Abstract
Background and Purpose
SUMO conjugation is a post-translational modification associated with many human diseases. Characterization of the SUMO-modified proteome is pivotal to defining the mechanistic link between SUMO conjugation and such diseases. This is particularly evident for SUMO2/3 conjugation, which is massively activated after brain ischemia/stroke, and is believed to be a protective response. The purpose of this study was to perform a comprehensive analysis of the SUMO3-modified proteome regulated by brain ischemia using a novel SUMO transgenic mouse.
Methods
To enable SUMO proteomics analysis in vivo, we generated transgenic mice conditionally expressing tagged SUMO1-3 paralogues. Transgenic mice were subjected to 10 minutes forebrain ischemia and 1 hour of reperfusion. SUMO3-conjugated proteins were enriched by anti-FLAG affinity purification and analyzed by LC-MS/MS.
Results
Characterization of SUMO transgenic mice demonstrated that all 3 tagged SUMO paralogues were functionally active, and expression of exogenous SUMOs did not modify the endogenous SUMOylation machinery. Proteomics analysis identified 112 putative SUMO3 substrates of which 91 candidates were more abundant in the ischemia group than the sham group. Data analysis revealed processes/pathways with putative neuroprotective functions, including glucocorticoid receptor signaling, RNA processing and SUMOylation-dependent ubiquitin conjugation.
Conclusions
The identified proteins/pathways modulated by SUMOylation could be the key to understanding the mechanisms linking SUMOylation to neuroprotection, and thus provide new promising targets for therapeutic interventions. The new transgenic mouse will be an invaluable platform for analyzing the SUMO-modified proteome in models of human disorders, and thereby help to mechanistically link SUMOylation to the pathological processes.
Keywords: SUMO, cerebral ischemia, proteomics, transgenic mice
Introduction
Small ubiquitin-like modifiers (SUMOs) are covalently conjugated to lysine residues of target proteins, and thereby modulate their function, stability, and localization 1, 2. SUMO1, SUMO2, and SUMO3 are widely expressed in mammalian tissues. SUMO2 and SUMO3 are almost identical and are often referred to as SUMO2/3. They are distinct from SUMO1, however, with about 50% homology. The SUMO conjugation (SUMOylation) is an energy-dependent process catalyzed by activating enzyme (SAE1/SAE2), conjugating enzyme (Ubc9), and ligating enzyme 1, 2. SUMOylation is a dynamic and reversible reaction, as SUMOylated proteins can be readily deconjugated by SUMO proteases 3.
SUMOylation modulates many cellular functions including DNA repair, genome maintenance, gene transcription, and protein degradation control 4–6, and plays key roles in many human diseases such as cerebral ischemia/stroke, cancer, and heart failure 1, 7–9. It is, therefore, of tremendous clinical interest to characterize the SUMO-modified proteome in these disorders. This is particularly evident for cerebral ischemia/stroke, because transient cerebral ischemia dramatically activates SUMOylation, and this is believed to be a neuroprotective stress response. It is, therefore, of key interest to identify the proteins SUMOylated after brain ischemia in order to uncover the mechanisms linking SUMOylation to neuroprotection. However, SUMO proteomics analysis is hampered by the low levels of SUMOylated proteins. A common strategy is, therefore, to perform proteomics analysis on purified SUMO-conjugated proteins from cells stably expressing tagged SUMOs. Recently, a His6-HA-SUMO1 knock-in mouse was generated, which allowed researchers, for the first time, to characterize the SUMO1-modified proteome in brains 10. In the present study, we generated a transgenic mouse (CAG-loxP-STOP-loxP-SUMO, hereafter referred to as CAG-SUMO) in which His-SUMO1, HA-SUMO2, and FLAG-SUMO3 are expressed in a Cre-dependent manner. Using this new mouse line, we report here the first profile of the SUMO3-modified proteome in mouse brain after ischemia, a pathological state associated with a dramatic activation of SUMO2/3 conjugation 8, 9.
Methods
Full details of the methods are provided in the online-only Supplemental Material.
Transgenic mice
Animal experiments were approved by the Duke University Animal Care and Use Committee. CAG-SUMO transgenic mice were generated by pronuclear injection of the transgene vector illustrated in Fig. 1A.
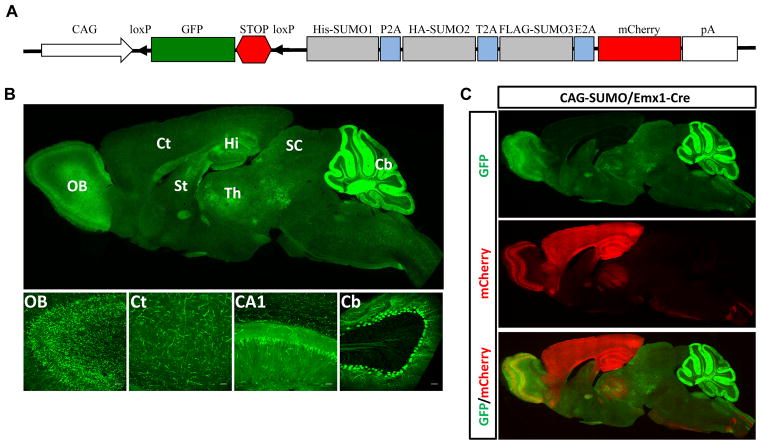
Figure 1.
Generation of CAG-SUMO transgenic mice. A, Scheme of the transgene construct. The transgene consists of the CMV early enhancer/chicken β-actin promoter (CAG), a fragment containing GFP and a transcriptional/translational STOP cassette (STOP) flanked by loxP sites, 3 tagged SUMOs linked by 2A sequences, mCherry, and a polyadenylation signal (pA). B, Expression patterns of native GFP fluorescence in a CAG-SUMO line 10 mouse brain. A sagittal brain section indicated widespread GFP expression (upper panel). Lower panel shows confocal images of different areas of the brain. C, A sagittal brain section of a double transgenic CAG-SUMO/Emx1-Cre mouse showing the mCherry expression, indicative of expression of tagged SUMOs, restricted to forebrain regions. OB, olfactory bulb; Ct, cortex; Hi, hippocampus; CA1, hippocampal CA1 subfield; St, striatum; Th, thalamus; SC, superior colliculus; Cb, cerebellum. Scale bars: 50 μm.
Animal surgery
Transient forebrain cerebral ischemia was performed as described previously, with minor modifications 8.
Sample preparation for proteomic analysis
Each sample for proteomic analysis was generated by FLAG pulldown of nuclear fraction from cortical tissues pooled from 4 mice. Three biological replicates were used for each group.
LC-MS/MS analysis and data analysis
All 9 samples derived from FLAG pulldown were separated on SDS-PAGE gel, and each lane was dissected into 14 slices. After in-gel digestion, peptides from each slice were analyzed by LC-MS/MS. Data were analyzed by various programs.
Results
Generation and characterization of CAG-SUMO mice
We designed a transgene vector based on Cre/lox recombination and 2A-mediated co-translational cleavage to express His-SUMO1, HA-SUMO2, and FLAG-SUMO3 from a single multicistronic transgene in a conditional manner (Fig. 1A). We used the cDNAs encoding precursor SUMO1-3, which allows the endogenous SUMO proteases to remove the extra 2A peptides 11, and expose the C-terminal di-glycine motif of SUMOs. GFP and mCherry were used as indicators of transgene expression before and after Cre recombination. We obtained 8 founder lines with varying levels and patterns of GFP expression. Due to ubiquitous expression of GFP, line 10 was chosen for the present study. To examine the global pattern of Cre-mediated transgene expression, mice were cross-bred with hemizygous β-Actin-Cre mice to generate double transgenic CAG-SUMO/β-Actin-Cre mice. CAG-SUMO line 10 exhibited strong GFP fluorescence in all organs examined including brain, heart, lung, kidney, and liver; those organs showed mCherry expression in CAG-SUMO/β-Actin-Cre mice. Expression of FLAG-SUMO3 was also confirmed in those organs (data not shown).
In the brain of CAG-SUMO line 10, GFP was ubiquitously expressed with strong signals in hippocampus and cerebellum (Fig. 1B). Since we were particularly interested in the SUMO-modified proteome regulated by forebrain ischemia, we mated hemizygous CAG-SUMO mice with homozygous Emx1Cre/Cre mice to generate double transgenic CAG-SUMO/Emx1-Cre as tagged SUMO-expressing mice and littermates Emx1Cre/+ as control mice. In line with a previous report 12, CAG-SUMO/Emx1-Cre mice showed strong mCherry expression in the cerebral cortex and hippocampus (Fig. 1C). For both CAG-SUMO and CAG-SUMO/Emx1-Cre mice, we did not find any obvious physical or behavioral abnormalities.
Post-ischemic SUMOylation in the brain of transgenic mice
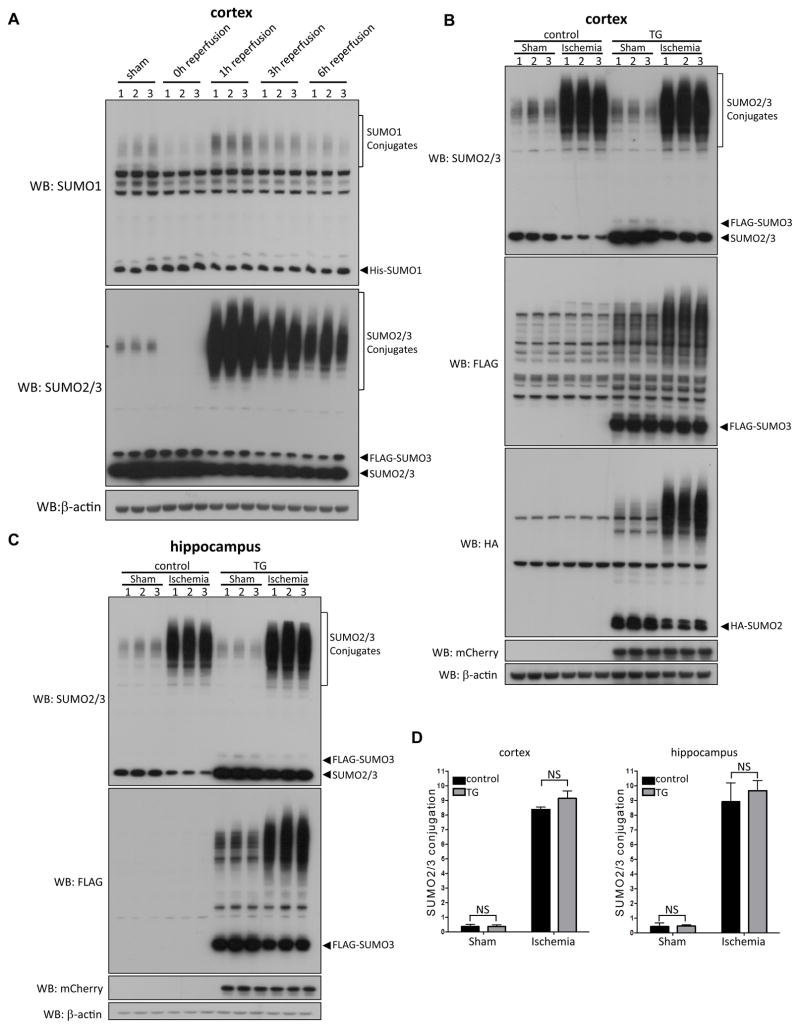
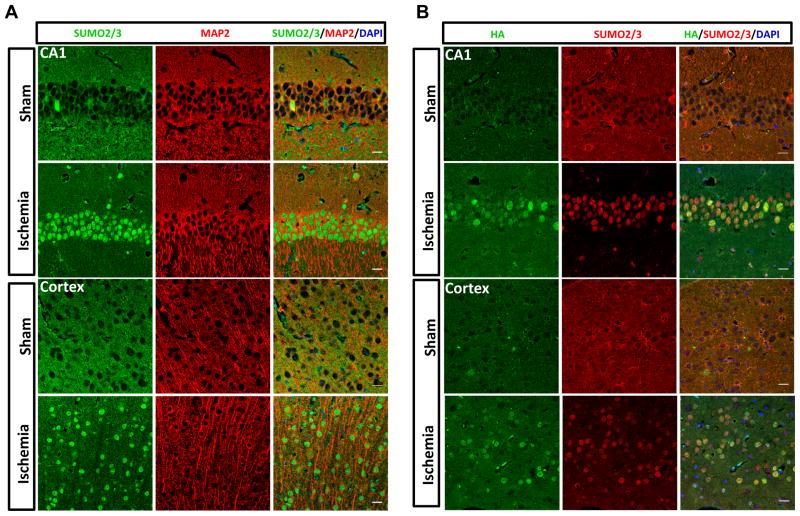
To extend our previous findings 8 and also demonstrate the suitability of CAG-SUMO/Emx1-Cre mice for in vivo SUMO studies, we investigated the temporal and spatial profiles of post-ischemic SUMOylation by endogenous and exogenous SUMOs in brains of Emx1Cre/+ control and CAG-SUMO/Emx1-Cre mice. First, the temporal profile studies indicated that SUMO1-3 conjugation was dramatically decreased during and rapidly activated after ischemia (Fig. 2A). Similar pattern was found for FLAG-SUMO3 (data not shown). Then, we carefully compared SUMOylation response to brain ischemia in Emx1Cre/+ and CAG-SUMO/Emx1-Cre mice to check for possible off-target effects that might be caused by overexpressing SUMOs. In both sham and ischemia groups, levels of SUMO1 and SUMO2/3 conjugates in the high-molecular-weight regions were comparable in controls and double transgenic mice, although there were substantially higher levels of unconjugated SUMOs in CAG-SUMO/Emx1-Cre mice due to expression of tagged SUMOs (Fig. 2B–D, data not shown). Finally, transient forebrain ischemia induced nuclear accumulation of SUMO2/3-conjugated proteins, and the same pattern was also observed for HA-SUMO2 and FLAG-SUMO3 (Fig. 3 and Supplemental Fig. IA).
Figure 2.
Effect of transient forebrain ischemia on SUMOylation. A, CAG-SUMO/Emx1-Cre mice were subjected to 10 minutes forebrain ischemia and 0, 1, 3, or 6 hours reperfusion (n = 3 per group). Sham-operated mice were used as control. SUMO conjugates in high-molecular-weight regions are marked by brackets. B–D, Comparison of SUMO2/3 conjugation in brains of Emx1Cre/+ (control) and CAG-SUMO/Emx1-Cre (TG) mice. Emx1Cre/+ and CAG-SUMO/Emx1-Cre mice were subjected to sham surgery or 10 minutes forebrain ischemia and 1 hour reperfusion (n = 3 per group). Protein samples were prepared from the cortex (B) and the hippocampus (C), and analyzed by Western blotting using indicated antibodies. D, The high-molecular-weight regions marked by brackets were used to quantify SUMO2/3 conjugates in the cortex and hippocampus. Films with shorter exposure times were used to measure intensities. Data were normalized to β-actin and presented as means ± SD. WB, Western blot; NS, not significant.
Figure 3.
Nuclear accumulation of SUMO2/3-conjugated proteins after ischemia. A–B, CAG-SUMO/Emx1-Cre mice were subjected to sham surgery or 10 minutes forebrain ischemia and 1 hour reperfusion. Brain sections were stained with antibodies against SUMO2/3, HA, and MAP2 as neuron-specific marker. Ischemia-induced nuclear accumulation of SUMO2/3- and HA-SUMO2-conjugated proteins was found both in cortex and hippocampus neurons. Scale bars: 20 μm.
SUMO3-modified proteome regulated by transient forebrain ischemia
In order to compare results to our previous SUMO3 proteomics analysis using an in vitro ischemia model 13, we focused on the SUMO3-modified proteome in this study. We chose 1 hour of reperfusion when SUMO2/3 conjugation was maximally activated (Fig. 2A). Furthermore, we used cortical tissues because we were interested in the neuroprotective role of SUMOylation, and the cortex is spared from damage in this ischemia model 14. For future studies, we also performed a small-scale HA pulldown to confirm enrichment of HA-SUMO2-conjugated proteins (Supplemental Fig. IB).
First, we optimized the FLAG pulldown procedure by using nuclear fractions as input for FLAG pulldown. This greatly enhanced specificity, since nuclear fractions were devoid of unconjugated FLAG-SUMO3, had markedly less unspecific bands on Western blots (Supplemental Fig. IA), and exhibited dramatically lower total protein levels (Supplemental Fig. IC). Indeed, FLAG-SUMO3-conjugated proteins were effectively immunoprecipitated from nuclear fractions (Supplemental Fig. ID). Interestingly, we did not notice a marked decrease in SUMO2/3 and HA signals in flow-through samples (Supplemental Fig. ID). This suggested that FLAG-SUMO3 represented only a small fraction of the total SUMO2/3 pool. We also found HA-SUMO2 in FLAG-SUMO3 pulldown eluates, and, notably, there was a shift toward higher molecular weights in the ischemic sample, implying increased length of SUMO2/3 chains (Supplemental Fig. ID).
For the large-scale SUMO3 proteomics study, 3 groups of mice were used: Emx1Cre/+ without surgery (control, to account for background binding to anti-FLAG beads), and CAG-SUMO/Emx1-Cre with sham (TG Sham) or ischemia surgery (TG Ischemia) (Fig. 4A). All 9 FLAG pulldown samples (n = 3/group) were confirmed by Western blotting (Supplemental Fig. IIA) and then separated on an SDS-PAGE gel (Fig. IIB). Fourteen gel slices per lane were cut for LC-MS/MS analysis (Supplemental Fig. IIB).
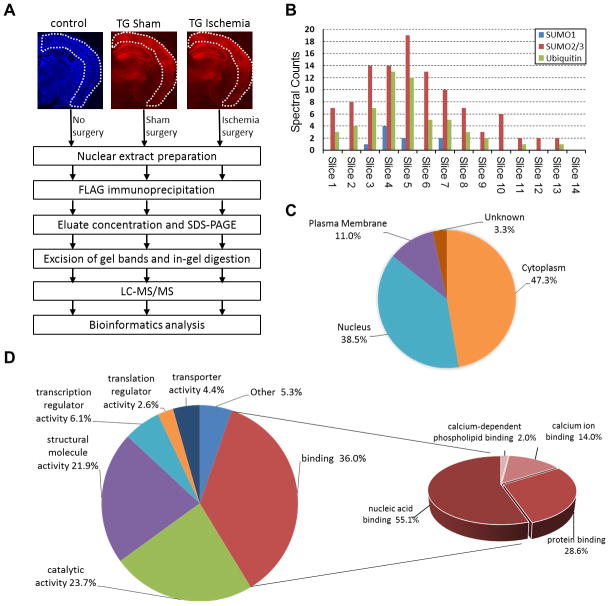
Figure 4.
Proteomics analysis of SUMO3-conjugated proteins in post-ischemic mouse brains. A, Overview of the workflow to identify FLAG-SUMO3-conjugates in the post-ischemic cerebral cortex. Coronal brain sections of Emx1Cre/+ (control; DAPI staining, blue) and CAG-SUMO/Emx1-Cre (TG; mCherry fluorescence, red) mice were shown to indicate cortical regions used in the study. B, Distribution of total spectral counts for SUMO1, SUMO2/3, and ubiquitin for each gel slice of ischemia samples from high (Slice 1) to low (Slice 14) molecular weights. C–D, Subcellular localization (C) and molecular functions (D) of the 91 putative SUMO3 substrates with upregulated SUMO3 conjugation state after ischemia were grouped by the PANTHER program.
Proteomics data showed that SUMO2/3 and ubiquitin shared a similar distribution of spectral counts (Fig. 4B), suggesting a marked post-ischemic activation of the cross-talk between these 2 post-translational modifications. Indeed, we found ubiquitin conjugation to be activated after ischemia, particularly pronounced in nuclei (Supplemental Fig. IIC). Based on selection criteria described in Supplemental Methods, 112 proteins were considered as putative SUMO3 substrates (Supplemental Table I), and 91 proteins (Supplemental Table I, asterisks) were considered as ischemia-upregulated candidates of which 46 candidates were found only in ischemia samples (Supplemental Table I, triangles), including the general transcription factor IIi (TFII-I/GTF2I), tripartite motif containing 33 (TRIM33), glucocorticoid receptor (GR/GCR), and B-cell lymphoma/leukemia 11B (CTIP2/BCL11B).
Gene Ontology annotation analysis by the PANTHER program indicated that 38.5% and 47.3% of the 91 candidates were predicted to have nuclear and cytoplasmic localization, respectively (Fig. 4C). We did not expect that most of the identified proteins would have predicted cytoplasmic localization, considering that nuclear fractions were used for proteomics analyses. A plausible explanation is that cytoplasmic SUMO3 target proteins were translocated to the nucleus after ischemia (for example, see below for GR). PANTHER analysis also revealed that most of the 91 proteins belonged to a group of binding proteins of which nucleic acid binding accounted for 55.1% (Fig. 4D). Ingenuity Pathway Analysis (IPA) core analysis of the 91 candidates revealed significant enrichment in the categories of neurological disease (46 targets) and cell death and survival (47 targets) (Supplemental Fig. IIIA,B). Strikingly, 27 proteins were grouped with functions in RNA processing with high confidence (IPA score = 72, Supplemental Fig. IIIC).
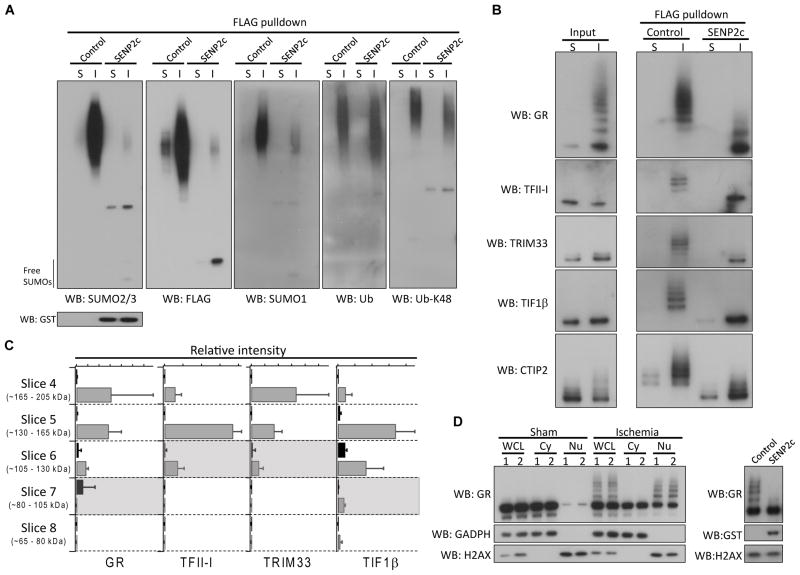
To verify proteomics analysis findings independently, a separate large-scale FLAG pulldown was performed using 3 CAG-SUMO/Emx1-Cre mice per group (sham and ischemia). First, we treated FLAG pulldown samples with SENP2c for de-SUMOylation. Western blot analysis confirmed that the strong high-molecular-weight smear of bands indeed represented SUMO-conjugated proteins (Fig. 5A). The presence of SUMO1 is consistent with the report that SUMO1-3 can form mixed chains 15. Interestingly, ubiquitinated proteins slightly shifted to lower molecular weight after SENP2c treatment (Fig. 5A). These data suggest that ubiquitin and SUMO conjugated to different lysine residues for a subset of post-ischemic SUMO3 conjugates. Similar findings were also reported in cells exposed to ischemia-like conditions or proteasome inhibitor MG132 13, 16.
Figure 5.
Verification of SUMO3 proteomics data. A–B, The FLAG pulldown samples (sham and ischemia) treated with/without SENP2c were analyzed by Western blotting with the indicated antibodies. SENP2c is GST tagged. C, Quantitative analysis of 4 identified SUMO3 substrates. The areas under the curve of two peptides from each protein were used to perform relative quantitation of sham (black) and ischemia (grey) samples as a function of molecular weight. The expected molecular weight of each unmodified protein was indicated by a shaded region. Data were presented as means ± SD (n = 3 per group). D, Whole cell lysates (WCL), cytoplasmic (Cy) and nuclear (Nu) fractions, prepared from cortical samples (n = 2/group, sham and ischemia) were analyzed by Western blotting (left panel) to evaluate SUMOylation of glucocorticoid receptor (GR). To confirm that slow migrating bands were indeed SUMOylated forms of GR, a post-ischemic nuclear fraction was treated with/without SENP2c. SENP2c treatment abolished almost all slow migrating bands (right panel). GADPH and H2AX were used as cytoplasmic and nuclear markers, respectively. S, sham; I, ischemia; WB, Western blot.; Ub, ubiquitin.
Then, we selected 5 candidate SUMO substrates for further verification, GR, TFII-I, TRIM33, TIF1β, and CTIP2. The unmodified forms of all 5 proteins were detected in sham and ischemia input samples (Fig. 5B). In contrast, only slower migrating bands were present in FLAG pulldown samples, with much stronger signals in the ischemia sample. After SENP2c treatment, almost all slower migrating bands disappeared, and bands representing unmodified forms appeared (Fig. 5B). These experiments convincingly confirmed that these 5 proteins are authentic SUMO3 substrates, and that SUMOylation of these proteins was markedly increased after ischemia (Fig. 5B).
We further determined the quantitative trend across molecular weight for 4 of these proteins (GR, TFII-I, TRIM33, and TIF1β) by performing a targeted data extraction from the LC-MS/MS analysis of each gel band (Fig. 5C). TFII-I and TRIM33 were virtually undetectable in sham samples, while GR and TIF1β were detectable in sham, but highly upregulated in ischemia samples. All 4 proteins showed the highest quantity in molecular weight regions that are significantly higher than the unmodified protein molecular weight (Fig. 5C), consistent with multiple SUMO modifications.
Finally, we further characterized GR SUMOylation. SUMO3-conjugated GR was already detected in post-ischemic nuclear fractions without immunoprecipitation (Fig. 5B, input), a remarkable observation considering that only a very small fraction of a given protein is usually SUMOylated. This suggests a dramatic post-ischemic activation of GR SUMOylation. Indeed, we found that ischemia/reperfusion triggered massive SUMO conjugation and nuclear accumulation of GR (Fig. 5D).
Discussion
Here, we presented a novel SUMO transgenic mouse model (CAG-SUMO) and the first study on tissue samples to uncover the SUMO-modified proteome in a pathological state. CAG-SUMO/Emx1-Cre mice did not show any obvious phenotype. This was expected since our data demonstrated that exogenous tagged SUMOs did not disturb global SUMOylation in the brain. Indeed, the tagged SUMOs were functionally intact and processed by the endogenous SUMOylation machinery in the same way as endogenous SUMOs (Fig. 2, 3 and Supplemental Fig. I). Moreover, expression of exogenous tagged SUMO1-3 did not induce an increase in levels of SUMO-conjugated proteins, although it resulted in a rise in levels of free SUMOs (Fig. 2). These data suggest that in the brain, SUMO conjugation is controlled by the activity of SUMOylation machinery and not by levels of free SUMOs. Together, our findings demonstrate that this new transgenic mouse model is well-suited for SUMO proteomics studies in vivo.
The new SUMO transgenic mouse model has several advantages over previously published approaches to investigate the SUMO2/3-modified proteome using tissue samples 17, 18. Protein sequences of SUMO2 and SUMO3 are almost identical and cannot be differentiated by available antibodies. Since SUMO2 and SUMO3 are expressed with different tags in this new transgenic mouse, it is possible for future studies to identify individually the SUMO2- and SUMO3-modified proteome. Furthermore, since tagged SUMOs are expressed in a conditional manner, they can be expressed in any cell/organ type for which the respective Cre mouse is available, thus making this new mouse model a universal tool for characterizing the SUMO-modified proteome.
Increased SUMOylation is believed to be a protective response that shields neurons from ischemia-induced damage 19,20. Transient forebrain ischemia triggered a massive increase in levels of SUMO2/3-conjugated proteins in neurons of the resistant cortex and vulnerable hippocampal CA1 subfield (Fig. 2 and 3). Whether this post-ischemic activation of SUMO2/3-conjugation is a stress response protecting neurons in both regions needs to be established in future studies. Recently, we have generated a novel SUMO knockdown transgenic mouse that will be well-suited for these studies 21. It also needs to be verified whether transient cerebral ischemia may activate SUMOylation in non-neuronal cells such as astrocytes and endothelial cells. Notably, SUMOylation of the liver X receptor in brain astrocytes blocks inflammatory responses 22, and inflammation is a major contributing factor to ischemia-induced brain damage. Crossing our CAG-SUMO mice with mice expressing Cre in non-neuronal brain cells will help to identify the post-ischemic SUMO-modified proteome in these cells and thereby clarify this important aspect.
We have identified 91 protein candidates that exhibit a post-ischemic upregulated SUMO3 conjugation state in cortex, a brain region relatively resistant to a short interruption of blood supply (Supplemental Table I, asterisks). This high stringency list includes many known SUMO substrates, such as TIF1β, hnRNPs, TFII-I, and GR 17, 23–25. When we compared the in vivo data presented here with a previous in vitro study 13, we found that 34 out of the 91 candidates (37%) were identified in both studies. Furthermore, we confidently confirmed 5 in vivo SUMO3 substrates regulated by ischemia (Fig. 5). Taken together, these analyses confirm the validity of the approach and new SUMO mouse model, and establish the credibility of our SUMO substrate list.
Our data revealed several potentially important processes modulated by SUMO3 conjugation that may play neuroprotective roles during reperfusion. First, we provided evidence that the cross-talk between ubiquitylation and SUMOylation was activated after ischemia. Our data suggested that ubiquitin and SUMO3 are conjugated to different lysine residues in a subset of SUMO3 substrates (Fig. 5A). This implies post-ischemic activation of the SUMO-dependent ubiquitin conjugation pathway that plays pivotal roles in DNA damage repair 26. Second, IPA analysis revealed that proteins involved in post-transcriptional RNA modification are highly enriched in post-ischemia FLAG-SUMO3 pulldown samples (Supplemental Fig. IIIC). These included various isoforms of hnRNPs, a group of proteins that are involved in mRNAs splicing, stability, and translation 27. Since transcription and translation processes are dramatically impaired during and following ischemia, the post-transcriptional regulation of existing mRNAs is crucial to determining the expression pattern in this pathological state. Therefore, we speculate that SUMOylation of such a large fraction of hnRNPs after ischemia is likely related to the decision of cell survival or death.
Finally, the glucocorticoid receptor (GR) was identified as a SUMO3 target in post-ischemia samples, at a level of SUMO conjugation so pronounced that the SUMOylated GR could be identified in homogenate even without enrichment (Fig. 5D). Stress-induced GR activation is associated with increased cell damage triggered by transient cerebral ischemia 28. GR function is modulated by post-translational modifications, and phosphorylation activates GR SUMOylation, resulting in repression of transcriptional activity 24. Taken together, these observations suggest that the massive post-ischemic activation of GR SUMOylation is a protective stress response.
In summary, we report here the first proteomics analysis of SUMO3-conjugated proteins in tissue samples from a pathological state, brain ischemia. We identified several pathways modulated by SUMOylation in the post-ischemic brain that warrant future investigations, since they could be therapeutic targets for neuroprotection in brain ischemia. Since a large portion of identified SUMO targets were nuclear proteins involved in gene expression, genome stability, and RNA processing, we expect activation of SUMOylation to have long-lasting effects on post-ischemic neurons. The new conditional SUMO transgenic mouse and the highly stringent purification approach developed in the present study provide an invaluable platform for in-depth analysis of the SUMO-modified proteome in vivo in physiological or pathological states under investigation.
Supplementary Material
Acknowledgments
The authors thank Laura Dubois and Meredith Turner for their excellent technical support, Dr. Christoph Harms and Dr. Matthew Foster for helpful discussions, and Kathy Gage for excellent editorial contribution.
Sources of Funding: This study was supported by a DREAM Innovation Grant from the Department of Anesthesiology and an American Stroke Association Scientist Development Grant 12SDG11950003 to W.Y., and NIH grants HL095552 and NS081299 to W.P.
Footnotes
Disclosures: None
References
- 1.Flotho A, Melchior F. Sumoylation: A regulatory protein modification in health and disease. Annu Rev Biochem. 2013;82:357–385. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- 2.Hay RT. Sumo: A history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 3.Hickey CM, Wilson NR, Hochstrasser M. Function and regulation of sumo proteases. Nat Rev Mol Cell Biol. 2012;13:755–766. doi: 10.1038/nrm3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ayaydin F, Dasso M. Distinct in vivo dynamics of vertebrate sumo paralogues. Mol Biol Cell. 2004;15:5208–5218. doi: 10.1091/mbc.E04-07-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson ES. Protein modification by sumo. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 6.Gareau JR, Lima CD. The sumo pathway: Emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol. 2010;11:861–871. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kho C, Lee A, Jeong D, Oh JG, Chaanine AH, Kizana E, et al. Sumo1-dependent modulation of serca2a in heart failure. Nature. 2011;477:601–605. doi: 10.1038/nature10407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang W, Sheng H, Warner DS, Paschen W. Transient global cerebral ischemia induces a massive increase in protein sumoylation. J Cereb Blood Flow Metab. 2008;28:269–279. doi: 10.1038/sj.jcbfm.9600523. [DOI] [PubMed] [Google Scholar]
- 9.Yang W, Sheng H, Warner DS, Paschen W. Transient focal cerebral ischemia induces a dramatic activation of small ubiquitin-like modifier conjugation. J Cereb Blood Flow Metab. 2008;28:892–896. doi: 10.1038/sj.jcbfm.9600601. [DOI] [PubMed] [Google Scholar]
- 10.Tirard M, Hsiao HH, Nikolov M, Urlaub H, Melchior F, Brose N. In vivo localization and identification of sumoylated proteins in the brain of his6-ha-sumo1 knock-in mice. Proc Natl Acad Sci USA. 2012;109:21122–21127. doi: 10.1073/pnas.1215366110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Felipe P, Luke GA, Hughes LE, Gani D, Halpin C, Ryan MD. E unum pluribus: Multiple proteins from a self-processing polyprotein. Trends Biotechnol. 2006;24:68–75. doi: 10.1016/j.tibtech.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 12.Gorski JA, Talley T, Qiu M, Puelles L, Rubenstein JL, Jones KR. Cortical excitatory neurons and glia, but not gabaergic neurons, are produced in the emx1-expressing lineage. J Neurosci. 2002;22:6309–6314. doi: 10.1523/JNEUROSCI.22-15-06309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang W, Thompson JW, Wang Z, Wang L, Sheng H, Foster MW, et al. Analysis of oxygen/glucose-deprivation-induced changes in sumo3 conjugation using silac-based quantitative proteomics. J Proteome Res. 2012;11:1108–1117. doi: 10.1021/pr200834f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheng H, Laskowitz DT, Mackensen GB, Kudo M, Pearlstein RD, Warner DS. Apolipoprotein e deficiency worsens outcome from global cerebral ischemia in the mouse. Stroke. 1999;30:1118–1124. doi: 10.1161/01.str.30.5.1118. [DOI] [PubMed] [Google Scholar]
- 15.Matic I, van Hagen M, Schimmel J, Macek B, Ogg SC, Tatham MH, et al. In vivo identification of human small ubiquitin-like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy. Mol Cell Proteomics. 2008;7:132–144. doi: 10.1074/mcp.M700173-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schimmel J, Larsen KM, Matic I, van Hagen M, Cox J, Mann M, et al. The ubiquitin-proteasome system is a key component of the sumo-2/3 cycle. Mol Cell Proteomics. 2008;7:2107–2122. doi: 10.1074/mcp.M800025-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Becker J, Barysch SV, Karaca S, Dittner C, Hsiao HH, Berriel DM, et al. Detecting endogenous sumo targets in mammalian cells and tissues. Nat Struct Mol Biol. 2013;20:525–531. doi: 10.1038/nsmb.2526. [DOI] [PubMed] [Google Scholar]
- 18.Bruderer R, Tatham MH, Plechanovova A, Matic I, Garg AK, Hay RT. Purification and identification of endogenous polysumo conjugates. EMBO Rep. 2011;12:142–148. doi: 10.1038/embor.2010.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Datwyler AL, Lattig-Tunnemann G, Yang W, Paschen W, Lee SL, Dirnagl U, et al. Sumo2/3 conjugation is an endogenous neuroprotective mechanism. J Cereb Blood Flow Metab. 2011;31:2152–2159. doi: 10.1038/jcbfm.2011.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee YJ, Mou Y, Maric D, Klimanis D, Auh S, Hallenbeck JM. Elevated global sumoylation in ubc9 transgenic mice protects their brains against focal cerebral ischemic damage. PLoS One. 2011;6:e25852. doi: 10.1371/journal.pone.0025852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang L, Rodriguiz RM, Wetsel WC, Sheng H, Zhao S, Liu X, et al. Neuron-specific SUMO1-3 knockdown in mice impairs episodic and fear memories. J Psychiatr Neurosci. doi: 10.1503/jpn.130148. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JH, Park SM, Kim OS, Lee CS, Woo JH, Park SJ, et al. Differential sumoylation of lxralpha and lxrbeta mediates transrepression of stat1 inflammatory signaling in ifn-gamma-stimulated brain astrocytes. Mol Cell. 2009;35:806–817. doi: 10.1016/j.molcel.2009.07.021. [DOI] [PubMed] [Google Scholar]
- 23.Li T, Evdokimov E, Shen RF, Chao CC, Tekle E, Wang T, et al. Sumoylation of heterogeneous nuclear ribonucleoproteins, zinc finger proteins, and nuclear pore complex proteins: A proteomic analysis. Proc Natl Acad Sci USA. 2004;101:8551–8556. doi: 10.1073/pnas.0402889101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davies L, Karthikeyan N, Lynch JT, Sial EA, Gkourtsa A, Demonacos C, et al. Cross talk of signaling pathways in the regulation of the glucocorticoid receptor function. Mol Endocrinol. 2008;22:1331–1344. doi: 10.1210/me.2007-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vassileva MT, Matunis MJ. Sumo modification of heterogeneous nuclear ribonucleoproteins. Mol Cell Biol. 2004;24:3623–3632. doi: 10.1128/MCB.24.9.3623-3632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prudden J, Pebernard S, Raffa G, Slavin DA, Perry JJ, Tainer JA, et al. Sumo-targeted ubiquitin ligases in genome stability. EMBO J. 2007;26:4089–4101. doi: 10.1038/sj.emboj.7601838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dreyfuss G, Kim VN, Kataoka N. Messenger-rna-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 28.Balkaya M, Prinz V, Custodis F, Gertz K, Kronenberg G, Kroeber J, et al. Stress worsens endothelial function and ischemic stroke via glucocorticoids. Stroke. 2011;42:3258–3264. doi: 10.1161/STROKEAHA.110.607705. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.