Abstract
Acat2 [gene name: sterol O-acyltransferase 2 (SOAT2)] esterifies cholesterol in enterocytes and hepatocytes. This study aims to identify repressor elements in the human SOAT2 promoter and evaluate their in vivo relevance. We identified TG-interacting factor 1 (Tgif1) to function as an important repressor of SOAT2. Tgif1 could also block the induction of the SOAT2 promoter activity by hepatocyte nuclear factor 1α and 4α. Women have ∼30% higher hepatic TGIF1 mRNA compared with men. Depletion of Tgif1 in mice increased the hepatic Soat2 expression and resulted in higher hepatic lipid accumulation and plasma cholesterol levels. Tgif1 is a new player in human cholesterol metabolism.
Keywords: hepatocyte nuclear factor 1α and 4α, liver, human, triglyceride
The gene encoding Acat2 is known by international convention as sterol O-acyltransferase 2 (SOAT2) (1). Acat2 expression is confined to enterocytes and hepatocytes (2, 3), where it is involved in intracellular cholesterol esterification for storage and secretion in the lipid core of VLDLs and chylomicrons. Soat2-deficient mice have lower plasma VLDL and LDL cholesterol levels (4, 5) and lower intestinal cholesterol absorption (6–8), are completely resistant to diet-induced hypercholesterolemia and gallstone formation (1), and do not develop atherosclerosis (5, 9). Additionally, these mice have reduced hepatic steatosis (10, 11), suggesting that SOAT2 also plays a role in TG metabolism. In humans, the products of SOAT2, such as cholesteryl palmitate and cholesteryl oleate, have been shown to predict intima media thickness, CVD mortality, and development of coronary artery disease in patients with acute coronary syndrome (12–14). Acat2 is, therefore, considered as a possible therapeutic target in prevention and treatment of atherosclerosis and the metabolic syndrome.
Hepatocyte nuclear factor (Hnf) 1α and 4α are two transcription factors that are expressed in various tissues, including liver, kidney, and pancreas, and play important roles in lipid and carbohydrate metabolism (15, 16). Mutations in the genes encoding these two transcription factors lead to two different forms of maturity onset diabetes of the young. We previously identified Hnf4α as a positive regulator of SOAT2 and found that subjects with mutations in the gene encoding Hnf4α have lower levels of esterified cholesterol in VLDL and LDL particles (17), which may partially be attributed to lower Acat2 activity in these subjects. In accordance, hepatic Hnf4α deficiency in mice reduced Soat2 mRNA expression, lipogenesis, de novo cholesterol synthesis, and VLDL secretion (18). Hnf4α is an upstream regulator of Hnf1α (19), and we earlier identified an important binding site for Hnf1 that serves as a positive regulator of the human SOAT2 gene and to which Hnf1α and Hnf1β can bind both in vivo and in vitro (20). In that study, we also found evidence for a potential repressor element, although this element was not characterized in detail. This study aims to identify repressor elements in the human SOAT2 promoter and evaluate their in vivo relevance.
MATERIALS AND METHODS
Materials
Probe and primers for human TG-interacting factor 1 (TGIF1) and GAPDH were purchased from Applied Biosystems (assay ID:Hs00820148_g1 and ID: Hs02758991_g1, respectively). All other primers were purchased from Cybergene AB (Stockholm, Sweden), and primer sequences are available upon request. Taqman and SYBR Green master mixes were purchased from Applied Biosystems (Stockholm, Sweden). All antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The human SOAT2 promoter region, −1,305 to +86 bp upstream of ATG start site, was previously cloned into pGL3 basic vector and used to create four nested deletion constructs: −1,196 to +86 (p-1196); −1,044 to +86p (p-1044); −782 to +86p (p-782); and −269 to +86p (p-269) (20). The human pCMV5 Flag Tgif1 expression vector was purchased from Addgene (http://www.addgene.org). The human Hnf1α expression vector was a generous gift from Prof. Pal R. Njølstad and Dr. Lise Bjørkhaug Gundersen, Haukeland University Hospital (Norway). The human Hnf4α expression vector was a generous gift from Dr. Theodore C. Simon, Washington University School of Medicine (St. Louis, MO). The human pCMV5 Flag myeloid ecotropic viral integration site 2d (Meis2d) expression vector was generated as described (21). Cholesterol and TG reagents were purchased from Roche Diagnostics (GmbH, Mannheim, Germany). Free cholesterol reagent was purchased from MTI Diagnostics (Idstein, Germany).
Methods
Subjects.
Patients undergoing laparoscopic cholecystectomy were included in the study after written informed consent had been given, and a liver biopsy (∼0.5 g) was collected from the edge of the right lobe during the operation. The subjects included 41 male patients (37 with gallstone disease and 4 with gallbladder polyps) and 58 female patients (47 with gallstone disease and 11 with gallbladder polyps). None of the subjects were subjected to any lipid-lowering therapies. Thirty-three of the females were premenopausal, and the rest were postmenopausal. The clinical characteristics of the patients are shown in Table 1. The study protocol was approved by the Ethical Committee at Ruijin Hospital, Shanghai Jiaotong University School of Medicine (2011, No. 47).
TABLE 1.
Clinical characteristics of the subjects included in this study
| Males (n = 41) | Females (n = 58) | |
| Age (years) | 54.2 ± 2.4 | 55.4 ± 1.5 |
| BMI (kg/m2) | 24.3 ± 0.5 | 24.9 ± 0.4 |
| Fasting glucose (mmol/l) | 5.59 ± 0.16 | 5.42 ± 0.13 |
| Total cholesterol (mmol/l) | 4.64 ± 0.17 | 4.88 ± 0.12 |
| TGs (mmol/l) | 1.45 ± 0.10 | 1.48 ± 0.10 |
| HDL (mmol/l) | 1.18 ± 0.04a | 1.46 ± 0.08 |
| LDL (mmol/l) | 3.15 ± 0.16 | 3.15 ± 0.12 |
| Apo AI (g/l) | 1.23 ± 0.03a | 1.37 ± 0.03 |
| Apo B (g/l) | 0.86 ± 0.04 | 0.84 ± 0.02 |
Data are expressed as mean ± SEM. Differences between males and females were tested using Student's t-tests.
P < 0.01 males versus females.
Animals.
Tgif1 null mice in a C57BL/6J strain background have been described previously (22, 23). Male mice (n = 6 per group) were maintained on a regular chow diet and euthanized at 9 to 15 weeks of age. The study was approved by the Animal Care and Use Committee of the University of Virginia.
Cell experiments, mutagenesis, and transfections.
HuH7 cells were cultured as described (20). Caco2 cells were grown in DMEM supplemented with 20% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin; the media were replaced every second day. Cells were maintained in 75 cm2 cell culture flasks and passaged when reaching ∼90% confluence. For all cell experiments, cells were plated out on 6-well tissue culture plates so that they reached ∼70% confluence after 24 h (i.e., ∼700,000 cells/well). Screening of promoter sequences was performed using the transcription factor TESS database (http://www.cbil.upenn.edu; University of Pennsylvania, Philadelphia, PA) and AliBaba 2.1 program (http://www.gene-regulation.com/pub/programs/alibaba2/index.html) to search for putative elements, and specific point mutations were generated as described (20).
Transfections of HuH7 cells were performed using 2 µg promoter (or mutated promoter) construct and 2 µg pSV-β-galactosidase control vector (Promega, Madison, WI), with or without expression vectors, using Lipofectin reagent (Invitrogen, Carlsbad, CA) at a ratio of 3:1 (Lipofectin-DNA). Transfections of Caco2 cells were performed using Lipofectamine LTX reagent (Invitrogen) at a ratio of 1.5:1 (Lipofectamine LTX-DNA) according to the manufacturer's recommendations. The pGL3 empty vector (Promega) was used to adjust for differences in the amount of DNA added to the cells. Twenty-four (Caco2) or 48 (HuH7) h after transfection, cell lysates were prepared in reporter lysis buffer (Promega). The β-galactosidase and luciferase activities were determined using β-galactosidase or luciferase assay kits, respectively, according to the manufacturer's instructions (Promega).
For gene expression analyses, Caco2 and HuH7 cells were transfected with 0, 0.5, 1, and 2 µg Tgif1 or Hnf4α, or cotransfected with both Tgif1 and Hnf4α, expression vectors using Lipofectamine LTX or Lipofectin reagent, respectively. Twenty-four or 48 h after transfection, respectively, RNA was extracted. Each experiment was performed in four replicates and repeated at least twice.
RNA preparation and real-time RT-PCR.
Total RNA was prepared using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. One microgram of RNA was transcribed into cDNA using Omniscript reverse transcriptase (Qiagen, Hilden, Germany) according to the manufacturer's protocol. Real-time RT-PCR was performed in triplicates, and arbitrary units were calculated by linearization of the CT values.
ChIP assay.
Chromatin immunoprecipitation (ChIP) assays were performed using ∼200 mg liver from a healthy donor, taken after written informed consent had been given, as described (20) (ethical permission approved by the ethics committee at Karolinska University Hospital in Huddinge). Chromatin was immunoprecipitated with antibodies against Tgif1 (sc-17800×), Meis2 (sc-10600×), or IgG (sc-2027). Ten microliters of purified, nonimmunoprecipitated chromatin was saved and used as input controls. Primers were designed to target specific binding sites in the human SOAT2 promoter. Relative binding was determined by the ΔΔCT method, normalizing the Tgif1- or Meis2-specific immunoprecipitations (IPs) to that in the nonspecific IgG IP and the input sample; relative enrichment = 2{[CT(input) − CT(specific IP)] − [CT(input) − CT(IgG IP)]}.
Plasma and hepatic lipids.
Plasma cholesterol and TGs were determined by enzymatic assays according to the manufacturer's protocols. Hepatic lipids were extracted as previously described (24, 25). In brief, ∼100 mg liver was extracted in chloroform-methanol (2:1, v/v) and solubilized in 1% TritonX-100 solution, and total and free cholesterol and TGs were determined by enzymatic assays. Esterified cholesterol concentrations were calculated by subtraction of free from total cholesterol, and then multiplied by 1.67 to convert the values into cholesteryl ester mass.
Statistical analysis.
Multiway ANOVA, followed by post hoc comparisons according to the Dunnett test, was used for differences in percent change of the SOAT2 promoter activity following mutation of putative repressor sites. Student's t-test was used for differences among patients. The Mann-Whitney U-test was used for differences between Tgif1 null and wild-type mice. The Kruskal-Wallis test was used for differences in SOAT2 gene expression following transfections with expression vectors (Statistica software; Stat Soft, Tulsa, OK).
RESULTS
Identification of Tgif1 as a repressor of the human SOAT2 promoter
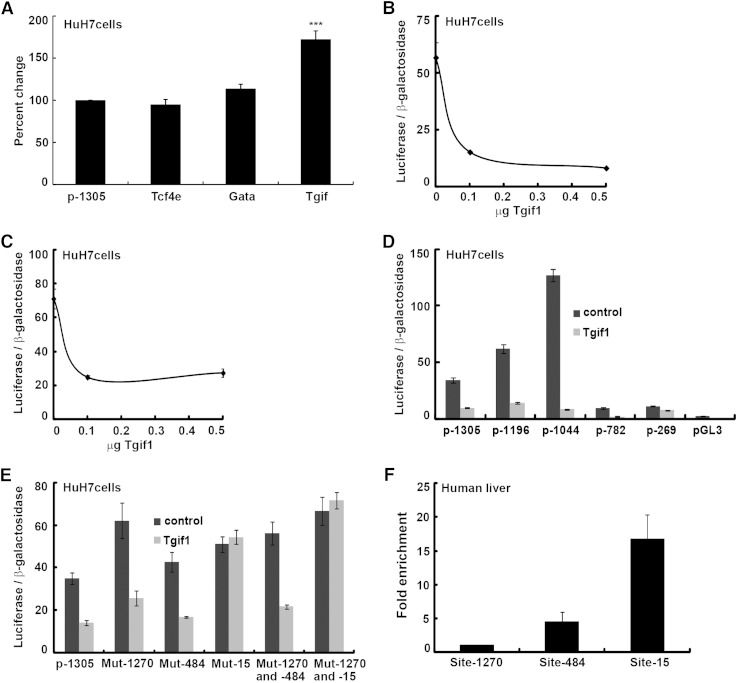
We have previously cloned a ∼1.4 kb construct of the human SOAT2 promoter region (−1,305 to +86 bp), which was used as template to create four deletion constructs: −1,196 to +86; −1,044 to +86p; −782 to +86p; and −269 to +86p (20). We found that the promoter activity increased >4-fold when comparing the full-length promoter with the −1,044 to +86 bp construct. The increased activity suggested the presence of potential repressor elements in the promoter region, which we aimed to identify here. We screened this region using TESS and AliBaba 2.1 and identified putative repressor elements, which displayed 100% match, for the transcription factors T-cell factor 4E, Gata, and Tgif. We mutated these sites in the human SOAT2 promoter and used these mutant constructs for transient transfections of human hepatoma HuH7 cells (Fig. 1A). Mutation of the putative binding site for Tgif, located −1,270 to −1,264 bp upstream of the transcription start site, significantly (P < 0.001) increased the basal promoter activity. To examine whether the transcription factor Tgif1, which recognizes this site, may function as a repressor of SOAT2, we transiently cotransfected HuH7 cells with the SOAT2 promoter and an expression vector for human Tgif1. As shown in Fig. 1B, Tgif1 overexpression dramatically decreased the promoter activity. However, transient cotransfections with the SOAT2 promoter construct in which the identified Tgif site was mutated and the expression vector for Tgif1 revealed that Tgif1 was still able to repress the promoter activity (Fig. 1C). Thus, Tgif1 either can exerts its repressive effect by also binding to other binding sites or to other DNA-bound proteins in the SOAT2 promoter.
Fig. 1.
Tgif1 is a repressor of SOAT2. (A) Putative repressor elements in the human SOAT2 promoter (p-1305) were mutated, and these mutated constructs [i.e., T-cell factor 4E (Tcf4e), Gata, and Tgif] were used for transient transfections of HuH7 cells. (B) The human SOAT2 promoter and an expression vector for Tgif1 were used to transiently cotransfect HuH7 cells. (C) Transient cotransfections with the SOAT2 promoter construct in which the identified Tgif site was mutated and the expression vector for Tgif1. (D) Transient cotransfections with deletion constructs of the SOAT2 promoter and the Tgif1 vector. (E) Three putative binding sites for Tgif, located at −1,270/−1,264 bp (Mut-1270), −484/−478 bp (Mut-484), and −15/−10 bp (Mut-15) upstream of the transcription start site in the SOAT2 promoter, were mutated individually or in combination with the −1,270/−1,264 bp site and used along with the Tgif1 vector. (F) Soluble chromatin, prepared from human liver, was immunoprecipitated with antibodies against either Tgif1 or IgG and amplified using primers designed to target the three putative Tgif binding sites in the human SOAT2 promoter. Data are expressed as mean ± SEM (n = 4). Differences were tested using the Dunnett test. *** P < 0.001.
Tgif1 exerts its repression by binding to a specific cis-element in the SOAT2 promoter
To investigate whether other Tgif binding sites were present in the SOAT2 promoter region, HuH7 cells were transiently cotransfected with deletion constructs of the SOAT2 promoter with or without the Tgif1 expression vector (Fig. 1D). Tgif1 repressed the promoter activity in all constructs, although the greatest effect was seen in the p-1044 bp construct. The entire promoter sequence (−1,305 to +86 bp) was therefore screened using TESS and AliBaba 2.1, and two additional putative binding sites for Tgif were identified, located at −484/−478 bp and at −15/−10 bp upstream of the transcription start site. These sites were mutated (see supplementary Fig. I) individually or in combination with the previously identified Tgif site (located at −1,270/−1,264 bp) and used for transient cotransfections of HuH7 cells together with the Tgif1 expression vector (Fig. 1E). Tgif1 was still able to repress the promoter activity when the −484/−478 bp site (Mut-484) was mutated alone or in combination with the −1,270/−1,264 bp Tgif site (Mut-1270 and -484). In contrast, mutation of the −15/−10 bp site (Mut-15) alone or in combination with mutation of the −1,270/−1,264 bp Tgif site (Mut-1270 and -15) completely abolished the repression by Tgif1. We next performed ChIP assays, using liver from a healthy donor, to determine the in vivo interaction of Tgif1 with the three identified Tgif sites in the SOAT2 promoter (Fig. 1F). Soluble chromatin was immunoprecipitated with a specific antibody against Tgif1 and amplified by PCR. A control antibody (IgG) was used to compare levels of specific DNA fragments. We found Tgif1 to be able to bind both to the −15/−10 bp site (Mut-15) and to the −484/−478 bp site (Mut-484). However, we observed no enrichment of the region −1,270/−1,264 bp (Mut-1270) consistent with the minimal effects seen when this site was mutated.
Tgif1 blocks the induction of SOAT2 promoter activity by Hnf1α and Hnf4α
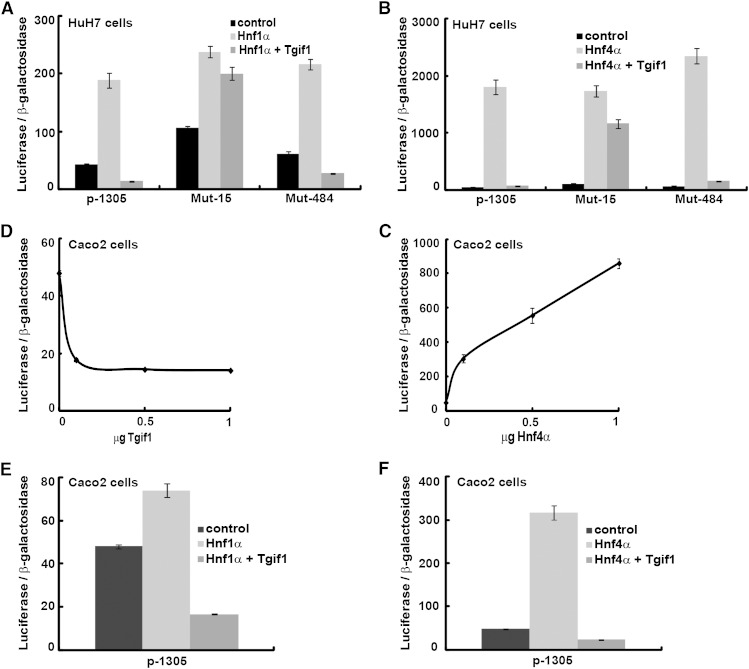
We have previously shown that Hnf1α (20) and Hnf4α (17) act as positive regulators of the human SOAT2 gene. Thus, we also investigated whether Tgif1 could block the stimulation by these two transcription factors of the SOAT2 promoter activity. Transient cotransfections of HuH7 cells with the SOAT2 promoter together with expression vectors for either Hnf1α or Hnf4α, with and without Tgif1, revealed that Tgif1 blocked the stimulation by Hnf1α (Fig. 2A) and Hnf4α (Fig. 2B). The inhibition of stimulation by Hnf1α and Hnf4α was exerted via binding of Tgif1 to the −15/−10 bp Tgif site because mutation of this site abolished these effects. Collectively, these results indicate that binding of Tgif1 to this site is crucial to determine the SOAT2 promoter activity.
Fig. 2.
Tgif1 blocks the induction by Hnf1α and Hnf4α of the SOAT2 promoter activity. HuH7 cells were transiently cotransfected with the SOAT2 promoter (p-1305) or with constructs of the promoter in which binding sites for Tgif were mutated along with vectors for Hnf1α (A) or Hnf4α (B), with and without Tgif1. Caco2 cells were transiently cotransfected with the SOAT2 promoter and vectors for Tgif1 (C) or Hnf4α (D). Caco2 cells were transiently cotransfected with the human SOAT2 promoter along with vectors for Hnf1α (E) or Hnf4α (F), with and without Tgif1. Data are expressed as mean ± SEM (n = 4).
As mentioned, Acat2 is also expressed in enterocytes. To investigate whether the effects of Tgif1 were cell specific, we performed transient cotransfections of intestinal Caco2 cells with the SOAT2 promoter and the Tgif1 expression vector. Interestingly, we found that Tgif1 was also able to function as a repressor of SOAT2 promoter activity in Caco2 cells (Fig. 2C). As expected, we identified Hnf4α as a strong positive regulator of intestinal SOAT2 promoter activity (Fig. 2D). Transient cotransfections of Caco2 cells with the SOAT2 promoter together with expression vectors for either Hnf1α or Hnf4α, with and without Tgif1, revealed that Tgif1 blocked the stimulation by Hnf1α (Fig. 2E) and Hnf4α (Fig. 2F). Thus, Tgif1 may also function as a repressor of intestinal SOAT2 expression, although the −15/−10 bp Tgif site does not appear to play such a dominant role in Caco2 cells (data not shown).
Effects of Meis2d on the SOAT2 promoter
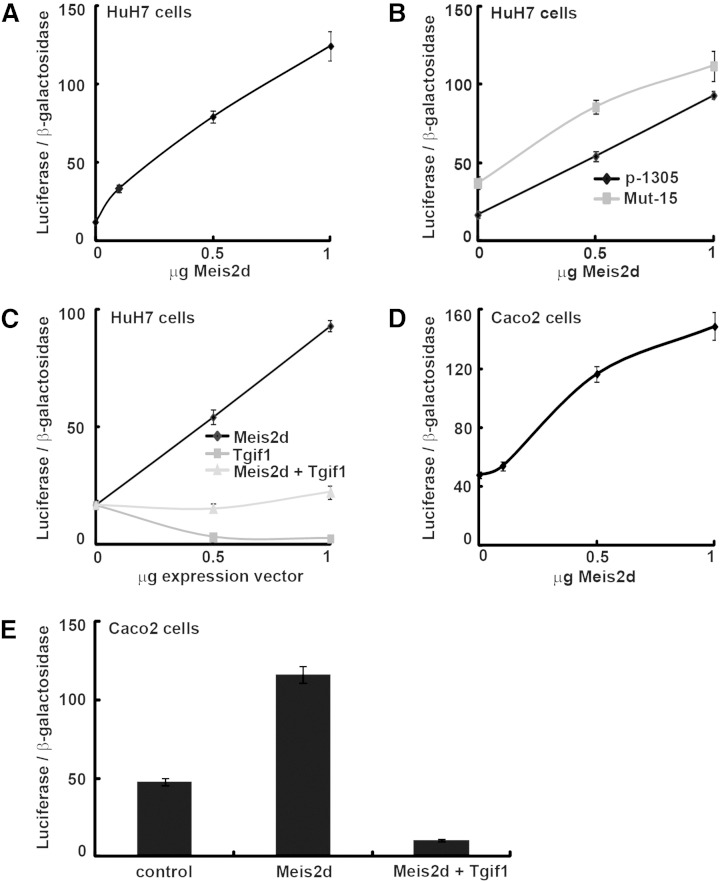
Tgif1 and Meis2 belong to the three amino acids loop extension (TALE) superfamily of homeodomains and act as a transcriptional repressor or activator, respectively (26). Five alternatively spliced Meis2 isoforms have been identified (Meis2a–e), all encoding functional proteins (26). Because Tgif1 and Meis2 bind to the same DNA sites, we used one of these isoforms, Meis2d, to investigate its role on the human SOAT2 promoter. Transient cotransfections of HuH7 cells revealed a dose-dependent increased transactivation by Meis2d overexpression (Fig. 3A). However, this transactivation was not exerted via the identified −15/−10 bp Tgif binding site (Mut-15) because Meis2d was still able to induce the promoter activity when this site was mutated (Fig. 3B). Transient cotransfections with both Tgif1 and Meis2d showed that these two transcription factors counteracted each other's effect on the SOAT2 promoter activity (Fig. 3C).
Fig. 3.
Effects of Meis2d on the SOAT2 promoter. Transient cotransfections of HuH7 cells with the human SOAT2 promoter (p-1305) (A) or with a construct of the promoter in which a binding site for Tgif was mutated (Mut-15) (B) along with a vector for Meis2d. (C) Transient cotransfections of HuH7 cells with the SOAT2 promoter and vectors for Meis2d and Tgif1. (D, E) Transient cotransfections of Caco2 cells with the SOAT2 promoter and vectors for Meis2d and Tgif1. Data are expressed as mean ± SEM (n = 4).
The cell specificity of these findings was investigated in Caco2 cells. Transient cotransfections of Caco2 cells with the human SOAT2 promoter and the expression vector for Meis2d revealed a dose-dependent increased transactivation by Meis2d overexpression (Fig. 3D). However, cotransfection with both Meis2d and Tgif1 showed a stronger repressing activity by Tgif1 on the human SOAT2 promoter in this model (Fig. 3E).
Plasma and hepatic lipids in Tgif1 null mice
To investigate the physiological effects of impaired Tgif1 function and to evaluate whether mice could function as a model for these studies, we aligned the human and mouse SOAT2 promoters (data not shown). The Tgif site responsible for the repression of SOAT2 (located at −15/−10 bp) was 100% conserved in mice and humans. This prompted us to investigate the effects of genetic depletion of Tgif1 in mice. As mentioned previously, Acat2 is involved in cholesterol esterification for storage and secretion in the lipid core of VLDL and chylomicrons and in intestinal cholesterol absorption (6, 8, 27). Genetic depletion of Tgif1 is thus expected to result in increased esterified cholesterol in the liver and in higher plasma cholesterol levels. We investigated this in male Tgif1 null mice fed a regular chow diet and euthanized at 9 to 15 weeks of age. Analysis of plasma lipids showed significantly higher plasma cholesterol levels (P < 0.05; Table 2) in Tgif1 null mice. No differences in plasma TG levels were observed. Liver analyses (Table 2) showed higher TG (P < 0.05) and esterified cholesterol (P < 0.01) mass in Tgif1 null mice compared with wild types. These data are in line with our initial predictions and further indicate a role for Tgif1 in the regulation of hepatic cholesterol metabolism.
TABLE 2.
Plasma and hepatic lipid analyses and mRNA expression levels in liver and proximal intestine in wild-type and Tgif1−/− mice
| Wild Types | Tgif1−/− | |
| Plasma lipids (mmol/l) | ||
| Cholesterol | 2.44 (2.10–2.49) | 2.78 (2.39–2.93)a |
| TGs | 0.76 (0.43–0.96) | 0.99 (0.51–1.21) |
| Hepatic lipid content (mg/g protein) | ||
| Esterified cholesterol | 15 (13–18) | 23 (20–29)b |
| Free cholesterol | 54 (51–59) | 51 (49–52) |
| TGs | 729 (485–1,302) | 1,220 (868–1,932)a |
| Hepatic mRNA expressions (AU) | ||
| Soat2 | 0.150 (0.117–0.198) | 0.223 (0.171–0.280)a |
| Soat1 | 0.094 (0.069–0.118) | 0.077 (0.062–0.143) |
| Tgif2 | 0.006 (0.004–0.009) | 0.005 (0.003–0.007) |
| Proximal intestine mRNA expressions (AU) | ||
| Soat2 | 2.338 (1.643–3.303) | 2.006 (1.676–2.518) |
| Soat1 | 1.097 (1.005–1.244) | 1.373 (0.575–1.900) |
| Tgif2 | 0.046 (0.015–0.079) | 0.099 (0.051–0.187)a |
Data are expressed as median and range in parentheses. Mann-Whitney U-tests were used to compare differences between wild-type and Tgif1−/− mice. AU, arbitrary units.
P < 0.05.
P < 0.01.
Gene expression in Tgif1 null mice
We also investigated the effects of genetic depletion of Tgif1 on gene expression in the liver and proximal intestine of these mice (Table 2). As expected, mice lacking Tgif1 had increased hepatic Soat2 mRNA expression (P < 0.05), but no effect was found on the intestinal Soat2 mRNA expression. Interestingly, we found significantly higher (P < 0.05) Tgif2 mRNA expression in the proximal intestine, but not in the liver, of Tgif1 null mice compared with wild types.
Sex-related differences in hepatic TGIF1 mRNA levels in humans
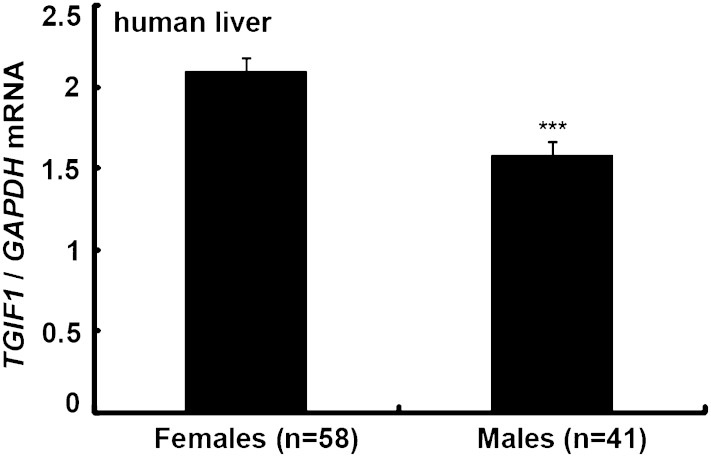
We have previously described a strong sex-related difference in hepatic Acat2 activity (28). Women have ∼70% lower Acat2 activity compared with men. We thus wanted to investigate whether sex-related differences also exist for the hepatic TGIF1 expression. Liver samples from Chinese normolipidemic gallstone patients (Table 1) were analyzed. Interestingly, we found that females had significantly higher TGIF1 mRNA levels compared with men (P < 0.001; Fig. 4).
Fig. 4.
Gene expression in humans. Hepatic TGIF1 mRNA levels were measured in 33 fertile and 24 postmenopausal female and 41 male normolipidemic Chinese subjects using real-time RT PCR. Gene expression was normalized to GAPDH mRNA, and data are expressed as mean ± SEM. Differences among patients were tested using multiway ANOVA, followed by post hoc comparisons according to the least significant difference test. *** P < 0.001.
DISCUSSION
In this study, we identified Tgif1 to function as an important transcriptional repressor of the gene encoding the cholesterol-esterifying enzyme Acat2.
Tgif1 is a homeodomain protein of the TALE superfamily, and loss-of-function mutations have been linked to holoprosencephaly, a developmental disease affecting craniofacial development (29). Tgif proteins interact with Sma- and Mad-related protein (Smad) 2 and 3 in response to transforming growth factor β signaling and repress Smad target gene expression (30) by displacement of coactivators and the recruitment of transcriptional corepressors. Tgif1 preferentially binds to the sequence CTGTCAA, with the underlined nucleotides being most important for the binding (31), but has also been shown to bind to retinoid X receptor (Rxr) binding sites and to the Rxr protein, suggesting that Tgif1 is a general repressor of retinoid signaling (22). Because Rxr is a common heterodimeric partner of many nuclear receptors, Tgif1 might repress other transcriptional pathways. There is evidence that liver X receptor/Rxr heterodimers are preferential targets for Tgif1 in mouse liver (32). Thus, Tgif1 may also regulate the pathways activated by sterols/lipids.
Previous studies regarding the transcriptional regulation of the human SOAT2 gene have identified Hnf1α, Hnf1β, and Hnf4α (17, 20) as positive regulators of its hepatic expression. Also, several sites in the SOAT2 promoter region have been reported to participate in its regulation: one binding site for Hnf1 located at −871/−866 bp (20) and four binding sites for Hnf4 located at −254/−247, −318/−311 bp, −1,006/−898 bp, and −38/−29 bp upstream of the translation start site (17, 33). Until now, no transcriptional repressor has been identified limiting therapeutic strategies to inhibit Acat2 protein expression. Here, we show that binding of Tgif1 to its cognate binding site at −15/−10 bp leads to a repression of the SOAT2 promoter activity (Fig. 1). Tgif1 is also able to block the induction of SOAT2 promoter activity by Hnf1α and Hnf4α, suggesting that Tgif1 participates in the metabolic control of SOAT2 gene expression. Binding of Tgif1 to the promoter region may thus function as a transcriptional “control point” of the human SOAT2 gene, determining its activity in human liver.
Previous studies have shown that Hnf1α and caudal type homeobox transcription factor 2 function as activators of the human SOAT2 promoter in intestinal Caco2 cells (34). For the first time, we identified Hnf4α as a strong activator of SOAT2 in the intestinal Caco2 cells (Fig. 2), and in accordance with the results in HuH7 cells, Tgif1 functions as a repressor of SOAT2 and is also able to block the stimulation by both Hnf1α and Hnf4α in these intestinal cells (Fig. 2). However, our transfection studies did not indicate that the −15/−10 bp Tgif site played such a dominant role in Caco2 cells. Unfortunately, we did not have access to human intestinal biopsies and were therefore not able to investigate the in vivo binding of Tgif1 to this or other putative binding sites. Thus, we cannot rule out a different mode of regulation of SOAT2 in intestinal cells, which may involve binding of Tgif1 to other DNA-bound factors.
Transfection of HuH7 and Caco2 cells with Tgif1 and/or Hnf4α expression vectors did not significantly alter the endogenous SOAT2 mRNA expression (supplementary Fig. II and data not shown). These findings may be due to relatively high endogenous levels of these transcription factors in HuH7 and Caco2 cells but may also be due to effects of Tgif1 and Hnf4α on other genes that in turn may affect the endogenous SOAT2 mRNA expression.
The studies in Tgif1 null mice were conducted to investigate the physiological role of Tgif1 in regulating Soat2 expression. However, major species differences in the cholesterol metabolism between humans and mice exist. For example, plasma cholesterol is mainly transported in LDL in humans and in HDL in rodents, humans have much lower cholesterol synthesis and hepatic LDL clearance (35), and humans (but not rodents) exhibit cholesteryl ester transfer protein activity in plasma. Despite these differences, we decided to use a mouse model because the Tgif site responsible for the repression of SOAT2 is conserved in mice and humans. We found that Tgif1 null mice had higher hepatic cholesteryl esters and TGs, higher hepatic Soat2 mRNA expression, and higher plasma cholesterol levels (Table 2). This further strengthens the importance of Tgif1 in regulating cholesterol metabolism. In the proximal intestine of Tgif1 null mice, we also found higher expression of the closely related and similarly functioning Tgif2 (Table 2). Thus, it is possible that Tgif2 may be able to compensate for the loss of Tgif1 in the proximal intestine as suggested by the lack of differences in Soat2 mRNA expression between Tgif1 and wild-type mice in this organ.
Here, we also showed for the first time that women have higher hepatic TGIF1 mRNA expression compared with men (Fig. 4). This finding is striking because the results here suggest that higher TGIF1 levels should be beneficial with respect to hepatic TG and cholesteryl ester accumulation (i.e., hepatic steatosis) and plasma cholesterol levels. Knowing the link between SOAT2 and hepatic steatosis or coronary heart disease, it would be interesting to study how levels of TGIF1 contribute to these diseases in humans.
Tgif1 is highly expressed in the human liver and weakly expressed in the small intestine (31), whereas Acat2 has the opposite tissue expression (2, 3, 28). Because Tgif1 functions as a repressor of SOAT2 in both hepatic and intestinal cells, it may, at least partly, be responsible for the different tissue expression levels of SOAT2 in humans.
Meis2 maps to human chromosome 15 (36), but, except for its involvement in developmental processes and cancer (37–39), not much is known about the function of this transcription factor. Although our transfection experiments revealed that Meis2d acts as a positive regulator of both the hepatic and intestinal SOAT2 expression (Fig. 3), the functional relevance of Meis2 is currently unclear and will be further investigated. We also performed ChIP assays with an antibody against Meis2 in human liver (data not shown), but we were not able to detect any binding by Meis2 to any of the identified Tgif sites in the SOAT2 promoter. However, the significance of this negative result is hard to interpret because we did not have a good positive control in our ChIP experiments.
In summary, for the first time we have identified Tgif1 as a transcriptional repressor of the human SOAT2 gene. Tgif1 is also able to oppose the induction of the promoter activity of SOAT2 by Hnf1α and Hnf4α and thereby control the hepatic and intestinal SOAT2 expression. Interestingly, sex-related differences in hepatic expression of TGIF1 are present in humans. Moreover, depletion of Tgif1 in mice increased the hepatic Soat2 expression and resulted in higher hepatic lipid accumulation and plasma cholesterol levels. Collectively, these results identify Tgif1 as a new player in cholesterol metabolism.
Supplementary Material
Acknowledgments
The authors thank Lilian Larsson for invaluable technical assistance.
Footnotes
Abbreviations:
- ChIP
- chromatin immunoprecipitation
- Hnf
- hepatocyte nuclear factor
- IP
- immunoprecipitation
- Rxr
- retinoid X receptor
- SOAT2
- sterol O-acyltransferase 2
- Tgif1
- TG-interacting factor 1
This work was supported by grants from the Swedish Research Council, the Swedish Medical Association, the Swedish Heart-Lung Foundation, the Albert and Gerda Svensson Foundation, the Swedish Diabetes Foundation, the Novo Nordisk Research Foundation, the Stockholm City Council, and the Karolinska Institutet, and by American Heart Association Grant 09GRNT2060150 (D.W.) and Natural Science Foundation of China Grant No. 81070367 (Z.J.).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two figures.
REFERENCES
- 1.Buhman K. K., Accad M., Novak S., Choi R. S., Wong J. S., Hamilton R. L., Turley S., Farese R. V., Jr 2000. Resistance to diet-induced hypercholesterolemia and gallstone formation in ACAT2-deficient mice. Nat. Med. 6: 1341–1347. [DOI] [PubMed] [Google Scholar]
- 2.Smith J. L., Rangaraj K., Simpson R., Maclean D. J., Nathanson L. K., Stuart K. A., Scott S. P., Ramm G. A., de Jersey J. 2004. Quantitative analysis of the expression of ACAT genes in human tissues by real-time PCR. J. Lipid Res. 45: 686–696. [DOI] [PubMed] [Google Scholar]
- 3.Parini P., Davis M., Lada A. T., Erickson S. K., Wright T. L., Gustafsson U., Sahlin S., Einarsson C., Eriksson M., Angelin B., et al. 2004. ACAT2 is localized to hepatocytes and is the major cholesterol-esterifying enzyme in human liver. Circulation. 110: 2017–2023. [DOI] [PubMed] [Google Scholar]
- 4.Brown J. M., Bell T. A., III, Alger H. M., Sawyer J. K., Smith T. L., Kelley K., Shah R., Wilson M. D., Davis M. A., Lee R. G., et al. 2008. Targeted depletion of hepatic ACAT2-driven cholesterol esterification reveals a non-biliary route for fecal neutral sterol loss. J. Biol. Chem. 283: 10522–10534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Willner E. L., Tow B., Buhman K. K., Wilson M., Sanan D. A., Rudel L. L., Farese R. V., Jr 2003. Deficiency of acyl CoA:cholesterol acyltransferase 2 prevents atherosclerosis in apolipoprotein E-deficient mice. Proc. Natl. Acad. Sci. USA. 100: 1262–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Repa J. J., Buhman K. K., Farese R. V., Jr, Dietschy J. M., Turley S. D. 2004. ACAT2 deficiency limits cholesterol absorption in the cholesterol-fed mouse: impact on hepatic cholesterol homeostasis. Hepatology. 40: 1088–1097. [DOI] [PubMed] [Google Scholar]
- 7.Turley S. D., Valasek M. A., Repa J. J., Dietschy J. M. 2010. Multiple mechanisms limit the accumulation of unesterified cholesterol in the small intestine of mice deficient in both ACAT2 and ABCA1. Am. J. Physiol. Gastrointest. Liver Physiol. 299: G1012–G1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nguyen T. M., Sawyer J. K., Kelley K. L., Davis M. A., Rudel L. L. 2012. Cholesterol esterification by ACAT2 is essential for efficient intestinal cholesterol absorption: evidence from thoracic lymph duct cannulation. J. Lipid Res. 53: 95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee R. G., Kelley K. L., Sawyer J. K., Farese R. V., Jr, Parks J. S., Rudel L. L. 2004. Plasma cholesteryl esters provided by lecithin:cholesterol acyltransferase and acyl-coenzyme a:cholesterol acyltransferase 2 have opposite atherosclerotic potential. Circ. Res. 95: 998–1004. [DOI] [PubMed] [Google Scholar]
- 10.Temel R. E., Lee R. G., Kelley K. L., Davis M. A., Shah R., Sawyer J. K., Wilson M. D., Rudel L. L. 2005. Intestinal cholesterol absorption is substantially reduced in mice deficient in both ABCA1 and ACAT2. J. Lipid Res. 46: 2423–2431. [DOI] [PubMed] [Google Scholar]
- 11.Alger H. M., Brown J. M., Sawyer J. K., Kelley K. L., Shah R., Wilson M. D., Willingham M. C., Rudel L. L. 2010. Inhibition of acyl-coenzyme a:cholesterol acyltransferase 2 (ACAT2) prevents dietary cholesterol associated steatosis by enhancing hepatic triglyceride mobilization. J. Biol. Chem. 285: 14267–14274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warensjo E., Sundstrom J., Vessby B., Cederholm T., Riserus U. 2008. Markers of dietary fat quality and fatty acid desaturation as predictors of total and cardiovascular mortality: a population-based prospective study. Am. J. Clin. Nutr. 88: 203–209. [DOI] [PubMed] [Google Scholar]
- 13.Ma J., Folsom A. R., Lewis L., Eckfeldt J. H. 1997. Relation of plasma phospholipid and cholesterol ester fatty acid composition to carotid artery intima-media thickness: the Atherosclerosis Risk in Communities (ARIC) Study. Am. J. Clin. Nutr. 65: 551–559. [DOI] [PubMed] [Google Scholar]
- 14.Miller C. D., Thomas M. J., Hiestand B., Samuel M. P., Wilson M. D., Sawyer J., Rudel L. L. 2012. Cholesteryl esters associated with acyl-CoA:cholesterol acyltransferase predict coronary artery disease in patients with symptoms of acute coronary syndrome. Acad. Emerg. Med. 19: 673–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka T., Jiang S., Hotta H., Takano K., Iwanari H., Sumi K., Daigo K., Ohashi R., Sugai M., Ikegame C., et al. 2006. Dysregulated expression of P1 and P2 promoter-driven hepatocyte nuclear factor-4alpha in the pathogenesis of human cancer. J. Pathol. 208: 662–672. [DOI] [PubMed] [Google Scholar]
- 16.Mendel D. B., Hansen L. P., Graves M. K., Conley P. B., Crabtree G. R. 1991. HNF-1 alpha and HNF-1 beta (vHNF-1) share dimerization and homeo domains, but not activation domains, and form heterodimers in vitro. Genes Dev. 5: 1042–1056. [DOI] [PubMed] [Google Scholar]
- 17.Pramfalk C., Karlsson E., Groop L., Rudel L. L., Angelin B., Eriksson M., Parini P. 2009. Control of ACAT2 liver expression by HNF4{alpha}: lesson from MODY1 patients. Arterioscler. Thromb. Vasc. Biol. 29: 1235–1241. [DOI] [PubMed] [Google Scholar]
- 18.Yin L., Ma H., Ge X., Edwards P. A., Zhang Y. 2011. Hepatic hepatocyte nuclear factor 4alpha is essential for maintaining triglyceride and cholesterol homeostasis. Arterioscler. Thromb. Vasc. Biol. 31: 328–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuo C. J., Conley P. B., Chen L., Sladek F. M., Darnell J. E., Jr, Crabtree G. R. 1992. A transcriptional hierarchy involved in mammalian cell-type specification. Nature. 355: 457–461. [DOI] [PubMed] [Google Scholar]
- 20.Pramfalk C., Davis M. A., Eriksson M., Rudel L. L., Parini P. 2005. Control of ACAT2 liver expression by HNF1. J. Lipid Res. 46: 1868–1876. [DOI] [PubMed] [Google Scholar]
- 21.Bjerke G. A., Hyman-Walsh C., Wotton D. 2011. Cooperative transcriptional activation by Klf4, Meis2, and Pbx1. Mol. Cell. Biol. 31: 3723–3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartholin L., Powers S. E., Melhuish T. A., Lasse S., Weinstein M., Wotton D. 2006. TGIF inhibits retinoid signaling. Mol. Cell. Biol. 26: 990–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartholin L., Melhuish T. A., Powers S. E., Goddard-Leon S., Treilleux I., Sutherland A. E., Wotton D. 2008. Maternal Tgif is required for vascularization of the embryonic placenta. Dev. Biol. 319: 285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bligh E. G., Dyer W. J. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 25.Ohshiro T., Matsuda D., Sakai K., Degirolamo C., Yagyu H., Rudel L. L., Omura S., Ishibashi S., Tomoda H. 2011. Pyripyropene a, an acyl-coenzyme a:cholesterol acyltransferase 2-selective inhibitor, attenuates hypercholesterolemia and atherosclerosis in murine models of hyperlipidemia. Arterioscler. Thromb. Vasc. Biol. 31: 1108–1115. [DOI] [PubMed] [Google Scholar]
- 26.Yang Y., Hwang C. K., D'Souza U. M., Lee S. H., Junn E., Mouradian M. M. 2000. Three-amino acid extension loop homeodomain proteins Meis2 and TGIF differentially regulate transcription. J. Biol. Chem. 275: 20734–20741. [DOI] [PubMed] [Google Scholar]
- 27.Turley S. D., Dietschy J. M. 2003. Sterol absorption by the small intestine. Curr. Opin. Lipidol. 14: 233–240. [DOI] [PubMed] [Google Scholar]
- 28.Jiang Z. Y., Jiang C. Y., Wang L., Wang J. C., Zhang S. D., Einarsson C., Eriksson M., Han T. Q., Parini P., Eggertsen G. 2009. Increased NPC1L1 and ACAT2 expression in the jejunal mucosa from Chinese gallstone patients. Biochem. Biophys. Res. Commun. 379: 49–54. [DOI] [PubMed] [Google Scholar]
- 29.Gripp K. W., Wotton D., Edwards M. C., Roessler E., Ades L., Meinecke P., Richieri-Costa A., Zackai E. H., Massague J., Muenke M., et al. 2000. Mutations in TGIF cause holoprosencephaly and link NODAL signalling to human neural axis determination. Nat. Genet. 25: 205–208. [DOI] [PubMed] [Google Scholar]
- 30.Wotton D., Lo R. S., Lee S., Massague J. 1999. A Smad transcriptional corepressor. Cell. 97: 29–39. [DOI] [PubMed] [Google Scholar]
- 31.Bertolino E., Reimund B., Wildt-Perinic D., Clerc R. G. 1995. A novel homeobox protein which recognizes a TGT core and functionally interferes with a retinoid-responsive motif. J. Biol. Chem. 270: 31178–31188. [DOI] [PubMed] [Google Scholar]
- 32.Melhuish T. A., Chung D. D., Bjerke G. A., Wotton D. 2010. Tgif1 represses apolipoprotein gene expression in liver. J. Cell. Biochem. 111: 380–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Z., Liu J., Xi Y., Yang R., Chen H., Li Z., Liu D., Liang C. 2012. Two novel cis-elements involved in hepatocyte nuclear factor 4alpha regulation of acyl-coenzyme A:cholesterol acyltransferase 2 expression. Acta Biochim. Biophys. Sin. (Shanghai). 44: 162–171. [DOI] [PubMed] [Google Scholar]
- 34.Song B. L., Wang C. H., Yao X. M., Yang L., Zhang W. J., Wang Z. Z., Zhao X. N., Yang J. B., Qi W., Yang X. Y., et al. 2006. Human acyl-CoA:cholesterol acyltransferase 2 gene expression in intestinal Caco-2 cells and in hepatocellular carcinoma. Biochem. J. 394: 617–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dietschy J. M., Turley S. D. 2002. Control of cholesterol turnover in the mouse. J. Biol. Chem. 277: 3801–3804. [DOI] [PubMed] [Google Scholar]
- 36.Smith J. E., Afonja O., Yee H. T., Inghirami G., Takeshita K. 1997. Chromosomal mapping to 15q14 and expression analysis of the human MEIS2 homeobox gene. Mamm. Genome. 8: 951–952. [DOI] [PubMed] [Google Scholar]
- 37.Larsen K. B., Lutterodt M. C., Laursen H., Graem N., Pakkenberg B., Mollgard K., Moller M. 2010. Spatiotemporal distribution of PAX6 and MEIS2 expression and total cell numbers in the ganglionic eminence in the early developing human forebrain. Dev. Neurosci. 32: 149–162. [DOI] [PubMed] [Google Scholar]
- 38.Oulad-Abdelghani M., Chazaud C., Bouillet P., Sapin V., Chambon P., Dolle P. 1997. Meis2, a novel mouse Pbx-related homeobox gene induced by retinoic acid during differentiation of P19 embryonal carcinoma cells. Dev. Dyn. 210: 173–183. [DOI] [PubMed] [Google Scholar]
- 39.Crijns A. P., de Graeff P., Geerts D., Ten Hoor K. A., Hollema H., van der Sluis T., Hofstra R. M., de Bock G. H., de Jong S., van der Zee A. G., et al. 2007. MEIS and PBX homeobox proteins in ovarian cancer. Eur. J. Cancer. 43: 2495–2505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.