Abstract
Determining how somatic copy-number alterations (SCNAs) promote cancer is an important goal. We characterized SCNA patterns among 4934 cancers from The Cancer Genome Atlas Pan-Cancer dataset. Whole-genome doubling, observed in 37% of cancers, was associated with higher rates of every other type of SCNA, TP53 mutations, CCNE1 amplifications, and alterations of the PPP2R complex. SCNAs that were internal to chromosomes tended to be shorter than telomere-bounded SCNAs, suggesting different mechanisms of generation. Significantly recurrent focal SCNAs were observed in 140 regions, including 102 without known oncogene or tumor suppressor gene targets and 50 with significantly mutated genes. Amplified regions without known oncogenes are enriched for genes involved in epigenetic regulation. When levels of genomic disruption were accounted for, 7% of region pairs anticorrelated, and these tended to encompass genes whose proteins physically interact, suggesting related functions. These results provide insights into mechanisms of generation and functional consequences of cancer SCNAs.
Introduction
Somatic copy-number alterations (SCNAs) affect a larger fraction of the genome in cancers than do any other type of somatic genetic alteration1–5. SCNAs play critical roles in activating oncogenes and inactivating tumor suppressors3,6–12 and an understanding of the biological and phenotypic effects of SCNAs has led to substantial advances in cancer diagnostics and therapeutics13–16.
A primary challenge in understanding SCNAs is to distinguish the driver events that contribute to oncogenesis and cancer progression from the passenger SCNAs that are acquired during cancer evolution but do not contribute towards it17–20. Positively selected SCNAs will tend to recur across cancers at elevated rates1,4,5. However, SCNAs may also recur in the absence of positive selection due to increased rates of generation or decreased negative selection21,22. For this reason, it is important to understand how mechanisms of SCNA generation, their temporal ordering, and negative selection shape the distribution of SCNAs genome-wide21–25.
A second challenge is to identify the oncogene and tumor suppressor gene targets of the driver SCNAs (which often encompass many genes) and elucidate the SCNA’s functional roles. The context of the SCNA can be informative. Positive correlations with other genetic events may indicate functional synergies, while anticorrelations may indicate functional redundancies because redundant events would not be required by the same cancer. Several approaches have been developed to determine functional effects of genetic events based on anticorrelation patterns26–28.
Here, we address these challenges through the analysis of 4934 cancer copy-number profiles across 11 cancer types, assembled through The Cancer Genome Atlas Project Pan-Cancer effort, enabling analysis of large numbers of cancers and comparison of patterns of copy-number change across cancer types. We have integrated rigorous statistical approaches into these analyses, including absolute allelic copy-number profiling29, as well as novel computational tools to determine individual SCNA events and their temporal ordering from these profiles, and to identify functionally relevant correlations between SCNAs.
Results
Cancer purities, ploidies, and rates of copy-number alteration within and across cancer types
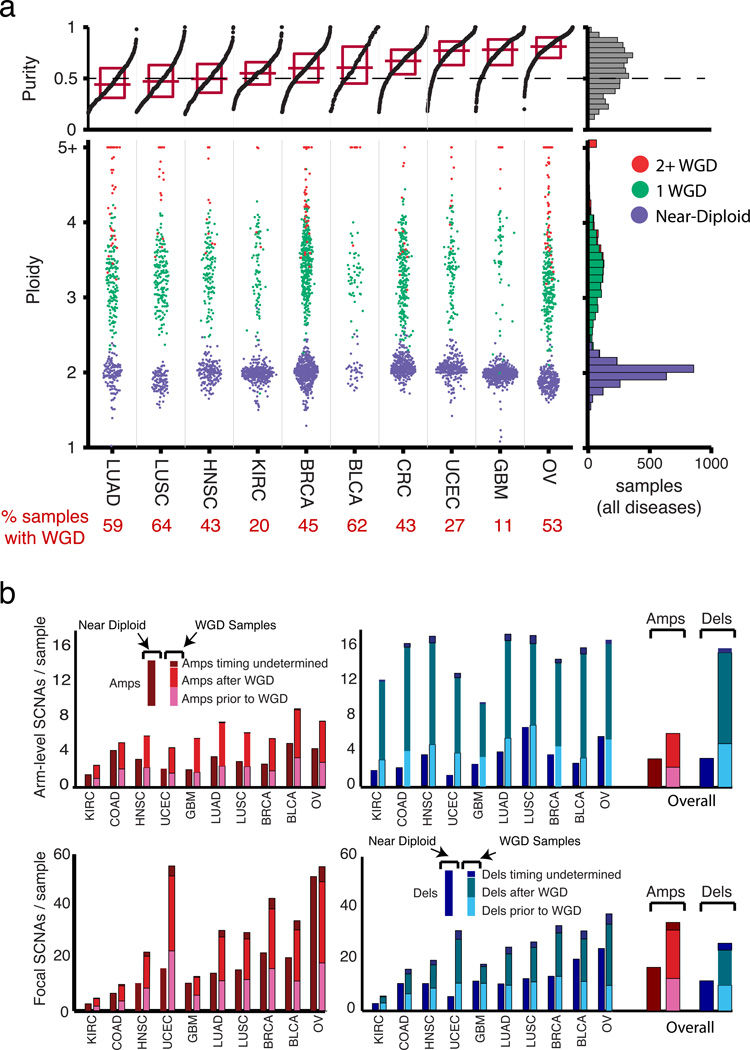
We analyzed the copy-number profiles of 4934 primary cancer specimens across 11 cancer types (minimum 136 for bladder cancer; maximum 880 samples for breast cancer; colon and rectal adenocarcinomas were combined; Supplementary Table 1). In each cancer, we determined copy-numbers at each of 1,559,049 loci relative to the median copy-number genome-wide, using Affymetrix SNP6 arrays and previously described algorithms1. For 3847 cancers, we also determined the purity, ploidy, and absolute allelic copy-number profiles29 of the malignant cells using SNP6 array data and, in 1069 cases, matched whole-exome sequencing data (Supplementary Table 1). In the other 1087 cases, purity and ploidy estimates were ambiguous and left uncalled. This included all cases of acute myeloid leukemias [LAMLs], which exhibit very few SCNAs.
We then inferred the sequence of somatic copy-alteration (SCNA) events that led to each copy-number profile, using the most parsimonious set of SCNAs that could generate the observed absolute allelic copy-numbers (Supplementary Fig. 1a, Methods). We determined the lengths, locations, and numbers of copies of change for each SCNA and, in many cases, their allelic structure (Supplementary Fig. 1b). We identified a total of 202,244 SCNAs, a median of 39 per cancer sample, comprising six categories: focal SCNAs that were shorter than one chromosome arm (a median of 11 amplifications and 12 deletions per sample); arm-level SCNAs that were chromosome-arm length or longer (a median of three amplifications and five deletions per sample); copy-neutral loss-of-heterozygosity events (cnLOHs), in which one allele had been deleted and the other amplified coextensively (a median of one per sample); and whole-genome duplications (WGDs, in 37% of cancers). By amplifications and deletions, we refer to copy-number gains and losses, respectively, of any length and amplitude.
Estimated purities and ploidies per cancer varied substantially within and across diseases (Fig. 1a). The purity estimates correlated with estimates derived from measurements of leukocyte and lymphocyte contamination using DNA methylation data from the same cancers (Supplementary Fig. 1c) (Shen et al, unpublished data)30, but tended to indicate lower purity, consistent with the presence of non-hematopoietic contaminating normal cells. Average ploidies within diseases mirrored their frequencies of WGD. The average estimated ploidy within samples that had undergone a single WGD was 3.31 (not four), suggesting that WGD events are associated with large amounts of genome loss. By contrast, samples that had not undergone WGD had an average estimated ploidy of 1.99.
Figure 1. Distribution of SCNAs across lineages.
(a) Sample purities (top panel) and ploidies (bottom panel) across lineages (see Supplementary Table 1 for a list of lineage abbreviations). Near-diploid samples are designated in purple; cancers that have undergone one or more than one WGD event are designated by green and red, respectively. Summarized data across all lineages are indicated on the right. (b) Numbers of arm-level (top) and focal (bottom) amplifications (left) and deletions (right) across lineages. For each lineage, near-diploid and WGD samples are indicated by bars on the left and right, respectively; events among WGD samples are resolved according to their timing relative to WGD.
Compared to the near-diploid cancers within each disease, cancers with WGD had higher rates of every other type of SCNA (Fig. 1b) and twice the rate of SCNAs overall. Across diseases, overall SCNA rates largely reflected rates of WGD (Supplementary Fig. 1d).
In cancers with WGD, most other SCNAs occurred after WGD (Fig. 1b, see Methods). The fractions of amplifications and deletions that were estimated to occur prior to WGD were highly correlated across diseases (R=0.64, Supplementary Fig. 1e), indicating a consistent estimate for the timing of WGD with respect to other SCNAs. WGD was inferred to occur earliest relative to focal SCNAs among diseases where WGD was common (ovarian, bladder, and colorectal cancers), and after most focal SCNAs in diseases in which WGD was least common (glioblastoma and kidney clear cell carcinoma).
SCNA lengths suggest varied mechanisms of generation
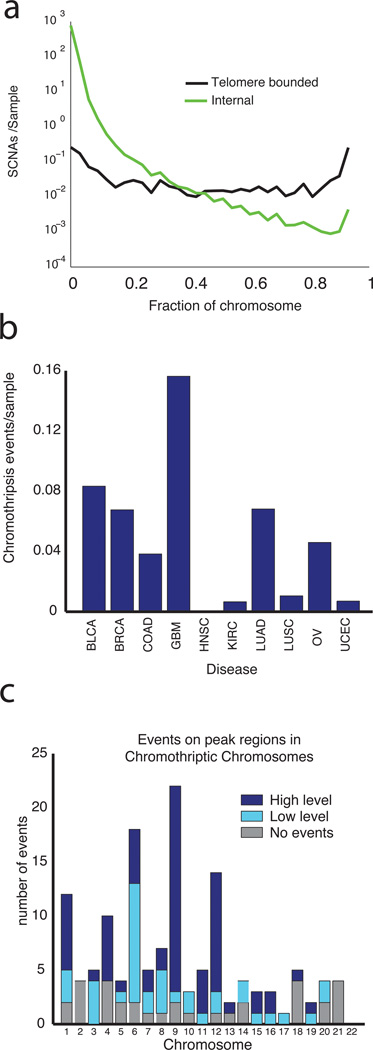
Focal SCNAs for which one boundary is the telomere (telomere-bounded) tend to be longer than SCNAs in which both boundaries are internal to a chromosome (median SCNA length: amplifications 19.6 Mb versus 0.9 Mb; deletions: 22.7 Mb versus 0.7 Mb, for telomere-bounded and internal events respectively). These differences reflect differences across the entire length distributions of internal and telomere-bounded events. Focal internal SCNAs were observed at frequencies inversely proportional to their lengths (Fig. 2a, Supplementary Fig. 2a–b), as noted previously1. However, telomere-bounded SCNAs tend to follow a superposition of 1/length and uniform length distributions. These distributions are the same whether measuring distance by kb, number of array markers, or number of genes, indicating that they do not result from variations in array resolution or gene density genome-wide (data not shown). Focal, telomere-bounded SCNAs also accounted for more SCNAs (12% and 26% of focal amplifications and deletions, respectively) than expected assuming random SCNA locations (p<0.0001). Both telomere-bounded and internal SCNAs are more likely to end within the centromere than expected given the centromere’s length (Supplementary Fig. 2c), but the differences in their length distributions remain when centromere-bounded events are excluded. Differences between telomere-bounded and internal SCNAs are even more marked for cnLOH events (Supplementary Fig. 2d).
Figure 2. Characteristics of different types of SCNA.
(a) The distribution of lengths of SCNAs originating at telomeres (black line) compared to SCNAs that are internal to the chromosome. (b) Rates of chromothripsis across lineages. (c) Rates of chromothripsis across chromosomes. Chromothripsis events that involved peak regions of amplification and deletion (see below) are indicated in blue (dark blue: amplifications >4.4 copies or deletions<−1; light blue: low-level events involving smaller changes); events that do not involve peak regions are indicated in grey.
We detected chromothripsis in 5% of samples, ranging from none of head and neck squamous cell carcinomas to 16% of glioblastomas (Fig. 2c; see Methods). The rate of chromothripsis was not related to overall rates of SCNA (r=0.13, p=0.3). As previously reported31, samples with chromothripsis were more likely to have chromothripsis on more than one chromosome (14/122 samples with chromothripsis had two to three such events, p=0.003).
Many chromothripsis events were concentrated in a few genomic regions, often associated with known driver events (Fig. 2d). In glioblastomas, chromothripsis events were concentrated in chromosomes 9 and 12 and corresponded respectively to homozygous loss of CDKN2A (20/22 samples) and coamplification of discontinuous regions containing CDK4 and MDM2 (9/12 samples). Across all cancers, 72% of chromothripsis events included a GISTIC peak region (see below).
Recurrent focal SCNAs
We identified 70 recurrently amplified and 70 recurrently deleted regions in a unified “Pan-Cancer” analysis across all lineages (Fig 3a, Supplementary Fig. 2e, Supplementary Table 2). SCNAs involving these regions included 21% of all focal amplifications and 23% of all focal deletions. Focal SCNAs within peak regions tended to be shorter than focal SCNAs elsewhere on the chromosome (median 12.2 Mb in peak regions vs 19.4 Mb genomewide, p<0.0001), and were more often high-amplitude events (p<0.0001). The number of focal SCNAs involving peak regions per sample tracked the total number of SCNAs (r=0.84, p<0.0001), ranging from 0.4 focal SCNAs in the typical acute myeloid leukemia to 12.3 focal SCNAs in the typical ovarian cancer (mean 5.2).
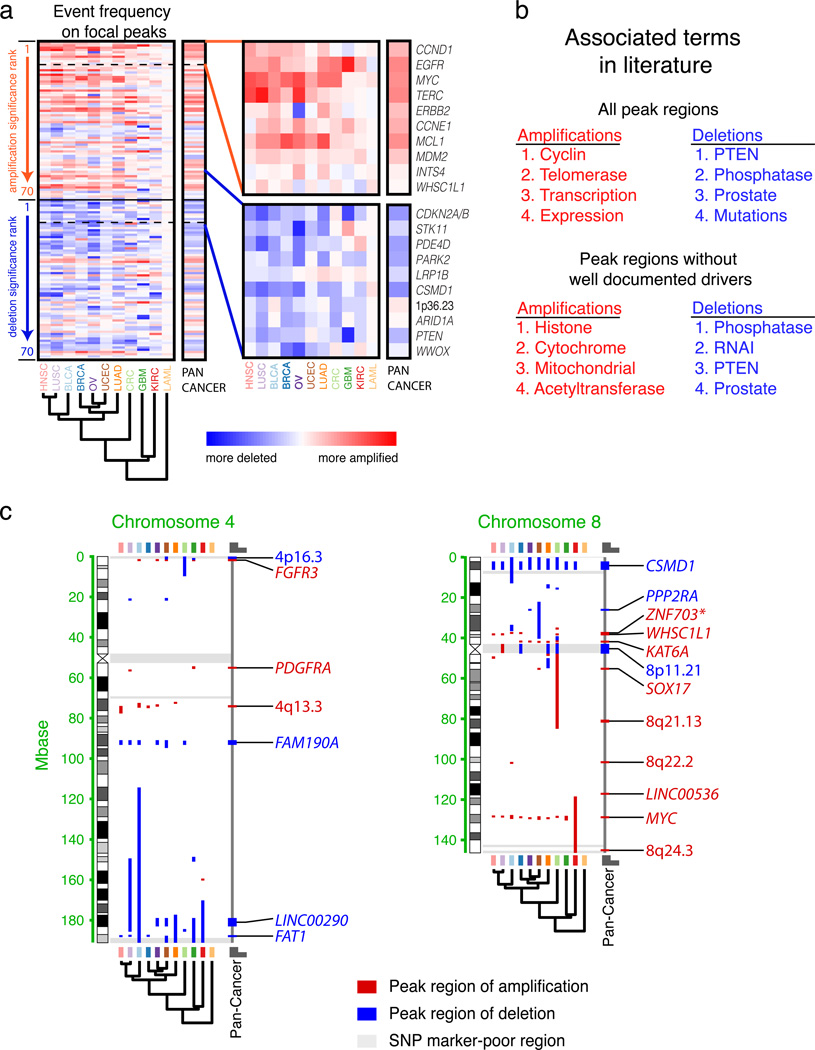
Figure 3. Significantly recurrent focal SCNAs.
(a) Frequencies of amplification minus frequencies of deletion (red and blue indicated propensity to amplifications and deletions, respectively) across lineages (x-axis; see Supplementary Table 1 for a list of lineage abbreviations) for all 84 significant peak regions of SCNA, arranged in order of significance (y-axis). The ordering of lineages reflects the results of unsupervised hierarchical clustering of these data. Magnified views of the values for the ten most significant amplification and deletion peaks, respectively, are shown to the right, alongside candidate targets for these regions. Criteria for selecting the indicated candidates are described in the Methods. (b) Associated terms in literature in peak regions containing fewer than 25 genes, according to a GRAIL analysis of (top) all peak regions and (bottom) peak regions without known cancer genes or large genes. (c) Illustration of locations of peak regions within chromosomes four and eight (other chromosomes are displayed in Supplementary Figure 3) across cancer types (designated by boxes on top and bottom colored according to the scheme in panel a) and the Pan-Cancer analysis (right-most column, denoted by a black line). Peaks are designated by candidate targets for each region, selected according to criteria described in the Methods.
Tissue types of similar lineages tended to have similar rates of amplification and deletion in peak SCNA regions (Fig. 3a). We observed clusters of squamous cell carcinomas (head and neck squamous cell carcinoma, lung squamous cell carcinoma and bladder cancer) and reproductive cancers (ovarian and endometrial cancer) with breast cancer.
The 70 peak regions of amplification contain a median of three genes each (including microRNAs), with 60 peaks containing fewer than 25 genes. Twenty-four of these peak regions contain an oncogene known to be activated by amplification (Supplementary Table 2), including seven of the top ten regions (CCND1, EGFR, MYC, ERBB2, CCNE1, MCL1, and MDM2). The ninth and tenth most significant regions (11q14.1 and 8p11.23, respectively) do not contain known oncogenes, but the latter contains the histone methyltransferase WHSC1L1 and is 18 kb away from the known amplified oncogene FGFR1. The fourth most significantly amplified peak region (3q26.2) contained TERC, which encodes the RNA substrate for the known oncogene TERT, which is itself in a peak region of amplification (5p15.33). Another peak with eight genes (9p13.3) contain RMRP, another TERT-associated RNA32.
The 70 peak regions of deletion contain a median of four genes (including microRNAs), with 52 peaks containing fewer than 25 genes. Twenty-two of these regions contain one of the 100 largest genes in the genome and 12 contain known tumor suppressors (Supplementary Table 2; two additional large regions contain the known tumor suppressors ATM and NOTCH1). Four others each contain a single gene (PPP2R2A, PTTG1IP, FOXK2, and LINC00290). We discuss PPP2R2A and its binding partner PPP2R1A (which is significantly mutated in the same set of cancers [Lawrence et al., unpublished data]33,34) in greater detail below. LINC00290 is a long non-coding RNA, a group whose role in cancer is increasingly being appreciated35,36. Two other regions contain suspected tumor suppressors (ERRFI137, and FOXC138).
The features most associated with genes in the amplification and deletion peak regions are known to be associated with cancer (Fig. 3b). We applied GRAIL39, which uses literature citations to find common features of genes in selected regions of the genome. We considered amplifications and deletions separately, and only peaks with fewer than 25 genes.
Among the 37 peak regions of amplification with fewer than 25 genes and without known targets (Supplementary Table 2), the most associated features were related to epigenetic and mitochondrial regulation: “Histone”, “Cytochrome”, “Mitochondrial”, and “Acetyltransferase” (Fig. 3b). Thirteen of these 37 regions contain chromatin-state and histone-modifying genes (Supplementary Table 2), reflecting significant enrichment (p<0.0001)40. Among these, five (BRD4, KAT6A, KAT6B, NSD1, and PHF1) are subject to recurrent rearrangements in leukemias, sarcomas, and midline carcinomas41–45. The BRD4 peak also contains NOTCH3, another potential oncogene46. Two others, KDM2A and KDM5A, are reported to regulate the activity of TP53 and RB1, respectively47,48. The finding that multiple peak regions of amplification contain epigenetic regulators is consistent with growing evidence suggesting epigenetic alterations and chromatin remodeling plays a critical role in many forms of cancer49–51. Ten regions contain genes encoding mitochronia-associated proteins (Supplementary Table 2); none of these are subject to recurrent rearrangements in cancer. The 21 peak regions of deletion with fewer than 25 genes and without known tumor suppressor or large genes were most associated with “Pten”, “Phosphatase”, “Leucine”, and “Prostate”.
Fifty of the 140 peak regions contain a significantly mutated gene, including 23 regions without known oncogene or tumor suppressor gene targets and 32 regions with fewer than 25 genes (Supplementary Table 2). We calculated the significance of mutations (including both point mutations and small insertion-deletion events identified in the paired sequencing data) for each gene in each region using the methods of [Lawrence et al, unpublished data]33,34 and corrected for multiple hypotheses reflecting the number of genes in the region. In three cases, there were two significantly mutated genes per peak, for a total of 35 significantly mutated genes. These 35 genes included eight of the 23 known amplification-activated oncogenes and all of the 12 known tumor suppressor genes in these peak regions (Supplementary Table 2). An additional two of the 35 genes (both in amplification peaks) are oncogenes known to be activated by mutations but not by amplifications.
Frame-shift and nonsense mutations that are likely to cause loss of function were significantly enriched in genes in deleted regions (p=0.0002), accounting for 19% of these mutations compared to 12% of mutations found in genes in amplified regions. We excluded regions with known oncogenes or tumor suppressor genes or more than 25 genes from this analysis. These findings are consistent with the prediction that deleted regions without known tumor suppressors are enriched for novel tumor suppressors or genes whose functions are non-essential.
Most peak regions in lineage-specific analyses intersected peak regions in other lineages, and indeed in the Pan-Cancer analysis (Fig. 3c, Supplementary Fig. 3). We obtained a median of 74 peak regions for each lineage (ranging from 25 in acute myeloid leukemia to 95 in endometrial cancer; 42% were amplification peaks and 58% were deletion peaks; Supplementary Table 3), resulting in a total of 770 peak regions. Of these, 84% intersected peak regions in at least one other lineage (p<0.0001), and 65% intersected peak regions in the Pan-Cancer analysis. Peak regions tended to be larger in the lineage-specific than the Pan-Cancer analyses (1.4 vs 0.7 Mb), indicating the improved resolution of the Pan-Cancer analysis.
Nevertheless, some significant SCNAs were identified in lineage-specific but not the Pan-Cancer analysis. Across all lineages, we identified 229 peaks not present in the Pan-Cancer analysis, including amplifications of the known amplified oncogenes MET, CCND2, ERBB3, and MYCN and deletions of the known tumor suppressor genes TP53 and CDKN2C.
Correlations reflect overall levels of genomic disruption
For each pair of peak regions, we looked for positive and negative correlations between focal SCNAs involving these regions (Fig. 4a). We compared the number of samples with SCNAs involving both regions between observed data and permuted data in which SCNAs were randomly assigned to samples while maintaining genomic positions and SCNA structure. We only permuted SCNAs within lineages (and sub-lineages when available) to avoid lineage-dependent confounders, and evaluated correlations between regions on different chromosomes to avoid correlations due to chromosomal structure (see Methods). We focused on peak regions with less than 25 genes.
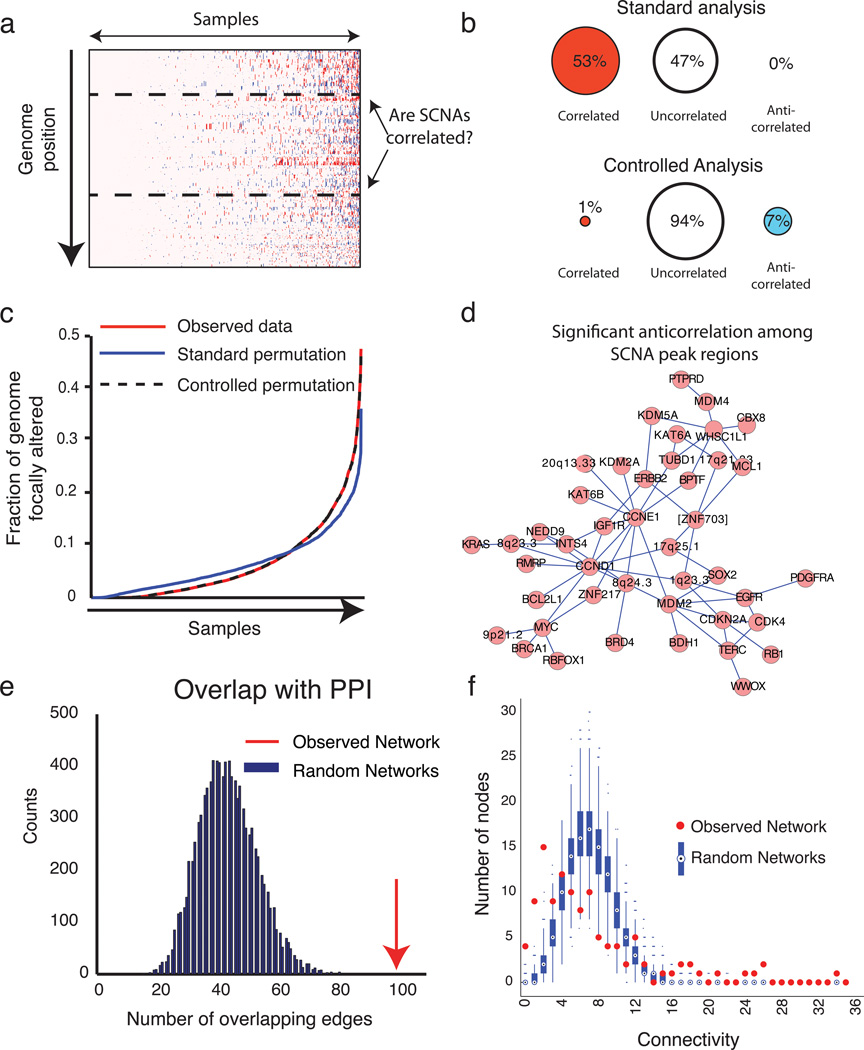
Figure 4. Correlations between SCNAs.
(a) Illustration of question, displaying a heatmap of copy-number profiles across 4934 cancers (x-axis), arranged in order of increasing genomic disruption. (b) Fraction of region pairs exhibiting significant positive correlation (left), negative correlation (right), or neither (middle), using standard analysis techniques (top) and after controlling for variations in genomic disruption (bottom). (c) Fraction of genome involved in focal SCNAs in samples displayed in panel (a) among observed data (red line), permutations generated by standard techniques (blue line) and permutations that maintain levels of genomic disruption (black dashed line). (d) Genetic interactome map for high-level SCNAs. Nodes represent peak regions with fewer than 25 genes and are connected by edges if focal high-level SCNAs (amplifications to >4.4 copies and deletions to <1 copy) are significantly anticorrelated. (e) The number of significant anticorrelations that overlap known protein-protein interactions in the observed genetic interactome network (red arrow) and permuted networks (blue bars). These results are from the analysis of all SCNAs; results from the high-level analysis are displayed in Supplementary Figure 4d. (f) Distribution of connectivity values (number of nodes to which each node is connected) for the observed genetic interactome network (red dots) and permuted networks (box plots) in the all-SCNAs analysis.
We identified significant positive correlations (q<0.25) between 53% of region pairs, but no significant anticorrelations (Fig. 4b). The high rate of positive correlations results from widely differing levels of genomic disruption across samples, which are not maintained in permuted datasets (Fig. 4c). Similar results are obtained with other standard statistical approaches such as Fisher’s exact tests (data not shown). These findings indicate that varying levels of overall genomic disruption confound analyses of functionally relevant correlations between SCNAs.
We therefore re-evaluated correlations between SCNAs after controlling for genomic disruption, by maintaining in the permuted data the fractions of the genome affected by each of amplifications and deletions in each sample (Fig. 4c, Supplementary Fig. 4a–b; Methods). We performed the analysis in two ways: evaluating all SCNAs (Supplementary Table 4), and evaluating only high-level amplifications and homozygous deletions (Supplementary Table 4; see Methods). In many cases, high-level amplification or homozygous deletion may be necessary to activate an oncogene or inactivate a tumor suppressor gene16 and in such cases, correlated features may be masked by noise in lower level events.
When evaluating all SCNAs, we identified significant positive correlations between <1% of region pairs (40 interactions, Supplementary Table 4) and anticorrelations between 7% of region pairs (396 interactions, Fig. 4b, Supplementary Table 4). Correcting for genomic disruption altered the estimated significance of these interactions and also changed the rank ordering of those significance estimates (Supplementary Fig. 4c). High-level amplifications and homozygous deletions are relatively rare, limiting our power to detect anticorrelations in the high-level analysis. Among the 1094 interactions we were powered to detect, we observed positive correlations between <1% of region pairs (3 interactions, Supplementary Table 4) and anticorrelations between 10% of region pairs (108 interactions, Fig. 4d, Supplementary Table 4). The three correlations included deletions of CDKN2A with amplifications of EGFR, amplifications of PDGFR with amplifications of CDK4, and deletions of PPP2RA with amplifications of 19p13.2.
We predicted that anticorrelated SCNAs would often indicate functional redundancies, and therefore genes in the affected regions would often be in similar pathways and interact physically. We tested this hypothesis by comparing networks representing significantly anticorrelated SCNAs (“anticorrelation networks”) with DAPPLE, a set of curated protein-protein interactions (PPIs)39 (see Methods).
Networks formed by our anticorrelations analyses and by PPIs significantly overlapped (p<0.0001 and p=0.006 for all-SCNA and high-level analyses, respectively, Fig. 4e, Supplementary Fig. 4d). For example, in the analysis of all SCNAs, we observed 100 overlapping edges, a 2-fold increase over the 43.4 overlapping edges expected by chance. This significance was not observed for correlated events (p=1 for both all-SCNA and high–level analyses). These results suggest that the observed anticorrelations are related to biological interactions.
The anticorrelations networks were enriched for both isolated nodes and highly connected “hub” regions (Fig. 4f). To analyze the structure of these networks, we generated control anticorrelation networks representing the most significant edges from permuted data in which we had randomized the SCNA sample assignments within lineage. In the all-SCNA analysis, 28 regions were anticorrelated with fewer than three other regions, relative to three isolated nodes in the average permutation (p<0.01).
The isolated nodes in the all-SCNA analysis were enriched for regions containing large genes (including 10 of 28 such regions; p=0.004). Conversely, they trended toward excluding regions with known oncogenes or tumor suppressors (five of 35 such regions; p=0.06). Most peak regions exhibit fewer anticorrelations in the high-level analysis, possibly due to decreased power. The most extreme exception was CDKN2A, which anticorrelated with 14 regions in the high-level analysis and only nine regions in the all-SCNA analysis. Consistent with these findings, CDKN2A is often inactivated by homozygous deletions.
We applied a similar analysis to identify events associated with WGD. We included both SCNAs and mutations, using the 200 most significantly mutated genes across the TCGA Pan-Cancer dataset [Lawrence et al, unpublished data34; see Methods). Three SCNA peak regions and two significantly mutated genes correlated with WGD (Supplementary Table 4). TP53 mutations and CCNE1 amplifications correlated with WGD; both have been functionally associated with tolerance of tetraploidy in experimental models52–55. Our findings indicate these associations apply to human tumors across multiple lineages. We also found that deletions of PPP2R2A and mutations of its binding partner PPP2R1A were correlated with WGD. These two genes belong to phospho-protein phosphatase complex 2 (PPP2), which regulates mitotic spindle formation and can lead to chromosomal missegregation and abnormal mitoses when depleted56,57.
Eleven genetic events anti-correlated with WGD, including two amplifications, five deletions and four mutations. (Supplementary Table 4). The deletions included CDKN2A, PTEN, and NF1, and three of the four mutations also involved genes known as or proposed to be tumor suppressors (CTCF58, MAP3K19, and ATM). The anticorrelations of these tumor suppressors may result from a greater difficulty in biallelically inactivating tumor suppressors in samples with extra copies subsequent to WGD29.
Portal for interactive viewing of results
Results from this study are available at http://www.broadinstitute.org/tcga, including segmented copy-number data (viewable using the Integrative Genomics Viewer59) and the frequency and significance of copy-number changes across and within cancer types.
Discussion
This study represents the largest analysis to date of high-resolution copy-number profiles generated using a single platform, and the first large-scale analysis of absolute allelic copy-number data across cancer types. We identified common patterns of SCNA across cancer types, including a tendency for telomeric events to be longer and more frequent than SCNAs within chromosomes, and for duplications of large regions of the genome (through WGD or polysomy) to lead to subsequent increases in numbers of SCNAs (especially deletions) in the duplicated regions. SCNAs also tend to reside in the same regions of the genome across different cancer types.
A primary challenge in the analysis of somatic genetic data is distinguishing between patterns of alteration that reflect mechanism by which those alterations are generated, positive selection, and negative selection. An underlying assumption of our analyses is that patterns of alteration that are observed across all chromosomes are likely to reflect mechanistic biases, whereas deviations from these patterns at individual loci are likely to reflect selective pressures.
The differences between telomere-bounded and internal SCNAs across all chromosomes suggest different mechanisms underlie their generation. Internal SCNAs have been proposed to occur as a result of apposition of their two breakpoints in three-dimensional space. Chromatin is arranged as a “fractal globule” during interphase60,61, in which the likelihood that two breakpoints would be apposed decreases proportional to the linear distance between them, implying a 1/length distribution. Conversely, SCNAs that start on the telomere may be related to telomere shortening and telomere crisis, and associated with a single double-strand break that could occur anywhere within the chromosome62.
Among the 140 peak regions in the Pan-Cancer analysis, only 35 contained known amplified oncogenes or tumor suppressor genes. SCNAs in some of the remaining regions may recur because these regions are subject to relatively small amounts of negative selection21 or due to mechanistic biases favoring the generation of SCNAs in these regions63, as has been suggested for deletions involving large genes1,5,64. Indeed, we found that SCNAs involving large genes often did not anticorrelate with any other genetic events, suggesting the genes in these regions may have limited functional roles in oncogenesis. However, it remains likely that many additional oncogenes and tumor suppressor genes are within these regions. Moreover, these 140 regions and the additional 229 peak regions identified in the lineage-specific analyses are likely to compose a subset of the regions that are significantly altered in cancer. Analyses of other cancer types have identified additional peak regions1,4, and the limited resolution of the array platform may have obscured detection of some SCNAs.
Varying levels of genomic disruption across cancers are likely to engender biases in analyses of correlations not only between SCNAs, but also between SCNAs and other features of these cancers. For example, increased genomic disruption has been associated with poor prognosis in multiple cancer types65,66. Poor prognosis is therefore likely to be associated with increased rates of SCNA across much of the genome. Controlling for this tendency will be required to identify SCNAs that are functionally associated with progression. It will also be important to account for other possible confounders, such as mechanistically linked events (e.g. chromothripsis or SCNAs that encompass multiple peak regions).
Whole-genome sequencing data can indicate the specific rearrangements that contributed to each SCNA11,24, and assessment of genetic heterogeneity within tumors can also distinguish early from late events23,29. Both of these are approaches are likely to inform the mechanisms by which SCNAs are generated and the selective pressures that shape them.
Online Methods
1. Generation of copy-number profiles
The pipeline used to generate relative copy-number estimates will described elsewhere (Tabak et al, unpublished data). In brief, probe-level signal intensities from Affymetrix SNP6 .CEL files were normalized to a uniform brightness across arrays and merged to form intensity values for each probeset using SNPFileCreator, a Java implementation of dChip67,68. These intensities were mapped to copy-number levels using Birdseed69 in the case of SNP markers, and on the basis of experiments with cell lines with varying dosage of X in the case of copy-number markers1. Recurrent germline copy-number variations (CNVs) were identified across all DNA samples from normal tissue and markers within these regions (representing ~15% of all markers) were removed from further analysis70. Noise was further reduced by application of Tangent normalization70 followed by Circular Binary Segmentation71,72. Quality control metrics were applied at various stages in the pipeline70, resulting in the removal of data representing 23 cancers out of 4957 primary cancers that had been profiled by SNP6 arrays.
HAPSEG73 and ABSOLUTE29, running on FireHose74, were applied to data from 4870 of these cancers, including both the SNP6 data and, when available, whole-exome sequencing data from the same cancers (1069 samples). Of these, purity and ploidy estimates and genome-wide absolute allelic copy-numbers were called in 3847 cancers (Supplementary Table 1). The 200 acute myeloid leukemia samples were not called by ABSOLUTE because they exhibited copy-number alterations across small fractions of their genomes, resulting in insufficient data for accurate calls by the algorithm.
2. Determination of SCNAs
We determined the most likely series of SCNAs that led to the copy-number profiles generated by ABSOLUTE for each homologous chromosome (henceforth, “allele”). Each SCNA was characterized by its length, amplitude, genomic position, and, when determinable, allele and the timing of its generation relative to neighboring segments. We deconstructed each chromosome individually in two sequential steps (to be described in greater detail in Zack et al, unpublished data):
Find a set of the most parsimonious arrangements of copy levels on the two parental alleles (allelic partitioning).
Find the most likely set of SCNA events that would give rise to these copy-number profile (allele deconstruction).
Allelic partitioning
Our data consist of integer copy-numbers of each allele at each locus. The data are segmented, with infrequent changes in copy-number between adjacent markers on the array (fewer than one breakpoint per 1000 markers). We start with no information about which copy levels or breakpoints belong on the same. The purpose of this section is to find a set of the most parsimonious partitions of copy levels between the two alleles.
There is some information inherent in the structure of the segmentation. Because breakpoints are rare, introducing breakpoints that are not necessary to explain our observations adds complexity to our model. There are only two situations in which this does not determine partitioning between the two alleles: 1) the two alleles are at the exact same copy level at a particular locus, or 2) both alleles have a breakpoint at the exact same SNP marker. The first situation is common; we expect the second situation to be rare. In either case, we lose the ability to confidently say whether segments preceding that position occurred on the same or opposite allele as segments subsequent to this position. We call these loci “flex-points” as we are free to swap segments between the two alleles only in these regions. We label regions between adjacent flex-points “contigs”, as the partitioning of these segments relative to one another is fixed. The total number of possible arrangements of a given chromosome is 2f where f is the number of flex-points on the chromosome.
If there are fewer than eight flex-points, we enumerate all possible permutations of the contigs across the two alleles. If there are eight or more flex-points, such enumeration is computationally prohibitive, and we focus on the most likely allelic partitions. We assume the most likely partitions will tend to assign unlikely copy-levels (which vary widely from the chromosome-wide average) to the same allele, so that they can be accounted for by a single unlikely event rather than requiring separate unlikely events on each allele.
Allele Deconstruction
Once the segments have been fixed to each allele, SCNA determination is performed in similar fashion to methods described previously1,75, which identify the combination of SCNAs that would result in the observed copy-number profile and have maximum likelihood of having occurred. The likelihood of an SCNA occurring is estimated according to the observed frequencies of SCNAs with similar lengths and amplitudes of copy-number change across the entire dataset.
Here, however, we consider absolute allelic copy-number levels, which are discrete numbers, whereas prior methods focused on continuous total copy ratios. The discretized data allow enumeration of more possible SCNA combinations (including multiple overlapping amplifications and deletions) than is computationally possible in continuous data. The absolute copy-numbers also require that we distinguish SCNA likelihoods in near-diploid samples from SCNA likelihoods in samples that have undergone WGD, which tend to have higher rates of other types of SCNA (Fig. 1b).
3. SCNA timing relative to WGD and chromosome duplication
We determined the temporal relations of individual SCNAs to WGD using different approaches for deletions and amplifications.
We considered deletions that involved a change from two copies to zero copies of an allele in WGD samples to have likely occurred prior to WGD. Similarly, deletions that involved a change from two copies to one copy of an allele were considered to have occurred after WGD. Other deletions were left uncalled because of ambiguities introduced by surrounding alterations. When determining timing of genome doubling, we did not include arm level or whole chromosome events, as the events of this size are too common to rule out two sequential events that appear to have the same breakpoints.
Amplifications are more ambiguous than deletions because the extra copies of DNA may end up elsewhere in the genome and be affected by subsequent events in those regions. However, because WGD affects the whole genome simultaneously, we expect estimates of WGD timing based on amplifications to be similar overall to estimates based on deletions. We called events with an even total copy change as occurring prior to WGD and events with odd copy change as occurring after WGD.
The same metrics were used to determine events before or after chromosome duplication (Figure 2b). Again, amplifications are more uncertain than deletions because they may involve disparate regions of the genome.
4. Chromothripsis detection
Chromothripsis results from different mechanisms to most focal events, and has a very different distribution across lineages31,76. We identified chromothripsis events in diploid samples based on three features that are observable in copy-number profiles and which have been associated with chromothripsis previously76:
A single chromosome exhibits an unexpectedly large number of SCNAs given the observed frequency of SCNAs within the sample.
SCNAs on this chromosome tend to abnormally closely spaced than we would expect by chance.
The SCNAs are non-overlapping (because they occurred simultaneously) and lead to copy-number changes of +1 or −1.
Prior estimates of rates of chromothripsis have been complicated by uncertainty as to the absolute numbers of copies of change. In our application of these criteria, we evaluated the absolute allelic copy-number data to identify chromosomes that contained more non-overlapping SCNAs that involved a single-copy change than we would expect by chance, given the number of SCNAs within the sample and using the binomial distribution. From these chromosomes, we applied the additional criterion that these SCNAs should be more tightly distributed within the chromosome than we would expect given a random selection of non-overlapping SCNAs within our dataset. If this criterion was not met, we applied a recursive algorithm to remove the SCNA furthest from the centroid location of the SCNAs potentially derived from chromothripsis, and recomputed these two statistics.
Further details of the method will be described separately (Zack et al, unpublished data).
5. Impurity-corrected GISTIC
In cases where we were able to estimate purity and ploidy from ABSOLUTE, we “corrected” total copy-ratios for signal dampening due to cancer cell impurity (i.e. contamination with normal DNA). We called this In-Silico Admixture Removal (ISAR).
The observed copy-ratio R(x) at locus x is a function of the purity α, cancer cell ploidy τ (representing the average copy-number genome-wide), and integer copy-number (in the cancer cells) q(x)29
where D represents the average ploidy across all cells in the cancer:
From this, we can determine q(x):
We assume that the functionally relevant number is the copy-ratio within cancer cells, representing the integer number of copies q(x) divided by the overall ploidy of the cell τ:
Use of R’(x) has the effect of amplifying the signal from low purity samples to be equivalent to higher purity samples. For samples for which ABSOLUTE calls were not available, we used R(x).
To determine significantly recurrent regions of SCNA, we used GISTIC 2.075 applied to the transformed copy-number data. We used a noise threshold of 0.3, a broad length cutoff of 0.5 chromosome arms, a confidence level of 95%, and a copy-ratio cap of 1.5.
For some lineage-specific analyses, dozens of regions on a single chromosome arm were identified as significant peaks due to the presence in many samples of discontinuous SCNAs (such as chromothripsis) on those chromosome arms. This phenomenon has been observed previously1. We narrowed these regions by applying in all lineage-specific analyses an “arm-level peel-off” correction that considers all SCNAs on a chromosome arm in a single sample to be part of a single event when determining whether multiple significantly recurrent events exist on that chromosome arm. This approach has also been used in prior analyses77.
The genes listed in each peak region include all protein-coding genes and microRNAs and additional non-coding RNAs as listed in the files refGene.txt, refLink.txt, refSeqStatus.txt, and wgRna.txt from the UCSC Golden Path database (ftp://hgdownload.cse.ucsc.edu/goldenPath/hg19/database/) as of 27 February 2012.
6. Significance of chromatin modifying genes among peak regions of amplification without known driver genes
To determine whether epigenetic regulators were enriched in peak regions, we compared the number of regions with epigenetic regulators (using a published list40) to permuted datasets in which each gene in each region was replaced by a gene randomly selected from elsewhere in the genome.
7. Correlation analysis
To determine the significance of SCNA co-occurrences, we compared the observed rate of co-occurrences to the rate of co-occurrences in 5000 permuted copy-number profiles for which we had randomized the sample assignment for each chromosome, while maintaining genomic position and lineage and sub-lineage assignments. We only considered SCNAs in different chromosomes to avoid confounding due to geographic proximity. This analysis generated the permuted distribution in Figure 4c (blue line) and Supplementary Figures 4a–b, and the FDR-corrected78 p-values in Figure 4b (top).
To control for variable rates of genomic disruption across samples, we modified the permutations so that they maintained both the numbers of amplified and deleted markers A0j and D0j in each sample j. After randomizing sample assignments for each chromosome as described above, we applied simulated annealing79,80 in which we picked a chromosome at random and swapped it between two randomly chosen samples within the same lineage at each step, and accepted the step with a probability 1− Etot, where:
and Atj and Dtj represent the numbers of amplified and deleted markers in sample j and step t. Tamp and Tdel are temperature factors that were slowly increased during the annealing, and the 1 in the denominator of each value is to avoid dividing by 0 in samples without any events. This approach generated the distributions shown in Figure 4c (dashed line) and the FDR-corrected78 p-values in 4b (bottom). This procedure was applied in two separates analyses: one in which we looked at all SCNAs that passed the noise thresholds we used for our GISTIC significance analyses (above), and one in which we only considered loci with copy-number <−1 or >4.4. The second analysis we termed our “high-level” analysis.
8. Intersection between mutual exclusivity network and Dapple network
To validate the functionality of our network, we looked at the overlap between our network and DAPPLE, a curated dataset of protein-protein interactions81 (PPIs). Of the >400,000 PPI pairs, we took only pairs with a score equal to 1 (indicating highest confidence). Two peak regions had an edge between them in the PPI network under two conditions;
A protein within the first peak was a direct interactor with a protein in the second peak.
A protein in the first peak had at least three distinct paths of length 2 in the PPI network to a protein in the second peak.
To improve specificity, we only tested regions containing fewer than 25 genes. We determined whether the similarity between the PPI network and the anticorrelation network was significant by comparing the extent of overlap to permutations in which the edges in the anticorrelation network were randomly reassigned while maintaining the overall connectivity of the graph (see Results). By comparing both observed and anticorrelation networks to the same PPI network, we controlled for the propensity of regions with many genes to map to more PPIs.
9. Somatic genetic correlates with WGD
To determine which of the 200 most significant somatic mutations correlate with WGD, we used the permmatswap function in the R82 package “vegan”83 with the “quasifit” handle [Lawrence et al., unpublished data]34 to produce a series of independent assignments for mutations on each gene within each sample. This function maintained the number of mutations per gene per lineage, as well as the number of the number of mutations per sample.
To determine which of the peak regions had SCNAs that correlate with WGD, we compared the number of times each SCNA was observed in WGD samples in our observed data to the number of times the SCNA was observed in WGD samples in the permutations created by our simulated annealing approach above.
10. Overlap of peak regions of SCNA
Two regions were considered to overlap if their 95% confidence intervals intersected. To determine significance of overlap, we compared the number of peak regions that overlapped across at least two lineages in the observed data to 100,000 permutations in which the locations of each peak region were randomly shuffled within its chromosome arm (disallowing extension past the telomere or centromere).
11. GRAIL analysis
We used GRAIL39 (www.broadinstitute.org/mpg/grail/) to find common functional terms in the literature for the genes in peak regions of SCNA. We used only PubMed abstracts through December 2006. We removed the following non-informative keywords from those GRAIL found most significant: "growth", "cancer", "cancers", "tumor", "tumors", "proliferation", "suppressor", "factors", "loss", "like", "rich", "cel", "cells", "yeast", "system", "family", "repeat", "deletions", "elegans", "national".
Supplementary Material
Acknowledgements
We would like to thank Abdul Shehata, Pratiti Bandopadhayay, Will Gibson, Ruben Ferrer, and Peleg Horowitz for their help and comments during construction of the paper. This work was conducted as part of The Cancer Genome Atlas Research Network, with funding from U24CA143867 (Genome Characterization Center: M.M., R.B., and G.G.) and U24CA143845 (Genome Data Analysis Center: G.G. and M.M.). Additional support was from NIH/NCI grants U54CA143798 (R.B.), U54HG003067 (S.G.), U24 CA143882 (P.W.L.), and the V Foundation and Pediatric Log-Grade Astrocytoma Foundation (R.B.).
Footnotes
Author Contributions
Travis Zack, Steven Schumacher, Scott Carter, Matthew Meyerson, Gad Getz, and Rameen Beroukhim conceived of study. Analytic methods were developed by Travis Zack, Steven Schumacher, Scott Carter, Barbara Tabak, Jeremiah Wala, Craig Mermel, Michael S. Lawrence, Gad Getz, and Rameen Beroukhim. Analyses were performed by Travis Zack, Steven Schumacher, Scott Carter, Andrew Cherniack, and Bryan Hernandez. The manuscript was written by Travis Zack, Steven Schumacher, Matthew Meyerson, Gad Getz, and Rameen Beroukhim.
References
- 1.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baudis M. Genomic imbalances in 5918 malignant epithelial tumors: an explorative meta-analysis of chromosomal CGH data. Bmc Cancer. 2007;7 doi: 10.1186/1471-2407-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim TM, et al. Functional genomic analysis of chromosomal aberrations in a compendium of 8000 cancer genomes. Genome Research. 2013;23:217–227. doi: 10.1101/gr.140301.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bignell GR, et al. Signatures of mutation and selection in the cancer genome. Nature. 2010;463 doi: 10.1038/nature08768. 893-U61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stephens PJ, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462 doi: 10.1038/nature08645. 1005-U60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weir BA, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450 doi: 10.1038/nature06358. 893-U22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammerman PS, et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas, N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490 doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xue W, et al. A cluster of cooperating tumor-suppressor gene candidates in chromosomal deletions. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:8212–8217. doi: 10.1073/pnas.1206062109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nik-Zainal S, et al. The Life History of 21 Breast Cancers. Cell. 2012;149 doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harada T, et al. Genome-wide DNA copy number analysis in pancreatic cancer using high-density single nucleotide polymorphism arrays. Oncogene. 2008;27:1951–1960. doi: 10.1038/sj.onc.1210832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsao MS, et al. Erlotinib in lung cancer - Molecular and clinical predictors of outcome. New England Journal of Medicine. 2005;353:133–144. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- 14.Cheang MCU, et al. Ki67 Index, HER2 Status, and Prognosis of Patients With Luminal B Breast Cancer. Journal of the National Cancer Institute. 2009;101:736–750. doi: 10.1093/jnci/djp082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim ES, et al. Gefitinib versus docetaxel in previously treated non-small-cell lung cancer (INTEREST): a randomised phase III trial. Lancet. 2008;372:1809–1818. doi: 10.1016/S0140-6736(08)61758-4. [DOI] [PubMed] [Google Scholar]
- 16.Lowe SW, et al. P53 STATUS AND THE EFFICACY OF CANCER-THERAPY IN-VIVO. Science. 1994;266:807–810. doi: 10.1126/science.7973635. [DOI] [PubMed] [Google Scholar]
- 17.Beroukhim R, et al. Assessing the significance of chromosomal aberrations in cancer: Methodology and application to glioma. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor BS, et al. Functional Copy-Number Alterations in Cancer. Plos One. 2008;3 doi: 10.1371/journal.pone.0003179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krasnitz A, Sun G, Andrews P, Wigler M. Target inference from collections of genomic intervals. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1306909110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mullighan CG, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 21.Solimini NL, et al. Recurrent Hemizygous Deletions in Cancers May Optimize Proliferative Potential. Science. 2012;337:104–109. doi: 10.1126/science.1219580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nijhawan D, et al. Cancer vulnerabilities unveiled by genomic loss. Cell. 2012;150:842–854. doi: 10.1016/j.cell.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landau DA, et al. Evolution and Impact of Subclonal Mutations in Chronic Lymphocytic Leukemia. Cell. 2013;152:714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang L, et al. Diverse mechanisms of somatic structural variations in human cancer genomes. Cell. 2013;153 doi: 10.1016/j.cell.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Notta F, et al. Evolution of human BCR-ABL1 lymphoblastic leukaemia-initiating cells. Nature. 2011;469 doi: 10.1038/nature09733. 362-+ [DOI] [PubMed] [Google Scholar]
- 26.Ciriello G, Cerami E, Sander C, Schultz N. Mutual exclusivity analysis identifies oncogenic network modules. Genome Research. 2012;22:398–406. doi: 10.1101/gr.125567.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaske CJ, et al. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics. 2010;26:i237–i245. doi: 10.1093/bioinformatics/btq182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vandin F, Upfal E, Raphael BJ. Algorithms for Detecting Significantly Mutated Pathways in Cancer. Journal of Computational Biology. 2011;18:507–522. doi: 10.1089/cmb.2010.0265. [DOI] [PubMed] [Google Scholar]
- 29.Carter SL, et al. Absolute quantification of somatic DNA alterations in human cancer. Nature Biotechnology. 2012;30 doi: 10.1038/nbt.2203. 413-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Synapse. DNA Methylation Based Purity. 2013 [Google Scholar]
- 31.Stephens PJ, et al. Massive Genomic Rearrangement Acquired in a Single Catastrophic Event during Cancer Development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maida Y, et al. An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature. 2009;461:230–235. doi: 10.1038/nature08283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lawrence MS SP, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, Kiezun A, Hammerman PS, McKenna A, Drier Y, Zou L, Ramos AH, Pugh TJ, Stransky N, Helman E, Kim J, Sougnez C, Ambrogio L, Nickerson E, Shefler E, Cortés ML, Auclair D, Saksena G, Voet D, Noble M, Dicara D, Lin P, Lichtenstein L, Heiman DI, Fennell T, Imielinski M, Hernandez B, Hodis E, Baca S, Dulak AM, Lohr J, Landau DA, Wu CJ, Melendez-Zajgla J, Hidalgo-Miranda A, Koren A, McCarroll SA, Mora J, Lee RS, Crompton B, Onofrio R, Parkin M, Winckler W, Ardlie K, Gabriel SB, Roberts CW, Biegel JA, Stegmaier K, Bass AJ, Garraway LA, Meyerson M, Golub TR, Gordenin DA, Sunyaev S, Lander ES, Getz G. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;489 doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Synapse. Pan-cancer significantly recurrent mutations. 2013 [Google Scholar]
- 35.Du Z, et al. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nature Structural & Molecular Biology. 2013;20 doi: 10.1038/nsmb.2591. 908-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheetham SW, Gruhl F, Mattick JS, Dinger ME. Long noncoding RNAs and the genetics of cancer. British Journal of Cancer. 2013;108:2419–2425. doi: 10.1038/bjc.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ying H, et al. Mig-6 controls EGFR trafficking and suppresses gliomagenesis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:6912–6917. doi: 10.1073/pnas.0914930107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Du J, et al. FOXC1, a target of polycomb, inhibits metastasis of breast cancer cells. Breast Cancer Research and Treatment. 2012;131:65–73. doi: 10.1007/s10549-011-1396-3. [DOI] [PubMed] [Google Scholar]
- 39.Raychaudhuri S, et al. Identifying Relationships among Genomic Disease Regions: Predicting Genes at Pathogenic SNP Associations and Rare Deletions. Plos Genetics. 2009;5 doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nature Reviews Drug Discovery. 2012;11:384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- 41.French CA, et al. Midline carcinoma of children and young adults with NUT rearrangement. Journal of Clinical Oncology. 2004;22:4135–4139. doi: 10.1200/JCO.2004.02.107. [DOI] [PubMed] [Google Scholar]
- 42.Borrow J, et al. The translocation t(8;l6)(p11, p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB binding protein. Nature Genetics. 1996;14:33–41. doi: 10.1038/ng0996-33. [DOI] [PubMed] [Google Scholar]
- 43.Champagne N, et al. Identification of a human histone acetyltransferase related to monocytic leukemia zinc finger protein. Journal of Biological Chemistry. 1999;274:28528–28536. doi: 10.1074/jbc.274.40.28528. [DOI] [PubMed] [Google Scholar]
- 44.Jaju RJ, et al. A novel gene, NSD1, is fused to NUP98 in the t(5;11)(q35;p15.5) in de novo childhood acute myeloid leukemia. Blood. 2001;98:1264–1267. doi: 10.1182/blood.v98.4.1264. [DOI] [PubMed] [Google Scholar]
- 45.Micci F, Panagopoulos I, Bjerkehagen B, Heim S. Consistent rearrangement of chromosomal band 6p21 with generation of fusion genes JAZF1/PHF1 and EPC1/PHF1 in endometrial stromal sarcoma. Cancer Research. 2006;66:107–112. doi: 10.1158/0008-5472.CAN-05-2485. [DOI] [PubMed] [Google Scholar]
- 46.Park JT, et al. Notch3 gene amplification in ovarian cancer. Cancer Research. 2006;66:6312–6318. doi: 10.1158/0008-5472.CAN-05-3610. [DOI] [PubMed] [Google Scholar]
- 47.Garkavtsev I, Kazarov A, Gudkov A, Riabowol K. Suppression of the novel growth inhibitor p33(ING1) promotes neoplastic transformation. Nature Genetics. 1996;14:415–420. doi: 10.1038/ng1296-415. [DOI] [PubMed] [Google Scholar]
- 48.Beshiri ML, et al. Coordinated repression of cell cycle genes by KDM5A and E2F4 during differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:18499–18504. doi: 10.1073/pnas.1216724109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gargalionis AN, Piperi C, Adamopoulos C, Papavassiliou AG. Histone modifications as a pathogenic mechanism of colorectal tumorigenesis. International Journal of Biochemistry & Cell Biology. 2012;44:1276–1289. doi: 10.1016/j.biocel.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 50.Berman BP, et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nature Genetics. 2012;44 doi: 10.1038/ng.969. 40-U62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fullgrabe J, Kavanagh E, Joseph B. Histone onco-modifications. Oncogene. 2011;30:3391–3403. doi: 10.1038/onc.2011.121. [DOI] [PubMed] [Google Scholar]
- 52.Andreassen PR, Lohez OD, Lacroix FB, Margolis RL. Tetraploid state induces p53-dependent arrest of nontransformed mammalian cells in G1. Molecular Biology of the Cell. 2001;12:1315–1328. doi: 10.1091/mbc.12.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rausch T, et al. Genome Sequencing of Pediatric Medulloblastoma Links Catastrophic DNA Rearrangements with TP53 Mutations. Cell. 2012;148:59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ho CC, Hau PM, Marxer M, Poon RYC. The requirement of p53 for maintaining chromosomal stability during tetraploidization. Oncotarget. 2010;1:583–595. doi: 10.18632/oncotarget.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dalton WB, Yu B, Yang VW. p53 suppresses structural chromosome instability after mitotic arrest in human cells. Oncogene. 2010;29:1929–1940. doi: 10.1038/onc.2009.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tang ZY, et al. PP2A is required for centromeric localization of sgol and proper chromosome segregation. Developmental Cell. 2006;10:575–585. doi: 10.1016/j.devcel.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 57.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nature Genetics. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 58.Filippova GN, et al. Tumor-associated zinc finger mutations in the CTCF transcription factor selectively alter its DNA-binding specificity. Cancer Research. 2002;62:48–52. [PubMed] [Google Scholar]
- 59.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings in Bioinformatics. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fudenberg G, Getz G, Meyerson M, Mirny LA. High order chromatin architecture shapes the landscape of chromosomal alterations in cancer. Nature Biotechnology. 2011;29 doi: 10.1038/nbt.2049. 1109-U75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lieberman-Aiden E, et al. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Artandi SE, et al. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406:641–645. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- 63.Yunis JJ, Soreng AL. CONSTITUTIVE FRAGILE SITES AND CANCER. Science. 1984;226:1199–1204. doi: 10.1126/science.6239375. [DOI] [PubMed] [Google Scholar]
- 64.Smith DI, Zhu Y, McAvoy S, Kuhn R. Common fragile sites, extremely large genes, neural development and cancer. Cancer Letters. 2006;232:48–57. doi: 10.1016/j.canlet.2005.06.049. [DOI] [PubMed] [Google Scholar]
- 65.Pinto AE, et al. DNA Ploidy is an Independent Predictor of Survival in Breast Invasive Ductal Carcinoma: A Long-term Multivariate Analysis of 393 Patients. Annals of Surgical Oncology. 2013;20:1530–1537. doi: 10.1245/s10434-012-2804-6. [DOI] [PubMed] [Google Scholar]
- 66.Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nature Genetics. 2006;38:1043–1048. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 67.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li CaWW. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biology. 2001;2 doi: 10.1186/gb-2001-2-8-research0032. research0032.1–research0032.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Korn JM, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nature Genetics. 2008;40:1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tabak B, Saksena G, Monti S. The Tangent copy-number inference pipeline for cancer genome analyses. Bioinformatics. doi: 10.1093/bioinformatics/btac586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 72.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–663. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- 73.Carter SL, Meyerson M, Getz G. Accurate estimation of homologue-specific DNA concentration ratios in cancer samples allows long-range haplotyping. 2011 Preprint at http://precedings.nature.com/documents/6494/version/1/ [Google Scholar]
- 74.Broad Institute. Broad Institute; 2011. FireHose. [Google Scholar]
- 75.Mermel CH, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biology. 2011;12 doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Korbel JO, Campbell PJ. Criteria for Inference of Chromothripsis in Cancer Genomes. Cell. 2013;152:1226–1236. doi: 10.1016/j.cell.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 77.Cancer Genome Atlas, N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Benjamini Y, Hochberg Y. CONTROLLING THE FALSE DISCOVERY RATE -A PRACTICAL AND POWERFUL APPROACH TO MULTIPLE TESTING. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289–300. [Google Scholar]
- 79.Kirkpatrick S, Gelatt CD, Vecchi MP. OPTIMIZATION BY SIMULATED ANNEALING. Science. 1983;220:671–680. doi: 10.1126/science.220.4598.671. [DOI] [PubMed] [Google Scholar]
- 80.Cerny V. THERMODYNAMICAL APPROACH TO THE TRAVELING SALESMAN PROBLEM - AN EFFICIENT SIMULATION ALGORITHM. Journal of Optimization Theory and Applications. 1985;45:41–51. [Google Scholar]
- 81.Rossin EJ, et al. Proteins Encoded in Genomic Regions Associated with Immune-Mediated Disease Physically Interact and Suggest Underlying Biology. Plos Genetics. 2011;7 doi: 10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Team RC. R: A language and Environment for Statistical Computing. 2012 [Google Scholar]
- 83.Oksanen Jari, Roeland FGB, Legendre Pierre, Minchin Peter R, O'Hara RB, Simpson Gavin L, Solymos Peter, Stevens M Henry H, Wagner Helene. Vegan: Community Ecology Package; 2012. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.