Abstract
The MEK/ERK and PI3K/AKT pathways are often concurrently activated by separate genetic alterations in colorectal cancer (CRC), which is associated with CRC progression and poor survival. However, how activating both pathways is required for CRC metastatic progression remains unclear. Our recent study showed that both ERK and AKT signaling are required to activate eIF4E-initiated cap-dependent translation via convergent regulation of the translational repressor 4E-BP1 for maintaining CRC transformation. Here, we identified that the activation of cap-dependent translation by cooperative ERK and AKT signaling is critical for promotion of CRC motility and metastasis. In CRC cells with coexistent mutational activation of ERK and AKT pathways, inhibition of either MEK or AKT alone showed limited activity in inhibiting cell migration and invasion, but combined inhibition resulted in profound effects. Genetic blockade of the translation initiation complex by eIF4E knockdown or expression of a dominant active 4E-BP1 mutant effectively inhibited migration, invasion and metastasis of CRC cells, whereas overexpression of eIF4E or knockdown of 4E-BP1 had the opposite effect and markedly reduced their dependence on ERK and AKT signaling for cell motility. Mechanistically, we found that these effects were largely dependent on the increase in mTORC1-mediated survivin translation by ERK and AKT signaling. Despite the modest effect of survivin knockdown on tumor growth, reduction of the translationally-regulated survivin profoundly inhibited motility and metastasis of CRC. These findings reveal a critical mechanism underlying the translational regulation of CRC metastatic progression, and suggest that targeting cap-dependent translation may provide a promising treatment strategy for advanced CRC.
Keywords: translational regulation, metastasis, ERK, AKT, survivin, colorectal cancer
INTRODUCTION
Colorectal cancer (CRC) is the second leading cause of cancer-related mortality in the United States.1 Although progress has been made in survival for earlier stage disease, only minimal improvement has been noted in patients with systemic metastases. Treatment options including traditional cytotoxic chemotherapy and newly targeted therapy with the inhibitors of the epidermal growth factor receptor and vascular endothelial growth factor are limited for patients with advanced metastatic CRC.2 Novel treatment strategies are therefore needed for these patients.
Mutational activation of mitogenic signaling is a frequent event in human cancers including CRC. The MAPK intracellular signaling cascade comprises RAS, RAF, MEK and ERK. Mutations in KRAS and BRAF that lead to hyperactivation of MEK/ERK signaling occur in 45% and 10% of CRC, respectively.3,4 In addition to ERK pathway activation, dysregulation of the PI3K/AKT signaling pathway, due to the activating mutations of the catalytic subunit of PI3K, p110α (PIK3CA) and the inactivating mutations or decreased function of the phosphatase PTEN, occurs at high frequency in CRC.4,5 Moreover, PIK3CA mutation is commonly coexisted with the KRAS or BRAF mutations in CRC.4 Uncontrolled activation of the ERK and AKT pathways in tumor cells is thought to play an important role in maintaining their proliferation, preventing apoptosis, and supporting processes required for the transformed and metastatic phenotypes. Several small molecule inhibitors targeting components of the RAF/MEK/ERK and PI3K/AKT pathways have been tested in a number of clinical and preclinical studies for the treatment of CRC but have shown only limited activity as a single agent.6–10
We and others recently showed that colon tumors with concurrent activation of the MEK/ERK and PI3K/AKT pathways by separate mutations are invariably resistant to inhibition of either pathway alone, but sensitive to combined inhibition of both pathways.6,8,9 We discovered that the resistance to inhibition of either pathway is associated with redundant activation of cap-dependent translation mediated by convergent phosphorylation and subsequent inhibition of the translational repressor 4E-BP1 function by the ERK and AKT pathways.8 We showed that combined inhibition of both pathways is required to effectively inhibit 4E-BP1 phosphorylation and cap-dependent translation, thereby suppressing CRC tumorigenesis in vivo.
The cap-dependent translation is a process by which most capped mRNAs are translated into proteins. Translation of certain key oncogenic mRNAs with a highly structured 5′-untranslated region (5′-UTR) has been shown to be strongly dependent on the mRNA cap-binding protein eIF-4E, the rate-limiting component of the translation initiation complex eIF4F, which also include the scaffolding protein eIF4G and the RNA helicase eIF4A.11 Consequently, these oncogenic mRNAs are preferentially and disproportionately affected by eIF4E availability and are sensitive to the alteration in the levels of eIF4E.12–14 The levels of free eIF4E can be increased substantially in cancer cells by a number of mechanisms, including increased eIF4E expression, decreased expression of eIF4E inhibitory binding proteins, 4E-BPs, and release of eIF4E from 4E-BPs by inactivating phosphorylation of 4E-BPs. Upregulation of eIF4E, reduction of 4E-BP expression, and hyperphosphorylation of 4E-BP resulting from the oncogenic activation of signaling pathways are significantly found in CRC and associated with CRC progression.15–18 The sum of these studies together with our previous findings8 suggest that the eIF4E-dependent translation represents a critical node of convergence for the oncogenic activation of the ERK and AKT signaling pathways, and the inappropriately high rates of translation initiation may be a mechanistic feature for driving metastatic progression of CRC. Here, we provide evidence that the effect of ERK and AKT signaling on metastatic progression of CRC is much mediated by their convergent activation of cap-dependent translation through selectively increasing translation of the survivin mRNA. Our data demonstrate that translational control is a critical step in CRC progression to metastasis and that a better understanding of the link between protein production in cancer cells and development of metastasis may lead to novel strategies to prevent and/or control advanced CRC.
RESULTS
ERK and AKT signaling are required to support migration and invasion of CRC cells with coexistent pathway activation
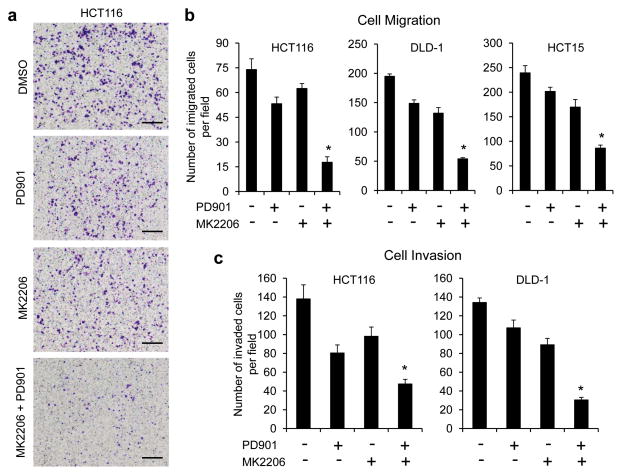
Migration and invasion are critical steps in initial progression of cancer that facilitate metastasis. Concurrent activation of the RAS/RAF/MEK/ERK and PI3K/AKT pathways occurs in a significant proportion of human CRCs.4 To ascertain the role of co-activation of both pathways in CRC metastasis, we first examined the effects of the MEK inhibitor PD0325901 (ref. 3) (hereafter referred to as PD901) and the AKT inhibitor MK2206 (ref. 19), alone and in combination, on CRC migration and invasion using Boyden chamber assays. We have previously shown that both PD901 and MK2206 can effectively inhibit phosphorylation of ERK and AKT and their downstream signaling, respectively, in a variety of cancers including CRC in tissue culture and in vivo.8,9,20 In CRC cells with coexistent KRAS and PIK3CA mutations (HCT116, DLD-1, HCT15), treatment with PD901 or MK2206 alone for 6 hours had only a modest effect on migration of the cells. However, a combination of both drugs was effective in profoundly inhibiting their migration (Figures 1a and b). Similar results were observed in the ability of these cells that invade through Matrigel 30 hours after drug exposure (Figure 1c), whereas cell cycle kinetics or cell viability were not affected within the same time interval.8,9 Collectively, these results suggest that the ERK and AKT signaling pathways cooperate to maintain migration and invasion of CRC cells in which both pathways are activated.
Figure 1.
Combined inhibition of MEK and AKT is required for effective inhibition of migration and invasion of CRC cells with coexistent KRAS and PIK3CA mutations. (a and b) Transwell migration analysis of HCT116, DLD-1 and HCT15 cells in the presence of 50 nM PD0325901 (PD901) and 1 μM MK2206, alone or in combination or DMSO as control for 6 hours. The results represent the mean number of migrated cells per field ± s.e.m. (n=3). Scale bar = 500 μm. (c) Transwell invasion analysis of HCT116 and DLD-1 cells in the presence of the drugs as indicated in (a and b) for 30 hours. The results represent the mean number of invaded cells per field ± s.e.m. (n=3). * P < 0.02 for combination of PD901 and MK2206 versus DMSO control, PD901 or MK2206.
ERK and AKT signaling regulate CRC cell migration and invasion through their convergent activation of cap-dependent translation
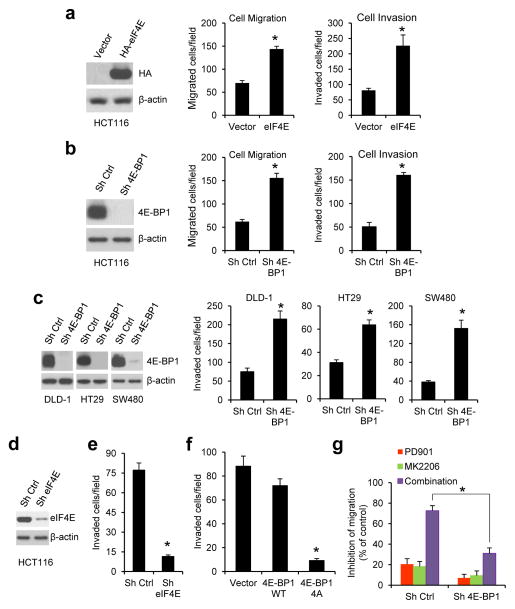
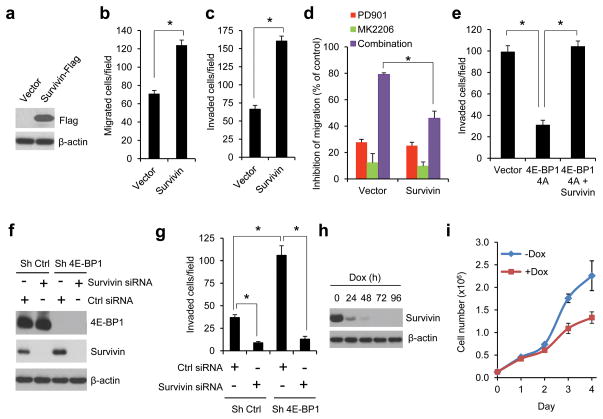
Our previous study showed that in CRC cells with concurrent activation of ERK and AKT signaling pathways, both pathways cooperate to maintain tumor growth by convergent activation of eIF4E-initiated cap-dependent translation.8 To determine whether the translational activation is also required for CRC cell migration and invasion, the cap-dependent translational activity was modulated by overexpression or knockdown of eIF4E and 4E-BP1. Boyden chamber assays showed that overexpression of eIF4E or knockdown of 4E-BP1 that activates cap-dependent translation markedly enhanced migration and invasion in HCT116 cells (Figures 2a and b). Similar results were obtained in three other CRC cell lines (DLD-1, HT29, SW480) with knockdown of 4E-BP1 expression (Figure 2c). By tracking single-cell movement with live cell imaging, we further confirmed that 4E-BP1 knockdown significantly increased migration of HCT116 cells (Supplementary Figure S1). Conversely, inhibition of cap-dependent translation by knocking down eIF4E or expression of mutant 4E-BP1-4A but not wild-type (wt) 4E-BP1 had the opposite effect (Figures 2d–f). As compared to 4E-BP1 wt, we showed previously that the mutant 4E-BP1-4A, in which its four known phosphorylation sites (T37, T46, S65, T70) were replaced with alanine, cannot be phosphorylated and binds constitutively to eIF4E, thus inhibiting cap-dependent translation8. Together, these data suggest that activation of cap-dependent translation plays a critical role in stimulation of CRC cell migration and invasion.
Figure 2.
Cap-dependent translation apparatus mediates the effect of ERK and AKT activation on CRC cell migration and invasion. (a–c) Overexpression of eIF4E (a) or silencing 4E-BP1 expression (b and c) promoted CRC cell migration and invasion. Exogenous eIF4E expression (a, left) and 4E-BP1 knockdown efficiency by shRNA (b and c, left) in the indicated cell lines were verified by immunoblot analysis. (d) HCT116 cells with stable expression of control (ctrl) shRNA or eIF4E shRNA were immunoblotted with the indicated antibodies. (e and f) Silencing eIF4E expression (e) or expression of 4E-BP1 4A but not 4E-BP1 WT (f) markedly inhibited invasion of HCT116 cells. (g) Silencing 4E-BP1 expression significantly reduced the inhibitory effect on cell migration induced by combined inhibition of MEK and AKT. Migration analysis of HCT116 cells with stable expression of control shRNA or 4E-BP1 shRNA was performed in the presence of 50 nM PD901 and 1 μM MK2206, alone or in combination or DMSO as control for 6 hours. The results are expressed as the inhibition of migration relative to the DMSO-treated controls. Data shown in graphs represent the mean ± s.e.m. (n=3). * P < 0.02 for eIF4E versus vector; 4E-BP1 4A versus 4E-BP1 WT or vector; sh eIF4E or sh 4E-BP1 versus sh Ctrl; and combination of PD901 and MK2206 in sh 4E-BP1 cells versus that in sh ctrl cells.
To test whether the cap-dependent translation apparatus mediates the effects of ERK and AKT activation on CRC motility, the effects of MEK and AKT inhibition, alone or in combination, were determined in HCT116 cells with stable knockdown of 4E-BP1 or expressing control shRNA. As shown in Figure 2g, combined inhibition of ERK and AKT kinases caused 73% inhibition of cell migration in control cells but had much less effect (31%) in cells in which 4E-BP1 expression was suppressed. Similar results were also observed in cell invasion assays (data not shown). These data suggest that 4E-BP1 or the eIF4E-initiated translation process is an important downstream effector of the ERK and AKT activation responsible for CRC cell migration and invasion.
Survivin expression is selectively regulated at the level of translation by ERK and AKT signaling
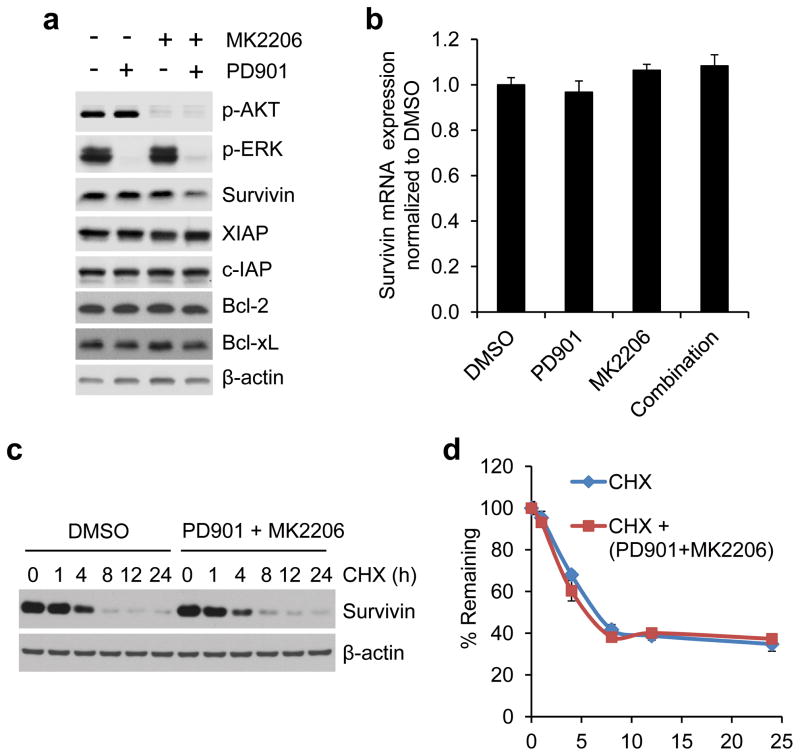
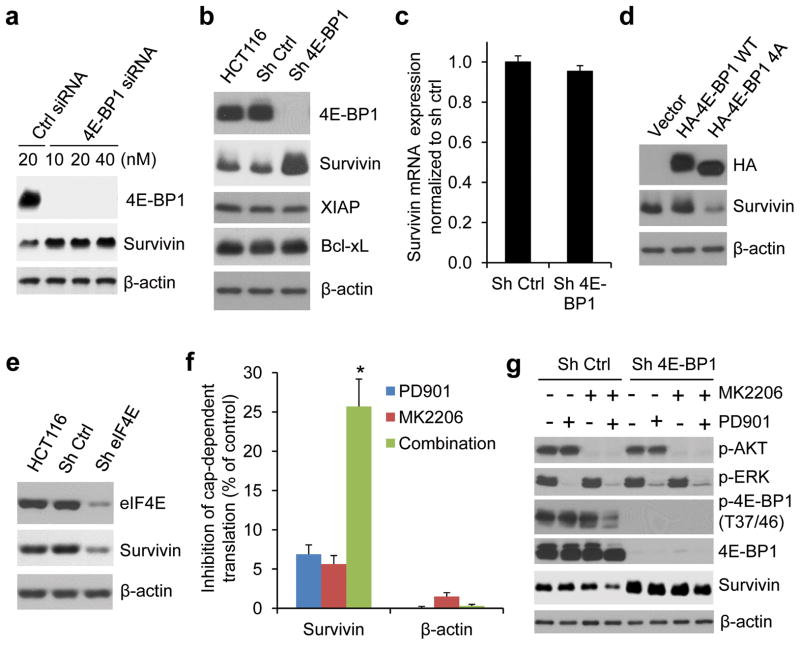
To understand how the translational activation by ERK and AKT signaling affects CRC progression, we determined the expression of several key proteins that are known to be involved in the regulation of apoptosis, proliferation and metastasis. Strikingly, we found that the anti-apoptotic and metastasis-related protein, survivin,21–25 was downregulated by combined inhibition of MEK and AKT kinases but not by inhibition of either kinase alone 12 hours after drug addition, in HCT116 and T84 CRC cells with coexistent KRAS and PIK3CA mutations (Figure 3a and Supplementary Figure S2). In contrast, expression of other anti-apoptotic proteins such as XIAP, c-IAP, Bcl-2 and Bcl-xL was unaffected even with combined inhibition. The downregulation of survivin expression was not associated with changes in the level of transcription or protein stability as analyzed by qRT-PCR or protein degradation rate using cycloheximide chase assay respectively (Figures 3b–d). Notably, we found that activation of cap-dependent translation by 4E-BP1 knockdown using siRNA or shRNA with different targeting sequences upregulated survivin protein expression, but its mRNA level remained unchanged in 4E-BP1 knockdown cells compared with the control cells (Figures 4a–c). Conversely, inhibition of cap-dependent translation by expression of 4E-BP1-4A but not 4E-BP1 wt, or by eIF4E knockdown resulted in a decrease of survivin protein (Figures 4d and e). Using a cap-dependent translation reporter luciferase (Luc) mRNA linked to the 5′-UTR of either survivin or β-actin for measurement of cap-dependent translation activity, we further found that inhibition of MEK or AKT kinase alone had modest inhibitory effects (5–7%) on the 5′survivin-Luc translation activity 12 hours after drug exposure, whereas combined inhibition markedly inhibited the activity (25%) but had no effect on 5′β-actin-Luc translation activity (Figure 4f). Most importantly, knockdown of 4E-BP1 expression prevented survivin reduction induced by combined inhibition of MEK and AKT (Figure 4g). In contrast, the combined inhibition effectively inhibited 4E-BP1 phosphorylation and downregulated survivin in the control cells (Figure 4g). Together, these data indicate that ERK and AKT signaling cooperate to specifically regulate survivin expression through the cap-dependent translation mechanism in CRC cells with coexistent pathway activation.
Figure 3.
Combined inhibition of MEK and AKT downregulates survivin expression at the level of protein but not total mRNA or protein stability. (a) HCT116 cells were treated with 50 nM PD901 and 1 μM MK2206, alone or in combination or DMSO as control for 12 hours. Cell lysates were immunoblotted with the indicated antibodies. (b) Quantitative RT-PCR analysis of mRNA expression of survivin relative to β-actin in HCT116 cells that were treated with the drugs as indicated in (a) for 12 hours (n=3). (c) HCT116 cells were treated with combination of 50 nM PD901 and 1 μM MK2206 or DMSO as control for 30 min, followed by addition of 20 μg/ml cycloheximide (CHX) for the indicated times. Cell lysates were immunoblotted with survivin and β-actin antibodies. (d) Immunoblots of survivin as shown in (c) were quantified using the FluoChem digital imaging system. The level of survivin remaining was obtained by normalizing β-actin level at each time, and the results are presented as mean ± s.e.m. (n=3).
Figure 4.
Survivin is translationally regulated by ERK and AKT signaling in CRC cells with coexistent pathway activation. (a) HCT116 cells were transfected with the indicated concentrations of 4E-BP1 siRNAs or control non-targeting siRNAs for 48 hours. Cell lysates were immunoblotted with the indicated antibodies. (b) Immunoblot analysis of HCT116 cells alone or with stable expression of control shRNA or 4E-BP1 shRNA with different targeting sequences used in (a). (c) Quantitative RT-PCR analysis of mRNA expression of survivin relative to β-actin in HCT116 cells with stable expression of control shRNA or 4E-BP1 shRNA (n=3). (d) Immunoblot analysis of HCT116 cells with stable expression of vector, HA-4E-BP1 WT or HA-4E-BP1 4A. (e) Immunoblot analysis of HCT116 cells alone or with stable expression of control shRNA or eIF4E shRNA. (f) HCT116 cells were transfected with a bicistronic luciferase reporter that detects cap-dependent translation of the Renilla luciferase gene linked to the 5′-UTR of either survivin or β-actin and cap-independent Polio IRES-mediated translation of the firefly luciferase gene. The transfected cells were treated with 50 nM PD0325901 and 1 μM MK2206, alone or in combination for 12 hours. Luciferase activities were measured by a dual-luciferase assay, and the Renilla/firefly luciferase luminescence ratio was calculated for cap-dependent translational activity. The results are expressed as the inhibition of cap-dependent translation relative to the DMSO-treated controls and presented as means ± s.e.m. (n=3). * P < 0.01 for combination of PD901 and MK2206 versus PD901 or MK2206. (g) Immunoblot analysis of HCT116 cells with stable expression of control shRNA or 4E-BP1 shRNA that were treated with 50 nM PD901 and 1 μM MK2206, alone or in combination for 12 hours.
Survivin mediates the effects of ERK and AKT activation on translational regulation of CRC cell migration and invasion
The importance of survivin downregulation in mediating the effects of combined MEK and AKT pathway inhibition was determined in HCT116 cells in which survivin protein was exogenously overexpressed (Figure 5a). These cells did not exhibit changes in cell proliferation (Supplementary Figure S3a) but showed a 2–3 fold increase in cell migration and invasion compared with the vector control cells (Figures 5b and c). Furthermore, in these cells, the effect of combined inhibition of MEK and AKT on suppression of cell migration was significantly reduced compared with that in the control cells (Figure 5d). In addition, overexpression of survivin could prevent inhibition of cell invasion induced by the cap-dependent translation inhibitor 4E-BP1-4A (Figure 5e). Conversely, silencing survivin expression 48 hours after transfection with siRNA profoundly inhibited cell invasion in both control and 4E-BP1 knockdown cells, although 4E-BP1 knockdown induced survivin expression and promoted cell invasion (Figures 5f and g). The inhibition of cell invasion by decreased survivin expression was not associated with the effects on cell viability and induction of apoptosis in the same time interval (Figures 5h and i and Supplementary Figure S3b). Knockdown of survivin expression by a tet-inducible shRNA only caused a slowing of cell growth but did not induce apoptosis in tissue culture after 48 hours of doxycycline exposure. Thus, these data suggest that survivin plays a prominent role in mediating the effects of ERK and AKT activation on translational control of CRC cell migration and invasion.
Figure 5.
Survivin is an important effector of ERK and AKT signaling responsible for translational control of CRC cell migration and invasion. (a) Immunoblot analysis of HCT116 cells with stable expression of vector or flag-tagged survivin. (b and c) Migration (b) or invasion (c) analysis of HCT116 cells with stable expression of vector or survivin. (d) Migration analysis of HCT116 cells with stable expression of vector or survivin in the presence of 50 nM PD901 and 1 μM MK2206, alone or in combination or DMSO as control for 6 hours. The results are expressed as the inhibition of migration relative to the DMSO-treated controls. (e) Invasion analysis of HCT116 cells with stable expression of vector, 4E-BP1 4A or 4E-BP1 4A with co-expressing survivin over 30 hours. (f and g) HCT116 cells with stable expression of control shRNA or 4E-BP1 shRNA were transfected with control siRNA or survivin siRNA for 48 hours, followed by immunoblot analysis with the indicated antibodies (f) and invasion analysis over 30 hours (g). (h and i) HCT116 cells with stable expression of tet-inducible survivin shRNA were treated with or without 50 ng/ml doxycycline (dox) for the indicated times, followed by immunoblot analysis with the indicated antibodies (h) and cell proliferation analysis (i). Data shown in graphs represent the mean ± s.e.m. (n=3). * P < 0.01.
mTORC1 largely mediates the effects of ERK and AKT activation on translational regulation of survivin and motility
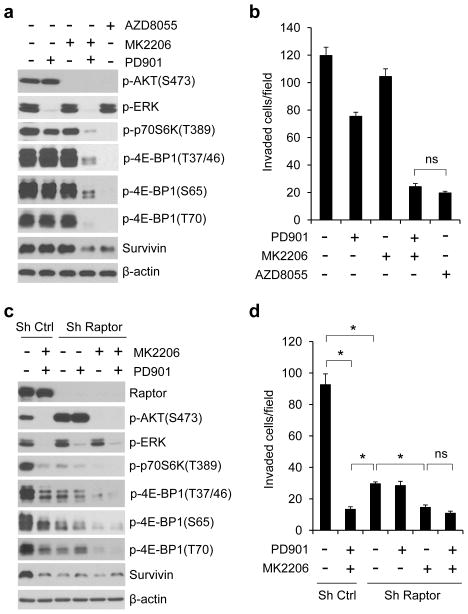
The mTOR kinase forms two distinct complexes, mTORC1 and mTORC2. They exert their actions by regulating other important kinases and substrates, such as the translational regulators p70S6 kinase (S6K) and 4E-BP1 by mTORC1 and AKT (on S473) by mTORC2.26 Both AKT and ERK have been shown to regulate mTORC1 activity via phosphorylation of TSC2.27,28 Thus, we explored whether mTORC1 mediates the effect of AKT and ERK activation on translational control of CRC cell motility. Consistent with our previous findings,8 inhibition of either AKT or MEK had no effect on phosphorylation of p70S6K or 4E-BP1 at any of its four phosphorylation sites in HCT116 and T84 cells with coexistent KRAS and PIK3CA mutations (Figure 6a and Supplementary Figure S2). However, combined inhibition of AKT and MEK profoundly inhibited phosphorylation of all these sites, suggesting that AKT and ERK cooperate to regulate mTORC1 activity. Rapamycin is a selective allosteric inhibitor of mTORC1, but it is much less effective than combined inhibition of AKT and MEK in inhibiting 4E-BP1 phosphorylation and cell invasion (Supplementary Figure S4). mTOR kinase inhibitors have been shown to be better mTORC1 inhibitors than rapamycin.29,30 As shown in Figures 6a and b, the mTOR kinase inhibitor AZD8055 (ref. 31) was as effective as combined inhibition of MEK and AKT in inhibiting 4E-BP1 phosphorylation, downregulating survivin expression, and repressing cell migration (data not shown) and invasion in HCT116 cells. However, AZD8055 also inhibited mTORC2 as indicated by loss of p-AKT on S473 (Figure 6a).
Figure 6.
The effects of ERK and AKT activation on translational regulation of survivin and cell invasion are largely mediated by mTORC1. (a and b) Immunoblot (a) and invasion (b) analyses of HCT116 cells that were treated with 50 nM PD901, 1 μM MK2206 and 500 nM AZD8055, alone or in combination for 12 hours (a) and 30 hours (b) respectively. (c and d) Immunoblot (c) and invasion (d) analyses of HCT116 cells with stable expression of control shRNA or raptor shRNA that were treated with 50 nM PD901 and 1 μM MK2206, alone or in combination for 12 hours (c) and 30 hours (d) respectively. Data shown in graphs represent the mean ± s.e.m. (n=3). *P < 0.02; ns, not significant.
To specifically determine the role of mTORC1, shRNAs were used to knockdown raptor, an obligatory component of mTORC1.32,33 Depletion of raptor in HCT116 cells markedly inhibited 4E-BP1 phosphorylation and survivin expression to a degree that equaled the effects of combined inhibition of MEK and AKT in control cells (Figure 6c). Moreover, raptor knockdown also profoundly suppressed cell invasion, although this effect was significantly less than that by inhibition of both MEK and AKT in control cells (Figure 6d). Interestingly, knockdown of raptor activated AKT by increasing p-AKT on S473, whereas inhibition of AKT elicited more profound inhibition of 4E-BP1 phosphorylation and cell invasion in raptor knockdown cells (Figures 6c and d). Collectively, these results suggest that the effects of AKT and ERK activation on translational regulation of survivin expression and CRC cell motility are mediated at least in a large part by the mTORC1-dependent mechanism.
Targeting cap-dependent translation or its dependent survivin expression suppresses CRC metastasis
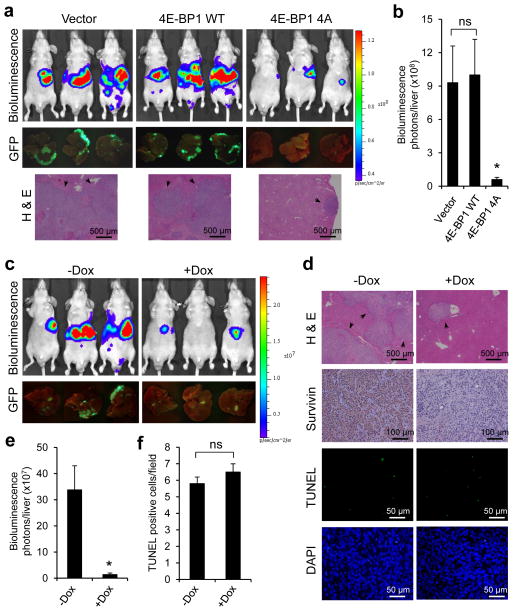
Our previous study showed that much of the biologic effect of ERK and AKT activation is mediated by inhibition of 4E-BP1 function through their convergent phosphorylation of 4E-BP1, and the non-phosphorylated mutant 4E-BP1-4A exerts similar inhibitory effects on cap-dependent translation and tumor growth as the combined inhibition of ERK and AKT in CRC.8 Our current data further suggest that 4E-BP1 and its translationally-regulated protein survivin, integrate the functions of ERK and AKT pathways in CRC cell migration and invasion. Thus, targeted inhibition of cap-dependent translation by 4E-BP1-4A or silencing survivin expression may significantly inhibit CRC metastasis. To explore this possibility, we used the experimental lung and liver metastasis models in vivo. Luciferase and GFP-labeled HCT116 cells with stable expression of 4E-BP1-4A or knockdown of survivin using a tet-inducible shRNA were injected intravenously or intrasplenically into athymic nude mice and formation of lung or liver metastasis was assessed by bioluminescent and fluorescent imaging and histological examination. As compared to wt 4E-BP1 and vector control, expression of 4E-BP1-4A profoundly suppressed lung and liver metastases in mice (Figures 7a–b and 8a–b) and survivin expression in metastatic cells (Supplementary Figure S5). Similar results were observed by silencing survivin expression in mice that were treated with doxycycline in drinking water for induction of survivin-targeting shRNAs (Figures 7c–e and 8c–e). Doxycycline effectively inhibited survivin expression but did not significantly induce apoptosis as shown by TUNEL staining and cleaved PARP expression, and had only a modest effect on tumor growth (Figures 7d, f–h, and 8d, f). These findings demonstrate that ERK/AKT-activated cap-dependent translation and its translational target survivin play crucial roles in establishment of CRC metastasis in vivo.
Figure 7.
Genetic blockade of cap-dependent translation or silencing survivin expression suppresses lung metastasis of CRC. (a) Bioluminescence, GFP images and H & E staining of lung metastasis in athymic nude mice that were injected intravenously with HCT116-Luc/GFP cells expressing vector, 4E-BP1 WT or 4E-BP1 4A at week 8 post-injection. (b) Quantitative analysis of bioluminescence in lung metastasis as shown in (a) (n=6 mice/group). (c) Bioluminescence and GFP images of lung metastasis in athymic nude mice that were injected intravenously with HCT116-Luc/GFP cells expressing tet-inucible survivin shRNA followed by maintenance with or without Dox (0.5 mg/ml) in the drinking water for 8 weeks. (d) Representative sections of lung metastasis as shown in (c) that are evaluated histologically for H & E, survivin, TUNEL and DAPI. (e) Quantitative analysis of bioluminescence in lung metastasis as shown in (c) (n=6 mice/group). (f) Scoring of TUNEL staining as shown in (d). The results represent the mean number of TUNEL positive tumor cells per field ± s.e.m. (n=4). (g) Mice (n=2) bearing established HCT116 xenografts expressing tet-inducible survivin shRNA were treated with Dox (0.5 mg/ml) in the drinking water for the indicated times. Tumor lysates were immunoblotted with the indicated antibodies. (h) Mice bearing established HCT116 xenografts expressing tet-inducible survivin shRNA were maintained with or without Dox (0.5 mg/ml) in the drinking water. The results represent the mean tumor volume ± s.e.m. (n=6 mice/group). * P < 0.03 for 4E-BP1 4A versus 4E-BP1 WT or vector, and + Dox versus − Dox; # P < 0.05; ns, not significant.
Figure 8.
Targeted inhibition of cap-dependent translation or silencing survivin expression suppresses liver metastasis of CRC. (a) Bioluminescence, GFP images and H & E staining of liver metastasis in athymic nude mice that were injected intrasplenically with HCT116-Luc/GFP cells expressing vector, 4E-BP1 WT or 4E-BP1 4A at week 3 post-injection. (b) Quantitative analysis of bioluminescence in liver metastasis as shown in (a) (n=5 mice/group). (c) Bioluminescence and GFP images of liver metastasis in athymic nude mice that were injected intrasplenically with HCT116-Luc/GFP cells expressing tet-inucible survivin shRNA followed by maintenance with or without Dox (0.5 mg/ml) in the drinking water for 3 weeks. (d) Representative sections of liver metastasis as shown in (c) that are evaluated histologically for H & E, survivin, TUNEL and DAPI. (e) Quantitative analysis of bioluminescence in liver metastasis as shown in (c) (n=5 mice/group). (f) Scoring of TUNEL staining as shown in (d). The results represent the mean number of TUNEL positive tumor cells per field ± s.e.m. (n=4). * P < 0.03 for 4E-BP1 4A versus 4E-BP1 WT or vector, and + Dox versus − Dox; ns, not significant.
DISCUSSION
A recent comprehensive genomic analysis shows that concurrent activation of both RAS/ERK and PI3K/AKT pathways by separate genetic alterations is a common feature of CRC associated with poor outcomes and metastatic progression.4 However, the selective advantage of activating both pathways in CRC progression and metastasis is unknown. In addition, the role of both ERK and AKT signaling pathways in cancer development has been studied mostly by focusing on the transcriptional and posttranslational mechanisms, whereas translational control of cancer development particularly in the process of metastasis has remained under-explored. In our previous study,8 we identified that the co-selection of mutational activation of both ERK and AKT signaling pathways is required to maintain transformed phenotype of CRC through convergent activation of cap-dependent translation. We found that inhibition of either pathway alone has a minor effect on cap-dependent translation and tumor growth, but combined inhibition of both pathways has a synergistic inhibitory effect. In this study, we further show that the cap-dependent translation also plays a critical role in mediating the effects of ERK and AKT activation on CRC cell migration and invasion in vitro and metastatic dissemination in vivo. Our study indicates that activation of cap-dependent translation is a crucial step in CRC progression to metastasis. Genetic inhibition of eIF4E-initiated translation markedly blocks migration, invasion and metastasis of CRC. Conversely, aberrant activation of translation by eIF4E amplification or 4E-BP1 reduction has the opposite effect and profoundly attenuates dependence of CRC cells on the ERK and AKT signaling for migration and invasion. Given that deregulation of cap-dependent translation due to overexpression of eIF4E or loss of 4E-BP1 function often occurs in CRC15–18 and has been shown to cause resistance to inhibition of upstream oncogenic signals,34–36 our findings suggest that directly targeting the convergence of ERK and AKT signaling on translation initiation may be an effective alternative to combination of both kinase inhibitors in advanced and disseminated CRC. Validation of this hypothesis requires additional clinical correlative studies and awaits the development of clinically effective inhibitions of translation initiation.
Metastasis is a complex process that requires the concerted action of numerous proteins, which impart tumor invasiveness, mediate angiogenesis, suppress apoptotic responses and cause proliferation in unrelated microenvironments. Unlike classic oncogenes and tumor suppressor genes, many genes that drive primary tumor progression to metastasis are not altered by mutation but are inappropriately expressed. Studies have primarily focused on the identification of deregulated gene expression at the level of transcription associated with the metastatic process.37 However, other studies have indicated that mRNA alterations do not always correlate precisely with expression of cognate proteins.38 Regulation of mRNA translation provides a direct and rapid means of regulating protein expression.13 A large body of evidence suggests that abundance of eIF4E does not limit overall translation rates but is especially important for translation of certain mRNAs with a highly structured 5′-UTR, such as proto-oncogenes and other growth factors.12–14 Thus, regulation of eIF4E function via upstream signals can provide an immediate level of expression control in the transcript-specific genes and alter the cellular phenotype. In this paper, we identified that survivin harboring a higher GC content (74%) with stable secondary structure in its 5′-UTR is specifically upregulated by ERK and AKT signaling via the eIF4E-dependent translation mechanism in CRC cells in which both signaling pathways are activated (Figure 4). Upregulation of survivin expression has been shown to be associated with CRC metastatic progression.25,39 The biologic effects of survivin are known through apoptotic inhibition, mitotic chromosomal alignment, and a recently characterized function: promotion of cellular motility and metastasis.21–24 Our data show that the translationally-upregulated survivin expression seems to be predominantly associated with promotion of CRC cell motility and metastasis, as noted by the following: 1) migration, invasion and metastasis were markedly compromised in CRC cells depleted of survivin under conditions not affecting apoptosis; 2) overexpression of survivin enhanced motility but not cell growth; and 3) reduction of survivin expression profoundly inhibited CRC metastasis but only caused a slowing of tumor growth. In addition, overexpression of survivin significantly rescued inhibition of cell motility induced by combined inhibition of ERK and AKT or by targeting inhibition of translation. Taken together, these data support the conclusion that the continuous translation of survivin by ERK and AKT signaling is critical for CRC progression to metastasis. Our results do not exclude the possibility that other common targets of ERK and AKT signaling contribute to the effects reported herein. How translationally-regulated survivin interacts with those targets to mediate metastatic progression of CRC is likely to be complex and a matter for further investigation.
mTORC1 is a downstream target of both AKT and ERK signaling27,28 and regulates cap-dependent translation through phosphorylation of 4E-BP1.40 It is thus reasonable to speculate that mTORC1 integrates the effect of ERK and AKT signaling pathways on translational control of CRC in which both pathways are mutationally activated. We show that combined inhibition of AKT and MEK is required to inhibit phosphorylation of 4E-BP1, survivin expression and cell invasion to a degree that is nearly equivalent to the effects of mTOR inhibition with the ATP-site inhibitor AZD8055. We identify that mTORC1 inhibition with depletion of raptor largely contributes to the effects of AZD8055 or combined inhibition of AKT and MEK, whereas inhibition of mTORC2 with knockdown of rictor does not affect the phosporylation of 4E-BP1 and survivin expression (data not shown). Furthermore, we have previously shown that targeted inhibition of mTORC1 via raptor knockdown exhibits a pronounced inhibitory effect on CRC metastasis.41 Thus, our data strongly suggest that mTORC1 functions as a key effector of AKT and ERK activation on translational regulation of CRC invasion and metastasis. These data support findings in a recent study showing that mTORC1 plays a critical role in translational regulation of a metastatic gene expression program for prostate cancer progression.42 Thus, targeting mTORC1 may provide a therapeutic benefit in blocking cancer invasion and metastasis. However, mTORC1 inhibition is known to activate negative feedback mechanisms leading to increased formation mTORC2 that phosphorylates and activates AKT,33,43,44 which may attenuate its therapeutic effects. Our finding that combined inhibition of mTORC1 and AKT activity or dual targeting of mTORC1 and mTORC2 with the mTOR kinase inhibitor induces more profound inhibition of 4E-BP1 phosphorylation and cell migration and invasion than mTORC1 inhibition alone in HCT116 cells is consistent with this hypothesis. Other studies also support this notion and demonstrate that mTOR kinase inhibitors or mTORC1 inhibition in combination with the dual PI3K/mTOR or AKT kinase inhibitors show greater activity in inhibiting the phosphorylation of 4E-BP1 and cap-dependent translation,42,45 and increase the efficacy of anticancer therapy.42,45–47
In summary, our study provides new insights into the biological and therapeutic relevance of translational control in CRC metastatic progression. Our findings reveal that the cap-dependent translation functions as a critical regulatory node that integrates signals from oncogenic activation of the ERK and AKT signaling pathways for promotion of cellular migration and invasion and metastasis of CRC. Mechanistically, we identify that survivin is a key translationally-regulated target of both pathways through convergence on the mTORC1/4E-BP1/eIF4E axis, and continuous translation of survivin by ERK and AKT signaling is crucial for CRC progression to metastasis. Thus, targeting cap-dependent translation that could simultaneously blocks upstream oncogenic signals and their downstream targets would hold potential as a future therapeutic strategy against the metastatic progression of CRC.
MATERIALS AND METHODS
Cell culture, plasmids, siRNA and shRNA
Human colon cancer cell lines were obtained from the American Type CultureCollection (ATCC, Manassas, VA, USA) and maintained in the appropriate medium with supplements as suggested by ATCC. PD0325901, MK2206 and AZD8055 were obtained from Selleck (Houston, TX, USA). The pCMV6-flag-myc-tagged human survivin expression plasmid was purchased from Origene (Rockville, MD, USA). HCT116 cells were transfected with the pCMV6-survivin or pCMV6 empty vector and selected by G418 (500 μg/ml) to generate stable transfectants. HCT116 cells with stable expression of HA-tagged 4E-BP1 and 4E-BP1-4A were previously generated.8 siRNA pool against human 4E-BP1 (L-003005), survivin (L-003459) or non-targeting control siRNA pool (D-001810-10) was from Dharmacon (Chicago, IL, USA). The human 4E-BP1, eIF4E and survivin shRNA expression plasmids were purchased from Open Biosystems (Lafayette, CO, USA). The human raptor and rictor shRNA expression plasmids were obtained from Addgene (Cambridge, MA, USA), and the specificity of the targeting sequences has been verified and described previously.33 For establishing stable transfectants with knockdown of specific protein expression, cell lines were lentivirally infected with the indicated shRNA construct followed by selection with puromycin (2 μg/ml) for 1 week. For bioluminescent tracking, cell lines were retrovirally infected with a reporter construct encoding firefly luciferase and green fluorescent protein (GFP).48 GFP-positive cells were enriched by fluorescence-activated cell sorting.
Migration and invasion assays
Migration and invasion assays were performed in Boyden chambers with coated collagen or Matrigel, respectively, as instructed by the manufacturer (BD Biosciences, San Jose, CA, USA) and described previously.41 Details are provided in the Supplementary Information.
Immunoblot analysis
Cells were lysed in NP-40 lysis buffer, and equal amounts of total protein were immunoblotted as described previously.8 All antibodies used are provided in the Supplementary Information.
Quantitative RT-PCR
Total cellular RNA was isolated using the RNeasy plus mini kit (Qiagen, Valencia, CA, USA). Equal amounts of RNA were used as templates for all reactions. Double-stranded cDNA was generated by using the SuperScript III First Strand Synthesis System (Invitrogen, Grand Island, NY, USA). Real-time PCR reactions were carried out with specific probes for human survivin (Hs00153353_m1) and β-actin (#4352935E) using the StepOne Real-Time PCR system (Applied Biosystems, Foster City, CA, USA).
Cycloheximide (CHX) chase assay
Cells were treated with CHX (20 μg/ml) and harvested at indicated time points. The cells were lysed in NP-40 lysis buffer and equal amounts of total protein were analyzed by immunoblot. To examine the effect of combination with PD0325901 and MK2206, one set of cells was pretreated with the combination of both drugs for 30 min before the addition of CHX.
Proliferation and apoptosis assays
The cell proliferation was analyzed by counting the number of viable cells in response to the treatment with the indicated drugs. The apoptosis assay was performed by flow cytometric analysis as described previously.8 Details are provided in the Supplementary Information.
Quantification of cap-dependent translation
The 5′-UTR cDNAs of human survivin and β-actin were obtained by RT-PCR from a HCT116 cDNA library using the primers as described in Supplementary Table S1. These 5′-UTR cDNAs were each inserted immediately upstream from the translation start codon of the renilla luciferase gene in the bicistronic luciferase reporter vector pcDNA3-rLuc-PolioIRES-fLuc, which directs cap-dependent translation of the renilla luciferase gene and cap-independent Polio IRES-mediated translation of the firefly luciferase gene.49 All sequences were verified by automated sequencing. Cells (80,000) were transfected with each the constructed bicistronic luciferase reporter plasmid (0.2 μg) in 12-well plates using X-tremeGENE Transfection Reagent (Roche Applied Science, Indianapolis, IN, USA). After 24 h transfection, cells were treated with kinase inhibitors for the indicated times, and cell lysates were assayed for renilla luciferase and firefly luciferase activities as described.8 Cap-dependent renilla activity was normalized against cap-independent firefly activity as the internal control. The renilla/firefly luciferase luminescence ratio was calculated for cap-dependent translational activity.
Animal studies
Male athymic nude mice (5–6 weeks old) were purchased from Taconic (Hudson, NY, USA) and maintained and treated under specific pathogen-free conditions. To established CRC xenografts, mice were subcutaneously injected with tumor cells (3×106/mouse) in a 1:1 mixture of media and Matrigel. Prior to initiation of treatment, mice were randomized among control and treated groups (n=6 per group). For induction of survivin shRNA expression, mice received doxycycline (0.5 mg/ml) in the drinking water. Tumor volume was measured and calculated as described previously.8 For the experimental lung and liver metastasis assays, cells with co-expression of firefly luciferase and GFP were injected into the tail vein (1×106/mouse) or spleen (5×106/mouse) of athymic nude mice (n=5 or 6 per group), respectively as described.50 To monitor metastasis, mice were imaged with luciferase signals using the IVIS Spectrum (Caliper Life Science, Hopkinton, MA, USA) and results were analyzed by Living Image 3.0 software.50 The lung and liver metastatic lesions were further examined by GFP imaging in paraffin-embedded sections stained with hematoxylin and eosin (H & E). Immunohistochemical analysis of survivin expression was performed using the Vectastain Elite ABC kit (Vector Laboratories, Burlingame, CA, USA). For the terminal deoxynucleotidyl transferase–mediated nick end labeling (TUNEL) apoptosis assay, the In Situ Cell Death Detection Kit (Roche Applied Science, Indianapolis, IN, USA) was used according to the manufacturer’s instruction as we have reported previously.51
Statistical analysis
The experiments were repeated at least twice. Results are expressed as mean ± SEM. as indicated. An independent Student’s t-test was performed to analyze the luciferase assay; a two-tailed Student’s t-test was used to compare the intergroup. P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
The authors thank Drs. Rina Plattner and Kathleen O’Connor and Ms. Donna Gilbreath for critical reading and editing of this manuscript, Dr. Ronald Blasberg for providing the SFG-FLuc-IRES2-GFP construct. Dr. Piotr Rychahou for the assistance in the animal experiments, and Ms. Dana Napier for the histologic tissue processing. This work was supported by grants from NCI (R01 CA175105 to Q-B She, P30 CA147886 and Gastrointestinal Cancer SPORE P20 CA 150343 to BM Evers), ACS (IRG 85-001-22 to QB She), NIH/NCATS UL1RR033173 (KL2RR0033171 to QB She), and the Markey Cancer Center Start-up fund (to QB She).
ABBREVIATIONS
- 4E-BP1
4E-binding protein 1
- CRC
colorectal cancer
- DMSO
dimethyl sulfoxide
- eIF4E
eukaryotic translation initiation factor 4E
- ERK
extracellular signal–regulated kinase
- MAPK
mitogen-activated protein kinase
- MEK
MAPK extracellular signal–regulated kinase
- mTOR
mammalian target of rapamycin
- mTORC1
mammalian target of rapamycin complex 1
- PI3K
phosphoinositide 3-kinase
- PIK3CA
p110α catalytic subunit of PI3K
- PTEN
phosphatase and tensin homolog
- UTR
untranslated region
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Chu E. An update on the current and emerging targeted agents in metastatic colorectal cancer. Clin Colorectal Cancer. 2012;11:1–13. doi: 10.1016/j.clcc.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 3.Sebolt-Leopold JS, Herrera R. Targeting the mitogen-activated protein kinase cascade to treat cancer. Nat Rev Cancer. 2004;4:937–947. doi: 10.1038/nrc1503. [DOI] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parsons DW, Wang TL, Samuels Y, Bardelli A, Cummins JM, DeLong L, et al. Colorectal cancer: mutations in a signalling pathway. Nature. 2005;436:792. doi: 10.1038/436792a. [DOI] [PubMed] [Google Scholar]
- 6.Wee S, Jagani Z, Xiang KX, Loo A, Dorsch M, Yao YM, et al. PI3K pathway activation mediates resistance to MEK inhibitors in KRAS mutant cancers. Cancer Res. 2009;69:4286–4293. doi: 10.1158/0008-5472.CAN-08-4765. [DOI] [PubMed] [Google Scholar]
- 7.Balmanno K, Chell SD, Gillings AS, Hayat S, Cook SJ. Intrinsic resistance to the MEK1/2 inhibitor AZD6244 (ARRY-142886) is associated with weak ERK1/2 signalling and/or strong PI3K signalling in colorectal cancer cell lines. Int J Cancer. 2009;125:2332–2341. doi: 10.1002/ijc.24604. [DOI] [PubMed] [Google Scholar]
- 8.She QB, Halilovic E, Ye Q, Zhen W, Shirasawa S, Sasazuki T, et al. 4E-BP1 is a key effector of the oncogenic activation of the AKT and ERK signaling pathways that integrates their function in tumors. Cancer Cell. 2010;18:39–51. doi: 10.1016/j.ccr.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halilovic E, She QB, Ye Q, Pagliarini R, Sellers WR, Solit DB, et al. PIK3CA Mutation Uncouples Tumor Growth and Cyclin D1 Regulation from MEK/ERK and Mutant KRAS Signaling. Cancer Res. 2010;70:6804–6814. doi: 10.1158/0008-5472.CAN-10-0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 11.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mamane Y, Petroulakis E, Martineau Y, Sato TA, Larsson O, Rajasekhar VK, et al. Epigenetic activation of a subset of mRNAs by eIF4E explains its effects on cell proliferation. PLoS One. 2007;2:e242. doi: 10.1371/journal.pone.0000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livingstone M, Atas E, Meller A, Sonenberg N. Mechanisms governing the control of mRNA translation. Phys Biol. 2010;7:021001. doi: 10.1088/1478-3975/7/2/021001. [DOI] [PubMed] [Google Scholar]
- 14.Silvera D, Formenti SC, Schneider RJ. Translational control in cancer. Nat Rev Cancer. 2010;10:254–266. doi: 10.1038/nrc2824. [DOI] [PubMed] [Google Scholar]
- 15.Rosenwald IB, Chen JJ, Wang S, Savas L, London IM, Pullman J. Upregulation of protein synthesis initiation factor eIF-4E is an early event during colon carcinogenesis. Oncogene. 1999;18:2507–2517. doi: 10.1038/sj.onc.1202563. [DOI] [PubMed] [Google Scholar]
- 16.Berkel HJ, Turbat-Herrera EA, Shi R, de Benedetti A. Expression of the translation initiation factor eIF4E in the polyp-cancer sequence in the colon. Cancer Epidemiol Biomarkers Prev. 2001;10:663–666. [PubMed] [Google Scholar]
- 17.Martin ME, Perez MI, Redondo C, Alvarez MI, Salinas M, Fando JL. 4E binding protein 1 expression is inversely correlated to the progression of gastrointestinal cancers. Int J Biochem Cell Biol. 2000;32:633–642. doi: 10.1016/s1357-2725(00)00007-8. [DOI] [PubMed] [Google Scholar]
- 18.Armengol G, Rojo F, Castellvi J, Iglesias C, Cuatrecasas M, Pons B, et al. 4E-binding protein 1: a key molecular “funnel factor” in human cancer with clinical implications. Cancer Res. 2007;67:7551–7555. doi: 10.1158/0008-5472.CAN-07-0881. [DOI] [PubMed] [Google Scholar]
- 19.Yap TA, Yan L, Patnaik A, Fearen I, Olmos D, Papadopoulos K, et al. First-in-man clinical trial of the oral pan-AKT inhibitor MK-2206 in patients with advanced solid tumors. J Clin Oncol. 2011;29:4688–4695. doi: 10.1200/JCO.2011.35.5263. [DOI] [PubMed] [Google Scholar]
- 20.Hanrahan AJ, Schultz N, Westfal ML, Sakr RA, Giri DD, Scarperi S, et al. Genomic complexity and AKT dependence in serous ovarian cancer. Cancer Discov. 2012;2:56–67. doi: 10.1158/2159-8290.CD-11-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altieri DC. Survivin, cancer networks and pathway-directed drug discovery. Nat Rev Cancer. 2008;8:61–70. doi: 10.1038/nrc2293. [DOI] [PubMed] [Google Scholar]
- 22.Mehrotra S, Languino LR, Raskett CM, Mercurio AM, Dohi T, Altieri DC. IAP regulation of metastasis. Cancer Cell. 2010;17:53–64. doi: 10.1016/j.ccr.2009.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKenzie JA, Liu T, Goodson AG, Grossman D. Survivin enhances motility of melanoma cells by supporting Akt activation and alpha 5 integrin upregulation. Cancer Res. 2010;70:7927–7937. doi: 10.1158/0008-5472.CAN-10-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang M, Coen JJ, Suzuki Y, Siedow MR, Niemierko A, Khor LY, et al. Survivin is a potential mediator of prostate cancer metastasis. Int J Radiat Oncol Biol Phys. 2010;78:1095–1103. doi: 10.1016/j.ijrobp.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chu XY, Chen LB, Wang JH, Su QS, Yang JR, Lin Y, et al. Overexpression of survivin is correlated with increased invasion and metastasis of colorectal cancer. J Surg Oncol. 2012;105:520–528. doi: 10.1002/jso.22134. [DOI] [PubMed] [Google Scholar]
- 26.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell. 2002;10:151–162. doi: 10.1016/s1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- 28.Ma L, Chen Z, Erdjument-Bromage H, Tempst P, Pandolfi PP. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell. 2005;121:179–193. doi: 10.1016/j.cell.2005.02.031. [DOI] [PubMed] [Google Scholar]
- 29.Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, Ruggero D, et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7:e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chresta CM, Davies BR, Hickson I, Harding T, Cosulich S, Critchlow SE, et al. AZD8055 is a potent, selective, and orally bioavailable ATP-competitive mammalian target of rapamycin kinase inhibitor with in vitro and in vivo antitumor activity. Cancer Res. 2010;70:288–298. doi: 10.1158/0008-5472.CAN-09-1751. [DOI] [PubMed] [Google Scholar]
- 32.Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 33.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 34.Dilling MB, Germain GS, Dudkin L, Jayaraman AL, Zhang X, Harwood FC, et al. 4E-binding proteins, the suppressors of eukaryotic initiation factor 4E, are down-regulated in cells with acquired or intrinsic resistance to rapamycin. J Biol Chem. 2002;277:13907–13917. doi: 10.1074/jbc.M110782200. [DOI] [PubMed] [Google Scholar]
- 35.Ilic N, Utermark T, Widlund HR, Roberts TM. PI3K-targeted therapy can be evaded by gene amplification along the MYC-eukaryotic translation initiation factor 4E (eIF4E) axis. Proc Natl Acad Sci U S A. 2011;108:E699–708. doi: 10.1073/pnas.1108237108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zindy P, Berge Y, Allal B, Filleron T, Pierredon S, Cammas A, et al. Formation of the eIF4F translation-initiation complex determines sensitivity to anticancer drugs targeting the EGFR and HER2 receptors. Cancer Res. 2011;71:4068–4073. doi: 10.1158/0008-5472.CAN-11-0420. [DOI] [PubMed] [Google Scholar]
- 37.Nguyen DX, Massague J. Genetic determinants of cancer metastasis. Nat Rev Genet. 2007;8:341–352. doi: 10.1038/nrg2101. [DOI] [PubMed] [Google Scholar]
- 38.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 39.Hernandez JM, Farma JM, Coppola D, Hakam A, Fulp WJ, Chen DT, et al. Expression of the antiapoptotic protein survivin in colon cancer. Clin Colorectal Cancer. 2011;10:188–193. doi: 10.1016/j.clcc.2011.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mamane Y, Petroulakis E, LeBacquer O, Sonenberg N. mTOR, translation initiation and cancer. Oncogene. 2006;25:6416–6422. doi: 10.1038/sj.onc.1209888. [DOI] [PubMed] [Google Scholar]
- 41.Gulhati P, Bowen KA, Liu J, Stevens PD, Rychahou PG, Chen M, et al. mTORC1 and mTORC2 regulate EMT, motility, and metastasis of colorectal cancer via RhoA and Rac1 signaling pathways. Cancer Res. 2011;71:3246–3256. doi: 10.1158/0008-5472.CAN-10-4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun SY, Rosenberg LM, Wang X, Zhou Z, Yue P, Fu H, et al. Activation of Akt and eIF4E survival pathways by rapamycin-mediated mammalian target of rapamycin inhibition. Cancer Res. 2005;65:7052–7058. doi: 10.1158/0008-5472.CAN-05-0917. [DOI] [PubMed] [Google Scholar]
- 44.O’Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mazzoletti M, Bortolin F, Brunelli L, Pastorelli R, Di Giandomenico S, Erba E, et al. Combination of PI3K/mTOR inhibitors: antitumor activity and molecular correlates. Cancer Res. 2011;71:4573–4584. doi: 10.1158/0008-5472.CAN-10-4322. [DOI] [PubMed] [Google Scholar]
- 46.Thomas HE, Mercer CA, Carnevalli LS, Park J, Andersen JB, Conner EA, et al. mTOR inhibitors synergize on regression, reversal of gene expression, and autophagy in hepatocellular carcinoma. Sci Transl Med. 2012;4:139ra184. doi: 10.1126/scitranslmed.3003923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Floc’h N, Kinkade CW, Kobayashi T, Aytes A, Lefebvre C, Mitrofanova A, et al. Dual Targeting of the Akt/mTOR Signaling Pathway Inhibits Castration-Resistant Prostate Cancer in a Genetically Engineered Mouse Model. Cancer Res. 2012;72:4483–4493. doi: 10.1158/0008-5472.CAN-12-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moroz E, Carlin S, Dyomina K, Burke S, Thaler HT, Blasberg R, et al. Real-time imaging of HIF-1alpha stabilization and degradation. PLoS One. 2009;4:e5077. doi: 10.1371/journal.pone.0005077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pause A, Belsham GJ, Gingras AC, Donze O, Lin TA, Lawrence JC, et al. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 50.Zaytseva YY, Rychahou PG, Gulhati P, Elliott VA, Mustain WC, O’Connor K, et al. Inhibition of Fatty Acid Synthase Attenuates CD44-Associated Signaling and Reduces Metastasis in Colorectal Cancer. Cancer Res. 2012;72:1504–1517. doi: 10.1158/0008-5472.CAN-11-4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.She QB, Chandarlapaty S, Ye Q, Lobo J, Haskell KM, Leander KR, et al. Breast tumor cells with PI3K mutation or HER2 amplification are selectively addicted to Akt signaling. PLoS ONE. 2008;3:e3065. doi: 10.1371/journal.pone.0003065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.