Abstract
Cholangiocytes, bile duct lining cells, actively adjust the amount of cholesterol and bile acids in bile through expression of enzymes and channels involved in transportation and metabolism of the cholesterol and bile acids. Herein, we report molecular mechanisms regulating bile acid biosynthesis in cholangiocytes. Among the cytochrome p450 (Cyp) enzymes involved in bile acid biosynthesis, sterol 27-hydroxylase (Cyp27) that is the rate-limiting enzyme for the acidic pathway of bile acid biosynthesis expressed in cholangiocytes. Expression of other Cyp enzymes for the basic bile acid biosynthesis was hardly detected. The Cyp27 expression was negatively regulated by a hydrophobic bile acid through farnesoid X receptor (FXR), a nuclear receptor activated by bile acid ligands. Activated FXR exerted the negative effects by inducing an expression of fibroblast growth factor 15/19 (FGF15/19). Similar to its repressive function against cholesterol 7α-hydroxylase (Cyp7a1) expression in hepatocytes, secreted FGF15/19 triggered Cyp27 repression in cholangiocytes through interaction with its cognate receptor fibroblast growth factor receptor 4 (FGFR4). The involvements of FXR and FGFR4 for the bile acid-induced Cyp27 repression were confirmed in vivo using knockout mouse models. Different from the signaling in hepatocytes, wherein the FGF15/19-induced repression signaling is mediated by c-Jun N-terminal kinase (JNK), FGF15/19-induced Cyp27 repression in cholangiocytes was mediated by p38 kinase. Thus, the results collectively suggest that cholangiocytes may be able to actively regulate bile acid biosynthesis in cholangiocytes and even hepatocyte by secreting FGF15/19. We suggest the presence of cholangiocyte-mediated intrahepatic feedback loop in addition to the enterohepatic feedback loop against bile acid biosynthesis in the liver.
Keywords: Cholangiocyte, Cyp27, p38 kinase
Introduction
Bile acids are synthesized in the liver from cholesterol and are secreted as an active ingredient of bile from the liver into the intestinal lumen to facilitate the absorption of hydrophobic lipid nutrients. Bile flows into gallbladder for storage and concentration by passing through bile ducts. Cholangiocytes are the epithelial cells lining the bile ducts [4]. We and others have found that cholangiocytes are reabsorptive/secretory epithelial cells that participate in a number of biological processes with pathophysiological consequences such as the modification of bile, secretory response to hepatotoxic substances, and cell proliferation [27]. For example, cholangiocytes (1) secrete several growth factors and pro-inflammatory molecules [27]; (2) mediate absorption of water, ions, and electrolytes [26]; (3) interact with immune cells for localized immune responses [7]; and (4) regulate cholesterol content in bile [32]. The importance of cholangiocytes becomes apparent in diseases caused by malfunction of cholangiocytes collectively called “cholangiopathies,” indicating that cholangiocytes are the primary cells for these pathological disorders [25, 27]. For example, obstruction of bile flow in the body (i.e., cholestasis) is often caused by gallstones formed in bile ducts as a result of increased cholesterol content in bile and increased hydrophobicity of bile acids [11]. It is not clear whether, and if so how, cholangiocytes regulate the content of hydrophobic bile acids in bile. Understanding the molecular mechanisms and transcription factors that regulate lipid and bile acid metabolism in cholangiocytes may provide therapeutic clues for the treatment of cholangiopathies.
Bile acids are potent ligands for the farnesoid X receptor (FXR, NR1H4), a nuclear receptor encoded by the NR1H4, which is highly expressed in the liver and intestine where bile acids are synthesized and recovered, respectively. Low FXR expression, polymorphisms in NR1H4, and FXR knockout all have been linked to gallstone disease in mouse models [18, 31]. Bile acids act through FXR to inhibit their own synthesis via distinct and redundant feedback loops. First, FXR induces the expression of small heterodimer partner (SHP, NR0B2) in hepatocytes [18, 30, 31]. SHP inhibits transactivation of liver receptor homolog-1 (LRH-1, NR5A2) through which to repress the expression of cytochrome p450 (Cyp) cholesterol 7α-hydroxylase 1 (Cyp7a1). Cyp7a1, in turn, catalyzes the rate-limiting step in cholesterol to bile acid conversion in hepatocytes [9]. FXR also induces the expression of human fibroblast growth factor (FGF) 19, and its mouse ortholog, FGF15, was identified as metabolic hormones in enterocytes and hepatocytes [23, 24]. FGF15/19 binds to fibroblast growth factor receptor 4 (FGFR4) which activates c-Jun N-terminal Kinase (JNK) and mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase 1/2 (ERK1/2) pathways resulting in repression of Cyp7a1 in hepatocytes [12, 13, 24].
Hydrophobic bile acids also inhibit the expression of the sterol 27-hydroxylase (Cyp27) in hepatocytes. Cyp27 encodes the rate-limiting enzyme for the alternative or acidic pathway of bile acid biosynthesis in hepatocytes [5, 8]. The acidic pathway is the predominant pathway of cholesterol to bile acid conversion in extrahepatic tissues [2]. However, underlying mechanisms of Cyp27 expression have not been clearly defined [21, 28, 29]. The nuclear receptor hepatocyte nuclear factor 4α (HNF4α, NR2A1) is known to be a master regulator of Cyp27 expression [5, 8] and is also required for optimal Cyp7a1 expression in hepatocytes [1, 14]. Bile acids negatively regulate HNF4α activity in rat liver and hepatoma cell lines via induction of SHP, which binds to HNF4α, or a SHP-independent mechanism wherein mRNA and protein levels of HNF4α are repressed [19, 34, 36].
In this study, we investigated the underlying mechanisms of Cyp27 expression in cholangiocytes. The bile acid-mediated repression of Cyp27 involved FXR-mediated induction of FGF15/19. FGF15/19 then acted in an autocrine fashion to activate its cognate receptor FGFR4, resulting in the repression of Cyp27. Unlike hepatocytes, in which FGF15/19 activities resulted in the inhibition of Cyp7a1 expression through JNK and MAPK, the inhibitory effect of FGF15/19 upon Cyp27 expression in cholangiocytes was mediated by p38 kinase. In addition to this, we found that the expression level of HNF4α, a master regulator of Cyp27 expression, was downregulated in cholangiocytes treated with bile acid. Our results indicate the presence of a novel regulatory mechanism through p38 kinase and HNF4α for the repression of bile acid biosynthesis in cholangiocytes. These results support the active participation by cholangiocytes in bile homeostasis and suggest a promising target mechanism for the development of drugs to cure diseases caused by the disruption of bile homeostasis.
Methods
Cells and reagents
Cholangiocytes and hepatocytes were isolated from rat liver and normal rat cholangiocytes (NRCs) were isolated and cultured as described [32]. Immortalized normal human cholangiocyte line (H69) was a gift from Dr. Greg Gores. The hepatoma cell line (HepG2) was obtained from ATCC. Chenodeoxycholic acid (CDCA), T0901317, GW501516, GW4064, the ERK inhibitor PD98059, water-soluble cholesterol, 27-hydrocholesterol, β-actin monoclonal antibodies, and other chemicals were obtained from Sigma. All cell culture reagents, the p38 kinase inhibitor, SB203580, and the JNK inhibitor, SP600125, were obtained from Invitrogen. Anti-p38 kinase antibody was obtained from Cell Signaling. Anti-CYP27 antibody was obtained from LSBio. The recombinant Human FGF19 Kit and Human FGF19 ELISA Kit were purchased from R&D Systems.
In vitro and in vivo models
For polarized culture, normal rat intrahepatic cholangiocytes and human cholangiocytes (H69) were used. These cells were maintained as previously described [32]. FXR [22] and FGFR4 [35] knockout mice were fed a standard rodent chow or a chow supplemented with 1 % (w/w) CDCA for 5 days. Bile ducts were then isolated from mouse livers using a laser microdissection system (Applied Biosystems). Mice were maintained under 12:12-hours light/night cycles and were fed ad libitum. Our animal protocols were approved by the Institutional Animal Care and Use Committee at University of Utah and Institute of Biosciences and Technology at Texas A&M Health Science Center.
RNA analysis
Total RNA was extracted by TRIzol reagent (Invitrogen) according to the manufacturer's instructions. For Northern blot analysis, 10 µg of RNA was denatured, electrophoresed, transferred to Zeta-Probe® Membrane (Bio-Rad), and probed with different cDNA probes. All the cDNA probes used in this study were prepared with reverse transcriptase using primers complementary to their target mRNA and were confirmed by sequencing. For real-time quantitative PCR (Q-PCR) analysis, 200 ng of purified total RNA was used to amplify the specific genes using a thermocycler.
Immunohistochemistry
For immunohistochemistry (IHC) analysis, paraffin-embedded rat liver sections (4 µm) were dewaxed, and antigens were retrieved. After blocking, slides were incubated with primary antibodies. EnVision Kit (Dako) was used for avidin–biotin complex method to visualize the signals [32].
Cholangiocytes culture on transwell inserts
NRCs, cultured confluently on a transwell insert (Corning), were used to measure cholesterol efflux. The radiolabeled cholesterol that is excreted from cholangiocytes was measured as described in our previous report [32]. Briefly, the NRCs were labeled with 0.5 µCi/mL [3H]-cholesterol (Amersham) and 50 µg/mL cholesterol for 24 h in a serum-free medium containing 0.2 % bovine serum albumin. After washing the cells with phosphate-buffered saline, the cholesterol-loaded NRCs were treated with ligands as indicated. Excreted radiolabeled cholesterol was captured with Apo-AI through both the apical and basolateral regions. Similarly, the paracrine and autocrine effects of FGF19 were determined by culturing human H69 cells on a transwell insert. H69 cells were treated with DMSO or 100 µM CDCA for 2 hours and then washed with the culture medium and transferred onto a well that contained cultured HepG2 cells on the bottom. After 24 hours of coculture, HepG2 cells were collected to analyze Cyp7a1 expression as described in the RNA analysis.
Western blot analysis
Cholangiocytes and hepatocytes were isolated from the rat liver. H69 cells were treated with 50 ng/mL FGF19 or 100 µM CDCA for 24 hours in the absence/presence of coincubation with the 50 µM of p38 kinase inhibitor, SB203580. Extracts were prepared from the hepatocytes and cholangiocytes and then separated by sodium dodecyl sulfate (SDS)–polyacryl-amide gel electrophoresis (PAGE). The separated proteins were transferred onto polyvinylidene difluouride membrane (Bio-Rad) and probed with antibodies.
Quantitative sandwich enzyme-linked immunosorbent assay
Excreted FGF19 in the medium used for human H69 cells, which were treated with 100 µM CDCA or DMSO for 24 hours, was quantitatively measured using the Sandwich ELISA Kit (R&D Systems) following the manufacturer's instructions. All the medium samples were assayed in duplicate.
Results
Expression of genes related to bile acid homeostasis in cholangiocytes
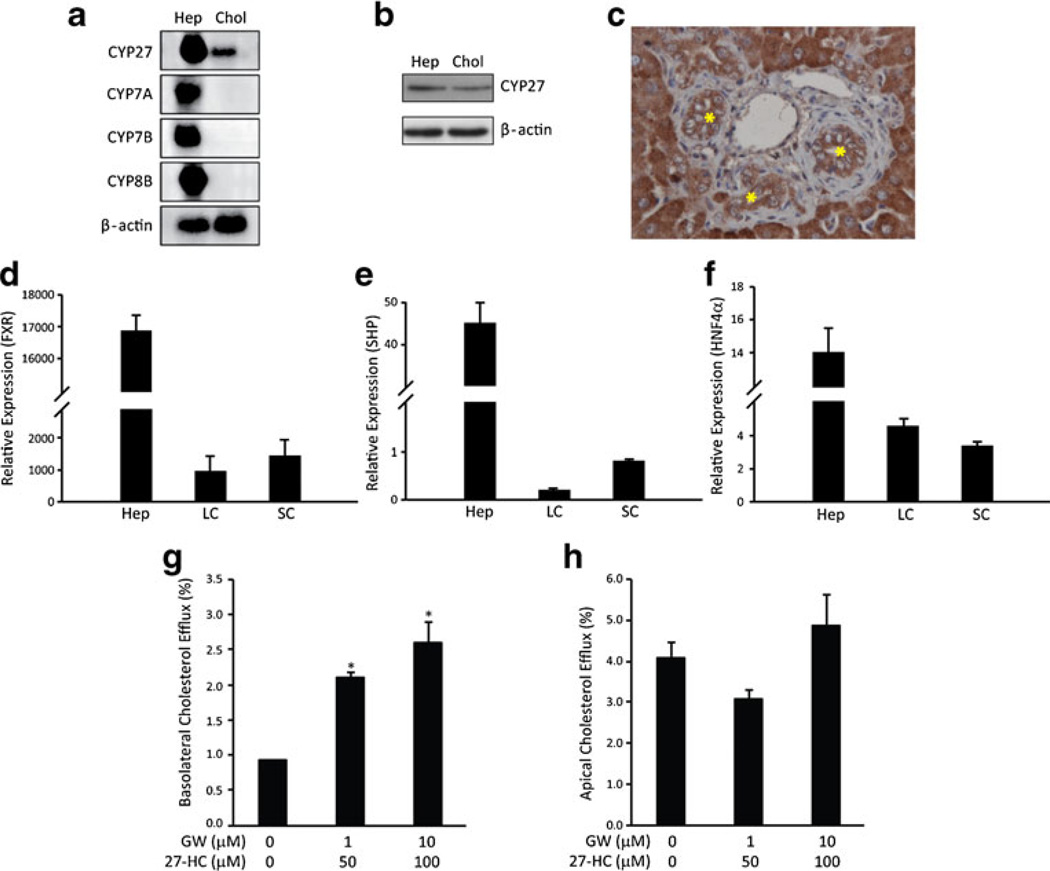
We first determined the expression levels of Cyp enzymes and nuclear receptors involved in bile acid homeostasis in cholangiocytes and hepatocytes. Among the Cyp enzymes that we analyzed, all were expressed by hepatocytes, but only Cyp27 was detected in cholangiocytes (Fig. 1a). Expression of Cyp27 in cholangiocytes was confirmed by detecting mRNA (Fig. 1a), protein (Fig. 1b), and protein expressed in liver tissue (Fig. 1c). FXR, SHP, and HNF4α, three nuclear receptors that play important roles in the regulation of cholesterol to bile acid conversion in hepatocytes, were also expressed in cholangiocytes, albeit at lower levels than those in hepatocytes (Fig. 1d–f).
Fig. 1.
Expression of genes related to bile acid homeostasis in cholangiocytes. Gene expression was analyzed using mRNA or total RNA purified fromfreshly isolated hepatocytes (Hep), large cholangiocyte (LC), and small cholangiocyte (SC) in rat. a Northern blot detected Cyp27 expression in cholangiocytes, whereas the other Cyp genes that are involved in bile acid synthesis could not be detected under the current experimental conditions. b Western blot detected CYP27 expression in hepatocytes and cholangiocytes from rats. c IHC detected CYP27 expression in the liver of rats. Strong cytoplasmic expression was shown in the hepatocytes and bile duct epithelia cells (star). d–f Quantitative PCR was used to quantify the expression of the nuclear receptors regulating bile acid homeostasis: FXR (d), SHP (e), and HNF4α (f). A LXR ligand 27- hydroxycholesterol (27-HC), the product of Cyp27, activated cholesterol efflux in cholangiocytes through the basolateral (g), but not the apical region (h) in NRCs. GW501516, a synthetic PPARδ ligand, enhanced the effect of 27-HC in the induction of efflux of cholesterol through the basolateral compartment (e). Error bars = S.D. (N=3). *P <0.01, statistical significance using Student's t test
Next, we tested whether 27-hydrocholesterol (27-HC), the enzymatic product of Cyp27, would also induce basolateral cholesterol efflux as a ligand to liver X receptor (LXR) since we reported previously that basolateral efflux of cholesterol from cholangiocytes was stimulated by coadministration of synthetic ligands that activate LXR or peroxisome proliferator-activated receptor delta (PPARδ) [32]. NRCs were cultured on a transwell insert and incubated with [3H]-labeled cholesterol for 24 h. Release of radiolabeled cholesterol was then determined after capture with delipidated Apo-AI. Similar to the combinatorial effect of a synthetic LXR ligand (T0901317) and a synthetic PPARδ ligand (GW501516), incubation of cholangiocytes with 27-HC and GW501516 induced basolateral cholesterol efflux (Fig. 1g). There was little effect on apical cholesterol efflux (Fig. 1h). Thus, cholangiocytes express Cyp27, and the enzymatic product of Cyp27 stimulates basolateral cholesterol efflux in cholangiocytes through LXR.
Bile acid-induced secretion of FGF15/19 in cholangiocytes
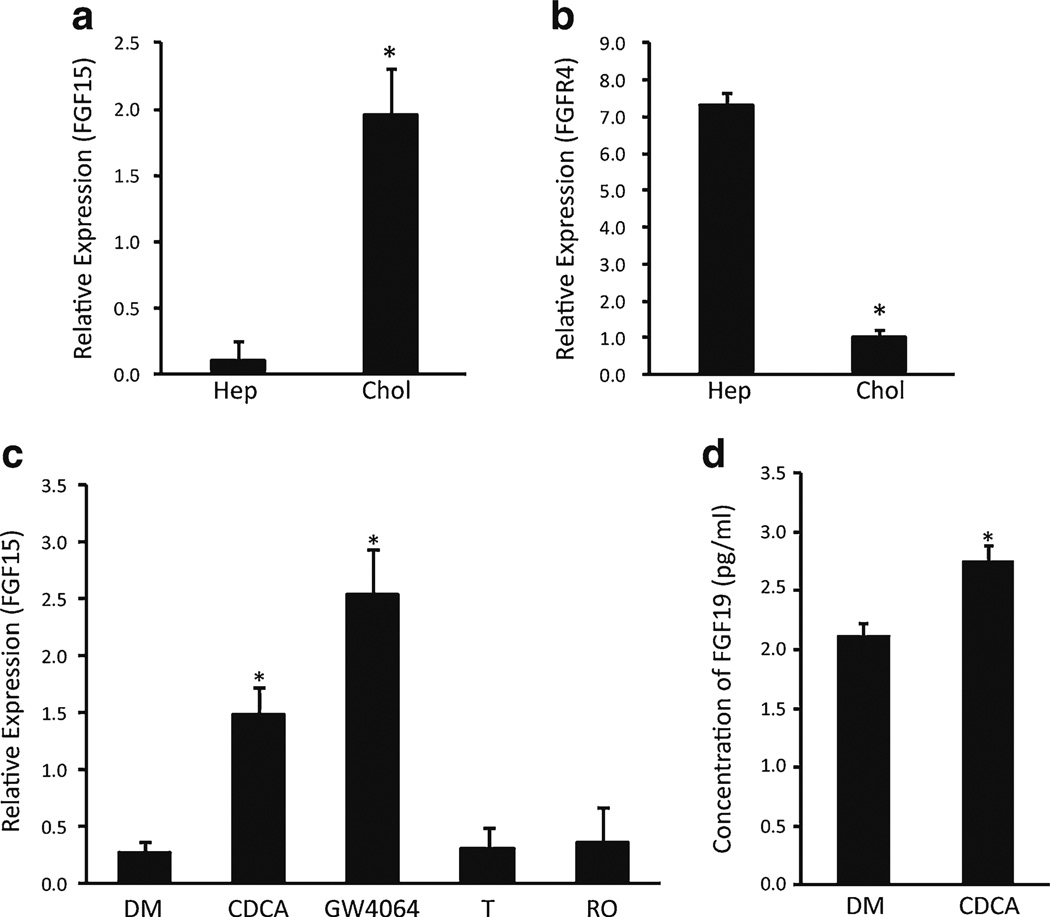
We next examined the expression of FGF15/19 in cholangiocytes. FGF19 and FGFR4 have been reported to be expressed in human gallbladder and bile duct epithelial cells [33, 37]. FGF15 mRNA was expressed at higher levels in cholangiocytes compared to hepatocytes (Fig. 2a); there were also FGFR4 mRNA in cholangiocytes, albeit at lower levels than those in the hepatocytes (Fig. 2b). Since FXR regulates FGF15/19 expression in the liver and intestine [6, 12, 22], we investigated whether similar regulation occurred in cholangiocytes. Treatment of rat cholangiocytes with the bile acid CDCA or the synthetic FXR ligand GW4064 increased FGF15 expression. Neither an LXR ligand (T0901317) nor a PPARγ ligand (rosiglitazone) yielded similar effects (Fig. 2c). Bile acids also induced an increase of secreted FGF19 in the medium of cultured human cholangiocytes (Fig. 2d). Thus, FXR was able to activate expression of FGF15/19 in cholangiocytes similar to that in enterocytes.
Fig. 2.
Bile acid-induced secretion of FGF15/19 in cholangiocytes. The level of gene expression was analyzed using mRNA purified from freshly isolated hepatocytes (Hep) and cholangiocytes (Chol), NRCs, and human cholangiocytes (H69). a Expression level of FGF15 in freshly isolated hepatocytes and cholangiocytes. b Expression level of FGFR4 in freshly isolated hepatocytes and cholangiocytes. c Expression level of FGF15 in cultured NRC line after treatment with DMSO (DM), CDCA, GW4064, T0901317 (T), or rosiglitazone (RO). d The protein concentration of secreted FGF19 in the medium of H69 cells treated with CDCA or DMSO for 24 hours was measured by Human FGF19 ELISA Kit. Error bars = S.D. (N =3). *P <0.01, statistical significance using Student's t test
Repression of Cyp27 and Cyp7a1 by bile acids and FGF15/19
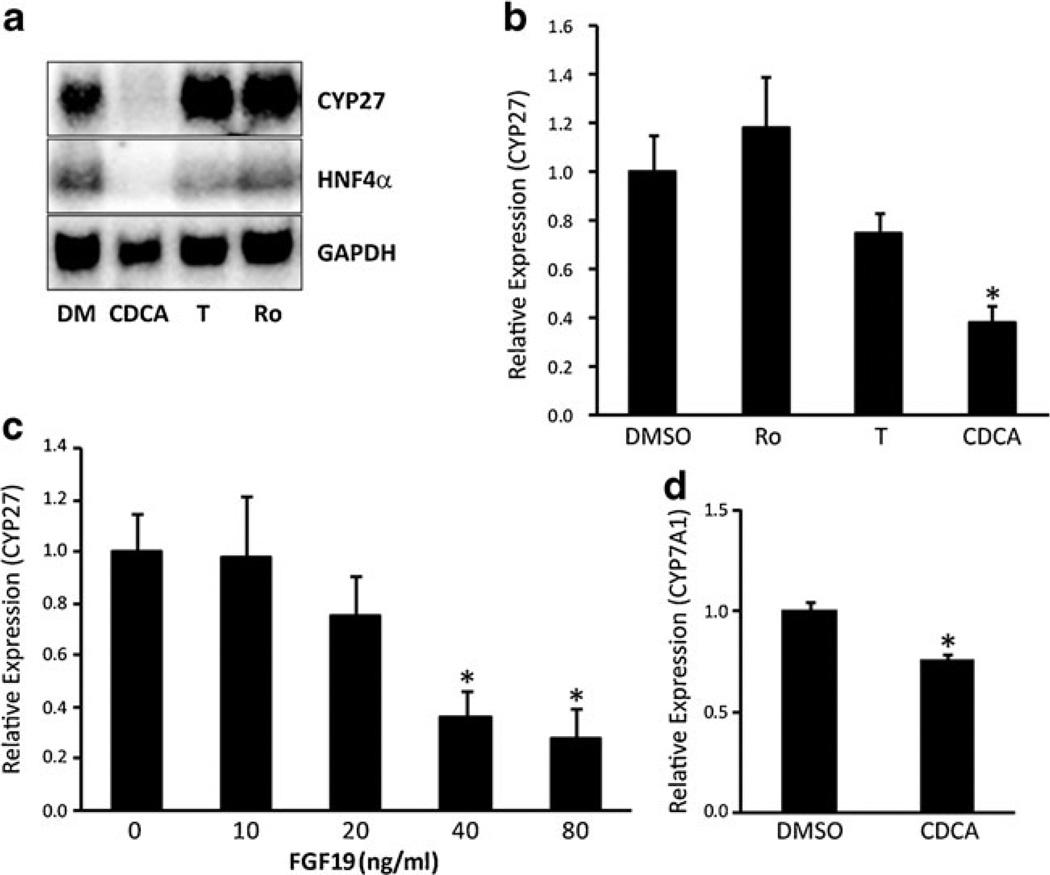
Since bile acids inhibit expression of Cyp27 and its master transcription factor HNF4α in hepatocytes, we determined whether similar effects would be observed in cholangiocytes. NRC treated with CDCA for 24 hours exhibited a marked decrease in Cyp27 and HNF4α expression (Fig. 3a, b). By contrast, ligands for LXR (T0901317) or PPARγ (rosiglitazone) had little effect. Recombinant FGF19 also elicited dose-dependent decreases in Cyp27 expression in the human cholangiocyte line, H69, implying that bile acid-dependent increases in FGF15/19 may be involved in this effect (Fig. 3c).
Fig. 3.
Repression of HNF4α, Cyp27, and Cyp7a1 by FGF15/19 signaling. The expression levels of HNF4α, Cyp27, and Cyp7a1 were analyzed using total RNA or mRNA purified from NRC or H69 treated with DMSO (DM), CDCA, T0901317 (T), rosiglitazone (RO), or FGF19. a The repression of Cyp27 and HNF4α in NRC cells were analyzed with Northern blot. b The repression of Cyp27 was confirmed using Q-PCR analysis in NRC cells. c FGF19 inhibited Cyp27 expression in a dose-dependent fashion in H69 cells. d Coincubation with H69 cells pretreated with CDCA significantly inhibited Cyp7a1 expression in HepG2 cells. Error bars = S.D. (N =3). *P <0.05, statistical significance using Student's t test
CDCA-induced secretion of FGF19 in the medium of cultured human cholangiocytes (Fig. 2d) led us to question whether bile acid-dependent increases in FGF15/19 secretion could elicit paracrine effects upon neighboring cells. To investigate this question, we used a transwell culture system to coculture a human cholangiocyte cell line (H69) and a hepatocyte cell line (HepG2). H69 cells cultured to confluence upon a transwell insert were treated with ± CDCA for 2 hours, washed three times with plain medium for 15 minutes each for complete removal of residual CDCA, and then transferred into a culture well that contained HepG2 cells grown on the bottom. Pretreatment of H69 cells with CDCA led to a significant inhibition of Cyp7a1 expression in the HepG2 cells in this system (P <0.05) (Fig. 3d). Thus, bile acids act on cholangiocytes to induce a secreted factor that represses the expression of Cyp7a1 in hepatocytes.
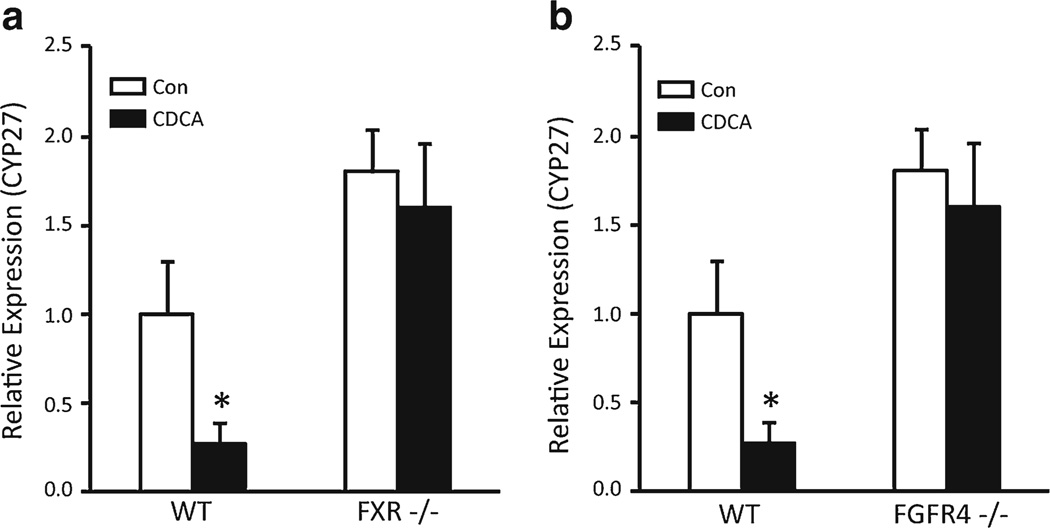
Next, we examined whether CDCA-induced Cyp27 repression can be observed in mouse cholangiocytes in vivo. Knockout models were used to determine whether this effect required FXR and FGFR4. The mice were fed on normal or 1 % CDCA-containing diet for 5 days, and bile ducts were isolated from livers using laser capture microdissection. Cyp27 mRNA expression was significantly inhibited in wild-type mice fed CDCA (Fig. 4a, b), but similar effects were not seen in FXR−/− (Fig. 4a) or FGFR4−/− mice (Fig. 4b). Thus, bile acids repress Cyp27 expression in cholangiocytes in vivo, and this effect requires FXR and the FGF15 receptor.
Fig. 4.
Confirmation of the FXR- and FGFR4-dependent repression of Cyp27 with CDCA in mouse models. Wild-type and knockout mice were fed normal diet or 1 % CDCA-containing diet for 5 days, and then, intrahepatic bile ducts were collected by a laser microdissection, and total RNA was purified. The expression of Cyp27 was analyzed using Q-PCR. Wild-type mice, which were fed the CDCA diet, showed decreased Cyp27 expression in bile ducts, whereas a FXR knockout (FXR−/−) mice and b FGFR4 knockout (FGFR4−/−)micemaintained a comparable level of Cyp27 expression even after feeding CDCAdiet. Error bars =S.D. (N =3). *P <0.05, statistical significance using Student's t test
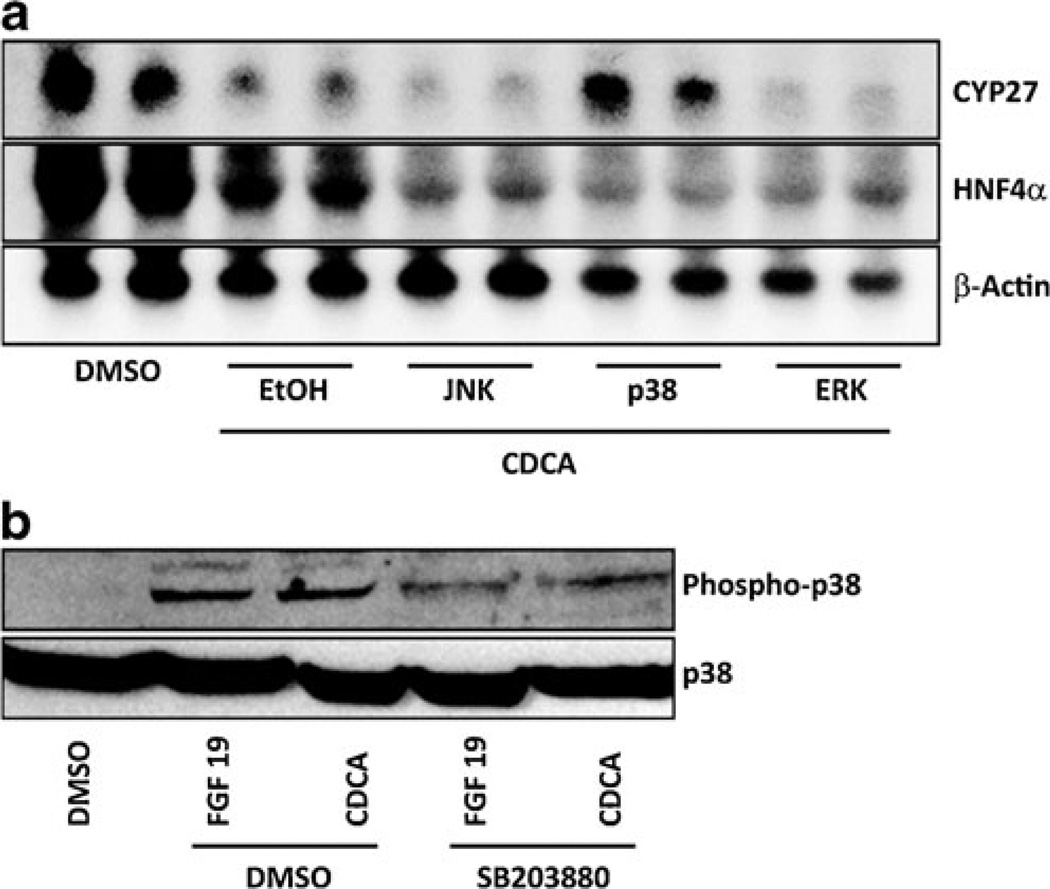
CDCA-dependent Cyp27 repression in cholangiocytes requires p38 kinase
We investigated intracellular signaling pathways that mediate CDCA-dependent repression of Cyp27 using small molecule inhibitors. Treatment of NRCs with small molecule inhibitors of MAPK kinase (MEK) (PD98059), JNK (SP600125), or p38 kinase (SB203580) revealed that CDCA-induced repression of Cyp27 was exclusively inhibited by p38 kinase inhibitor SB203580 (Fig. 5a).Western blot analysis confirmed that p38 kinase phosphorylation, which is indicative of kinase activation [20], was increased after treatment with CDCA or FGF15/19 and decreased by the presence of p38 kinase inhibitor SB203880 (Fig. 5b). CDCA treatment also inhibited HNF4α expression in cholangiocytes (Fig. 5a). However, this effect was not reversed by the treatment of cholangiocytes with the p38 kinase inhibitor, SB203880 (Fig. 5a). These results suggest that p38 kinase mediates a signal from the FGFR4–FGF15/19 complex to downregulates Cyp27 expression in cholangiocytes, whereas p38 kinase signaling is not responsible for the CDCA-induced HNF4α repression in cholangiocytes.
Fig. 5.
Two mechanisms for Cyp27 repression by CDCA in cholangiocytes. NRCs were treated with CDCA together with ethanol (EtOH), a JNK inhibitor (SP600125), an ERK inhibitor (PD98059), or a p38 kinase inhibitor (SB203580). Expression levels of Cyp27 and HNF4α were analyzed using Northern blot. a CDCA treatment with vehicle (EtOH) decreased Cyp27 and HNF4α, but the decrease was inhibited by cotreatment with a p38 kinase inhibitor. b Phosphorylation of p38 kinase was increased after treatment with CDCA or FGF15, which was inhibited in the presence of a p38 kinase inhibitor
Discussion
Evidences indicate that cholangiocytes are actively involved in bile homeostasis rather than just lining cells of the bile conduit [4]. In the current study, we demonstrated that bile acids exerted inhibitory effects upon Cyp27 expression in cholangiocytes. Expression profiles of genes for bile acid synthesis in cholangiocytes were different from that observed in hepatocytes. There was low expression of Cyp7a and Cyp8b, which encode enzymes involved in the classic/neutral bile acid biosynthesis processing, yet there was a considerable expression of Cyp27, which encodes an enzyme that catalyzes the first step of the alternative/acidic bile acid biosynthesis. The absence of Cyp7a and Cyp8b expressions suggested that cholangiocytes might have, like other somatic cells, a lesser role in the maintenance of bile homoeostasis; however, we hypothesized that cholangiocytes could regulate bile homeostasis based on their physical proximity both to bile flow and the neighboring hepatocytes. Our current data support our hypothesis that cholangiocytes produce acidic bile acid through expression of Cyp27 and also give an influence toward the bile acid biosynthesis in neighboring hepatocytes through secretion of FGF15/19.
Loss of CDCA-induced repression of Cyp27 in bile ducts isolated from the livers of the FXR−/− or the FGFR4−/− mice confirmed the essential role of FXR- and FGF15/19-mediated signaling for the Cyp27 repression in cholangiocytes in vivo. Interestingly, in contrast with JNK- or MAPK/ERK-mediated repression of Cyp7a1 in hepatocytes, FGF15/FGFR4 signaling downregulated Cyp27 expression through activation of p38 kinase in cholangiocytes. In hepatocytes, the FGF15/19 signaling to repress Cyp7a1 expression has been known to be mediated by SHP and AP-1 [12, 13, 24]. SHP inhibits the function of LRH-1, the master transcription factor required for expression of Cyp7a1, and JNK activates SHP function via phosphorylation [9, 10]. In our study, SHP-mediated repression of LRH-1 does not seem to be involved in Cyp27 repression in cholangiocytes since SHP is neither highly expressed (Fig. 1e) nor induced by bile acid treatment in cholangiocytes (data not shown). Instead, we found that SHP-independent HNF4α was a pathway regulating the expression of Cyp27 in cholangiocytes. Supporting our findings, previous studies performed to analyze Cyp27 promoter identified that transcription factors such as Sp1, Sp3, and HNF4α cooperated to induce the Cyp27 expression [8], and a HNF4α-binding site was identified in the promoter of the Cyp27 gene [5]. Bile acid treatment downregulated the expression and DNA-binding affinity of HNF4α [19]. Similar to the previous results, we found that bile acid signaling negatively regulated HNF4α activity by downregulating HNF4α expression in cholangiocytes.
FXR activation with selective agonists has been investigated as a therapy for biliary diseases such as cholesterol gallstone disease [16, 18] and cholestasis [3, 15, 17] with promising results. Cholesterol gallstone formation is a prevalent disease in the USA primarily caused by altered metabolism and homeostasis of cholesterol and increased hydrophobicity of bile acids in bile. Our data suggest that cholangiocytes are a promising target to prevent cholesterol gallstone formation because their physical location enables them to alter bile flow directly and quickly. We have previously reported that cholangiocytes were involved in cholesterol homeostasis in bile through activation of the nuclear receptors, LXRβ and PPARδ; activation of which induced cholesterol efflux from the bile to the liver [32]. Given that feeding of a high cholesterol and cholic acid diet induced cholesterol gallstone disease in mice, possibility of cholangiocytes responding to the changes in the bile induced by this diet suggests that targeted activation of the nuclear receptors in cholangiocytes holds a future potential as a therapeutic option for preventing or even curing cholesterol gallstone diseases by restoring homeostasis of cholesterol and bile acids in the bile of patients with elevated bile acid hydrophobicity or altered cholesterol metabolism.
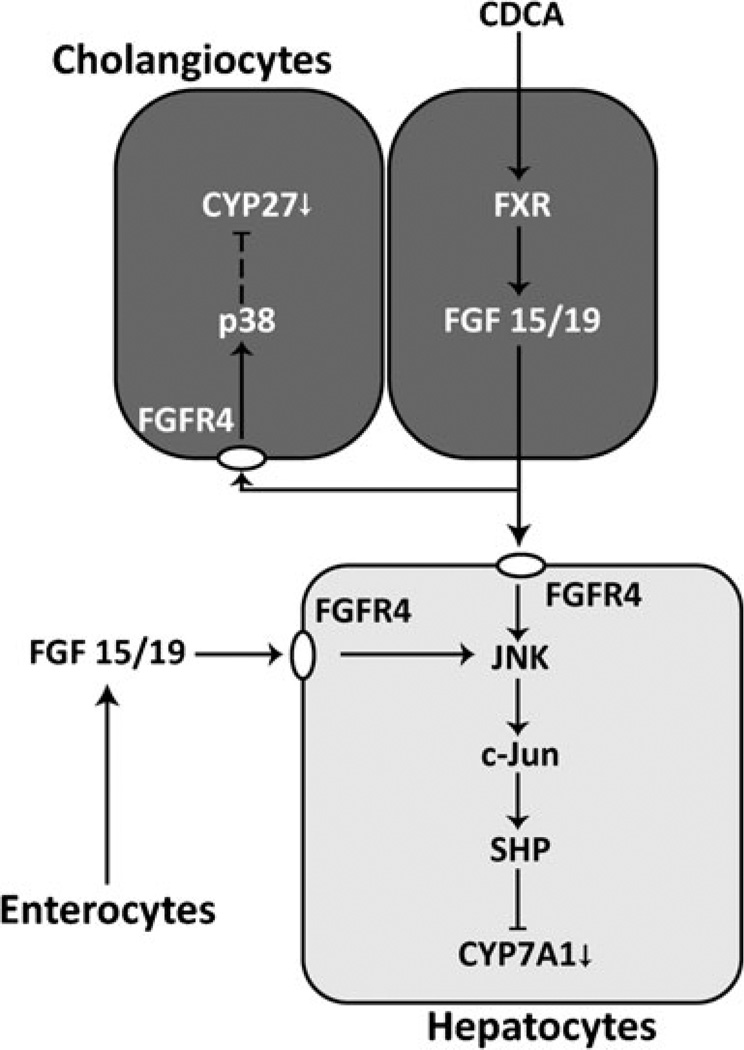
In this study, we have shown that Cyp27 expression in cholangiocytes can be negatively regulated by bile acid signaling via FXR or HNF4α. FXR signaling is mediated by FXR-induced FGF15/19, which represses Cyp27 expression through binding to FGFR4 and inducing p38 kinase signaling. Bile acid treatment decreases HNF4α expression by still unknown mechanisms. A hormonal effect of FGF15 via enterohepatic circulation in mouse has been reported. Cyp7a1 is repressed in the liver by induced FGF15 in enterocytes by bile acid-activated FXR or by intravenous administration of FGF15 [13]. In addition to the enterohepatic feedback loop, our results indicate the presence of FGF15/19-mediated intrahepatic feedback loop through cholangiocytes against bile acid biosynthesis in the liver. Although it is still immature, the schematic representation summarizes the feedback regulations against bile acid biosynthesis in hepatocytes and cholangiocytes (Fig. 6). Discovery of the p38 kinase target proteins and of a mechanism for the bile acid-induced repression of HNF4α remains to be open questions, and we will pursue the answers to these questions in our future work.
Fig. 6.
Schematic description of FGF15/19-mediated repression of Cyp27 and Cyp7a1 in cholangiocytes and hepatocytes. In cholangiocytes or enterocytes, activated FXR induces expression of FGF15/19, which affects neighboring cholangiocytes and hepatocytes through paracrine signaling. FGF15/19 binds to FGFR4, initiating a signaling to repress Cyp27 expression in cholangiocytes through p38 kinase or Cyp7a1 expression in hepatocytes through JNK
Acknowledgments
This work was supported by grants from the National Institutes of Health (R01 DK53366 to D.D.M., RC4 DK90849 to P.W., R01 DK54208 to G.D.L., R01 DK080440 to L.W., R01 DK062975, and DK76898), from the Dr. Nicholas C. Hightower Centennial Chair of Gastroenterology from Scott & White Hospital to G.A., and from the National Natural Science Foundation of China (no. 81270868 to X.X.).
Footnotes
Conflict of interest None.
Contributor Information
Dongju Jung, Department of Molecular and Cellular Biology, Baylor College of Medicine, Houston, TX 77030, USA.
J. Philippe York, Center for Genomic Medicine and Diabetes Research, Houston Methodist Research Institute, Weill Cornell School of Medicine, Weill Cornell Medical College, 6670 Bertner Street, Houston, TX 77030, USA.
Li Wang, Departments of Medicine and Oncological Sciences, University of Utah School of Medicine, Salt Lake City, UT 84112, USA.
Chaofeng Yang, Center for Cancer and Stem Cell Biology, Institute of Biosciences and Technology, Texas A&M Health Science Center, Houston, TX 77030, USA.
Aijun Zhang, Center for Genomic Medicine and Diabetes Research, Houston Methodist Research Institute, Weill Cornell School of Medicine, Weill Cornell Medical College, 6670 Bertner Street, Houston, TX 77030, USA.
Heather L. Francis, Department of Research, Central Texas Veterans Health Care System, Temple, TX 77807, USA Scott & White Digestive Disease Research Center, Division of Gastroenterology, Department of Medicine College of Medicine, Research Education Scott & White and Texas A&M Health Science Center, Temple, TX 77807, USA.
Paul Webb, Center for Genomic Medicine and Diabetes Research, Houston Methodist Research Institute, Weill Cornell School of Medicine, Weill Cornell Medical College, 6670 Bertner Street, Houston, TX 77030, USA.
Wallace L. McKeehan, Center for Cancer and Stem Cell Biology, Institute of Biosciences and Technology, Texas A&M Health Science Center, Houston, TX 77030, USA
Gianfranco Alpini, Department of Research, Central Texas Veterans Health Care System, Temple, TX 77807, USA; Scott & White Digestive Disease Research Center, Division of Gastroenterology, Department of Medicine College of Medicine, Research Education Scott & White and Texas A&M Health Science Center, Temple, TX 77807, USA.
Gene D. LeSage, Department of Internal Medicine, Quillen College of Medicine, East Tennessee State University, Johnson City, TN 37614, USA
David D. Moore, Department of Molecular and Cellular Biology, Baylor College of Medicine, Houston, TX 77030, USA
Xuefeng Xia, Email: xxia2@tmhs.org, Center for Genomic Medicine and Diabetes Research, Houston Methodist Research Institute, Weill Cornell School of Medicine, Weill Cornell Medical College, 6670 Bertner Street, Houston, TX 77030, USA; The Third Affiliated Hospital, Guangzhou Medical University, Guangzhou, Guangdong, China.
References
- 1.Abrahamsson A, Gustafsson U, Ellis E, Nilsson LM, Sahlin S, Bjorkhem I, Einarsson C. Feedback regulation of bile acid synthesis in human liver: importance ofHNF-4alpha for regulation of CYP7A1. Biochem Biophys Res Commun. 2005;330(2):395–399. doi: 10.1016/j.bbrc.2005.02.170. [DOI] [PubMed] [Google Scholar]
- 2.Babiker A, Andersson O, Lund E, Xiu RJ, Deeb S, Reshef A, Leitersdorf E, Diczfalusy U, Bjorkhem I. Elimination of cholesterol in macrophages and endothelial cells by the sterol 27-hydroxylase mechanism, Comparison with high density lipoprotein-mediated reverse cholesterol transport. J Biol Chem. 1997;272(42):26253–26261. doi: 10.1074/jbc.272.42.26253. [DOI] [PubMed] [Google Scholar]
- 3.Barbier O, Torra IP, Sirvent A, Claudel T, Blanquart C, Duran-Sandoval D, Kuipers F, Kosykh V, Fruchart JC, Staels B. FXR induces the UGT2B4 enzyme in hepatocytes: a potential mechanism of negative feedback control of FXR activity. Gastroenterology. 2003;124(7):1926–1940. doi: 10.1016/s0016-5085(03)00388-3. [DOI] [PubMed] [Google Scholar]
- 4.Bogert PT, LaRusso NF. Cholangiocyte biology. Curr Opin Gastroenterol. 2007;23(3):299–305. doi: 10.1097/MOG.0b013e3280b079fb. [DOI] [PubMed] [Google Scholar]
- 5.Chen W, Chiang JY. Regulation of human sterol 27-hydroxylase gene (CYP27A1) by bile acids and hepatocyte nuclear factor 4alpha (HNF4alpha) Gene. 2003;313:71–82. doi: 10.1016/s0378-1119(03)00631-0. [DOI] [PubMed] [Google Scholar]
- 6.Chiang JY. Regulation of bile acid synthesis: pathways, nuclear receptors, and mechanisms. J Hepatol. 2004;40(3):539–551. doi: 10.1016/j.jhep.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 7.Fava G, Glaser S, Francis H, Alpini G. The immunophysiology of biliary epithelium. Semin Liver Dis. 2005;25(3):251–264. doi: 10.1055/s-2005-916318. [DOI] [PubMed] [Google Scholar]
- 8.Garuti R, Croce MA, Piccinini L, Tiozzo R, Bertolini S, Calandra S. Functional analysis of the promoter of human sterol 27-hydroxylase gene in HepG2 cells. Gene. 2002;283(1–2):133–143. doi: 10.1016/s0378-1119(01)00874-5. [DOI] [PubMed] [Google Scholar]
- 9.Goodwin B, Jones SA, Price RR, Watson MA, McKee DD, Moore LB, Galardi C, Wilson JG, Lewis MC, Roth ME, Maloney PR, Willson TM, Kliewer SA. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell. 2000;6(3):517–526. doi: 10.1016/s1097-2765(00)00051-4. [DOI] [PubMed] [Google Scholar]
- 10.Gupta S, Stravitz RT, Dent P, Hylemon PB. Down-regulation of cholesterol 7alpha-hydroxylase (CYP7A1) gene expression by bile acids in primary rat hepatocytes is mediated by the c-Jun N-terminal kinase pathway. J Biol Chem. 2001;276(19):15816–15822. doi: 10.1074/jbc.M010878200. [DOI] [PubMed] [Google Scholar]
- 11.Hay DW, Carey MC. Pathophysiology and pathogenesis of cholesterol gallstone formation. Semin Liver Dis. 1990;10(3):159–170. doi: 10.1055/s-2008-1040470. [DOI] [PubMed] [Google Scholar]
- 12.Holt JA, Luo G, Billin AN, Bisi J, McNeill YY, Kozarsky KF, Donahee M, Wang DY, Mansfield TA, Kliewer SA, Goodwin B, Jones SA. Definition of a novel growth factor-dependent signal cascade for the suppression of bile acid biosynthesis. Genes Dev. 2003;17(13):1581–1591. doi: 10.1101/gad.1083503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inagaki T, Choi M, Moschetta A, Peng L, Cummins CL, McDonald JG, Luo G, Jones SA, Goodwin B, Richardson JA, Gerard RD, Repa JJ, Mangelsdorf DJ, Kliewer SA. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2(4):217–225. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 14.Jung D, Kullak-Ublick GA. Hepatocyte nuclear factor 1 alpha: a key mediator of the effect of bile acids on gene expression. Hepatology. 2003;37(3):622–631. doi: 10.1053/jhep.2003.50100. [DOI] [PubMed] [Google Scholar]
- 15.Liu YP, Binz J, Numerick MJ, Dennis S, Luo GZ, Desai B, Mac-Kenzie KI, Mansfield TA, Kliewer SA, Goodwin B, Jones SA. Hepatoprotection by the farnesoid X receptor agonist GW4064 in rat models of intra- and extrahepatic cholestasis. J Clin Investig. 2003;112(11):1678–1687. doi: 10.1172/JCI18945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Modica S, Gadaleta RM, Moschetta A. Deciphering the nuclear bile acid receptor FXR paradigm. Nucl Recept Signal. 2010;8:e005. doi: 10.1621/nrs.08005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Modica S, Petruzzelli M, Bellafante E, Murzilli S, Salvatore L, Celli N, Di Tullio G, Palasciano G, Moustafa T, Halilbasic E, Trauner M, Moschetta A. Selective activation of nuclear bile acid receptor FXR in the intestine protects mice against cholestasis. Gastroenterology. 2012;142(2):355–U269. doi: 10.1053/j.gastro.2011.10.028. [DOI] [PubMed] [Google Scholar]
- 18.Moschetta A, Bookout AL, Mangelsdorf DJ. Prevention of cholesterol gallstone disease by FXR agonists in a mouse model. Nat Med. 2004;10(12):1352–1358. doi: 10.1038/nm1138. [DOI] [PubMed] [Google Scholar]
- 19.Popowski K, Eloranta JJ, Saborowski M, Fried M, Meier PJ, Kullak-Ublick GA. The human organic anion transporter 2 gene is transactivated by hepatocyte nuclear factor-4 alpha and suppressed by bile acids. Mol Pharmacol. 2005;67(5):1629–1638. doi: 10.1124/mol.104.010223. [DOI] [PubMed] [Google Scholar]
- 20.Raingeaud J, Gupta S, Rogers JS, Dickens M, Han J, Ulevitch RJ, Davis RJ. Pro-inflammatory cytokines and environmental stress cause p38mitogen-activated protein kinase activation by dual phosphorylation on tyrosine and threonine. J Biol Chem. 1995;270(13):7420–7426. doi: 10.1074/jbc.270.13.7420. [DOI] [PubMed] [Google Scholar]
- 21.Rao YP, Vlahcevic ZR, Stravitz RT, Mallonee DH, Mullick J, Avadhani NG, Hylemon PB. Down-regulation of the rat hepatic sterol 27-hydroxylase gene by bile acids in transfected primary hepatocytes: possible role of hepatic nuclear factor 1alpha. J Steroid Biochem Mol Biol. 1999;70(1–3):1–14. doi: 10.1016/s0960-0760(99)00099-0. [DOI] [PubMed] [Google Scholar]
- 22.Sinal CJ, Tohkin M, Miyata M, Ward JM, Lambert G, Gonzalez FJ. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell. 2000;102(6):731–744. doi: 10.1016/s0092-8674(00)00062-3. [DOI] [PubMed] [Google Scholar]
- 23.Sinha J, Chen F, Miloh T, Burns RC, Yu Z, Shneider BL. beta-Klotho and FGF-15/19 inhibit the apical sodium-dependent bile acid transporter in enterocytes and cholangiocytes. Am J Physiol Gastrointestin Liver Physiol. 2008;295(5):G996–G1003. doi: 10.1152/ajpgi.90343.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song KH, Li T, Owsley E, Strom S, Chiang JY. Bile acids activate fibroblast growth factor 19 signaling in human hepatocytes to inhibit cholesterol 7alpha-hydroxylase gene expression. Hepatology. 2009;49(1):297–305. doi: 10.1002/hep.22627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strazzabosco M, Fabris L, Spirli C. Pathophysiology of cholangiopathies. J Clin Gastroenterol. 2005;39(4) Suppl 2:S90–S102. doi: 10.1097/01.mcg.0000155549.29643.ad. [DOI] [PubMed] [Google Scholar]
- 26.Tietz PS, Chen XM, Gong AY, Huebert RC, Masyuk A, Masyuk T, Splinter PL, LaRusso NF. Experimental models to study cholangiocyte biology. World J Gastroenterol. 2002;8(1):1–4. doi: 10.3748/wjg.v8.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tietz PS, Larusso NF. Cholangiocyte biology. Curr Opin Gastroenterol. 2006;22(3):279–287. doi: 10.1097/01.mog.0000218965.78558.bc. [DOI] [PubMed] [Google Scholar]
- 28.Twisk J, de Wit EC, Princen HM. Suppression of sterol 27-hydroxylase mRNA and transcriptional activity by bile acids in cultured rat hepatocytes. Biochem J. 1995;305(Pt 2):505–511. doi: 10.1042/bj3050505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vlahcevic ZR, Jairath SK, Heuman DM, Stravitz RT, Hylemon PB, Avadhani NG, Pandak WM. Transcriptional regulation of hepatic sterol 27-hydroxylase by bile acids. Am J Physiol. 1996;270(4 Pt 1):G646–G652. doi: 10.1152/ajpgi.1996.270.4.G646. [DOI] [PubMed] [Google Scholar]
- 30.Wang L, Lee YK, Bundman D, Han Y, Thevananther S, Kim CS, Chua SS, Wei P, Heyman RA, Karin M, Moore DD. Redundant pathways for negative feedback regulation of bile acid production. Dev Cell. 2002;2(6):721–731. doi: 10.1016/s1534-5807(02)00187-9. [DOI] [PubMed] [Google Scholar]
- 31.Wittenburg H, Lyons MA, Li R, Churchill GA, Carey MC, Paigen B. FXR and ABCG5/ABCG8 as determinants of cholesterol gallstone formation from quantitative trait locus mapping in mice. Gastroenterology. 2003;125(3):868–881. doi: 10.1016/s0016-5085(03)01053-9. [DOI] [PubMed] [Google Scholar]
- 32.Xia X, Jung D, Webb P, Zhang A, Zhang B, Li L, Ayers SD, Gabbi C, Ueno Y, Gustafsson JA, Alpini G, Moore DD, Lesage GD. Liver X receptor beta and peroxisome proliferator-activated receptor delta regulate cholesterol transport in murine cholangiocytes. Hepatology. 2012;56(6):2288–2296. doi: 10.1002/hep.25919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie MH, Holcomb I, Deuel B, Dowd P, Huang A, Vagts A, Foster J, Liang J, Brush J, Gu Q, Hillan K, Goddard A, Gurney AL. FGF-19, a novel fibroblast growth factor with unique specificity for FGFR4. Cytokine. 1999;11(10):729–735. doi: 10.1006/cyto.1999.0485. [DOI] [PubMed] [Google Scholar]
- 34.Yang Y, Zhang M, Eggertsen G, Chiang JY. On the mechanism of bile acid inhibition of rat sterol 12alpha-hydroxylase gene (CYP8B1) transcription: roles of alpha-fetoprotein transcription factor and hepatocyte nuclear factor 4alpha. Biochim Biophys Acta. 2002;1583(1):63–73. doi: 10.1016/s1388-1981(02)00186-5. [DOI] [PubMed] [Google Scholar]
- 35.Yu C, Wang F, Kan M, Jin C, Jones RB, Weinstein M, Deng CX, McKeehan WL. Elevated cholesterolmetabolism and bile acid synthesis inmice lacking membrane tyrosine kinase receptor FGFR4. J Biol Chem. 2000;275(20):15482–15489. doi: 10.1074/jbc.275.20.15482. [DOI] [PubMed] [Google Scholar]
- 36.Zhang M, Chiang JY. Transcriptional regulation of the human sterol 12alpha-hydroxylase gene (CYP8B1): roles of hepatocyte nuclear factor 4alpha in mediating bile acid repression. J Biol Chem. 2001;276(45):41690–41699. doi: 10.1074/jbc.M105117200. [DOI] [PubMed] [Google Scholar]
- 37.Zweers SJ, Booij KA, Komuta M, Roskams T, Gouma DJ, Jansen PL, Schaap FG. The human gallbladder secretes fibroblast growth factor 19 into bile: towards defining the role of fibroblast growth factor 19 in the enterobiliary tract. Hepatology. 2012;55(2):575–583. doi: 10.1002/hep.24702. [DOI] [PubMed] [Google Scholar]