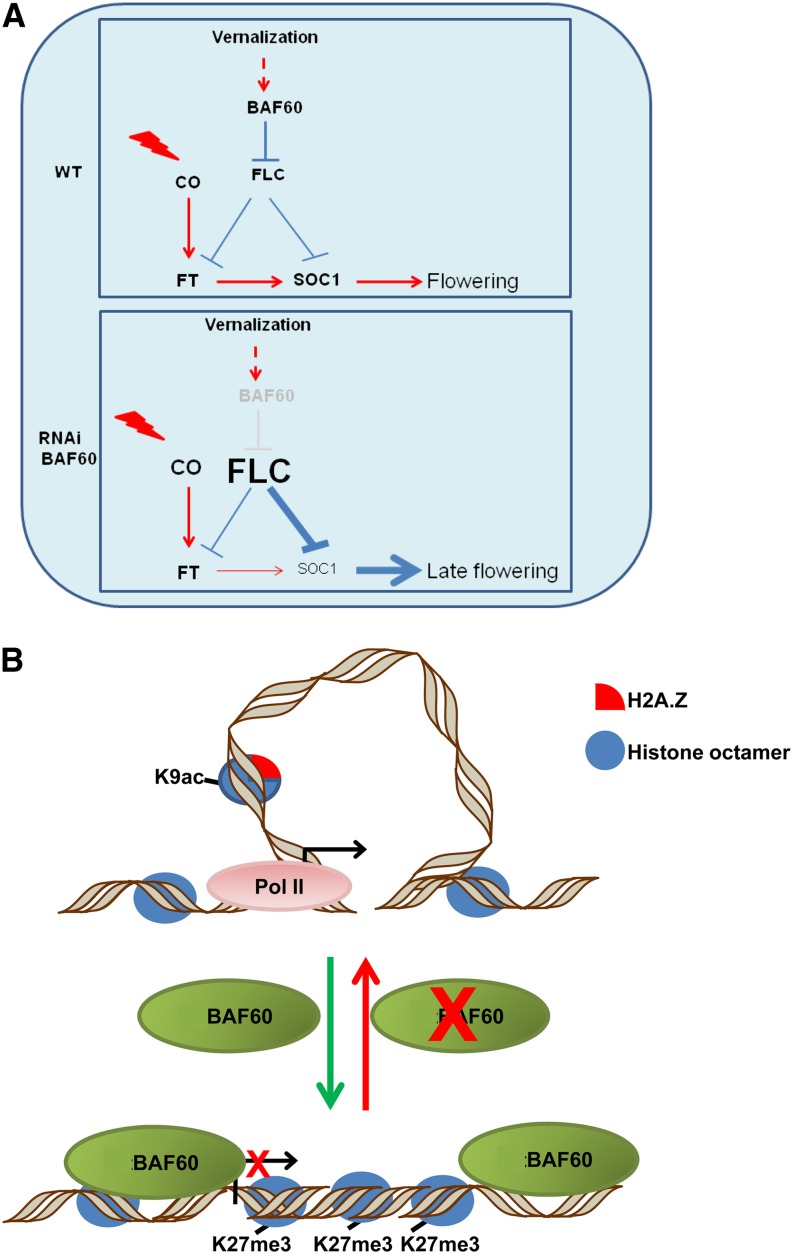
It is shown that BAF60, a subunit of the chromatin-remodeling complex SWI/SNF, induces a change in the floral repressor FLOWERING LOCUS C at the high-order chromatin level, thereby repressing the photoperiod flowering pathway in Arabidopsis. Specifically, BAF60 modulates histone density, composition, and posttranslational modification, thereby controlling gene loop formation at FLOWERING LOCUS C.
Abstract
SWI/SNF complexes mediate ATP-dependent chromatin remodeling to regulate gene expression. Many components of these complexes are evolutionarily conserved, and several subunits of Arabidopsis thaliana SWI/SNF complexes are involved in the control of flowering, a process that depends on the floral repressor FLOWERING LOCUS C (FLC). BAF60 is a SWI/SNF subunit, and in this work, we show that BAF60, via a direct targeting of the floral repressor FLC, induces a change at the high-order chromatin level and represses the photoperiod flowering pathway in Arabidopsis. BAF60 accumulates in the nucleus and controls the formation of the FLC gene loop by modulation of histone density, composition, and posttranslational modification. Physiological analysis of BAF60 RNA interference mutant lines allowed us to propose that this chromatin-remodeling protein creates a repressive chromatin configuration at the FLC locus.
INTRODUCTION
DNA/histone complexes generate a barrier, thereby reducing the accessibility of transcription factors and the general transcriptional machinery to DNA. Among the mechanisms that have evolved to overcome this barrier is chromatin remodeling. Chromatin remodelers, which have been referred to as chromatin-remodeling machines (CRMs), are multiple-subunit complexes that use the energy of ATP hydrolysis to modify DNA–histone interactions and alter the location or conformation of nucleosomes (Clapier and Cairns, 2009). These multiple-protein complexes, with unique subunit compositions, regulate access to chromatin DNA by influencing the structure, the type of histone variants, and nucleosome positioning. Hence, these complexes mediate a remarkably broad spectrum of biological processes involving chromatin. The distinctive feature of all CRMs is the presence of a central ATPase, belonging to the SWI2/SNF2 family (Bork and Koonin, 1993; Eisen et al., 1995). The Arabidopsis thaliana genome encodes 41 Snf2-related proteins distributed into six superfamilies, among which the Snf2-like family encompasses the motor subunits for SWI/SNF complexes (Knizewski et al., 2008).
The SWI/SNF complex is recruited to the promoter region of a target gene by specific transcription factors in order to facilitate or repress nearby transcription (Weissman and Knudsen, 2009). The structure of this complex is highly conserved, and SWI/SNF subunits can be subclassified into three categories: (1) the above-mentioned enzymatic ATPase, (2) core subunits, and (3) accessory subunits. In mammals, the chromatin-remodeling activity of the SWI/SNF complex is essential for developmental processes, hormone responses, and tumor suppression, and the accessory subunits are hypothesized to dictate the specificity of SWI/SNF complex function (Wu, 2012).
In plants, experimental analyses and database surveys indicate the probable existence of SWI/SNF-type complexes, but no complete plant CRM has been isolated and characterized so far (Jerzmanowski, 2007). Nevertheless, several components of these complexes are evolutionarily conserved in plants, such as the SWI3 subunits (Sarnowski et al., 2005), the SNF5/BSH subunit (Brzeski et al., 1999), the nuclear actin-related protein ARP4 (Kandasamy et al., 2005), BRAHMA (BRM), or SPLAYED (SYD). Both of these latter proteins are ATPases of Arabidopsis SWI/SNF complexes and have been shown to participate in the control of flower development and flowering time (Wagner and Meyerowitz, 2002; Farrona et al., 2004; Hurtado et al., 2006; Bezhani et al., 2007; Fornara et al., 2010; Wu, 2012). Likewise, the SWI3B protein interacts with the flowering regulator FCA (Sarnowski et al., 2005). Among plant developmental processes, flowering is one of the most studied. Research over the last 15 years, mainly in Arabidopsis, has provided a good understanding of how different environmental and endogenous cues are integrated to cause a developmental switch from the shoot apical meristem to a flower (He, 2009). Floral induction is regulated by exogenous (daylength and temperature) and endogenous signals; for example, in the case of Arabidopsis, flowering is induced by long days and cold treatment (vernalization), but plants maintained in short-day (SD) conditions will eventually flower, owing to the autonomous pathway of flowering time regulation. Two central flowering regulators, CONSTANS (CO) and FLOWERING LOCUS C (FLC), antagonistically regulate flowering (Putterill et al., 1995; Samach et al., 2000): the CO gene, encoding a zinc finger protein, acts as a floral activator and mediates the photoperiod pathway in long-day (LD) conditions, whereas the FLC gene, encoding a MADS box protein, acts as a floral repressor and mediates the autonomous pathway as well as the vernalization pathway. In turn, CO and FLC regulate the expression of downstream genes, the so-called flowering pathway integrators FLOWERING LOCUS T (FT), SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1), and LEAFY. These three genes integrate signals from multiple flowering pathways, and their expression levels determine flowering time (Simpson and Dean, 2002; Parcy, 2005). In SD conditions, the CO pathway does not seem to be involved in the control of flowering, since the CO protein is not being stably produced (Valverde et al., 2004). Under this condition, induction of SOC1 is mediated by gibberellic acid signaling, as evidenced by the gibberellic acid biosynthetic mutant ga1-3, which fails to flower under SD conditions (Wilson et al., 1992).
Analyses of mutations that activate or silence the expression of FLC have revealed a conserved regulatory pathway that modulates transcription via chromatin regulation by histone posttranslational modifications. For instance, early-flowering mutants with reduced levels of FLC expression display elevated levels of trimethylation on lysine-27 of histone H3 (H3K27me3) at this locus (Doyle and Amasino, 2009), whereas FLC expression correlates positively with acetylation on lysine-9 of histone H3 (H3K9Ac; Wang et al., 2006).
A series of early-flowering mutants identified a major role for the SWR1 complex in FLC expression. The chromatin remodeling SWR1/SRCAP complex belongs to a different class of chromatin remodelers than SWI/SNF complexes and relies on the activity of ATPases from the SWR1-like family, comprising INO80 and SWR1 (Knizewski et al., 2008). This complex is involved in the substitution of histone H2A by the histone variant H2A.Z (March-Díaz and Reyes, 2009). Indeed, mutations in PIE1, ARP6 (also known as SUF3 and ESD1 in Arabidopsis), and SEF/AtSW6C show early flowering and reduced FLC expression (Noh and Amasino, 2003; Choi et al., 2005, 2007; Deal et al., 2005; Martin-Trillo et al., 2006; March-Díaz et al., 2007; Lázaro et al., 2008), and ARP6 and PIE1 were found to be required for H2A.Z deposition at FLC (Deal et al., 2007). Finally, it has been shown that disruption of BRM results in high FLC expression and that the brm mutation restores high accumulation of FLC mRNA in pie and sef mutants, suggesting that SWR1/SCRAP and SWI/SNF complexes play antagonistic roles in FLC regulation (Farrona et al., 2011). Whether BRM acts by modulating H2A.Z incorporation remains to be established, but expression of FLC in the brm mutant does not require the incorporation of H2A.Z, suggesting that this may be the case. H2A.Z is associated with both gene activation and gene repression and is frequently enriched at the 5′ and 3′ ends of genes (Marques et al., 2010). In the case of FLC, incorporation of H2A.Z at this locus would lead to relative nucleosome instability, thereby facilitating nucleosome displacement by the transcription machinery.
More recently, Crevillén et al. (2013) have used quantitative chromosome conformation capture (3C) in a range of Arabidopsis genotypes and showed that higher order chromatin structure is probably involved in FLC regulation. They detected a gene loop at the FLC locus, reflecting interaction between sequences in the FLC 5′ flanking region with sequences in the 3′ flanking region, and clearly demonstrated that this loop structure could be efficiently disrupted during vernalization. This loop did not reform after transfer of plants back to warm conditions. However, the mechanism involved in the formation and/or stabilization of this structure remains an open question.
As stated above, several components of the Arabidopsis SWI/SNF complexes are involved in the control of flowering. In addition, SWI/SNF complexes have been shown to produce in vitro DNA loops in a nucleosome-dependent manner (Zhang et al., 2006). This prompted us to investigate the function of another putative subunit of SWI/SNF complexes in the control of flowering. The SWP73 component of the yeast (Saccharomyces cerevisiae) SWI/SNF complex has a functional role in transcriptional activation (Cairns et al., 1996). Orthologs of SWP73, called BAP60 in Drosophila melanogaster and BAF60 in human, have been extensively characterized biochemically and genetically (Tang et al., 2010). The Arabidopsis genome encodes two highly similar homologs of SWP73: At5g14170 (also named At BAF60, At SWP73B, or CHC1 according to the Chromatin Consortium Database [http://chromdb.org]) and At3g01890 (also named At SWP73A). Although BAF60 has been shown to be involved in Agrobacterium tumefaciens–mediated root transformation (Crane and Gelvin, 2007), the repair of UV-B light–damaged DNA (Campi et al., 2012), and probably cell cycle regulation (Jégu et al., 2013), its molecular role in chromatin remodeling and plant development remains unclear.
In this work, we investigated the function of Arabidopsis BAF60 at the chromatin level and in the regulation of flowering time. We show that BAF60 is required for the repression of FLC expression. Downregulation of BAF60 results in a late-flowering phenotype in LD conditions, mainly through the increase of FLC expression in correlation with the decrease of its downstream targets. Furthermore, BAF60 regulates histone mark modifications, H2A.Z deposition, chromatin condensation, and loop formation via a direct binding at the FLC locus. BAF60 thus regulates the expression of FLC by creating a repressive chromatin configuration at this locus and plays a key role in the control of chromatin structure.
RESULTS
H2A.Z Deposition and Chromatin Condensation Are Impaired in BAF60 RNA Interference Lines
To perform a functional analysis of BAF60 in Arabidopsis, we first searched for loss-of-function mutants in publicly available mutant databases. Unfortunately, all T-DNA insertion lines showed insertions in the promoter or 5′ untranslated region, and mRNA accumulation remained unchanged in homozygous lines (Supplemental Figure 1). Therefore, we used two independent RNA interference (RNAi) lines described previously (Crane and Gelvin, 2007) for which the extent of silencing was documented by RT-PCR at the consortium database (http://chromdb.org). Quantitative real-time RT-PCR (qRT-PCR) confirmed the down-regulation of BAF60 in these lines (Supplemental Figure 2).
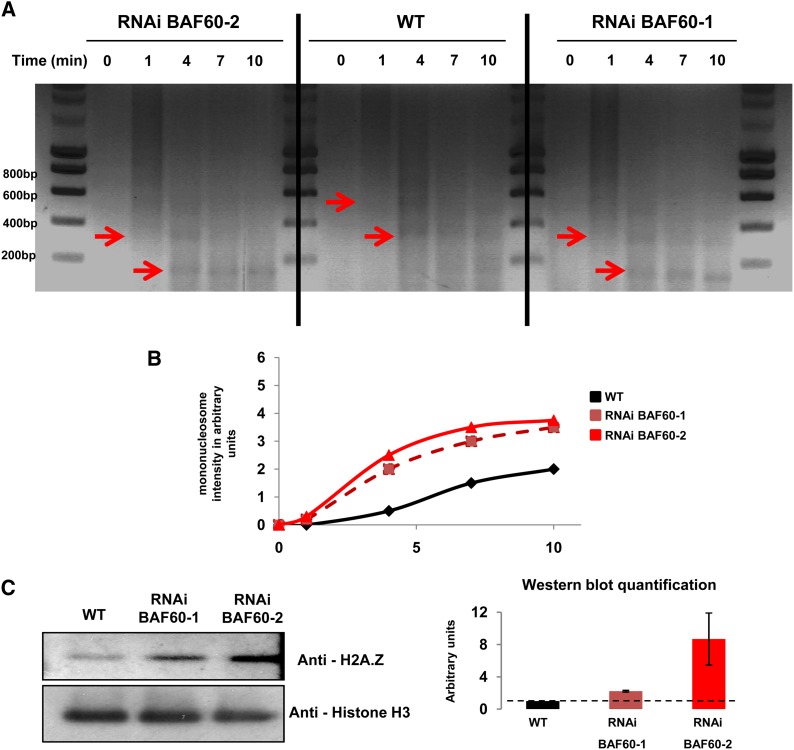
ATP-dependent chromatin-remodeling complexes may act in concert with various histone-modifying enzymes to modulate chromatin structure (Hargreaves and Crabtree, 2011). The global pattern of different histone modifications was thus analyzed by immunoblot assays in BAF60 RNAi lines, but no obvious difference was found. We next asked whether global chromatin condensation could be modified in BAF60 RNAi lines. Micrococcal nuclease (MNase) digestion provides a useful tool for analyzing chromatin structure because it cleaves the linker DNA between adjacent nucleosomes. Depending on the extent of digestion, chromatin fragments with different sizes corresponding to nucleosome number are generated and form a specific DNA ladder reflecting the level of chromatin compaction. To test the consequences of BAF60 down-regulation on chromatin structure, we performed a limited MNase digestion assay. The patterns of permeability to nuclease digestion (Figures 1A and 1B; Supplemental Figure 3) indicated a nuclease hypersensitivity of the BAF60 RNAi line chromatin compared with the wild type.
Figure 1.
BAF60 Is Involved in H2A.Z Deposition and Chromatin Condensation.
(A) Nuclei from wild-type or RNAi BAF60 cells were isolated and analyzed in an MNase digestion assay. The reaction was stopped after 1, 4, 7, and 10 min, and equal amounts of digested DNA were loaded on agarose gels and stained with ethidium bromide. The red arrows indicate nucleosomes.
(B) Curves representing the intensity of the bands corresponding to mononucleosomes as a function of time.
(C) Nuclear protein fractions extracted from wild-type or BAF60 RNAi plants were analyzed by immunoblotting using an anti-H2A.Z antibody. The relative quantifications (right panel) were normalized using an antibody raised against histone H3. Error bars refer to sd of three biological replicates.
H2A.Z histone variants are known to facilitate intramolecular folding of nucleosomal arrays while simultaneously inhibiting the formation of highly condensed structures resulting from intermolecular associations (Fan et al., 2002). Previous studies suggest that BRM, the ATPase subunit hypothesized to function in the same complex as BAF60, might be involved in H2A.Z deposition (Farrona et al., 2011). Therefore, we tested H2A.Z deposition in the BAF60 knockdown lines. Immunoblot analysis revealed the higher accumulation of H2A.Z in both RNAi lines (Figure 1C). The specificity of the H2A.Z antibody used was demonstrated using the double mutant hta9 hta11, in which two of the three genes encoding H2A.Z variants are inactivated. In our conditions, H2A.Z accumulation was undetectable in this mutant (Supplemental Figure 4). To confirm these results, we performed an immunostaining experiment on the wild type and both RNAi lines. Quantification of the H2A.Z fluorescent signal revealed a higher accumulation of H2A.Z in both RNAi lines, confirming our protein gel blot analysis (Supplemental Figure 5). In order to determine whether this global increase was due to an enhanced expression of H2A.Z variants, quantitative analysis of the expression of genes encoding H2A.Z was performed. The Arabidopsis genome encodes 13 H2A histones, 3 of which are H2A.Z variants (Deal et al., 2007). Quantitative PCR showed that in Arabidopsis, H2A.Z mRNA was mainly transcribed from the HTA9 gene (Supplemental Figure 6) and that no increase in the expression of this gene was detected in BAF60 RNAi lines, suggesting that H2A.Z accumulation is regulated at the protein level. As non-chromatin-bound histones are rapidly degraded by phosphorylation and ubiquitylation-dependent proteolysis (Singh et al., 2009), our results support the hypothesis that BAF60 is involved in negatively regulating H2A.Z deposition into chromatin, which leads to H2A.Z stabilization when BAF60 is downregulated.
BAF60 Influences Flowering Time and CO, FT, SOC1, and FLC Expression in Long and Short Days
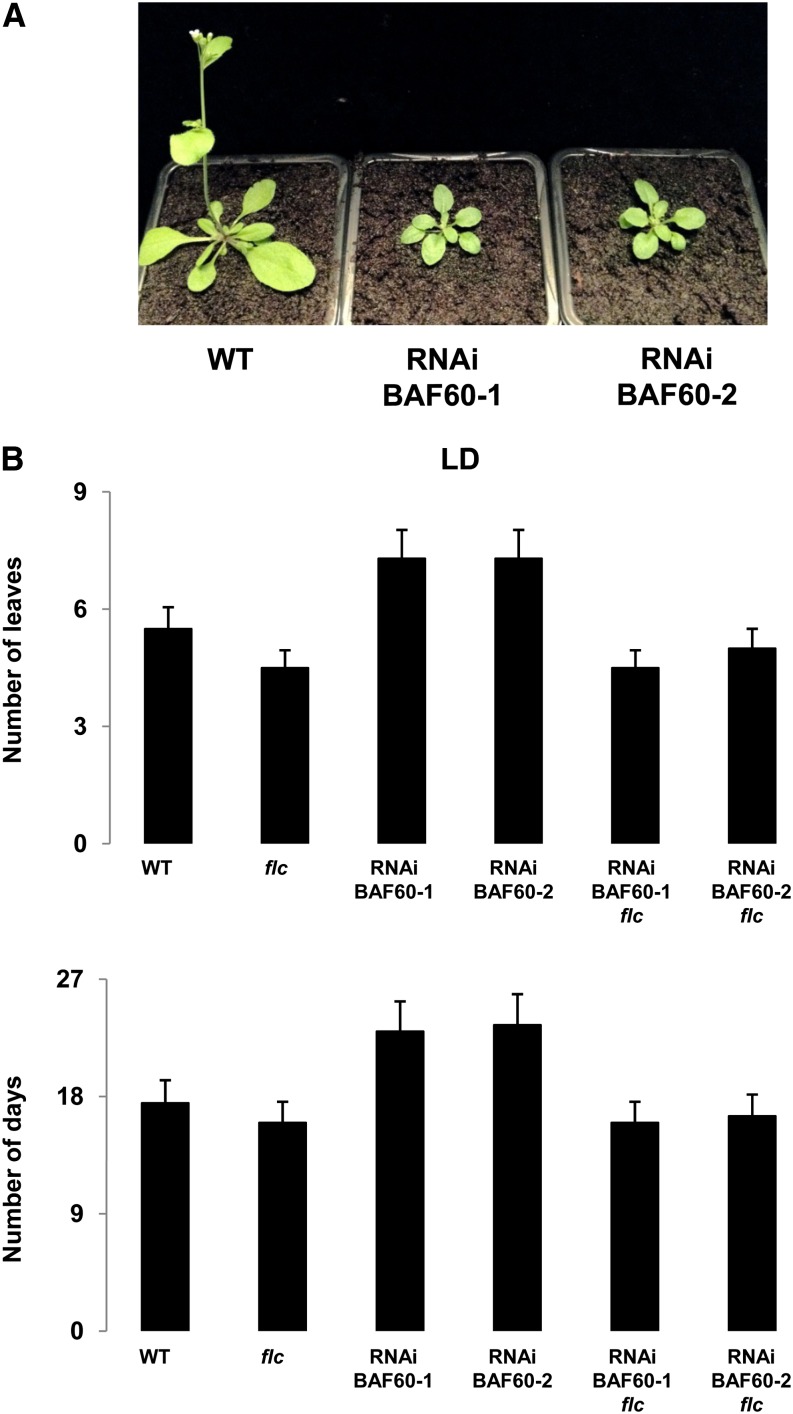
Among genes known to be regulated via H2A.Z incorporation into chromatin, several are involved in the control of flowering time (Deal et al., 2007; Farrona et al., 2011). Hence, we compared the flowering time of BAF60 RNAi lines and Wassilewskija control plants in two independent experiments. Both RNAi lines, grown under normal growth conditions (LD photoperiod), displayed a late-flowering phenotype measured by the number of days to bolting and the rosette leaf number at flowering (Figures 2A and 2B). Wild-type plants bolted nearly 17 d after sowing, whereas 23 d were required for the RNAi lines, which also had increased number of rosette leaves in comparison with the wild type (Figure 2B). This phenotype was associated with a decrease in leaf size, as shown in Figure 2A.
Figure 2.
BAF60 Regulates Flowering Time.
(A) Twenty-day-old wild-type plant and two independent BAF60 RNAi lines grown under LD conditions.
(B) Flowering time of plants grown under LD photoperiod. Values shown are averages ± sd of days (top) or number of rosette leaves (bottom) at flowering of at least 30 plants.
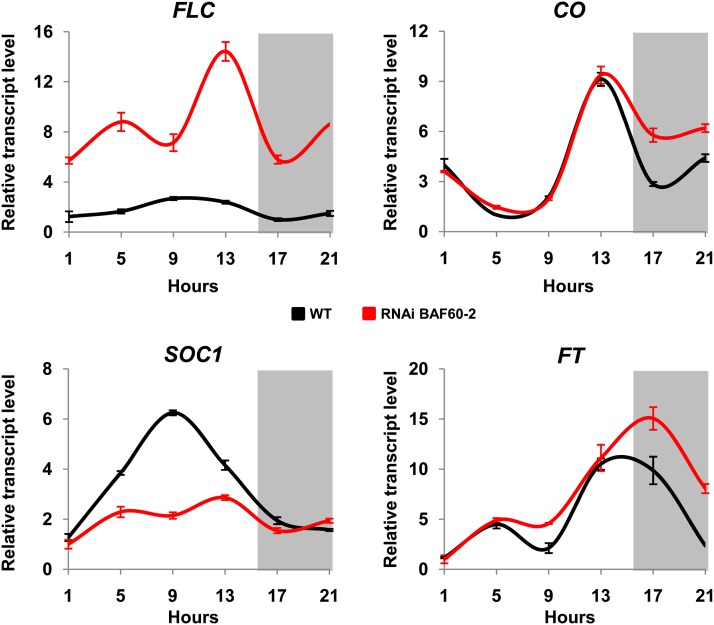
This late-flowering phenotype observed in LD conditions suggests that BAF60 could be involved in the photoperiod pathway of flowering. This pathway is mediated by CO, a transcription factor required to activate the expression of FT under LD conditions. FT moves from leaves to the apical meristem to trigger a cascade of events that lead to plant flowering, including the activation of SOC1 expression (Fornara et al., 2010). To address the molecular mechanisms underlying the late-flowering phenotype of BAF60 RNAi lines, we quantified the expression of these different regulators in these plants. Because CO, FT, and SOC1 mRNA levels show a circadian rhythm, their expression was examined during a 24-h period. In both RNAi lines, the accumulation of CO mRNA was only slightly upregulated compared with wild-type plants (Figure 3; Supplemental Figure 7). Consistently, a slight overaccumulation of FT mRNA, in contrast to a 3-fold reduction of SOC1 mRNA, was detected in BAF60 RNAi lines compared with the control (Figure 3). To correlate the late-flowering phenotype observed in the RNAi lines and SOC1 downregulation, we performed a time-course analysis of SOC1 expression in wild-type and RNAi plants growing under LD conditions. In the wild type, we observed that SOC1 expression reached a peak 19 d after sowing, whereas this peak was observed more than 23 d after sowing in the RNAi lines (Supplemental Figure 8). Hence, SOC1 misregulation was consistent with the late-flowering phenotype.
Figure 3.
BAF60 Controls the Expression of Flowering Genes.
qRT-PCR data show the relative expression of the indicated genes in wild-type and BAF60 RNAi plants in LD conditions. Total RNA samples were collected every 4 h from 14-d-old plants during a 24-h period. mRNA abundance was quantified by qRT-PCR and expressed relative to the abundance of UBQ10 transcripts. Gray areas behind the traces represent night periods. Average relative quantity ± sd is shown for each sample.
Knowing that FT and SOC1 are targets of FLC, we then assayed the expression of FLC, which was 7-fold upregulated in the RNAi lines. Taken together, these results provide evidence that genes involved in the control of flowering time are deregulated in BAF60 RNAi lines. The resulting balance between the action of activators and repressors of flowering favors the inhibition of flowering via upregulation of FLC and downregulation of SOC1.
To further explore this hypothesis, the flowering time of the BAF60 RNAi lines introgressed into the flc background was compared with that of the flc mutant and the wild-type control plants. In two independent experiments, the flc BAF60 RNAi lines, grown under normal growth conditions (LD photoperiod), displayed an early-flowering phenotype, measured by the days to bolting and the rosette leaf number at flowering (Figure 2B). These results demonstrate that the late-flowering phenotype observed in the RNAi lines was due to the overexpression of FLC. Since BAF60 affects FLC expression, we also asked whether BAF60 could be involved in the autonomous pathway regulating flowering time. Under SD conditions, considering the number of days to bolting, the flowering time was the same for all genotypes. However, the number of leaves at bolting was lower in both BAF60 RNAi lines compared with the control (Supplemental Figure 9). CO and FT were initially identified as mutants that flower late in LD conditions but not in SD conditions (Koornneef et al., 1991). However, ectopic expression of CO leads to early flowering, irrespective of daylength (Onouchi et al., 2000), showing that transcriptional regulation of CO expression is an important determinant in the control of flowering. Therefore, we examined the expression of CO, FT, and SOC1 by qRT-PCR in SD conditions. The CO mRNA level was 4-fold upregulated in correlation with a 5-fold increase in accumulation of FT mRNA and increased SOC1 mRNA levels in both RNAi lines compared with the wild type (Supplemental Figure 10). Although we also observed an increase in the FLC mRNA level, this seemed insufficient to repress FT and SOC1 (Supplemental Figure 10). Taken together, these results suggest that the upregulation of CO could induce the early flowering observed in SD conditions (Supplemental Figure 11).
BAF60 Influences the Expression of FLC by Regulating Histone Modifications and H2A.Z Deposition
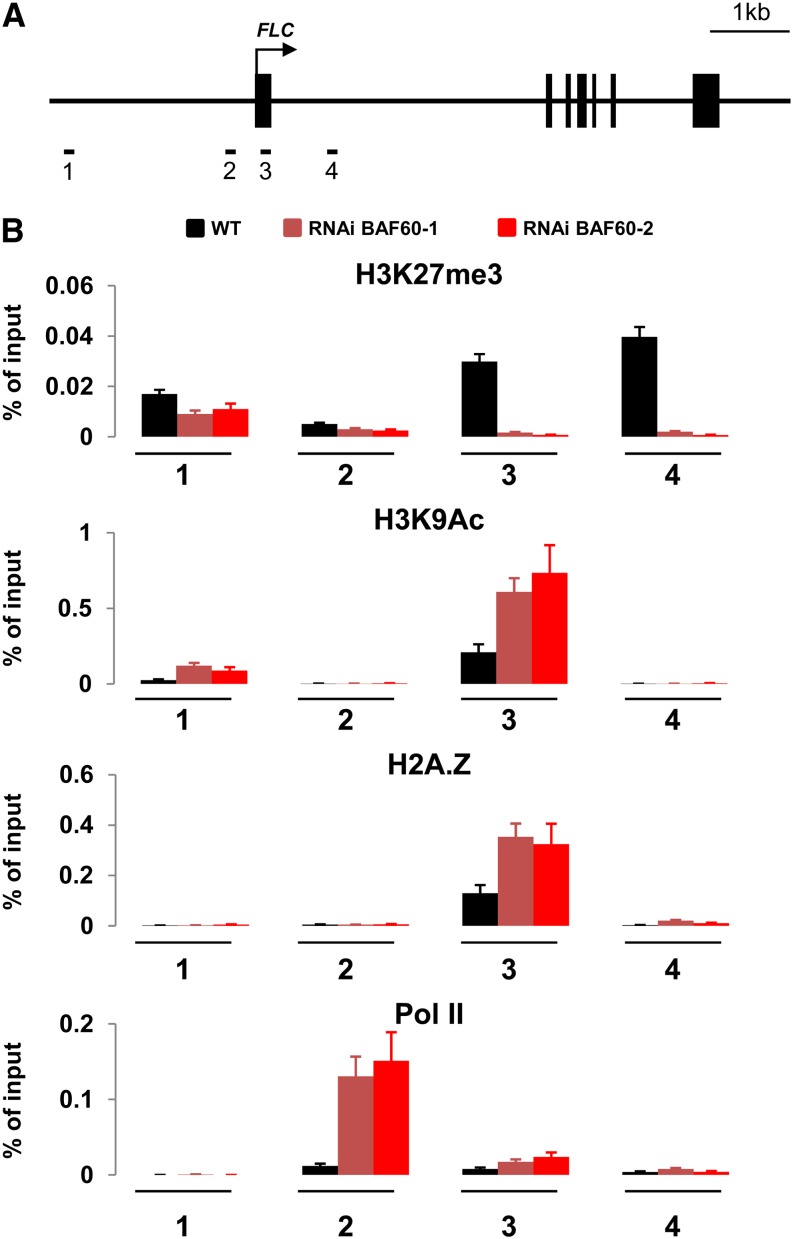
FLC repressors and activators may act on chromatin structure at the FLC locus either via modification of specific histones or via H2A.Z deposition (Crevillén and Dean, 2011). Since BAF60 may alter the chromatin configuration of its target genes, we analyzed how the absence of BAF60 affects histone modifications at the FLC locus. Both a transcription-repressing mark (H3K27me3) and a transcription-activating mark (H3K9Ac) were assessed by chromatin immunoprecipitation (ChIP) assays using primers specific for three regions of the FLC locus, covering the FLC promoter (distal and proximal regions), the first exon, and the first intron of the gene (Figure 4A).
Figure 4.
BAF60 Controls Histone Modification State and RNA Polymerase II Occupancy at the FLC Locus.
(A) Schematic representation of the regions of the FLC locus analyzed. Black boxes correspond to exons, the arrow indicates the site of translation initiation, and numbers indicate the positions of primer pairs used.
(B) Quantification data of the ChIP results. Nuclei were extracted from 14-d-old seedlings grown under LD conditions, and immunoprecipitation was performed with antibodies specific for H3K27me3, H3K9Ac, H2A.Z, and RNA polymerase II. Average relative quantity ± sd is shown for each sample (two biological replicates).
In the BAF60 RNAi line, H3K27me3 was decreased while H3K9Ac was increased in the 5′ part of the FLC locus (Figure 4B). These results are consistent with the observed FLC upregulation caused by the downregulation of BAF60. We next examined the possibility that BAF60 could be involved in H2A.Z deposition at the FLC locus. ChIP assays were performed on wild-type and BAF60 RNAi lines using an H2A.Z antibody. In both genetic backgrounds, chromatin H2A.Z incorporation was restricted to the FLC exons 1, 6, and 7, and this incorporation was higher in the RNAi lines (Figure 4B; Supplemental Figure 12).
H2A.Z-containing nucleosomes facilitate RNA polymerase II passage by affecting Pol II elongation complexes and favoring efficient nucleosome remodeling over the gene (Santisteban et al., 2011). To assess the effect of H2A.Z enrichment on RNA polymerase II occupancy, ChIP assays were performed using an antibody raised against RNA polymerase II. We observed that RNA polymerase II was present on the proximal promoter of FLC in both genotypes, with a 12-fold increase in RNA polymerase II occupancy in the BAF60 RNAi line (Figure 4B). Altogether, our results suggest that BAF60 inhibits H3K9Ac, H2A.Z, and RNA polymerase II deposition at the FLC locus to promote its repression.
BAF60 Regulates the FLC Loop
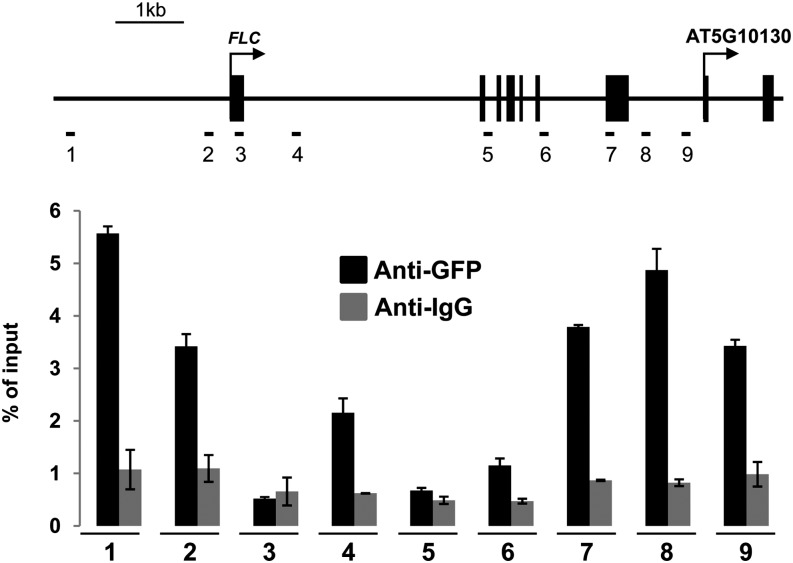
Taken together, our results show that BAF60 downregulation results in a general increase in H2A.Z incorporation into chromatin, notably at the FLC locus. To determine whether FLC is a direct target of BAF60, we first generated plants overexpressing a cyan fluorescent protein (CFP)–tagged version of BAF60. This allowed us to check that the protein accumulates in the nucleus (Supplemental Figure 13). We then examined the ability of BAF60 to bind to the FLC locus by ChIP using an anti-GFP antibody. The same assay using antibodies against IgG was used as a negative control. As shown in Figure 5, we observed a specific association of BAF60 with the promoter region of the FLC gene, the first intron, and the 3′ part of the locus.
Figure 5.
BAF60 Binds to the FLC Locus.
Top, schematic representation of the FLC locus. The position of each primer pair used for ChIP-quantitative PCR is indicated. Arrows indicate the positions of the transcription start sites. Exons are represented as black boxes and introns as black lines. Bottom, quantification data of ChIP results. Nuclei were extracted after cross-linking from 14-d-old seedlings expressing the BAF60-CFP transgene and sonicated, and chromatin/protein complexes were immunoprecipitated with antibodies specific for GFP or IgG. Error bars refer to sd of three biological replicates.
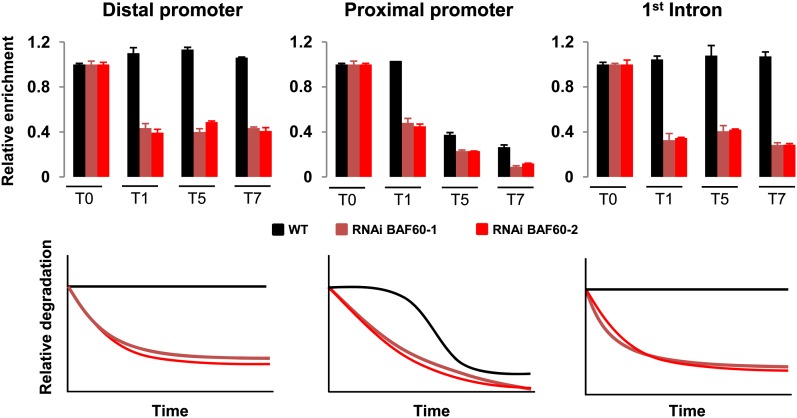
Considering the binding of BAF60 on FLC and the higher incorporation of the H2A.Z variant at this locus in the BAF60 RNAi lines, we wondered whether BAF60 could influence chromatin condensation in this particular genomic region. In order to measure DNA accessibility and to obtain a quantitative assessment of chromatin condensation at the FLC locus, we performed a limited MNase digestion followed by real-time PCR. As shown in Figure 6, in the wild type, we observed a nuclease-hypersensitive site in the proximal part of the FLC promoter, whereas no obvious difference was noticed in the two other parts of the genomic region tested. This result is in accordance with a previous analysis showing an apparent nucleosome-free region immediately upstream of the transcription start site (Shivaswamy et al., 2008). However, in BAF60 RNAi plants, we observed nuclease hypersensitivity for the three genomic regions tested, reflecting a reduction in chromatin condensation and thus probably in nucleosome occupancy at the FLC locus in BAF60 knockdown lines.
Figure 6.
BAF60 Is Required for Chromatin Condensation at the FLC Locus.
Quantification of MNase sensitivity was measured using qRT-PCR at the FLC locus after a limited MNase digestion with four time point kinetics. Data were normalized to the control without MNase digestion, and graphical representation of the data is presented below.
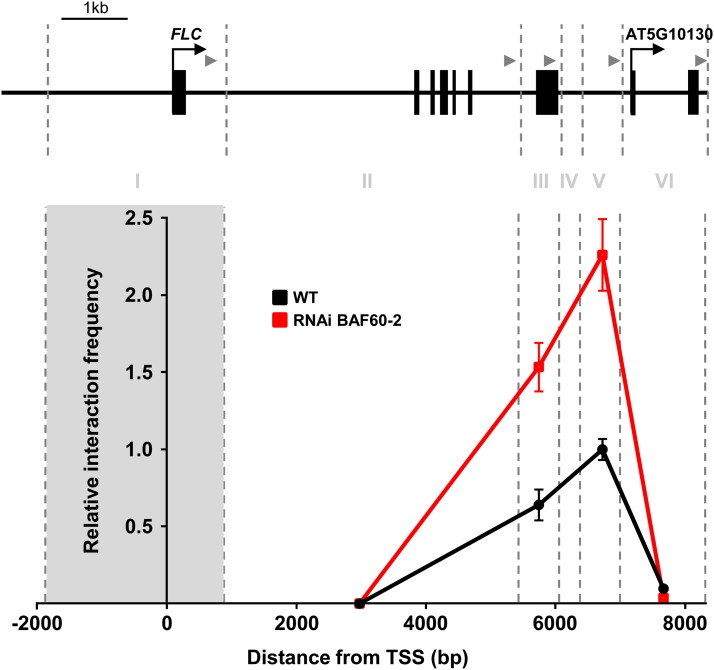
Recently, Crevillén et al. (2013) demonstrated that the FLC locus could make a loop that plays a role in its transcriptional regulation. Since histone depletion facilitates chromatin loops (Diesinger et al., 2010), we performed 3C experiments to explore whether BAF60 could regulate the maintenance of this high-order chromatin structure. First, we confirmed the results showing a chromatin loop at the FLC locus in the wild type and compared it with BAF60 RNAi lines (Figure 7; Supplemental Figure 14). As a control, we verified that no PCR product was observed when our plant material was not cross-linked or ligase was not added in the 3C reaction. Interestingly, we observed that twice as many FLC loops were present and that this structure was more stable in the BAF60 RNAi context. These results suggest that BAF60 plays a negative role in loop formation and/or stabilization, at least at the FLC locus, and allowed us to elucidate the molecular mechanisms governing chromatin conformation involved in the control of flowering.
Figure 7.
BAF60 Regulates Gene Loop Formation.
Quantitative 3C of the FLC locus is shown using FI as the anchor region in 14-d-old wild-type and RNAi BAF60-2 seedlings. Relative interaction frequencies were calculated as described in Methods. Data are averages of three biological replicates each with three technical replicates. In the graph, BamHI and BglII restriction sites are indicated with vertical dotted lines, the analyzed FLC regions are numbered with Roman numerals, and the anchor region is highlighted with gray shading. A schematic representation of the FLC locus is shown above, with the positions of primers used for the 3C analysis represented by gray arrowheads. TSS, transcription start site.
BAF60 Is Necessary for Efficient Disruption of the FLC Loop after Vernalization
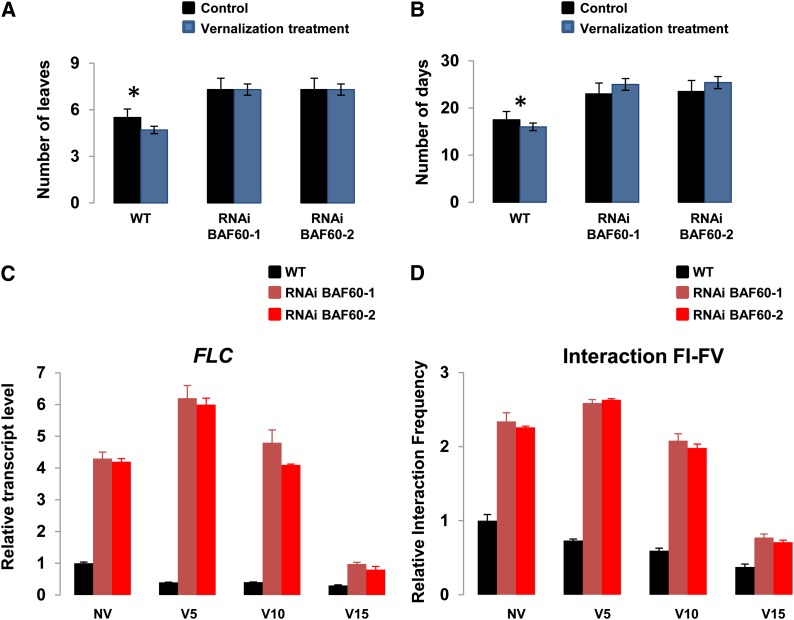
Since a major conclusion from Crevillén et al. (2013) was that the FLC gene loop could be disrupted by vernalization, we analyzed the flowering phenotype of both RNAi lines after vernalization. The vernalization treatment efficiently reduced flowering time in the wild type: as expected, plants displayed a significant reduction of the rosette leaf number at flowering (Student’s t test, P = 0.05) and of the number of days to bolting (Student’s t test, P = 0.05; Figures 8A and 8B). However, we observed that the vernalization did not have any effect on the two RNAi lines regarding the number of days to bolting and the rosette leaf number at flowering (Figures 8A and 8B).
Figure 8.
BAF60 Regulates Gene Loop Dynamics after Vernalization Treatment.
(A) Flowering time measured as days to bolting after vernalization of wild-type plants and BAF60 RNAi lines. Vernalization significantly reduced flowering time in the wild type but not in RNAi lines.
(B) Flowering time measured as number of leaves at the time of bolting after vernalization of wild-type plants and BAF60 RNAi lines. The obtained results were the same as in (A).
(C) qRT-PCR analysis of FLC expression in the wild type (black bars) and BAF60 RNAi lines (red bars). FLC expression was always higher in BAF60 RNAi lines than in the wild type, and it only started to decrease 10 d after vernalization instead of 5 d after vernalization in the wild type.
(D) 3C assay of loop formation on the FLC locus after vernalization. Interaction between regions I and V decreased in the wild type after vernalization (black bars). By contrast, this association was more stable in the two RNAi lines (red bars) and started to decrease only 10 d after vernalization.
Having established that BAF60 was necessary for the vernalization response, we investigated the relationship between the phenotype observed, the expression of FLC, and the loop frequency. qRT-PCR analysis revealed that in the wild type, the decrease in FLC expression started 5 d after vernalization treatment, whereas in both RNAi lines, a clear decrease was observed only 15 d after vernalization treatment (Figure 8C). These results correlate with the phenotype displayed by the two RNAi lines and suggested that BAF60 could be a regulator of the vernalization pathway through its action on FLC expression.
Then, knowing that BAF60 was required for the loop destabilization and/or inhibition of its formation in control conditions, we hypothesized that during vernalization, the phenotype observed in both RNAi lines could be a consequence of the presence of a more stable FLC loop. As reported previously, loop formation was reduced by vernalization in the wild type (Crevillén et al., 2013). Interestingly, we observed that the FLC loop was more stable after vernalization in both RNAi lines (Figure 8D), and these observations were in agreement with the expression data and the phenotype observed. Altogether, these results suggested that BAF60 is necessary for disrupting the loop after vernalization treatment to reduce FLC expression, thereby inducing flowering.
DISCUSSION
Despite an intensive study of ATP-dependent chromatin-remodeling complexes in yeast, a single-celled organism, the function of these complexes in developmental processes in higher eukaryotes remains largely unknown. Even though a SWI/SNF complex has not yet been isolated from plant tissues, homologs of most of its components are present in Arabidopsis. In order to gain further insight regarding the role of these complexes, we analyzed the molecular function of one accessory subunit, At BAF60, a homolog of the yeast SWP73 protein. We show that BAF60 is a positive regulator of chromatin condensation, probably by its negative effect on H2A.Z incorporation into chromatin, and, more specifically, that it controls the formation of a chromatin regulatory loop involving a crucial flowering gene, FLC, thereby modulating flowering time and the vernalization response.
We showed that BAF60 RNAi lines displayed a late-flowering phenotype only in LD conditions, together with upregulation of the FLC gene and downregulation of SOC1. Analysis of flc BAF60 RNAi lines confirmed that the late-flowering phenotype was attributable to FLC misregulation. Under SD conditions, the flowering time of the RNAi lines was not significantly different from the control in terms of number of days, although the number of leaves was decreased compared with the wild type. In this case, this phenotype was correlated with an upregulation of CO, FT, and SOC1, which could not be prevented by FLC activation. Hence, loss of BAF60 function has opposing effects on regulators of flowering time, resulting in the simultaneous upregulation of repressors and activators both in LD and SD conditions. The resulting balance between these regulators leads to delayed flowering in LD conditions and early flowering in SD conditions.
Our data indicate that BAF60 is a regulator of the photoperiod via direct targeting of FLC. Therefore, the regulation of FT and SOC1 by BAF60 might take place in at least two different ways: through CO expression and/or independently of CO. The late-flowering phenotype of the knockdown BAF60 RNAi lines in LD conditions can be explained by the higher levels of expression of FLC, which lead to a downregulation of SOC1 (Figure 9A).
Figure 9.
Model for the Role of BAF60 in the Control of Flowering Time.
(A) Genetic model. Under LD conditions and after a vernalization treatment in wild-type plants, FLC is repressed by BAF60 to promote flowering. In BAF60 RNAi lines, a high level of FLC expression induces a repression of SOC1 and leads to a late-flowering phenotype.
(B) Molecular model. Under LD conditions in wild-type plants, an equilibrium exists between the formation and the release of the FLC loop in which BAF60 is involved. This equilibrium could be destabilized by the knockdown of BAF60.
Although in SD conditions the CO pathway does not seem to be involved in the control of flowering (Valverde et al., 2004), overexpression of either CO or FT causes photoperiod-independent early flowering (Kardailsky et al., 1999; Kobayashi et al., 1999; Onouchi et al., 2000). In addition, high levels of FLC do not inhibit the early flowering of CO-overexpressing plants (Hepworth et al., 2002). Hence, the upregulation of CO observed under SD conditions in BAF60 RNAi plants is likely to account for the observed early-flowering phenotype and probably cannot be overcome by the simultaneous upregulation of FLC. Altogether, our results suggest that BAF60 is a repressor of CO mainly in SD conditions (Supplemental Figure 11). How this photoperiod specificity occurs needs to be determined in the future.
Although BRM, like BAF60, is a repressor of FLC and CO, the SWI/SNF ATPase mutants brm and syd display precocious activation of FT both in LD and SD conditions (Wagner and Meyerowitz, 2002; Farrona et al., 2004, 2011; Hurtado et al., 2006). SYD and BRM may have common targets or participate in common processes (Bezhani et al., 2007; Wu, 2012). However, their interaction with BAF60 has not been reported yet, and other partners to which BAF60 could be associated remain unknown. The observation that both BRM and BAF60 repress the expression of FLC and CO strongly suggests that these two proteins function in the same complex. The opposing outcomes of the brm mutation and BAF60 downregulation with respect to flowering time in LD conditions is puzzling. These contrasting effects of mutations predicted to affect the same complex suggest distinctive activities of the various subunits even though they regulate the same general process: chromatin remodeling. Indeed, downregulation of BAF60 and BRM or SYD results in a different balance between activators and repressors of the floral transition, leading to opposite flowering phenotypes. This observation highlights the complexity of fine-tuning mechanisms that govern the floral transition and suggests that several subcomplexes differing by their composition and target genes may be at work to modulate the balance of flowering regulators (Srikanth and Schmid, 2011).
From a molecular point of view, we showed that BAF60 could directly repress FLC transcription by modulation of its chromatin structure. Genome topology has emerged as a key player in genome functions (Cavalli and Misteli, 2013). A contribution of local genome chromatin loops in transcription has long been appreciated (reviewed in Hou and Corces, 2012). Chromatin loops can regulate transcription via a variety of mechanisms, for example, by bringing a distant enhancer into contact with a promoter (Tolhuis et al., 2002). On the contrary, they can associate several polycomb response elements to target sites with the polycomb machinery and repress gene expression (Tiwari et al., 2008). Other types of chromatin loops involve insulator binding proteins (Yang and Corces, 2012). In the case of the FLC locus, the association between the 5′ and 3′ ends of the gene could allow efficient recycling of RNA polymerase II and other general transcription factors inside the loop, which are released after the termination of transcription (Ansari and Hampsey, 2005). Surprisingly, the FLC loop was not disturbed by mutations that affect several chromatin regulatory pathways and lead to different expression levels (Crevillén et al., 2013), indicating that expression of FLC can be modulated independently of the spatial organization at this locus. However, the formation of this loop may mediate the vernalization pathway for FLC regulation, since its formation was disrupted during vernalization, concomitant with the downregulation of FLC. We showed here that BAF60 interacts with the 5′ and 3′ parts of the FLC locus, and its downregulation induces the formation and/or stabilization of the loop structure, expression of FLC, and deposition of active epigenetic marks in this locus. Altogether, our results suggest that BAF60 could regulate FLC by negatively affecting the loop dynamics through the modulation of histone density, composition, and posttranslational modification (Figure 6B). Consistently, BAF60 RNAi lines are insensitive to vernalization, providing evidence for the physiological relevance of the involvement of BAF60 in the regulation of FLC loop stability or formation.
Another mechanism accounting for the role of BAF60 as a negative regulator of FLC expression is the incorporation of H2A.Z into chromatin. Indeed, we showed that BAF60 is involved in chromatin condensation at the FLC locus in part via the modulation of H2A.Z incorporation into chromatin. This may be consistent with the ability of S. cerevisiae SWI/SNF RSC to remodel nucleosomes, thereby creating “inactive” nucleosomal states at the locus. Among the players involved in the control of the nucleosomal state, the H2A.Z variant plays important roles both in activating transcription and antagonizing the spread of heterochromatin into euchromatic regions (Marques et al., 2010). Previously, data obtained from Drosophila and humans showed that H2A.Z levels positively correlate with gene expression (Barski et al., 2007; Mavrich et al., 2008). However, H2A.Z variants may not participate directly in the activation of transcription but rather commit to a chromatin state competent for activation by other factors or prevent its silencing by DNA methylation (Zilberman et al., 2008). In Arabidopsis, recent results have shown that H2A.Z is required for gene activation in response to warmer temperature (Kumar and Wigge, 2010). Our data show that deposition of the histone variant H2A.Z over the FLC locus is correlated with an increase in its transcription, as evidenced by increased mRNA accumulation and higher polymerase II binding on its proximal promoter and the stabilization/formation of a loop. This is in agreement with recent results showing that the loss of H2A.Z from chromatin in arp6 and pie1 mutants results in reduced FLC expression and premature flowering, indicating that this histone variant is correlated with high expression levels of FLC. BRM represses FLC expression, pointing to an antagonistic role of the SWI/SNF and SWR1/SRCAP complexes, on H2A.Z deposition at the FLC locus. Under such a hypothesis, FLC expression levels would result from the balance between these opposing activities on its chromatin. In agreement with this model, analysis of genetic interactions between BRM and the histone H2A.Z deposition machinery demonstrates that brm mutations overcome the requirement of H2A.Z for FLC activation. In the absence of BRM, a constitutively open chromatin conformation may render H2A.Z dispensable (Farrona et al. 2011). Regarding specifically the loop structure, Crevillén et al. (2013) demonstrated that the loop was present in the arp6 mutant, suggesting that incorporation of H2A.Z alone could not explain its formation. MNase sensitivity assays revealed that the FLC locus was more accessible to digestion in BAF60 RNAi lines, suggesting that nucleosomes are more distant in the absence of BAF60. Whether this role of BAF60 is mediated by H2A.Z incorporation or also results from a direct effect on nucleosome positioning remains to be established. More generally, our results show a role for BAF60 in the modulation of H2A.Z incorporation into chromatin, which is not confined to the FLC locus but observed at the whole-genome scale, as evidenced by the reduced condensation of the genome in BAF60 RNAi lines.
The discovery of SWI/SNF-related complexes possessing histone deacetylase activity, a function shown to repress transcription, suggests that nucleosome-remodeling activities can be coupled with other chromatin-modifying activities to induce transcriptional repression (Knoepfler and Eisenman, 1999; Lai and Wade, 2011). Our results show that depletion of BAF60 also modifies the deposition of two important histone marks, H3K27me3 and H3K9Ac, involved in the repression and activation of the transcriptional state, respectively. These results support a model whereby a combination of specific histone modifications associated with particular histone variants could modulate the dynamics of gene loop formation, although we cannot rule out the possibility that some of the changes in histone modifications we observed could be a consequence of altered gene expression rather than its cause. In conclusion, BAF60 functions in the control of flowering time by regulating the expression of genes through the incorporation of H2A.Z into chromatin, induction of histone modification, and control of gene loop formation. Our work reveals an important role for BAF60 in gene loop formation in plant chromatin control and raises the possibility that SWI/SNF complexes, notably those containing BAF60, are key factors governing the equilibrium between the formation and dissociation of the gene loop in a developmental context such as the control of flowering.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana seeds of both RNAi lines, BAF60-1 (CS30982) and BAF60-2 (CS23961), in the Wassilewskija (wild-type) background, were obtained from the Nottingham Arabidopsis Stock Centre. The flc-3 mutant line has been described previously (Guyomarc’h et al. 2006). Plants were grown in chambers at 20°C on soil or on sterile half-strength Murashige and Skoog medium and 0.8% agar in SD (8 h of light at 20°C, 16 h of darkness at 18°C) or LD (16 h of light at 20°C, 8 h of darkness at 18°C) conditions. Seeds were surface-sterilized by treatment with bayrochlore for 20 min, washed, and imbibed in sterile water for 2 to 4 d at 4°C to obtain homogeneous germination. To vernalize, seeds were grown for 7 d at standard warm-growing conditions (16 h of light at 20°C, 8 h of darkness at 18°C) before being transferred to cold (16 h of light and 8 h of darkness at 5°C) conditions for 2 weeks and then returned to standard warm-growing conditions.
Production of Plants Overexpressing a CFP-Tagged Version of BAF60
Total RNA was extracted from seedlings or leaves of Arabidopsis ecotype Columbia with the Nucleospin RNA II kit (Macherey-Nagel) according to the manufacturer’s instructions. First-strand cDNA was synthesized from 2 µg of total RNA using Improm-II reverse transcriptase (A3802; Promega) according to the manufacturer’s instructions. A 1604-bp BAF60 cDNA from the start codon, deleted of the last base of the stop codon, was amplified by PCR from the cDNA pool using the following primers: BAF60-f (5′-GGGGTACCATGTCTGGTAACAACAACAATC-3′) and BAF60r (5′-CCGCTCGAGCACCAACTCCCTGGCCCATCGTTC-3′). The PCR product was cloned into the pGEM-T vector and sequenced. The cDNA was then excised via a KpnI/XhoI restriction and subcloned into the pEntr1C vector. Then, to obtain CFP translational fusions (35S:BAF60-CFP), the BAF60 cDNA in pEntr1C vector was recombined according to the Gateway procedure (Invitrogen) downstream of the CaMV35S promoter in the binary vector pB7CWG2 (Plant Systems Biology, Vlaams Interuniversitair Instituut voor Biotechnologie, Ghent University). The expression vectors were introduced into Agrobacterium tumefaciens strain GV3101 by electroporation, and transgenic lines were generated by the floral dip method. Ten independent lines from the T1 to T6 generations were successively selected on sand supplemented with Basta (10 mg/L phosphinothricin).
RNA Extraction and qRT-PCR Analysis
Total RNA was extracted from seedlings with the RNeasy MiniPrep kit (Qiagen) according to the manufacturer’s instructions. First-strand cDNA was synthesized from 2 μg of total RNA using Improm-II reverse transcriptase (A3802; Promega) according to the manufacturer’s instructions. Four percent of the synthesized cDNA was mixed with 100 nM of each primer and LightCycler 480 Sybr Green I master mix (Roche Applied Science) for quantitative PCR analysis. Products were amplified and fluorescent signals acquired with the LightCycler 480 detection system. The specificity of amplification products was determined by melting curves. UBQ10 was used as an internal control for signal normalization. Exor4 relative quantification software (Roche Applied Science) automatically calculates relative expression levels of the selected genes with algorithms based on the ΔΔCt method. Data were from duplicates of at least three biological replicates. The sequences of all primers used are given in Supplemental Table 1.
MNase Sensitivity Assay
Five grams of 14-d-old seedlings was ground, and nuclei were isolated with 1% Triton X-100, filtered through a 63-µm filter, and incubated in MN buffer (20 mM Tris-HCl, pH 7.5, 70 mM NaCl, 20 mM KCl, 5 mM MgCl2, and 3 mM CaCl2). One hundred units of MNase (Invitrogen) was added to initiate the kinetics. Adding EGTA and EDTA to a final concentration of 2 mM stopped the reaction. DNA was extracted with chloroform, precipitated with isopropanol, resuspended in ultrapure water, and quantified with Qubit. Sensitivity to MNase was then analyzed by visualizing the DNA digestion pattern by agarose gel electrophoresis using 1 µg of DNA or by qRT-PCR using 6 ng of digested DNA.
Immunoblot and ChIP Analyses
Nuclear proteins were extracted from 14-d-old in vitro–grown seedlings. After quantification with the Bradford method, identical amounts of proteins were resolved by SDS-PAGE and then transferred to a polyvinylidene difluoride membrane (Bio-Rad) using a Mini-Protean 3 Cell (Bio-Rad). Immunoblot analysis was performed using 1 µg/mL primary polyclonal antibodies raised against histone H2A.Z (Abcam) and histone H3 (Abcam) and then with secondary antibodies conjugated to alkaline phosphatase. Antibody complexes were detected by chemiluminescence using the Immun-Start AP Substrate kit (Bio-Rad).
ChIP assays were performed on 14-d-old in vitro–grown seedlings using anti-GFP (Clontech), anti-IgG (Millipore), anti-H3K27me3 (Millipore), anti-H3K9Ac (Millipore), H2A.Z (Abcam), and RNA polymerase II (Santa Cruz) antibodies, using a method modified from Gendrel et al. (2005). Briefly, after plant material fixation in 1% (v/v) formaldehyde, the tissues were homogenized and the nuclei were isolated and lysed. Cross-linked chromatin was sonicated using a water bath Bioruptor UCD-200 (Diagenode; 15-s on/15-s off pulses, 15 times). The complexes were immunoprecipitated with antibodies overnight at 4°C with gentle shaking and incubated for 1 h at 4°C with 50 μL of Dynabeads Protein A (Invitrogen). Immunoprecipitated DNA was then recovered using the IPure kit (Diagenode) and analyzed by qRT-PCR. An aliquot of untreated sonicated chromatin was processed in parallel and used as the total input DNA control.
3C
Two grams of 14-d-old seedlings was cross-linked in 1% (v/v) formaldehyde at room temperature for 20 min. The cross-linked plantlets were ground, and nuclei were isolated and treated with 0.3% SDS at 65°C for 40 min. SDS was sequestered with 2% Triton X-100. Digestions were performed overnight at 37°C with 400 units of BamHI (Fermentas) and 400 units of BglII (Fermentas). The restriction enzymes were inactivated by the addition of 1.6% SDS and incubation at 65°C for 20 min. SDS was sequestered with 1% Triton X-100. DNA was ligated by incubation at 22°C for 5 h in a 5-mL volume using 100 units of T4 DNA ligase (Fermentas). Reverse cross-linking was performed by overnight treatment at 65°C. DNA was recovered after proteinase K treatment by phenol/chloroform extraction and ethanol precipitation. Relative interaction frequencies were calculated by qRT-PCR using 15 ng of DNA. A region uncut by BamHI and BglII in the proximal promoter was used to normalize the amount of DNA.
Confocal Imaging
Plantlets were mounted in 5% glycerol and directly imaged on a TCS-SP2 upright microscope (Leica Microsystems) with 488/543-nm excitation, 488/543-nm beamsplitter filter, and 515/615-nm (green channel) and 610/625-nm (red channel) detection windows. Transmitted light was also collected.
Immunofluorescence
Seedlings were fixed in 4% paraformaldehyde in PHEM (60 mM PIPES, 25 mM HEPES, 10 mM EGTA, and 2 mM MgCl2, pH 6.9) for 1 h at room temperature (vacuum was applied for 20 min to facilitate uptake of the fixation solution). Seedlings were washed 5 min in PHEM and 5 min in PBS, pH 6.9, and chopped in a Petri dish in PBS supplemented with 0.1% (w/v) Triton X-100. The mixture was filtered (50 µm) and centrifuged for 10 min at 2000g. The supernatant was carefully removed, and the pellet was washed once with PBS and gently resuspended in 20 μL of PBS, and a drop was placed on a poly-Lys slide and air-dried. The slides were rehydrated with PBS and permeabilized two times by 10 min of incubation in PBST (PBS plus 0.1% [v/v] Tween 20). The slides were placed in a moist chamber and incubated overnight at 4°C with primary anti-histone H2A.Z antibody (Abcam) in PBST supplemented with 3% (w/v) BSA. The slides were washed 5 × 10 min in PBST (at room temperature) and incubated 1 h at room temperature in the dark with the secondary antibody (A11037; Invitrogen; Alexa Fluor goat anti-rabbit) diluted 1:400 (v/v) in PBST and 3% BSA. The slides were washed 5 × 10 min in PBST. The slides were then mounted with a drop of Vectashield with 4′,6-diamidino-2-phenylindole and observed with the suitable cube fluorescence filters (BP340-380, DS400, and BP450-490 for 4′,6-diamidino-2-phenylindole, BP570-590, DS595, and BP605-655 for A594).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: BAF60 (AT5G14170), FLC (AT5G10140), SOC1 (AT2G45660), FT (AT1G65480), and CO (AT5G15840).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. qRT-PCR Data Showing Relative Expression of BAF60 (AT5G14170) in Two Available T-DNA Insertion Lines.
Supplemental Figure 2. qRT-PCR Data Showing Relative Expression of BAF60 in Wild-Type and BAF60 RNAi Plants Cultivated under LD or SD Conditions.
Supplemental Figure 3. BAF60 Is Involved in Chromatin Condensation.
Supplemental Figure 4. Specificity of H2A.Z Antibody.
Supplemental Figure 5. Immunofluorescence Detection and Quantification of H2A.Z in Isolated Nuclei of the Wild Type, RNAi BAF60-1, and RNAi BAF60-2.
Supplemental Figure 6. qRT-PCR Data Showing Relative Expression of HTA8, HTA9, and HTA11 in Wild-Type and BAF60 RNAi Plants.
Supplemental Figure 7. qRT-PCR Data Showing Relative Expression of the Indicated Genes in Wild-Type and BAF60 RNAi Plants in SD Conditions.
Supplemental Figure 8. qRT-PCR Data Showing Relative Expression of SOC1 in Wild-Type and BAF60 RNAi Plants in LD Conditions.
Supplemental Figure 9. Flowering Time of Plants Grown under an SD Photoperiod.
Supplemental Figure 10. qRT-PCR Data Showing Relative Expression of the Indicated Genes in Wild-Type and BAF60 RNAi Plants in SD Conditions.
Supplemental Figure 11. Genetic Model for the Role of BAF60 in Flowering Time Regulation under SD Conditions.
Supplemental Figure 12. BAF60 Controls Histone H2A.Z Incorporation at the FLC Locus.
Supplemental Figure 13. BAF60 Is Localized in Nuclei.
Supplemental Figure 14. Quantitative 3C of the FLC Locus Using FI as the Anchor Region in 14-d-Old Wild-Type and RNAi BAF60-2 Seedlings.
Supplemental Table 1. Sequences of Primers Used in This Study (5′ to 3′).
Supplementary Material
Acknowledgments
We thank Philip Wigge for generously providing the double mutant hta9-hta11.
AUTHOR CONTRIBUTIONS
T.J. designed and performed research. D.L., M.D., S.D., and F.A. performed research. C.B. generated new genetic tools. C.R., H.H., and M.C. wrote the article. M.B. designed the research and wrote the article.
Glossary
- CRM
chromatin-remodeling machine
- LD
long-day
- SD
short-day
- 3C
chromosome conformation capture
- RNAi
RNA interference
- qRT-PCR
quantitative real-time RT-PCR
- MNase
micrococcal nuclease
- ChIP
chromatin immunoprecipitation
Footnotes
Online version contains Web-only data.
References
- Ansari A., Hampsey M. (2005). A role for the CPF 3′-end processing machinery in RNAP II-dependent gene looping. Genes Dev. 19: 2969–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. (2007). High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837. [DOI] [PubMed] [Google Scholar]
- Bezhani S., Winter C., Hershman S., Wagner J.D., Kennedy J.F., Kwon C.S., Pfluger J., Su Y., Wagner D. (2007). Unique, shared, and redundant roles for the Arabidopsis SWI/SNF chromatin remodeling ATPases BRAHMA and SPLAYED. Plant Cell 19: 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P., Koonin E.V. (1993). An expanding family of helicases within the ‘DEAD/H’ superfamily. Nucleic Acids Res. 21: 751–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzeski J., Podstolski W., Olczak K., Jerzmanowski A. (1999). Identification and analysis of the Arabidopsis thaliana BSH gene, a member of the SNF5 gene family. Nucleic Acids Res. 11: 2393–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns B.R., Levinson R.S., Yamamoto K.R., Kornberg R.D. (1996). Essential role of Swp73p in the function of yeast Swi/Snf complex. Genes Dev. 10: 2131–2144. [DOI] [PubMed] [Google Scholar]
- Campi M., D’Andrea L., Emiliani J., Casati P. (2012). Participation of chromatin-remodeling proteins in the repair of ultraviolet-B-damaged DNA. Plant Physiol. 158: 981–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli G., Misteli T. (2013). Functional implications of genome topology. Nat. Struct. Mol. Biol. 20: 290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K., Kim S., Kim S.Y., Kim M., Hyun Y., Lee H., Choe S., Kim S.G., Michaels S., Lee I. (2005). SUPPRESSOR OF FRIGIDA3 encodes a nuclear ACTIN-RELATED PROTEIN6 required for floral repression in Arabidopsis. Plant Cell 17: 2647–2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K., Park C., Lee J., Oh M., Noh B., Lee I. (2007). Arabidopsis homologs of components of the SWR1 complex regulate flowering and plant development. Development 134: 1931–1941. [DOI] [PubMed] [Google Scholar]
- Clapier C.R., Cairns B.R. (2009). The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78: 273–304. [DOI] [PubMed] [Google Scholar]
- Crane Y.M., Gelvin S.B. (2007). RNAi-mediated gene silencing reveals involvement of Arabidopsis chromatin-related genes in Agrobacterium-mediated root transformation. Proc. Natl. Acad. Sci. USA 104: 15156–15161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crevillén P., Dean C. (2011). Regulation of the floral repressor gene FLC: The complexity of transcription in a chromatin context. Curr. Opin. Plant Biol. 14: 38–44. [DOI] [PubMed] [Google Scholar]
- Crevillén P., Sonmez C., Wu Z., Dean C. (2013). A gene loop containing the floral repressor FLC is disrupted in the early phase of vernalization. EMBO J. 32: 140–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal R.B., Kandasamy M.K., McKinney E.C., Meagher R.B. (2005). The nuclear actin-related protein ARP6 is a pleiotropic developmental regulator required for the maintenance of FLOWERING LOCUS C expression and repression of flowering in Arabidopsis. Plant Cell 17: 2633–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal R.B., Topp C.N., McKinney E.C., Meagher R.B. (2007). Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A.Z. Plant Cell 19: 74–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diesinger P.M., Kunkel S., Langowski J., Heermann D.W. (2010). Histone depletion facilitates chromatin loops on the kilobasepair scale. Biophys. J. 99: 2995–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle M.R., Amasino R.M. (2009). A single amino acid change in the Enhancer of Zeste ortholog CURLY LEAF results in vernalization-independent, rapid flowering in Arabidopsis. Plant Physiol. 151: 1688–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen J.A., Sweder K.S., Hanawalt P.C. (1995). Evolution of the SNF2 family of proteins: Subfamilies with distinct sequences and functions. Nucleic Acids Res. 23: 2715–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J.Y., Gordon F., Luger K., Hansen J.C., Tremethick D.J. (2002). The essential histone variant H2A.Z regulates the equilibrium between different chromatin conformational states. Nat. Struct. Biol. 3: 172–176. [DOI] [PubMed] [Google Scholar]
- Farrona S., Hurtado L., Bowman J.L., Reyes J.C. (2004). The Arabidopsis thaliana SNF2 homolog AtBRM controls shoot development and flowering. Development 131: 4965–4975. [DOI] [PubMed] [Google Scholar]
- Farrona S., Hurtado L., March-Díaz R., Schmitz R.J., Florencio F.J., Turck F., Amasino R.M., Reyes J.C. (2011). Brahma is required for proper expression of the floral repressor FLC in Arabidopsis. PLoS ONE 6: e17997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F., de Montaigu A., Coupland G. (2010). SnapShot: Control of flowering in Arabidopsis. Cell 141: 550. [DOI] [PubMed] [Google Scholar]
- Gendrel A.-V., Lippman Z., Martienssen R., Colot V. (2005). Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods 2: 213–218. [DOI] [PubMed] [Google Scholar]
- Guyomarc’h S., Benhamed M., Lemonnier G., Renou J.P., Zhou D.X., Delarue M. (2006). MGOUN3: Evidence for chromatin-mediated regulation of FLC expression. J. Exp. Bot. 57: 2111–2119. [DOI] [PubMed] [Google Scholar]
- Hargreaves D.C., Crabtree G.R. (2011). ATP-dependent chromatin remodeling: Genetics, genomics and mechanisms. Cell Res. 21: 396–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y. (2009). Control of the transition to flowering by chromatin modifications. Mol. Plant 2: 554–564. [DOI] [PubMed] [Google Scholar]
- Hepworth S.R., Valverde F., Ravenscroft D., Mouradov A., Coupland G. (2002). Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J. 21: 4327–4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou C., Corces V.G. (2012). Throwing transcription for a loop: Expression of the genome in the 3D nucleus. Chromosoma 121: 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado L., Farrona S., Reyes J.C. (2006). The putative SWI/SNF complex subunit BRAHMA activates flower homeotic genes in Arabidopsis thaliana. Plant Mol. Biol. 62: 291–304. [DOI] [PubMed] [Google Scholar]
- Jégu T., et al. (2013). Multiple functions of Kip-related protein5 connect endoreduplication and cell elongation. Plant Physiol. 161: 1694–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerzmanowski A. (2007). SWI/SNF chromatin remodeling and linker histones in plants. Biochim. Biophys. Acta 1769: 330–345. [DOI] [PubMed] [Google Scholar]
- Kandasamy M.K., Deal R.B., McKinney E.C., Meagher R.B. (2005). Silencing the nuclear actin-related protein AtARP4 in Arabidopsis has multiple effects on plant development, including early flowering and delayed floral senescence. Plant J. 41: 845–858. [DOI] [PubMed] [Google Scholar]
- Kardailsky I., Shukla V.K., Ahn J.H., Dagenais N., Christensen S.K., Nguyen J.T., Chory J., Harrison M.J., Weigel D. (1999). Activation tagging of the floral inducer FT. Science 286: 1962–1965. [DOI] [PubMed] [Google Scholar]
- Knizewski L., Ginalski K., Jerzmanowski A. (2008). Snf2 proteins in plants: Gene silencing and beyond. Trends Plant Sci. 10: 557–565. [DOI] [PubMed] [Google Scholar]
- Knoepfler P.S., Eisenman R.N. (1999). Sin meets NuRD and other tails of repression. Cell 99: 447–450. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y., Kaya H., Goto K., Iwabuchi M., Araki T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962. [DOI] [PubMed] [Google Scholar]
- Koornneef M., Hanhart C.J., van der Veen J.H. (1991). A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 229: 57–66. [DOI] [PubMed] [Google Scholar]
- Kumar S.V., Wigge P.A. (2010). H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140: 136–147. [DOI] [PubMed] [Google Scholar]
- Lai A.Y., Wade P.A. (2011). Cancer biology and NuRD: A multifaceted chromatin remodelling complex. Nat. Rev. Cancer 11: 588–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lázaro A., Gómez-Zambrano A., López-González L., Piñeiro M., Jarillo J.A. (2008). Mutations in the Arabidopsis SWC6 gene, encoding a component of the SWR1 chromatin remodelling complex, accelerate flowering time and alter leaf and flower development. J. Exp. Bot. 59: 653–666. [DOI] [PubMed] [Google Scholar]
- March-Díaz R., García-Domínguez M., Florencio F.J., Reyes J.C. (2007). SEF, a new protein required for flowering repression in Arabidopsis, interacts with PIE1 and ARP6. Plant Physiol. 143: 893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- March-Díaz R., Reyes J.C. (2009). The beauty of being a variant: H2A.Z and the SWR1 complex in plants. Mol. Plant 4: 565–677. [DOI] [PubMed] [Google Scholar]
- Marques M., Laflamme L., Gervais A.L., Gaudreau L. (2010). Reconciling the positive and negative roles of histone H2A.Z in gene transcription. Epigenetics 5: 267–272. [DOI] [PubMed] [Google Scholar]
- Martin-Trillo M., Lázaro A., Poethig R.S., Gómez-Mena C., Piñeiro M.A., Martinez-Zapater J.M., Jarillo J.A. (2006). EARLY IN SHORT DAYS 1 (ESD1) encodes ACTIN-RELATED PROTEIN 6 (AtARP6), a putative component of chromatin remodelling complexes that positively regulates FLC accumulation in Arabidopsis. Development 133: 1241–1252. [DOI] [PubMed] [Google Scholar]
- Mavrich T.N., et al. (2008). Nucleosome organization in the Drosophila genome. Nature 453: 358–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh Y.-S., Amasino R.M. (2003). PIE1, an ISWI family gene, is required for FLC activation and floral repression in Arabidopsis. Plant Cell 15: 1671–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onouchi H., Igeño M.I., Périlleux C., Graves K., Coupland G. (2000). Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. Plant Cell 12: 885–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy F. (2005). Flowering: A time for integration. Int. J. Dev. Biol. 49: 585–593. [DOI] [PubMed] [Google Scholar]
- Putterill J., Robson F., Lee K., Simon R., Coupland G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847–857. [DOI] [PubMed] [Google Scholar]
- Samach A., Onouchi H., Gold S.E., Ditta G.S., Schwarz-Sommer Z., Yanofsky M.F., Coupland G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616. [DOI] [PubMed] [Google Scholar]
- Santisteban M.S., Hang M., Smith M.M. (2011). Histone variant H2A.Z and RNA polymerase II transcription elongation. Mol. Cell. Biol. 31: 1848–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnowski T.J., Ríos G., Jásik J., Swiezewski S., Kaczanowski S., Li Y., Kwiatkowska A., Pawlikowska K., Koźbiał M., Koźbiał P., Koncz C., Jerzmanowski A. (2005). SWI3 subunits of putative SWI/SNF chromatin-remodeling complexes play distinct roles during Arabidopsis development. Plant Cell 17: 2454–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaswamy S., Bhinge A., Zhao Y., Jones S., Hirst M., Iyer V.R. (2008). Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol. 6: e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson G.G., Dean C. (2002). Arabidopsis, the Rosetta stone of flowering time? Science 296: 285–289. [DOI] [PubMed] [Google Scholar]
- Singh R.K., Kabbaj M.H., Paik J., Gunjan A. (2009). Histone levels are regulated by phosphorylation and ubiquitylation-dependent proteolysis. Nat. Cell Biol. 11: 925–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth A., Schmid M. (2011). Regulation of flowering time: All roads lead to Rome. Cell. Mol. Life Sci. 68: 2013–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L., Nogales E., Ciferri C. (2010). Structure and function of SWI/SNF chromatin remodeling complexes and mechanistic implications for transcription. Prog. Biophys. Mol. Biol. 102: 122–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari V.K., McGarvey K.M., Licchesi J.D., Ohm J.E., Herman J.G., Schübeler D., Baylin S.B. (2008). PcG proteins, DNA methylation, and gene repression by chromatin looping. PLoS Biol. 6: 2911–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B., Palstra R.J., Splinter E., Grosveld F., de Laat W. (2002). Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell 10: 1453–1465. [DOI] [PubMed] [Google Scholar]
- Valverde F., Mouradov A., Soppe W., Ravenscroft D., Samach A., Coupland G. (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006. [DOI] [PubMed] [Google Scholar]
- Wagner D., Meyerowitz E.M. (2002). SPLAYED, a novel SWI/SNF ATPase homolog, controls reproductive development in Arabidopsis. Curr. Biol. 12: 85–94. [DOI] [PubMed] [Google Scholar]
- Wang J., Tian L., Lee H.S., Chen Z.J. (2006). Nonadditive regulation of FRI and FLC loci mediates flowering-time variation in Arabidopsis allopolyploids. Genetics 173: 965–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman B., Knudsen K.E. (2009). Hijacking the chromatin remodeling machinery: Impact of SWI/SNF perturbations in cancer. Cancer Res. 69: 8223–8230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R.N., Heckman J.W., Somerville C.R. (1992). Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol. 100: 403–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.I. (2012). Diverse functions of ATP-dependent chromatin remodeling complexes in development and cancer. Acta Biochim. Biophys. Sin. (Shanghai) 44: 54–69. [DOI] [PubMed] [Google Scholar]
- Yang J., Corces V.G. (2012). Insulators, long-range interactions, and genome function. Curr. Opin. Genet. Dev. 22: 86–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Smith C.L., Saha A., Grill S.W., Mihardja S., Smith S.B., Cairns B.R., Peterson C.L., Bustamante C. (2006). DNA translocation and loop formation mechanism of chromatin remodeling by SWI/SNF and RSC. Mol. Cell 24: 559–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D., Coleman-Derr D., Ballinger T., Henikoff S. (2008). Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature 456: 125–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.