Abstract
The methyltetrahydrofolate (CH3-H4folate) corrinoid-iron-sulfur protein (CFeSP) methyltransferase (MeTr) catalyzes transfer of the methyl group of CH3-H4folate to cob(I)amide. This key step in anaerobic CO and CO2 fixation is similar to the first half-reaction in the mechanisms of other cobalamin-dependent methyltransferases. Methyl transfer requires electrophilic activation of the methyl group of CH3-H4folate, which includes proton transfer to the N5 group of the pterin ring and poises the methyl group for reaction with the Co(I) nucleophile. The structure of the binary CH3-H4folate/MeTr complex (revealed here) lacks any obvious proton donor near the N5 group. Instead, an Asn residue and water molecules are found within H-bonding distance of N5. Structural and kinetic experiments described here are consistent with the involvement of an extended H-bonding network in proton transfer to N5 of the folate that includes an Asn (Asn-199 in MeTr), a conserved Asp (Asp-160), and a water molecule. This situation is reminiscent of purine nucleoside phosphorylase, which involves protonation of the purine N7 in the transition state and is accomplished by an extended H-bond network that includes water molecules, a Glu residue, and an Asn residue. In MeTr, the Asn residue swings from a distant position to within H-bonding distance of the N5 atom upon CH3-H4folate binding. An N199A variant exhibits only ~20-fold weakened affinity for CH3-H4folate but a much more marked 20,000–40,000-fold effect on catalysis, suggesting that Asn-199 plays an important role in stabilizing a transition state or high energy intermediate for methyl transfer.
The methyltetrahydrofolate (CH3-H4 folate)5 corrinoid-iron-sulfur protein (CFeSP) methyltransferase (MeTr) from Moorella thermoacetia catalyzes the methyl group transfer from CH3-H4folate to cob(I)amide (Reaction 1) in the Wood-Ljungdahl pathway of anaerobic CO and CO2 fixation (1, 2) to generate an organometallic methyl-Co species on the CFeSP. The methylated CFeSP then reacts with carbon monoxide and CoA to generate acetyl-CoA in a reaction catalyzed by the bifunctional NiFeS protein, carbon monoxide dehydrogenase/acetyl-CoA synthase (2). The MeTr reaction is similar to a variety of cobalamin-dependent methylation reactions, including methyl transfer from CH3-H4folate to the bound cobalamin cofactor of methionine synthase, which subsequently transfers its methyl group from methylcobalamin to homocysteine (3, 4).
| REACTION 1 |
The MeTr reaction mechanism consists of several steps (5). The first two steps include a pH-dependent conformational change followed by binding of CH3-H4folate. Equilibrium dialysis studies shows that MeTr has a high affinity binding site for CH3-H4folate with a Kd of 10 μm (6). CH3-H4folate binds to MeTr in the unprotonated state and then undergoes rapid protonation (5, 6). This proton transfer step generates a positive charge at the N5 position of the pterin ring, resulting in electrophilic activation of the methyl group of CH3-H4folate (3). This activation step is important because H4folate is a very poor leaving group, but it is not rate-limiting. Next, the CFeSP binds, and its Co(I) center catalyzes an SN2 displacement of the N5 methyl group of CH3-H4folate to form the methylated CFeSP. There is evidence that proton transfer to the pterin occurs in a tertiary complex with CH3-H4folate and cob(I)alamin for the cobalamin-dependent methionine synthase (MetH) (7) and in the ternary complex with CH3-H4folate and homocysteine for the enzyme cobalamin-independent methionine synthase (8).
Interestingly, the crystal structure of MeTr (in the absence of substrate) reveals no obvious proton donor within H-bonding distance of the N5 position of CH3-H4folate, which was modeled into the structure (9). The only amino acid that located near enough to N5 to participate in H-bonding is the side chain of Asn-199; however, this group is not chemically suitable to act as a proton donor. Thus, the mechanism of proton transfer leading to electrophilic activation of the methyl group is unclear. To solve this paradox, one might posit that the proton donor moves into place after CH3-H4folate binds; however, the recent crystal structure of the CH3-H4folate-MetH complex shows that there is no acidic group positioned near the N5 group of the pterin and, like the MeTr structure, Asn-199 (MeTr numbering) is within H-bonding distance to N5 (10) (Fig. 1).
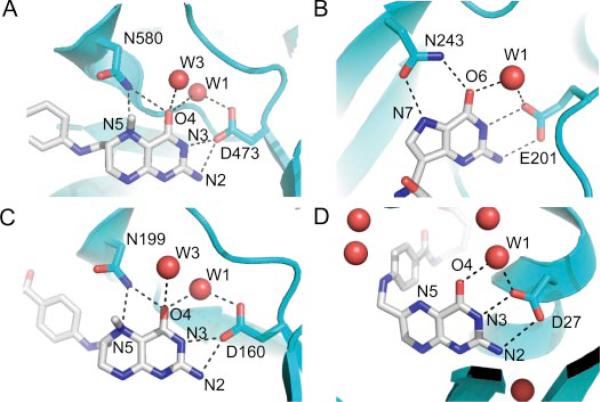
FIGURE 1. Similarity of H-bonding networks around substrates.
A, MetH (carbon, blue) with bound CH3-H4folate (carbon, gray) (PDB code 1Q8J). B, PNP (carbon, blue) with bound transition state ImmH (carbon, gray) (PDB code 1B8O). C, MeTr (carbon, blue) with bound CH3-H4folate (carbon, gray). D, DHFR (carbon, blue) with bound folate (carbon, gray) (PDB code 7DFR).
The lack of discernible proton delivery systems in the crystal structures of enzymes that catalyze proton transfer is surprisingly common. Besides methyltransferases, this situation is encountered in a number of enzymes, including dihydrofolate reductase (DHFR) (11) and purine nucleoside phosphorylase (PNP) (12). The latter catalyzes the reversible phosphorolysis of purine nucleoside to form the corresponding purine base and ribose 1-phosphate. Fig. 1 compares the H-bonding patterns in MeTr, MetH, and DHFR near the N5 of CH3-H4folate (or folate in DHFR) with PNP in the vicinity of N7 of the transition state analog immucillin-H (ImmH). In addition to the structural homology, there are mechanistic similarities of the methyltransferases with PNP as well. To facilitate their bond cleavage reactions, both sets of enzymes appear to require substrate protonation and subsequent stabilization of a protonated species in the transition state. Evidence that N7 is protonated in the transition state of the PNP reaction comes from a series of elegant studies by Schramm and co-workers using transition state analogs (13, 14). They find a difference in affinity of 107 (10.1 kcal/mol) between ImmH, an analog that is an H-bond donor at N7 (always protonated) and 4-aza-3-deaza-ImmH, an analog that is an H-bond acceptor at N7 (never protonated), suggesting that analogs that are protonated at N7 mimic the transition state. Available to accept the H-bond from N7 of substrate is Asn-243 (12, 13). The oxygen of the carboxamide side chain of Asn-243 is 3.3 Å away from N7 with an unprotonated substrate bound and moves closer (2.8–2.9 Å) when protonated substrate analogs are bound (12, 13). NMR studies show that a single H-bond between Asn-243 and N7 of substrate is only partly responsible for the full 10.1 kcal/mol difference between ImmH and 4-aza-3-deaza-ImmH binding affinity (13, 14); however, this bond appears to be enmeshed within an extended H-bonding network (Fig. 1) that is responsible for this transition state stabilization. The equivalent residue to Asn-243 in MeTr, Asn-199, is conserved in all methyltransferases. Given the provocative location of Asn-199 in MeTr and its potential role in transition state stabilization in the transmethylation reaction, we performed site-directed mutagenesis, kinetic, and structural studies to evaluate the contribution of this residue to catalysis.
EXPERIMENTAL PROCEDURES
General DNA Manipulations
DNA isolation and manipulation were performed using standard techniques (15). Plasmid DNA was purified with a QIAprep spin miniprep kit (Qiagen, Valencia, CA). DNA fragments were purified from agarose gels using a QIAquick gel extraction kit (Qiagen). The DNA sequences of all PCR-generated DNA fragments were confirmed by automated sequencing of both strands by the Genomics Core Research Facility (University of Nebraska-Lincoln, NE) with a Beckman Coulter CEQ2000XL 8-capillary DNA sequencer using dye terminator chemistry.
Site-directed Mutagenesis of MeTr
Asn-199 was mutated to Ala using the QuikChange site-directed mutagenesis protocol from Stratagene (La Jolla, CA) using the wild-type pET3a-MeTr plasmid (9) as a template. The PCR product was digested with DpnI and transformed into Escherichia coli Top10 (Invitrogen) cells. The N199A mutant plasmid was then isolated and transformed into B834(DE3)pLysS(met–) (Novagen) E. coli cells.
Enzyme Purification
E. coli cells transformed with pET3a-MeTr (N199A mutant or wild-type) were grown in 3 or 4 liters of Terrific Broth essentially as described earlier (9). The cells were induced with 1 mm isopropylthio-β-d-galactoside when the OD at 600 nm reached 0.5–0.8, grown for 3 h at 37 °C or 11 h at 25 °C, and then harvested by centrifugation at 5,000 rpm for 30 min. The cells (12 or 16 g) were suspended in 50 or 40 ml of sonication buffer (50 mm Tris, pH 7.6, containing 2 mm 1,4-dithio-dl-threitol (DTT), 1 mg/ml lysozyme, 1 units/ml DNase I, and 0.1 mg/ml phenylmethylsulfonyl fluoride) and disrupted by sonication for 10 min at 0 °C. After centrifugation at 32,000 rpm for 90 min at 4 °C with a Beckman Ti35 rotor, the super-natant fraction was recovered as cell-free extract. The cell-free extract was heated at 70 °C for 40 min and then centrifuged at 10,000 rpm for 15 min at 4 °C. MeTr was then purified from the cell-free extract using a 150-ml phenyl-Sepharose column and buffer-exchanged and concentrated by ultrafiltration into 50 mm Tris, pH 7.6, containing 0.1 m NaCl and 2 mm DTT. The concentration of protein was determined with the rose bengal method (16). A typical yield from one liter of culture was 50 mg of MeTr.
Activity Measurements
For steady-state assays, MeTr activity was determined in the reverse direction by following transfer of the methyl group of methylcobalamin to H4folate as described earlier (17). Briefly, the assay was performed at pH 8.4 and 55 °C using 66 μm methylcobalamin, 300 μm H4folate, and the decrease in absorbance at 520 nm from methyl-Co(III) was measured. The forward reaction was measured essentially as described earlier (17) at 25 °C in a reaction mixture containing 0.1 m potassium succinate, pH 5.0, 60 mm NaCl, 3.6 μm CFeSP, 87 μm CH3-H4folate, MeTr (4.1 nm monomer for the wild-type or 8.3 μm monomer for the N199A variant) and 165 μm Ti(III)-citrate to reduce Co(II) to the Co(I) state. The decrease in absorbance at 390 nm (Co(I) decay) and increase at 450 (methyl-Co(III) formation) were measured. The absorbance changes for the forward and reverse reactions were monitored using an OLIS Cary-14-converted spectrometer (Bogart, GA).
Presteady-state kinetic experiments for the wild-type MeTr and N199A variant were performed at 25 °C under anaerobic conditions. For both experiments, 300 μm 6S-CH3-H4folate, MeTr (40 μm of monomer), 1 mm Ti(III)citrate, 0.1 m MES, pH 5.1, 0.1 m NaCl was rapidly mixed with 10 μm CFeSP, 1 mm Ti(III)citrate (to reduce the CFeSP to the Co(I) state), 0.1 m MES, pH 5.1, 0.1 m NaCl. The decrease in absorption of Co(I) at 390 and the increase in absorption of the methylated Co(III) at 450 nm were recorded. For the CFeSP, the Δε between cob(I)amide and methylcob(III)amide at 390 and 450 nm is 17 and 6 mm–1 cm–1, respectively (18). The experiment with the wild-type protein was performed with a DX.17MV sequential stopped-flow ASVD spectrophotometer from Applied Photo-physics (Leatherbarrow, England), whereas for the N199A variant MeTr, the same experiments were performed at room temperature using an S2000 miniature fiber optic spectrometer (Ocean Optics, Inc., Dunedin, FL) inside a Vacuum Atmospheres (Hawthorne, CA) anaerobic chamber.
Binding Studies
Equilibrium dialysis experiments were performed as described (6) using a lower solution containing 40 μm N199A-MeTr monomer and an upper solution containing varying concentrations of (6R,S)-14CH3-H4folate (4–75 μm). Both solutions contained 0.1 mm MES, pH 6.1, and 1 mm DTT.
Fluorescence quenching experiments were performed at pH 7.6 by titrating 2.8 μm MeTr monomer with CH3-H4folate as described (6). We could not measure the Kd at low pH values because under these conditions, the fluorescence of CH3-H4folate was much more intense than that of MeTr.
Isothermal titration calorimetry (ITC) experiments for wild-type and N199A MeTr were performed with a MicroCal VPITC isothermal titration calorimeter (MicroCal Inc., Northampton, MA) at pH 5.1 and 7.6. Before the experiment, the enzyme solutions were dialyzed against the buffer with appropriate pH, i.e. 0.1 m MES, pH 5.1, or 50 mm Tris, pH 7.6, containing 0.1 m NaCl and 2 mm DTT if needed for anaerobiosis. The reaction cell was filled with a 0.05 mm (as monomer) MeTr solution. The titrated ligand [6S]-5-CH3-H4folate (1.5 mm) was dissolved in the same buffer used for dialysis of the enzyme. The stirring speed was 310 rpm, and the thermal power of 29 injections of 10 μl was recorded every 120 s. Thermogram analysis was performed using the Origin 7.0 software supplied with the instrument.
Crystal Growth
Crystals of MeTr with CH3-H4folate (CH3-H4folate/MeTr) and of N199A with CH3-H4folate (CH3-H4folate/N199A) were produced at room temperature using a hanging drop setup and equal amounts of protein and precipitant (typically 2 μl of protein to 2 μl of precipitant). The protein solutions were kept in an anaerobic bottle before crystallization. Small aliquots were taken by syringe for the experiments. Typical protein solution contained 15–25 mg/ml MeTr in 50 mm Tris pH 7.6, 100 mm NaCl, 2 mm DTT. For the complexes with CH3-H4folate, a 3-fold molar excess of the 6-S-monoglutarate substrate (Schricks Laboratories, Jona, Switzerland) was added to the crystallization mixture. A precipitant stock solution contained 8–15% (w/v) polyethylene glycol monomethyl ether 5000, 20–50 mm calcium acetate, 50 mm HEPES, pH 7.5, and 20% glycerol. To obtain single crystals, this stock solution was diluted 50–100-fold in 20% glycerol prior to crystallization. Typically, 4 μl of the stock mixture were added to either 196 μl or 396 μl of 20% glycerol to make the working crystallization solution. Crystals formed two-dimensional plates and were very fragile and difficult to manipulate. All crystals were cryo-frozen directly from the drops into the cryostream. The 20% glycerol in the crystallization condition acted as cryoprotectant.
Data Collection
The CH3-H4folate/MeTr data set was obtained at the National Synchrotron Light Source (NSLS, Brookhaven National Laboratory) X12-C line on a B4 Brandeis CCD 4 cell detector. The CH3-H4folate/N199A data set was also collected at NSLS on the X25 beam line with a MAR345 image plate detector. Diffraction data were integrated with MOSFLM (19) and scaled and merged using SCALA (20) and TRUNCATE (21) programs in the CCP4 suite (22). Data statistics are shown in Table 1.
TABLE 1.
Data collection and refinement statistics
| Wild-type with CH3-H4 folate | N199A with CH3-H4 folate | |
|---|---|---|
| Data | ||
| Wavelength (Å) | 1.3000 | 0.91938 |
| Space group | P212121 | P212121 |
| Unit cell dimensions (Å) | 50.16 78.55 135.62 | 50.08 78.17 135.98 |
| Resolution (Å) | 30.92-2.20 (2.26-2.20)a | 46.73-2.30 (2.36-2.30) |
| Reflections total/unique | 103105/26806 (6074/1750) | 130051/24151 (8478/1752) |
| Completeness | 96.2 (87.9) | 99.1 (99.3) |
| Multiplicity | 3.8 (3.5) | 5.4 (4.8) |
| Rsymb (%) | 0.086 (0.206) | 0.134 (0.385) |
| Mean (I)/sd(I) | 11.5 (5.3) | 11.1 (4.3) |
| Wilson B factor (Å2) | 14.8 | 17.2 |
| Refinement | ||
| Resolution (Å) | 30.88-2.20 (2.26-2.20) | 40.19-2.30 (2.36-2.30) |
| Rcryst/Rfreec (%) | 16.26/22.40 (17.4/25.0) | 16.77/22.37 (17.40/24.11) |
| r.m.s.d. bonds (Å) | 0.007 | 0.007 |
| r.m.s.d. angle (degree) | 1.049 | 1.078 |
| Reflections in working/test set | 24177/2586 (1571/165) | 21756/2369 (1588/158) |
| Nonhydrogen protein atoms | 4485 | 4358 |
| Nonhydrogen heteroatoms | 2 Ca + 64 in CH3-H4folate | 2 Ca + 64 in CH3-H4folate |
| Waters | 426 | 357 |
| Average B-factor, all atoms (Å2) | 9.38 | 11.36 |
| CH3-H4folate | 13.4 | 19.5 |
| Correlation coefficientd | 0.947/0.903 | 0.946/0.905 |
Values in parentheses are for the highest resolution bin
Rsym = math where Ii is the intensity of the measured reflection and Im is the mean intensity of all symmetry related reflections.
Rcryst, Rfree = math where the working (cryst) and the free R-factors are calculated using the working and the test (free) reflection sets, respectively. The test reflections were held aside throughout the refinement.
Fo — Fc/Fo — Fcfree.
Structure Determination
The crystal structure of CH3-H4folate/MeTr was solved by molecular replacement using the structure of MeTr in space group P212121 (PDB code 1F6Y) as the search model. In particular, a MeTr dimer model with all protein atoms and no water molecules was used for the initial molecular replacement in the program EPMR (23), yielding a correct solution with a correlation coefficient CC = 0.538 and a residual factor r = 0.466. Unaccounted for electron density in the TIM barrel was modeled as CH3-H4folate with restraints generated from the HIC-Up server. Electron density fitting was performed initially in XtalView (24) and later in COOT (25). The structure was refined with TLS restrained refinement parameters without a σ cutoff and with loose NCS restraints as implemented in REFMAC (26, 27). Since the publication of the apo-MeTr structure, a sequence revision was published for the residues 224–225 (Met-224–Ser-225 to Asp-224–Ala-225). These changes have been incorporated into the current structures. The model also contains a cis-peptide bond between residues Asn-96 and Ser-97 in both subunits and another one between Asn-188 and Pro-189 in the ordered loop of subunit A. In addition, a 6.5-σ electron density peak suggests the presence of a metal ion near residue Asp-224. A metal ion modeled into this electron density is in position to interact with Asp-224, five water molecules, and the carboxyl group of Gly-222, with distances ranging from 2.38 to 2.75 Å. Based on these oxygen-rich interactions, the bond lengths, and the σ of the electron density peak, a divalent cation such as calcium (present in the crystallization conditions) is a reasonable assignment. In subunit A, the putative calcium ion is well ordered, but in subunit B, it has much weaker density and was modeled at 50% occupancy. The final model for CH3-H4folate/MeTr has excellent stereochemistry (Table 1), and no residues in the disallowed region of the Ramachandran plot (440 of 469 non-glycine and non-proline residues were in the most favored region, 29 were in the additional allowed region, 0 were in the generously allowed region, and 0 were in the disallowed region). The loop after the sixth helix (184–188) in subunit B is disordered, and these residues were omitted from the final model. The final model has 519 protein residues of MeTr, two calcium atoms, two CH3-H4folate molecules, and 426 ordered water molecules.
The CH3-H4folate/N199A structure was solved to 2.3 Å resolution using rigid body refinement and TLS restrained refinement in REFMAC starting from the CH3-H4folate/MeTr model and keeping analogous reflections in the test set for the Rfree reference. The model was modified by changing Asn-199 to alanine. A Ramachandran plot shows that 440 of 469 non-glycine and non-proline residues are in the most favored region, 28 are in additional allowed region, 1 is in generously allowed region (leucine 162, subunit A), and 0 is in the disallowed region. The final model has 519 protein residues of MeTr, two calcium atoms, two CH3-H4folate molecules, and 357 ordered water molecules. As in the CH3-H4folate/MeTr model, the loop after the sixth helix (184–188) in subunit B is disordered, and these residues were omitted from the final model. Composite omit maps calculated in CNS (28) were used to check both the CH3-H4-folate/MeTr and the CH3-H4-folate/N199A structures.
RESULTS
Effect of Asn-199 on CH3-H4folate Binding
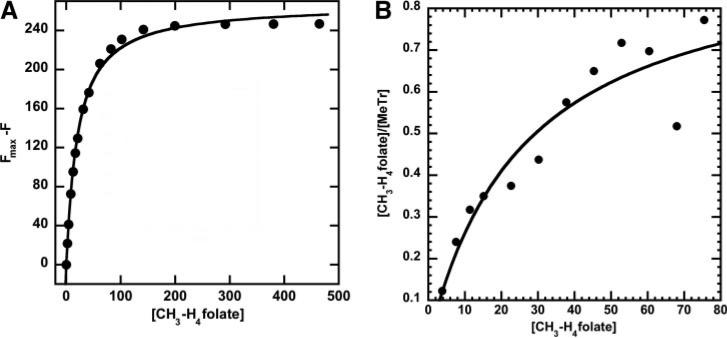
Since Asn-199 is in hydrogen-bonding contact with N5 of CH3-H4folate, we expected that the N199A mutation would decrease affinity of MeTr for this substrate. However, we do not observe a large decrease in affinity. Based on equilibrium dialysis experiments with the N199A variant and 6S-CH3-H4folate (Fig. 2A), the Kd value is 26.5 ± 8.8 μm, and the enzyme reaches a maximum ratio of 0.95 ± 0.13 mol of CH3-H4folate bound per mol of MeTr at pH 6.1. With the wild-type protein, the Kd value was determined by equilibrium dialysis to be pH-independent between pH 4.9 and 8.5 and is about 2.6-fold lower (10 ± 1 μm) (6).
FIGURE 2. Binding of CH3-H4 folate to the N199A variants.
A, 2.8μm N199A-MeTr monomer was titrated with varying concentrations of [6S]-CH3-H4folate, and the data were plotted as fluorescence decrease versus concentration of substrate. B, 40 μm N199A-MeTr (monomer) was reacted with varying concentrations of (6R,S)-14CH3-H4folate in equilibrium dialysis experiments. The [CH3-H3folate] on the y axis refers to bound substrate. For both curves, the data were fit to a single-site binding isotherm.
Tryptophan residue(s) undergo fluorescence quenching when MeTr binds CH3-H4folate (Fig. 2B) (6, 17). The Kd value obtained from tryptophan fluorescence quenching experiments is 21.3 ± 1.0 μm for the N199A variant at pH 7.6, which compares with a value between 0.64 μm (17) and 2.1 μm (6), which were measured earlier for the wild-type protein at pH 7.6.
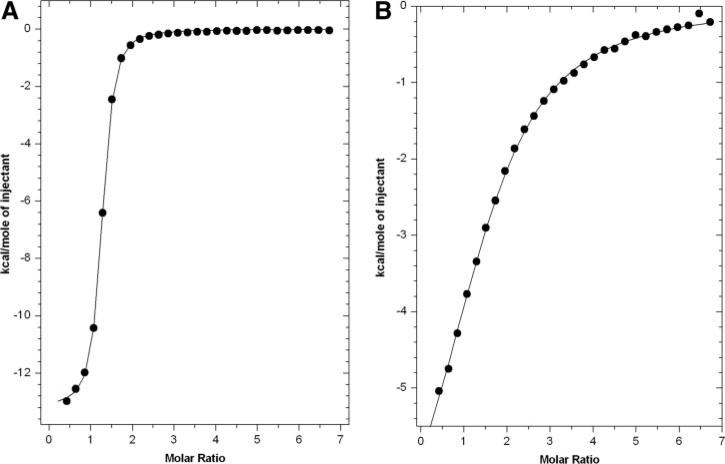
ITC experiments were performed for the wild-type protein and the N199A variant. With the N199A variant, the Kd value for 6S-CH3-H4folate is 33.2 ± 1.3 μm at pH 5.1 (Fig. 3A). This is similar to the values determined by equilibrium dialysis and by fluorescence quenching (Table 2). For the wild-type protein, the Kd value for 6S-CH3-H4folate is 0.79 ± 0.02 μm at pH 7.6 (Fig. 3B), which is similar to the values measured by ITC at pH 5.1 (0.29 μm, not shown). These Kd values for the wild-type enzyme are also similar to those measured by tryptophan fluorescence quenching experiments, 0.64 (17) to 2.1 μm (6). From the values of ΔH and ΔS, binding can be considered to be enthalpy-driven with ΔG values of –6.0 kcal/mol for the mutant and –8.3 kcal/mol for the wild-type protein.
FIGURE 3. ITC measurements with wild-type MeTr and N199A variant.
The experiments were performed by titrating 50μm MeTr (as monomer) in 0.1 m buffer, 0.1 m NaCl, 1 mm DTT with 1.5 mm 6S-CH3-H4folate in the same buffer. For wild-type and N199A MeTr, the buffers were MES, pH 5.1, and Tris-HCl, pH 7.6. A, for the N199A variant (measured at pH 5.1), the fit parameters are n = 1.2, K = 1.26 (± 0.04) × 106 m, ΔH = –13200 ± 40 cal/mol, ΔS = –17.1 cal/mol-K. B, for wild-type MeTr (measured at pH 7.6), the fit parameters are n = 1.5, K = 3.01 (± 0.13) × 104 m, ΔH = –8187 ± 209 cal/mol, ΔS = –7.44 cal/mol-K.
TABLE 2.
Comparison of wild-type MeTr versus the N199A variant
| Value | Wild-type | N199A |
|---|---|---|
| Kd | ||
| Equilibrium dialysis | 10 ± 1 μM (pH 4.85-8.5) (6)a | 26.5 ± 8.8 μM (pH 6.1) |
| Tryptophan fluorescence quenching | 0.6 ± 0.2 μM (17) | 21.3 ± 1.0 μM (pH 7.6) |
| 2.1 ± 0. 2 μM (6) (pH 7.6) | ||
| Isothermal calorimetry | 0.79 ± 0.02 μM (pH 7.6) | 33.2 ± 1.3 μM (pH 5.1) |
| 0.29 μM (pH 5.1) | ||
| Steady-state kinetic parameters | ||
| Reverse Rate (with methylcobalamin) | 0.05 s-1 | 0.00003 s-1 |
| Forward Rate (with CFeSP) | 1.1 s-1 | 0.0005 s-1 |
| kcat/Km (forward) | 0.5 × 106M-1s-1 | 20 M-1 s-1 |
| Rel kcat/Km | 2.5 × 104 | 1 |
| Presteady-state kinetic parameters | ||
| kobs (forward reaction with CFeSP) | 7.2 s-1 (pH5.1) | 0.0077 s-1 (pH 5.1) |
| kobs/Kd | 4.8 × 106M-1s-1 | 260 M-1 s-1 |
| Rel kobs/Kd | 1.85 × 104 | 1 |
Kd values varied from 2.1 to 6.2 over the pH range of 4.85-8.5 but with no obvious trend as a function of pH (6).
In summary, for binding of CH3-H4folate by the N199A variant, the Kd values determined by ITC (33.2 μm), equilibrium dialysis (26.5 μm), and fluorescence quenching (21.3 μm) agree quite well (Table 2). The Kd values for the wild-type protein vary fairly widely, from 0.29 μm (ITC) and 0.64–2.1 μm (fluorescence quenching) to 10 μm (equilibrium dialysis). The Km value for CH3-H4folate for the wild-type protein is similar (2.0 μm) (5). Even if we disregard the fairly high Kd value (10 μm) obtained by equilibrium dialysis, the binding affinity of MeTr for CH3-H4folate is compromised only ~20-fold (30/1.5) by alteration of Asn-199 to Ala. This loss in affinity would contribute to a total energy cost of only about 2 kcal/mol at 50 °C (the growth temperature for M. thermoacetica), indicating that substrate binding is minimally affected by alteration of Asn-199.
Effect of Asn-199 on Steady-state and Presteady-state Kinetics
We compared steady-state and presteady-state kinetic studies to determine the role of Asn-199 in catalysis. Because the N199A variant is severely compromised in catalysis, ~500-fold more enzyme is required in each reaction to accurately measure the reaction velocity.
Since the MeTr reaction is reversible (Keq = 14 (5)), the steady-state velocity was measured in the forward (methylation of Co(I)) and reverse (methylation of H4folate) directions. The steady-state velocity for the methyl transfer from methylcobalamin to H4folate for wild-type MeTr is 84.4 nmol min–1 mg–1, which is similar to that measured earlier under similar conditions (18), whereas for the N199A variant, the velocity is 1700-fold slower, 0.05 nmol min–1 mg–1 (not shown). For the forward reaction, methylation of the CFeSP by CH3-H4folate, the steady-state velocity is 2.0 μmol min–1 mg–1 (1.1 s–1) for the wild-type protein. For the N199A mutant, the velocity is 2200-fold slower, 0.8 nmol min–1 mg–1. In the forward and reverse reactions, the absorbance changes due to the formation/decay of Co(I) and methyl-Co(III) were both monitored, and the observed rates were nearly identical.
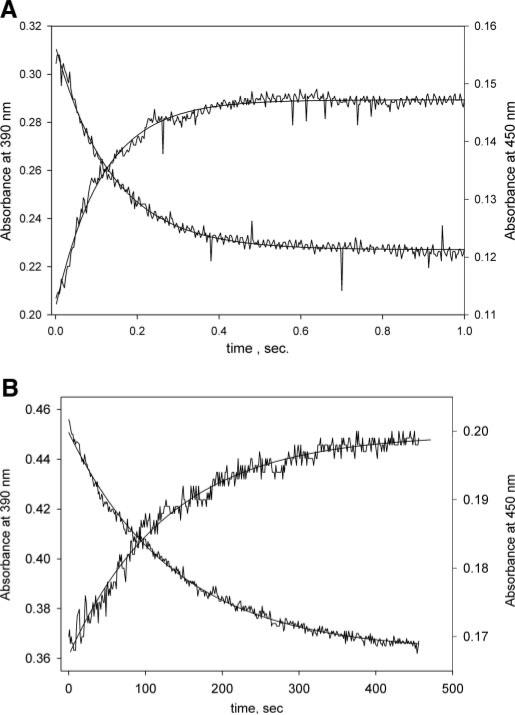
Presteady-state kinetic studies of the methylation of H4folate by the methylated CFeSP were performed at pH 5.1 under saturating substrate conditions using wild-type MeTr and the N199A variant (Fig. 4). The rate constant for wild-type MeTr was 7.2 s–1, whereas the kobs under identical conditions for the N199A variant was 7.70 × 10–3 s–1. Based on the amplitudes at 390 (ε = 17 mm–1 cm–1) and 450 nm, 5 μm CFeSP (100%) underwent methylation. This ~1,000-fold difference in kobs by substituting Asn-199 with Ala (above) is comparable with the 2,000-fold decrease in the steady-state Vmax value. Using the Kd values described above, the kobs/Kd value for the wild-type MeTr at pH 5.1 is 4.8 × 106 m–1 s–1, which is 18,500-fold larger than the value of 260 m–1 s–1 measured for the N199A variant. A similar calculation using the Kd values measured above and the ~2,000-fold decrease in steady-state rates indicates that substitution of Asn-199 by Ala lowers the kcat/Kd in both forward and reverse directions by ~40,000-fold (~2,000 × 30/1.5). Based on the relative steady-state values of kcat/Km, a similar catalytic penalty of 25,000-fold is calculated for the N199A substitution (Table 2). Thus, Asn-199 stabilizes the transition state for methylation of H4folate by the methylated CFeSP by 6.3–6.9 kcal/mol.
FIGURE 4. Presteady-state kinetics of the wild-type and N199A variant.
In these experiments performed at 25 °C and pH 5.1, a solution containing 300 μm [6S]-CH3-H4folate, and 40μm MeTr (monomer) was rapidly mixed with a solution containing 10 μm CFeSP and 1 mm Ti(III) (see “Experimental Procedures” for details).A,forthewild-typeprotein,thefitparametersforthe450nmabsorbance increase are: amplitude, 0.031; rate constant, 7.2 s–1; absorbance end point, 0.143. For decay of the 390 nm absorbance, the parameters are: amplitude, 0.084; rateconstant,7.2s–1; and absorbance end point, 0.227. B, for the N199A mutant, the fit parameters for the 450 nm absorbance increase were as follows. The amplitude is 0.032, the rate constant is 0.0077 s–1, and the absorbance end point is 0.199. For the 390 nm absorbance decay, the amplitude is 0.086, the rate constant is 0.0074 s–1, and the absorbance end point is 0.364.
Overall Structure
The structures of the binary complexes of CH3-H4folate/MeTr and CH3-H4folate/N199A were determined to 2.2 Å resolution with 16.3 and 22.4 R/Rfree factors and 2.3 Å resolution with 16.8 and 22.4% R/Rfree, respectively (Table 1). Each monomer of MeTr contains eight β strands and eight helices, forming a typical TIM barrel fold with the binding site for CH3-H4folate located at the C termini of the β strands. The overall structures of the complexes and the apoenzymes are very similar with no major loop movements observed. The r.m.s.d. values between the 485&-carbons for the dimer of CH3-H4folate/MeTr when compared with the apo-MeTr dimer is 0.46 Å. The individual monomers of apo-MeTr are also very similar to the monomers of CH3-H4folate/MeTr with r.m.s.d. values under 0.39 Å for 236–241 common &-carbons. Comparing the CH3-H4folate-binding domains of MetH and MeTr reveals 30% sequence identity, resulting in a 3.61 Å r.m.s.d. for 421 α-carbons for the dimers and r.m.s.d. values from 1.58 to 1.81 Å for individual monomers with 187–201 common α-carbons.
CH3-H4-folate Binding in the Active Site
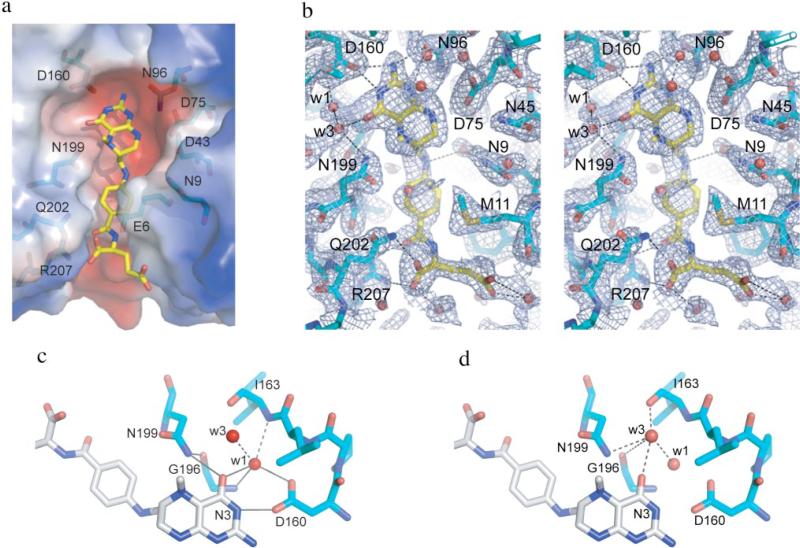
CH3-H4folate binds in the negatively charged TIM barrel cavity as expected based on the apo-MeTr structure (Fig. 5A) (9) and as described in the structure of the ternary complex of the Thermotoga maritima methionine synthase with homocysteine and CH3-H4folate (10). The CH3-H4folate molecule in MeTr has a similar conformation to that in MetH, except with respect to the orientation of the glutamate tail. There are slight differences in the conformations of the folate glutamate tails in the two subunits of MeTr as well, indicating that this part of the binding site is quite flexible. The presence of the folate-binding pocket at the surface of the protein allows for this conformational flexibility and the accommodation of different length polygluta-mate tails of CH3-H4folate. The p-aminobenzoic acid ring and the glutamate tail of CH3-H4folate extend to fill a gap on the side of the TIM barrel created by a short eighth β strand and a short loop. p-Aminobenzoic acid is oriented almost parallel to the pterin ring, allowing for a long conformation of the CH3-H4 folate substrate. The methyl group of CH3-H4folate projects above the pterin plane at ~60 degrees.
FIGURE 5. MeTr active site.
a, active site with CH3H4folate bound (carbon in yellow) and labels on the polar residues (protein carbons in light blue). Protein surface charge distribution, as calculated in PyMOL, is depicted as follows: red, negative charges, and blue, positive charges. b, stereo view of active site. Blue mesh indicates a 1-σ 2Fo – Fc electron density omit map density. Potential H-bonds are shown as dashed lines. c, H-bonding network involving water molecule W1. Solid lines represent the H-bonding networks described under “Results,” whereas dashed lines represent addition H-bonds made by W1. d, H-bonds involving water molecule W3 (dashed lines). This figure was created in PyMOL.
The pterin-binding site is conserved in a variety of enzymes that utilize H4folate or tetrahydromethanopterin, including MeTr, methyltetrahydromethanopterin:cob(I)alamin methyltransferase (MtrH) in the sodium ion translocation step in methanogenesis, MetH, and dihydropteroate synthases (DHPS). The following residues are part of this conserved binding site: Asp-43/-358/-45/-65, Asp-75/-390/-84/-101, Asn-96/-411/-103/-127, Asp-160/-473/-167/-194 in MeTr(1F6Y)/MetH(1Q8J)/DHPS(1AD4)/MtrH (29). The majority of these residues contribute to the negatively charged character of that cavity and create a complementary binding surface for CH3-H4folate. All polar atoms in the folate ring have an H-bonding partner, either an amino acid side chain or a well ordered water molecule. This H-bonding pattern is conserved in the binding of CH3-H4folate to MetH and in the binding of dihydropterin pyrophosphate to DHPS (30, 31). The major difference between the methyltransferases and DHPSs is in the nature of the hydrogen bond to the N5 position of the pterin. In the methyltransferases, the H-bonding partner is a conserved Asn (Asn-199/-508/-228 for MeTr/MetH/MtrH), whereas in the DHPSs, it is a conserved Lys. Another intriguing interaction between protein and folate is made by Asp-160, the so-called pterin hook (9). This residue hydrogen bonds to two atoms of the pterin (Fig. 5B) as well as to a water molecule described below and has been implicated in activation of CH3-H4folate for methyl transfer (see “Discussion”).
Electron density at an interesting position inside the CH3-H4folate-binding pocket was modeled as a water molecule (Fig. 5C, W1). This water is in position to hydrogen bond with the O4 of CH3-H4folate (2.89/2.73 Å in subunits A/B of MeTr and 2.95/2.95 Å in subunits A/B of the N199A variant) and with the carboxyl side chain of Asp-160 (2.80/2.77 Å in subunits A/B of MeTr and 2.72/2.72 Å subunits A/B of the N199A variant). Water W1 also makes close contact with the amide nitrogens of residues Ile-163 (3.10/3.20 Å), Gly-196 (2.93/2.88 Å), as well as another water molecule (Fig. 5C, W3). Interestingly, W1 is a part of two H-bonding networks: (a) an oxygen-rich ring in the plane of the pterin ring with five polar atoms, one carboxyl oxygen of Asp-160 to W1, W1 to O4 folate, N3 folate to the other carboxyl oxygen of Asp-160; (b) another network including the backbone oxygen and nitrogen of Gly-196, the side-chain amide of Asn-199, and O4 of the pterin (Fig. 5C). A water molecule in this position is also found in DHFR, MetH (Fig. 1), and DHPS, and thus, seems to be integral for binding pterin within a TIM barrel structure.
Another conserved interaction in the CH3-H4folate-binding pocket is the water above O4 (Fig. 5D, W3). Distances from W3 to O4 of CH3-H4folate are 2.67/2.76 Å in A (2.87/2.82 Å in B); distances to the carboxyl oxygen of Ile-163 are 2.71/2.77 Å in A (2.74/2.68 Å in B); and distances to the side-chain amide of Asn-199 are 3.16/3.50 Å for subunits A and B in the CH3-H4folate/MeTr structure. A water molecule in a similar location is observed in one of the wild-type apo-MeTr subunits and in subunit A of the MetH structure (Fig. 1). For MetH, the distances of this water to neighboring atoms are similar to those observed in the MeTr structures (O4 (2.70 Å), carboxyl oxygen of Val-476 (Ile-163 equivalent) (2.73 Å); side-chain amide of Asn-508 (Asn-199 equivalent) (3.48 Å)).
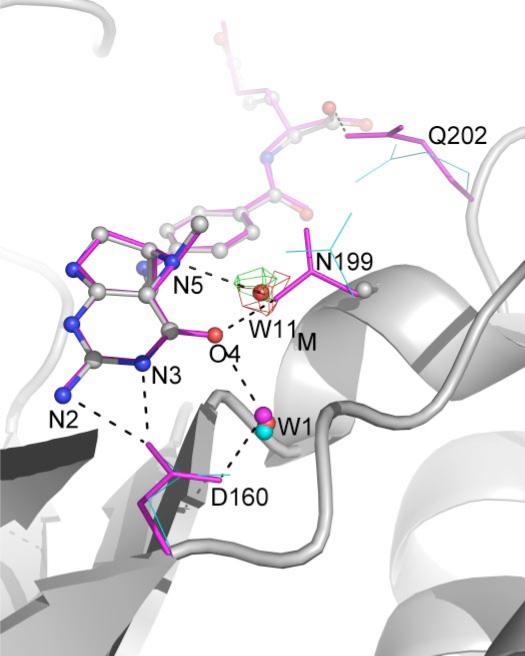
Position of Asn-199 in the Apo versus Folate-bound Structures
In the wild-type apo-MeTr structure (1F6Y), the side-chain conformation of Asn-199 is quite different from that observed when CH3-H4folate is present. Although the backbone α carbon of Asn-199 moves only 0.25 Å, the amide nitrogen of the side chain of Asn-199 swings 4.0 Å (Cγ moves 2.3 Å and OD1 by 2.5 Å) toward the N5 and O4 positions of CH3-H4folate and “locks” the substrate into place. Fig. 6 illustrates the change in position of the Asn-199 side chain as well as the similar movement of the Gln-202 side chain. In the “swung-in” position, Asn-199 can hydrogen-bond to both N5 (as mentioned previously) and O4 of the folate. An analogous Asn is found in the structure of the folate-binding domains of MetH (Fig. 1). For both MetH and MeTr, the bridging Asn is located almost equidistant between O4 and N5 (~2.8–2.9 Å). Although the resolution of the structures prohibits identification of oxygen from nitrogen, the bridging atom of Asn has been assigned in both enzyme structures as NH2 based on the close distance to O4, a distance that would be repulsive for two non-protonated oxygen atoms. This orientation of the Asn side chain suggests that N5 of folate is not protonated since folate protonated at N5 would be expected to contact the Asn residue in a similar manner to that observed in PNP (with oxygen of the carboxamide serving as the H-bond acceptor from substrate N5-H and the amide of the carboxamide serving as H-bond donor to substrate O4). Again, resolution prohibits analysis of the protonation state of the bound folate; however, biochemical studies of MetH suggest that N5 of folate is only protonated in the ternary complex with cobalamin, so N5 should not be protonated in the MetH structure (7). In the unprotonated form, both N5 and O4 of folate are H-bond acceptors, and a bridging Asn-NH2 could serve as an H-bond donor to both atoms. In contrast, biochemical studies indicate that MeTr does not require a ternary complex with cobalamin for N5 protonation. CH3-H4folate binds in an unprotonated state, but rapid protonation then occurs (5). Given the nature and similarities of the H-bonding to N5 of folate in the MeTr and MetH structures (Fig. 1), it is likely both folates are in the same protonation state (i.e. N5 is unprotonated). The simplest explanation is that the rapid protonation observed in solution (5) does not occur under our crystallization conditions.
FIGURE 6. Comparison of folate-binding sites in wild-type and N199A MeTr.
This view includes H-bonding interactions (dashed lines) between folate and Asp-160, conserved water W1 (spheres), and Gln-202. Wild-type apo-MeTr is in cyan (thin lines); CH3-H4folate/N199A is in gray (ball and stick), and wild-type CH3-H4folate/MeTr is in magenta (thin sticks).
Comparison of Wild-type and N199A Structures
The structure of the N199A variant shows the same level of hydrogen bonding to the folate and very similar folate conformations (r.m.s.d. values of 0.14 Å). Interestingly, a water molecule (Fig. 6, W11M) is recruited to bind to N5 and O4, replacing the interaction of the side-chain amide from Asn-199 in the wild-type protein. The displacement of the water from that position occupied by the side-chain amide of Asn-199 in the wild-type protein is 0.84 and 0.67 Å for subunits A and B. The distances between this water molecule and N5 of folate are 2.75 and 2.67 Å in the two subunits, indicating that W11M should be a good binding partner of N5. However, the distances to O4 of the folate are slightly longer: 3.36 and 3.09 Å. Thus, the almost perfect bridge between folate N5 and O4 atoms created by Asn-199 is disrupted in the mutant protein. At a resolution of 2.2–2.3 Å, it is difficult to know how significant such distance changes are; however, it is interesting that the Asn to Ala mutation shows such a dramatic change in kcat when the H-bonding network is so well conserved between mutant and the wild-type proteins.
DISCUSSION
Cobalamin-dependent methyltransferases play an important role in redox homeostasis and amino acid metabolism in many organisms (including humans) as well as in one-carbon metabolism and CO2 fixation in anaerobic microbes (3). Substrates that can be used by the methyltransferases include methanol, methylated amines, methylated thiols, methoxylated aromatics, and CH3-H4folate along with its methanogenic analog, methyltetrahydromethano-pterin. In all these reactions, a methyltransferase analogous to MeTr binds the methyl donor and transfers the methyl group to a cobalamin-containing protein. Subsequently, the methyl group is transferred from protein-bound methylcobalamin to an acceptor, which is acetyl-CoA synthase in organisms that perform the Wood-Ljungdahl pathway, coenzyme M in the process of methanogenesis, or homocysteine in methionine synthase. An important question in all these systems is how the methyl group of the methyl donor undergoes activation. In the transfer of the methyl group from methanol catalyzed by methanol:coenzyme M methyltransferase, it appears that a zinc ion acts as a Lewis acid to donate positive charge on oxygen (32), whereas in the methyl transfer from CH3-H4folate, protonation of the N5 group on the pterin appears to be the key step in activation. Generation of a positive charge at N5 would activate the methyl group, enhancing its reactivity toward nucleophilic attack by cob(I)alamin. Here, we have used kinetics, site-directed mutagenesis, and crystallography to provide insight into the mechanism of activation of the N5-methyl group.
Comparison of the structures of MeTr and MetH show significant similarity in the binding interactions of CH3-H4folate (Fig. 1), suggesting that the mechanisms of CH3-H4folate activation will be the same for these two methyltransferases. Neither structure shows an obvious proton transfer pathway to N5. In both structures, the amino acid closest to N5 is an Asn residue, a residue that is involved in an elaborate H-bonding network (Fig. 5, B–D). Mutation of this Asn residue to Ala affects kcat much more than Km, consistent with a minor role (2 kcal/mol) of Asn-199 in substrate binding and a key role (6.3–6.9 kcal/mol) in transition state stabilization. Taken together, the structural and kinetic data presented here, along with previous studies, suggest a mechanism for CH3-H4folate activation by MeTr outlined below.
The first steps in the MeTr mechanism involve a pH-dependent conformational change followed by the binding of unprotonated CH3-H4folate. Although in solution, binding of folate to MeTr is followed by rapid protonation (5), we believe that the structure presented here is of the unprotonated form. As discussed above, crystallography at this resolution cannot be used to determine protonation states; however, the similarity of the H-bonding pattern near folate N5 of the unprotonated CH3-H4folate bound to MetH suggests that the folate bound to MeTr is also unprotonated. When folate binds to MeTr, we observe a change in the position of Asn-199. It swings into the active site and helps lock the folate into its binding pocket. The NH2 of the Asn side chain is in position to serve as an H-bond donor to both N5 and O4 of folate (Fig. 7). Other important contacts to folate include Asp-160 and a series of conserved water molecules.
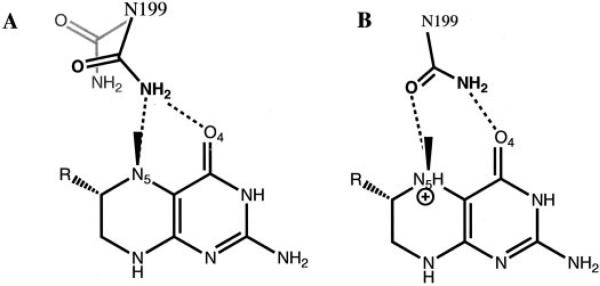
FIGURE 7. Schematic of two observed and one hypothetical conformation of Asn-199.
A, observed change in the position of Asn-199 upon binding of unprotonated CH3-H4folate. The swung-out position of Asn-199, found in the apo structure, is shown in gray. The bridging donor conformation of Asn-199 is shown in black. B, hypothetical PNP-like conformation of Asn-199 when protonated CH3-H4folate is bound to MeTr.
The next step in the mechanism is proton transfer to N5. Here, structural comparisons of MeTr with MetH, DHFR, and PNP suggest the players in the pathway. Based on mutagenesis and computational studies of DHFR, the corresponding Asp-27 to Asp-160 and the corresponding water to W1 are proposed to be the source for protonation of N5 via the nearby O4 group of the pterin (33). Mutating this residue to a neutral Asn or Ser in DHFR greatly reduces activity toward dihydrofolate at physiological pH, but the activity could be rescued at very acidic pHs (34). Although the details of the proton transfer pathway have been disputed in the literature (11, 35–37), the involvement of Asp-27 and the conserved water molecules is not in question. Kinetic data indicate that the analogous Asp-473 residue in MetH from E. coli plays a crucial role in catalysis (38). Likewise for PNP, structural and biochemical studies of substrate analogs suggest that Glu-207, the corresponding residue to Asp-160 (MeTr numbering), as well as corresponding water molecules (Fig. 1), are important for proton transfer to substrate in that system (12, 14). Although it has not been suggested for PNP that Asn-243 is a direct part of the proton transfer pathway, NMR data suggest that mutation of Asn-243 affects the extended H-bonding network and may indirectly affect proton transfer.
Following protonation of N5, stabilization of the protonated substrate would be expected to be important in catalysis since a positive charge at N5 would enhance the reactivity of the methyl group toward nucleophilic attack by cob(I)alamin. The fact that mutagenesis of Asn-199 affects kcat so much more than Km suggests that the rate-determining step in the reaction has been impaired. Since the rate-determining step is the methyl transfer and not protonation, Asn-199 is implicated in the stabilization of a transition state or a high energy intermediate in methyl transfer. The most obvious source for such stabilization is through a direct H-bond from the Asn side chain to N5-H. It is likely that our structure does not display this exact interaction. We speculate that the conformation of Asn that exists in the transition state resembles that of the Asn-243 in PNP with transition state analog ImmH bound (Fig. 1). In this conformation, the carboxamide oxygen is the H-bond acceptor from N7-H, and the carboxamide nitrogen is the H-bond donor to O6. A similar orientation of Asn-199 in MeTr would serve the same purpose; the carboxamide oxygen would accept the H-bond from N5-H, and the carboxamide nitrogen would donate an H-bond to O4 (Fig. 7). This dual H-bonding function could explain the rationale behind the placement of Asn at this structural position in the methyltransferase enzymes. Initially, we expected a residue in this position to be capable of serving as a source of protons (an acid to protonate N5), and the presence of Asn was challenging to explain. Given its pKa, Asn is not a candidate to serve as a catalytic acid. If, however, as we now believe, the role of this residue is to serve as an H-bond acceptor of the N5 proton and the H-bond donor to O4, then the choice of Asn can be rationalized. Asn is one of two residues (Gln being the other) with a branched side chain that has, at all pHs, an H-bond acceptor on one end and donor on the other. Although backbone carbonyls and amide nitrogens could serve this function (acceptor from N5-H and donor to O4), the backbone is less flexible, and the mobility of the Asn side chain seems to be important. In this mechanistic proposal, Asn must have three conformations: “swung-out” to allow folate binding, “bridging donor” to donated H-bonds to both N5 (unprotonated) and O4, and finally, “PNP-like” where both atoms of the carboxamide interact with folate N5-H and O4 (Fig. 7). The flexibility of the Asn side chain at this position in the structure would allow for these three states to be realized.
Along these lines, it is interesting to consider why a water molecule could not serve as the stabilizing H-bond acceptor and donor to folate. In the structure of N199A in complex with CH3-H4folate, a water molecule fills in the site emptied by the N199A mutation. This water molecule is in position to H-bond to both N5 and O4 of folate, yet catalytic efficiency is down 20,000-fold (Fig. 6). There are several possible explanations for why a water molecule is less ideal than Asn for the functions required. First, there is an entropic cost associated with water binding to MeTr. Second, although water molecule W11M appears to be a reasonable mimic of the carboxamide NH2 in the bridging donor position of Asn-199, it may not be able to successfully rearrange once folate is protonated to serve as an acceptor to N5-H and donor to O4. In fact, the ability of water to serve as both donor and acceptor of H-bonds may be a negative influence here. A water molecule locked into the wrong orientation might even hinder protonation of N5 through H-bond donation. The carboxamide oxygen of Asn, in contrast, is only able to accept H-bonds.
Finely tuned H-bonding networks appear to be the common theme for MeTr, MetH, PNP, and DHFR. Changes in these networks are tolerated (turnover is observed in the case of several of the mutant proteins), but the fine-tuning is damaged (14, 34, 38, 39). Instead of placing a residue capable of being a catalytic acid directly above the final site of protonation, these enzymes appear to employ an elaborate H-bonding network for proton transfer. Also, with the exception of DHFR, which does not need to stabilize a protonated intermediate in the transition state, they use a residue not typically thought of as catalytic, Asn, for that stabilization.
Acknowledgments
Molecular graphics images were produced using PyMOL. Use of the NSLS, Brookhaven National Laboratory, was supported by National Institutes of Health Grants GM39451 (to S. W. R.) and GM69857 (to C. L. D) and Center Grant 1P20RR17675 to the University of Nebraska Redox Biology Center and by a grant from the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-98CH10886. We thank Joe Spicha for preliminary kinetic work on the N199A variant and Dr. Javier Seravalli for helpful discussions.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
This article was selected as a Paper of the Week.
This paper is dedicated to Professor Martha Ludwig, who passed away on November 27, 2006. Martha was a thoughtful, thorough, and innovative scientist who made extremely important contributions to our understanding of the structure and function of cobalamin-dependent methyltransferases. Martha was a crystallographer who was a faculty member in the Biological Chemistry Department at the University of Michigan for almost 40 years and a member of the National Academy of Sciences and of the Institute of Medicine. She also was a caring friend who we wish could have seen this paper in print.
The atomic coordinates and structure factors (codes 2E7F and 2OGY) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The abbreviations used are: CH3-H4folate, methyltetrafolate; MetH, cobalamin-dependent methionine synthase; MeTr, CH3-H4folate:corrinoid iron-sulfur protein methyltransferase; MtrH, methyltetrahydromethanopterin: cob(I)alamin methyltransferase; DHFR, dihydrofolate reductase; PNP, purine nucleoside phosphorylase; DHPS, dihydropteroate synthases; ImmH, immucillin-H; PDB, Protein Data Bank; CFeSP, corrinoid-iron-sulfur protein; DTT, dithiothreitol; MES, 4-morpholineethanesulfonic acid; ITC, isothermal titration calorimetry; r.m.s.d., root mean square deviation.
REFERENCES
- 1.Ragsdale SW. In: Encyclopedia of Catalysis. Horvath IT, Iglesia E, Klein MT, Lercher JA, Russell AJ, Stiefel EI, editors. John Wiley and Sons, Inc.; New York: 2003. pp. 452–467. [Google Scholar]
- 2.Ragsdale SW. CRC Crit. Rev. Biochem. Mol. Biol. 2004;39:165–195. doi: 10.1080/10409230490496577. [DOI] [PubMed] [Google Scholar]
- 3.Banerjee R, Ragsdale SW. Ann. Rev. Biochem. 2003;72:209–247. doi: 10.1146/annurev.biochem.72.121801.161828. [DOI] [PubMed] [Google Scholar]
- 4.Matthews RG. Acc. Chem. Res. 2001;34:681–689. doi: 10.1021/ar0000051. [DOI] [PubMed] [Google Scholar]
- 5.Seravalli J, Zhao S, Ragsdale SW. Biochemistry. 1999;38:5728–5735. doi: 10.1021/bi982473c. [DOI] [PubMed] [Google Scholar]
- 6.Seravalli J, Shoemaker RK, Sudbeck MJ, Ragsdale SW. Biochemistry. 1999;38:5736–5745. doi: 10.1021/bi9824745. [DOI] [PubMed] [Google Scholar]
- 7.Smith AE, Matthews RG. Biochemistry. 2000;39:13880–13890. doi: 10.1021/bi001431x. [DOI] [PubMed] [Google Scholar]
- 8.Taurog RE, Matthews RG. Biochemistry. 2006;45:5092–5102. doi: 10.1021/bi060052m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doukov T, Seravalli J, Stezowski J, Ragsdale SW. Structure (Camb.) 2000;8:817–830. doi: 10.1016/s0969-2126(00)00172-6. [DOI] [PubMed] [Google Scholar]
- 10.Evans JC, Huddler DP, Hilgers MT, Romanchuk G, Matthews RG, Ludwig ML. Proc. Natl. Acad. Sci. U. S. A. 2004;101:3729–3736. doi: 10.1073/pnas.0308082100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rod TH, Brooks CL., III J. Am. Chem. Soc. 2003;125:8718–8719. doi: 10.1021/ja035272r. [DOI] [PubMed] [Google Scholar]
- 12.Fedorov A, Shi W, Kicska G, Fedorov E, Tyler PC, Furneaux RH, Hanson JC, Gainsford GJ, Larese JZ, Schramm VL, Almo SC. Biochemistry. 2001;40:853–860. doi: 10.1021/bi002499f. [DOI] [PubMed] [Google Scholar]
- 13.Lewandowicz A, Ringia EA, Ting LM, Kim K, Tyler PC, Evans GB, Zubkova OV, Mee S, Painter GF, Lenz DH, Furneaux RH, Schramm VL. J. Biol. Chem. 2005;280:30320–30328. doi: 10.1074/jbc.M505033200. [DOI] [PubMed] [Google Scholar]
- 14.Kicska GA, Tyler PC, Evans GB, Furneaux RH, Shi W, Fedorov A, Lewandowicz A, Cahill SM, Almo SC, Schramm VL. Biochemistry. 2002;41:14489–14498. doi: 10.1021/bi026636f. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: a Laboratory Manual. 2nd Edition Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 16.Elliott JI, Brewer JM. Arch. Biochem. Biophys. 1978;190:351–357. doi: 10.1016/0003-9861(78)90285-0. [DOI] [PubMed] [Google Scholar]
- 17.Zhao SY, Ragsdale SW. Biochemistry. 1996;35:2476–2481. doi: 10.1021/bi952310d. [DOI] [PubMed] [Google Scholar]
- 18.Zhao S, Roberts DL, Ragsdale SW. Biochemistry. 1995;34:15075–15083. doi: 10.1021/bi00046a013. [DOI] [PubMed] [Google Scholar]
- 19.Leslie AG. Acta Crystallogr. Sect. D Biol. Crystallogr. 2006;62:48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- 20.Evans PR. Protein Crystallogr. 1997;33:22–24. [Google Scholar]
- 21.French GS, Wilson KS. Acta Crystallogr. Sect. A. 1978;34:517–534. [Google Scholar]
- 22.Collaborative Computational Project Number 4 Acta Crystallogr. Sect. D Biol. Crystallogr. 1994;50:760–763. [Google Scholar]
- 23.Kissinger CR, Gehlhaar DK, Fogel DB. Acta Crystallogr. Sect. D Biol. Crystallogr. 1999;55:484–491. doi: 10.1107/s0907444998012517. [DOI] [PubMed] [Google Scholar]
- 24.McRee DE. J. Struct. Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 25.Emsley P, Cowtan K. Acta Crystallogr. Sect. D Biol. Crystal-logr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 26.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr. Sect. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 27.Winn MD, Isupov MN, Murshudov GN. Acta Crystal-logr. Sect. D Biol. Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 28.Brunger AT. J. Mol. Biol. 1988;203:803–816. doi: 10.1016/0022-2836(88)90211-2. [DOI] [PubMed] [Google Scholar]
- 29.Hippler B, Thauer RK. FEBS Lett. 1999;449:165–168. doi: 10.1016/s0014-5793(99)00429-9. [DOI] [PubMed] [Google Scholar]
- 30.Achari A, Somers DO, Champness JN, Bryant PK, Rosemond J, Stammers DK. Nat. Struct. Biol. 1997;4:490–497. doi: 10.1038/nsb0697-490. [DOI] [PubMed] [Google Scholar]
- 31.Babaoglu K, Qi J, Lee RE, White SW. Structure (Camb.) 2004;12:1705–1717. doi: 10.1016/j.str.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 32.Sauer K, Thauer RK. Eur. J. Biochem. 1997;249:280–285. doi: 10.1111/j.1432-1033.1997.t01-1-00280.x. [DOI] [PubMed] [Google Scholar]
- 33.Cummins PL, Gready JE. J. Am. Chem. Soc. 2001;123:3418–3428. doi: 10.1021/ja0038474. [DOI] [PubMed] [Google Scholar]
- 34.Howell EE, Villafranca JE, Warren MS, Oatley SJ, Kraut J. Science. 1986;231:1123–1128. doi: 10.1126/science.3511529. [DOI] [PubMed] [Google Scholar]
- 35.Casarotto MG, Basran J, Badii R, Sze KH, Roberts GC. Biochemistry. 1999;38:8038–8044. doi: 10.1021/bi990301p. [DOI] [PubMed] [Google Scholar]
- 36.Cannon WR, Garrison BJ, Benkovic SJ. J. Mol. Biol. 1997;271:656–668. doi: 10.1006/jmbi.1997.1173. [DOI] [PubMed] [Google Scholar]
- 37.Ferrer S, Silla E, Martı S, Moliner V. J. Phys. Chem. B. 2003;107:14036–14041. [Google Scholar]
- 38.Smith AE. Ph.D. thesis. University of Michigan; Ann Arbor, MI: 2002. The Folate Active Site of Methionine Synthase: Binding and Activation of Methyltetrahydrofolate. [Google Scholar]
- 39.Stoeckler JD, Poirot AF, Smith RM, Parks RE, Jr., Ealick SE, Takabayashi K, Erion MD. Biochemistry. 1997;36:11749–11756. doi: 10.1021/bi961971n. [DOI] [PubMed] [Google Scholar]