Abstract
High mobility group box 1 (HMGB1) is a central mediator in inflammation and immunity. Recently, it was shown that different redox states of the three cysteines of HMGB1 endow it with mutually exclusive activities, such as inducing chemotaxis or the transcription of cytokines and chemokines, via the interaction with different receptors. The different HMGB1 redox forms can be identified by mass spectrometry in body fluids of patients and may hold promise as biomarkers. We propose here a systematic nomenclature of the different redox forms of HMGB1 and related proteins, to replace the conflicting names used so far by different laboratories.
High mobility group box 1 (HMGB1) is a 25-kDa nuclear protein that has attracted increasing interest as a mediator of tissue damage and inflammation (1). HMGB1 is released passively after cell death and is secreted actively by macrophages and many other cell types during infection, injury or severe stress (such as ischemia/reperfusion). Extracellular HMGB1 can act both as a chemoattractant for leukocytes, stem cells and other cells involved in tissue repair and as a proinflammatory mediator to induce both recruited leukocytes and parenchymal cells to release tumor necrosis factor (TNF)-α, interleukin (IL)-1, IL-6 and other cytokines. Extracellular HMGB1 interacts with several receptors and partner proteins, including receptor for advanced glycation end-products (RAGE), Toll-like receptors (TLRs) and the C-X-C motif chemokine 12( CXCL12), with which it forms a complex that binds the C-X-C chemokine receptor type 4 (CXCR4).
Recently, with the application of high-resolution and sensitive proteomic mass spectrometry, it has become apparent that the different activities of HMGB1 depend on multiple redox forms of three cysteine residues within the protein (2,3). Indeed, the different HMGB1 redox forms interact with different receptors, and there is increasing attention to which redox form of HMGB1 is responsible for specific pathophysiological action. Moreover, there is evidence that HMGB1, which is predominantly fully reduced inside the cell, can be oxidized in the cytoplasm after ROS production (4,5). Thus, HMGB1 can now no longer be conceptualized as a single protein species. At present, different laboratories refer to the different HMGB1 redox forms in different ways. A unified nomenclature is thus required to (a) facilitate interactions between various laboratories, (b) expedite literature searches and (c) avoid confusion, in particular, in sourcing the individual redox forms from commercial providers. Even more importantly, a unified nomenclature will be important in the clinics: the different redox forms can be identified in the body fluids of patients with several different complaints and might be used as biomarkers for diagnosis, outcome prediction and rationally based therapeutic decisions (6). Participants in the Merinoff World Congress on HMGB1, held at the Feinstein Institute for Medical Research on October 9–11, 2013, thus decided to introduce a unified nomenclature.
Cysteine represents a potent nucleophile, and the chemistry of sulfur in protein cysteine residues is rich in complexity: the sulfur atom can exist in multiple redox forms that can form bonds to another sulfur atom or to hydrogen, nitrogen, oxygen and carbon atoms. Furthermore, the identification of the chemical form of cysteine sulfur in proteins often involves the formation of chemical bonds with specific reagents that form characteristic adducts; an effective nomenclature also requires the facile identification of such adducts. We therefore propose the adoption of a nomenclature that is based on the name of the protein (HMGB1 or one its family members, HMGB2, -3 and -4) and the amino acid number of the cysteine residue in the polypeptide chain (for example, C23), followed by a suffix identifying the atom(s) or chemical group bonded to the cysteine sulfur atom (for example, “h” to indicate the formation of a single bond to a hydrogen atom to generate the sulfhydryl moiety, “se” for the formation of a chemical bond to one oxygen atom to form a sulfenic group, “so” for the formation of chemical bonds to three oxygen atoms to form a sulfonic group, or “nem” to indicate the formation of a chemical bond to nitrogen in the N-ethylmaleimide group, which is used to tag reduced cysteines in polypeptides). Similarly, iodoacetamide has been used as a tool to chemically characterize HMGB1 redox states by adding the carboxyamidomethyl group to the sulfur in the protein; the suffix “iodo” can indicate the formation of this chemical adduct.
This nomenclature is reminiscent of and compatible with the nomenclature of posttranslational modifications of histones (7) and other proteins (including HMGB1 itself) on noncysteine amino acid residues. Indeed, HMGB1 can be acetylated or methylated on lysines (K) and phosphorylated on serine/threonines (S/T). A special case, not readily accommodated by the current nomenclature of posttranslational modifications, is the formation of a disulfide bond to another cysteine, because the unequivocal identification of that specific cysteine requires the indication of its residue number as well. We propose the suffix “-” (hyphen) connecting the cysteines involved in the disulfide bond. We also propose that the absence of a specific indication unequivocally indicates the absence of a posttranslational modification, which in the case of cysteines would otherwise be indicated by the “h” suffix.
Fully reduced HMGB1 is thus identified as “HMGB1C23hC45hC106h,” or more simply “HMGB1,” since the protein is not modified posttranslationally. HMGB1 containing a single intramolecular disulfide bond between C23 and C45 and no posttranslational modification on C106 is to be identified as “HMGB1C23-C45C106h,” or more simply “HMGB1C23-C45,” since the C106 residue is not modified posttranslationally. HMGB1 terminally oxidized to contain sulfonyl groups on all cysteines should be identified as HMGB1C23soC45soC106so.
This nomenclature also accommodates HMGB1 adducts obtained after reaction with specific chemical species, such as HMGB1C23-C45C106nem or HMGB1C23-C45C106iodo, which are obtained after reaction of the sulfhydryl moiety on C106 within HMGB1C23-C45 with N-ethylmaleimide or iodoacetamide, respectively. This nomenclature could also accommodate chemical species not currently identified in vivo or in vitro, such as reaction products of cysteine sulfur with nitrogen oxide (for review of cysteine S-nitrosylation, see [8]).
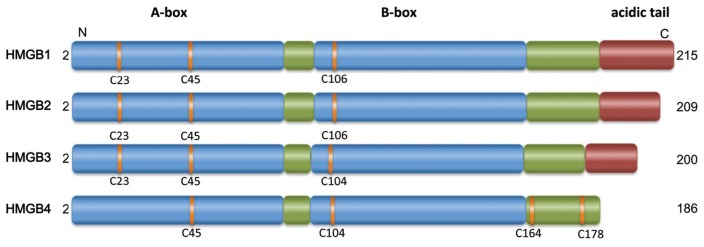
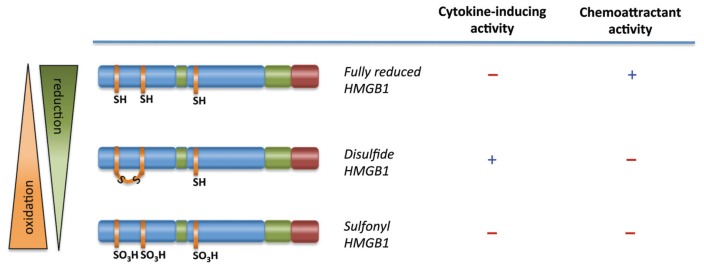
Figure 1 indicates the domain structure of HMGB1 and its family members; we expect HMGB2 to undergo the same posttranslational modification as HMGB1, although so far these modifications have not been identified or even been sought after. Table 1 shows a partial list of atoms or groups that can be bound to cysteine 23 of HMGB1; cysteines 45 and 106 follow the same rules. Table 2 lists the redox forms of HMGB1 that have been identified so far in vivo and the biochemical and physiological activities that have been attributed to them, and Figure 2 provides a pictorial representation of the various forms.
Figure 1.
Schematic structures of HMGB1, -2, -3 and -4. HMGB1 is a 25-kDa nuclear protein of 215 amino acids. The first methionine is cleaved off posttranslationally. HMGB1 comprises two DNA-binding domains (blue), boxes A and B, and a negatively charged C-terminal tail (red). The green boxes are connecting segments. The critical cysteines are highlighted in yellow. Box A, when isolated from the rest of the molecule, acts as a specific antagonist of HMGB1. HMGB2, HMGB3 and HMGB4 have a similar overall domain organization, but are products of different genes. No alternative protein products from alternative splicing are known.
Table 1.
Suggested abbreviations for cysteine modifications.
| Abbreviation of modification | Modification | Description |
|---|---|---|
| C23h | R-SH | Thiol |
| C23-C45 | R-S-S-R | Disulfide bond with cysteine 45 |
| C23so | R-SO3− | Sulfonate modification |
| C23nem | R-S-NEM | Adduct after reaction with NEM |
| C23iodo | R-S-carboxy-amidomethyl | Adduct after reaction with iodoacetamide |
The modification of cysteine 23 in HMGB1 is given as an example.
Table 2.
Redox forms of HMGB1 identified so far.
| Complete notation | Simple identification | Description |
|---|---|---|
| HMGB1C23hC45hC106h | Fully reduced HMGB1 | HMGB1 with no posttranslational modifications; predominant form in nucleus and cytoplasm in basal conditions; forms a heterocomplex with CXCL12 that interacts with receptor CXCR4 to promote cell migration |
| HMGB1C23-C45C106h | Disulfide HMGB1 | HMGB1 with a disulfide bond connecting C23 and C45; interacts with receptor TLR4 to induce cytokine and chemokine transcription and secretion |
| HMGB1C23soC45soC106so | Sulfonyl HMGB1 | Formed after exposure to ROS; has no activity for cell migration or cytokine induction |
Figure 2.
The functional activity of HMGB1 is regulated by its redox status. In its fully reduced form (HMGB1C23hC45hC106h), HMGB1 has cell migration–inducing activity but no cytokine-inducing activity. In its partly oxidized form, disulfide HMGB1 (HMGB1C23-C45C106h) has no cell migration–induced activity but possesses cytokine-inducing activity. The terminally oxidized form, sulfonyl HMGB1 (HMGB1C23soC45soC106so), has neither cell migration–inducing activity nor cytokine-inducing activity.
Recombinant box A and box B (Figure 1) are important molecular research tools to dissect the various functions of the HMGB1 domains. Although the importance of the redox state of these molecules has not been fully investigated to date, the nomenclature system we propose can be applied to the individual recombinant boxes as well. For example, fully reduced box A or box B is identified as “BoxAC23hC45h” and “BoxBC106h,” or more simply box A or box B, since the cysteine residue is not modified posttranslationally. Furthermore, as with the full-length protein, an intramolecular disulfide bond between C23 and C45 in box A would be identified as BoxAC23-C45. Box A or box B terminally oxidized to contain sulfonyl groups on all cysteines should be identified as BoxAC23soC45so and BoxBC106so.
We propose to phase out all currently used identifications of HMGB1 that do not conform to this nomenclature and thus might generate confusion. For use in short abstracts or titles, where space is at a premium, the terms “fully reduced HMGB1,” “disulfide HMGB1” or “sulfonyl HMGB1” are acceptable, but the systematic identification should be used in the full text. For oral presentations, we similarly propose to use the terms “fully reduced HMGB1,” “disulfide HMGB1” or “sulfonyl HMGB1” in speech, but at the same time use the systematic identification in the projected images.
As we continue to move forward with a consensus and understanding of HMGB1 functions, we are confident that the proposed nomenclature will bring clarity to the entire burgeoning field of HMGB1 biology, and immunity in general.
Footnotes
Online address: http://www.molmed.org
DISCLOSURE
The authors declare that they have no competing interests as defined by Molecular Medicine, or other interests that might be perceived to influence the results and discussion reported in this paper.
REFERENCES
- 1.Andersson U, Tracey KJ. HMGB1 is a therapeutic target for sterile inflammation and infection. Annu Rev Immunol. 2011;29:139–62. doi: 10.1146/annurev-immunol-030409-101323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Venereau E, et al. Mutually exclusive redox forms of HMGB1 promote cell recruitment or proinflammatory cytokine release. J Exp Med. 2012;209:1519–28. doi: 10.1084/jem.20120189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang H, et al. Redox modification of cysteine residues regulates the cytokine activity of HMGB1. Mol Med. 2012;18:250–9. doi: 10.2119/molmed.2011.00389. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Kazama H, et al. Induction of immunological tolerance by apoptotic cells requires caspase-dependent oxidation of high-mobility group box-1 protein. Immunity. 2008;29:21–32. doi: 10.1016/j.immuni.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang D, et al. Endogenous HMGB1 regulates autophagy. J Cell Biol. 2010;190:881–92. doi: 10.1083/jcb.200911078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dear JW, Antoine DJ. Stratification of paracetamol overdose patients using new toxicity biomarkers: current candidates and future challenges. Expert Rev Clin Pharmacol. 2014;7:181–9. doi: 10.1586/17512433.2014.880650. [DOI] [PubMed] [Google Scholar]
- 7.Turner BM. Reading signals on the nucleosome with a new nomenclature for modified histones. Nat Struct Mol Biol. 2005;12:110–12. doi: 10.1038/nsmb0205-110. [DOI] [PubMed] [Google Scholar]
- 8.Gould N, Doulias PT, Tenopoulou M, Raju K, Ischiropoulos H. Regulation of protein function and signaling by reversible cysteine S-nitrosylation. J Biol Chem. 2013;288:26473–9. doi: 10.1074/jbc.R113.460261. [DOI] [PMC free article] [PubMed] [Google Scholar]