Abstract
Objective
Inappropriate transcriptional activation of innate immunity is a pathologic feature of several cardio-metabolic disorders but little is known of inflammatory modulation of long intergenic non-coding RNAs (lincRNAs) in disease-relevant human tissues.
Approach and Results
We applied deep RNA sequencing (RNA-seq; >500 million filtered reads per sample) to blood and adipose during low-dose experimental endotoxemia (LPS) in a healthy human, with targeted replication in separate individuals undergoing endotoxemia (n=6), to identify inflammatory lincRNAs. A subset of these lincRNAs was examined for expression in adipocytes and monocytes, modulation in adipose in obesity, and overlap with genome-wide association study signals for inflammatory and cardio-metabolic traits. Of a stringent set of 4,284 lincRNAs, about 11–22% were expressed with 201 and 56 lincRNAs modulated by LPS in blood or adipose, respectively. Tissue-specific expression of a subset of six LPS-lincRNAs was replicated with LPS-modulation confirmed for all three expressed in blood and two of four expressed in adipose. The broader generalizabilty of findings in blood of subject A was confirmed by RNA-seq in seven additional subjects. We confirmed adipocytes and monocytes as potential cell sources of selective LPS-regulated lincRNAs and two of these, linc-DMRT2 (P=0.002) and linc-TP53I13 (P=0.01), were suppressed in adipose in obesity. Finally, we provide examples of LPS-modulated lincRNAs that overlap SNPs that are associated with cardiometabolic traits.
Conclusion
Our findings provide novel insights into tissue-level, inflammatory transcriptome regulation in cardio-metabolic diseases. These are complementary to more usual approaches limited to interrogation of DNA variations.
Keywords: Genomics, cardio-metabolic disease, RNA-seq, alternative splicing, linc-RNA
INTRODUCTION
A portion of the genetic basis of complex diseases is apparent from recent genome-wide association studies (GWAS). It is evident, however, that scanning the genome can identify only a portion of the genomic influence on many heritable disorders1. Transcriptomics provides a promising avenue for a more complete understanding of the functional elements of the genome that mediate tissue-specific disease manifestations. Indeed, the advent of high-throughput RNA sequencing (RNA-seq) has demonstrated the existence of a far greater and tissue-specific transcriptomic complexity than previously estimated2–7.
The conventional view of the genome as simply containing structural and protein-coding genes is evolving rapidly with the realization of the diversity of RNA species in mammalian cells4–6, 8. Regions previously thought to be silent are transcribed into a wide-range of functional transcripts broadly known as non-coding RNAs (ncRNAs). One class of such ncRNAs loosely known as long (>200nt) noncoding RNAs (lncRNAs) are distinct from more studied microRNAs (miRs) and as yet are poorly understood5–7. Although debate continues regarding the proportion of lncRNAs that are likely to be functional6, 7, many have messenger RNA (mRNA)-like features (e.g., polyA tail and exons/introns)7 and several important examples have critical functions in cell biology (e.g., the cis-regulatory Xist9, Hotair10, and Mistral11) including ANRIL12, the likely causal trans-regulatory “gene” at the common 9p21 vascular disease locus. Because lncRNAs are poorly characterized in terms of evolution, conservation, secondary structure and functions5, 7, there is a dearth of understanding of lncRNAs in human physiology and disease. To simplify their study separately from other RNA species, recent focus often has been on the subset of lncRNAs known as long “intergenic” (or intervening) lncRNA (i.e., lincRNAs) that do not overlap with exons of protein-coding genes5, 7, 8.
Inappropriate and sustained tissue-specific transcriptional activation of innate immunity by diet and lifestyle factors in genetically prone individuals is a pathologic feature of several cardio-metabolic disorders including atherosclerosis, obesity and insulin resistance. Although early data on lncRNA modulation during activation of innate immunity are emerging13, 14, little is known in human cells and tissues. We utilize low-dose experimental endotoxemia (lipopolysaccharide, or LPS) to activate toll-like receptor 4 (TLR4) signaling in humans as a model to study in vivo inflammatory cardio-metabolic responses15–20. Epidemiological21, 22, clinical23 and experimental19 studies in human as well as disease modeling in rodents24 support the relevance of this model. In our prior studies, we reported endotoxemia-induced metabolic dyslipidemia18 and systemic insulin resistance19 coincident with adipose inflammation19, 25. Prior studies also suggest cell-specific transcriptome16, 25 modulation, at least of protein-coding genes, during endotoxemia.
Recently, a reciprocal pattern of inflammatory lincRNA induction with subsequent negative feedback on immune signaling has been described13, 14, suggesting a functional role of specific lincRNAs in regulating innate immunity. Here, we applied very-deep RNA-seq to whole blood and adipose during experimental endotoxemia (LPS 1 ng/kg intravenous bolus) in a participant enrolled in the Genetics of Evoked-responses to Niacin and Endotoxemia (GENE) study26, with targeted replication in additional GENE subjects, in order to identify inflammatory lincRNAs that may modulate complex cardio-metabolic disorders. We report novel tissue-specific, inflammatory regulation of specific lincRNAs, examine their genomic features, probe potential cellular origins and explore their potential relevance in human cardio-metabolic diseases.
RESULTS
Overview, clinical characteristics and RNA-seq data alignment and filtering
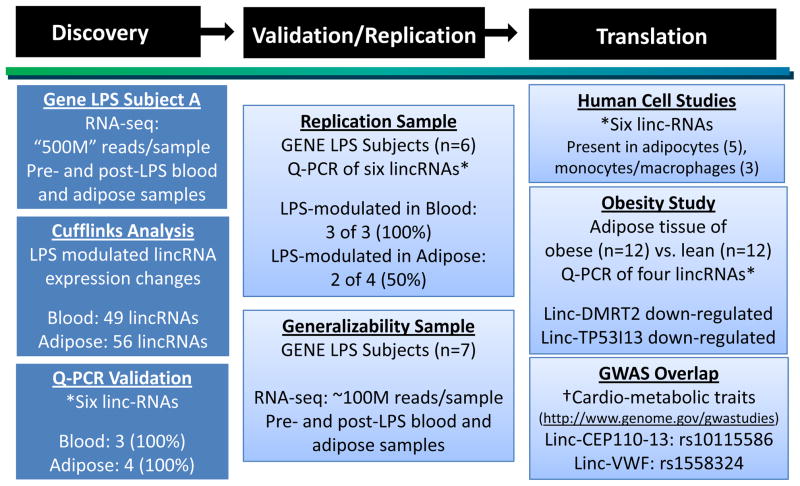
An overview of study design and results is shown in Figure 1. The GENE study is a single-center U.Penn-based National Institute of Health-sponsored endotoxemia protocol (n=284, 33% African Americans, age 18–45; NIH.gov clinical trial NCT00953667)26. Whole blood RNA samples were collected at baseline and multiple time points post-LPS. Gluteal subcutaneous fat tissue was biopsied and snap-frozen pre- and post-LPS as described19, 25. In this report, we focus initially on a self-reported European Ancestry (EA) individual (subject A) for deep RNA-seq. To enrich discovery, subject A was chosen from those with above-median inflammatory response (Table 1). Our primary analysis was of whole blood and adipose RNA-seq data; RNA-seq data was generated also in peripheral blood mononuclear cells (PBMCs) to compare to findings for whole blood. Selective LPS-modulated events in blood and adipose of subject A were assessed for replication in independent GENE participants (n=6, subjects B-G) also chosen from those with above-average inflammatory response (Table 1). The broader generalizabilty of findings in subject A was examined by RNA-seq in seven additional GENE subjects (Suppl Table I). Based on previous mRNA profiling19, 25 we selected baseline and 2-hour blood samples, and baseline and 4-hour adipose samples for RNA-seq.
Figure 1. Overview of study design and results.
Discovery (subject A) and replication subjects (B–G) were selected from those participants in the Genetics of Evoked-response to Niacin and Endotoxemia (GENE) study with above median inflammatory cytokine responses during endotoxemia (Table 1). Deep RNA-sequencing generated >500 million (“500M”) reads per sample.
*Of six LPS-regulated lincRNAs selected for follow-up, three were expressed in blood and four expressed in adipose. Quantitative real-time PCR (qRT-PCR) assays were used for validation (subject A), replication (subjects B-G), human cell studies and a pilot case-control study of obesity (the four adipose lincRNAs were tested). †All LPS-regulated lincRNAs (49 in blood and 56 in adipose) were tested relative to expressed non-LPS modulated lincRNAs (182 blood and 413 adipose) for enrichment with SNPs that have genome-wide significant association with inflammatory and cardio-metabolic traits in the NHGRI GWAS catalogue (http://www.genome.gov/gwastudies). Linc-CEP110-13, induced by LPS, overlaps a SNP (rs10115586) that associates with N-Glycosylation of circulating IgG. Linc-VWF, a LPS-induced lincRNA, contained a SNP (rs1558324) associated with mean platelet volume, a predictor of cardiovascular disease.
Table 1.
Clinical characteristics of study subjects (A–G) and all GENE study European ancestry participants (A) at baseline and (B) during endotoxemia
| (A) | Full Gene Sample | |||
|---|---|---|---|---|
| Male EA N=99 Mean (SD) |
Female EA N=93 Mean (SD) |
Subject A* (Female) | Subjects B-G† Mean (SD) (3 Females & 3 Males) |
|
| Age (Years) | 25.6 (6.7) | 23.9 (4.0) | ||
| Total Cholesterol (mg/dL) | 151 (33) | 150 (26) | 147 | 141 (26) |
| HDL-Cholesterol (mg/dL) | 47 (10) | 55 (13) | 66 | 61 (17) |
| Triglycerides (mg/dL) | 84 (45) | 78 (33) | 63 | 65 (21) |
| LDL-Cholesterol (mg/dL) | 87 (30) | 79 (24) | 68 | 67 (22) |
| Systolic Blood Pressure (mmHg) | 113 (11) | 109 (10) | 112 | 111 914) |
| Diastolic Blood Pressure (mmHg) | 65 (9.0) | 62 (8.0) | 66 | 64 (6.4) |
| Glucose (mg/dL) | 87 (13) | 82 (16) | 79 | 85 (5.9) |
| (B) | Male EA Mean (SD) |
Female EA Mean (SD) |
Subject A* | Subjects B-G† Mean (SD) |
|
|---|---|---|---|---|---|
| IL-6 (pg/ml) | Baseline | 2.5 (1.5) | 3.2 (1.7) | 3.9 | 2.1 (2.1) |
| Peak (2hour) | 152 (142) | 198 (181) | 294 | 292 (280) | |
| Fold Change | 60 | 62 | 76 | 218 | |
| Percentile of peak | 84th | ||||
| TNFα (pg/ml) | Baseline | 1.3 (0.8) | 1.3 (0.8) | 0.8 | 1.3 (0.9) |
| Peak (2 hour) | 50 (35) | 58 (52) | 50 | 77 (30) | |
| Fold Change | 38 | 44 | 65 | 79 | |
| Percentile of peak | 64th | ||||
| CRP (mg/L) | Baseline | 1.1 (3.6) | 1.5 (2.6) | 0.8 | 0.5 (0.4) |
| Peak (24 hour) | 18 (9.0) | 16 (8.1) | 20 | 18 (5.8) | |
| Percentile of peak | 67th | ||||
| SAA (mg/L) | Baseline | 7.1 (32) | 7.3 (31) | 2.6 | 3.2 (1.3) |
| Peak (24 hour) | 95 (43) | 82 (46) | 81 | 87 (30) | |
| Percentile of peak | 52nd | ||||
| Temp. (°F) | Baseline | 97.6 (0.4) | 97.9 (0.5) | 98.6 | 97.5 (0.5) |
| Peak | 99.4 (0.9) | 99.5 (0.8) | 100.2 | 99.3 (0.5) | |
| Percentile of peak | 87th |
GENE=Genetics of Evoked-responses to Niacin and Endotoxemia; HDL=high-density lipoprotein; LDL=low density lipoprotein; IL=interleukin; TNF=tumor necrosis factor; CRP=C reactive protein; SAA=serum amyloid A. TEMP=temperature.
We obtained 1,400 million (M)/976M reads and 912M/1,040M reads for the pre-/post-LPS blood and adipose samples, respectively. The data were aligned and filtered as described in Methods. We obtained a high mapping rate with 95% and 98% of reads mapped to the reference genome for pre- and post-LPS blood samples and 85% and 82% of reads pre- and post-LPS adipose samples. Of these, 60% and 63% of reads in blood and 53% and 49% of reads in adipose were uniquely mapped and properly filtered (Suppl Table II A). For all blood RNA-seq samples (subject A and others), we eliminated hemoglobin gene sequences (located on chromosomes 11 and 16) because these sequences are uninformative (for LPS modulation) yet dominate the blood RNA-seq data due to their high abundance. In our analysis, we only considered reads from autosomal and sex chromosomes, and this left 586/881M filtered reads for pre-/post-LPS blood samples and 482M/505M filtered reads for pre-/post-LPS adipose samples, respectively. For ease of notation, we denote these four datasets as 500M RNA-seq data. In lower-depth RNA-Seq of subject A, we obtained 116M/138M and 67M/65M reads for the pre-/post-LPS blood and adipose samples, respectively (~50M or “low” RNA-seq depth data) (Suppl Table II A). The FPKM expression levels for the deeply sequenced 500M samples from subject A were highly correlated (correlation coefficients 0.92 to 0.98) with the corresponding “low” depth replicate samples. For PBMCs of subject A, we obtained 102M and 129M reads before and after LPS respectively (Suppl Table II A). In RNA-seq of blood of seven additional GENE subjects, we generated 152M–230M reads (Suppl Table II B).
RNA-seq of lincRNAs in blood and adipose
In order to limit spurious findings, we focused on the “stringent” subset of 4,284 lincRNAs with at least one isoform reconstructed in at least two human tissues or by two assemblers in the same tissue from the full dataset of 8,179 lincRNAs identified through RNA-seq by Cabili et al8. For consistency across tissues, we included lincRNAs that were expressed at FPKM >0.
Sequencing depth for analysis of human lincRNAs
Recently, we analyzed sequencing depth requirements for detection of differentially expressed (DE) protein-coding genes in human adipose and found that substantially greater depth was required to detect DE than simple basal expression27. Because expression of lincRNAs is generally lower than protein-coding genes, our first step was to determine by empirical evaluations the sequence depth required to assess DE (e.g., LPS-modulation). Briefly, treating the 500M adipose datasets as the gold standard, we evaluated percentages of expressed lincRNAs and DE lincRNAs detected at lower sequencing depths (using randomly selected reads with incrementally lower sequencing depths). Although ~75% of expressed lincRNAs can be detected at sequencing depths of 300M, we found that 400M read-depth was required to detect DE and even higher depth to detect DE of lincRNAs with lower expression, e.g., 450M depth for lincRNAs < 25th percentile expression (Suppl Figure I). Thus, our deeply-sequenced 500M datasets provided substantial depth for detection of DE lincRNAs even for those with low expression. By contrast, in subject A “low” depth RNA-seq replicates, only 13% of DE lincRNAs in adipose were detected relative to 500M data.
LincRNA expression in blood and adipose
Of the 4,284 human lincRNAs, ~22% were expressed (at FPKM>0 and “OK” status in cuffdiff analysis)28 in blood and ~11% in adipose of 500M subject A samples; 931 and 453, respectively, prior to LPS and 894 and 445, respectively, post-LPS. In the “low” RNA-seq data for subject A, only 409 and 402 and 101 and 100 lincRNAs were expressed pre- and post-LPS in blood and adipose, respectively. For protein-coding genes, we detected 14,564 and 11,464, respectively, prior to LPS and 14,521 and 11,418, respectively, post-LPS in blood and adipose.
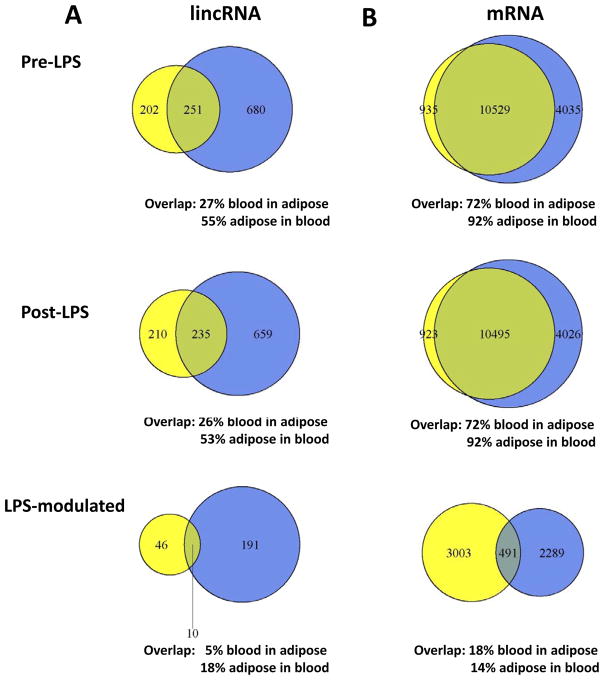
Cabili et al.8 and others5–7 suggest that human lincRNAs have more tissue-specific expression than protein-coding genes. For lincRNAs and protein coding mRNAs expressed in either blood or adipose of subject A, we counted the number that overlapped between the two tissues (Figure 2). Among 931 in blood and 453 lincRNAs in adipose prior to LPS, 251 overlapped (27% for blood; 55% for adipose) (Figure 2A). Of 894 lincRNAs in blood and 445 in adipose after LPS, 235 overlapped (26% for blood; 53% for adipose). Compared to protein-coding genes (Figure 2B), the percentages of blood-adipose overlapping lincRNAs were significantly lower (e.g., pre-LPS blood, 27% vs. 73%, Fisher’s exact test P< 2.2×10−16; and adipose 55% vs. 89%, P< 2.2×10−16), suggesting greater tissue-specificity for lincRNAs than protein-coding genes. The differences were less pronounced but still significant when analysis was restricted to protein-coding genes with low expression levels, similar to lincRNAs (e.g., pre-LPS, blood 27% vs. 55%, Fisher’s exact test P< 2.2×10−16; adipose 55% vs. 61%, P=0.018).
Figure 2.
Venn diagram for expression, tissue overlap and LPS-modulation of (A) lincRNAs and (B) protein-coding genes in adipose (yellow, left) and blood (blue, right) of subject A.
LPS-modulation of lincRNAs
In subject A, we detected a modest number of LPS-regulated lincRNAs, 201 and 56 in blood and adipose, respectively (Figure 2A, details in Suppl Table III A & B). The majority was down-regulated in both blood (n=150, 75%) and adipose (n=38, 68%). This pattern of greater LPS down-regulation was similar to that for LPS-modulated protein coding genes (n=1,996, 72% in blood and 3,242, 93% in adipose, respectively) and is consistent with prior published data 16.
One concern when examining apparent LPS-modulation of gene expression in tissues with mixed cell content (e.g., whole blood) is that LPS-modulation of lincRNAs might be due to change in cell composition rather than due to actual LPS-regulation of lincRNA expression 29, 30. We addressed this concern indirectly for whole blood findings by generating RNA-seq data before and after LPS in PBMCs of subject A at the same time points as the whole blood RNA-seq (pre- and 2-hours post-LPS). If LPS-driven change in blood cell composition was the predominant basis for LPS-modulation in whole blood, then we would expect much less LPS-modulation of lincRNAs in the PBMC samples because of PBMCs more homogenous, monocyte-rich, cell composition. However, this was not the case. We observed LPS-modulation of lincRNAs in PBMCs to a degree that was very similar to that observed in whole blood (18.4% and 21.6% in PBMCs and whole blood, respectively) (Suppl Table IV). Indeed, there was substantial overlap (81%, 82% and 48%) of pre- and post-LPS lincRNA expression and LPS-modulation respectively in PBMCs and whole blood.
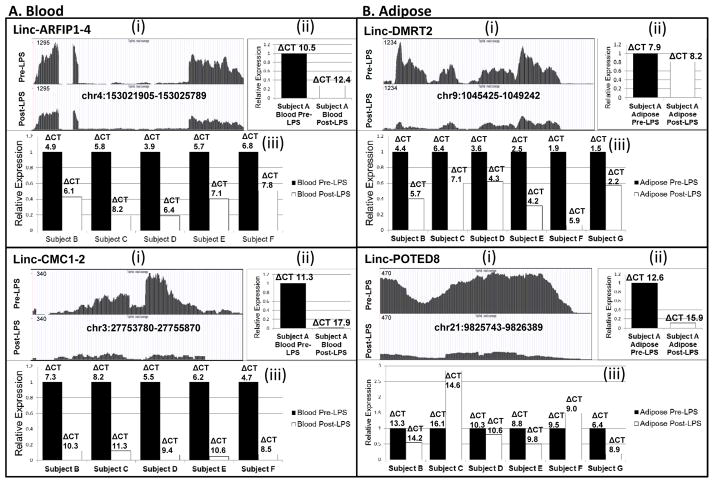
In preliminary translation, we selected six LPS-regulated lincRNAs in subject A for validation by qRT-PCR. These were chosen to represent a range of FPKM expression and statistical significance of LPS modulation as well as presence either in blood, adipose or both; three were expressed in blood and four were expressed in adipose. We confirmed tissue patterns of expression and validated the direction of LPS-modulation for all six lincRNAs in subject A blood and adipose. (Panels (i) and (ii), Figure 3 and Suppl Figure II).
Figure 3. Validation and replication for LPS modulated lincRNAs.
Data for (A) blood lincRNAs, linc-ARFIP1-4 and linc-CMC1-2 as well as (B) adipose lincRNAs, linc-DMRT2 and linc-POTED8 are shown. (i) Genome browser views of RNA-seq data before and after LPS in subject A. (ii) Quantitative real-time PCR (qRT-PCR) validation of lincRNA in tissue of subject A. (iii) qRT-PCR replication of lincRNA in tissues of subject B-G (subject G blood sample failed). For (ii) and (iii) expression data are presented as bar-graphs of relative fold-change compared with pre-LPS. The delta CT’s represent the median cycle threshold for lincRNAs relative to B-actin mRNA as the reference in each sample.
This same subset of lincRNAs was examined by qRT-PCR for replication of LPS-modulation in separate GENE study subjects (n=6) (Table 1). Tissue-specific expression was confirmed for these lincRNAs in all subjects and the direction of LPS-modulation replicated for three of three blood-lincRNAs (linc-ARFIP1-4, linc-CMC1-2, linc-TP53I13) and two of four adipose-lincRNAs (linc-DMRT2, linc-POTED8) (Panel (iii), Figure 3 and Suppl Figure II). This suggests generalizability of LPS-modulation, particularly in blood, for a subset of LPS-regulated lincRNAs identified in subject A.
Tissue-specificity of LPS-modulated lincRNAs
For subject A LPS-regulated lincRNAs (Suppl Table III), there was minor overlap between blood and adipose (Figure 2A) (10 lincRNAs; 5% of blood in adipose, 18% of adipose in blood; linc-TP53I13, linc-POTED-8; linc-ARIH2, linc-STIM2-1, linc-TACC3, linc-ACSL1-1, linc-HEATR2-2, linc-CCND1-4, lincCGNL1-2, linc-TMEM105-2). This pattern of tissue-specificity was generally similar to LPS-modulated protein-coding genes (Figure 2B). Compared to such protein-coding genes, the percentages of blood-adipose overlapping LPS-regulated lincRNAs were slightly lower (e.g., 5% vs. 18% of blood in adipose, Fisher’s exact test P = 3.7×10−7), but similar when compared to a subset of LPS-modulated protein coding genes with a similar (low) level of expression (e.g., 5% vs. 8% of blood in adipose, P = 0.14). In further support of tissue specificity of LPS-modulated lincRNAs, we found no correlation between blood and adipose in the degree of LPS modulation of all lincRNAs (expressed either as fold-change or absolute change) (data not shown). Thus, LPS-regulated lincRNAs are tissue-specific in a manner generally consistent with that observed for LPS-modulated protein-coding genes.
Generalizability of RNA-seq findings for lincRNAs
In order to address the broader generalizability of RNA-seq patterns in subject A, we generated in seven additional GENE study participants (Suppl Table I) additional RNA-seq data (Suppl Table II B) for the same time points pre-and post-LPS in blood and compared findings to that in subject A. These analyses of additional subjects revealed; (a) substantial overlap between detectable lincRNAs to that in subject A (~94% and 93%) before and after LPS respectively and (b) substantial overlap in LPS-modulated lincRNAs to that in subject A (~79%) (Table 2). These RNA-seq data provide support for the broader generalizability of lincRNA expression and LPS-modulation despite the confounding influence of lower RNA-seq depth in the additional blood samples and an expected heterogeneity in LPS clinical and transcriptomic response across humans16, 26.
Table 2.
Numbers of whole blood expressed and LPS-modulated lincRNAs at RNA-seq in GENE study subject A compared to additional GENE study subjects (n=7).
| Subject A* | Additional Subjects (n=7)* | Overlap | # Percent Overlap | |
|---|---|---|---|---|
| Pre-LPS | 931 | 504 | 472 | 94% of 7-subject in subject A |
| Post-LPS | 894 | 504 | 472 | 93% of 7-subject in subject A |
| LPS-modulated | 201 | 24 | 19 | 79% of 7-subject in subject A |
RNA-seq depth was substantially deeper in whole blood of subject A (“500M” data, see Suppl Table 2A) than in whole blood of the additional GENE subjects (~81–165M data, Suppl T2B). Thus, as expected, the number of lincRNAs detected and in particular LPS-modulated lincRNAs (see Suppl Figure 1) in the additional subject whole blood is lower than in subject A.
Of lincRNAs detected and LPS-modulated in the whole blood of the seven additional GENE subjects (“7-subjects”), there was substantial overlap with those detected and LPS-modulated in the whole blood of subject A.
Conservation of LPS-regulated lincRNAs
Recent data suggest rapid evolution and limited sequence conservation of lincRNAs5, 8. This questions the functional importance for poorly conserved lincRNAs but also raises the possibility of functions not dependent on primary sequence7. Using primate phastCons scores from 46-way vertebrate genome alignment31 (available in conservation track, UCSC genome browser; http://genome.ucsc.edu/), expressed lincRNAs were markedly less conserved than protein-coding genes while, in adipose but not blood, lincRNAs were slightly more conserved than intergenic regions (Suppl Figure III). LPS-regulated lincRNAs were not more conserved than non-LPS-regulated (data not shown).
Transcription factor binding sites of lincRNAs
We hypothesized that if LPS modulation of lincRNAs reflects evolutionary conserved functions down-stream of canonical TLR4 signaling, then LPS-regulated (vs. non-LPS-regulated) lincRNAs would more likely harbor upstream sequences with active promoters and that these sequences would be enriched for NFKB transcription factor binding. To test this hypothesis, we obtained the motifs that correspond to NF-κB family members NFKB1, NFKB2, RELA, RELB and REL from the TRANSFAC database (version 2012.3) 32. Sequences encompassing 2.5kb upstream and 300bp downstream of a lincRNA’s start site were searched for motif occurrences using FIMO33. Motifs for NFKB1 were enriched in LPS-regulated lincRNAs in blood (Fisher’s exact test P = 0.005). We also calculated the number of motifs in the 2.8kb region for each lincRNA that contained at least one motif. For the combination of all NF-κB family transcription factors, LPS-regulated lincRNAs had more motifs than non-LPS-regulated lincRNAs in blood (one-sided two-sample t-test P=0.037).
Translational and genetic studies of LPS-regulated linc-RNAs
Cell source and modulation in obesity
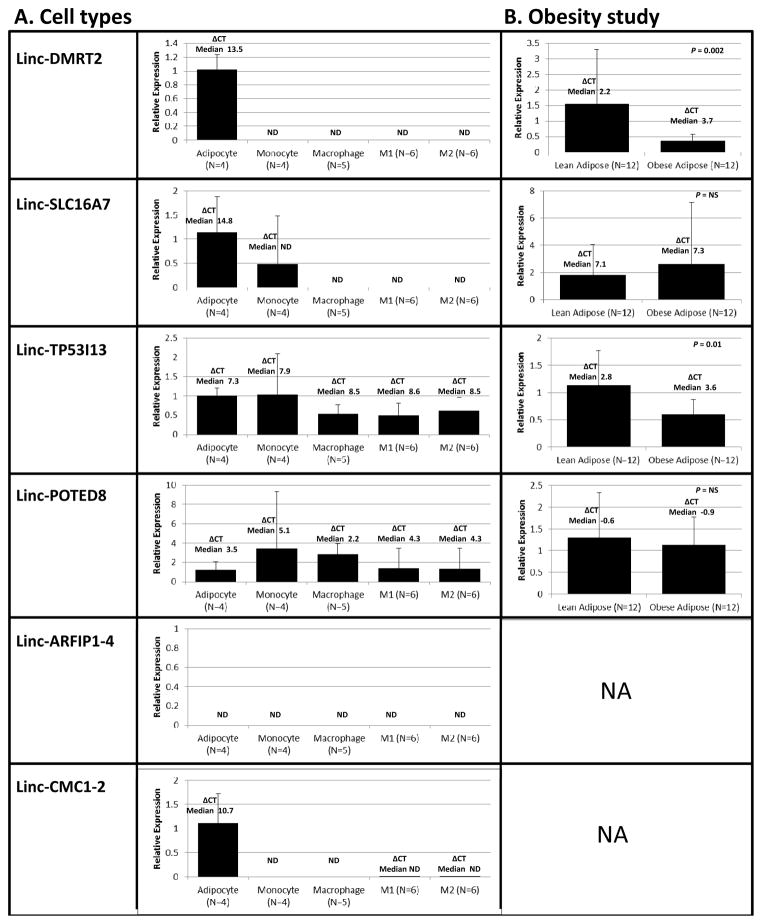
For LPS-modulated lincRNAs selected for validation, we tested expression in vitro in human cell-types (adipocytes, monocytes and macrophages) that are functionally implicated in cardio-metabolic diseases. Adipocytes expressed relatively high levels of several lincRNAs (linc-DMRT2, linc-SLC16A7, linc-CMC1-2). Consistent with expression in both blood and adipose, linc-TP53I13 and linc-POTED8 were found in monocytes and macrophages as well as in adipocytes (Figure 4A). Linc-ARFIP1-4 and linc-CMC1-2 were not expressed in monocytes suggesting an alternative blood-cell source (e.g., platelets or neutrophils) for in vivo expression in blood.
Figure 4. Translational studies of selective LPS-modulated lincRNAs.
(A) Human cells; expression of linc-DMRT2, linc-SLC16A7, linc-TP53I1, linc-POTED8, linc-ARFIP1-4, linc-CMC1-2 (by qRT-PCR), in adipocytes, primary human monocytes, macrophages, M1-type macrophages (M1), and M-2 type macrophages (M2). LincRNA expression data are presented as bar graphs of the mean (SD) fold-change in in monocytes, macrophages and M1 or M2 macrophages relative to adipocyte expression; the delta CT’s represent the median cycle threshold for lincRNAs relative to B-actin mRNA as the reference in each sample; (B) Adipose of lean vs. obese subjects; expression (by qRT-PCR) of linc-DMRT2, linc-SLC16A7, linc-TP53I1, linc-POTED8 in subcutaneous adipose of obese (n=12) compared to lean (n=12) humans. LincRNA expression data are presented as bar graphs of the mean (SD) fold-change in obese vs. lean adipose tissue. Linc-DMRT2 (P=0.002) and linc-TP53I1 (P=0.01) expression are suppressed in obesity (Mann Whitney U non-parametric test on the delta CTs). ND=not detected. NA=not assessed.
We explored disease relevance in obesity of the four selected adipose-expressed lincRNAs and found that linc-DMRT2 (P=0.002) and linc-TP53I13 (P=0.01) were down-regulated in obese adipose (Figure 4B).
Overlap of LPS-modulated lincRNAs with GWAS loci for cardio-metabolic traits
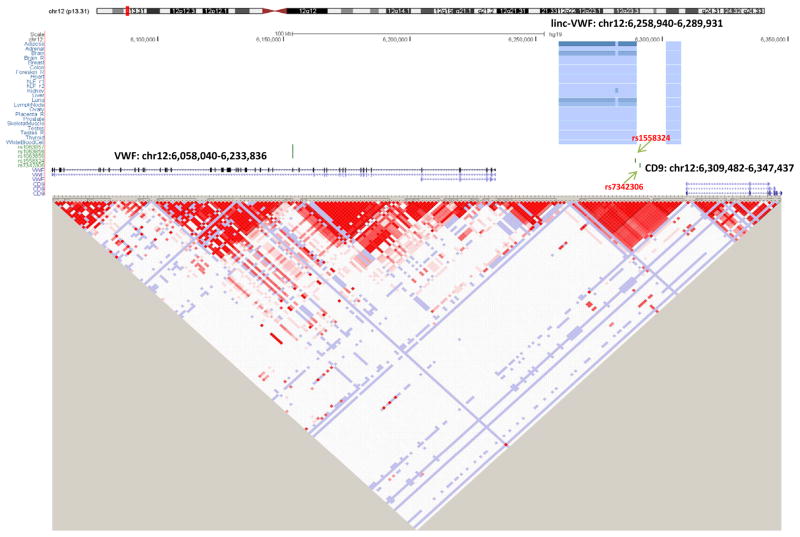
In order to illustrate the potential for RNA-seq to reveal lincRNA that may underlie GWAS disease associations, we interrogated established SNPs for inflammatory and cardio-metabolic traits using the NHGRI GWAS catalogue (http://www.genome.gov/gwastudies; Suppl Table V) for their overlap with LPS-regulated lincRNAs. First, we identified overlap between linc-CEP110-13 (chr9: 120521860-120639328) and rs10115586, a SNP in intron 1 of linc-CEP110-13 that is associated with N-Glycosylation of immunoglobulin G (IgG) 34. Linc-CEP110-13 is LPS-induced in adipose (P=0.0028, FDR adjusted P=0.016; Suppl Table III B) with a similar trend in blood (P=0.016, FDR adjusted P=0.068). rs10115586 lies 79kb downstream of TLR4, the LPS receptor, but is not in LD (r2 <0.2; http://www.sph.umich.edu/csg/abecasis/MACH/download/1000G.2012-03-14.html, 1000 genome data) with SNPs in TLR4. We also found overlap of linc-VWF (chr12: 6258940-6289931), with rs1558324 a SNP in intron 1 of linc-VWF that is associated with mean platelet volume (P=1.55×10−21, Gieger et al35), a biomarker of platelet function that predicts MI36. A second SNP (rs7342306) near linc-VWF (912 bp 5′) had a strong signal for platelet count (P=4.29×10−11, Gieger et al35). Linc-VWF is induced in adipose (Suppl Table III B) but not expressed in our blood RNA-seq data. Notably, these linc-VWF SNPs have no linkage disequilibrium (r2 <0.2) (Figure 5) with SNPs (e.g., rs1063856 or rs1063857) in VWF (located ~105 kb upstream) that are associated with circulating VWF levels37 or with SNPs in CD9 which lies ~20kb 3′ of linc-VWF.
Figure 5. (A) Genome browser view and (B) linkage disequilibrium plot for the chromosome 12 linc-VWF and VWF regions in European Ancestry.
Highlighted linc-VWF SNPs (rs1558324, rs7342306) have genome-wide association with platelet traits in Gieger et al.
DISCUSSION
Activation of innate immunity is a pathologic feature of several cardio-metabolic disorders. We utilized low-dose human endotoxemia to perform genome-wide discovery of inflammatory lincRNAs as candidates for complex diseases characterized by disturbance of innate immunity. We report substantial and tissue-specific change in expression of blood and adipose lincRNAs, confirm monocytes, macrophages and adipocytes as potential cell-sources for selective LPS-regulated lincRNAs. We provide evidence that some LPS-regulated adipose lincRNAs are expressed in adipocytes and suppressed in adipose of obese humans (linc-DMRT2 and linc-TP53I13). Finally, we found that LPS-induced linc-CEP110-13, which lies downstream of TLR4, contains a SNP associated with N-Glycosylation of IgG34 while linc-VWF, also LPS-induced, harbors SNPs for platelet traits35 that predict CVD events36.
Knowledge of lncRNA species and biology is emerging rapidly but current classifications are likely to include diverse non-coding RNA subsets with distinct genomic, evolutionary and functional characteristics5–8. Many lincRNAs have basic mRNA-like features and several important examples (e.g., Xist, Hotair, Mistral) have emerged with critical roles in cell differentiation, growth and apoptosis9–11. It is likely that a lncRNA, ANRIL, is the causal “gene” at the 9p21 vascular disease locus38. Yet, it is probable that many lincRNAs lack function and bioinformatic strategies for identifying functionality are deficient7. Thus, there is limited knowledge of lincRNA regulation in human tissues or their role in human disease.
We found that the majority of lincRNAs, as well as protein-coding RNAs, were down-regulated in blood and adipose by LPS. Although this is a novel finding for lincRNAs and of uncertain significance, Calvano et al. reported similar results for protein coding mRNAs in circulating leukocytes during endotoxemia 16. Indeed like Calvano, we found in blood that LPS up-regulated mRNAs largely reflect the expected innate immune and stress response to endotoxemia whereas down-regulated mRNAs were enriched for genes involved in mitochondrial processes. In adipose down-regulated mRNAs were enriched for distinct metabolic and biosynthetic pathways (not shown). Overall, LPS-induced pathways in blood and adipose converge on inflammatory and stress responses whereas LPS-suppressed genes and pathways may be more tissue-specific incorporating the more specialized functions of these tissues. This is consistent with the concept of energy modulation in response to inflammation in both tissues 16, 30.
One feature of protein-coding genes is evolutionary conservation of functional domains. Current evidence, however, suggests rapid sequence evolution and an apparent lack of conservation for lincRNAs. Similar to Cabili et al8, we found that the subset of lincRNAs present in blood and adipose is much less conserved than protein-coding genes. Indeed, lincRNAs appear to be only slightly more conserved than intergenic non-protein coding, non-lincRNA regions. However, current whole-genome alignment methods using conservations scores31 may not detect short conserved functional domains as noted for several functional lincRNAs (e.g., Xist, Megamind and Miat)7. A subset of lincRNAs demonstrates synteny (conserved neighboring protein coding genes) without apparent primary sequence conservation, suggesting conserved local cis-regulatory functions, while some lincRNA functions may not depend on primary sequence5, 7, 8. In our analysis, promoter regions of LPS-modulated lincRNAs in blood were enriched in binding sites for the NF-κB family. Thus, despite lack of primary sequence conservation, some LPS-modulated lincRNAs are transcriptional targets of TLR4 canonical signaling increasing the likelihood of specific regulatory functions.
Recently, specific lincRNAs have been implicated in regulation of immune and inflammatory signaling as in vitro13, 14. Carpenter et al. demonstrated Tlr2 and Tlr4 induction of numerous lincRNAs in mouse bone marrow-derived macrophages and showed that one, lincRNA-Cox2, mediated activation and repression of distinct classes of immune genes via nuclear ribonucleoproteins13. Rapicavoli et al. identified a set of pseudogene lncRNAs induced by TNFα in an NFKB manner in mouse cells and identified one, Lethe, that functioned in negative feedback signaling to NFKB14. Although these rodent lincRNAs are apparently not conserved in human, the pattern of inflammatory lincRNA induction with subsequent negative feedback on inflammatory cascades argues for functional roles for some LPS-modulated lincRNAs.
Our translational studies suggest clinical relevance of LPS-regulated lincRNAs. First, we found that some were expressed differentially in human adipocytes, monocytes or differentiated/polarized macrophages. This suggests potential functions in cells with prominent roles in cardio-metabolic homeostasis. Second, of a small number of adipose LPS-regulated lincRNAs prioritized for follow up, linc-DMRT2 and linc-TP53I13 were down-regulated in adipose of obese humans. Third, although we have yet to establish effects of disease SNPs on lincRNA functions, we did find overlap of LPS-induced lincRNAs with GWAS SNPs for complex inflammatory traits. Linc-CEP110-13 overlaps a SNP (rs10115586) associated with N-Glycosylation of circulating IgG. Such post-translational glycomic modifications are emerging as mechanistically important in inflammatory and cardio-metabolic pathophysiologies34, 39–41. Linc-VWF, induced by LPS and highly expressed in endothelial cells8, contains a SNPs (rs1558324) associated with mean platelet volume35, a biomarker of platelet function that predicts CVD36. Linc-VWF is located ~105 kb from VWF, an endothelial and platelet-derived plasma glycoprotein that plays a central role in hemostasis and thrombosis 42. There is no LD between linc-VWF rs1558324 and any SNPs in VWF that have been associated with circulating VWF levels37. Functional studies are required to directly implicate these lincRNAs in SNP-disease association. It is possible, however, that inflammatory and genetic regulation of specific lincRNAs converge in human inflammatory cardio-metabolic disorders.
Our study has several unique strengths and also limitations. This is the first in vivo profiling of blood and adipose lincRNAs during inflammation in human and utilizes an experimental model of proven clinical relevance in cardio-metabolic and inflammatory diseases15–20 that controls temporality limiting spurious findings due to reverse causation or confounding. We applied very deep RNA-seq facilitating identification of LPS-modulation of transcripts with low expression, a typical feature of lincRNAs. Further, we estimated the depth of coverage requirements and, like others6, note that many contemporary studies use sequencing depths that will detect modulation only for very abundant lincRNAs. Although our discovery was limited to one individual, we replicated several specific findings in independent subjects undergoing endotoxemia. Separately, we established broader generalizability of lincRNA expression and LPS-modulation through RNA-seq of additional GENE participants. Relative to subject A, a lower proportion of lincRNAs were LPS-modulated in the additional subjects but this is not surprising given the lower RNA-seq depth in additional sequencing and the expected heterogeneity in LPS response across humans 16, 26; we note that of LPS modulated lincRNAs in the additional subjects, almost 80% were also LPS modulated in subject A suggesting reasonable reproducibility in LPS modulation in blood data. We detected LPS-modulation of lincRNAs in PBMCs to a very similar degree to that in whole blood suggesting that modulation of lincRNAs in whole blood is not simply due to change in blood cell composition after LPS. Further, in our large GENE study (~300 participants) it was impractical in terms of time and cost to separate each cell population in all subjects while we also have concerns that cell separation can lead to variable degrees of cell activation. We recognize that while our studies in whole blood, PBMCs and adipose identify transcriptomic events of interest, these findings require cell-specific validation and functional follow-up. Importantly, we did perform preliminary translational experiments in cells and disease tissues and exploited GWAS resources to demonstrate disease relevance.
As evidence for the pervasive nature of human lncRNAs continues to emerge5, 6, our focus on a “stringent” set of human lincRNAs8 may have limited the breadth of discovery. Indeed, in our blood RNA-seq data, we did observe ~2-fold LPS-modulation of MIAT, the ncRNA containing SNPs associated with MI in Japanese43. However, MIAT was excluded from our lincRNA dataset based on its proximity to protein-coding genes. Similarly, ANRIL, also known as CDKN2B-AS1, the lncRNA at the 9p21 vascular disease locus38 was excluded from our analysis because it overlaps exons of a protein coding mRNA, CDKN2B. Parenthetically, we retrieved all reads for this 9p21 region in our datasets and found almost no evidence for expression (minimal or absent reads; all FPKM <0.05) of ANRIL exons that did not overlap CDKN2B suggesting that ANRIL has low expression in blood, PBMCs and adipose. On balance, in this work we favor a conservative approach while we pursue RNA-seq studies with larger lincRNAs sets and de-novo lincRNA assembly. We acknowledge that our disease associations are preliminary, do not provide evidence for lincRNAs as causal at the locus and require follow-up. Functional validations are ongoing but not trivial and are outside the scope of this report.
Our study provides a unique view of in vivo tissue-specific inflammatory modulation of lincRNAs in humans. We reveal LPS-regulation of distinct blood and adipose lincRNAs and provide evidence for roles of specific lincRNAs in cardio-metabolic disease. Findings are novel and our experimental approach has specific potential for clinical and functional translation.
Supplementary Material
Significance.
Inflammation is a key feature of cardio-metabolic diseases, and is modulated by expression of innate immunity genes. Recent studies suggest pervasive transcription of the genome with long intergenic non-coding RNAs (lincRNAs) representing a major transcript class with emerging roles in human biology. LincRNAs may modulate immune signaling, however their specific involvement in inflammation and cardio-metabolic pathophysiology has not been addressed. Using a model of human inflammation (endotoxemia; LPS) in combination with deep RNA-sequencing, we profiled LPS-modulation of >4000 lincRNAs in blood and adipose. We report tissue-specific, inflammatory regulation of specific lincRNAs and confirm monocytes, macrophages and adipocytes as cell-sources for selective LPS-regulated lincRNAs. We found that some LPS-regulated adipose lincRNAs are suppressed in human obesity and that specific LPS-induced lincRNAs harbor SNPs that have genome wide association with cardio-metabolic traits. Thus, we reveal tissue-specific LPS-regulation of distinct lincRNAs and roles of specific LPS-modulated lincRNAs in inflammatory cardio-metabolic pathophysiology.
Acknowledgments
Sources of Funding: This project was funded, in part, by R01HL113147 from the NIH and by the Penn Genome Frontiers Institute under a grant with the Pennsylvania Department of Health (which disclaims responsibility for any analyses, interpretations or conclusions). M.P.R. is also supported by R01-DK-090505, R01-DK071224, U01-HL108636 and K24-HL107643, M.L. is also supported by R01HG004517 and R01HG005854 from the NIH. J.F.F is supported by a postdoctoral fellowship grant from the American Heart Association (12POST11840017).
Non-standard Abbreviations
- RNA-seq
RNA sequencing
- LPS
lipopolysaccharide
- TLR4
toll-like receptor 4
- GENE
Genetics of Evoked-Responses to Niacin and Endotoxemia
- LincRNA
long intergenic non-coding RNA
- AS
alternative splicing
- qRT-PCR
quantitative real-time PCR
- FPKM
Fragments Per Kilobase of transcript per Million mapped reads
Footnotes
DATA AVAILABILITY
RNA-seq data have been deposited in the Gene Expression Omnibus (GEO) database (accession number GSE50792).
Disclosures: The authors have no disclosures related to this work.
References
- 1.Yang J, Manolio TA, Pasquale LR, et al. Genome partitioning of genetic variation for complex traits using common snps. Nature genetics. 2011;43:519–525. doi: 10.1038/ng.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Z, Gerstein M, Snyder M. Rna-seq: A revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Consortium EP, Dunham I, Kundaje A, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derrien T, Johnson R, Bussotti G, et al. The gencode v7 catalog of human long noncoding rnas: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hangauer MJ, Vaughn IW, McManus MT. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding rnas. PLoS genetics. 2013;9:e1003569. doi: 10.1371/journal.pgen.1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ulitsky I, Bartel DP. Lincrnas: Genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding rnas reveals global properties and specific subclasses. Genes & development. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for xist in × chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 10.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding rna as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bertani S, Sauer S, Bolotin E, Sauer F. The noncoding rna mistral activates hoxa6 and hoxa7 expression and stem cell differentiation by recruiting mll1 to chromatin. Molecular cell. 2011;43:1040–1046. doi: 10.1016/j.molcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM. Molecular interplay of the noncoding rna anril and methylated histone h3 lysine 27 by polycomb cbx7 in transcriptional silencing of ink4a. Molecular cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, Byron M, Monks B, Henry-Bezy M, Lawrence JB, O’Neill LA, Moore MJ, Caffrey DR, Fitzgerald KA. A long noncoding rna mediates both activation and repression of immune response genes. Science. 2013;341:789–792. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rapicavoli NA, Qu K, Zhang J, Mikhail M, Laberge RM, Chang HY. A mammalian pseudogene lncrna at the interface of inflammation and anti-inflammatory therapeutics. eLife. 2013;2:e00762. doi: 10.7554/eLife.00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suffredini AF, Fromm RE, Parker MM, Brenner M, Kovacs JA, Wesley RA, Parrillo JE. The cardiovascular response of normal humans to the administration of endotoxin. N Engl J Med. 1989;321:280–287. doi: 10.1056/NEJM198908033210503. [DOI] [PubMed] [Google Scholar]
- 16.Calvano SE, Xiao W, Richards DR, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 17.Hudgins LC, Parker TS, Levine DM, Gordon BR, Saal SD, Jiang XC, Seidman CE, Tremaroli JD, Lai J, Rubin AL. A single intravenous dose of endotoxin rapidly alters serum lipoproteins and lipid transfer proteins in normal volunteers. Journal of lipid research. 2003;44:1489–1498. doi: 10.1194/jlr.M200440-JLR200. [DOI] [PubMed] [Google Scholar]
- 18.McGillicuddy FC, de la Llera Moya M, Hinkle CC, Joshi MR, Chiquoine EH, Billheimer JT, Rothblat GH, Reilly MP. Inflammation impairs reverse cholesterol transport in vivo. Circulation. 2009;119:1135–1145. doi: 10.1161/CIRCULATIONAHA.108.810721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehta NN, McGillicuddy FC, Anderson PD, Hinkle CC, Shah R, Pruscino L, Tabita-Martinez J, Sellers KF, Rickels MR, Reilly MP. Experimental endotoxemia induces adipose inflammation and insulin resistance in humans. Diabetes. 2010;59:172–181. doi: 10.2337/db09-0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agwunobi AO, Reid C, Maycock P, Little RA, Carlson GL. Insulin resistance and substrate utilization in human endotoxemia. J Clin Endocrinol Metab. 2000;85:3770–3778. doi: 10.1210/jcem.85.10.6914. [DOI] [PubMed] [Google Scholar]
- 21.Ridker PM. Clinical application of c-reactive protein for cardiovascular disease detection and prevention. Circulation. 2003;107:363–369. doi: 10.1161/01.cir.0000053730.47739.3c. [DOI] [PubMed] [Google Scholar]
- 22.Sarwar N, Butterworth AS, Freitag DF, et al. Interleukin-6 receptor pathways in coronary heart disease: A collaborative meta-analysis of 82 studies. Lancet. 2012;379:1205–1213. doi: 10.1016/S0140-6736(11)61931-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Cromphaut SJ, Vanhorebeek I, Van den Berghe G. Glucose metabolism and insulin resistance in sepsis. Current pharmaceutical design. 2008;14:1887–1899. doi: 10.2174/138161208784980563. [DOI] [PubMed] [Google Scholar]
- 24.Michelsen KS, Wong MH, Shah PK, Zhang W, Yano J, Doherty TM, Akira S, Rajavashisth TB, Arditi M. Lack of toll-like receptor 4 or myeloid differentiation factor 88 reduces atherosclerosis and alters plaque phenotype in mice deficient in apolipoprotein e. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:10679–10684. doi: 10.1073/pnas.0403249101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shah R, Lu Y, Hinkle CC, McGillicuddy FC, Kim R, Hannenhalli S, Cappola TP, Heffron S, Wang X, Mehta NN, Putt M, Reilly MP. Gene profiling of human adipose tissue during evoked inflammation in vivo. Diabetes. 2009;58:2211–2219. doi: 10.2337/db09-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferguson JF, Patel PN, Shah RY, Mulvey CK, Gadi R, Nijjar PS, Usman HM, Mehta NN, Shah R, Master SR, Propert KJ, Reilly MP. Race and gender variation in response to evoked inflammation. J Transl Med. 2013;11:63. doi: 10.1186/1479-5876-11-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, Ferguson JF, Xue C, Silverman IM, Gregory B, Reilly MP, Li M. Evaluating the impact of sequencing depth on transcriptome profiling in human adipose. PLoS ONE. 2013;8:e66883. doi: 10.1371/journal.pone.0066883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by rna-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cobb JP, Mindrinos MN, Miller-Graziano C, et al. Application of genome-wide expression analysis to human health and disease. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:4801–4806. doi: 10.1073/pnas.0409768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haimovich B, Zhang Z, Calvano JE, Calvano SE, Kumar A, Macor MA, Corbett S, Coyle SM, Lowry SF. Cellular metabolic regulators: Novel indicators of low-grade inflammation in humans. Annals of surgery. 2013 doi: 10.1097/SLA.0b013e31829a4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siepel A, Bejerano G, Pedersen JS, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matys V, Kel-Margoulis OV, Fricke E, et al. Transfac and its module transcompel: Transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grant CE, Bailey TL, Noble WS. Fimo: Scanning for occurrences of a given motif. Bioinformatics (Oxford, England) 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lauc G, Huffman JE, Pucic M, et al. Loci associated with n-glycosylation of human immunoglobulin g show pleiotropy with autoimmune diseases and haematological cancers. PLoS genetics. 2013;9:e1003225. doi: 10.1371/journal.pgen.1003225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gieger C, Radhakrishnan A, Cvejic A, et al. New gene functions in megakaryopoiesis and platelet formation. Nature. 2011;480:201–208. doi: 10.1038/nature10659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chu SG, Becker RC, Berger PB, Bhatt DL, Eikelboom JW, Konkle B, Mohler ER, Reilly MP, Berger JS. Mean platelet volume as a predictor of cardiovascular risk: A systematic review and meta-analysis. J Thromb Haemost. 2010;8:148–156. doi: 10.1111/j.1538-7836.2009.03584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Desch KC, Ozel AB, Siemieniak D, et al. Linkage analysis identifies a locus for plasma von willebrand factor undetected by genome-wide association. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:588–593. doi: 10.1073/pnas.1219885110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holdt LM, Hoffmann S, Sass K, et al. Alu elements in anril non-coding rna at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks. PLoS genetics. 2013;9:e1003588. doi: 10.1371/journal.pgen.1003588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reilly MP, Li M, He J, et al. Identification of adamts7 as a novel locus for coronary atherosclerosis and association of abo with myocardial infarction in the presence of coronary atherosclerosis: Two genome-wide association studies. Lancet. 2011;377:383–392. doi: 10.1016/S0140-6736(10)61996-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang H, Mooney CJ, Reilly MP. Abo blood groups and cardiovascular diseases. International journal of vascular medicine. 2012;2012:641917. doi: 10.1155/2012/641917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lauc G, Essafi A, Huffman JE, et al. Genomics meets glycomics-the first gwas study of human n-glycome identifies hnf1alpha as a master regulator of plasma protein fucosylation. PLoS genetics. 2010;6:e1001256. doi: 10.1371/journal.pgen.1001256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sadler JE. Von willebrand factor: Two sides of a coin. J Thromb Haemost. 2005;3:1702–1709. doi: 10.1111/j.1538-7836.2005.01369.x. [DOI] [PubMed] [Google Scholar]
- 43.Ishii N, Ozaki K, Sato H, Mizuno H, Saito S, Takahashi A, Miyamoto Y, Ikegawa S, Kamatani N, Hori M, Saito S, Nakamura Y, Tanaka T. Identification of a novel non-coding rna, miat, that confers risk of myocardial infarction. Journal of human genetics. 2006;51:1087–1099. doi: 10.1007/s10038-006-0070-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.