Summary
Platensimycin (PTM) and platencin (PTN) are potent inhibitors of bacterial fatty acid synthases and have emerged as promising antibacterial drug leads. We previously characterized the PTM and PTN biosynthetic machineries in the Streptomyces platensis producers. We now identify two mechanisms for PTM and PTN resistance in the S. platensis producers - the ptmP3 or ptnP3 gene within the PTM-PTN or PTN biosynthetic cluster and the fabF gene within the fatty acid synthase locus. PtmP3/PtnP3 and FabF confer PTM and PTN resistance by target replacement and target modification, respectively. PtmP3/PtnP3 also represents an unprecedented mechanism for fatty acid biosynthesis in which FabH and FabF are functionally replaced by a single condensing enzyme. These findings challenge the current paradigm for fatty acid biosynthesis and should be considered in future development of effective therapeutics targeting fatty acid synthase.
Introduction
The rapid development of antibiotics beginning over sixty years ago was one of the most important advancements in the history of treating human disease. Pathogenic bacteria, however, remain a threat to human health, and the inevitable tendency of bacteria to evolve toward resistance to present therapeutics is well recognized and cause for concern. The discovery of platensimycin (PTM) and platencin (PTN), the second entirely new class of natural antibiotics with a novel mode of action discovered in the past four decades, represents an encouraging step toward addressing a pharmaceutical pipeline that is inadequately producing a sufficient number of drugs to treat newly emergent and resistant pathogenic strains (Taubes, 2008; Wang et al., 2006; Wang et al., 2007). It has been reported recently that PTM is also a potent and selective inhibitor of mammalian fatty acid synthase (Wu et al., 2011).
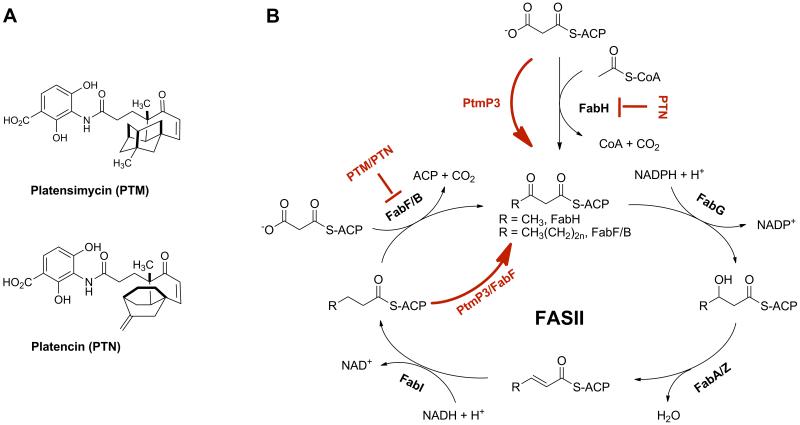
PTM and PTN act by inhibiting bacterial fatty acid synthase (FASII) (Fig. 1). PTM selectively inhibits β-ketoacyl-acyl carrier protein (ACP) synthases (KAS) I/II, or FabB/F, while PTN is a dual inhibitor of both FabF and KAS III, or FabH (Wang et al., 2006; Wang et al., 2007). These FASII enzymes catalyze unique condensation reactions, representing essential steps for chain initiation (by FabH) and elongation (by FabB/F) in fatty acid biosynthesis (Fig. 1B). Importantly, both PTM and PTN show broad spectrum antibacterial activity, do not exhibit cross-resistance to clinically relevant pathogens, and have shown in vivo efficacy without toxicity (Wang et al., 2006; Wang et al., 2007). While poor pharmacokinetic profiles have limited development of these compounds into clinically significant therapeutics, synthetic strategies toward PTM and PTN analogs and isolation of natural congeners from native and engineered producing strains aim to overcome these limitations (Martens and Demain, 2011; Saleem et al., 2011; Shen et al., 2009; Smanski et al., 2012; Yu et al., 2010; Yu et al., 2013; Zhang et al., 2011).
Fig. 1.
(A) Structures of platensimycin (PTM) and platencin (PTN) and (B) the bacterial fatty acid synthesis cycle (FASII). Highlighted in red are (i) PTM inhibiting FabF/B, the elongation step of FASII, and PTN dually inhibiting FabF and FabH, the initiation step FASII, and (ii) the two complementary mechanisms of PTM and PTN resistance in S. platensis by target replacement (i.e., FabF and FabH by PtmP3) and target modification (i.e., FabF by a PTM-insensitive variant). See also Figure S1.
Among our studies on the biosynthetic gene clusters responsible for PTM and PTN production in Streptomyces platensis strains (Smanski et al., 2011), we sought to identify the self-resistance mechanism(s) in these producing organisms. Characterization of the PTM and PTN biosynthetic machineries revealed that S. platensis MA7327 is a PTM and PTN dual producer and therefore must maintain resistance to both PTM and PTN. S. platensis MA7339 is an exclusive PTN producer and must minimally maintain resistance to PTN, but not necessarily to PTM; however, the dual inhibitory nature of PTN suggests that a resistance element for PTN could also confer resistance to PTM. In addition, we previously introduced the “PTM cassette”, a 5.4-kb DNA fragment consisting of five genes present within the PTM-PTN dual-producing cluster but absent within the PTN cluster (Fig. S1A) into the MA7339 strain. This afforded a PTM and PTN dual-producing recombinant strain, confirming that the MA7339 strain maintains resistance to both PTN and PTM (Smanski et al., 2011). Bioinformatics analysis of the open reading frames within both the PTM-PTN dual-producing cluster (S. platensis MA7327) and the PTN cluster (S. platensis MA7339) revealed the same set of four genes – ptmP1/ptmP2/ptmP3/ptmP4 or ptnP1/ptnP2/ptnP3/ptnP4 in the PTM-PTN and PTN clusters, respectively – without apparent roles in biosynthesis or regulation, which might encode elements conferring resistance to PTM, PTN, or both (Fig. S1A and Table S1) (Smanski et al., 2011).
Development of resistance in pathogenic bacteria has widely been attributed to horizontal gene transfer from nonpathogenic bacteria with one potential source being antibiotic-producing bacteria that developed highly effective mechanisms to avoid suicide (D’Costa et al., 2006; Hopwood, 2007). This hypothesis is supported by examples in vancomycin and aminoglycoside clinical resistance mechanisms (Benveniste and Davies, 1973; Marshall et al., 1997; Pootoolal et al., 2002). Understanding self-resistance mechanisms within PTM and PTN producing organisms therefore is imperative for predicting, determining, and thereby managing, potential resistance that could develop with any future use of PTM, PTN, or their derivatives in the clinic.
Here we report identification of PtmP3/PtnP3 as the major self-resistance conferring element in the native PTM-PTN dual producer S. platensis MA7327 and the PTN producer S. platensis MA7339. We demonstrate that PtmP3 can functionally replace both the FabF and FabH enzymes of the housekeeping FASII in Streptomyces species. Investigation of the FabF and FabH enzymes within the FASII locus of the S. platensis strains further reveals that FabF is also resistant to PTM, providing a second form of self-resistance to PTM, while FabH remains sensitive to PTN. PtmP3 and FabF in the PTM and PTN producing S. platensis species therefore confer PTM and PTN resistance by target replacement (i.e., FabF and FabH by PtmP3) and target modification (i.e., FabF by a PTM-insensitive variant), respectively (Fig. 1B). PtmP3 represents an unprecedented mechanism for fatty acid biosynthesis in which FabH and FabF are functionally replaced by a single condensing enzyme. These findings challenge the current paradigm for fatty acid biosynthesis and should be considered in future development of effective therapeutics targeting FASII.
Results
Bioinformatics analysis reveals candidates for self-resistance genes
Four genes within the PTM and PTN biosynthetic clusters – ptmP1/ptmP2/ptmP3/ptmP4 or ptnP1/ptnP2/ptnP3/ptnP4 – were previously ruled out as contributors to, or regulators of, the biosynthetic reactions leading to production of PTM and PTN (Smanski et al., 2011). Bioinformatics analysis assigned these genes putative roles in self-resistance based on their homology to enzymes of known functions (Fig. S1A and Table S1) (Smanski et al., 2011). The N-terminal regions of PtmP1/PtnP1 show sequence similarity to KAS III condensing enzymes; however, while this region retains asparagine and histidine in a conserved Cys-Asn-His catalytic triad, it lacks the cysteine residue essential in forming the acyl-enzyme intermediate during the condensation reaction (White et al., 2005). PtmP2/PtnP2 show sequence similarity to acetyl-CoA acetyltransferases, belonging to the thiolase family of enzymes. Interestingly, structural features, including an N-terminal region containing a cysteine residue (Cys85) and a C-terminal region containing a histidine residue (His338), align with conserved active site regions in both thiolases and KAS condensing enzymes. However, PtmP2/PtnP2 lack additional key active site residues, critical for either thiolase or condensing enzyme activity: a C-terminal catalytic cysteine residue for type II thiolase activity or a second C-terminal catalytic His/Asn residue for condensing enzyme activity (Huang et al., 1998; Kursula et al., 2002). PtmP3/PtnP3 are FabF (KAS II) homologs, retaining the three key residues in a Cys-His-His catalytic triad conserved among KAS I/II enzymes (Fig. S2) (White et al., 2005). The similarity of PtmP1-P3/PtnP1-P3 to condensing enzymes, the targets of PTM and PTN, led to their classification as putative resistance elements. Finally, PtmP4/PtnP4 show high similarity to members of the major facilitator superfamily, suggesting a role in efflux activity and possible resistance (Kumar and Schweizer, 2005).
Streptomyces species are sensitive to both PTM and PTN
In order to identify a suitable naïve host for resistance studies, the susceptibility of various Streptomyces strains to PTM and PTN was determined using a standard disk diffusion assay (Supplementary Experimental Procedures). Clear zones of growth inhibition were observed around discs impregnated with PTM or PTN on R2YE agar plates inoculated with Streptomyces albus J1074 and Streptomyces avermitilis SUK-A4, while faint zones of inhibition were observed on plates inoculated with Streptomyces lividans K4-114 and Streptomyces coelicolor CH999 (Fig. S3). S. albus J1074 was selected as a suitable naïve host. Both producing strains of S. platensis MA7327 and MA7339 were confirmed to be highly resistant to PTM and PTN. No zones of inhibition were observed around discs impregnated with high concentrations of either molecule (Fig. S3).
Expression of ptmP3 in naïve S. albus J1074 confers resistance to both PTM and PTN
We used an in vivo approach to screen whether predicted self-resistance elements could confer resistance to PTM and/or PTN in a sensitive Streptomyces host. Each candidate resistance element was cloned from S. platensis MA7327 genomic DNA into the shuttle vector pBS12015 and introduced into S. albus J1074 by conjugation (Supplementary Experimental Procedures). This resulted in stable recombinant S. albus strains where the expression vectors containing candidate resistance elements were fully integrated into the Streptomyces chromosome (Table S2). The susceptibility of these recombinant Streptomyces strains to PTM and PTN was determined using the disk diffusion assay described above, and minimum inhibitory concentrations (MIC) (Table 1) were determined using a 96-well plate assay format (Fig. S4). S. albus harboring ptmP3 (S. albus SB12011) showed high levels of resistance to both PTM and PTN, whereas strains harboring ptmP1, ptmP2, or ptmP4 showed identical levels of sensitivity compared with the naïve S. albus J1074 host (Fig. 2A).
Table 1.
PTM and PTN susceptibility of S. platensis and S. albus J1074 wild-type and recombinant strains as measured by minimum inhibitory concentrations (MIC) (μg/mL). See also Figures S4 and S5.
| MIC (μg/mL) |
|||
|---|---|---|---|
| Strain Name | Genotype | PTM | PTN |
| S. platensis (S.p.): | |||
| MA7327 | wild-type | >256 | >64 |
| MA7339 | wild-type | >256 | >64 |
| SB12021 | MA7327/ΔptmU4-P4 | >256 | 32 |
| S. albus (S.a.): | |||
| J1074 | wild-type | 64 | 0.125 |
| SB12011 | pBS12018 (ptmP3) | >256 | >64 |
| SB12017 | pBS12028 [ptmP3(C162L)] | 64 | <0.0625 |
| SB12013 | pBS12020 (S.p. fabF) | >256 | 32 |
| SB12014 | pBS12021 (S.p. fabH) | 32 | <0.0625 |
| SB12018 | pBS12029 (S.a. fabF) | 32 | <0.0625 |
| SB12019 | pBS12030 (S.a. fabH) | 128 | 0.125 |
Fig. 2.
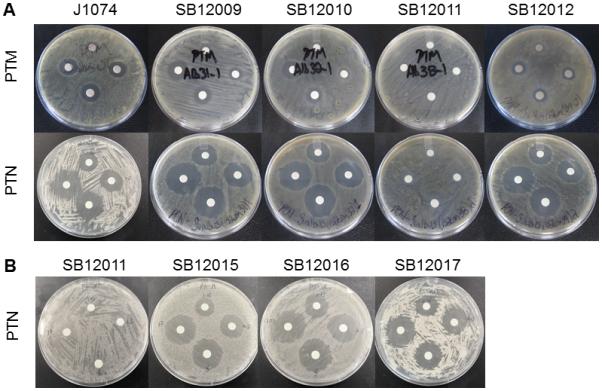
Disk diffusion assays for PTM and PTN susceptibility of recombinant Streptomyces albus J1074 strains that carry varying expression constructs. (A) PTM and PTN susceptibility: J1074 (wild-type), SB12009 (J1074/pBS12016 and ptmP1 expressing), SB12010 (J1074/pBS12017 and ptmP2 expressing), SB12011 (J1074/pBS12018 and ptmP3 expressing), and SB12012 (J1074/pBS12019 and ptmP4 expressing), challenged with 5, 10, 15, and 20 μg of PTM or 1.25, 2.5, 5, and 10 μg of PTN (clockwise from top filter, respectively). (B) PTN susceptibility: SB12011 (J1074/pBS12018 and ptmP3 expressing); SB12015 [J1074/pBS12026 and ptmP3 (C162A) expressing], SB12016 [J1074/pBS12027 and ptmP3 (C162Q) expressing], and SB12017 (J1074/pBS12028 and ptmP3 (C162L) expressing], challenged with 1.25, 2.5, 5, and 10 μg of PTN (clockwise from top filter, respectively). See also Figure S2 and Table S1.
Substitutions of Cys162 in PtmP3 abolish its ability to confer resistance
PtmP3 is homologous to the KAS I/II enzymes of FabB/F, targets of PTM and PTN (Fig. 1B). Sequence alignments with characterized bacterial FabB/FabF enzymes confirm that PtmP3/PtnP3 retain the three residues (Cys162, His300, and His334) in a conserved catalytic triad (Fig. S2). We hypothesized that PtmP3/PtnP3 contribute to resistance by carrying out the FabB/F condensation reaction inhibited by PTM and PTN. Mutating the apparent catalytic cysteine residue in PtmP3 would eliminate its KAS II catalytic activity and thus its resistance to PTM and PTN. Site-directed mutagenesis of PtmP3 gave mutants C162A, C162L and C162Q (Supplementary Methods). All three substitutions completely abolished PTM/PTN resistance and exhibited wild-type host sensitivity (Table 1 and Fig. 2B). These results indicate that Cys162 is essential for conferring resistance to PTM and PTN and support the hypothesis that PtmP3/PtnP3 is a resistant variant of a KAS II that is competent to constitute a functional FASII for fatty acid biosynthesis in Streptomyces.
Deletion of a locus spanning ptmU4-ptmP4 introduces PTN susceptibility, but not PTM susceptibility, in S. platensis MA7327
To assess the essential nature of PtmP3/PtnP3 for PTM/PTN resistance in the producing strains and explore the possibility of additional resistance elements located within the biosynthetic gene cluster or elsewhere within the S. platensis genome, a mutant strain was created in the S. platensis MA7327 background (Supplementary Experimental Procedures). A 6.6-kb locus containing the genes ptmU4, ptmA3, ptmP3, and ptmP4 was deleted to yield S. platensis SB12021 mutant strain, abolishing its ability to produce both PTM and PTN (Figs. S1 and S5A). The susceptibility of SB12021 to PTM and PTN was determined using the 96-well assay. Interestingly, the deletion mutant is slightly more susceptible to PTN, but retains full resistance to PTM (Table 1 and Fig. S4). These data confirm that PtmP3 is both sufficient and necessary for resistance to PTN in the producing strains, but that an additional element capable of providing the strain with PTM resistance, in the absence of PtmP3, also exists.
Expression of the native fabF from S. platensis MA7327 in a naïve Streptomyces host confers resistance to PTM
The fabF gene from the housekeeping FASII locus in S. platensis MA7327 was a likely candidate for contributing to the PTM resistance observed in the ΔptmU4-ptmP4 deletion mutant strain SB12021. The S. platensis fabF was cloned, sequenced, and utilized to construct S. albus SB12013, a recombinant S. albus J1074 strain overexpressing the S. platensis fabF. S. albus SB12018, a recombinant S. albus J1074 strain overexpressing the S. albus fabF was similarly constructed as a control to rule out potential gene dosage effect (Supplementary Experimental Procedures). These strains were assayed for PTM/PTN susceptibility as compared with the naïve S. albus J1074 host. While S. albus SB12018 exhibited similar susceptibility as the naïve S. albus J1074 host to both PTM and PTN, S. albus SB12013 showed full resistance to PTM, but remained sensitive to PTN at a concentration of 32 μg/mL (Table 1 and Fig. S3). These data confirmed that the native fabF in PTM/PTN-producing strains of S. platensis is inherently resistant to PTM (fully) and PTN (partially) and is responsible for a second form of resistance to PTM/PTN in the natural producers. However, while the S. platensis native FabF and PtmP3 are both sufficient to confer natural resistance levels to PTM (>256 μg/mL), PtmP3 confers resistance to PTN at a higher level than S. platensis FabF.
Expression of the native fabH from S. platensis MA7327 in a naïve Streptomyces host does not confer resistance to either PTM or PTN
FabH from the housekeeping FASII locus in S. platensis was similarly investigated. The S. platensis fabH gene was cloned, sequenced, and utilized to construct S. albus SB12014, a recombinant S. albus J1074 strain overexpressing S. platensis fabH, and a recombinant S. albus J1074 strain, SB12019, overexpressing S. albus fabH was also constructed as a control (Supplementary Experimental Procedures). These strains were assayed for PTM/PTN susceptibility as compared with the naïve S. albus J1074 host. We observed that overexpression of fabH was insufficient to confer resistance to either PTM or PTN in the naïve host (Table 1 and Fig. S4).
Either fabF or fabH within the housekeeping FASII locus can be deleted in PTM-PTN producing S. platensis strains
We sought further confirmation that the resistance element, PtmP3, can functionally replace both FASII condensing enzymes, FabF and FabH, as a mechanism of resistance to their inhibition by PTM/PTN in the producing S. platensis strains. To delete fabF or fabH in S. platensis MA7327, a cosmid, pBS12014, harboring the conserved Streptomyces FASII locus (fabD-fabH-acpP-fabF) was isolated from a previously constructed S. platensis MA7327 genomic library (Fig. S1) (Smanski et al., 2009). pBS12014 was modified by λRED-mediated PCR-targeted gene replacement of fabF or fabH with an apramycin resistance cassette to yield cosmids pBS12020 and pBS12021, respectively. To insure constitutive expression of ptmP3, pBS12022, an integrative plasmid in which the expression of ptmP3 is under the control of the ermE* promoter, was first introduced into S. platensis MA7327, to yield S. platensis SB12020. Introduction of cosmids, pBS12020 or pBS12021, into the S. platensis MA7327 wild-type or the SB12020 recombinant strain by conjugation, followed by screening for double-crossover recombinants, yielded deletion of fabF or fabH from the chromosomal FASII locus in S. platensis (Figs. S5E and S5G). Successful isolation of ΔfabF and ΔfabH recombinant S. platensis strains, SB12022, SB12023, SB12024 and SB12025, respectively, demonstrated that fabF and fabH can be readily deleted from the chromosome of PTM-PTN producing strains.
Discussion
PTM and PTN dual producers have adapted two complementary mechanisms to confer PTM and PTN resistance
Acquired resistance to an antibiotic by a microorganism generally falls into one of four major mechanisms: antibiotic modification or destruction, target modification, metabolic pathway circumvention, or modification of the uptake and/or efflux of the antibiotic (Wright, 2011). Target modification impacts the effectiveness of an antimicrobial agent and ultimately provides the means for developing high-level resistance to a specific agent. Point mutations introduced into target genes, which provide a selective advantage, are fundamental target modifications that can confer resistance and a simple single point mutation arising in a target gene can provide clinically relevant resistance (Wehrli, 1993). Typically, more advanced modification, involving several advantageous point mutations or variation through homologous recombination with foreign DNA, is required to alter the antibiotic-target interaction to provide high-level resistance. Other target-mediated mechanisms of resistance include overexpression of the target gene or acquisition of a replacement target gene that has a lower affinity for the antibiotic.
Antibiotic sequestration, as in the case of bleomycin and BlmA (Galm et al., 2005), is another possible mechanism of resistance. The point mutations of Cys162 in PtmP3 were designed to distinguish between target replacement and antibiotic sequestration mechanisms. It has been reported that mutation of the catalytic cysteine to glutamine in E. coli FabF resulted in a 50-fold increase in PTM binding (Wang et al., 2006). Therefore, a sequester mechanism would introduce PTM/PTN susceptibility in the PtmP3 C162A and C162L mutants while retaining resistance in the C162Q mutant. In fact, the C162A, C162L, and C162Q mutants all lost resistance, inferring PtmP3 acts as a target replacement, and not a sequestration protein (Fig. 2B). With our finding that S. platensis FabF is inherently resistant to PTM and PTN, we revealed that the PTM and PTN producing organisms adapted two complementary mechanisms to confer PTM and PTN resistance: target replacement (i.e., FabF and FabH by PtmP3/PtnP3) and target modification (i.e., FabF by a PTM-insensitive variant).
PtmP3/PtnP3 is the major self-resistance element, acting as a functional replacement for both FabF and FabH of FASII in Streptomyces
Bioinformatics analysis of the gene clusters responsible for PTM and PTN biosynthesis revealed four candidate genes encoding self-resistance to the producers. The results from this study exclude PtmP1/PtnP1, PtmP2/PtnP2, and PtmP4/PtnP4 from playing significant roles in PTM and PTN resistance, and their functions or possible contributions to the production of PTM/PTN remain unclear. On the other hand, we confirmed PtmP3/PtnP3 as the major resistance element, conferring both PTM (targeting FabF) and PTN (targeting both FabF and FabH) resistance by complementing the target condensing enzymes during inhibition. These findings are consistent with the fact that while PtmP1/PtnP1, PtmP2/PtnP2, and PtmP3/PtnP3 all show sequence similarity to KAS condensing enzymes, PtmP1/PtnP1 and PtmP2/PtnP2 lack key active site residues known to be essential for function. Only PtmP3/PtnP3 retains a catalytic triad conserved among KAS I/II enzymes (Fig. S2) and was hypothesized as the best candidate for conferring resistance in the form of an insensitive copy of the native target of PTM or PTN. Our results confirmed this hypothesis, although the dual capability of PtmP3/PtnP3 to provide resistance to both antibiotics and apparently carry out the disparate functions of both the FabF (KAS II) and the FabH (KAS III) enzymes in Streptomyces was unexpected (Fig. 1B).
Evolutionary relationship between the two resistance mechanisms provided by PtmP3/PtnP3 and FabF in S. platensis
Construction of the S. platensis MA7327 ΔptmU4-ptmP4 deletion mutant strain, SB12021, revealed that upon deletion of ptmP3, the strain becomes sensitive to PTN, but remains resistant to PTM. This established the presence of another resistance element to PTM in S. platensis. Lacking any obvious candidates remaining within the PTM/PTN biosynthetic gene cluster prompted investigation of the S. platensis native genes within the housekeeping FASII locus, namely fabF, the target of PTM. Expression of the native S. platensis fabF in a naïve host conferred high-level resistance to PTM, as well as partial resistance to PTN, and implied its likely responsibility for the residual PTM resistance observed in the SB12021 strain.
PtmP3 (ACS13710) shows high sequence homology to other FabF enzymes, including the S. platensis MA7327 FabF (KF543053) (51% identity and 67% similarity) and the E. coli FabF (CAA84431) (38% identity and 55% similarity). S. platensis MA7327 FabF shows an average of 81% sequence identity and 90% similarity to other Streptomyces spp. FabF enzymes, including S. coelicolor (NP_626636), S. griseus (YP_001826617), S. avermitilis (NP_826962), and S. albus (AGI908332), and relatively low sequence holomogy to FabF from E. coli (39% identity and 57% similarity) (Fig. S2). Point mutations arising in the S. platensis FabF, reducing its affinity for PTM, may explain its resistance to PTM. However, this essential and highly conserved primary metabolism gene would have a very low tolerance for mutation, as illustrated by the high sequence conservation to and among other Streptomyces spp. FabF enzymes. Acquisition of a second fabF gene from the environment, or more likely, duplication of its own fabF gene near the PTM/PTN biosynthetic cluster, provides a gene locus capable of genetic variation (without affecting primary metabolism and growth) and allows coevolution, with the production of PTM and PTN, of a highly efficient resistance element such as PtmP3. Furthermore, having a resistance element such as PtmP3 residing within the PTM-PTN biosynthetic cluster (as opposed to within a conserved region of the chromosome such as the FASII locus), provides the advantage of horizontal cluster transfer among organisms to afford other PTM, PTN, or PTM-PTN dual producers.
Implications of PtmP3/PtnP3 as a dual FabF and FabH enzyme of FASII on fatty acid biosynthesis
PtmP3/PtnP3 is sufficient to confer resistance in a naïve Streptomyces host to both PTM, an inhibitor of FabF (KAS II), and PTN, a dual inhibitor of FabF (KAS II) and FabH (KAS III) (Fig. 1B). Furthermore, isolation of viable S. platensis strains containing ΔfabF or ΔfabH deletion in backgrounds expressing ptmP3 suggest the ability of PtmP3 to functionally complement the activities of each of these enzymes essential for fatty acid biosynthesis. KAS II/III are a group of enzymes that ultimately perform the same chemical reaction (decarboxylative Claisen condensation), but its various members are defined by their substrate preferences. FabH (KAS III) utilizes acetyl-CoA and initiates the fatty acid biosynthetic pathway by catalyzing the first decarboxylative condensation with malonyl-ACP, while FabF (KAS II) catalyzes the same decarboxylative condensation reaction, but condenses acyl-ACP substrates of varying lengths with malonyl-ACP to carry out the consecutive rounds of elongation (Fig. 1B). FabH has been shown to be essential for viability in E. coli, Streptomyces coelicolor, and Lactococcus lactis, demonstrating FabH is the key contributor for initiating fatty acid biosynthesis (Lai and Cronan, 2003; Li et al., 2005; Revill et al., 2001). That PtmP3, a KAS II homolog, can apparently replace both the functions of FabF (KAS II) and FabH (KAS III) in Streptomyces is unexpected, but plausible. In L. lactis, a fabH bypass mutant strain can be fully complemented by engineering overproduction of the L. lactis FabF (Morgan-Kiss and Cronan, 2008). Based on previously observed activities of E. coli FabF/B enzymes in vitro (comparable detailed mechanistic studies of the L. lactis enzymes in vitro have not been reported), it has been proposed that in the absence of acyl-ACP substrate, FabF decarboxylates the lone malonyl-ACP substrate to acetyl-ACP, which becomes an available substrate for condensation with malonyl-ACP to yield acetoacetyl-ACP, the FabH product (Alberts et al., 1972; McGuire et al., 2000; McGuire et al., 2001; Morgan-Kiss and Cronan, 2008). The reported example in L. lactis is likely a product of the evolutionary tendency of L. lactis toward genome minimization and metabolic simplification (Makarova et al., 2006; Morgan-Kiss and Cronan, 2008). Nonetheless, it is a significant precedent for a bacterium evolving towards the ability to survive with only a single functional KAS enzyme and adding complexity to the current understanding of fatty acid synthesis in bacteria. PtmP3/PtnP3 apparently exploits a similar evolutionary pathway towards metabolic simplification in fatty acid biosynthesis, combining the unique functions of two enzymes (each a target) into one functional enzyme. This strategy yields a unique enzyme for primary metabolism and provides traditional insensitivity toward an antibiotic. Further mechanistic studies will provide insight on how PtmP3/PtnP3 balances its apparent expanded substrate preferences while maintaining catalytic efficiency.
Impact of the PtmP3/PtnP3 resistance mechanism on potential emergence of PTM and PTN resistance in the clinical setting
Bacterial fatty acid synthesis is an essential pathway, and FASII represents an attractive target for clinically useful antibiotics. Although the effectiveness of targeting FASII enzymes has been debated (Balemans et al., 2010; Brinster et al., 2009; Brinster et al., 2010; Parsons and Rock, 2011), there are examples, such as isoniazid and triclosan, of successful FASII inhibitor use (Lu and Tonge, 2008; Russell, 2004). The limited clinical use of antibiotics targeting FASII may partially account for why PTM and PTN exhibit no cross-resistance to other key antibiotic-resistant pathogens (Wang et al., 2006; Wang et al., 2007). However, the unavoidable and highly efficient self-resistance mechanisms represent potential sources for clinical resistance to antibiotics of natural product origin. PtmP3/PtnP3, identified as the major natural resistance gene to PTM and PTN in the producing organism S. platensis, is a potential source of clinical resistance. Its identity as an apparent “hybrid” KAS II/III isoform capable of complementing both native FabF (KAS II) and FabH (KAS III) enzymes in Streptomyces represents a clever resistance mechanism developed by a producer to counteract an inhibitor (PTN) of two separate essential targets. The potential for PtmP3/PtnP3 to confer resistance to other clinically relevant FabF/FabH inhibitors as well as its ability to complement FabF/FabH and/or confer resistance in other organisms and pathogens should be investigated further. These studies will guide the future development of new and effective therapeutics targeting FASII.
Significance
PTM and PTN are potent inhibitors of bacterial fatty acid synthesis – PTM specifically inhibits the elongation condensing enzymes FabB/F, and PTN dually inhibits FabF and the initiation condensing enzyme FabH. PTM and PTN have emerged as promising antibacterial drug leads. We previously characterized the PTM and PTN biosynthetic machineries in the PTM-PTN dual producer S. platensis MA7327 and the PTN producer S. platensis MA7339, respectively. We now identify two mechanisms for PTM and PTN resistance in the producers. The ptmP3 or ptnP3 gene within the PTM-PTN or PTN biosynthetic cluster was identified as the major resistance conferring element. Expression of ptmP3 in a naïve Streptomyces host conferred full resistance to both PTM and PTN, while site-directed mutagenesis of the active site Cys162 of PtmP3 completely abolished its ability to confer resistance. The fabF gene within the housekeeping fatty acid synthase locus was identified as the second resistance conferring element. Deletion of ptmP3 alone was insufficient to eliminate PTM resistance from the producer, and expression of S. platensis fabF in a naïve Streptomyces host conferred full resistance to PTM. PtmP3 and FabF in the PTM and PTN producers therefore confer PTM and PTN resistance by target replacement (i.e., FabF and FabH by PtmP3) and target modification (i.e., FabF by a PTM-insensitive variant), respectively. PtmP3 also represents an unprecedented mechanism for fatty acid biosynthesis in which FabH and FabF are functionally replaced by a single condensing enzyme. These findings challenge the current paradigm for fatty acid biosynthesis and should be considered in future development of effective therapeutics targeting fatty acid synthase.
Experimental Procedures
Bacterial strains, plasmids, primers, and culture conditions
For a complete list of bacterial strains, plasmids, and PCR primers used in this study, see Tables S2 and S3. Escherichia coli DH5α was used for routine cloning (Sambrook and Russel, 2001). Vectors pGEM-T Easy (Promega) and pCR2.1-TOPO (Invitrogen) were obtained from commercial sources. PTM and PTN producers, Streptomyces platensis MA7327 and Streptomyces platensis MA7339, respectively, were kindly provided by Dr. Sheo B. Singh at Merck Research Laboratories (Rahway, NJ). E. coli carrying plasmids were grown in Luria-Bertani (LB) medium and selected using appropriate antibiotics (Sambrook and Russel, 2001). Standard media and protocols were used for Streptomyces growth and sporulation (Kieser, et al., 2000). Media components and all other chemicals were obtained from standard commercial sources.
DNA isolation and manipulation
Plasmid preparation from E. coli and gel extraction was carried out with commercial kits (Qiagen and Omega Bio-Tek). Total Streptomyces DNA isolation, restriction endonuclease digestions, and ligations were performed according to standard procedures (Kieser, et al., 2000; Sambrook and Russel, 2001). Automated DNA sequencing was performed at the University of Wisconsin Biotechnology Center (Madison, WI), or by GENEWIZ and oligonucleotide primer synthesis was completed by Integrated DNA Technologies.
Platensimycin (PTM) and Platencin (PTN)
The PTM and PTN utilized in this study was isolated and purified from fermentations of S. platensis SB12002 and S. platensis SB12001, respectively, as previously described (Smanski, et al., 2009).
Disk diffusion assay
A standard disk diffusion assay was used to determine susceptibility of wild-type and recombinant Streptomyces strains to PTM and PTN. In brief, quantities of PTM (5-200 μg) or PTN (1.25-40 μg) in methanol were applied to 7 mm paper disks (Whatman) and dried. The disks were placed onto solid R2YE agar plates applied with dense Streptomyces liquid culture (~48 h). These plates were incubated overnight at 30 °C and the zones of inhibition were subsequently observed.
96-Well plate assay for measuring the MIC values
Molten R2YE agar was prepared and contained either 0, 0.25, 0.5, 1, 2, 4, 8, 16, 32, 64, 128, or 256 μg/mL of PTM, or 0, 0.0625, 0.125, 0.25, 0.5, 1, 2, 4, 8, 16, 32, or 64 μg/mL of PTN. These mixtures were applied to sterile 96-well plates; each well contained 100 μL of agar media. Each well was inoculated with 10 μL of sterile water containing ~103 Streptomyces spores. Plates were incubated overnight at 30 °C. Growth in wells containing PTM or PTN was compared with control wells (no antibiotics added).
Cloning and constructing the expression plasmids of the putative resistance elements
Putative resistance elements ptmP1, ptmP2, ptmP3, and ptmP4 were amplified from S. platensis MA7327 genomic DNA using LA Taq with GC Buffer II (Clontech Laboratories, Inc.) according to the manufacturer’s instructions. The resulting PCR products were cloned into pGEM-T Easy or pCR2.1-TOPO. These constructs were used for subcloning individual elements into pBS12015, a modified Streptomyces integrating vector containing the apramycin resistance cassette and the constitutively active promoter, ermE*, upstream of the multiple cloning site to ensure a high level of expression of the insert (Bierman, et al., 1992). The expression plasmids pBS12016, pBS12017, pBS12018, and pBS12019 (containing ptmP1, ptmP2, ptmP3, and ptmP4, respectively) were constructed for expression of individual elements in S. albus J1074.
Site-directed mutagenesis of ptmP3
Point mutations in ptmP3 were generated through site-directed mutagenesis by overlap extension using PCR (Ho, et al., 1989). The mutagenized fragments were cloned into pGEM-T Easy vector and subcloned into pBS12015 to yield pBS12026 (C162A), pBS12027 (C162Q), and pBS12028 (C162L).
Construction and isolation of recombinant Streptomyces strains
After transformation of each pSET152ermE*-derived expression construct (Table S2) into E. coli ET12567/pUZ8002 electrocompetent cells, intergeneric conjugation between E. coli and Streptomyces was performed according to standard protocol (Kieser, et al., 2000). This resulted in stable, recombinant strains of Streptomyces with the various expression constructs fully integrated into the Streptomyces chromosome. For a complete list of recombinant strains, see Table S2.
Construction of the ΔptmU4-ptmP4 mutant S. platensis SB12021
Cosmid pBS12005 was modified by PCR targeting, using standard protocols (Gust, et al., 2003), to generate the construct, pBS12023. Briefly, primers RLDC2for and RLDC2rev were used to amplify the aac(3)IV cassette from pIJ773. Introduction of the amplified fragment into E. coli BW25113/pIJ790/pBS12005 resulted in the replacement of the locus spanning ptmU4-ptmP4 with the apramycin resistance cassette on pBS12005 to generate pBS12023. The construct was introduced into S. platensis MA7327 by intergeneric conjugation using E. coli ET12567/pUZ8002 as the donor strain. Apramycin-resistant exconjugants were picked and replica-plated to identify apramycin-resistant, kanamycin-sensitive clones in which homologous recombination on each side of the locus of gene replacement had taken place, resulting in S. platensis SB12021. Double crossover mutants were verified by PCR and Southern analysis (Figs. S5B and S5D).
Cloning and sequencing the fabF and fabH genes within the FASII locus from S. platensis MA7327 and S. albus J1074
Sequence data for the native fab genes in S. platensis MA7327 and MA7339 strains were unavailable. Genome sequences of other Streptomyces spp. were analyzed for the homologous fab cluster containing fabD, fabH, acpP, and fabF. Primers for fabF amplification were designed based on highly conserved regions found in S. coelicolor, S. avermitilis, and S. griseus (Table S3). Fragments containing the fabF or fabH genes were amplified from S. platensis MA7327 and S. platensis MA7339 genomic DNA. These fragments were cloned into pGEM-T Easy for sequencing and further subcloned into the Streptomyces integrating vector, pBS12015, to yield pBS12020 and pBS12021. Similar procedures were followed for the construction of pBS12029 and pBS12030, the integrating vector pBS12015 containing S. albus fabF and fabH, respectively.
Construction of the fabF or fabH deletion mutants S. platensis SB12022 and SB12023
Cosmids containing the S. platensis fab cluster were isolated from a previously constructed S. platensis MA7327 genomic library (Smanski, et al., 2009). A 10 μL culture containing the genomic library was diluted to 106 cells/mL and spread onto LB plates containing 50 μg/mL of kanamycin. PCR primers FabHscreenFor and FabHscreenRev were used to screen resulting single colonies. Colonies yielding the expected 432 bp band were isolated and their respective cosmids purified using standard methods. Cosmid pBS12014, isolated from the S. platensis MA7327 cosmid library and verified to contain both fabF and fabH, was transformed into E. coli BW25113/pIJ790 to generate ΔfabF and ΔfabH single knockout mutants using the λRED-mediated PCR-targeting mutagenesis method (Gust, et al., 2003). The modified cosmids were introduced to S. platensis MA7327 by intergeneric conjugation using E. coli ET12567/pUZ8002 as the donor strain. Apramycin-resistant exconjugants were selected and these putative single-crossover recombinants were replica plated to identify apramycin-resistant, kanamycin-sensitive clones in which homologous recombination on each side of the locus of gene replacement had taken place. Single and double crossover mutants were verified by PCR and Southern analysis. Strains S. platensis SB12022 and SB12024 were confirmed to be ΔfabF::aac(3)IV mutants, while S. platensis SB12023 and SB12025 were confirmed to be ΔfabH::aac(3)IV mutants. For Southern analysis, digoxigenin labeling of DNA probes, hybridization, and detection were performed according to the manufacturer’s protocols for DIG-High Prime (Roche Diagnostics).
Supplementary Material
Highlights.
PTM and PTN are potent inhibitors of bacterial fatty acid synthases
Two mechanisms for PTM and PTN resistance are discovered in Streptomyces platensis
PtmP3/PtnP3 confer PTM and PTN resistance by target replacement
FabF confers PTM and PTN resistance by target modification
Acknowledgments
We thank Dr. Sheo B. Singh, Merck Research Laboratories, Rahway, NJ, for providing S. platensis MA7327 and S. platensis MA7339 wild-type strains and the John Innes Center, Norwich, UK, for providing the REDIRECT technology kit. This work was supported in part by NIH Grant AI079070. MJS was supported in part by NIH Predoctoral Training Grant GM08505.
Footnotes
Accession Numbers The S. platensis MA7327 fabF and fabH genes were deposited in the GenBank database with Accession Nos. KF543053 and KF543054, respectively.
Supplemental Information Supplemental Information includes five supplemental figures, three supplemental tables, and can be found online at http://dx.doi.org/10.1016/j.chembiol.xxxx.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alberts AW, Bell RM, Vagelos PR. Acyl carrier protein. XV. β-Ketoacyl-acyl carrier protein synthetase. J. Biol. Chem. 1972;247:3190–3198. [PubMed] [Google Scholar]
- Balemans W, Lounis N, Gilissen R, Guillemont J, Simmen K, Andries K, Koul A. Essentiality of FASII pathway for Staphylococcus aureus. Nature. 2010;463:E3–E4. doi: 10.1038/nature08667. [DOI] [PubMed] [Google Scholar]
- Benveniste R, Davies J. Aminoglycoside antibiotic-inactivating enzymes in actinomycetes similar to those present in clinical isolates of antibiotic-resistant bacteria. Proc. Natl. Acad. Sci. USA. 1973;70:2276–2280. doi: 10.1073/pnas.70.8.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierman M, Logan R, O’Brien K, Seno ET, Rao RN, Schoner B,E. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- Brinster S, Lamberet G, Staels B, Trieu-Cuot P, Gruss A, Poyart C. Type II fatty acid synthesis is not a suitable antibiotic target for Gram-positive pathogens. Nature. 2009;458:83–86. doi: 10.1038/nature07772. [DOI] [PubMed] [Google Scholar]
- Brinster S, Lamberet G, Staels B, Trieu-Cuot P, Gruss A, Poyart C. Essentiality of FASII pathway for Staphlococcus aureus. Brinster et al. reply. Nature. 2010;463:E4–E5. [Google Scholar]
- D’Costa VM, McGrann KM, Hughes DW, Wright GD. Sampling the antibiotic resistome. Science. 2006;311:374–377. doi: 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- Galm G, Hager MH, Van Lanen SG, Ju J, Thorson JS, Shen B. Antitumor antibiotics: bleomycin, enediynes, and mitomycin. Chem. Rev. 2005;105:739–758. doi: 10.1021/cr030117g. [DOI] [PubMed] [Google Scholar]
- Gust B, Challis GL, Fowler K, Kieser T, Chater KF. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA. 2003;100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Hopwood DA. How do antibiotic-producing bacteria ensure their self-resistance before antibiotic biosynthesis incapacitates them? Mol. Microbiol. 2007;63:937–940. doi: 10.1111/j.1365-2958.2006.05584.x. [DOI] [PubMed] [Google Scholar]
- Huang W, Jia J, Edwards P, Dehesh K, Schneider G, Lindqvist Y. Crystal structure of β-ketoacyl-acyl carrier protein synthase II from E. coli reveals the molecular architecture of condensing enzymes. EMBO J. 1998;17:1183–1191. doi: 10.1093/emboj/17.5.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. The John Innes Foundation; Norwich, UK: 2000. [Google Scholar]
- Kumar A, Schweizer HP. Bacterial resistance to antibiotics: active efflux and reduced uptake. Adv. Drug Delivery Rev. 2005;57:1486–1513. doi: 10.1016/j.addr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Kursula P, Ojala J, Lambeir A-M, Wierenga RK. The catalytic cycle of biosynthetic thiolase: a conformational journey of an acetyl group through four binding modes and two oxyanion holes. Biochemistry. 2002;41:15543–15556. doi: 10.1021/bi0266232. [DOI] [PubMed] [Google Scholar]
- Lai C-Y, Cronan JE. β-Ketoacyl-acyl carrier protein synthase III (FabH) is essential for bacterial fatty acid synthesis. J. Biol. Chem. 2003;278:51494–51503. doi: 10.1074/jbc.M308638200. [DOI] [PubMed] [Google Scholar]
- Li Y, Florova G, Reynolds KA. Alteration of the fatty acid profile of Streptomyces coelicolor by replacement of the initiation enzyme 3-ketoacyl acyl carrier protein synthase III (FabH) J. Bacteriol. 2005;187:3795–3799. doi: 10.1128/JB.187.11.3795-3799.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Tonge PJ. Inhibitors of FabI, an enzyme drug target in the bacterial fatty acid biosynthesis pathway. Acc. Chem. Res. 2008;41:11–20. doi: 10.1021/ar700156e. [DOI] [PubMed] [Google Scholar]
- Makarova K, et al. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. USA. 2006;103:15611–15616. doi: 10.1073/pnas.0607117103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CG, Broadhead G, Leskiw BK, Wright GD. D-Ala-D-Ala ligases from glycopeptide antibiotic-producing organisms are highly homologous to the enterococcal vancomycin-resistance ligases VanA and VanB. Proc. Natl. Acad. Sci. USA. 1997;94:6480–6483. doi: 10.1073/pnas.94.12.6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens E, Demain AL. Platensimycin and platencin: promising antibiotics for future application in human medicine. J. Antibiot. 2011;64:705–710. doi: 10.1038/ja.2011.80. [DOI] [PubMed] [Google Scholar]
- McGuire KA, McGuire JN, von Wettstein-Knowles P. Acyl carrier protein (ACP) inhibition and other differences between β-ketoacyl synthase (KAS) I and II. Biochem. Soc. Trans. 2000;28:607–610. doi: 10.1042/0300-5127:0280607. [DOI] [PubMed] [Google Scholar]
- McGuire KA, Siggaard-Andersen M, Bangera MG, Olsen JG, von Wettstein-Knowles P. β-Ketoacyl-[acyl carrier protein] synthase I of Escherichia coli: Aspects of the condensation mechanism revealed by analyses of mutations in the active site pocket. Biochemistry. 2001;40:9836–9845. doi: 10.1021/bi0105577. [DOI] [PubMed] [Google Scholar]
- Morgan-Kiss RM, Cronan JE. The Lactococcus lactis FabF fatty acid synthetic enzyme can functionally replace both the FabB and FabF proteins of Escherichia coli and the FabH protein of Lactococcus lactis. Arch. Microbiol. 2008;190:427–437. doi: 10.1007/s00203-008-0390-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons JB, Rock CO. Is bacterial fatty acid synthesis a valid target for antibacterial drug discovery? Curr. Opinion Microbiol. 2011;14:544–549. doi: 10.1016/j.mib.2011.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pootoolal J, Neu J, Wright GD. Glycopeptide antibiotic resistance. Ann. Rev. Pharmacol. Toxicol. 2002;42:381–408. doi: 10.1146/annurev.pharmtox.42.091601.142813. [DOI] [PubMed] [Google Scholar]
- Revill WP, Bibb MJ, Scheu A-K, Kieser HJ, Hopwood DA. β-Ketoacyl acyl carrier protein synthase III (FabH) is essential for fatty acid biosynthesis in Streptomyces coelicolor A3(2) J. Bacteriol. 2001;183:3526–3530. doi: 10.1128/JB.183.11.3526-3530.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AD. Whither triclosan? J. Antimicrob. Chemother. 2004;53:693–695. doi: 10.1093/jac/dkh171. [DOI] [PubMed] [Google Scholar]
- Saleem M, Hussain H, Ahmed I, van Ree T, Krohn K. Platensimycin and its relatives. A recent story in the struggle to develop new naturally derived antibiotics. Nat. Prod. Rep. 2011;28:1534–1579. doi: 10.1039/c1np00010a. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russel D. Molecular cloning: A Laboratory Manual. 3rd Ed Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2001. [Google Scholar]
- Shen HC, et al. Synthesis and biological evaluation of platensimycin analogs. Bioorg. Med. Chem. Lett. 2009;19:1623–1627. doi: 10.1016/j.bmcl.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Smanski MJ, Casper J, Peterson RM, Yu Z, Rajski SR, Shen B. Expression of the platencin biosynthetic gene cluster in heterologous hosts yielding new platencin congeners. J. Nat. Prod. 2012;75:2158–2167. doi: 10.1021/np3005985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smanski MJ, Peterson RM, Rajski SR, Shen B. Engineered Streptomyces platensis strains that overproduce antibiotics platensimycin and platencin. Antimicrob. Agents Chemother. 2009;53:1299–1304. doi: 10.1128/AAC.01358-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smanski MJ, Yu Z, Casper J, Lin S, Peterson RM, Chen Y, Wendt-Pienkowski E, Rajski SR, Shen B. Dedicated ent-kaurene and ent-atiserene synthases for platensimycin and platencin biosynthesis. Proc. Natl. Acad. Sci. USA. 2011;108:13498–13503. doi: 10.1073/pnas.1106919108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubes G. The bacteria fight back. Science. 2008;321:356–361. doi: 10.1126/science.321.5887.356. [DOI] [PubMed] [Google Scholar]
- Yu Z, Rateb ME, Smanski MJ, Peterson RM, Shen B. Isolation and structural elucidation of glucoside congeners of platencin from Streptomyces platensis SB12600. J. Antibiot. 2013;66:291–294. doi: 10.1038/ja.2013.1. [DOI] [PubMed] [Google Scholar]
- Yu Z, Smanski MJ, Peterson RM, Marchillo K, Andes D, Rajski SR, Shen B. Engineering of Streptomyces platensis MA7339 for overproduction of platencin and congeners. Org. Lett. 2010;12:1744–1747. doi: 10.1021/ol100342m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, et al. Platensimycin is a selective FabF inhibitor with potent antibiotic properties. Nature. 2006;441:358–361. doi: 10.1038/nature04784. [DOI] [PubMed] [Google Scholar]
- Wang J, et al. Discovery of platencin, a dual FabF and FabH inhibitor with in vivo antibiotic properties. Proc. Natl. Acad. Sci. USA. 2007;104:7612–7616. doi: 10.1073/pnas.0700746104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrli W. Rifampin: mechanisms of action and resistance. Rev. Infect. Dis. 1983;5:S407–411. doi: 10.1093/clinids/5.supplement_3.s407. [DOI] [PubMed] [Google Scholar]
- White SW, Zheng J, Zhang Y-M, Rock CO. The structural biology of type II fatty acid biosynthesis. Ann. Rev. Biochem. 2005;74:791–831. doi: 10.1146/annurev.biochem.74.082803.133524. [DOI] [PubMed] [Google Scholar]
- Wright GD. Molecular mechanisms of antibiotic resistance. Chem. Commun. 2011;47:4055–4061. doi: 10.1039/c0cc05111j. [DOI] [PubMed] [Google Scholar]
- Wu M, et al. Antidiabetic and antisteatotic effects of the selective fatty acid synthase (FAS) inhibiotr platensimycin in mouse models of diabetes. Proc. Natl. Acad. Sci. USA. 2011;108:5378–5383. doi: 10.1073/pnas.1002588108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, et al. Platensimycin and platencin congeners from Streptomyces platensis. J. Nat. Prod. 2011;74:329–340. doi: 10.1021/np100635f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.