Abstract
Large amounts of manure have been applied to arable soils as fertilizer worldwide. Manure is often contaminated with veterinary antibiotics which enter the soil together with antibiotic resistant bacteria. However, little information is available regarding the main responders of bacterial communities in soil affected by repeated inputs of antibiotics via manure. In this study, a microcosm experiment was performed with two concentrations of the antibiotic sulfadiazine (SDZ) which were applied together with manure at three different time points over a period of 133 days. Samples were taken 3 and 60 days after each manure application. The effects of SDZ on soil bacterial communities were explored by barcoded pyrosequencing of 16S rRNA gene fragments amplified from total community DNA. Samples with high concentration of SDZ were analyzed on day 193 only. Repeated inputs of SDZ, especially at a high concentration, caused pronounced changes in bacterial community compositions. By comparison with the initial soil, we could observe an increase of the disturbance and a decrease of the stability of soil bacterial communities as a result of SDZ manure application compared to the manure treatment without SDZ. The number of taxa significantly affected by the presence of SDZ increased with the times of manure application and was highest during the treatment with high SDZ-concentration. Numerous taxa, known to harbor also human pathogens, such as Devosia, Shinella, Stenotrophomonas, Clostridium, Peptostreptococcus, Leifsonia, Gemmatimonas, were enriched in the soil when SDZ was present while the abundance of bacteria which typically contribute to high soil quality belonging to the genera Pseudomonas and Lysobacter, Hydrogenophaga, and Adhaeribacter decreased in response to the repeated application of manure and SDZ.
Introduction
The use of animal manure for fertilization of agricultural soils has a long tradition in many parts of the world and is generally assumed to be ecologically more friendly and sustainable than mineral fertilizer. In particular organic and bio-dynamic farms depend on manure or compost as sources for fertilization. However, industrial husbandries with typically large numbers of animals sharing limited space depend on the prophylactic and therapeutic use of antibiotics. In addition, in several parts of the world antibiotics are still being used as growth promoters. Although antibiotics as growth promoters were banned in many European countries, considerable amounts of antibiotics such as tetracyclines, β-lactams and sulfonamides are still used in animal husbandries [1]. Depending on their physicochemical properties, many antibiotics such as sulfonamides are to a large extent excreted via urine or feces and are not or only to a low extent degraded during manure storage [2], [3]. Thus, spreading manure on agricultural soils does not only introduce nutrients required for maintaining the soil fertility but also antibiotics, their metabolites and antibiotic resistant bacteria. Indeed, antibiotics have been detected in the environment due to the use of manure for soil fertilization or direct deposition via dung and urine of animals grazing on pastures [4], [5].
The rapid sequestration of most antibiotics in soils, as e.g. observed for the sulfonamide antibiotic SDZ [6], leads usually to low concentrations of bioavailable SDZ, which are far below the minimal inhibitory concentrations after a single application of manure. Most data published indicated only short-term effects on the microbial community after a single application of manure spiked with antibiotics followed by a fast regeneration of the community structure [7], [8] and its function [9]–[11]. However, agricultural management implies in most cases a repeated application of manure mainly during the vegetation period to keep the level of nutrients needed for best possible plant growth. Surprisingly, little is known of the effects of repeated application of manure containing antibiotics on microbial communities in soil which might cause cumulative effects. Despite a rapid dissipation and sequestration of SDZ in soil [3], [6], [12] an increased abundance of sul1 and sul2 resistance genes and their transferability was observed in soils treated with manure containing SDZ compared to control manure under microcosm, mesocosm and field conditions [13]–[15], suggesting that manure containing antibiotics enhanced the spreading of the antibiotic resistance genes in soils. In particular the repeated application of manure in combination with antibiotics may set the ground for an increased abundance of resistant bacteria as recently reported by Heuer et al. [16] and thus might stimulate the spreading of antibiotic resistance genes and mobile genetic elements in agricultural ecosystems. Antibiotic resistance genes localized on mobile genetic elements can be captured by human and veterinary pathogens and thus pose a threat to the treatment of bacterial diseases [17]–[19].
In the present microcosm experiment, the effect of repeated application of manure containing antibiotics on the soil bacterial community was investigated by 454 sequencing of 16S rRNA gene fragments amplified from total community (TC)-DNA. We have chosen the sulfonamide SDZ as a model compound as it is still frequently used in pig husbandry [4] and highly persistent in manure [20]. Manure unspiked and spiked with SDZ in two concentrations was applied three times, and the bacterial community composition was monitored over a period of 193 days. We hypothesized that repeated application of manure spiked with SDZ to soil would increase the degree of disturbance and reduce the resilience of soil bacterial communities compared to a single application of manure spiked with SDZ or soils treated with manure not containing antibiotics. Resilience was measured as the stability of bacterial diversity after changes in soil properties ([21]; here manure addition). To differentiate between short and long-term effects, the soil was sampled 3 and 60 days after each manure application, respectively.
Materials and Methods
Experimental design
The experimental design is described in detail by Heuer et al. [16]. In short, topsoil samples (Ap horizon) of a silt loam soil (Orthic Luvisol) with no history of manure application were used. Pots were filled with 100 g of 2 mm-sieved soils and treated with 4 g of manure from healthy pigs (approximately 30 m3/ha) spiked with SDZ or not to achieve a SDZ concentration in soil of 10 mg/kg, 100 mg/kg, or no SDZ (Sigma-Aldrich, Germany), which corresponded to samples S10, S100, and S0, respectively. Soil samples without manure application served as controls (U). To avoid effects based on different manure composition at the different time points of application, manure was taken before the experiment and frozen in aliquots at −20°C. For each time point of application, an aliquot of the manure was thawn and the corresponding amount of SDZ was directly applied before mixing the manure into the soil. The SDZ concentration S10 was chosen in accordance to maximal inputs of sulfonamides detected in agricultural soils [3], [22]. Manure loads were applied on days 0, 63 and 133. The loosely covered pots were incubated at 15°C in the dark. Water was added to the microcosms twice a week to compensate for weight losses and to maintain a soil moisture of about 55% of the soil's water-holding capacity. For each treatment and time point 5 replicates were prepared and treated individually for the subsequent analysis. Samples were taken every 3rd and 60th day after manure application (days 3, 60, 66, 123, 136 and 193) and kept at −80°C until further analysis. No specific permissions were required for collecting the manure and soil and the study did not involve endangered or protected species.
DNA extraction
Extraction of total community DNA from soil samples was performed according to Griffith et al. [23]. Briefly, 0.4 g soil per replicate were added to a lysing matrix tube (MP Biomedicals, Germany) and submitted to phenol∶chloroform extraction starting with homogenization for 30 seconds at 5.5 m S-1 (Precellys, PeqLab, Germany). Extracted DNA was finally resolved in 50 µl DNase-free water. Quality and quantity of DNA extracts were checked using a spectrophotometer (Nanodrop, PeqLab, Germany). For the extraction of total community DNA from manure, 10 ml manure were centrifuged at about 6500 g for 10 min, the pellet was homogenized and DNA was extracted from 0.5 g manure pellet using the FastDNA SPIN kit for soil (MP Biomedicals, Heidelberg, Germany), followed by a purification step using the Geneclean spin kit (MP Biomedicals, Heidelberg, Germany), according to the manufacturer's instructions.
Preparation of the 16S rRNA gene amplicons, pyrosequencing and data processing
To generate the amplicon library for Bacteria, 16S primers 926-F (5′-AAACTYAAAKGAATTGACGG-3′), Escherichia coli position 907–926 and 630-R (5′-CAKAAAGGAGGTGATCC-3′), E. coli position 1528–1544 [24], were selected. Fusion primers were designed according to the guidelines of ROCHE (www.my454.com), by extending the specific primers with a 10 base multiplex identifier (MID), a 4 base key and the respective sequencing primers A or B for bidirectional sequencing. The optimal PCR conditions were determined by gradient PCRs with annealing temperatures ranging from 50°C to 60°C. Amplicons were finally generated using 50 ng of extracted DNA, an annealing temperature of 50°C and 22 PCR cycles. The PCR products were purified with AMPure Beads (Agencourt, Beckman Coulter, Krefeld, Germany) according to the Amplicon Library Preparation Method Manual (www.my454.com) and pooled in equimolar amounts.
Sequencing of the 16S rRNA genes was performed on a second-generation pyrosequencer (454 GS FLX Titanium, ROCHE, Germany) following the manufacturer's protocol for amplicon sequencing (www.my454.com). The number of replicates per treatment included in the final sequencing run can be found in Table S1 in File S1.
The automatic amplicon pipeline of the GS Run Processor (ROCHE) was used to perform an initial quality filtering of the pyrosequencing raw reads in order to remove failed reads, low quality reads and adaptor sequences. Sequence files were submitted to the NCBI Sequence Read Archive (www.ncbi.nlm.nih.gov/sra/) and are available with the study accession number SRP038712.
Sequence analysis
The two data sets acquired by the forward and reverse primers were analyzed separately to avoid the systematical bias of individual primers. The analyses were mainly performed according to Ding et al. [25]. Only those sequences with a length above 200 bp after removing the barcode, primer and unpaired regions were subjected to further analysis. The unpaired regions were truncated based on a standalone BlastN analysis against a bacterial database described by Pruesse et al. [26]. Sequences were grouped into operational taxonomic units (OTUs>97% sequence identity) using software package mothur (v1.14.0). The classification of sequences was performed using software RDP MultiClassifier at >80% confidence [27]. A taxonomic OTU report with each row representing one OTU containing taxonomic positions (from phylum to genus) and the number of sequences for each sample was constructed based on the OTU assignment and on the classification.
Analyses based on the taxonomic OTU report were done with R (version 2.14). To allow the comparison of samples with different number of reads, all algorithms selected rely on relative abundance of taxonomic groups. To check whether the forward and reverse primers revealed a similar taxonomic composition or not, the percent of Bray-Curtis similarity of taxonomic composition between two data sets was calculated for each sample based on the percent of classified taxa at phylum, class, order, family, and genus level using the R add-on package “vegan”. To evaluate the influence of repeated application of manure containing SDZ on the bacterial community compositions, the pairwise Pearson dissimilarities between S0 and S10 or between S0 and S100 for day 193 were calculated. To compare the microbial community composition between samples, non-metric multidimensional scaling (NMDS) analysis was carried out based on the Pearson dissimilarity matrix using R package MASS. The 3-dimensional plots of NMDS analysis were created with the R package “rgl” in conjunction with the Linux command “import”. Tukey's honest significance tests under a generalized linear model via a logistic function for binomial data with the package multcomp [28] was applied to identify the discriminative taxa between treatments using a Bonferroni adjusted p value<0.05.
Results
Effects of the repeated application of manure containing SDZ on the composition of soil bacterial communities
In this study, bacterial 16S rRNA gene fragments amplified from TC-DNA extracted from 50 composite soil samples were sequenced using forward (926F) and reverse (630R) primers. A total of 132,278 sequences for 50 samples with 790–5685 high quality sequences per sample were obtained with the forward primer, and 162,446 sequences for 49 samples with 1105–7053 high quality sequences per sample with the reverse primer.
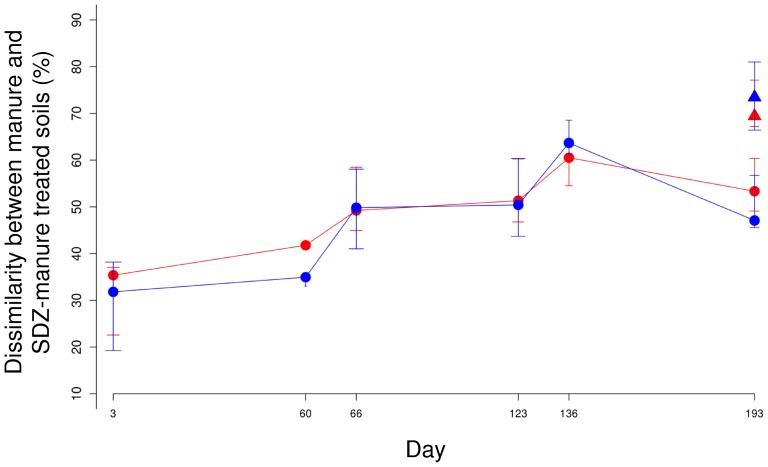
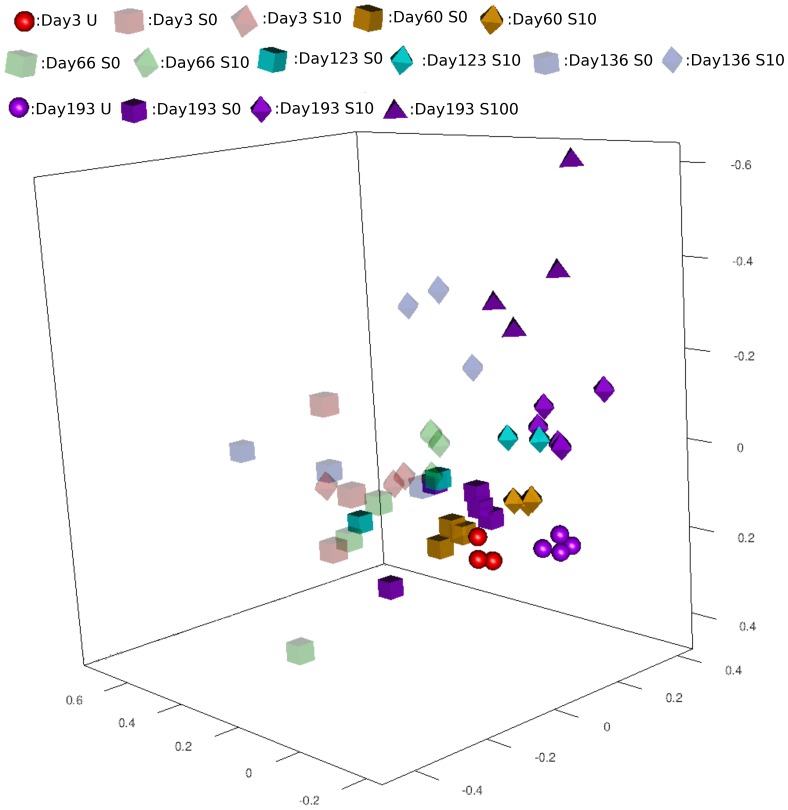
To study the effect of repeated applications of manure containing SDZ on the soil bacterial community composition, 16S rRNA gene pyrosequencing data sets acquired from TC-DNA of soils treated with SDZ manure or control manure were compared. Sequences obtained with the forward and reverse primers were treated independently and grouped into OTUs (>97% sequence identity). Both datasets revealed similar changes of bacterial community composition after manure application (Figure 1). In general, the Pearson dissimilarities of the bacterial community compositions between the S0 and S10 soils increased with repeated manure application (Figure 1). Shortly after each manure application the dissimilarity between the bacterial community composition of S0 and S10 soils was strongly increased. Sixty days after the first and second manure applications the dissimilarity remained at the same level as shortly after manure application, while 60 days after the third application a decrease in dissimilarity between S0 and S10 soil was observed. Compared with the S10 soils on day 193, the S100 soils were less similar to S0 soils in the bacterial community composition, indicating that a higher concentration of SDZ caused more pronounced changes of the soil bacterial communities (Figure 1). Non-metric multidimensional scaling based on the OTU reports acquired for the forward (Figure S1 in File S1) and reverse (Figure 2) data set confirmed the findings presented in Figure 1 as it showed that the bacterial community composition of S10 soils increasingly deviated from the S0 soils with time, and this difference was even more pronounced for S100 soils. Furthermore, the faint color symbols in Figure 2 representing the bacterial community composition shortly after manure application strongly deviated from the community composition of untreated soil suggesting a pronounced disturbance of the system. In contrast to S10 and S100 soils, the bacterial community composition of S0 soils (indicated by cubes) 60 days after the third manure application still grouped in the vicinity of the untreated soils (Figures 2 and S1) suggesting a stronger resilience of the soil bacterial community towards manure when no antibiotic was present.
Figure 1. Dissimilarity between soils treated with manure (S0) and manure spiked with SDZ (S10 indicated by circles; S100 indicated by triangles) at different sampling times.
Red symbols: results based on the data set acquired by the forward primer; blue symbols: results based on the data set acquired by the reverse primer. Error bars indicate the first and third quartiles.
Figure 2. Non metric multidimensional scaling based on OTU reports acquired by the reverse primer sequencing.
Balls: untreated soils; cubes: S0; octahedron: S10; pyramid: S100.
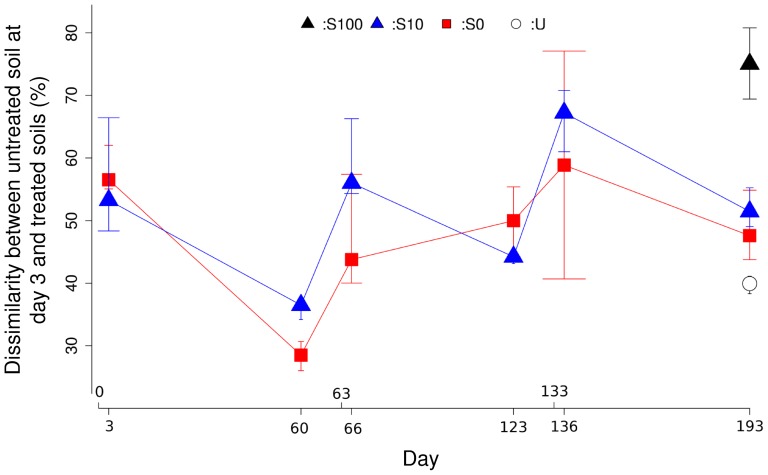
The difference between bacterial communities of the initial soil (day 3 untreated) and of the treated soil was calculated based on Pearson dissimilarity (Figure 3). A pronounced increase in difference was observed for soils shortly after each manure application (S0, S10). The dissimilarity increased with the repeated applications of manure containing SDZ for samples collected 60 days after each manure application. The highest deviation was observed for soil treated three times with manure containing the high concentration (100 mg/kg) of SDZ. At the same sampling time, the least difference was found for the untreated control soils.
Figure 3. Dissimilarity between soils treated with manure (S0, S10 and S100) or not (U) and the untreated initial soils (Day3 U) at different sampling times.
Dissimilarity was calculated from the data acquired by the reverse primer. Error bars indicate the first and third quartiles.
Taxa with significantly altered relative abundance in response to repeated application of manure spiked with SDZ or unspiked
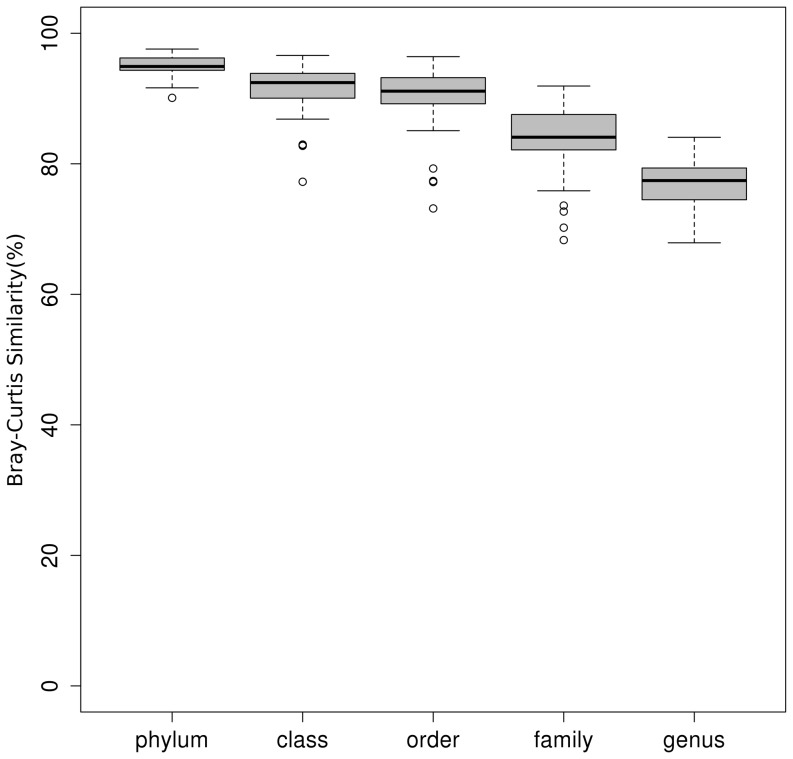
About 86% of the sequences obtained with the forward primer could be assigned to 21 phyla while 90% of the reverse primer derived sequences were affiliated to 19 phyla. The percent Bray-Curtis similarities of the taxonomic composition between the data sets acquired with the forward and reverse primers, respectively, were calculated for each sample based on the percent of the classified taxa at different taxonomic ranks (from phylum to genus level). Although from the phylum to the genus level the similarity of the taxonomic composition decreased as shown in Figure 4, the overall taxonomic composition of both data sets was very similar.
Figure 4. Box plot showing the similarity of taxonomic composition at different ranks between data acquired by the forward and reverse primers.
To identify the taxa which significantly (Bonferroni adjusted p value<0.05) responded to the repeated application of manure, comparisons were made between soils treated with S0 or not (U). All taxa with significantly different relative abundance (Bonferroni adjusted p value<0.05) were considered. In soil samples taken three days after the addition of manure short-term responders were identified (summarized in Table S2 for the reverse primer derived sequence set and in Table S3 in File S1 for the sequence set obtained with the forward primer). Based on these data (Table S2 and S3) we could mainly identify genera belonging to the Proteobacteria such as Delftia (Betaproteobacteria), Pseudomonas, Pseudoxanthomonas, Stenotrophomonas (Gammaproteobacteria), Bacillus, Lactobacillus, Streptocococcus, Clostridiales (Firmicutes) and Arthrobacter (Actinobacteria) which were significantly increased in relative abundance in S0 soils compared to the untreated soil as short time responders to the manure application. The increase in the relative abundance of Firmicutes (Table S4 in File S1) is likely related to the high abundance of these organisms in the manure (data not shown). In turn the relative abundance of different genera of the Acidobacteria and Planctomycetaceae but also Gemmatimonadetes, the genus Methylibium (Betaproteobacteria) decreased in S0 soils compared to the untreated soils (Table S2 and S3 in File S1). An increased relative abundance of the genera Devosia, Clostridium, Peptostreptococcus and Adheribacter in response to the manure application was observed by comparing U and S0 soil on day 193 (Table S5 and S6 in File S1). In contrast, Myxococcales seemed to be negatively affected by manure. On day 193 the relative abundance of Acidobacteria was still significantly decreased in all manure treatments compared to untreated soil with the exception of GP10 which showed a higher relative abundance in S10. Some taxa, such as the family Planctomycetaceae, were not affected by manure application.
With repeated application of manure the number of taxa with significantly altered relative abundance in S10 compared to S0 soil increased (summarized in Table 1 for the reverse primer and in Table S7 in File S1 for the forward primer). An increased abundance in S10 soils was observed in particular after repeated manure application for the genera Devosia, Sphingobium, Gp10 and Gemmatimonas. Shortly after manure application a significantly increased abundance was observed in S10 compared to the S0 soils for the phylum Chloroflexi, the class of the Deltaproteobacteria and the orders of Clostridales and Actinomycetales. Acidobacterial genera Gp4, Gp6 and Gp10 with one exception showed always higher relative abundances in S10 but only in some cases these differences were found to be significant (Table 1). The relative abundances of Pseudomonas, Lysobacter and Adheribacter were lower in all S10 compared to S0 soils indicating that the presence of SDZ exerted a negative effect on these taxa.
Table 1. Taxa with significantly different relative abundance between manured soils with or without SDZ treatments shortly after manure addition as revealed by barcoded pyrosequencing using the reverse primer (Bonferroni adjusted p value<0.05).
| Phylum | Class | Order | Family | Genus | Day 3 | Day 66 | Day136 | |||
| S0 | S10 | S0 | S10 | S0 | S10 | |||||
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Hyphomicrobiaceae | Devosia | 0.2±0 | 0.3±0 | 0.5±0 | 1.5±0 | 1.1±0 | 2.4±0 |
| Sphingomonadales | Sphingomonadaceae | Sphingobium | 0.1±0 | 0.1±0 | 0.1±0 | 0.3±0 | 0.1±0 | 0.4±0 | ||
| Betaproteobacteria | Burkholderiales | Comamonadaceae | 3.8±3 | 1.6±0 | 3.3±1 | 2.1±0 | 2.6±1 | 3±1 | ||
| Oxalobacteraceae | Herminiimonas | 0.1±0 | 0.1±0 | 0.5±0 | 0±0 | 3±4 | 0±0 | |||
| Deltaproteobacteria | 1.1±0 | 1.2±0 | 1.4±0 | 1.3±0 | 3.1±1 | 4.4±2 | ||||
| Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | Pseudomonas | 0.8±1 | 0.2±0 | 2.6±4 | 0.2±0 | 0.6±1 | 0±0 | |
| Xanthomonadales | Xanthomonadaceae | Lysobacter | 2.1±1 | 1.2±0 | 4.1±1 | 0.8±1 | 1.7±1 | 0.2±0 | ||
| Stenotrophomonas | 1.6±2 | 0.1±0 | 0±0 | 0±0 | 0.1±0 | 0.3±0 | ||||
| Acidobacteria | Acidobacteria_Gp10 | Gp10 | 1±0 | 1.5±0 | 1.7±0 | 3±1 | 1±0 | 2.1±0 | ||
| Acidobacteria_Gp4 | Gp4 | 3.3±0 | 4.7±1 | 4.8±1 | 5.3±0 | 4.2±2 | 4.1±2 | |||
| Acidobacteria_Gp6 | Gp6 | 8.7±1 | 10.5±2 | 12.1±2 | 16.1±1 | 8.7±4 | 9.4±2 | |||
| Firmicutes | Bacilli | Lactobacillales | 5.4±1 | 5.3±2 | 3.9±1 | 6.3±2 | 2.8±1 | 3.8±1 | ||
| Clostridia | Clostridiales | 5.9±2 | 6.4±1 | 6±1 | 8.2±1 | 7.5±1 | 10.5±1 | |||
| Actinobacteria | Actinobacteria | Actinomycetales | 7.1±1 | 5.8±2 | 2.9±1 | 3.3±1 | 4.7±5 | 5.9±6 | ||
| Bacteroidetes | Sphingobacteria | Sphingobacteriales | Cytophagaceae | Adhaeribacter | 1.5±0 | 0.7±0 | 2.3±1 | 0.6±0 | 1.5±0 | 0.4±0 |
| Chloroflexi | 3.3±1 | 3.6±0 | 1.9±1 | 3.8±1 | 4.5±0 | 6.2±1 | ||||
| Gemmatimonadetes | Gemmatimonadetes | Gemmatimonadales | Gemmatimonadaceae | Gemmatimonas | 0.2±0 | 0.3±0 | 0.3±0 | 0.5±0 | 0.4±0 | 1±1 |
Bold numbers indicate taxa with significantly higher relative abundance in SDZ treated or non treated soils. Soils were collected three days after each treatment.
Samples taken 60 days after the first, second and third manure applications mainly revealed the same taxa with increased or decreased relative abundance in S10 compared to S0 as in the samples taken shortly after manure application (shown in Table 2 for the reverse primer and in Table S8 in File S1 for the forward primer). With repeated application of manure (S0, S10) the number of taxa with significantly different relative abundance increased from 3 (60 days after the first manure application) to 11 (60 days after the third manure application). Although not significant at all time points the genera Devosia, Gemmatimonas and Acidobacteria Gp6 and Gp10 showed an increased relative abundance in S10 compared to S0 soils. In S100 soils analyzed only for the samples taken 60 days after the third manure application (day 193) the relative abundances of Devosia and Gemmatimonas were increased compared to their abundance in S10 and S0 soils clearly indicating that bacteria belonging to these genera take profit from high concentrations of SDZ. A higher relative abundance in S100 soils was also observed for Anerolineaceae (Chloroflexi), Microbacteriaceae and the genus Clostridium. Most striking was the selection of Stenotrophomonas in S100 soils as sequences of this genus were not detected in S0 and in very low relative abundance in S10 soils on day 193. Also acidobacterial genera were identified as responders to SDZ. However, the different genera of Acidobacteria responded differently to SDZ manure. The S10 soils had a higher abundance of Acidobacteria belonging to the genera Gp10 and Gp6 (except day 60) compared to S0. On day 193, the relative abundance of Gp4, Gp6 and Gp10 was lower in S100 than in S10 soil. The negative effect of SDZ on Pseudomonas, Lysobacter and Adheribacter was also detected in S100 but there was no significant difference in their relative abundance between S10 and S100.
Table 2. Taxa with significantly different relative abundance between manured soils with or without SDZ treatments as revealed by barcoded pyrosequencing using the reverse primer (Bonferroni adjusted p value<0.05).
| Phylum | Class | Order | Family | Genus | Day 60 | Day 123 | Day 193 | ||||
| S0 | S10 | S0 | S10 | S0 | S10 | S100 | |||||
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Hyphomicrobiaceae | Devosia | 0.4±0 | 1.5±0 | 1.8±1 | 2.2±0 | 1.5±0b | 2.6±1a | 3.5±1a |
| Rhizobiaceae | 0.4±0 | 0.3±0 | 0.3±0 | 0.5±0 | 0.6±0a | 0.4±0ab | 0.4±0b | ||||
| Rhodospirillales | Rhodospirillaceae | Magnetospirillum | 0.4±0 | 0.4±0 | 0.3±0 | 0.6±0 | 0.8±0ab | 0.9±0a | 0.5±0b | ||
| Sphingomonadales | Sphingomonadaceae | 4±1 | 3.4±1 | 4±1 | 3.4±1 | 5.4±1a | 3.6±0b | 3.4±1b | |||
| Betaproteobacteria | Burkholderiales | Alcaligenaceae | 0.1±0 | 0.1±0 | 0.1±0 | 0.1±0 | 0.1±0b | 0.1±0b | 0.8±0a | ||
| Burkholderiales_incertae_sedis | Methylibium | 1.8±0 | 1.3±0 | 1.2±0 | 1.3±0 | 2±1a | 1.8±1a | 0.9±0b | |||
| Comamonadaceae | Hydrogenophaga | 0.1±0 | 0.1±0 | 0.6±0 | 0.2±0 | 0.5±0a | 0.1±0b | 0±0b | |||
| Oxalobacteraceae | Herminiimonas | 0±0 | 0±0 | 1.2±0 | 0±0 | 0.5±0a | 0±0ab | 0±0b | |||
| Methylophilales | Methylophilaceae | 0.1±0 | 0.1±0 | 0.3±0 | 0.1±0 | 0.3±0a | 0.1±0ab | 0±0b | |||
| Rhodocyclales | Rhodocyclaceae | Shinella | 0±0 | 0±0 | 0±0 | 0.1±0 | 0±0b | 0.2±0b | 1.2±1a | ||
| Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | Pseudomonas | 0.5±1 | 0.1±0 | 0.1±0 | 0±0 | 1.2±2a | 0.2±0b | 0±0b | |
| Xanthomonadales | Xanthomonadaceae | Lysobacter | 1.6±0 | 0.7±0 | 1.7±1 | 0.6±0 | 0.9±0a | 0.2±0b | 0.1±0b | ||
| Pseudoxanthomonas | 0.7±1 | 0.4±0 | 1.9±1 | 0.4±0 | 0.7±0b | 0.4±0b | 2±2a | ||||
| Rhodanobacter | 0±0 | 0±0 | 0.3±0 | 0.3±0 | 0.3±0b | 0.9±1a | 0.2±0b | ||||
| Stenotrophomonas | 0±0 | 0±0 | 0.1±0 | 0.3±0 | 0±0b | 0.2±0b | 3.5±3a | ||||
| Deltaproteobacteria | Myxococcales | Haliangiaceae | Haliangium | 0.5±0 | 0.1±0 | 0.8±0 | 0.1±0 | 0.8±0a | 0.1±0b | 0.1±0b | |
| Nannocystaceae | 0.3±0 | 0±0 | 0.2±0 | 0±0 | 0.3±0a | 0.2±0ab | 0±0b | ||||
| Polyangiaceae | 0.2±0 | 0.3±0 | 0.2±0 | 0.3±0 | 0.3±0b | 0.2±0b | 0.8±1a | ||||
| Acidobacteria | Acidobacteria_Gp10 | Gp10 | 2.8±0 | 4.3±1 | 2±0 | 5.4±1 | 1.6±0c | 4.4±0a | 2.5±1b | ||
| Acidobacteria_Gp4 | Gp4 | 7.9±1 | 8.6±1 | 4.4±0 | 5±1 | 7.5±2a | 5.8±2a | 4.5±1b | |||
| Acidobacteria_Gp6 | Gp6 | 18.2±2 | 22.1±3 | 14.2±2 | 17.8±0 | 13.6±3b | 15.8±3a | 11.1±2c | |||
| Firmicutes | Bacilli | Lactobacillales | 0.1±0 | 0.1±0 | 0±0 | 0.4±0 | 0.1±0b | 0.2±0ab | 0.5±0a | ||
| Clostridia | Clostridiales | Clostridiaceae | Clostridium | 0.5±0 | 0.4±0 | 1.2±1 | 1.1±0 | 1.4±0b | 1.7±0b | 3.7±1a | |
| Clostridia | Peptostreptococcaceae | Peptostreptococcus | 0.1±0 | 0.1±0 | 0.1±0 | 0.5±0 | 0.3±0b | 0.4±0ab | 0.8±0a | ||
| Bacteroidetes | Sphingobacteria | Sphingobacteriales | Cytophagaceae | Adhaeribacter | 1.7±0 | 0.9±0 | 1.5±0 | 0.7±0 | 2.1±1a | 0.6±0b | 0.4±0b |
| Actinobacteria | Actinobacteria | Actinomycetales | Microbacteriaceae | Leifsonia | 0±0 | 0.1±0 | 0±0 | 0.1±0 | 0±0b | 0.2±0b | 0.4±0a |
| Chloroflexi | Anaerolineae | Anaerolineales | Anaerolineaceae | 1±0 | 1.1±0 | 1.9±0 | 1.9±0 | 1.5±0b | 1.8±1ab | 2.2±1a | |
| Gemmatimonadetes | Gemmatimonadetes | Gemmatimonadales | Gemmatimonadaceae | Gemmatimonas | 1.1±1 | 1.6±0 | 0.5±0 | 1.7±2 | 0.7±0c | 1.6±1b | 2.5±1a |
| Planctomycetes | Planctomycetacia | Planctomycetales | Planctomycetaceae | 2.7±0 | 2.6±1 | 2.5±0 | 2.4±0 | 2.6±1a | 2.5±0a | 1.7±0b | |
| Verrucomicrobia | Subdivision3 | Subdivision3_genera_incertae_sedis | 0.4±0 | 0.3±0 | 0.6±0 | 0.5±0 | 0.4±0a | 0.5±0a | 0.1±0b | ||
Bold numbers indicate taxa with significantly higher relative abundance in SDZ treated or non treated soils; letters a, b and c indicate taxa with significant difference in relative abundance between S0, S10 and S100 treated soils collected at day 193. Soils were collected 60 days after each manure application.
Discussion
The importance of the soil microbiome for various ecosystem services such as nutrient cycling, soil fertility, degradation of pollutants, and plant growth promotion is well recognized [29]. However, antibiotics introduced into agricultural fields via manure might alter the ability of soil microbes to fulfill crucial ecosystem services, changing the diversity and activity of key functional groups by enhancing antibiotic resistant populations while decreasing the abundance of sensitive populations in soils [16], [30]. In the present study, the effects of repeated application of SDZ manure on soil bacterial communities were explored by barcoded pyrosequencing of 16S rRNA gene amplicons from TC-DNA. More pronounced changes of bacterial communities were observed for soils that were repeatedly treated with SDZ manure. The strongest effects were observed for soils treated three times with manure containing a high concentration of SDZ (Figures 2 and 4). For the first time, taxonomic groups affected by the presence of SDZ were systematically identified, and numerous taxa were found with significantly altered relative abundance (Tables 1 and 2). These findings largely extended our understanding of the influence of SDZ introduced via manure on soil bacterial communities.
Although the bioavailable fraction of SDZ in soil rapidly decreased after each manure application [16], cumulative inputs of SDZ via manure still affected bacterial communities. Also in the study by Byrne-Bailey et al. [31], the proportion of sulfachloropyridazine resistant bacteria increased in soils after treatment with manure containing antibiotics. The repeated application of SDZ manure to grassland soil was found to affect nitrite oxidizing bacteria [32]. The influence of SDZ on soil bacterial communities is more likely to be detected when additional nutrients (root exudates, manure) are added to stimulate bacterial growth [33], [34] as SDZ affects only growing bacteria and bacterial activities are generally low in oligotrophic environments such as soils.
The amendment of soil with SDZ manure resulted in a significantly increased relative abundance of numerous Gram-negative and Gram-positive taxa such as Devosia, Shinella, Stenotrophomonas, Clostridium, Peptostreptococcus, Leifsonia, Gemmatimonas, suggesting that the presence of SDZ provided a selective advantage for these taxa. Mainly the increase in bacteria affiliated to Clostridia detected shortly after the addition of manure likely resulted from manure-derived bacteria. In our study more than 60% of sequences in manure could be affiliated to Clostridium (data not shown), which confirms data from previous studies where the dominance of Clostridium in manure has been reported [35]–[37]. A steep decrease of Clostridia was observed 60 days after the application. Only in S100 soils the relative abundance of Clostridia was found to be significantly higher than in the other soils on day 193, suggesting that the high concentration of SDZ slowed down the decline of Clostridium populations. Whether the enhanced abundance of Clostridium in S100 soils was due to the selection of SDZ resistant populations deserves further research.
Although the resistance to SDZ was not reported yet for most of these taxa, it is likely that some of the populations enriched in the present study might be resistant to SDZ either due to the presence of resistance genes or due to intrinsic resistance. Isolates belonging to Stenotrophomonas and Clostridium were previously reported to be resistant to other sulfonamide antibiotics [38], [39]. Based on the same TC-DNA, Heuer et al. [16] found an accumulation of sul gene carrying populations in the soil repeatedly treated with SDZ manure. Sul genes located on mobile genetic elements such as class 1 integrons or plasmids might have spread among different taxa [40], [41]. The presence of antibiotics, even at very low concentrations could accelerate the genetic exchanges between bacteria [42], [43]. The enhanced transferability of resistant genes was recently also reported by Jechalke et al. [15] in a field experiment using manure from SDZ-treated pigs. However, we cannot exclude that some of the taxa were intrinsically resistant to, or even able to mineralize SDZ as a SDZ degrader identified as Microbacterium lacus was recently isolated from the same Merzenhausen soil [44]. Interestingly, in the present study an OTU affiliated to Leifsonia, which is phylogenetically closely related to Microbacterium was found to be enriched in S100 soils.
In this study, the consensus sequences of an enriched OTU in soils treated with manure shared high similarity with the 16S rRNA gene of S. maltophilia which is also known as a human pathogen [38]. However, these enriched bacteria are not necessarily human pathogens as Stenotrophomonas was also found in different environments such as rhizosphere or soils [45], [46]. Nevertheless, the enrichments of Shinella, Stenotrophomonas, Clostridium and Peptostreptococcus in soil which was continually treated with manure containing a high concentration of SDZ still urge for further investigation due to potential implications for public health. The enrichment of these bacteria, which are phylogenetically closely related to human pathogens, may improve the chance of transferring antibiotic resistance genes to human pathogens, since horizontal gene transfer is more prevalent between closely related organisms than between those distantly related (reviewed by Boto [47]). Soil particles carrying viable bacteria can be transported over long distances and might contribute to the spreading of antibiotic resistant bacteria over wide geographic ranges [48]. The ecological role of taxa such as Devosia, Leifsonia and Gemmatimonas has not been fully understood and thus the effect of their changed abundance on soil functions remains to be explored. Few studies suggested that some members of Devosia [49], [50] and Gemmatimonas [51] might be associated with nitrogen cycling.
The relative abundances of several other taxa such as Pseudomonas, Lysobacter, Hydrogenophaga, Haliangium, and Adhaeribacter were found to be significantly lower in the soils treated by SDZ manure. Several members of Lysobacter or Pseudomonas are known as biological control agents against soilborne phytopathogens such as Rhizoctonia solani, Thielaviopsis basicola [52], [53]. Several studies suggested that these beneficial bacteria belonging to Pseudomonas or Lysobacter probably play an important role for soil suppressiveness and plant growth [45], [54]. In the present study, the consensus sequences of an OTU with decreased relative abundance in soil treated with SDZ manure shared 100% similarities with 16S rRNA gene of the strain of P. brassicacearum (NCBI accession number: NR_074834) which is a beneficial root-associated bacterium [55]. The decline of Pseudomonas and Lysobacter in arable soils might increase the susceptibility towards fungal pathogens. However, in general the application of manure to soils is one strategy to suppress soilborne diseases. Our data showed that the relative abundance of Pseudomonas was much higher in soils treated with manure than in untreated soils on days 3 and 193. The presence of SDZ in manure likely reduces the beneficial effects of manure on improving soil health as the abundance of Pseudomonas was reduced. However, resilience of these beneficial populations might still occur in the rhizosphere as decreased concentrations of SDZ were recently observed in the rhizosphere of maize and grass compared to bulk soil [6]. Little is known about the ecological roles of Hydrogenophage, Haliangium, Adhaeribacter in soil. Hydrogenophaga spp. were reported as main biphenyl degraders in the rhizosphere of horseradish (Armoracia rusticana) contaminated with polychlorinated biphenyls [56].
The present study is the first to explore the influence of veterinary antibiotics entering soil via manure on the diversity and abundance of Acidobacteria. The results revealed that Gp4 was negatively affected by repeated application of manure containing the high concentration of SDZ. Low concentrations of SDZ in manure seemed to favor Gp6 and Gp10, while high concentrations of SDZ could deprive the effects. Acidobacteria are known to be adapted to an oligotrophic lifestyle. Indeed, in the present study the relative abundance of Acidobacteria was significantly higher in the untreated soil than in the soil amended with manure. Due to their high abundance and ubiquitous distribution in soils, Acidobacteria might play an important role in terrestrial ecosystems. In combination with the analysis of three genomes of acidobacterial isolates, Ward et al. [57] suggested that Acidobacteria might significantly contribute to the terrestrial carbon cycle.
Along with nutrients, a large amount of manure bacteria was also introduced into soil which could have also contributed to the dramatically altered bacterial community compositions observed in the present study shortly after each manure application. However, the majority of these introduced manure bacteria might not be very well adapted to soil, allowing the resilience of the indigenous soil bacteria community. In the present study we showed that repeated application of manure spiked with SDZ to soil increased the degree of disturbance and reduced the resilience of soil bacterial communities compared to a single application of manure spiked with SDZ or soils treated with manure without antibiotics. In a recent study by Poulsen et al. [58], the taxonomic composition of bacterial communities based on the abundance of phyla and proteobacterial classes were highly similar between soils with amendments of different organic wastes such as manure. In the present study, we observed clear effects of manure on the soil bacterial community composition as the NMDS analysis revealed separated groups for S0 and untreated soils on day 193 (Figure 3). In contrast to several subgroups of the Acidobacteria, Myxococcales (Deltaproteobacteria), numerous taxa belonging to different phyla (Proteobacteria, Firmicutes and Bacteriodetes) were observed with significantly higher relative abundance in the soils amended with manure than in the untreated control 60 days after the third application. Very likely the added nutrients favored taxa with a more copiotrophic lifestyle. In contrast to the field experiment analyzed by Poulsen et al. [58], the conditions in the present experiment were more controlled and the time period after the last manure amendment might have been shorter although this was not specified in the Poulsen study.
In summary, repeated input of SDZ into the Merzenhausen soil via manure, especially at a high concentration, caused pronounced changes in soil bacterial communities. The presence of SDZ provided a selective advantage for species affiliated to the genera Devosia, Shinella, Stenotrophomonas, Clostridium, Peptostreptococcus, Leifsonia, Gemmatimonas, while suppressing Pseudomonas, Lysobacter, Hydrogenophage, Haliangium, Adhaeribacter. We could show that SDZ-containing manure caused a more pronounced disturbance of the soil bacterial community compared to control manure and reduced resilience at least for the time period studied. However, whether the findings obtained can be generalized to other soil types with different soil properties remains an open question which needs further attention. Furthermore, mainly the long-term consequences of repeated manure application for soil ecosystem functions and human health demand further investigations.
Supporting Information
Supporting Files. Table S1. The number of replicates per treatment included in the final sequencing run using the reverse primer. The numbers in brackets indicate the number of samples sequenced using the forward primer. Table S2. Taxa with significantly different relative abundance between soils on day 3 using the reverse primer (Bonferroni adjusted p value<0.05). Table S3. Taxa with significantly different relative abundance between soils on day 3 using the forward primer (Bonferroni adjusted p value<0.05). Table S4. Relative abundance of detected phyla based on the average of the forward and reverse dataset. Table S5. Taxa with significantly different relative abundance between soils on day 193 using the reverse primer (Bonferroni adjusted p value<0.05). Table S6. Taxa with significantly different relative abundance between soils on day 193 using the forward primer (Bonferroni adjusted p value<0.05). Table S7. Taxa with significantly different relative abundance between manured soils with or without SDZ treatments as revealed by barcoded pyrosequencing using the forward primer (Bonferroni adjusted p value<0.05). Table S8. Taxa with significantly different relative abundance between manured soils with or without SDZ treatments as revealed by barcoded pyrosequencing using the forward primer (Bonferroni adjusted p value<0.05). Figure S1. Non metric multidimensional scaling based on OTU reports acquired by the forward primer sequencing. Balls: untreated soils; cubes: S0; octahedron: S10; pyramid: S100.
(DOC)
Funding Statement
This work was funded by the Deutsche Forschungsgemeinschaft (DFG) in the framework of the Research Unit FOR 566 “Veterinary medicines in soil: Basic research for risk analysis” (SM59/5-3). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kools SAE, Moltmann JF, Knacker T (2008) Estimating the use of veterinary medicines in the European union. Regulatory Toxicology and Pharmacology 50: 59–65. [DOI] [PubMed] [Google Scholar]
- 2. Lamshöft M, Sukul P, Zühlke S, Spiteller M (2007) Metabolism of C-14-labelled and non-labelled sulfadiazine after administration to pigs. Anal Bioanal Chem 388: 1733–1745. [DOI] [PubMed] [Google Scholar]
- 3. Heuer H, Focks A, Lamshöft M, Smalla K, Matthies M, et al. (2008) Fate of sulfadiazine administered to pigs and its quantitative effect on the dynamics of bacterial resistance genes in manure and manured soil. Soil Biol Biochem 40: 1892–1900. [Google Scholar]
- 4. Sarmah AK, Meyer MT, Boxall ABA (2006) A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment. Chemosphere 65: 725–759. [DOI] [PubMed] [Google Scholar]
- 5. Sittig S, Kasteel R, Groeneweg J, Vereecken H (2012) Long-term sorption and sequestration dynamics of the antibiotic sulfadiazine: a batch study. J Environ Qual 41: 1497–1506. [DOI] [PubMed] [Google Scholar]
- 6. Rosendahl I, Siemens J, Groeneweg J, Linzbach E, Laabs V, et al. (2011) Dissipation and sequestration of the veterinary antibiotic sulfadiazine and its metabolites under field conditions. Environ Sci Technol 45: 5216–5222. [DOI] [PubMed] [Google Scholar]
- 7. Hammesfahr U, Heuer H, Manzke B, Smalla K, Thiele-Bruhn S (2008) Impact of the antibiotic sulfadiazine and pig manure on the microbial community structure in agricultural soils. Soil Biol Biochem 40: 1583–1591. [Google Scholar]
- 8. Hammesfahr U, Bierl R, Thiele-Bruhn S (2011) Combined effects of the antibiotic sulfadiazine and liquid manure on the soil microbial-community structure and functions. J of Plant Nutr Soil Sci 174: 614–623. [Google Scholar]
- 9. Schauss K, Focks A, Heuer H, Kotzerke A, Schmitt H, et al. (2009) Analysis, fate and effects of the antibiotic sulfadiazine in soil ecosystems. Trac-Trends Anal Chem 28: 612–618. [Google Scholar]
- 10. Kotzerke A, Sharma S, Schauss K, Heuer H, Thiele-Bruhn S, et al. (2008) Alterations in soil microbial activity and N-transformation processes due to sulfadiazine loads in pig-manure. Environ Pollut 153: 315–322. [DOI] [PubMed] [Google Scholar]
- 11. Kotzerke A, Hammesfahr U, Kleineidam K, Lamshoft M, Thiele-Bruhn S, et al. (2011) Influence of difloxacin-contaminated manure on microbial community structure and function in soils. Biol Fertil Soils 47: 177–186. [Google Scholar]
- 12. De Gelder L, Ponciano JM, Abdo Z, Joyce P, Forney LJ, et al. (2004) Combining mathematical models and statistical methods to understand and predict the dynamics of antibiotic-sensitive mutants in a population of resistant bacteria during experimental evolution. Genetics 168: 1131–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Heuer H, Smalla K (2007) Manure and sulfadiazine synergistically increased bacterial antibiotic resistance in soil over at least two months. Environ Microbiol 9: 657–666. [DOI] [PubMed] [Google Scholar]
- 14. Kopmann C, Jechalke S, Rosendahl I, Groeneweg J, Krögerrecklenfort E, et al. (2013) Abundance and transferability of antibiotic resistance as related to the fate of sulfadiazine in maize rhizosphere and bulk soil. FEMS Microbiol Ecol 83: 125–134. [DOI] [PubMed] [Google Scholar]
- 15. Jechalke S, Kopmann C, Rosendahl I, Groeneweg J, Weichelt V, et al. (2013) Increased abundance and transferability of resistance genes after field application of manure from sulfadiazine-treated pigs. Appl Environ Microbiol 79: 1704–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Heuer H, Solehati Q, Zimmerling U, Kleineidam K, Schloter M, et al. (2011) Accumulation of sulfonamide resistance genes in arable soils due to repeated application of manure containing sulfadiazine. Appl Environ Microbiol 77: 2527–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Heuer H, Schmitt H, Smalla K (2011) Antibiotic resistance gene spread due to manure application on agricultural fields. Curr Opin Microbiol 14: 236–243. [DOI] [PubMed] [Google Scholar]
- 18. Gaze WH, Krone SM, Larsson DGJ, Li X, Robinson JA, et al. (2013) Influence of humans on evolution and mobilization of environmental antibiotic resistome. Emerg Infect Dis 19 Article 871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Forsberg KJ, Reyes A, Wang B, Selleck EM, Sommer MOA, et al. (2012) The shared antibiotic resistome of soil bacteria and human pathogens. Science 337: 1107–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sukul P, Lamshoft M, Zuhlke S, Spiteller M (2008) Sorption and desorption of sulfadiazine in soil and soil-manure systems. Chemosphere 73: 1344–1350. [DOI] [PubMed] [Google Scholar]
- 21. Griffiths BS, Philippot L (2013) Insights into the resistance and resilience of the soil microbial community. FEMS Microbiol Rev 37: 112–129. [DOI] [PubMed] [Google Scholar]
- 22. Lamshöft M, Sukul P, Zühlke S, Spiteller M (2010) Behaviour of C-14-sulfadiazine and C-14-difloxacin during manure storage. Sci Total Environ 408: 1563–1568. [DOI] [PubMed] [Google Scholar]
- 23. Griffiths RI, Whiteley AS, O'Donnell AG, Bailey MJ (2000) Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl Environ Microbiol 66: 5488–5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Timmers RA, Rothballer M, Strik D, Engel M, Schulz S, et al. (2012) Microbial community structure elucidates performance of Glyceria maxima plant microbial fuel cell. Appl Microbiol Biotechnol 94: 537–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ding G-C, Heuer H, Smalla K (2012) Dynamics of bacterial communities in two unpolluted soils after spiking with phenanthrene: soil type specific and common responders. Front Microbiol 3 doi:10.3389/fmicb.2012.00290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, et al. (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35: 7188–7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naïve bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hothorn T, Bretz F, Westfall P (2008) Simultaneous inference in general parametric models. Biometrical J 50: 346–363. [DOI] [PubMed] [Google Scholar]
- 29. Torsvik V, Øvreås L (2002) Microbial diversity and function in soil: from genes to ecosystems. Curr Opin Microbiol 5: 240–245. [DOI] [PubMed] [Google Scholar]
- 30. Zhu YG, Johnson TA, Su JQ, Qiao M, Guo GX, et al. (2013) Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc Natl Acad Sci U S A 110: 3435–3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Byrne-Bailey KG, Gaze WH, Kay P, Boxall ABA, Hawkey PM, et al. (2009) Prevalence of sulfonamide resistance genes in bacterial isolates from manured agricultural soils and pig slurry in the United Kingdom. Antimicrob Agents Chemother 53: 696–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ollivier J, Schacht D, Kindler R, Groeneweg J, Engel M, et al. (2013) Effects of repeated application of sulfadiazine-contaminated pig manure on the abundance and diversity of ammonia and nitrite oxidizers in the root-rhizosphere complex of pasture plants under field conditions. Front Microbiol 4 Article 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brandt KK, Sjøholm OR, Krogh KA, Halling-Sørensen B, Nybroe O (2009) Increased pollution-induced bacterial community tolerance to sulfadiazine in soil hotspots amended with artificial root exudates. Environ Sci Technol 43: 2963–2968. [DOI] [PubMed] [Google Scholar]
- 34. Zielezny Y, Groeneweg J, Vereecken H, Tappe W (2006) Impact of sulfadiazine and chlorotetracycline on soil bacterial community structure and respiratory activity. Soil Biol Biochem 38: 2372–2380. [Google Scholar]
- 35. Doornbos RF, van Loon LC, Bakker P (2012) Impact of root exudates and plant defense signaling on bacterial communities in the rhizosphere. A review. Agron Sustain Dev 32: 227–243. [Google Scholar]
- 36. Peu P, Brugere H, Pourcher AM, Kerouredan M, Godon JJ, et al. (2006) Dynamics of a pig slurry microbial community during anaerobic storage and management. Appl Environ Microbiol 72: 3578–3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yi J, Zheng R, Li FG, Chao Z, Deng CY, et al. (2012) Temporal and spatial distribution of Bacillus and Clostridium histolyticum in swine manure composting by fluorescent in situ hybridization (FISH). Appl Microbiol Biotechnol 93: 2625–2632. [DOI] [PubMed] [Google Scholar]
- 38. Brooke JS (2012) Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin Microbiol Rev 25: 2–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Neikirk RL, Krieg NR (1964) Sulfonamide resistance of Clostridium sporogenes . J Bacteriol 87: 1526–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Heuer H, Kopmann C, Binh CTT, Top EM, Smalla K (2009) Spreading antibiotic resistance through spread manure: characteristics of a novel plasmid type with low %G plus C content. Environ Microbiol 11: 937–949. [DOI] [PubMed] [Google Scholar]
- 41. Heuer H, Smalla K (2012) Plasmids foster diversification and adaptation of bacterial populations in soil. FEMS Microbiol Rev 36: 1083–1104. [DOI] [PubMed] [Google Scholar]
- 42. Davies J, Spiegelman GB, Yim G (2006) The world of subinhibitory antibiotic concentrations. Curr Opin Microbiol 9: 445–453. [DOI] [PubMed] [Google Scholar]
- 43. Martinez JL (2009) Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ Pollut 157: 2893–2902. [DOI] [PubMed] [Google Scholar]
- 44. Tappe W, Herbst M, Hofmann D, Koeppchen S, Kummer S, et al. (2013) Degradation of sulfadiazine by Microbacterium lacus strain SDZm4, isolated from lysimeters previously manured with slurry from sulfadiazine-medicated pigs. Appl Environ Microbiol 79: 2572–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hayward AC, Fegan N, Fegan M, Stirling GR (2010) Stenotrophomonas and Lysobacter: ubiquitous plant-associated gamma-proteobacteria of developing significance in applied microbiology. J Appl Microbiol 108: 756–770. [DOI] [PubMed] [Google Scholar]
- 46. Ryan RP, Monchy S, Cardinale M, Taghavi S, Crossman L, et al. (2009) The versatility and adaptation of bacteria from the genus Stenotrophomonas . Nat Rev Microbiol 7: 514–525. [DOI] [PubMed] [Google Scholar]
- 47. Boto L (2010) Horizontal gene transfer in evolution: facts and challenges. Proc Biol Sci 277: 819–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Prospero JM, Blades E, Mathison G, Naidu R (2005) Interhemispheric transport of viable fungi and bacteria from Africa to the Caribbean with soil dust. Aerobiologia 21: 1–19. [Google Scholar]
- 49. Vanparys B, Heylen K, Lebbe L, De Vos P (2005) Devosia limi sp. nov., isolated from a nitrifying inoculum. Int J Syst Evol Microbiol 55: 1997–2000. [DOI] [PubMed] [Google Scholar]
- 50. Rivas R, Velazquez E, Willems A, Vizcaino N, Subba-Rao NS, et al. (2002) A new species of Devosia that forms a unique nitrogen-fixing root-nodule symbiosis with the aquatic legume Neptunia natans (L.f.) druce. Appl Environ Microbiol 68: 5217–5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yang Y, Wang X, Shi JF, Li JJ (2012) The influence of the discharging sewage on microbial diversity in sludge from Dongting Lake. World J Microb Biot 28: 421–430. [DOI] [PubMed] [Google Scholar]
- 52. Sullivan RF, Holtman MA, Zylstra GJ, White JF, Kobayashi DY (2003) Taxonomic positioning of two biological control agents for plant diseases as Lysobacter enzymogenes based on phylogenetic analysis of 16S rDNA, fatty acid composition and phenotypic characteristics. J Appl Microbiol 94: 1079–1086. [DOI] [PubMed] [Google Scholar]
- 53. Weller DM (2007) Pseudomonas biocontrol agents of soilborne pathogens: looking back over 30 years. Phytopathology 97: 250–256. [DOI] [PubMed] [Google Scholar]
- 54. Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, et al. (2011) Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332: 1097–1100. [DOI] [PubMed] [Google Scholar]
- 55. Ortet P, Barakat M, Lalaouna D, Fochesato S, Barbe V, et al. (2011) Complete genome sequence of a beneficial plant root-associated bacterium, Pseudomonas brassicacearum . J Bacteriol 193: 3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Uhlik O, Jecna K, Mackova M, Vlcek C, Hroudova M, et al. (2009) Biphenyl-metabolizing bacteria in the rhizosphere of horseradish and bulk soil contaminated by polychlorinated biphenyls as revealed by stable isotope probing. Appl Environ Microbiol 75: 6471–6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ward NL, Challacombe JF, Janssen PH, Henrissat B, Coutinho PM, et al. (2009) Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl Environ Microbiol 75: 2046–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Poulsen PHB, Abu Al-Soud W, Bergmark L, Magid J, Hansen LH, et al. (2013) Effects of fertilization with urban and agricultural organic wastes in a field trial - prokaryotic diversity investigated by pyrosequencing. Soil Biol Biochem 57: 784–793. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Files. Table S1. The number of replicates per treatment included in the final sequencing run using the reverse primer. The numbers in brackets indicate the number of samples sequenced using the forward primer. Table S2. Taxa with significantly different relative abundance between soils on day 3 using the reverse primer (Bonferroni adjusted p value<0.05). Table S3. Taxa with significantly different relative abundance between soils on day 3 using the forward primer (Bonferroni adjusted p value<0.05). Table S4. Relative abundance of detected phyla based on the average of the forward and reverse dataset. Table S5. Taxa with significantly different relative abundance between soils on day 193 using the reverse primer (Bonferroni adjusted p value<0.05). Table S6. Taxa with significantly different relative abundance between soils on day 193 using the forward primer (Bonferroni adjusted p value<0.05). Table S7. Taxa with significantly different relative abundance between manured soils with or without SDZ treatments as revealed by barcoded pyrosequencing using the forward primer (Bonferroni adjusted p value<0.05). Table S8. Taxa with significantly different relative abundance between manured soils with or without SDZ treatments as revealed by barcoded pyrosequencing using the forward primer (Bonferroni adjusted p value<0.05). Figure S1. Non metric multidimensional scaling based on OTU reports acquired by the forward primer sequencing. Balls: untreated soils; cubes: S0; octahedron: S10; pyramid: S100.
(DOC)