Abstract
Oxidation of LDL by the myeloperoxidase (MPO)-H2O2-chloride system is a key event in the development of atherosclerosis. The present study aimed at investigating the interaction of MPO with native and modified LDL and at revealing posttranslational modifications on apoB-100 (the unique apolipoprotein of LDL) in vitro and in vivo. Using amperometry, we demonstrate that MPO activity increases up to 90% when it is adsorbed at the surface of LDL. This phenomenon is apparently reflected by local structural changes in MPO observed by circular dichroism. Using MS, we further analyzed in vitro modifications of apoB-100 by hypochlorous acid (HOCl) generated by the MPO-H2O2-chloride system or added as a reagent. A total of 97 peptides containing modified residues could be identified. Furthermore, differences were observed between LDL oxidized by reagent HOCl or HOCl generated by the MPO-H2O2-chloride system. Finally, LDL was isolated from patients with high cardiovascular risk to confirm that our in vitro findings are also relevant in vivo. We show that several HOCl-mediated modifications of apoB-100 identified in vitro were also present on LDL isolated from patients who have increased levels of plasma MPO and MPO-modified LDL. In conclusion, these data emphasize the specificity of MPO to oxidize LDL.
Keywords: inflammation, myeloperoxidase activity, hypochlorous acid, 3-chlorotyrosine, epitope mapping, low density lipoprotein, apolipoprotein B-100
Over the past several years, considerable evidence has been obtained in support of the hypothesis that oxidants generated by the heme enzyme myeloperoxidase (MPO, EC1.11.2.2) play a key role in oxidation reaction of the artery wall. The enzyme, abundantly present in neutrophils and, to a lesser extent, in monocytes, is released during inflammatory activation of immune cells. MPO produces hypochlorous acid (HOCl) by the reaction of hydrogen peroxide (H2O2) and chloride ions (Cl−), and this potent oxidant contributes to the antimicrobial activity of phagocytes (1, 2). However, evidence has emerged that either chronic or prolonged production of HOCl by the MPO-H2O2-Cl− system contributes to tissue damage and the initiation and propagation of vascular diseases (3). HOCl-modified epitopes were present in acute and chronic vascular inflammatory diseases where staining was found to be associated with monocytes/macrophages, smooth muscle cells, and endothelial cells (4–6). As human atherosclerotic lesions contain elevated levels of MPO, the enzyme may act as a catalyst for LDL oxidation (7). Furthermore, the oxidation of LDL/apoB-100, leading to species called “oxidized LDLs (OxLDLs),” plays a crucial role in the pathogenesis of atherosclerosis (8–10). Fingerprints for in vivo modifications of apoB-100 by the MPO-H2O2-Cl− system were observed by immunohistological analyses (4, 11) and GC-MS (12). Observations that an increasing oxidant/LDL molar ratio of HOCl-modified apoB-100 is paralleled by a decreased ligand interaction by the classical LDL receptor (13) suggested that scavenger receptors on macrophages mediate the uptake of HOCl-modified LDL (HOCl-LDL) (14, 15). In addition to its capacity to promote lipid accumulation in monocytes/macrophages (16), HOCl-LDL adversely affects biological properties of smooth muscle cells and endothelial cells (13), thus favoring progression of atherosclerosis. Furthermore, LDL oxidized by the MPO-H2O2-Cl− system (MPO-LDL) accumulates in macrophages and exerts proinflammatory effects on monocytes and endothelial cells (17, 18).
Modification by reagent HOCl alters the lipid moiety of LDL but primarily leads to amino acid oxidation favoring posttranslational modification (PTM) of the protein moiety. Lysine (Lys), histidine (His), and the N-terminal α-amino group may form reactive chloramine species, which may lead to secondary oxidation processes (13). Methionine (Met) can be converted into sulfoxide form while tyrosine (Tyr) may be converted into 3-chlorotyrosine (Cl-Tyr), a specific marker for the MPO-H2O2-Cl− system-mediated oxidation in vivo (12, 19, 20) and in vitro (19). Furthermore, it has been reported that MPO, probably due to its charge, can bind LDL (21, 22). That binding, which seems to be mediated via the protein moiety of LDL (23), may result in localized formation of oxidants and hence side-specific damages (22, 24). ApoB-100, the unique protein of LDL, is a highly hydrophobic protein with 4,536 amino acid residues (molecular mass 550 kDa). Furthermore, apoB-100 contains a high number of amino acid residues prone to be modified by HOCl.
In the present work, we have studied the impact of adsorption of MPO on native and HOCl-modified LDL and on its structural and enzymatic features. Using high-resolution MS, we then performed a comprehensive survey of PTMs on apoB-100 treated with HOCl added as reagent or generated enzymatically. Numerous modifications were identified including methionine sulfoxide (O-Met), (di)-oxidized tryptophan [(di-)OxTrp], and Cl-Tyr. Finally, we compared these in vitro findings with oxidation patterns of LDL that has been isolated from hemodialysis patients who have increased levels of MPO and MPO-LDL (25).
EXPERIMENTAL SECTION
Materials and reagents
Ammonium bicarbonate, formic acid (FA), NaCl, K2HPO4, methanol, and acetonitrile were from Merck (Darmstadt, Germany). Trichloroacetic acid, butylated hydroxytoluene (BHT), dithiothreitol, iodoacetamide, 13C9-tyrosine, 13C15N-lysine, thioglycolic acid, 2,2,2-trifluoroethanol, tosyl phenylalanyl chloromethyl ketone-treated trypsin (from bovine pancreas), and peptide-N-glycosidase (PNGase) F (from Elizabethkingia meningoseptica) were from Sigma-Aldrich (St. Louis, MO). NaOCl solution was from Carl Roth GmbH + Co. KG (Karlsruhe, Germany). PD-10 desalting columns were from GE Healthcare (Fairfield, CT). Agilent Si-SCX 1 ml 100 mg solid phase extraction cartridges, HCl, phenol, and benzoic acid were from VWR (Leuven, Belgium). RapiGest SF was from Waters (Milford, MA). Water was purified using a Milli-Q purification system (Millipore, Bedford, MA). MPO isolated from human neutrophils was from Planta (Vienna, Austria), and human recombinant MPO was expressed in CHO cells (26).
Isolation and oxidation of LDL
Isolation of LDL.
LDL was isolated by ultracentrifugation from healthy blood donors at the A. Vesale Hospital (Charleroi, Belgium) (27). Protein content was measured using the Lowry technique (28).
Chemical oxidation of LDL (HOCl-LDL).
LDL (1 mg) was dispersed in 1 ml PBS (150 mM Cl− and 10 mM phosphate, pH 7.4) and incubated for 2 h at 37°C with NaOCl (added as a single addition with gentle vortexing) at oxidant/LDL molar ratios in between 25:1 and 333:1. When needed, 1 mM Met was added to HOCl-LDL preparations to quench the reaction at the end.
Enzymatic oxidation of LDL (MPO-LDL).
LDL (1 mg) was dispersed in 1 ml PBS (pH 7.4) and incubated with MPO and H2O2 for 4 h at 37°C. Two soft oxidations by MPO were performed using 110 nM of MPO and 50 or 100 µM of H2O2. Assuming quantitative conversion of H2O2, this results in formation of almost 50 and 100 µM HOCl [93% of conversion of H2O2 in HOCl was measured by comparing taurine chlorination by HOCl or the MPO system (29, 30)], equivalent to a molar ratio of oxidant/lipoprotein of 25:1 and 50:1, respectively. Controls were performed in the absence of MPO.
A more intensive enzymatic oxidation was performed as described previously (11). Briefly, MPO-LDL was generated by mixing 8 µl of HCl 1 M (final concentration: 4 mM), 45 µl of MPO (final concentration: 250 nM), a volume containing 1.6 mg LDL (final concentration: 0.8 mg/ml in PBS, pH 7.4), and 40 µl of H2O2 50 mM (final concentration: 1 mM). The volume was adjusted to 2 ml with PBS (pH 7.4) containing 1 g/l of EDTA. In this oxidation condition, the oxidant/lipoprotein molar ratio is 625:1.
Copper oxidation.
Briefly, LDL (1 mg/ml) in PBS was incubated with 10 µM CuSO4 for 24 h at 37°C (18). The oxidation was stopped by the addition of 25 µM BHT and incubation on ice for 1 h.
Isolation of apoB-100 from LDL
ApoB-100 was isolated from LDL as described previously (31). Briefly, LDL was precipitated with 500 µl trichloroacetic acid (10%, v/v) and centrifuged for 10 min at 4,500 g. The supernatant was discarded, and the precipitate was treated one more time with trichloroacetic acid. Pellets were then mixed with 1.1 ml of water-methanol-diethyl ether (1:3:7, v/v/v) to remove lipids. The mixture was sonicated for 30 min and then centrifuged for 10 min at 4500 g. The supernatant was discarded, and the procedure was repeated.
Isolation of apoB-100 from patients
Alternatively, LDL was isolated from patients on chronic maintenance hemodialysis therapy (n = 9) or healthy volunteers (n = 9). Briefly, blood (5 ml) was drawn before dialysis in patients or in volunteers. Four hundred microliters of LDL solution from each patient/volunteer was treated as described previously. This study conforms to the Declaration of Helsinki, and its protocol was approved by the Ethics Committee of the ISPPC (“Intercommunale de Santé Publique du Pays de Charleroi”) Hospital. Finally, all subjects gave their written informed consent.
Amperometric measurements of MPO activity
The chlorination activity of MPO (100 nM) in the absence or presence of indicated lipoprotein concentrations was measured by continuously monitoring H2O2 consumption by amperometry using a combined platinum/reference electrode, which was covered with a hydrophilic and a dialysis membrane (cutoff 3,600 Da), fitted to the Amperometric Biosensor Detector 3001 (Universal Sensors Inc.). The applied electrode potential at pH 7.4 was 650 mV, and the electrode filling solution was freshly prepared half-daily (32). The electrode was calibrated against known H2O2 concentrations. In detail, consumption of H2O2 (100 µM) in the presence of 100 mM Cl− and 10 mM PBS (pH 7.4) was followed for 5 min at 25°C. The presence of LDL preparations did not affect the amperometric measurement of H2O2. In the presence of H2O2, Cl−, and LDL, no H2O2 consumption occurred within the 5 min experiment as long as MPO was absent. Reactions were started by addition of MPO either free or preincubated for 5 min with LDL or HOCl-LDL. One unit of chlorination activity is defined as the consumption of 1 µmol of H2O2 per minute at 25°C in the presence of 100 mM Cl−.
The peroxidase activity of MPO was measured in 10 mM PBS (pH 7.4) using 100 µM of Tyr as the one-electron donor. One unit of peroxidase activity is defined as the consumption of 1 µmol H2O2 per minute at 25°C in the presence of 100 µM Tyr.
Circular dichroism spectrometry
Circular dichroism (CD) spectra were performed on a Chirascan spectrometer equipped with a thermostatic cell holder (Applied Photophysics, Leatherhead, UK). For recording far-UV spectra (190–260 nm), conditions were as follows: path length, 1 mm; spectral bandwidth, 3 nm; step size, 1 nm; scan time by nanometer, 10 s; protein concentrations, 0.1 mg/ml MPO and 0.1 mg/ml LDL or HOCl-LDL (oxidant/LDL molar ratio of 50:1). In the near-UV visible region (260–480 nm), conditions were as follows: path length, 10 mm; spectral bandwidth, 1 nm; step size, 1 nm; scan time by nanometer, 8 s; protein concentrations, 0.5 mg/ml MPO and 0.5 mg/ml LDL or HOCl-LDL (oxidant/LDL ratio = 50:1). All CD measurements were performed in 5 mM PBS (pH 7.4) at 25°C (33). Each spectrum was automatically corrected with the baseline to remove birefringence of the cell.
ApoB-100 hydrolysis for amino acid analysis
ApoB-100 protein pellets (obtained from isolated native or oxidized LDL samples) were hydrolyzed using a StartS microwave oven and a protein hydrolysis reactor according to the manufacturer's protocol (Milestone, Italy). Briefly, apoB-100 (1 mg) was placed into the vial, 200 µl of acid mixture [6 M HCl supplemented with 10% (v/v) thioglycolic acid, 0.1% (m/v) phenol, and 0.1% (m/v) benzoic acid] and internal standards were added, and hydrolysis was carried out for 40 min at 160°C. 13C9-Tyr and 13C15N-Lys were used as internal standards. Samples were thereafter purified using Si-SCX solid phase extraction cartridges. Briefly, columns were flushed twice with 1 ml methanol and then equilibrated with 2 ml FA (0.2 M). After hydrolysis, the samples were loaded and washed with 2 ml FA (0.2 M). Amino acids were eluted with 2 ml methanol containing 5% (w/v) NH4OH. Samples were evaporated to dryness using a vacuum centrifuge and finally dissolved in 50 µl water before injection of 10 µl into the LC-MS system.
The LC system was a rapid resolution LC (RRLC) 1200 series using a Zorbax Eclipse XDB Phenyl RR column [4.6 × 150 mm inner diameter (ID), 3.5 µM particle size] and coupled to an electrospray ion source (ESI) in positive mode quadrupole TOF (QTOF) 6520 series mass spectrometer from Agilent Technologies. Amino acid residues were separated by an acetonitrile gradient, and amino acids of interest were analyzed by MS/MS using Mass Hunter Acquisition® and Qualitative Analysis® (Agilent Technologies).
Identification of PTMs of ApoB-100
Digestion.
Protein pellets from native or oxidized LDL preparations were treated using an optimized method that ensures an optimal protein recovery (31). Briefly, apoB-100 was unfolded using 250 µl of RapiGest SF (Waters) 0.2% (w/w) in 50 mM ammonium bicarbonate buffer (pH 7.8), reduced using dithiothreitol (20 mM) at 37°C during 30 min, and finally, alkylated with iodoacetamide (60 mM) for 30 min in the dark. The solution was heated at 100°C (5 min), and apoB-100 was then digested by trypsin (enzyme-protein = 1:10, w/w) at 37°C for 24 h. The reaction was then stopped by heating the sample at 100°C for 30 min. Deglycosylation of tryptic peptides was performed with PNGase F (10 U/mg of protein) during 24 h at 37°C. The sample was then adjusted to 0.5% FA, incubated (30 min, 37°C), and centrifuged (10 min, 13,000 g). The supernatant was evaporated to dryness in a centrifugal vacuum evaporator. Finally, peptides were dissolved in 50 µl FA [0.1% (v/v) in water] before analysis.
Additionally, unfolding of protein was performed by 2,2,2-trifluoroethanol. Briefly, to 1 mg protein, 50 µl of 2,2,2-trifluoroethanol, 50 µl of ammonium bicarbonate buffer (100 mM, pH 7.8), and 2 µl of 200 mM dithiothreitol were added. The sample was heated at 60°C (1 h) and then cooled to 20°C. Eight microliters of 200 mM iodoacetamide was added, and the sample was kept in the dark at 20°C (1 h). Finally, 2 µl of 200 mM dithiothreitol was added, and the sample was kept at 20°C (1 h) in the dark before dilution with 600 µl of water and 200 µl of bicarbonate buffer (100 mM). ApoB-100 was then digested at 37°C (24 h) by adding 50 µl of trypsin (enzyme-protein = 1:10, w/w). Trypsin was then inactivated by heating the sample (100°C, 30 min). After cooling to 20°C, deglycosylation was performed at 37°C (24 h) by adding 10 units of PNGase F. Finally, the sample was acidified by addition of 2 µl of FA (100%) and evaporated to dryness in a centrifugal vacuum evaporator. Peptides were dissolved in 50 µl of FA [0.1% (v/v) in water] before analysis.
LC-MS/MS process, data acquisition, and analysis.
Ten microliters of the resulting samples was injected into the LC system and analyzed as described previously (31). Briefly, analyses were performed with the same RRLC 1200 series mentioned previously. Peptides were separated on a Fused Core Ascentis® Express C18 column (100 × 2.1 mm ID, 2.7 µm particle size) from Supelco (Bellefonte, PA) using a 105 min gradient of FA and acetonitrile. The ESI-QTOF mass spectrometer mentioned previously was used for the MS/MS analyses. Auto-MS/MS spectra were acquired in positive and high-resolution acquisition mode (4 GHz) (31). Data were acquired by the Mass Hunter Acquisition® software and analyzed by the Mass Hunter Qualitative Analysis with Bioconfirm® and by Spectrum Mill® software (Agilent Technologies). Peptide identification and validation were based on mass error (ppm), peptide scores, and % score peak intensity (SPI), which are essential to validate peptide mapping as previously described (31).
To improve the sensitivity for the analysis of the samples from patients, a nanoLC system coupled to the QTOF-MS was used. Tryptic peptides were separated on an Agilent Technologies nanoLC Chip Cube II system using a Polaris HR nanochip column. This consists of a 360 nl enrichment column and a 75 µm × 150 mm separation column, both packed with Polaris C18 phasis 3 µm particle size, 180 Å pore size. Mobile phases and gradient were identical to those used for the RRLC separation mentioned previously. Samples were loaded on the enrichment column of the chip using the capillary pump in 97% of the aqueous mobile phase at a flow rate of 1.5 µl/min. The nanoflow pump was used to generate the analytical gradient with a flow of 0.40 µl/min. MS/MS parameters were identical to those used for RRLC analyses.
Statistical analysis
Data were analyzed using the SigmaPlott® 12.0 software (Systat®, San Jose, CA). Differences were considered statistically significant with a two-tailed P < 0.05. Comparisons were made using one-way ANOVA and a Dunnett's post hoc test.
RESULTS
Activity and structure of MPO at the surface of LDL
Measurement of MPO activity.
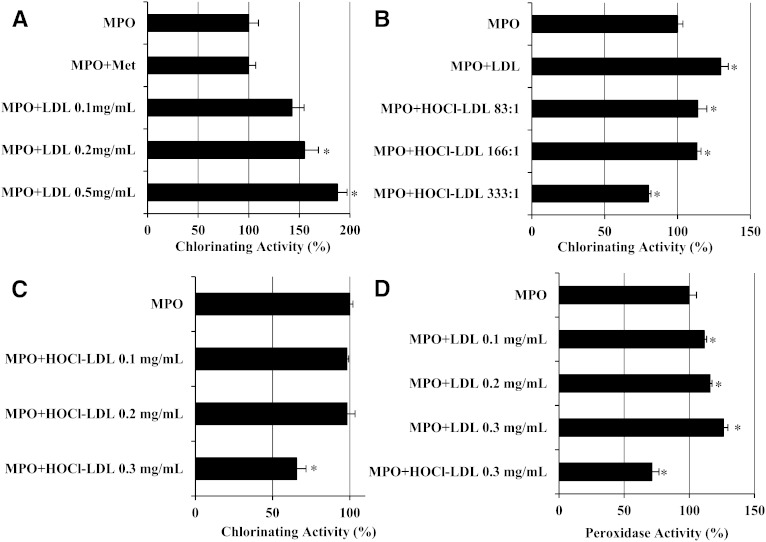
MPO interacts preferentially with the protein moiety of native LDL (23, 34). It is likely that the enzyme may interact in a similar manner with modified LDL; we therefore studied whether this interaction is paralleled by changes in the activity of MPO. Figure 1A shows that the chlorinating activity of MPO increases as a function of increasing LDL concentration; at the highest LDL concentration, the chlorinating activity of MPO increased by 90%. In order to investigate whether an increased consumption of H2O2 could result from scavenging of HOCl by LDL (which would protect MPO from inhibition by its own product), Met was added. However, the presence of this HOCl scavenger had no impact on the observed kinetics when compared with samples containing MPO only.
Fig. 1.
Measurement of MPO activity by amperometric monitoring of H2O2 consumption. Chlorinating activity of MPO (100 nM) was measured in the presence of 100 mM Cl− and (A) Met as HOCl scavenger or indicated native-LDL concentrations, (B) 0.3 mg/ml native LDL or HOCl-LDL (at indicated oxidant/lipoprotein molar ratio), or (C) indicated concentrations of HOCl-LDL (oxidant/lipoprotein molar ratio = 333:1). (D) Peroxidase activity of MPO (100 nM) was measured in the presence of 100 µM Tyr and indicated concentrations of native LDL or 0.3 mg/ml HOCl-LDL (oxidant/lipoprotein molar ratio = 333:1). MPO and lipoproteins were incubated 5 min at 25°C before measurement. MPO or MPO-lipoprotein solutions were added to start the reaction. Values are given as percentages of activity of free MPO (100% activity). One unit of activity is defined as the consumption of 1 µmol H2O2 per minute at 25°C in the presence of 100 µM electron donor (Cl− or Tyr). Results represent mean ± SD of at least three experiments. * P < 0.05 (significant difference versus MPO).
Second, the effect of HOCl-LDL on H2O2 consumption by MPO was investigated. Compared with native LDL (0.3 mg/ml), the chlorinating activity of MPO incubated with HOCl-LDL at identical concentrations was lower (Fig. 1B). Although at low oxidant/lipoprotein molar ratios (83:1 and 166:1) the chlorinating activity of MPO was significantly higher (∼20%) compared with MPO alone, a high oxidant/lipoprotein molar ratio (333:1) significantly decreased MPO activity by ∼20%.
Third, we tested whether the chlorinating activity of MPO is dependent on the concentration of HOCl-LDL at a given oxidant/lipoprotein molar ratio (333:1). Preincubation of MPO with HOCl-LDL did not affect the chlorinating activity at low concentrations of HOCl-LDL, whereas consumption of H2O2 by MPO was decreased by ∼35% at the highest lipoprotein concentration (Fig. 1C).
Next, we investigated whether the observed behavior was independent of an electron donor and thus independent of the nature of the oxidation product released by MPO. Peroxidase activity in the presence of Tyr showed similar effects on H2O2 consumption by MPO, namely increased activity in the presence of native LDL but decreased peroxidase activity in the presence of highly modified LDL (Fig. 1D).
These data illustrate that native LDL enhances its ability to be oxidized by MPO apparently due to a specific interaction between MPO and nonmodified apoB-100. In contrast, HOCl-LDL may decrease the activity of MPO, suggesting that oxidation of apoB-100 by HOCl may modulate the MPO-apoB-100 interaction.
Structural features of MPO adsorbed on LDL.
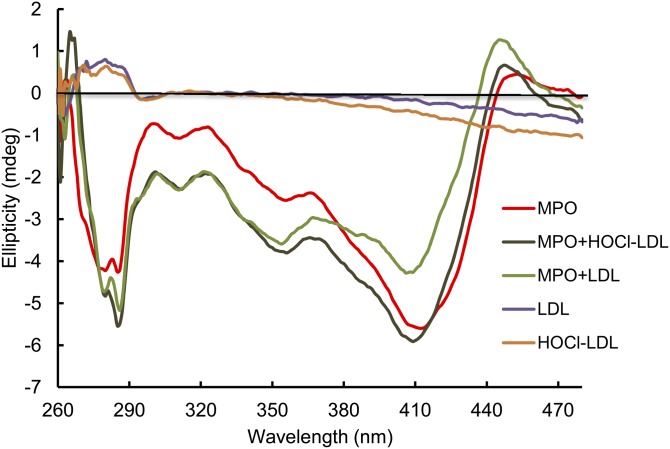
Next, we investigated whether altered activity of MPO after preincubation with native LDL or HOCl-LDL (Fig. 1) could also be reflected by changes on a spectroscopic basis. Electronic CD spectroscopy was chosen because it allows analysis of both the overall secondary structure and the (asymmetric) environment of the prosthetic heme group. In the far-UV region, the spectra of MPO in the presence of LDL or HOCl-LDL exactly matched the sum of spectra of individual lipoproteins at the same protein concentrations (data not shown). This suggests that the structural changes must be local and that the overall content of the secondary structure of MPO (which is mainly α-helical) is not affected.
However, this observation seems to be in contrast to the Soret region. Native MPO has a Soret minimum at 410 nm, whereas the ellipticity of native LDL and HOCl-LDL is negligible at this wavelength region (Fig. 2). Upon incubation of MPO with native LDL, ellipticity is lost in the Soret region, whereas in the near-UV range (260–350 nm) the difference was very small. This observation is indicative for structural changes in the immediate surroundings of the prosthetic group. The effect on the CD of the heme group was smaller when MPO was incubated with HOCl-LDL. The Soret minimum shifted to 411 nm with native MPO-like ellipticity, whereas between 300 and 350 nm ellipticity was lost. This indicates that HOCl-LDL has some local impact on the tertiary structure of MPO but not on the architecture of the active site. In any case, the observed changes in the enzymatic activity in the presence of native- or HOCl-LDL are reflected by structural changes at the active site of the peroxidase with native LDL having a stronger impact than HOCl-LDL.
Fig. 2.
Structural investigation of MPO by CD. CD near-UV visible spectra of 0.5 mg/ml free MPO (red), 0.5 mg/ml LDL (mauve), HOCl-LDL (oxidant/lipoprotein molar ratio = 50:1, orange), or 0.5 mg/ml MPO incubated with 0.5 mg/ml LDL (light green) or with 0.5 mg/ml HOCl-LDL (oxidant/lipoprotein molar ratio = 50:1, dark green) in 5 mM PBS (pH 7.4) at 25°C. For details, see the Experimental Section. mdeg, millidegree.
MS analysis of apoB-100 modified chemically or enzymatically
Total hydrolysis of apoB-100.
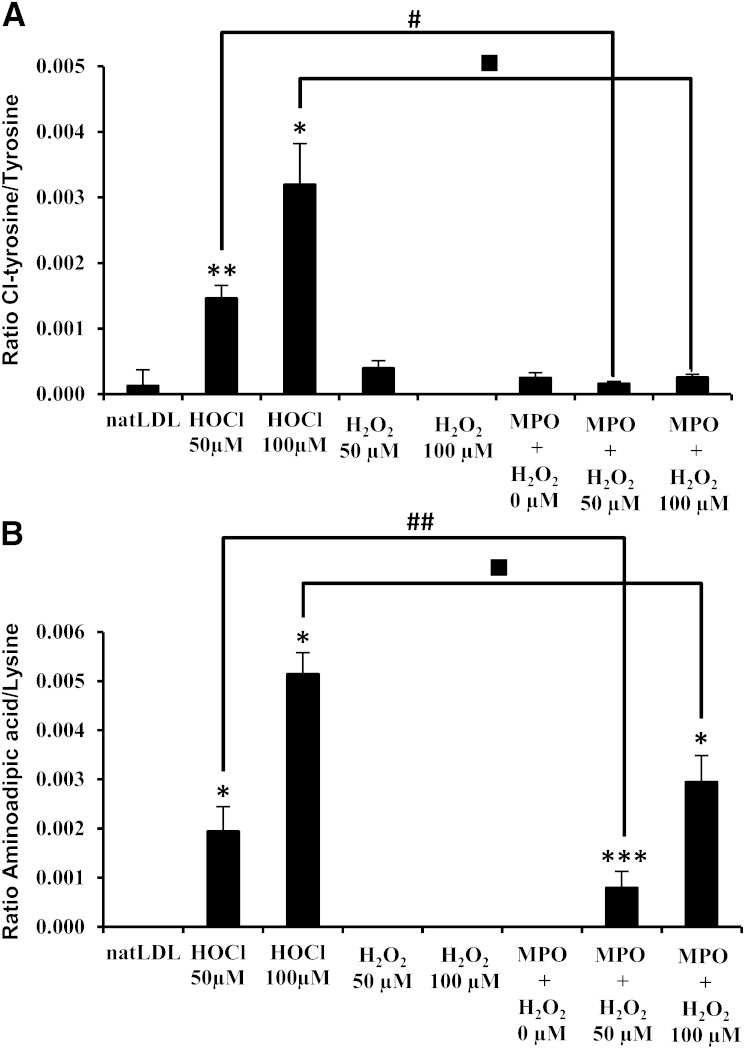
In order to investigate whether modifications of apoB-100 differed when LDL was oxidized under several conditions, we investigated the formation of Cl-Tyr, O-Met, (di-)OxTrp, and aminoadipic acid, a degradation product of N-chloramine Lys (35, 36). Figure 3 shows that the ratios of aminoadipic acid/Lys and Cl-Tyr/Tyr (Fig. 3A, B, respectively) increased as a function of increasing concentrations of HOCl (added as a reactant), although oxidation of apoB-100 by HOCl (generated by the MPO-H2O2-Cl− system) resulted in formation of Cl-Tyr only. No increase in the aminoadipic acid/Lys ratio was found when apoB-100 was oxidized enzymatically or when H2O2 and Cl− were added in the absence of MPO (Fig. 3).
Fig. 3.
Aminoadipic acid and Cl-Tyr quantification after in vitro oxidation. Amino acid residues of apoB-100 from native LDL (natLDL), HOCl-LDL (50 or 100 µM of HOCl), and LDL oxidized by H2O2 (50 or 100 µM) or by the MPO-H2O2-Cl− system (0, 50, or 100 µM H2O2 considering almost equimolar formation of HOCl from H2O2) were analyzed after total acid hydrolysis. Ratios of (A) aminoadipic acid/Lys and (B) Cl-Tyr/Tyr were calculated for each oxidation condition. Results represent mean ± SEM of three experiments. * P < 0.001, ** P < 0.005, and *** P < 0.05 versus natLDL; # P < 0.001 and ## P < 0.05 versus HOCl-LDL 50 µM; and ■ P < 0.001 versus HOCl-LDL 100 µM.
Next, we focused on oxidation of Met and tryptophan (Trp). Independent of whether apoB-100 was native or modified (either chemically or enzymatically), Met was widely oxidized. Formation of OxTrp and di-OxTrp was found in all LDL/apoB-100 samples with no statistical difference even if there was a tendency to increased levels of di-OxTrp under both oxidative conditions (twice the level observed in native LDL).
To confirm that chlorination is specific for HOCl treatment, apoB-100 was oxidized by copper (18) and then subjected to hydrolysis. No Cl-Tyr formation was observed, while low levels of (di-)OxTrp may also occur, data that parallel previous findings (37).
Mapping of in vitro modifications on apoB-100.
Next, we analyzed the location of PTMs on apoB-100. Instead of RapiGest SF (31), 2,2,2-trifluoroethanol was also used as the unfolding agent to recover apoB-100 with the same results on protein sequence recovery (79%) or even higher.
Analyses revealed that few residues (Met4, Met785, Met1873, Met2015, Trp2468, Trp3943, and Met4192) were already oxidized in native LDL. The latter residues seem thus to be highly sensitive to oxidation.
Oxidation of apoB-100 was first performed by reagent HOCl under soft conditions (oxidant/lipoprotein molar ratio = 25:1 or 50:1). Table 1 summarizes all PTMs identified in the corresponding tryptic peptides. A total of 43 modified peptides have been identified, and modifications (n = 46) include formation of O-Met (n = 33), OxTrp (n = 8), di-OxTrp (n = 3), and Cl-Tyr (n = 2). In most peptides, a single amino acid modification (primarily O-Met but also OxTrp, di-OxTrp, or Cl-Tyr) was found, while in three peptides two modifications were observed (either two O-Met residues or the combination of O-Met and OxTrp).
TABLE 1.
List of apoB-100-derived tryptic peptides carrying modifications generated by reagent HOCl
| Peptide | Error (ppm) | Peptide Score | % SPI | Z | Sequence with Fragmentation | Modified Amino Acid |
| 1–14 | 1.9 | 17.8 | 91 | 2 | (-)E E/E m/L/E/N/V|S/L/V|C/P K(D) | O-Met4 |
| 91–101 | 0.5 | 12.9 | 71 | 2 | (K)N S|E|E|F/A/A/A/m/S/R(Y) | O-Met99 |
| 203–220 | 3.3 | 17.8 | 92 | 2 | (K)G m/T R\P L\S T L|I|S|S|S Q|S|C|Q\Y(T) | O-Met204 |
| 249–260 | 2.1 | 18.0 | 85 | 2 | (K)Y G/m|V|A|Q|V|T/Q/T L/K(L) | O-Met251 |
| 279–287 | 2.0 | 13.6 | 93 | 2 | (K)m/G|L|A/F/E/S T/K(S) | O-Met279 |
| 401–407 | 3.0 | 10.2 | 79 | 2 | (R)E I|F/N/m/A/R(D) | O-Met405 |
| 464–480 | 3.0 | 15.2 | 76 | 2 | (R)V/I/G/N m|G Q/T m E Q/L/T|P E/L/K(S) | O-Met468, 472 |
| 486–498 | 3.5 | 11.6 | 74 | 3 | (K)C/V|Q/S T K/P S L m I Q/K(A) | O-Met495 |
| 532–540 | 1.3 | 15.5 | 85 | 2 | (R)L/A|A|Y|L/m L/m/R(S) | O-Met537, 539 |
| 550–563 | 1.9 | 17.5 | 82 | 3 | (K)I V|Q\I|L/P\w/E/Q/N E Q V/K(N) | OxTrp556 |
| 550–563 | 1.2 | 14.0 | 84 | 2 | (K)I V|Q|I|L|P w E Q N E Q V/K(N) | di-OxTrp556 |
| 594–604 | 2.3 | 15.7 | 92 | 2 | (K)E S/Q|L|P/T V/m/D/F R(K) | O-Met601 |
| 691–705 | 0.5 | 14.9 | 80 | 2 | (K)A L/Y|w|V|n/G Q/V|P D G V|S K(V) | OxTrp694 |
| 691–705 | 1.1 | 16.3 | 92 | 2 | (K)A L|Y|w|V|n G Q/V|P D G V/S K(V) | di-OxTrp694 |
| 706–715 | 1.9 | 8.3 | 71 | 3 | (K)V/L|V/D H F G y T/K(D) | Cl-Tyr713 |
| 771–777 | 3.0 | 11.5 | 86 | 2 | (K)L/L|L/m/G A/R(T) | O-Met774 |
| 778–791 | 2.7 | 13.1 | 86 | 2 | (R)T L|Q|G|I|P Q m I G E V I/R(K) | O-Met785 |
| 837–846 | 1.4 | 10.1 | 89 | 2 | (K)L E\V\A N m\Q\A|E|L(V) | O-Met842 |
| 870–891 | 3.9 | 19.0 | 83 | 3 | (R)S G/V|Q|m/N/T/N/F F/H E S G L E A H V/A/L/K(A) | O-Met874 |
| 1,029–1,051 | 3.4 | 17.9 | 97 | 2 | (R)q S m T\L\S\S E\V|Q/I|P D F/D V/D L/G T I/L/R(V) | O-Met1031 |
| 1,119–1,132 | 0.6 | 20.4 | 70 | 2 | (K)L L|L|Q/m/D S/S/A T A/Y/G S/T V/S/K(R) | O-Met1123 |
| 1,162–1,175 | 2.2 | 15.7 | 93 | 2 | (K)m T|S/N/F|P V/D/L/S D/Y/P K(S) | O-Met1162 |
| 1,176–1,183 | 1.2 | 11.3 | 62 | 2 | (K)S/L/H|m/Y/A N/R(L) | O-Met1179 |
| 1,203–1,213 | 1.1 | 18.2 | 87 | 2 | (K)L I|V|A|m/S/S/W/L/Q/K(A) | O-Met1207 |
| 1,203–1,213 | 2.3 | 12.0 | 81 | 2 | (K)L/I|V|A/m S S w/L Q/K(A) | O-Met1207 OxTrp1210 |
| 1,676–1,697 | 0.4 | 23.7 | 94 | 2 | (K)A A L T/E\L|S|L/G|S/A/Y/Q/A m/I/L/G/V/D/S K(N) | O-Met1690 |
| 1,871–1,876 | 1.1 | 8.2 | 91 | 1 | (R)S V|m|A|P\F(T) | O-Met1873 |
| 1,890–1,901 | 3.8 | 14.1 | 91 | 2 | (K)L A|L|W/G E H T G Q|L\y(S) | Cl-Tyr1901 |
| 1,986–2,016 | 3.2 | 18.0 | 82 | 3 | (R)T L A D/L T L L\D S P I K V|P L\L\L|S/E|P I/N I|I/D A L/E/m R(D) | O-Met2015 |
| 2,214–2,228 | 0.3 | 11.7 | 63 | 2 | (K)S G S S T/A|S/w/I|Q/N V/D|T K(Y) | OxTrp2221 |
| 2,321–2,333 | 0.8 | 19.4 | 81 | 2 | (R)Y E|V|D Q/Q/I|Q/V|L/m/D K(L) | O-Met2331 |
| 2,462–2,470 | 0.8 | 10.2 | 74 | 2 | (K)I/T|L/I I N w L Q(E) | OxTrp2468 |
| 2,565–2,582 | 2.2 | 22.7 | 93 | 2 | (K)T I|L/G T/m|P A/F/E/V/S L/Q/A/L/Q K(A) | O-Met2570 |
| 2,979–2,993 | 4.6 | 12.9 | 73 | 3 | (K)G/m|A|L|F G E/G K/A E/F T G R(H) | O-Met2980 |
| 3,122–3,128 | 1.5 | 6.6 | 64 | 2 | (K)D F|S/L w/E K(T) | OxTrp3126 |
| 3,237–3,249 | 2.2 | 21.6 | 90 | 2 | (F)T/I|E|m/S/A/F/G Y/V|F/P K(A) | O-Met3240 |
| 3,250–3,264 | 2.7 | 21.1 | 93 | 2 | (K)A V|S|m|P/S/F/S I/L|G/S D/V R(V) | O-Met3253 |
| 3,423–3,434 | 2.6 | 12.1 | 80 | 3 | (K)S K P T\V|S|S/S m E F/K(Y) | O-Met3431 |
| 3,926–3,946 | 1.6 | 15.1 | 75 | 3 | (R)D F|S|A/E|Y/E E/D G K Y E/G L/Q E w E G K(A) | OxTrp3943 |
| 4,080–4,094 | 0.0 | 16.0 | 72 | 2 | (R)N L/Q|N N|A/E/w/V/Y/Q/G A/I/R(Q) | OxTrp4087 |
| 4,080–4,094 | 2.1 | 19.4 | 88 | 2 | (R)N/L|Q|N N\A|E\w/V|Y/Q/G A/I/R(Q) | di-OxTrp4087 |
| 4,187–4,195 | 2.9 | 14.9 | 89 | 2 | (R)E E|L/C/T/m/F/I/R(E) | O-Met4192 |
| 4,235–4,243 | −0.1 | 11.2 | 80 | 2 | (K)L I|D|V|I/S/m/Y R(E) | O-Met4241 |
LDL was modified by HOCl added as a reactant (50 or 100 µM) (oxidant/LDL molar ratio = 25:1 and 50:1). ApoB-100 mapping was carried out by RRLC-MS/MS. Each modified peptide is characterized by its peptide score, % SPI, mass error, fragmentation, and position(s) of modified residue(s).
Next, PTMs generated via the MPO-H2O2-Cl− system also under soft conditions (oxidant/lipoprotein molar ratio = 25:1 and 50:1 assuming almost equimolar conversion of H2O2 to HOCl) were identified. Under these conditions, several modifications already observed with reagent HOCl were found. However, 13 additional peptides specific for MPO-mediated modification (Table 2) were found. Again, O-Met is the most abundant PTM, while only a single Cl-Tyr-containing peptide was found to be specific for MPO treatment.
TABLE 2.
List of apoB-100-derived tryptic peptides carrying modifications generated only by the MPO-H2O2-Cl− system under soft conditions (molar ratios = 25:1 and 50:1)
| Peptide | Error (ppm) | Peptide Score | % SPI | Z | Sequence with Fragmentation | Modified Residue |
| 74–83 | 2.7 | 15.0 | 75 | 2 | (K)E V|y/G/F/N/P E/G K(A) | Cl-Tyr76 |
| 716–733 | 3.3 | 10.9 | 83 | 3 | (K)D D/K H E Q D\m V|N|G I/m L S V/E/K(L) | O-Met723, 728 |
| 1,074–1,088 | 4.1 | 15.8 | 75 | 2 | (K)I/T/E|V|A/L/m G H/L|S C D|T\K(E) | O-Met1080 |
| 1,189–1,197 | 2.8 | 14.5 | 79 | 2 | (R)V/P|Q/T/D m T F/R(H) | O-Met1194 |
| 1,282–1,290 | 5.0 | 7.9 | 64 | 2 | (R)D/L/K/m/L\E/T V/R(T) | O-Met1285 |
| 1,711–1,723 | 1.6 | 14.9 | 80 | 2 | (K)L/S|N D|m m/G S/Y/A/E/m/K(F) | O-Met1715, 1716, 1722 |
| 2,392–2,396 | 0.8 | 6.8 | 75 | 3 | (K)F/L/D m L I/K/K(L) | O-Met2395 |
| 2,810–2,826 | 1.2 | 11.6 | 81 | 3 | (R)T E H G S E m\L|F|F/G N A I E/G K(S) | O-Met2816 |
| 3,393–3,403 | 2.5 | 10.4 | 89 | 2 | (K)N/m|E|V|S V A/T T T K(A) | O-Met3394 |
| 3,411–3,422 | 2.4 | 11.3 | 78 | 3 | (R)m/N/F/K\Q E\L/N/G N T K(S) | O-Met3411 |
| 3,435–3,447 | 3.3 | 15.0 | 78 | 2 | (K)Y D|F|n|S/S m L Y/S/T/A/K(G) | O-Met3441 |
| 4,151–4,158 | −0.5 | 17.3 | 85 | 2 | (R)V/T|Q/E/F H|m/K(V) | O-Met4157 |
| 4,515–4,526 | −0.8 | 9.5 | 83 | 2 | (K)L/Q|S T/T/V m N/P Y m/K(L) | O-Met4521, 4525 |
LDL was modified by HOCl generated by the MPO-H2O2-Cl− system using 50 or 100 µM of H2O2 (oxidant/LDL molar ratio = 25:1 and 50:1, considering almost equimolar formation of HOCl). ApoB-100 mapping was carried out by RRLC-MS/MS. Peptides carrying a modification observed in Table 1 were omitted, indicating PTMs to be MPO specific. Each modified peptide is characterized by its peptide score, % SPI, mass error, fragmentation, and position(s) of modified residue(s).
Furthermore, oxidation of LDL was performed by the MPO-H2O2-Cl− system under more drastic conditions (oxidant/lipoprotein molar ratio = 625:1) previously used to raise monoclonal antibodies to detect MPO-LDL in vivo and in vitro (11). In addition to oxidative modifications listed in Table 1, 41 tryptic peptides carrying PTMs (n = 46) were identified (Table 3). These peptides further include 3 residues of 3-hydroxytyrosine (HO-Tyr), 5 O-Met residues, 10 OxTrp residues, 18 di-OxTrp residues, and 10 Cl-Tyr residues, respectively. Again, in most peptides a single amino acid modification (primarily di-OxTrp in this case) was found, while few peptides included either two (O-Met and HO-Tyr or O-Met and Cl-Tyr) or three modifications (two O-Met and HO-Tyr). These findings indicate that a more pronounced enzymatic oxidation of apoB-100 further targets Trp and Tyr residues.
TABLE 3.
List of apoB-100-derived tryptic peptides carrying modifications generated by the MPO-H2O2-Cl− system under drastic conditions (molar ratio = 625:1)
| Peptide | Error (ppm) | Peptide Score | % SPI | Z | Sequence with Fragmentation | Modified Amino Acid |
| 102–105 | 1.9 | 3.1 | 80 | 2 | (R)y/E/L K(L) | Cl-Tyr102 |
| 748–751 | 3.7 | 3.0 | 84 | 2 | (R)A/y/L R(I) | Cl-Tyr749 |
| 1,108–1,118 | −0.5 | 15.0 | 76 | 2 | (R)S/E|I|L|A|H|w/S|P A K(L) | OxTrp1114 |
| 1,108–1,118 | −0.6 | 12.0 | 81 | 2 | (R)S E|I|L|A/H w|S/P A K(L) | di-OxTrp1114 |
| 1,176–1,183 | 3.2 | 12.6 | 87 | 3 | (K)S/L/H/m y|A/N/R(L) | O-Met1179 HO-Tyr1180 |
| 1,573–1,583 | 1.0 | 14.1 | 71 | 2 | (R)S E|y|Q|A|D/Y/E/S L/R(F) | Cl-Tyr1575 |
| 1,682–1,694 | 0.1 | 10.8 | 63 | 2 | (L)S L|G S\A|y Q A m I/L/G/V/D/S K(N) | O-Met1690 Cl-Tyr1687 |
| 1,743–1,750 | 2.3 | 6.3 | 82 | 2 | (K)L D/N\I/y/S S D K F Y/K(Q) | Cl-Tyr1747 |
| 1,890–1,901 | 0.4 | 10.2 | 71 | 2 | (K)L A|L/w|G E H T G Q|L\Y(S) | OxTrp1893 |
| 1,890–1,901 | 0.9 | 13.5 | 79 | 2 | (K)L A|L|w|G E H T G Q|L|Y(S) | di-OxTrp1893 |
| 1,941–1,955 | −0.5 | 10.4 | 70 | 3 | (K)V S A L L|T|P A E Q T/G T w K(L) | OxTrp1954 |
| 1,941–1,955 | 1.8 | 13.4 | 80 | 2 | (K)V S A|L|L|T|P A E/Q T G T w/K(L) | di-OxTrp1954 |
| 2,086–2,106 | 1.7 | 18.2 | 94 | 3 | (R)A A|L|G K\L|P Q Q A N D\Y|L|N/S/F/N w/E R(Q) | di-OxTrp2104 |
| 2,214–2,228 | −2.9 | 15.8 | 74 | 2 | (K)S G S S T/A/S/w|I|Q/N/V|D/T/K(Y) | di-OxTrp2221 |
| 2,334–2,341 | 1.8 | 11.9 | 73 | 2 | (K)L/V|E/L/A/H Q y(K) | Cl-Tyr2341 |
| 2,402–2,415 | −0.7 | 5.1 | 64 | 3 | (K)S F|D/y H Q F V/D E T N D K(I) | Cl-Tyr2405 |
| 2,462–2,478 | 0.3 | 8.5 | 74 | 2 | (K)I T L\I I\N w\L Q E\A\L|S\S A/S L(A) | OxTrp2468 |
| 2,462–2,478 | −1.6 | 10.8 | 83 | 2 | (K)I T L\I\I N w\L\Q E\A\L|S|S A/S L(A) | di-OxTrp2468 |
| 2,496–2,507 | 0.7 | 12.0 | 80 | 2 | (R)m y/Q/m D I|Q/Q/E L/Q/R(Y) | O-Met2496, 2499 HO-Tyr2497 |
| 2,542–2,548 | 2.0 | 12.5 | 77 | 2 | (Y)S/I|Q/D/w/A/K(R) | OxTrp2546 |
| 2,533–2,548 | −0.1 | 15.0 | 70 | 2 | (K)N L\T D|F/A/E|Q/Y/S I/Q/D w/A/K(R) | di-OxTrp2546 |
| 2,731–2,736 | 2.0 | 6.2 | 73 | 2 | (K)L/y/S I/L/K(I) | Cl-Tyr2732 |
| 2,889–2,899 | 3.6 | 11.0 | 73 | 3 | (K)A G H\I A\w|T S/S/G/K(G) | di-OxTrp2894 |
| 3,051–3,065 | 0.1 | 17.9 | 80 | 2 | (F)L/S/P S A/Q/Q|A|S w/Q/V/S A/R(F) | OxTrp3060 |
| 3,051–3,065 | 1.3 | 13.9 | 73 | 2 | (F)L/S/P S A/Q|Q/A/S/w/Q V/S A R(F) | di-OxTrp3060 |
| 3,122–3,128 | 2.0 | 10.7 | 73 | 2 | (K)D F|S L/w/E K(T) | di-OxTrp3126 |
| 3,511–3,520 | 0.9 | 16.2 | 85 | 2 | (K)I/D|D|I|w/N L/E/V/K(E) | OxTrp3515 |
| 3,511–3,520 | 1.5 | 16.2 | 84 | 2 | (K)I/D|D|I|w|N L/E/V/K(E) | di-OxTrp3515 |
| 3,532–3,541 | 0.3 | 16.9 | 77 | 2 | (R)I/Y|S|L/w/E/H|S T/K(N) | OxTrp3536 |
| 3,532–3,541 | 1.7 | 13.6 | 83 | 3 | (R)I/Y/S L/w|E H/S T K(N) | di-OxTrp3536 |
| 3,604–3,611 | 1.7 | 4.5 | 68 | 3 | (K)I R w|K/N E/V R(I) | di-OxTrp3606 |
| 3,647–3,655 | −4.4 | 5.0 | 78 | 2 | (K)N I|I/L/P V y D K(S) | Cl-Tyr3653 |
| 3,656–3,662 | 1.3 | 6.8 | 78 | 2 | (K)S L|w/D F L K(L) | OxTrp3658 |
| 3,656–3,662 | 7.5 | 10.3 | 72 | 2 | (K)S/L|w/D/F L/K(L) | di-OxTrp3658 |
| 3,860–3,876 | 0.1 | 18.3 | 85 | 3 | (R)F E\V D S\P\V|Y/n/A/T/w/S A/S L K(N) | OxTrp3871 |
| 3,860–3,876 | 0.7 | 17.2 | 80 | 2 | (R)F E|V|D/S|P V/Y/n/A T w/S A/S L K(N) | di-OxTrp3871 |
| 3,926–3,946 | −0.4 | 14.2 | 70 | 3 | (R)D F|S|A/E|Y|E E D G K Y E/G L/Q E w E G K(A) | di-OxTrp3943 |
| 4,106–4,120 | 1.4 | 10.6 | 69 | 3 | (K)A/A|S/G/T/T/G T y Q E W K D K(A) | HO-Tyr4114 |
| 4,106–4,120 | −0.1 | 11.2 | 64 | 3 | (K)A/A|S/G/T/T/G T Y Q E w/K D K(A) | di-OxTrp4117 |
| 4,235–4,243 | 0.2 | 10.5 | 87 | 2 | (K)L I|D|V|I|S/m y R(E) | O-Met4241 Cl-Tyr4242 |
| 4,359–4,372 | 1.2 | 15.3 | 74 | 2 | (R)E E Y|F/D|P S I|V/G w/T V/K(Y) | di-OxTrp4369 |
LDL was modified by HOCl generated by the MPO-H2O2-Cl− system using drastic conditions (oxidant/LDL molar ratio = 625:1 considering equimolar formation of HOCl). ApoB-100 mapping was carried out by RRLC-MS/MS, and listed modifications are the ones not recovered with the softest conditions described in Table 1 and Table 2. Each modified peptide is characterized by its peptide score, % SPI, mass error, fragmentation, and position(s) of modified residue(s).
Mapping of MPO-mediated PTMs of apoB-100 isolated from patients undergoing hemodialysis or healthy volunteers.
To confirm whether our in vitro findings may occur also in vivo, LDL was isolated from patients (n = 9) who have high levels of MPO (38, 39) and circulating MPO-LDL (25) and compared with healthy volunteers (n = 9). As levels of oxidized LDL in patients with hemodialysis are still low when compared with levels of native LDL, apoB-100 was analyzed using a nano-LC-MS/MS with high-resolution analytical column (Polaris CHIP), which is more sensitive than an LC system.
In sum, 90 PTMs were identified over patients and volunteers. Again, O-Met (n = 31) is a prominent modification compared with HO-Tyr (n = 30), OxTrp (n = 16), and di-OxTrp (n = 12), while only one Cl-Tyr was observed. Most of the modifications match those found when apoB-100 was treated in vitro, whereas several were newly detected, such as HO-Tyr76, 249, 425, 693, 713, 1579, 1753, 1747, 1965, 1972, 2189, 2362, 2405, 2732, 3206, 3268, 3533, 3653, 3744, 4055, 4057, 4184, 4205, 4242, 4424, 4464, Cl-Tyr125, O-Met507, 1022, 1162, 1239, 1560, 2481, OxTrp936, 4063, and di-OxTrp936. Among PTMs already detected in vitro, O-Met4 was described as MPO specific (22). In our experiments, O-Met4 was observed in apoB-100 of all patients but also all volunteers, showing that this site is highly sensitive and is not relevant as a biomarker. The same results were observed for (di)-OxTrp1114 and 3536, which were previously described (37).
In contrast, several less prominent PTMs seemed interesting, such as O-Met2499, which was observed only in patients (n = 2); OxTrp2894, observed in five patients and only one volunteer; OxTrp3606, observed in four patients and only one volunteer; and finally, Cl-Tyr125, observed in only one patient. Furthermore, O-Met2499, OxTrp2894, and OxTrp3606 are identical to those observed when LDL/apoB-100 was treated with the MPO-H2O2-Cl− system under conditions using the molar ratio 625:1 (Table 3). Cl-Tyr125 might also be interesting as it is specific to the MPO-H2O2-Cl− system and was never detected before.
DISCUSSION
Methodologies for detecting and identifying protein modifications have become increasingly important as the roles of modified (lipo)proteins in disease etiology and biomarkers are becoming more apparent (40). Because OxLDL impairs the physiological functioning of various cells (13, 17) and plays a causal role in atheroma plaque formation, both the nature of oxidants as well as the modified entity of the lipoprotein particle are of importance. Oxidation of LDL can be carried out by, among others, transition metals, hemoglobin, lipoxygenases, and reactive oxygen species generated by vascular endothelium or phagocytes, and HOCl can modify LDL at the lipid and the protein moieties in vitro and/or in vivo (13). Modified lipids were preferentially identified by MALDI-TOF-MS and 31P NMR spectroscopy (40–42), while MALDI-TOF-MS, MS/MS, and LC-MS/MS are suitable techniques to identify the respective protein modifications basically after tryptic digestion (31, 40). Here, we have used an LC-MS/MS method with a high-resolution MS (a QTOF) to identify PTMs on tryptic peptides from delipidated apoB-100 samples (31) treated with HOCl added as reagent or generated enzymatically. Furthermore, oxidative PTMs occurring in vitro were compared with those occurring under in vivo conditions. Finally, we were interested in whether the extent of HOCl modification of apoB-100 can in turn modulate the activity of MPO to generate oxidants.
ApoB-100 is probably the most difficult protein for structural analysis because of its huge size and its insolubility in aqueous buffer after delipidation (43). However, studying modifications of apoB-100 in relation to cardiovascular diseases is of major importance (40). Yang and colleagues (44) were the first to identify 88% of the sequence of native apoB-100 by HPLC analysis of tryptic peptides coupled to an automatic sequencer (different than the MS technique). Oxidation of LDL with HOCl produced a diverse array of 2,4-dinitrophenylhydrazine-reactive peptides, with little indication of selectivity (24). HOCl treatment of apoB-100 resulted in modification at cysteine (Cys), Trp, Met, and Lys. Thirteen out of 14 modified tryptic peptides identified (corresponding to position 53–66[Cys61]; 113–129[Lys120]; 181–187[Cys185]; 228–235[Cys234]; 428–463[Cys451]; 1,108–1,118[Trp1114]; 1,203–1,213[Trp1210]; 1,890–1,903[Trp1893]; 3,560–3,600[Trp3567, 3569]; 3,710–3,735[Cys3734]; 3,877–3,908[Cys3890]; 3,879–3,908[Cys3890]; and 4,187–4,195[Cys4190] of apoB-100) were found to reside at the external surface (the so-called trypsin-releasable fraction) of LDL (10). However, MPO-catalyzed oxidation of LDL produced a much different pattern of 2,4-dinitrophenylhydrazine-reactive products. Two peptides (corresponding to position 3,710–3,735[Met3719] and 4,187–4,195[Met4192] of apoB-100) were detected at low abundance, while the most prominent peptide (position 53–66) was modified at Cys61 (22).
To analyze PTMs on apoB-100, we used an RRLC system coupled to high-resolution MS/MS detection. This methodology is extremely efficient because LC enables high resolution of tryptic peptides by C18 column, while high-resolution MS/MS provides high accuracy of m/z values and a fragmentation pattern of the compounds (i.e., peptides). The latter technique enables both mapping of peptides and detection of untargeted PTMs. Here, we are the first to present the most comprehensive pattern of tryptic peptides of apoB-100 treated by reagent HOCl. In sum, 97 tryptic peptides carrying at least one PTM could be identified. Although others (10) have tried to differentiate between trypsin-releasable, nontrypsin-releasable, and mixed fractions of apoB-100, we directly performed delipidation of total LDL prior to trypsin digestion. Using two different concentrations of reagent HOCl (50 or 100 µM), 46 modified tryptic peptides were found.
Although some modifications (n = 15) were also present when LDL was modified by the MPO-H2O2-Cl− system (Table 2), the pattern of enzymatic modification differs from that using reagent HOCl. Among the residues found exclusively in enzymatically mediated oxidation are peptides 74–83[Cl-Tyr76]; 716–733[O-Met723/728]; 1,074–1,088[O-Met1080]; 1,189–1,197[O-Met1194]; 1,282–1,290[O-Met1285]; 1,711–1,723[O-Met1715/1716/1722]; 2,392–2,396[O-Met2395]; 2,810–2,826[O-Met2816]; 3,393–3,403[O-Met3394]; 3,411–3,422[O-Met3411]; 3,435–3,447[O-Met3441]; 4,151–4,158[O-Met4157]; and 4,515–4,526[O-Met4521/4525]. Tyr76, which was chlorinated by the MPO-H2O2-Cl− system, was also reported to act as a specific target for LDL nitration (45). Met, apparently the first target for oxidation, is highly reactive toward HOCl-mediated attack (13), and Met residues may protect proteins from critical oxidative damage (46). In addition to Met3719 and Met4192, Met4 and Cys61, 3734, 4190 were reported to be specific MPO-mediated modifications of apoB-100 (22, 24). Our results suggest that O-Met4 and O-Met4192 are already present on non-in vitro oxidized LDL isolated from healthy volunteers. These data also reflect the high sensitivity of those residues to oxidative damages. Modifications of residues Met3719 and Cys61/3734/4190 were not found under our experimental conditions. Cys residues have been reported to rapidly react with HOCl (47, 48). Despite careful analysis of possibly oxidized Cys residues, none were detected under our experimental conditions.
We further show that oxidation of apoB-100 by HOCl (added as a reagent) leads to a higher content of aminoadipic acid compared with MPO-mediated oxidation. On the other hand, no statistical difference in chemical and enzymatic oxidation was seen for Trp and Met residues, a fact underlining their high sensitivity toward oxidation. So again, oxidation patterns are dependent on the oxidant, suggesting that reagent HOCl does not exactly mimic the respective MPO enzymatic oxidation.
Both the extent of modification as well as the difference in the charge of amino acid side chains may alter the binding properties of MPO to the respective LDL particle in vivo or in vitro. Adsorption of MPO on native LDL increased both chlorinating and peroxidase activities (Fig. 1), and this was reflected by structural changes in the heme cavity using CD spectroscopy (Fig. 2). These findings are in accordance with Sokolov et al. (49) who demonstrated that the addition of LDL (but not HDL) protects MPO from inhibition by ceruloplasmin. This observation favors a specific interaction between LDL and MPO that differs from MPO interaction with high-density lipoprotein (34). An increase in the activity of MPO at the surface of LDL might indicate that native LDL enhances its own oxidation by MPO and that MPO activity could be underestimated in vitro and/or in vivo. On the other hand, MPO activity decreased with increasing HOCl/LDL molar ratios and HOCl-LDL concentration. The more wild-type-like Soret CD spectrum of MPO in the presence of HOCl-LDL is further support. Apparently, oxidative LDL modifications (e.g., O-Met, Cl-Tyr, OxTrp, or N-chloramine Lys) alter the interaction between apoB-100 and MPO and, at least at higher concentrations, diminish MPO activity to some extent.
Recently, monoclonal antibodies were raised against LDL that had been modified by MPO under conditions using the drastic molar ratio 625:1 (11) as performed in the present study. The respective epitope identified by one of the antibodies corresponded to a 66 kDa fragment of apoB-100 starting with Gly1612 (11). Using this oxidation protocol, a total of 41 peptides carrying at least one PTM (e.g., primarily modified Trp and Tyr residues but also O-Met residues) were detected (Table 3). In that particular 66 kDa fragment, six modifications (Met1690, Tyr1687/1747, and Trp1893/1954/2104) are probably the likely recognition sites for the antibody.
Cl-Tyr, a protein modification specific for MPO-catalyzed oxidation, has previously been identified on LDL isolated from human plaque material or LDL plasma samples from patients with cardiovascular disease (12). Although several Tyr residues are prone to be modified either enzymatically or nonenzymatically by HOCl, we are the first to identify Cl-Tyr residues on apoB-100 at positions Tyr76, 102, 749, 1575, 1687, 1747, 1901, 2341, 2405, 2732, 3653, and 4242. An increased level of Cl-Tyr, apparently a consequence of high plasma MPO levels (50, 51), was also reported in plasma samples from dialysis patients (38, 39). This observation prompted us to follow PTMs on apoB-100 isolated from plasma of hemodialysis patients who have high plasma MPO-LDL levels (25) and compared with apoB-100 of healthy volunteers. Among the observed PTMs, O-Met and OxTrp residues were detected, as well as only one Cl-Tyr. This suggests that Cl-Tyr formation on apoB-100 is rare, but Tyr125 is an interesting PTM for future studies in MPO. However, only one patient among nine carried this MPO-specific modification. Furthermore, O-Met2499 was only observed in patients and so is an interesting PTM that should be investigated in future studies.
Although we could not find PTM in the close vicinity of the LDL receptor binding domain [between residues 3,346 and 3,369 of apoB-100 (52, 53)], the remaining PTMs, markers of oxidative stress and/or lipoprotein abnormalities (54), could still contribute to impaired binding of MPO-LDL to the LDL receptor favoring scavenger receptor-mediated uptake as reported for nitrated LDL (45). Furthermore, Trp3606 is close to the binding domain and might interact with the latter when oxidized, and OxTrp3606 was observed in more patients (n = 4) than volunteers (n = 1).
Summarizing, using LC-MS/MS we further identified in vitro a series of modified tryptic peptides (n = 97) that are indicative of modification by HOCl, added as reagent or generated enzymatically. Among these modifications, several have been identified in vivo from patients suffering from kidney failure and undergoing hemodialysis therapy where high MPO levels are causally linked with LDL modifications and abnormalities in clearance of the modified LDL particles. Our data also highlight that, when studying LDL oxidation, HOCl added as a reagent does not completely mimic enzymatic modification via the MPO-H2O2-Cl− system and that MPO activity could be misestimated under in vitro or in vivo experiments due to the fact that MPO/apoB-100 interaction is specific and changes the enzymatic activity. Future experiments mapping PTMs of LDL in different cardiovascular diseases are also of importance to link individual oxidative modifications with disease severity.
Acknowledgments
The authors thank the members of the Laboratory of Pharmaceutics and Biopharmaceutics and the Laboratory of Pharmacology (Université Libre de Bruxelles) for their scientific support.
Footnotes
Abbreviations:
- CD
- circular dichroism
- Cl-Tyr
- 3-chlorotyrosine
- di-OxTrp
- dioxidized tryptophan
- FA
- formic acid
- HOCl-LDL
- HOCl-modified LDL
- HOCl
- hypochlorous acid
- HO-Tyr
- 3-hydroxytyrosine
- MPO
- myeloperoxidase
- MPO-LDL
- LDL modified by the MPO-H2O2-chloride system
- O-Met
- methionine sulfoxide
- OxTrp
- oxidized tryptophan
- PTM
- posttranslational modification
- QTOF
- quadrupole TOF
- RRLC
- rapid resolution LC
- SPI
- score peak intensity
This study was supported by the Belgian Fund for Scientific Research (FRS-FNRS) Grant 34553.08, a grant from the FER 2007 (Université Libre de Bruxelles), and the Austrian Science Fund Grants FWF F3007 and W1226-B18. C. Delporte is a postdoctoral researcher funded by the FRS-FNRS, C. Noyon is a research fellow of the FRS-FNRS, and L. Vanhamme is Research Director of the FRS-FNRS.
REFERENCES
- 1.Klebanoff S. J. 2005. Myeloperoxidase: friend and foe. J. Leukoc. Biol. 77: 598–625. [DOI] [PubMed] [Google Scholar]
- 2.Arnhold J., Flemmig J. 2010. Human myeloperoxidase in innate and acquired immunity. Arch. Biochem. Biophys. 500: 92–106. [DOI] [PubMed] [Google Scholar]
- 3.Davies M. J., Hawkins C. L., Pattison D. I., Rees M. D. 2008. Mammalian heme peroxidases: from molecular mechanisms to health implications. Antioxid. Redox Signal. 10: 1199–1234. [DOI] [PubMed] [Google Scholar]
- 4.Hazell L. J., Arnold L., Flowers D., Waeg G., Malle E., Stocker R. 1996. Presence of hypochlorite-modified proteins in human atherosclerotic lesions. J. Clin. Invest. 97: 1535–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malle E., Waeg G., Schreiber R., Grone E. F., Sattler W., Grone H. J. 2000. Immunohistochemical evidence for the myeloperoxidase/H2O2/halide system in human atherosclerotic lesions: colocalization of myeloperoxidase and hypochlorite-modified proteins. Eur. J. Biochem. 267: 4495–4503. [DOI] [PubMed] [Google Scholar]
- 6.Malle E., Buch T., Grone H. J. 2003. Myeloperoxidase in kidney disease. Kidney Int. 64: 1956–1967. [DOI] [PubMed] [Google Scholar]
- 7.Daugherty A., Dunn J. L., Rateri D. L., Heinecke J. W. 1994. Myeloperoxidase, a catalyst for lipoprotein oxidation, is expressed in human atherosclerotic lesions. J. Clin. Invest. 94: 437–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daugherty A., Roselaar S. E. 1995. Lipoprotein oxidation as a mediator of atherogenesis: insights from pharmacological studies. Cardiovasc. Res. 29: 297–311. [PubMed] [Google Scholar]
- 9.Itabe H. 2009. Oxidative modification of LDL: its pathological role in atherosclerosis. Clin. Rev. Allergy Immunol. 37: 4–11. [DOI] [PubMed] [Google Scholar]
- 10.Yang C. Y., Gu Z. W., Yang H. X., Yang M., Gotto A. M., Jr, Smith C. V. 1997. Oxidative modifications of apoB-100 by exposure of low density lipoproteins to HOCL in vitro. Free Radic. Biol. Med. 23: 82–89. [DOI] [PubMed] [Google Scholar]
- 11.Moguilevsky N., Zouaoui Boudjeltia K., Babar S., Delree P., Legssyer I., Carpentier Y., Vanhaeverbeek M., Ducobu J. 2004. Monoclonal antibodies against LDL progressively oxidized by myeloperoxidase react with ApoB-100 protein moiety and human atherosclerotic lesions. Biochem. Biophys. Res. Commun. 323: 1223–1228. [DOI] [PubMed] [Google Scholar]
- 12.Hazen S. L., Heinecke J. W. 1997. 3-Chlorotyrosine, a specific marker of myeloperoxidase-catalyzed oxidation, is markedly elevated in low density lipoprotein isolated from human atherosclerotic intima. J. Clin. Invest. 99: 2075–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malle E., Marsche G., Arnhold J., Davies M. J. 2006. Modification of low-density lipoprotein by myeloperoxidase-derived oxidants and reagent hypochlorous acid. Biochim. Biophys. Acta. 1761: 392–415. [DOI] [PubMed] [Google Scholar]
- 14.Marsche G., Weigle B., Sattler W., Malle E. 2007. Soluble RAGE blocks scavenger receptor CD36-mediated uptake of hypochlorite-modified low-density lipoprotein. FASEB J. 21: 3075–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marsche G., Zimmermann R., Horiuchi S., Tandon N. N., Sattler W., Malle E. 2003. Class B scavenger receptors CD36 and SR-BI are receptors for hypochlorite-modified low density lipoprotein. J. Biol. Chem. 278: 47562–47570. [DOI] [PubMed] [Google Scholar]
- 16.Hazell L. J., Stocker R. 1993. Oxidation of low-density lipoprotein with hypochlorite causes transformation of the lipoprotein into a high-uptake form for macrophages. Biochem. J. 290: 165–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boudjeltia K. Z., Legssyer I., Van Antwerpen P., Kisoka R. L., Babar S., Moguilevsky N., Delree P., Ducobu J., Remacle C., Vanhaeverbeek M., et al. 2006. Triggering of inflammatory response by myeloperoxidase-oxidized LDL. Biochem. Cell Biol. 84: 805–812. [DOI] [PubMed] [Google Scholar]
- 18.Calay D., Rousseau A., Mattart L., Nuyens V., Delporte C., Van Antwerpen P., Moguilevsky N., Arnould T., Boudjeltia K. Z., Raes M. 2010. Copper and myeloperoxidase-modified LDLs activate Nrf2 through different pathways of ROS production in macrophages. Antioxid. Redox Signal. 13: 1491–1502. [DOI] [PubMed] [Google Scholar]
- 19.Hazen S. L., Crowley J. R., Mueller D. M., Heinecke J. W. 1997. Mass spectrometric quantification of 3-chlorotyrosine in human tissues with attomole sensitivity: a sensitive and specific marker for myeloperoxidase-catalyzed chlorination at sites of inflammation. Free Radic. Biol. Med. 23: 909–916. [DOI] [PubMed] [Google Scholar]
- 20.Zheng L., Nukuna B., Brennan M. L., Sun M., Goormastic M., Settle M., Schmitt D., Fu X., Thomson L., Fox P. L., et al. 2004. Apolipoprotein A-I is a selective target for myeloperoxidase-catalyzed oxidation and functional impairment in subjects with cardiovascular disease. J. Clin. Invest. 114: 529–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carr A. C., Myzak M. C., Stocker R., McCall M. R., Frei B. 2000. Myeloperoxidase binds to low-density lipoprotein: potential implications for atherosclerosis. FEBS Lett. 487: 176–180. [DOI] [PubMed] [Google Scholar]
- 22.Yang C. Y., Gu Z. W., Yang M., Lin S. N., Garcia-Prats A. J., Rogers L. K., Welty S. E., Smith C. V. 1999. Selective modification of apoB-100 in the oxidation of low density lipoproteins by myeloperoxidase in vitro. J. Lipid Res. 40: 686–698. [PubMed] [Google Scholar]
- 23.Sokolov A. V., Chekanov A. V., Kostevich V. A., Aksenov D. V., Vasilyev V. B., Panasenko O. M. 2011. Revealing binding sites for myeloperoxidase on the surface of human low density lipoproteins. Chem. Phys. Lipids. 164: 49–53. [DOI] [PubMed] [Google Scholar]
- 24.Yang C., Wang J., Krutchinsky A. N., Chait B. T., Morrisett J. D., Smith C. V. 2001. Selective oxidation in vitro by myeloperoxidase of the N-terminal amine in apolipoprotein B-100. J. Lipid Res. 42: 1891–1896. [PubMed] [Google Scholar]
- 25.Vaes M., Zouaoui Boudjeltia K., Van Antwerpen P., Babar S., Deger F., Neve J., Vanhaerverbeek M., Ducobu J. 2006. Low-density lipoprotein oxidation by myeloperoxidase occurs in the blood circulation during hemodialysis (Abstract). Atheroscler. Suppl. 7: 525. [Google Scholar]
- 26.Moguilevsky N., Garcia-Quintana L., Jacquet A., Tournay C., Fabry L., Pierard L., Bollen A. 1991. Structural and biological properties of human recombinant myeloperoxidase produced by Chinese hamster ovary cell lines. Eur. J. Biochem. 197: 605–614. [DOI] [PubMed] [Google Scholar]
- 27.Havel R. J., Eder H. A., Bragdon J. H. 1955. The distribution and chemical composition of ultracentrifugally separated lipoproteins in human serum. J. Clin. Invest. 34: 1345–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peterson G. L. 1977. A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal. Biochem. 83: 346–356. [DOI] [PubMed] [Google Scholar]
- 29.Kettle A. J., Winterbourn C. C. 1988. The mechanism of myeloperoxidase-dependent chlorination of monochlorodimedon. Biochim. Biophys. Acta. 957: 185–191. [DOI] [PubMed] [Google Scholar]
- 30.Van Antwerpen P., Moreau P., Zouaoui Boudjeltia K., Babar S., Dufrasne F., Moguilevsky N., Vanhaeverbeek M., Ducobu J., Neve J. 2008. Development and validation of a screening procedure for the assessment of inhibition using a recombinant enzyme. Talanta. 75: 503–510. [DOI] [PubMed] [Google Scholar]
- 31.Delporte C., Van Antwerpen P., Zouaoui Boudjeltia K., Noyon C., Abts F., Metral F., Vanhamme L., Reye F., Rousseau A., Vanhaeverbeek M., et al. 2011. Optimization of apolipoprotein-B-100 sequence coverage by liquid chromatography-tandem mass spectrometry for the future study of its posttranslational modifications. Anal. Biochem. 411: 129–138. [DOI] [PubMed] [Google Scholar]
- 32.Furtmüller P. G., Jantschko W., Regelsberger G., Jakopitsch C., Arnhold J., Obinger C. 2002. Reaction of lactoperoxidase compound I with halides and thiocyanate. Biochemistry. 41: 11895–11900. [DOI] [PubMed] [Google Scholar]
- 33.Furtmüller P. G., Zederbauer M., Jantschko W., Helm J., Bogner M., Jakopitsch C., Obinger C. 2006. Active site structure and catalytic mechanisms of human peroxidases. Arch. Biochem. Biophys. 445: 199–213. [DOI] [PubMed] [Google Scholar]
- 34.Sokolov A. V., Ageeva K. V., Cherkalina O. S., Pulina M. O., Zakharova E. T., Prozorovskii V. N., Aksenov D. V., Vasilyev V. B., Panasenko O. M. 2010. Identification and properties of complexes formed by myeloperoxidase with lipoproteins and ceruloplasmin. Chem. Phys. Lipids. 163: 347–355. [DOI] [PubMed] [Google Scholar]
- 35.Temple A., Yen T. Y., Gronert S. 2006. Identification of specific protein carbonylation sites in model oxidations of human serum albumin. J. Am. Soc. Mass Spectrom. 17: 1172–1180. [DOI] [PubMed] [Google Scholar]
- 36.Sell D. R., Strauch C. M., Shen W., Monnier V. M. 2007. 2-aminoadipic acid is a marker of protein carbonyl oxidation in the aging human skin: effects of diabetes, renal failure and sepsis. Biochem. J. 404: 269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang C., Gu Z. W., Yang M., Lin S. N., Siuzdak G., Smith C. V. 1999. Identification of modified tryptophan residues in apolipoprotein B-100 derived from copper ion-oxidized low-density lipoprotein. Biochemistry. 38: 15903–15908. [DOI] [PubMed] [Google Scholar]
- 38.Himmelfarb J., McMenamin M. E., Loseto G., Heinecke J. W. 2001. Myeloperoxidase-catalyzed 3-chlorotyrosine formation in dialysis patients. Free Radic. Biol. Med. 31: 1163–1169. [DOI] [PubMed] [Google Scholar]
- 39.Delporte C., Franck T., Noyon C., Dufour D., Rousseau A., Madhoun P., Desmet J. M., Serteyn D., Raes M., Nortier J., et al. 2012. Simultaneous measurement of protein-bound 3-chlorotyrosine and homocitrulline by LC-MS/MS after hydrolysis assisted by microwave: application to the study of myeloperoxidase activity during hemodialysis. Talanta. 99: 603–609. [DOI] [PubMed] [Google Scholar]
- 40.Spickett C. M., Reis A., Pitt A. R. 2013. Use of narrow mass-window, high-resolution extracted product ion chromatograms for the sensitive and selective identification of protein modifications. Anal. Chem. 85: 4621–4627. [DOI] [PubMed] [Google Scholar]
- 41.Panasenko O. M., Spalteholz H., Schiller J., Arnhold J. 2003. Myeloperoxidase-induced formation of chlorohydrins and lysophospholipids from unsaturated phosphatidylcholines. Free Radic. Biol. Med. 34: 553–562. [DOI] [PubMed] [Google Scholar]
- 42.Lessig J., Schiller J., Arnhold J., Fuchs B. 2007. Hypochlorous acid-mediated generation of glycerophosphocholine from unsaturated plasmalogen glycerophosphocholine lipids. J. Lipid Res. 48: 1316–1324. [DOI] [PubMed] [Google Scholar]
- 43.Yang C. Y., Kim T. W., Pao Q., Chan L., Knapp R. D., Gotto A. M., Jr, Pownall H. J. 1989. Structure and conformational analysis of lipid-associating peptides of apolipoprotein B-100 produced by trypsinolysis. J. Protein Chem. 8: 689–699. [DOI] [PubMed] [Google Scholar]
- 44.Yang C. Y., Gu Z. W., Weng S. A., Kim T. W., Chen S. H., Pownall H. J., Sharp P. M., Liu S. W., Li W. H., Gotto A. M., Jr, et al. 1989. Structure of apolipoprotein B-100 of human low density lipoproteins. Arteriosclerosis. 9: 96–108. [DOI] [PubMed] [Google Scholar]
- 45.Hamilton R. T., Asatryan L., Nilsen J. T., Isas J. M., Gallaher T. K., Sawamura T., Hsiai T. K. 2008. LDL protein nitration: implication for LDL protein unfolding. Arch. Biochem. Biophys. 479: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levine R. L., Berlett B. S., Moskovitz J., Mosoni L., Stadtman E. R. 1999. Methionine residues may protect proteins from critical oxidative damage. Mech. Ageing Dev. 107: 323–332. [DOI] [PubMed] [Google Scholar]
- 47.Pattison D. I., Davies M. J. 2001. Absolute rate constants for the reaction of hypochlorous acid with protein side chains and peptide bonds. Chem. Res. Toxicol. 14: 1453–1464. [DOI] [PubMed] [Google Scholar]
- 48.Pattison D. I., Davies M. J., Hawkins C. L. 2012. Reactions and reactivity of myeloperoxidase-derived oxidants: differential biological effects of hypochlorous and hypothiocyanous acids. Free Radic. Res. 46: 975–995. [DOI] [PubMed] [Google Scholar]
- 49.Sokolov A. V., Ageeva K. V., Pulina M. O., Cherkalina O. S., Samygina V. R., Vlasova I. I., Panasenko O. M., Zakharova E. T., Vasilyev V. B. 2008. Ceruloplasmin and myeloperoxidase in complex affect the enzymatic properties of each other. Free Radic. Res. 42: 989–998. [DOI] [PubMed] [Google Scholar]
- 50.Rutgers A., Heeringa P., Kooman J. P., van der Sande F. M., Cohen Travaert J. W. 2003. Peripheral blood myeloperoxidase activity increases during hemodialysis. Kidney Int. 64: 760. [DOI] [PubMed] [Google Scholar]
- 51.Madhusudhana Rao A., Apoorva R., Anand U., Anand C. V., Venu G. 2012. Effect of hemodialysis on plasma myeloperoxidase activity in end stage renal disease patients. Indian J. Clin. Biochem. 27: 253–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Borén J., Ekström U., Ågren B., Nilsson-Ehle P., Innerarity T. L. 2001. The molecular mechanism for the genetic disorder familial defective apolipoprotein B100. J. Biol. Chem. 276: 9214–9218. [DOI] [PubMed] [Google Scholar]
- 53.Rutledge A. C., Su Q., Adeli K. 2010. Apolipoprotein B100 biogenesis: a complex array of intracellular mechanisms regulating folding, stability, and lipoprotein assembly. Biochem. Cell Biol. 88: 251–267. [DOI] [PubMed] [Google Scholar]
- 54.Pawlak K., Mysliwiec M., Pawlak D. 2013. Oxidized low-density lipoprotein (oxLDL) plasma levels and oxLDL to LDL ratio - are they real oxidative stress markers in dialyzed patients? Life Sci. 92: 253–258. [DOI] [PubMed] [Google Scholar]