-B*57-KF11, Mamu-A*01-CM9 and HLA-B*42/81-TL9 (Table 2). However, it should be emphasized that, just as the expression of HLA-B*57 does not necessarily translate into immune control of HIV infection, the generation of Gag-specific responses also does not confer universal protection against disease progression. Likewise, targeting of an identical epitope in the context of different class I alleles results can result in very different mutational escape, which likely impacts T cell efficacy (Leslie et al., 2006). However the data presented above suggest that, like expression of HLA-B*57, a broad Gag-specific response in general is a factor that is likely to contribute to improved control of HIV infection.
Influence of epitopes outside Gag
Studies in the macaque-SIV model have, in the main, supported the role of Gag-specificity and the impact of escape mutants on viral RC in CD8+ T cell-mediated immune control of immunodeficiency virus infection. However, the work on Indian rhesus elite controllers of SIV has also highlighted the fact that effective CD8+ T cell responses can be found outside of Gag. Among Mamu-B*08 and Mamu-B*17 elite controller macaques the epitopes mediating control are not in Gag, but in Nef or Vif (Maness et al., 2008; Valentine et al., 2009). Mamu-B*08 animals infected with SIVmac239 containing mutants in the key non-Gag epitopes were unable to contain viraemia as successfully as those infected with wildtype virus (Valentine and Watkins, 2008). Mamu-B*08 animals vaccinated with constructs expressing the 3 critical epitopes in Nef and Vif showed markedly improved control of viraemia compared with unvaccinated animals (Mudd and Watkins, 2012). Moreover the mechanism implicated here does not appear to involve the cost to viral RC of escape mutants. Likewise, in HIV infection there is evidence of effective antiviral pressure, as measured by effective inhibition of virus replication in vitro, directed against epitopes other than Gag, including Env and Nef (Chen et al., 2012a; Chen et al., 2011; Yang et al., 1997).
Thus, although the epitopes that form the dominant CD8+ T cell responses may play an important part in contributing to immune control because of features relating to protein specificity, the MHC class I molecule itself evidently also performs a critical role in establishing the effectiveness of particular CD8+ T cell responses. The absence to date of an identified HLA class I molecule that protects against HIV disease progression and yet does not have a dominant Gag-specific response continues to obscure the precise role of epitope and HLA in immune control.
The special case of HLA-C
One aspect of HLA-mediated control of HIV that still remains quite unclear is the role of HLA-C. Several GWAS in Caucasians have shown that a SNP 35kb upstream of the HLA-C locus is strongly associated with viral set point (Fellay et al., 2009; Fellay et al., 2007; Pereyra et al., 2010). The protective CC variant is linked with high HLA-C expression (Thomas et al., 2009), and variation at −35 marks a polymorphism within the 3’UTR of HLA-C that regulates binding of a microRNA to its target site, which has been shown to regulate HLA-C expression (Kulkarni et al., 2011). Strong binding of this microRNA (hsa-miR-148) results in relatively low expression of HLA-C, whereas alleles that do not bind this microRNA have high HLA-C expression. Of note, however, the −35 C variant is in strong LD with HLA-C alleles that are themselves in strong LD with protective HLA-B alleles in Caucasian cohorts (Corrah et al., 2011). Examples here are HLA-Cw*02:02 and HLA-B*27:05, HLA-Cw*06:02 and HLA-B*57:01, HLA-Cw*0801/02 and HLA-B*14, HLA-Cw*12:02 and HLA-B*52. The −35 T variant is in LD with HLA alleles such as HLA-Cw*07:01, which is in tight LD with the risk allele HLA-B*08:01. The fact that the −35 SNP is also present in African American cohorts but does not correlate with viral set point in those populations (McLaren et al., 2012; Pelak et al., 2010; Shrestha et al., 2009) might relate to the different patterns of LD between HLA-B and HLA-C alleles referred to above. The protective HLA-B alleles in African cohorts include HLA-B*57:03 and HLA-B*58:01, which are both in LD with HLA-Cw*07:01, a low expressing allele, and the principal risk allele HLA-B*58:02, which is in strong LD with HLA-Cw*06:02, a high expressing allele. The precise role of HLA-C alleles in control of HIV is likely to be facilitated and clarified by further studies including direct measurement of expression levels of different HLA-C alleles and involving different study populations.
Conclusions and Future Directions
The discovery more than 15 years ago that HLA-B*57 and HLA-B*27 are associated with improved HIV control (Kaslow et al., 1996), followed by the association of HLA-B*35 with worse outcome (Carrington et al., 1999), has been consistently validated, and the major impact of these and other HLA class I alleles on disease progression has been further revealed as HLA typing methods have improved and cohort sizes have expanded. Initially, the evidence appeared to suggest that the epitope presented by such HLA class I molecules was key to immune control. Loss of the critical epitope meant loss of the protective effect associated with the HLA allele, and there is strong support for this for multiple but by no means all epitopes, particularly those in Gag. More recent data from studies in humans and in the SIV-macaque model, however, also indicate an important component of HLA-associated immune control of HIV infection is related to the HLA molecule itself, either through specific characteristics of peptide binding or through allele-specific interactions with HLA ligands including TCRs on CD8 T cells, KIRs on NK cells or LILRs on dendritic cells.
It is likely, also, that the precise mechanism underlying the protective effects of MHC class I molecules may vary from one to another. For example, there is evidence that the beneficial effects of HLA-B*57 and HLA-B*27 appear to differ in their timing during the course of HIV infection (Gao et al., 2005). Resolution of the relative contributions of these different effects, and identification of the precise mechanisms governing them, will require large population based studies, ideally longitudinal studies from the time of acute infection, and will likely reveal multifactorial impacts on viral control.
The concepts of HLA molecules driving the evolution of HIV, together with differential impacts of HLA alleles on disease outcome, raise important questions as the quest for an HIV vaccine continues. Despite HLA-specific associations with disease outcome, it is important to note that even among persons with protective alleles such as B*27 and B*57, the majority of persons with these alleles progress, whereas some persons with no protective alleles remain asymptomatic, suggesting that host genetics are not an absolute constraint, and that HLA alone is poorly predictive of outcome. This begs the question as to what other factors modulate the HLA effect (Figure 2). Indeed, it is estimated that less than 20% of viral setpoint variation can be explained by HLA and other genetic polymorphisms (Carrington and Walker, 2012).
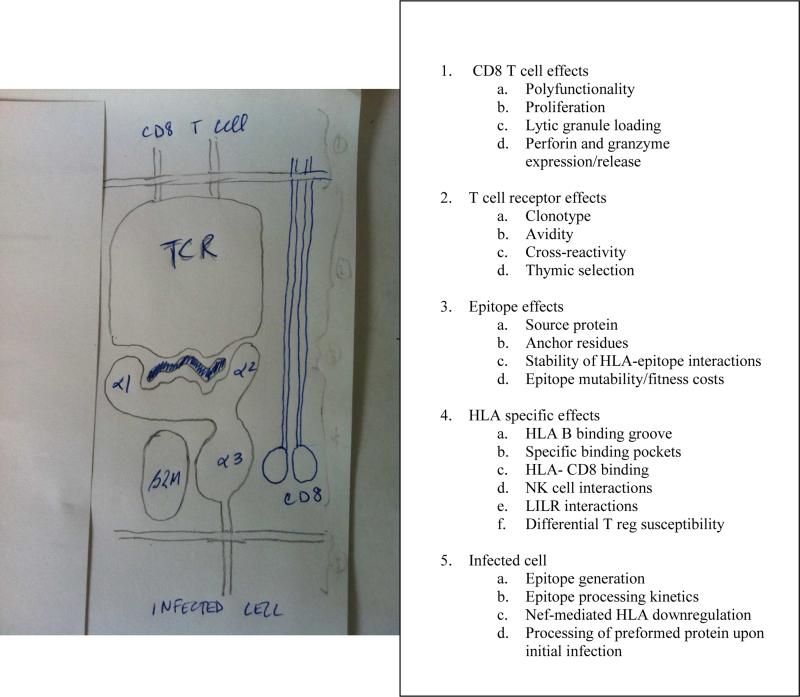
Figure 2.
Potential mechanisms of HLA-associated impact on viral control. The HLA class I molecule is a heterodimer consisting of three alpha helices and β-2 microglobulin. The alpha 1 and alpha 2 domains together form the peptide binding groove, which presents viral peptides at the surface of an infected cell, and this complex is in turn recognized by the T cell receptor on CD8+ T cells, through interactions with both the TCR and CD8 receptor (shown). The HLA molecule or peptide-HLA complex may also bind KIR and LILR (not shown). The mechanisms by which HLA and its interactions with CD8+ T cells, TCR, epitope, and infected cells, as well as HLA-specific mechanisms, have been shown to modulate the efficacy of HIV control are listed in the adjacent table.
Basic concepts outlined in this review provide multiple potential paths forward to better define those components required for effective durable control of HIV. The fitness landscape of HIV needs to be defined, as efforts to harness immune responses to impair viral replication capacity offer hope for enhanced control. The importance of Gag targeting is clear, but definition of other non-Gag epitopes that can be beneficially targeted, particularly those that are associated with a fitness cost, need to be a priority. The specific properties of HLA ligands in enhancing immune control need to be better defined, and offer opportunities to improve immune control through TCR, KIR and LILR interactions. And a better definition of the properties of the CD8 T cells that mediate infected cell killing, and how to enhance this function, remains a priority. The path toward an HIV vaccine has not been easy, and the ongoing evolution of HIV under HLA-associated immune selection pressure is a significant challenge. However, the prevalence and extent of HIV infection, together with readily measureable clinical parameters of disease severity, offer an unprecedented opportunity to dissect the multiple parameters that contribute to immune control, which will have benefit to harnessing the immune system well beyond HIV.
Acknowledgements
Support for this work came from the NIH (BDW and PJRG), the Wellcome Trust (PJRG), the Collaboration for Vaccine Discovery of the Bill and Melinda Gates Foundation (BDW), the Harvard University Center for AIDS Research (BDW), and the Mark and Lisa Schwartz Foundation (BDW).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- Almeida JR, Price DA, Papagno L, Arkoub ZA, Sauce D, Bornstein E, Asher TE, Samri A, Schnuriger A, Theodorou I, et al. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. The Journal of experimental medicine. 2007;204:2473–2485. doi: 10.1084/jem.20070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter G, Heckerman D, Schneidewind A, Fadda L, Kadie CM, Carlson JM, Oniangue-Ndza C, Martin M, Li B, Khakoo SI, et al. HIV-1 adaptation to NK-cell-mediated immune pressure. Nature. 2011;476:96–100. doi: 10.1038/nature10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altfeld M, Goulder P. ‘Unleashed’ natural killers hinder HIV. Nature genetics. 2007;39:708–710. doi: 10.1038/ng0607-708. [DOI] [PubMed] [Google Scholar]
- Altfeld M, Kalife ET, Qi Y, Streeck H, Lichterfeld M, Johnston MN, Burgett N, Swartz ME, Yang A, Alter G, et al. HLA Alleles Associated with Delayed Progression to AIDS Contribute Strongly to the Initial CD8(+) T Cell Response against HIV-1. PLoS medicine. 2006;3:e403. doi: 10.1371/journal.pmed.0030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammaranond P, van Bockel DJ, Petoumenos K, McMurchie M, Finlayson R, Middleton MG, Davenport MP, Venturi V, Suzuki K, Gelgor L, et al. HIV immune escape at an immunodominant epitope in HLA-B*27-positive individuals predicts viral load outcome. J Immunol. 2011;186:479–488. doi: 10.4049/jimmunol.0903227. [DOI] [PubMed] [Google Scholar]
- Bailey JR, Brennan TP, O'Connell KA, Siliciano RF, Blankson JN. Evidence of CD8+ T-cell-mediated selective pressure on human immunodeficiency virus type 1 nef in HLA-B*57+ elite suppressors. Journal of virology. 2009;83:88–97. doi: 10.1128/JVI.01958-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MS, Ng HL, Ali A, Yang OO. Cross-clade detection of HIV-1-specific cytotoxic T lymphocytes does not reflect cross-clade antiviral activity. The Journal of infectious diseases. 2008;197:390–397. doi: 10.1086/525281. [DOI] [PubMed] [Google Scholar]
- Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. The foreign antigen binding site and T cell recognition regions of class I histocompatibility antigens. Nature. 1987a;329:512–518. doi: 10.1038/329512a0. [DOI] [PubMed] [Google Scholar]
- Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. 1987b;329:506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- Blanco-Gelaz MA, Suarez-Alvarez B, Gonzalez S, Lopez-Vazquez A, Martinez-Borra J, Lopez-Larrea C. The amino acid at position 97 is involved in folding and surface expression of HLA-B27. Int. Immunol. 2006;18:211–220. doi: 10.1093/intimm/dxh364. [DOI] [PubMed] [Google Scholar]
- Carlson JM, Listgarten J, Pfeifer N, Tan V, Kadie C, Walker BD, Ndung'u T, Shapiro R, Frater J, Brumme ZL, et al. Widespread impact of HLA restriction on immune control and escape pathways of HIV-1. Journal of virology. 2012;86:5230–5243. doi: 10.1128/JVI.06728-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington M, Martin MP, van Bergen J. KIR-HLA intercourse in HIV disease. Trends Microbiol. 2008;16:620–627. doi: 10.1016/j.tim.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington M, Nelson GW, Martin MP, Kissner T, Vlahov D, Goedert JJ, Kaslow R, Buchbinder S, Hoots K, O'Brien SJ. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science (New York, N.Y. 1999;283:1748–1752. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- Carrington M, Walker BD. Immunogenetics of spontaneous control of HIV. Annual review of medicine. 2012;63:131–145. doi: 10.1146/annurev-med-062909-130018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DY, Balamurugan A, Ng HL, Cumberland WG, Yang OO. Epitope targeting and viral inoculum are determinants of Nef-mediated immune evasion of HIV-1 from CTLs. Blood. 2012a doi: 10.1182/blood-2012-02-409870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DY, Balamurugan A, Ng HL, Yang OO. Antiviral activity of human immunodeficiency virus type 1 Gag-specific cytotoxic T lymphocyte targeting is not necessarily intrinsically superior to envelope targeting. Journal of virology. 2011;85:2474–2478. doi: 10.1128/JVI.01726-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Ndhlovu ZM, Liu D, Porter LC, Fang JW, Darko S, Brockman MA, Miura T, Brumme ZL, Schneidewind A, et al. TCR clonotypes modulate the protective effect of HLA class I molecules in HIV-1 infection. Nature immunology. 2012b;13:691–700. doi: 10.1038/ni.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Piechocka-Trocha A, Miura T, Brockman MA, Julg BD, Baker BM, Rothchild AC, Block BL, Schneidewind A, Koibuchi T, et al. Differential neutralization of human immunodeficiency virus (HIV) replication in autologous CD4 T cells by HIV-specific cytotoxic T lymphocytes. Journal of virology. 2009;83:3138–3149. doi: 10.1128/JVI.02073-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen GB, Gandhi RT, Davis DM, Mandelboim O, Chen BK, Strominger JL, Baltimore D. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity. 1999;10:661–671. doi: 10.1016/s1074-7613(00)80065-5. [DOI] [PubMed] [Google Scholar]
- Collins KL, Chen BK, Kalams SA, Walker BD, Baltimore D. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature. 1998;391:397–401. doi: 10.1038/34929. [DOI] [PubMed] [Google Scholar]
- Corrah TW, Goonetilleke N, Kopycinski J, Deeks SG, Cohen MS, Borrow P, McMichael A, Brackenridge S. Reappraisal of the relationship between the HIV-1-protective single-nucleotide polymorphism 35 kilobases upstream of the HLA-C gene and surface HLA-C expression. Journal of virology. 2011;85:3367–3374. doi: 10.1128/JVI.02276-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford H, Lumm W, Leslie A, Schaefer M, Boeras D, Prado JG, Tang J, Farmer P, Ndung'u T, Lakhi S, et al. Evolution of HLA-B*5703 HIV-1 escape mutations in HLA-B*5703-positive individuals and their transmission recipients. The Journal of experimental medicine. 2009;206:909–921. doi: 10.1084/jem.20081984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmasso C, Carpentier W, Meyer L, Rouzioux C, Goujard C, Chaix ML, Lambotte O, Avettand-Fenoel V, Le Clerc S, de Senneville LD, et al. Distinct genetic loci control plasma HIV-RNA and cellular HIV-DNA levels in HIV-1 infection: the ANRS Genome Wide Association 01 study. PloS one. 2008;3:e3907. doi: 10.1371/journal.pone.0003907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bakker PI, McVean G, Sabeti PC, Miretti MM, Green T, Marchini J, Ke X, Monsuur AJ, Whittaker P, Delgado M, et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nature genetics. 2006;38:1166–1172. doi: 10.1038/ng1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinges WL, Richardt J, Friedrich D, Jalbert E, Liu Y, Stevens CE, Maenza J, Collier AC, Geraghty DE, Smith J, et al. Virus-specific CD8+ T-cell responses better define HIV disease progression than HLA genotype. Journal of virology. 2010;84:4461–4468. doi: 10.1128/JVI.02438-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty PC, Zinkernagel RM. A biological role for the major histocompatibility antigens. Lancet. 1975;1:1406–1409. doi: 10.1016/s0140-6736(75)92610-0. [DOI] [PubMed] [Google Scholar]
- Draenert R, Le Gall S, Pfafferott KJ, Leslie AJ, Chetty P, Brander C, Holmes EC, Chang SC, Feeney ME, Addo MM, et al. Immune selection for altered antigen processing leads to cytotoxic T lymphocyte escape in chronic HIV-1 infection. The Journal of experimental medicine. 2004;199:905–915. doi: 10.1084/jem.20031982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards BH, Bansal A, Sabbaj S, Bakari J, Mulligan MJ, Goepfert PA. Magnitude of functional CD8+ T-cell responses to the gag protein of human immunodeficiency virus type 1 correlates inversely with viral load in plasma. Journal of virology. 2002;76:2298–2305. doi: 10.1128/jvi.76.5.2298-2305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elahi S, Dinges WL, Lejarcegui N, Laing KJ, Collier AC, Koelle DM, McElrath MJ, Horton H. Protective HIV-specific CD8+ T cells evade Treg cell suppression. Nature medicine. 2011;17:989–995. doi: 10.1038/nm.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emu B, Sinclair E, Hatano H, Ferre A, Shacklett B, Martin JN, McCune JM, Deeks SG. HLA class I-restricted T-cell responses may contribute to the control of human immunodeficiency virus infection, but such responses are not always necessary for long-term virus control. Journal of virology. 2008;82:5398–5407. doi: 10.1128/JVI.02176-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerberg T, Cerottini JC, Michielin O. Structural prediction of peptides bound to MHC class I. J Mol Biol. 2006;356:521–546. doi: 10.1016/j.jmb.2005.11.059. [DOI] [PubMed] [Google Scholar]
- Feeney ME, Tang Y, Roosevelt KA, Leslie AJ, McIntosh K, Karthas N, Walker BD, Goulder PJ. Immune escape precedes breakthrough human immunodeficiency virus type 1 viremia and broadening of the cytotoxic T-lymphocyte response in an HLA-B27-positive long-term-nonprogressing child. Journal of virology. 2004;78:8927–8930. doi: 10.1128/JVI.78.16.8927-8930.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellay J, Ge D, Shianna KV, Colombo S, Ledergerber B, Cirulli ET, Urban TJ, Zhang K, Gumbs CE, Smith JP, et al. Common genetic variation and the control of HIV-1 in humans. PLoS Genet. 2009;5:e1000791. doi: 10.1371/journal.pgen.1000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellay J, Shianna KV, Ge D, Colombo S, Ledergerber B, Weale M, Zhang K, Gumbs C, Castagna A, Cossarizza A, et al. A whole-genome association study of major determinants for host control of HIV-1. Science (New York, N.Y. 2007;317:944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Villanueva PO, Yunis EJ, Delgado JC, Vittinghoff E, Buchbinder S, Leung JY, Uglialoro AM, Clavijo OP, Rosenberg ES, Kalams SA, et al. Control of HIV-1 viremia and protection from AIDS are associated with HLA-Bw4 homozygosity. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5140–5145. doi: 10.1073/pnas.071548198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Bashirova A, Iversen AK, Phair J, Goedert JJ, Buchbinder S, Hoots K, Vlahov D, Altfeld M, O'Brien SJ, Carrington M. AIDS restriction HLA allotypes target distinct intervals of HIV-1 pathogenesis. Nature medicine. 2005;11:1290–1292. doi: 10.1038/nm1333. [DOI] [PubMed] [Google Scholar]
- Gao X, Nelson GW, Karacki P, Martin MP, Phair J, Kaslow R, Goedert JJ, Buchbinder S, Hoots K, Vlahov D, et al. Effect of a single amino acid change in MHC class I molecules on the rate of progression to AIDS. The New England journal of medicine. 2001;344:1668–1675. doi: 10.1056/NEJM200105313442203. [DOI] [PubMed] [Google Scholar]
- Geldmacher C, Currier JR, Herrmann E, Haule A, Kuta E, McCutchan F, Njovu L, Geis S, Hoffmann O, Maboko L, et al. CD8 T-cell recognition of multiple epitopes within specific Gag regions is associated with maintenance of a low steady-state viremia in human immunodeficiency virus type 1-seropositive patients. Journal of virology. 2007;81:2440–2448. doi: 10.1128/JVI.01847-06. genome.gov/26525384, h.w. National Human Genome Research Project. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulder PJ, Brander C, Tang Y, Tremblay C, Colbert RA, Addo MM, Rosenberg ES, Nguyen T, Allen R, Trocha A, et al. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature. 2001;412:334–338. doi: 10.1038/35085576. [DOI] [PubMed] [Google Scholar]
- Goulder PJ, Phillips RE, Colbert RA, McAdam S, Ogg G, Nowak MA, Giangrande P, Luzzi G, Morgan B, Edwards A, et al. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nature medicine. 1997;3:212–217. doi: 10.1038/nm0297-212. [DOI] [PubMed] [Google Scholar]
- Hersperger AR, Martin JN, Shin LY, Sheth PM, Kovacs CM, Cosma GL, Makedonas G, Pereyra F, Walker BD, Kaul R, et al. Increased HIV-specific CD8+ T-cell cytotoxic potential in HIV elite controllers is associated with T-bet expression. Blood. 2011;117:3799–3808. doi: 10.1182/blood-2010-12-322727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton H, Frank I, Baydo R, Jalbert E, Penn J, Wilson S, McNevin JP, McSweyn MD, Lee D, Huang Y, et al. Preservation of T cell proliferation restricted by protective HLA alleles is critical for immune control of HIV-1 infection. J Immunol. 2006;177:7406–7415. doi: 10.4049/jimmunol.177.10.7406. [DOI] [PubMed] [Google Scholar]
- Huang J, Burke PS, Cung TD, Pereyra F, Toth I, Walker BD, Borges L, Lichterfeld M, Yu XG. Leukocyte Immunoglobulin-Like Receptors maintain unique antigen-presenting properties of circulating myeloid dendritic cells in HIV-1 elite controllers. Journal of virology. 2010 doi: 10.1128/JVI.01009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Goedert JJ, Sundberg EJ, Cung TD, Burke PS, Martin MP, Preiss L, Lifson J, Lichterfeld M, Carrington M, Yu XG. HLA-B*35-Px-mediated acceleration of HIV-1 infection by increased inhibitory immunoregulatory impulses. The Journal of experimental medicine. 2009;206:2959–2966. doi: 10.1084/jem.20091386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Nei M. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature. 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- Iglesias MC, Almeida JR, Fastenackels S, van Bockel DJ, Hashimoto M, Venturi V, Gostick E, Urrutia A, Wooldridge L, Clement M, et al. Escape from highly effective public CD8+ T-cell clonotypes by HIV. Blood. 2011;118:2138–2149. doi: 10.1182/blood-2011-01-328781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii H, Kawada M, Tsukamoto T, Yamamoto H, Matsuoka S, Shiino T, Takeda A, Inoue M, Iida A, Hara H, et al. Impact of vaccination on cytotoxic T lymphocyte immunodominance and cooperation against simian immunodeficiency virus replication in rhesus macaques. Journal of virology. 2012;86:738–745. doi: 10.1128/JVI.06226-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaslow RA, Carrington M, Apple R, Park L, Munoz A, Saah AJ, Goedert JJ, Winkler C, O'Brien SJ, Rinaldo C, et al. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nature medicine. 1996;2:405–411. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- Kawada M, Tsukamoto T, Yamamoto H, Iwamoto N, Kurihara K, Takeda A, Moriya C, Takeuchi H, Akari H, Matano T. Gag-specific cytotoxic T-lymphocyte-based control of primary simian immunodeficiency virus replication in a vaccine trial. Journal of virology. 2008;82:10199–10206. doi: 10.1128/JVI.01103-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawada M, Tsukamoto T, Yamamoto H, Takeda A, Igarashi H, Watkins DI, Matano T. Long-term control of simian immunodeficiency virus replication with central memory CD4+ T-cell preservation after nonsterile protection by a cytotoxic T-lymphocyte-based vaccine. Journal of virology. 2007;81:5202–5211. doi: 10.1128/JVI.02881-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima Y, Kuse N, Gatanaga H, Naruto T, Fujiwara M, Dohki S, Akahoshi T, Maenaka K, Goulder P, Oka S, Takiguchi M. Long-term control of HIV-1 in hemophiliacs carrying slow-progressing allele HLA-B*5101. Journal of virology. 2010;84:7151–7160. doi: 10.1128/JVI.00171-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima Y, Pfafferott K, Frater J, Matthews P, Payne R, Addo M, Gatanaga H, Fujiwara M, Hachiya A, Koizumi H, et al. Adaptation of HIV-1 to human leukocyte antigen class I. Nature. 2009;458:641–645. doi: 10.1038/nature07746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiepiela P, Leslie AJ, Honeyborne I, Ramduth D, Thobakgale C, Chetty S, Rathnavalu P, Moore C, Pfafferott KJ, Hilton L, et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–775. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- Kiepiela P, Ngumbela K, Thobakgale C, Ramduth D, Honeyborne I, Moodley E, Reddy S, de Pierres C, Mncube Z, Mkhwanazi N, et al. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nature medicine. 2007;13:46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- Klein MR, van Baalen CA, Holwerda AM, Kerkhof Garde SR, Bende RJ, Keet IP, Eeftinck-Schattenkerk JK, Osterhaus AD, Schuitemaker H, Miedema F. Kinetics of Gag-specific cytotoxic T lymphocyte responses during the clinical course of HIV-1 infection: a longitudinal analysis of rapid progressors and long-term asymptomatics. The Journal of experimental medicine. 1995;181:1365–1372. doi: 10.1084/jem.181.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloverpris HN, Harndahl M, Leslie AJ, Carlson JM, Ismail N, van der Stok M, Huang KH, Chen F, Riddell L, Steyn D, et al. HIV control through a single nucleotide on the HLA-B locus. Journal of virology. 2012 doi: 10.1128/JVI.01020-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler RN, Walsh AM, Saathoff E, Tovanabutra S, Arroyo MA, Currier JR, Maboko L, Hoelscher M, Robb ML, Michael NL, et al. Class I HLA-A*7401 is associated with protection from HIV-1 acquisition and disease progression in Mbeya, Tanzania. The Journal of infectious diseases. 2010;202:1562–1566. doi: 10.1086/656913. [DOI] [PubMed] [Google Scholar]
- Kosmrlj A, Read EL, Qi Y, Allen TM, Altfeld M, Deeks SG, Pereyra F, Carrington M, Walker BD, Chakraborty AK. Effects of thymic selection of the T-cell repertoire on HLA class[thinsp]I-associated control of HIV infection. Nature. 2010;465:350–354. doi: 10.1038/nature08997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni S, Savan R, Qi Y, Gao X, Yuki Y, Bass SE, Martin MP, Hunt P, Deeks SG, Telenti A, et al. Differential microRNA regulation of HLA-C expression and its association with HIV control. Nature. 2011;472:495–498. doi: 10.1038/nature09914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaro E, Godfrey SB, Stamegna P, Ogbechie T, Kerrigan C, Zhang M, Walker BD, Le Gall S. Differential HIV epitope processing in monocytes and CD4 T cells affects cytotoxic T lymphocyte recognition. The Journal of infectious diseases. 2009;200:236–243. doi: 10.1086/599837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaryan A, Song W, Lobashevsky E, Tang J, Shrestha S, Zhang K, McNicholl JM, Gardner LI, Wilson CM, Klein RS, et al. The influence of human leukocyte antigen class I alleles and their population frequencies on human immunodeficiency virus type 1 control among African Americans. Hum Immunol. 2011;72:312–318. doi: 10.1016/j.humimm.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gall S, Stamegna P, Walker BD. Portable flanking sequences modulate CTL epitope processing. The Journal of clinical investigation. 2007;117:3563–3575. doi: 10.1172/JCI32047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie A, Matthews PC, Listgarten J, Carlson JM, Kadie C, Ndung'u T, Brander C, Coovadia H, Walker BD, Heckerman D, Goulder PJ. Additive contribution of HLA class I alleles in the immune control of HIV-1 infection. Journal of virology. 2010 doi: 10.1128/JVI.00320-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie A, Price DA, Mkhize P, Bishop K, Rathod A, Day C, Crawford H, Honeyborne I, Asher TE, Luzzi G, et al. Differential selection pressure exerted on HIV by CTL targeting identical epitopes but restricted by distinct HLA alleles from the same HLA supertype. J Immunol. 2006;177:4699–4708. doi: 10.4049/jimmunol.177.7.4699. [DOI] [PubMed] [Google Scholar]
- Lichterfeld M, Kaufmann DE, Yu XG, Mui SK, Addo MM, Johnston MN, Cohen D, Robbins GK, Pae E, Alter G, et al. Loss of HIV-1-specific CD8+ T cell proliferation after acute HIV-1 infection and restoration by vaccine-induced HIV-1-specific CD4+ T cells. The Journal of experimental medicine. 2004;200:701–712. doi: 10.1084/jem.20041270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichterfeld M, Yu XG. The emerging role of leukocyte immunoglobulin-like receptors (LILRs) in HIV-1 infection. J Leukoc Biol. 2012;91:27–33. doi: 10.1189/jlb.0811442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limou S, Le Clerc S, Coulonges C, Carpentier W, Dina C, Delaneau O, Labib T, Taing L, Sladek R, Deveau C, et al. Genomewide Association Study of an AIDS-Nonprogression Cohort Emphasizes the Role Played by HLA Genes (ANRS Genomewide Association Study 02). The Journal of infectious diseases. 2009;199:419–426. doi: 10.1086/596067. [DOI] [PubMed] [Google Scholar]
- Madden DR, Garboczi DN, Wiley DC. The antigenic identity of peptide-MHC complexes: a comparison of the conformations of five viral peptides presented by HLA-A2. Cell. 1993;75:693–708. doi: 10.1016/0092-8674(93)90490-h. [DOI] [PubMed] [Google Scholar]
- Maness NJ, Yant LJ, Chung C, Loffredo JT, Friedrich TC, Piaskowski SM, Furlott J, May GE, Soma T, Leon EJ, et al. Comprehensive immunological evaluation reveals surprisingly few differences between elite controller and progressor Mamu-B*17-positive Simian immunodeficiency virus-infected rhesus macaques. Journal of virology. 2008;82:5245–5254. doi: 10.1128/JVI.00292-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh SGE, Parham P, Barber LD. The HLA factsbook. Academic Press; San Diego: 2000. [Google Scholar]
- Martinez-Picado J, Prado JG, Fry EE, Pfafferott K, Leslie A, Chetty S, Thobakgale C, Honeyborne I, Crawford H, Matthews P, et al. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. Journal of virology. 2006;80:3617–3623. doi: 10.1128/JVI.80.7.3617-3623.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masemola A, Mashishi T, Khoury G, Mohube P, Mokgotho P, Vardas E, Colvin M, Zijenah L, Katzenstein D, Musonda R, et al. Hierarchical targeting of subtype C human immunodeficiency virus type 1 proteins by CD8+ T cells: correlation with viral load. Journal of virology. 2004;78:3233–3243. doi: 10.1128/JVI.78.7.3233-3243.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matano T, Kobayashi M, Igarashi H, Takeda A, Nakamura H, Kano M, Sugimoto C, Mori K, Iida A, Hirata T, et al. Cytotoxic T lymphocyte-based control of simian immunodeficiency virus replication in a preclinical AIDS vaccine trial. The Journal of experimental medicine. 2004;199:1709–1718. doi: 10.1084/jem.20040432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews PC, Adland E, Listgarten J, Leslie A, Mkhwanazi N, Carlson JM, Harndahl M, Stryhn A, Payne RP, Ogwu A, et al. HLA-A*7401-mediated control of HIV viremia is independent of its linkage disequilibrium with HLA-B*5703. J Immunol. 2011;186:5675–5686. doi: 10.4049/jimmunol.1003711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren PJ, Ripke S, Pelak K, Weintrob AC, Patsopoulos NA, Jia X, Erlich RL, Lennon NJ, Kadie CM, Heckerman D, et al. Fine-mapping classical HLA variation associated with durable host control of HIV-1 infection in African Americans. Hum Mol Genet. 2012 doi: 10.1093/hmg/dds226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza D, Johnson SA, Peterson BA, Natarajan V, Salgado M, Dewar RL, Burbelo PD, Doria-Rose NA, Graf EH, Greenwald JH, et al. Comprehensive analysis of unique cases with extraordinary control over HIV replication. Blood. 2012;119:4645–4655. doi: 10.1182/blood-2011-10-381996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migueles SA, Laborico AC, Shupert WL, Sabbaghian MS, Rabin R, Hallahan CW, Baarle DV, Kostense S, Miedema F, McLaughlin M, et al. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nature immunology. 2002;3:1061–1068. doi: 10.1038/ni845. [DOI] [PubMed] [Google Scholar]
- Migueles SA, Osborne CM, Royce C, Compton AA, Joshi RP, Weeks KA, Rood JE, Berkley AM, Sacha JB, Cogliano-Shutta NA, et al. Lytic granule loading of CD8+ T cells is required for HIV-infected cell elimination associated with immune control. Immunity. 2008;29:1009–1021. doi: 10.1016/j.immuni.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migueles SA, Sabbaghian MS, Shupert WL, Bettinotti MP, Marincola FM, Martino L, Hallahan CW, Selig SM, Schwartz D, Sullivan J, Connors M. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:2709–2714. doi: 10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura T, Brockman MA, Brumme ZL, Brumme CJ, Pereyra F, Trocha A, Block BL, Schneidewind A, Allen TM, Heckerman D, Walker BD. HLA-associated alterations in replication capacity of chimeric NL4-3 viruses carrying gag-protease from elite controllers of human immunodeficiency virus type 1. Journal of virology. 2009a;83:140–149. doi: 10.1128/JVI.01471-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura T, Brockman MA, Schneidewind A, Lobritz M, Pereyra F, Rathod A, Block BL, Brumme ZL, Brumme CJ, Baker B, et al. HLA-B57/B*5801 human immunodeficiency virus type 1 elite controllers select for rare gag variants associated with reduced viral replication capacity and strong cytotoxic T-lymphocyte [corrected] recognition. Journal of virology. 2009b;83:2743–2755. doi: 10.1128/JVI.02265-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura T, Brumme CJ, Brockman MA, Brumme ZL, Pereyra F, Block BL, Trocha A, John M, Mallal S, Harrigan PR, Walker BD. HLA-associated viral mutations are common in human immunodeficiency virus type 1 elite controllers. Journal of virology. 2009c;83:3407–3412. doi: 10.1128/JVI.02459-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudd PA, Ericsen AJ, Walsh AD, Leon EJ, Wilson NA, Maness NJ, Friedrich TC, Watkins DI. CD8+ T cell escape mutations in simian immunodeficiency virus SIVmac239 cause fitness defects in vivo, and many revert after transmission. Journal of virology. 2011;85:12804–12810. doi: 10.1128/JVI.05841-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudd PA, Watkins DI. Nature. 2012 In press. [Google Scholar]
- Ngumbela KC, Day CL, Mncube Z, Nair K, Ramduth D, Thobakgale C, Moodley E, Reddy S, de Pierres C, Mkhwanazi N, et al. Targeting of a CD8 T cell env epitope presented by HLA-B*5802 is associated with markers of HIV disease progression and lack of selection pressure. AIDS research and human retroviruses. 2008;24:72–82. doi: 10.1089/aid.2007.0124. [DOI] [PubMed] [Google Scholar]
- Norstrom MM, Buggert M, Tauriainen J, Hartogensis W, Prosperi MC, Wallet MA, Hecht FM, Salemi M, Karlsson AC. Combination of immune and viral factors distinguish low-risk versus high-risk HIV-1 disease progression in HLA-B*5701 subjects. Journal of virology. 2012 doi: 10.1128/JVI.01165-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitsky V, Gilbert P, Peter T, McLane MF, Gaolekwe S, Rybak N, Thior I, Ndung'u T, Marlink R, Lee TH, Essex M. Association between virus-specific T-cell responses and plasma viral load in human immunodeficiency virus type 1 subtype C infection. Journal of virology. 2003;77:882–890. doi: 10.1128/JVI.77.2.882-890.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg GS, Jin X, Bonhoeffer S, Dunbar PR, Nowak MA, Monard S, Segal JP, Cao Y, Rowland-Jones SL, Cerundolo V, et al. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science (New York, N.Y. 1998;279:2103–2106. doi: 10.1126/science.279.5359.2103. [DOI] [PubMed] [Google Scholar]
- Parsons MS, Wren L, Isitman G, Navis M, Stratov I, Bernard NF, Kent SJ. HIV infection abrogates the functional advantage of natural killer cells educated through KIR3DL1/HLA-Bw4 interactions to mediate anti-HIV antibody-dependent cellular cytotoxicity. Journal of virology. 2012;86:4488–4495. doi: 10.1128/JVI.06112-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne RP, Kloverpris H, Sacha JB, Brumme Z, Brumme C, Buus S, Sims S, Hickling S, Riddell L, Chen F, et al. Efficacious early antiviral activity of HIV Gag- and Pol-specific HLA-B 2705-restricted CD8+ T cells. Journal of virology. 2010;84:10543–10557. doi: 10.1128/JVI.00793-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelak K, Goldstein DB, Walley NM, Fellay J, Ge D, Shianna KV, Gumbs C, Gao X, Maia JM, Cronin KD, et al. Host determinants of HIV-1 control in African Americans. The Journal of infectious diseases. 2010;201:1141–1149. doi: 10.1086/651382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereyra F, Addo MM, Kaufmann DE, Liu Y, Miura T, Rathod A, Baker B, Trocha A, Rosenberg R, Mackey E, et al. Genetic and immunologic heterogeneity among persons who control HIV infection in the absence of therapy. The Journal of infectious diseases. 2008;197:563–571. doi: 10.1086/526786. [DOI] [PubMed] [Google Scholar]
- Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PI, Walker BD, Ripke S, Brumme CJ, Pulit SL, Carrington M, et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science (New York, N.Y. 2010;330:1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riviere Y, McChesney MB, Porrot F, Tanneau-Salvadori F, Sansonetti P, Lopez O, Pialoux G, Feuillie V, Mollereau M, Chamaret S, et al. Gag-specific cytotoxic responses to HIV type 1 are associated with a decreased risk of progression to AIDS-related complex or AIDS. AIDS research and human retroviruses. 1995;11:903–907. doi: 10.1089/aid.1995.11.903. [DOI] [PubMed] [Google Scholar]
- Robinson J, Mistry K, McWilliam H, Lopez R, Parham P, Marsh SG. The IMGT/HLA database. Nucleic acids research. 2011;39:D1171–1176. doi: 10.1093/nar/gkq998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacha JB, Chung C, Rakasz EG, Spencer SP, Jonas AK, Bean AT, Lee W, Burwitz BJ, Stephany JJ, Loffredo JT, et al. Gag-specific CD8+ T lymphocytes recognize infected cells before AIDS-virus integration and viral protein expression. J Immunol. 2007a;178:2746–2754. doi: 10.4049/jimmunol.178.5.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacha JB, Chung C, Reed J, Jonas AK, Bean AT, Spencer SP, Lee W, Vojnov L, Rudersdorf R, Friedrich TC, et al. Pol-specific CD8+ T cells recognize simian immunodeficiency virus-infected cells prior to Nef-mediated major histocompatibility complex class I downregulation. Journal of virology. 2007b;81:11703–11712. doi: 10.1128/JVI.00926-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sáez-Cirión A, Lacabaratz C, Lambotte O, Versmisse P, Urrutia A, Boufassa F, Barré-Sinoussi F, Delfraissy J-F, Sinet M, Pancino G, Venet A. HIV controllers exhibit potent CD8 T cell capacity to suppress HIV infection ex vivo and peculiar cytotoxic T lymphocyte activation phenotype. Proceedings of the National Academy of Sciences. 2007;104:6776–6781. doi: 10.1073/pnas.0611244104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidewind A, Brockman MA, Sidney J, Wang YE, Chen H, Suscovich TJ, Li B, Adam RI, Allgaier RL, Mothe BR, et al. Structural and Functional Constraints Limit Options for Cytotoxic T-Lymphocyte Escape in the Immunodominant HLA-B27-Restricted Epitope in Human Immunodeficiency Virus Type 1 Capsid. J. Virol. 2008;82:5594–5605. doi: 10.1128/JVI.02356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidewind A, Brockman MA, Yang R, Adam RI, Li B, Le Gall S, Rinaldo CR, Craggs SL, Allgaier RL, Power KA, et al. Escape from the dominant HLA-B27-restricted cytotoxic T-lymphocyte response in Gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. Journal of virology. 2007;81:12382–12393. doi: 10.1128/JVI.01543-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha S, Aissani B, Song W, Wilson CM, Kaslow RA, Tang J. Host genetics and HIV-1 viral load set-point in African-Americans. AIDS (London, England) 2009;23:673–677. doi: 10.1097/QAD.0b013e328325d414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidney J, Peters B, Frahm N, Brander C, Sette A. HLA class I supertypes: a revised and updated classification. BMC immunology. 2008;9:1. doi: 10.1186/1471-2172-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steers NJ, Currier JR, Kijak GH, di Targiani RC, Saxena A, Marovich MA, Kim JH, Michael NL, Alving CR, Rao M. Cell type-specific proteasomal processing of HIV-1 Gag-p24 results in an altered epitope repertoire. Journal of virology. 2011;85:1541–1553. doi: 10.1128/JVI.01790-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Jones GB, Gillespie G, Overton IM, Kaul R, Roche P, McMichael AJ, Rowland-Jones S, Jones EY. Structures of three HIV-1 HLA-B*5703-peptide complexes and identification of related HLAs potentially associated with long-term nonprogression. J Immunol. 2005;175:2459–2468. doi: 10.4049/jimmunol.175.4.2459. [DOI] [PubMed] [Google Scholar]
- Streeck H, Lichterfeld M, Alter G, Meier A, Teigen N, Yassine-Diab B, Sidhu HK, Little S, Kelleher A, Routy JP, et al. Recognition of a defined region within p24 gag by CD8+ T cells during primary human immunodeficiency virus type 1 infection in individuals expressing protective HLA class I alleles. Journal of virology. 2007;81:7725–7731. doi: 10.1128/JVI.00708-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Malhotra R, Song W, Brill I, Hu L, Farmer PK, Mulenga J, Allen S, Hunter E, Kaslow RA. Human leukocyte antigens and HIV type 1 viral load in early and chronic infection: predominance of evolving relationships. PloS one. 2010;5:e9629. doi: 10.1371/journal.pone.0009629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R, Apps R, Qi Y, Gao X, Male V, O'HUigin C, O'Connor G, Ge D, Fellay J, Martin JN, et al. HLA-C cell surface expression and control of HIV/AIDS correlate with a variant upstream of HLA-C. Nature genetics. 2009;41:1290–1294. doi: 10.1038/ng.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyer RM, McNevin J, Liu Y, Zhang SC, Krizan RW, Abraha A, Tebit DM, Zhao H, Avila S, Lobritz MA, et al. Variable fitness impact of HIV-1 escape mutations to cytotoxic T lymphocyte (CTL) response. PLoS pathogens. 2009;5:e1000365. doi: 10.1371/journal.ppat.1000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto T, Takeda A, Yamamoto T, Yamamoto H, Kawada M, Matano T. Impact of cytotoxic-T-lymphocyte memory induction without virus-specific CD4+ T-Cell help on control of a simian immunodeficiency virus challenge in rhesus macaques. Journal of virology. 2009;83:9339–9346. doi: 10.1128/JVI.01120-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine LE, Loffredo JT, Bean AT, Leon EJ, MacNair CE, Beal DR, Piaskowski SM, Klimentidis YC, Lank SM, Wiseman RW, et al. Infection with “escaped” virus variants impairs control of simian immunodeficiency virus SIVmac239 replication in Mamu-B*08-positive macaques. Journal of virology. 2009;83:11514–11527. doi: 10.1128/JVI.01298-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine LE, Watkins DI. Relevance of studying T cell responses in SIV-infected rhesus macaques. Trends Microbiol. 2008;16:605–611. doi: 10.1016/j.tim.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang OO, Kalams SA, Rosenzweig M, Trocha A, Jones N, Koziel M, Walker BD, Johnson RP. Efficient lysis of human immunodeficiency virus type 1-infected cells by cytotoxic T lymphocytes. Journal of virology. 1996;70:5799–5806. doi: 10.1128/jvi.70.9.5799-5806.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang OO, Kalams SA, Trocha A, Cao H, Luster A, Johnson RP, Walker BD. Suppression of human immunodeficiency virus type 1 replication by CD8+ cells: evidence for HLA class I-restricted triggering of cytolytic and noncytolytic mechanisms. Journal of virology. 1997;71:3120–3128. doi: 10.1128/jvi.71.4.3120-3128.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang OO, Nguyen PT, Kalams SA, Dorfman T, Gottlinger HG, Stewart S, Chen IS, Threlkeld S, Walker BD. Nef-mediated resistance of human immunodeficiency virus type 1 to antiviral cytotoxic T lymphocytes. Journal of virology. 2002;76:1626–1631. doi: 10.1128/JVI.76.4.1626-1631.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang OO, Sarkis PT, Ali A, Harlow JD, Brander C, Kalams SA, Walker BD. Determinant of HIV-1 mutational escape from cytotoxic T lymphocytes. The Journal of experimental medicine. 2003;197:1365–1375. doi: 10.1084/jem.20022138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkernagel RM, Doherty PC. Restriction of in vitro T cell-mediated cytotoxicity in lymphocytic choriomeningitis within a syngeneic or semiallogeneic system. Nature. 1974;248:701–702. doi: 10.1038/248701a0. [DOI] [PubMed] [Google Scholar]
- Zuniga R, Lucchetti A, Galvan P, Sanchez S, Sanchez C, Hernandez A, Sanchez H, Frahm N, Linde CH, Hewitt HS, et al. Relative dominance of Gag p24-specific cytotoxic T lymphocytes is associated with human immunodeficiency virus control. Journal of virology. 2006;80:3122–3125. doi: 10.1128/JVI.80.6.3122-3125.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]