Peroxisomes are dynamic single-membrane organelles in eukaryotes. Peroxins are essential proteins required for peroxisome biogenesis and play multiple roles in development and growth. DAU/APEM9, a recently identified peroxin, plays a critical role in peroxisome biogenesis, morphology, and function during pollen maturation and germination in planta.
Abstract
Pollen undergo a maturation process to sustain pollen viability and prepare them for germination. Molecular mechanisms controlling these processes remain largely unknown. Here, we report an Arabidopsis thaliana mutant, dayu (dau), which impairs pollen maturation and in vivo germination. Molecular analysis indicated that DAU encodes the peroxisomal membrane protein ABERRANT PEROXISOME MORPHOLOGY9 (APEM9). DAU is transiently expressed from bicellular pollen to mature pollen during male gametogenesis. DAU interacts with peroxisomal membrane proteins PEROXIN13 (PEX13) and PEX16 in planta. Consistently, both peroxisome biogenesis and peroxisome protein import are impaired in dau pollen. In addition, the jasmonic acid (JA) level is significantly decreased in dau pollen, and the dau mutant phenotype is partially rescued by exogenous application of JA, indicating that the male sterility is mainly due to JA deficiency. In addition, the phenotypic survey of peroxin mutants indicates that the PEXs most likely play different roles in pollen germination. Taken together, these data indicate that DAU/APEM9 plays critical roles in peroxisome biogenesis and function, which is essential for JA production and pollen maturation and germination.
INTRODUCTION
The life cycle of flowering plants alternates between a diploid, sporophytic generation and a haploid, gametophytic generation. During evolution, key innovations associated with the male gametophyte occurred, including the reversal of microspore polarity, the conversion from motile to immotile gametes, and the emergence of siphonogamy (Rudall and Bateman, 2007). In Arabidopsis thaliana anthers, microspores undergo an asymmetric division to produce a large vegetative cell and a small generative cell. Subsequently, the generative cell goes through a symmetric division to generate two sperm cells. The tricellular pollen grains have gained pollination competence from this moment though still in undehisced anthers (Kandasamy et al., 1994). The tricellular grains undergo a maturation process prior to release to prepare themselves for survival in terrestrial environment (Taylor and Hepler, 1997). The mature pollen grains released from the dehiscent anthers are greatly desiccated and metabolically dormant, which maintains pollen viability (Swanson et al., 2004). Jasmonic acid (JA) is an essential signal regulating anther development and pollen maturation (Turner et al., 2002; Browse, 2009). Anther dehiscence–defective phenotypes have been reported in mutants defective in the JA biosynthetic pathway, including fatty acid desaturation (McConn and Browse, 1996), defective in anther dehiscence1 (dad1) (Ishiguro et al., 2001), allene oxide synthase (Park et al., 2002), 12-oxophytodienoic acid reductase3 (opr3) (Stintzi and Browse, 2000), delayed-dehiscence1 (dde1), and dde2 (Sanders et al., 2000; von Malek et al., 2002). Among these mutants, phenotypic analysis of pollen was only conducted in opr3 and dad1 (Stintzi and Browse, 2000; Ishiguro et al., 2001). Pollen grains from opr3 and dad1 mutants develop normally up to the tricellular stage but are sterile after release. The dad1 grains are unable to germinate when manually laid on the stigmas. These findings indicate that JA is also involved in pollen maturation and germination, but the underlying molecular mechanisms are poorly understood.
Peroxisomes are single membrane–bound organelles that function in diverse metabolic pathways. Recent advances have largely boosted our knowledge of peroxisome biogenesis although far from fully elucidating this process (Hu et al., 2012; Kim and Mullen, 2013). So far, several models have been proposed, including the ER vesiculation model, the autonomous peroxisome growth and division model, and the recent ER semiautonomous peroxisome model (Mullen and Trelease, 2006; Hu et al., 2012). Generally, peroxisome biogenesis consists of three steps: biogenesis of the peroxisomal membrane, import of peroxisome matrix proteins, and peroxisome division. Proteins involved in peroxisome biogenesis are termed PEROXINS (PEXs). In yeast and mammalian cells, peroxisomal membrane proteins PEX3 and PEX16 as well as a farnesylated, mostly cytosolic protein PEX19 are required for the formation of nascent peroxisomes and import of peroxisomal membrane proteins (Götte et al., 1998; Ghaedi et al., 2000; Kim et al., 2006; Matsuzaki and Fujiki, 2008). PEX16 localized both in the endoplasmic reticulum (ER) and peroxisome is involved in early peroxisome biogenesis (Karnik and Trelease, 2005; Kim et al., 2006; Mullen and Trelease, 2006). Peroxisome matrix proteins, tagged by PTS1 or PTS2 peptide signals, are synthesized on free polyribosomes and imported into peroxisomes posttranslationally. PTS1- and PTS2-containing proteins are first recognized by receptors PEX5 and PEX7, respectively, in the cytosol (Dammai and Subramani, 2001; Hayashi et al., 2005; Singh et al., 2009; Ramón and Bartel, 2010). Next, the receptor-cargo complex docks onto the peroxisomal membrane proteins PEX13 and PEX14 (Hayashi et al., 2000; Mano et al., 2006; Singh et al., 2009), and then the cargoes are released in the peroxisome and the receptors are recycled back to the cytosol. The recycling machinery requires the RING-finger E3 ligase complex composed of PEX2, PEX10, and PEX12 (Dammai and Subramani, 2001; Schumann et al., 2003; Sparkes et al., 2003; Fan et al., 2005; Nito et al., 2007; Kaur et al., 2013), the ubiquitin-conjugating enzyme PEX4 anchored to the membrane by PEX22 (Zolman et al., 2005; Nito et al., 2007), and the APEM9-tethered AAA-ATPase PEX1-PEX6 complex (Grou et al., 2009; Goto et al., 2011).
Peroxisomes in plants display profound metabolic plasticity manifested by their diverse function and morphology. They are the site of fatty acid β-oxidation in plant cells and involved in the generation of phytohormones JA and indole-3-acetic acid as well as in other metabolic and signaling pathways. OPR3 converts the chloroplast-produced 12-oxophytodienoic acid to OPC8:0, which is converted to JA after three rounds of β-oxidation in the peroxisome (Turner et al., 2002; Hu et al., 2012). Consistently, the male sterility of opr3 can be rescued by exogenous JA but not 12-oxophytodienoic acid (Stintzi and Browse, 2000).
The biogenesis and function of peroxisomes in reproduction are largely unknown in plants. Recently, it was reported that ABSTINENCE BY MUTUAL CONSENT (AMC), a putative ortholog of PEX13, is involved in male-female gametophyte recognition, but the mechanism remains unknown (Boisson-Dernier et al., 2008). Here, we identified a male gametophytic mutant dayu (dau) in Arabidopsis, which is defective in pollen maturation and germination in planta. DAU encodes a peroxisomal membrane protein recently identified as APEM9 (Goto et al., 2011). Peroxisome biogenesis/function and matrix protein import were both impaired in dau pollen, and the male sterility of dau/DAU plants was partially restored by the exogenous application of JA. A similar phenotype was observed in pex13 but not in pex10, pex12, pex14, or pex16 mutants, suggesting that peroxins likely play different roles in pollen. We also found that DAU is a dual transmembrane protein that interacts with PEX13 and PEX16 in plants. Together, we showed that DAU/APEM9, which is required for peroxisome biogenesis and function, plays a critical role during pollen maturation/germination.
RESULTS
Isolation of the dau Mutant
To understand mechanisms controlling pollen development, we performed a genetic screen for mutants with a distorted Mendelian segregation from our Arabidopsis gene/enhancer trap lines (Sundaresan et al., 1995; Page and Grossniklaus, 2002). A gene trap line, designated as dayu (dau, after the Chinese legendary hero), exhibited a kanamycin-resistant (Kanr) to kanamycin-sensitive (Kans) ratio of 1.28:1 (370:289). Further reciprocal crosses between dau/DAU and wild-type plants showed a Kanr:Kans ratio of 0.88:1 (191:216) in F1 progenies when dau/DAU plants were used as the female and of 0.13:1 (55:415) when dau/DAU plants as the male. These data suggest that the dau mutation causes severe defects in the male gametophyte. In addition, no homozygous dau mutant was obtained, indicating that the mutation might cause embryo lethality. Therefore, we examined the embryo development from the self-pollinated dau/DAU plants and found that ∼17.10% of embryos (n = 1158) displayed obvious abortion. The mutant embryos were arrested at the heart stage and were ultimately shrunk (Supplemental Figure 1).
Pollen Maturation Is Impaired in the dau Mutant
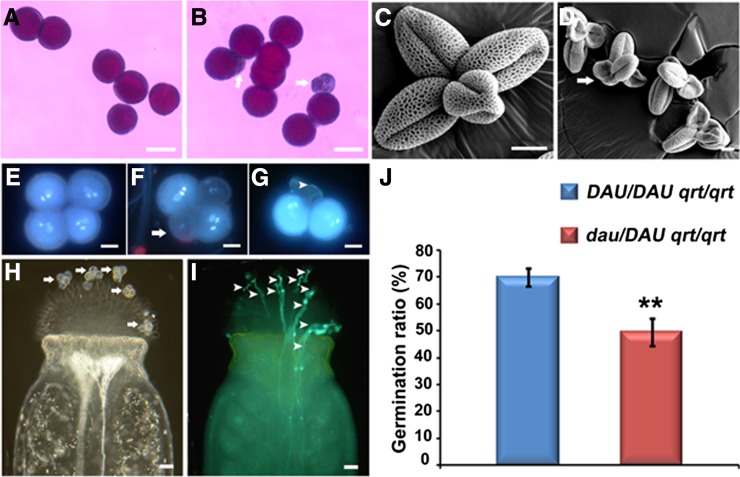
Since the function of the male gametophyte is impaired in the dau mutant, the viability of mature pollen was investigated with Alexander’s stain (Alexander, 1969). The viable wild-type pollen were stained red purple (Figure 1A), while a few aborted pollen from dau/DAU plants were not stained (Figure 1B). Statistical analysis indicates that the wild-type pollen have an abortion rate of 0.90% (n = 2116), while dau/DAU plants grown in the same conditions have a pollen abortion rate of 4.80% (n = 8536) (Student’s t test, 0.01 < P < 0.05). We further examined the pollen morphology by scanning electron microscopy. Quartet pollen grains (Preuss et al., 1994; Copenhaver et al., 2000) from DAU/DAU qrt/qrt and dau/DAU qrt/qrt plants were collected for the scanning electron microscopy analysis (Figures 1C and 1D). Compared with the wild-type pollen (Figure 1C) with an abortion ratio of 0.61% (n = 653), the majority of pollen grains from dau/DAU qrt/qrt plants were morphologically normal except for 4.01% pollen (n = 873, Student’s t test, P < 0.01), which were small and shrunken (Figure 1D). These data indicate that pollen viability is slightly affected in the dau mutant.
Figure 1.
Phenotype of dau/DAU Mature Pollen Released from Anthers.
(A) and (B) Alexander staining showing viable wild-type pollen (A) and dau/DAU mutant pollen with a few aborted grains (arrows) (B).
(C) and (D) Scanning electron micrograph of DAU/DAU qrt/qrt pollen (C) and dau/DAU qrt/qrt mutant pollen with a few aborted grains (arrow) (D).
(E) to (G) DAPI staining of mature pollen grains from dau/DAU qrt/qrt plants showing normal grains with three clearly stained nuclei (E), grains with faint nuclear staining (arrow) (F), and collapsed grains (arrowhead) (G).
(H) and (I) Hand pollination of five dau/DAU qrt/qrt quartet pollen (20 grains) on wild-type stigma showing positions of the pollen grains (arrows) (H) and 10 germinated pollen tubes (arrowheads) (I).
(J) Statistical comparison of in vivo pollen germination ratios between DAU/DAU qrt/qrt and dau/DAU qrt/qrt. Data presented are mean values from three independent experiments (n > 300). **Student’s t test, P < 0.01.
Bars = 50 µm in (A) and (B), 10 µm in (C) to (G), and 25 µm in (H) and (I).
To assess whether the dau mutation impairs pollen development, pollen grains from dau/DAU qrt/qrt plants were stained with 4′,6-diamidino-2-phenylindole (DAPI) to check cell cycle progression. At stages of pollen mitosis I and II, the quartet pollen grains from dau/DAU qrt/qrt plants, which have two wild-type grains and two dau grains, displayed a similar nuclear appearance (Supplemental Figure 2), indicating that pollen development is normal at these stages. Among the mature quartet pollen released from dau/DAU qrt/qrt anthers (n = 1104), 81.70% contained three clearly stained nuclei (Figure 1E), 12.41% showed three visible nuclei with faint staining (Figure 1F), and 5.89% contained totally disrupted nuclei (Figure 1G). In comparison, the mature pollen from DAU/DAU qrt/qrt anthers had an abortion ratio of 0.88% (n = 1026). These data indicate that the pollen grains from dau/DAU qrt/qrt plants develop normally up to the tricellular stage; thereafter, a small fraction of the mutant pollen is disrupted during maturation. Since the tricellular pollen in undehisced anthers have gained pollination competence (Kandasamy et al., 1994), we wondered whether the dau tricellular pollen within anthers were functional. When tricellular pollen from dau/DAU qrt/qrt undehisced anthers were manually pollinated onto mature wild-type stigmas, the average Kanr:Kans ratio of the F1 progeny increased to 0.36:1 (668:1842) (Student’s t test, P < 0.01), nearly 3 times that of naturally released dau pollen. This suggests that the dau tricellular pollen germinate more efficiently than mature dau pollen. Taken together, these data indicate that dau mutation impairs the pollen maturation process.
The dau Mutation Impairs in Vivo Pollen Germination
To check whether the dau mutation affects in vivo pollen germination and tube growth, stigmas of wild-type flowers were pollinated with limited pollen grains shed from DAU/DAU qrt/qrt and dau/DAU qrt/qrt plants. Four hours after pollination, the pollinated pistils were collected for pollen tube analysis. When five quartets (20 pollen grains) from dau/DAU qrt/qrt plants were pollinated on the wild-type stigma, only 10 grains produced pollen tubes (Figures 1H and 1I). Statistically, the pollen germination ratio assayed this way was around 69.98% for DAU/DAU qrt/qrt pollen and 49.51% for dau/DAU qrt/qrt pollen (Figure 1J). This suggests that the dau mutation inhibits pollen germination on the stigma. Moreover, no obvious defects in pollen tube growth or guidance in vivo were detected, suggesting that the male sterility is mainly caused by the impaired pollen germination on the stigma.
Peroxisome Biogenesis Is Disrupted in dau Pollen
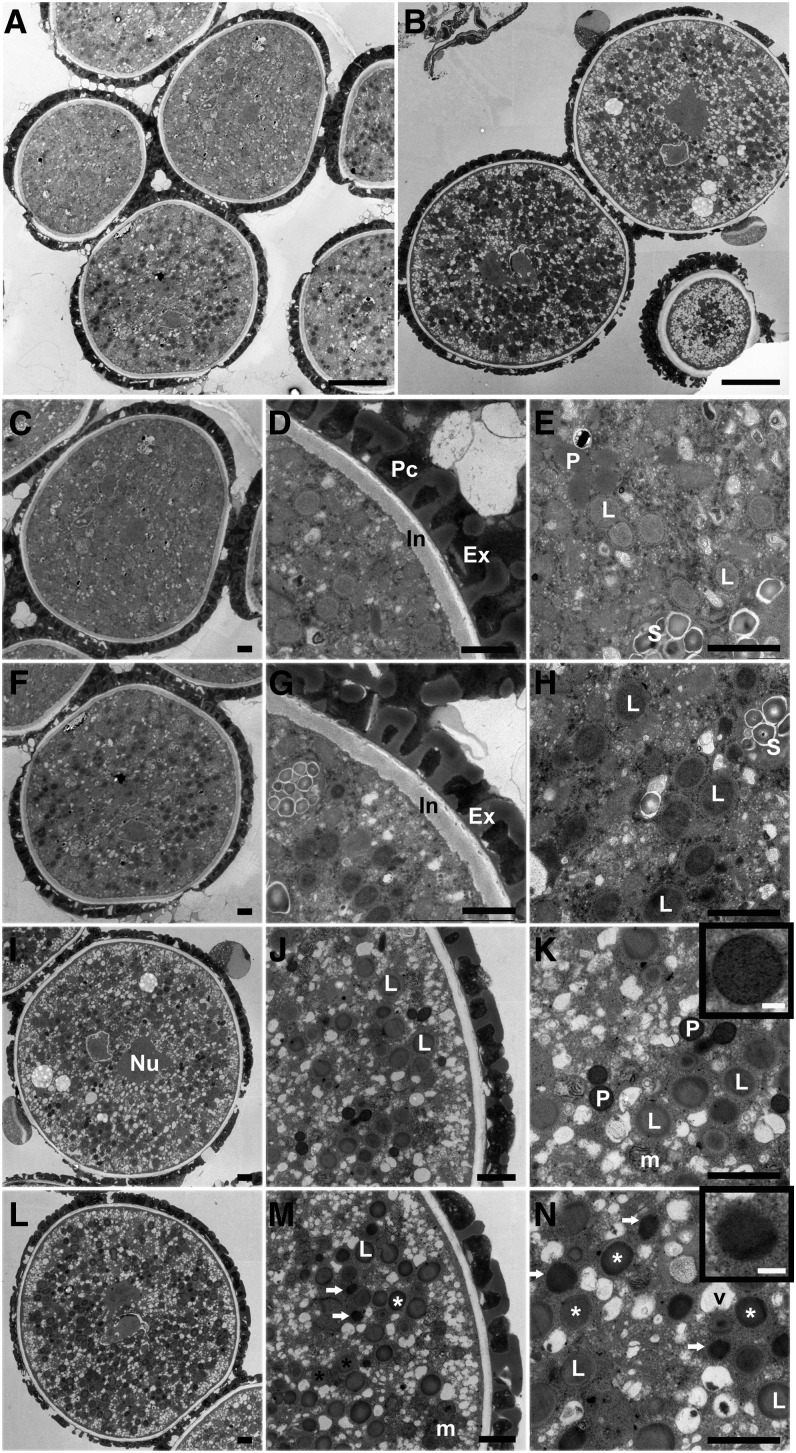
To unveil what caused the defect in pollen germination, transmission electron microscopy (TEM) was performed to explore the cytological abnormality in dau pollen. In wild-type tricellular pollen from undehisced anthers, nuclei of sperm cells and the vegetative cell were visible (Figures 2A and 2C), and the pollen coat contained a large amount of tryphine (Figure 2D). The cytoplasm of the vegetative cell was rich in mitochondria, lipid bodies, peroxisomes, starch granules, and small vesicles (Figure 2E). In dau grains at the same stage, nuclei of sperm cells and the vegetative cell were also visible. The pollen coat and pollen wall were also morphologically normal (Figures 2F and 2G). However, no typical peroxisome was observed in dau pollen grains, and lipid bodies were densely stained (Figure 2H), compared with the wild-type grains (Figure 2E).
Figure 2.
TEM Analysis of dau Pollen Compared with the Wild Type.
(A) dau/DAU qrt/qrt tricellular pollen grains in undehisced anthers.
(B) dau/DAU qrt/qrt mature grains, stained with DAB.
(C) A wild-type tricellular pollen grain in undehisced anthers.
(D) A portion of (C) showing wild-type pollen cell wall.
(E) A magnification of (C) showing peroxisomes, lipid bodies, and the starch granules as indicated.
(F) A dau mature pollen grain in undehisced anthers.
(G) A portion of (F) showing dau mutant pollen cell wall.
(H) A magnification of (F) showing darkly stained lipid bodies.
(I) to (N) TEM micrographs of mature pollen released from dehiscent anthers with DAB staining.
(I) A wild-type mature pollen grain. The nucleus of one sperm cell is indicated.
(J) A magnified region of (I) showing gray lipid bodies.
(K) Detail of (I) showing heavily stained peroxisomes with clear boundary (inset), lipid bodies, and mitochondria. Mitochondrial cristae were also stained by DAB.
(L) An overview of a dau mature pollen grain.
(M) A magnified region of (L). Note DAB-stained peroxisome-like structure (arrow) and two types of lipid body (star).
(N) A magnification of (L) showing darkly stained peroxisome-like structures (arrow and inset) and stained (white star) and nonstained (dark star) lipid bodies.
Ex, exine; In, intine; L, lipid body; Nu, nucleus; m, mitochondria; P, peroxisome; Pc, pollen coat; S, starch granule. Bars = 5 µm in (A) and (B), 1 µm in (C) to (N), and 0.1 µm in the insets of (K) and (N).
To visualize peroxisomes, we performed 3,3′-diaminobenzidine tetrahydrochloride (DAB) staining for catalase activity, which marks the peroxisome (Lorenzo et al., 1990). In mature wild-type pollen released from dehiscent anthers (Figures 2B, 2I, to 2K), peroxisomes were intensively stained and displayed clear round structure of single-layered membrane (Figure 2K). While in dau pollen at the same stage (Figures 2B and 2L to 2N), the DAB-stained structures lacking clear membrane, designated as peroxisome-like structures, were present (Figure 2N). These observations suggest that peroxisome biogenesis and membrane integrity are partially disrupted in dau pollen.
DAU Encodes the Peroxisomal Membrane Protein APEM9
To isolate the gene disrupted in the mutant, Ds flanking sequences were obtained using thermal asymmetric interlaced PCR (Liu et al., 1995). Sequence analysis indicated that the Ds element is inserted at +218 bp in the first intron of the At3G10572 gene and caused an 8-bp nucleotide duplication at the insertion site. Southern hybridization using Ds-5′ probes and the At3G10572 fragment further confirmed that a single Ds element is inserted in the mutant genome.
To verify whether the dau phenotype is caused by Ds insertion into At3G10572, a complementation assay was performed. The construct containing a 2.8-kb genomic DNA fragment of At3G10572 was introduced into dau/DAU plants by Agrobacterium tumefaciens–mediated infiltration (Bechtold and Pelletier, 1998). Six independent transgenic lines were obtained. The Kanr:Kans ratios of T2 plants were raised to 2.15:1 (n = 2058), compared with 1.28:1 in dau/DAU plants. In addition, several T3 plants homozygous for the Ds insertion and the transgene were obtained. These data demonstrated that the male sterility in dau/DAU is indeed caused by the loss of At3G10572 gene function.
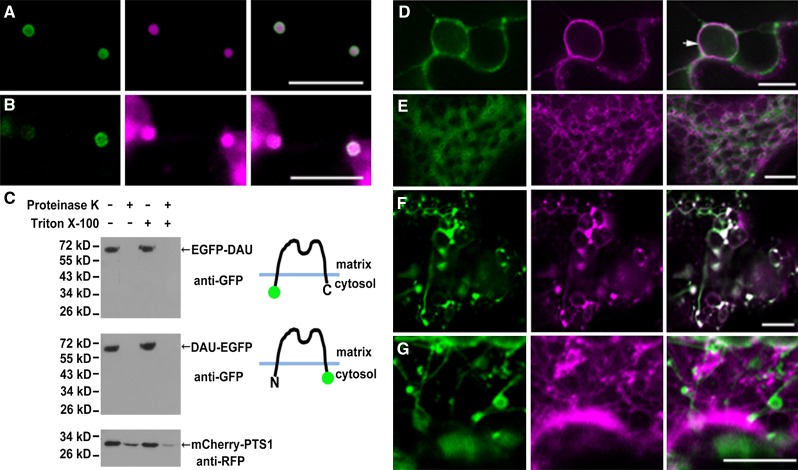
Recently, an allelic mutation of dau, designated as aberrant peroxisome morphology9 (apem9), was reported to disrupt peroxisome morphology and protein import in Arabidopsis (Goto et al., 2011). It was shown that DAU/APEM9 encodes a peroxisomal membrane protein. Secondary structure prediction suggests that DAU/APEM9 has one or two putative transmembrane domains (TMDs) (Goto et al., 2011). To determine the membrane topology of DAU/APEM9, we performed protease protection assays (Lisenbee et al., 2003) with peroxisomes purified from tobacco (Nicotiana benthamiana) leaves transiently expressing DAU tagged with enhanced green fluorescent protein (EGFP). To determine the localization of N- and C-terminal EGFP-tagged DAU, the EGFP-DAU or DAU-EGFP construct, along with the peroxisome marker mCherry-PTS1 (Nelson et al., 2007), was transiently coexpressed in tobacco leaves. EGFP-DAU was mainly localized on the peroxisomal membrane (Figure 3A), which is consistent with the previous observation that GFP-APEM9 was targeted to the peroxisomal membrane (Goto et al., 2011). DAU-EGFP was targeted to the peroxisomal membrane and also to the perinuclear ER (Figures 3B and 3D), and its overexpression impaired the import of mCherry-PTS1 and PTS2-mCherry (Supplemental Figures 3A and 3B). However, in DAU-EGFP–overexpressing cells, PEX12, PEX13, and PEX16 are able to target to peroxisomes (Supplemental Figures 3C to 3E). In addition, the DAU-EGFP transgene fully complemented the dau mutation when the ProDAU:DAU-EGFP construct was transformed into dau/DAU plants. Among the 12 independent transgenic lines, the ratio of Kanr to Kans of their progeny was raised significantly to 2.54:1 on average, compared with the 1.28:1 ratio in dau/DAU plants (Supplemental Table 1). Consistently, dau/dau plants were obtained in T3 plants. These results indicated that the DAU-EGFP fusion protein is functional. Protease protection assays on purified peroxisomes expressing EGFP-DAU and DAU-EGFP showed that both the N- and C-terminal tagged EGFP were sensitive to protease digestion with or without Triton X-100 treatment (Figure 3C), indicating that both the N and C terminus of DAU are exposed to the cytosol. As a control, peroxisome matrix protein mCherry-PTS1 was protected from protease digestion, unless the peroxisomal membrane was solubilized with Triton X-100 (Figure 3C). Taken together, we conclude that DAU most likely contains two transmembrane domains with both termini facing the cytosol. There is a caveat, although unlikely, that the GFP fusion might disrupt the membrane topology of DAU.
Figure 3.
Topology of DAU and Localization of DAU Truncated Protein.
(A) Coexpression of EGFP-DAU and the peroxisomal marker mCherry-PTS1 in tobacco leaves, showing peroxisomal membrane-localized EGFP-DAU and peroxisomal matrix-localized mCherry-PTS1.
(B) Coexpression of DAU-EGFP and peroxisomal marker mCherry-PTS1 in tobacco leaves, showing that DAU-EGFP is targeted to the peroxisomal membrane and mCherry-PTS1 is localized in the peroxisomal matrix and cytosol.
(C) Determination of DAU topology through the proteinase protection assay. Both the N terminus and C terminus of DAU face the cytosol. Peroxisomes from tobacco leaves expressing EGFP-DAU, DAU-EGFP, and mCherry-PTS1 were subjected to proteinase K treatment in the presence or absence of Triton X-100. Treated samples were subjected to SDS-PAGE and immunoblot analysis.
(D) Coexpression of DAU-EGFP and the ER marker mCherry-HDEL in perinuclear ER in tobacco leaves. Arrow indicates the nuclear membrane.
(E) Coexpression of EGFP-DAU(1-115) and the ER marker mCherry-HDEL in tobacco leaves.
(F) Coexpression of DAU(267-333)-EGFP and mCherry-PTS1 in tobacco leaves. Note the formation of tubular peroxisomes.
(G) Coexpression of DAU(267-333)-EGFP and mCherry-HDEL in tobacco leaves. Bars = 10 µm.
To further explore the topology of DAU, we generated DAU truncations containing either the N- or C-terminal TMD and monitored their localization. The truncated protein EGFP-DAU(1-115) containing the N-terminal TMD was not localized to the peroxisome or ER, but accumulated in the cytosol (Figure 3E). The DAU(267-333)-EGFP containing the C-terminal TMD was targeted to the peroxisome (Figure 3F). These results indicate that the peroxisomal membrane targeting signal of DAU is located in or adjacent to the C-terminal TMD, whereas the N terminus alone is not sufficient for peroxisomal membrane targeting. Intriguingly, DAU(267-333)-EGFP expression caused peroxisome elongation or tubulation (Figure 3F), and the tubular structures were not part of the ER (Figure 3F). Taken together, these data imply that DAU most likely plays a role in peroxisome biogenesis and morphology.
DAU Is Expressed during the Later Stages of Pollen Development
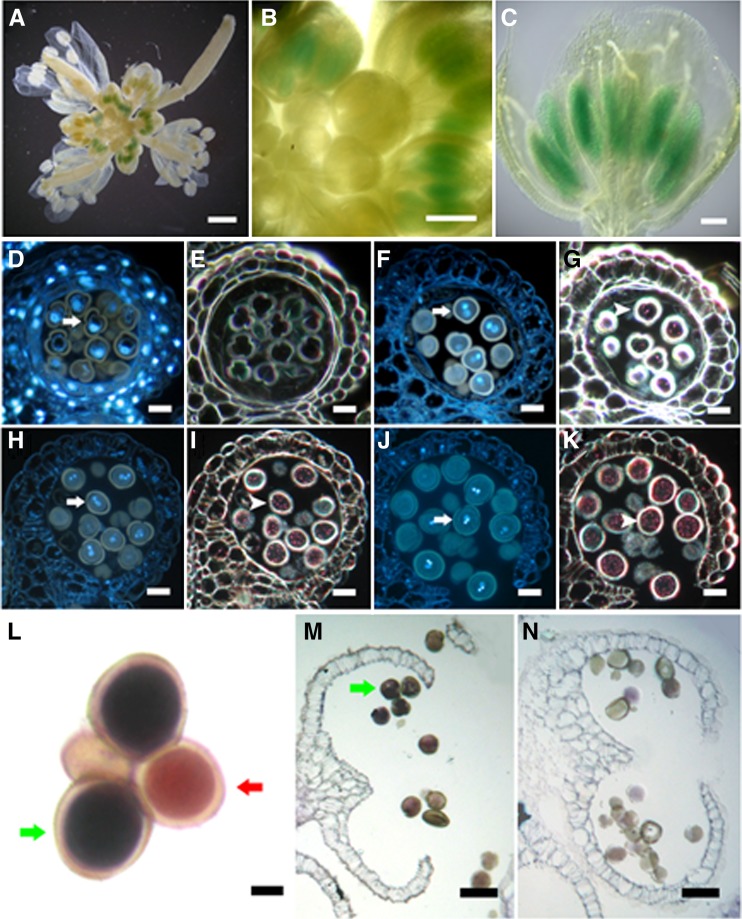
Quantitative RT-PCR analysis showed that APEM9 is expressed in various tissues, with a higher level in buds and flowers (Goto et al., 2011). We used a ProDAU:GUS (for β-glucuronidase) reporter system to monitor DAU expression during pollen development. GUS signals were detected in the anthers of the transgenic plants (Figures 4A to 4C). To further observe the GUS activity in pollen, the GUS-stained anthers were sectioned and the semithin sections were stained with DAPI to determine their developmental stage. No GUS signal was observed in microspores before and at the stage of pollen mitosis I (Figures 4D and 4E). GUS signals were detected in bicellular, tricellular, and mature pollen grains (Figures 4F to 4K), indicating that DAU is expressed after the first mitosis during pollen development.
Figure 4.
Expression Pattern of DAU Gene in Flowers.
(A) to (K) GUS staining of ProDAU:GUS transgenic plants showing GUS activity in inflorescence (A), flower buds (B), and anthers (C).
(D) to (K) The semithin sections of a GUS-stained transgenic inflorescence showing nucleus visualized by DAPI (arrow) and GUS signal in dark (arrowhead). Note no GUS signal in one-nucleate pollen grains ([D] and [E]). GUS signals were detected in bicellular pollen ([F] and [G]), tricellular pollen ([H] and [I]), and mature pollen at anther dehiscence ([J] and [K]).
(L) to (N) DAU expression detected by RNA in situ hybridization.
(L) In mature dau/DAU qrt/qrt quartet pollen, DAU was detected in wild-type grains (green arrow) but not in dau mutant grains (red arrow).
(M) DAU was detected in wild-type pollen grains (green arrow) at anther dehiscence.
(N) Negative control hybridized with DAU sense probe.
Bars = 1 mm in (A) to (C), 20 µm in (D) to (K), 5 µm in (L), and 40 µm in (M) to (N).
To validate that DAU is expressed in the mature pollen grains after anther dehiscence, RNA in situ hybridization was performed on dau/DAU qrt/qrt pollen (Figures 4L to 4N). The signal was detected in mature wild-type pollen (Figures 4L and 4M) but not in dau mutant pollen, suggesting that dau is a null mutant (Figure 4L). No signal was observed in the control when the sense RNA probe was used (Figure 4N). Together, these results show that DAU is expressed during the later stages of pollen development.
DAU Interacts with PEX13 and PEX16
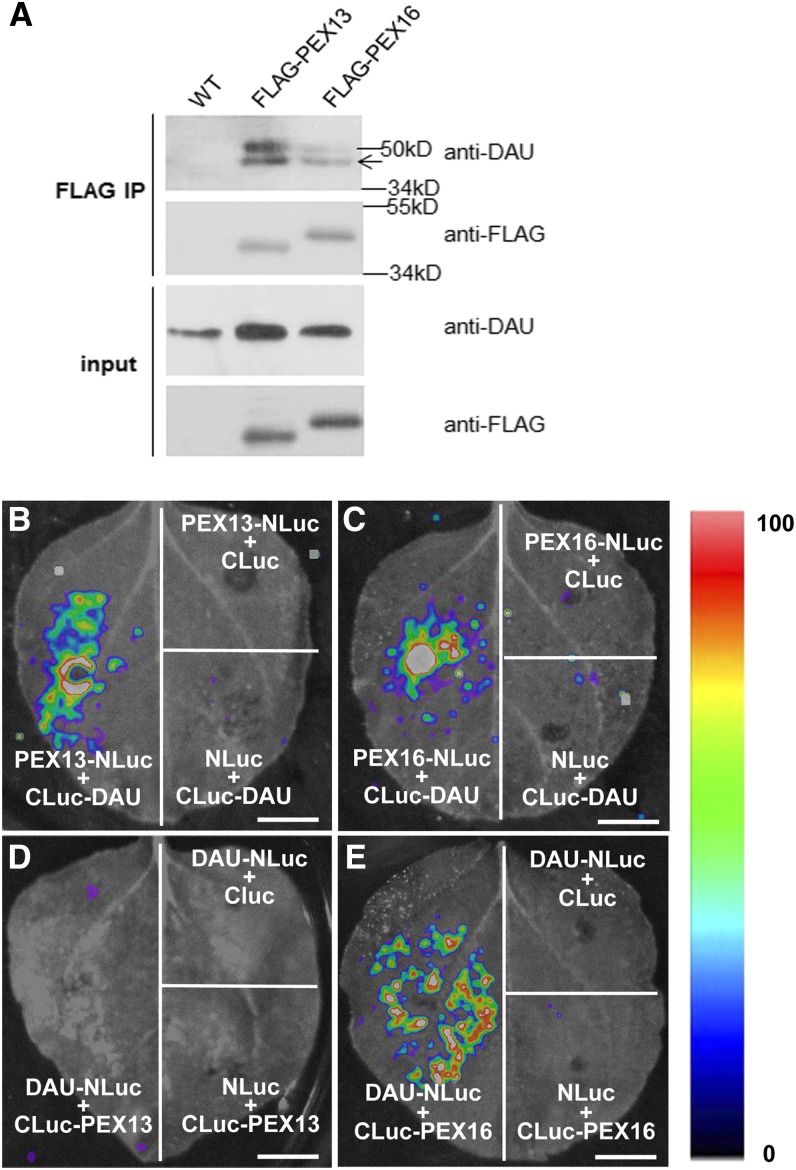
We next tested whether DAU could interact with other peroxins in vivo. Antibody against DAU/APEM9 was prepared, and the DAU antibody specifically recognized the target proteins in Arabidopsis and transiently transformed tobacco leaves (Supplemental Figure 4). We also generated transgenic plants carrying both the Pro35S:FLAG-PEX13 and Pro35S:FLAG-PEX16 constructs. A coimmunoprecipitation assay showed that DAU interacted with PEX13 and PEX16 (Figure 5A). In addition, a firefly luciferase complementation imaging assay (Chen et al., 2008) was performed. As shown in Figures 5B and 5C, combinations of CLuc-DAU with PEX13-NLuc and PEX16-NLuc showed strong LUC activity, suggesting that the N-terminal domain of DAU interacts with PEX13 and PEX16. Moreover, a combination of DAU-NLuc and CLuc-PEX13 did not show LUC complementation (Figure 5D), while the interaction between DAU-NLuc and CLuc-PEX16 was detected (Figure 5E), indicating that the C-terminal of DAU can interact with PEX16 but not PEX13. This suggests that the N and C termini of PEX13 may face to peroxisomal matrix and cytosol, respectively, while both termini of PEX16 may face the cytosol, as DAU does. Taken together, our data showed that DAU physically interacts with PEX13 and PEX16, both in a luciferase complementation imaging assay and in planta.
Figure 5.
Interactions between DAU and PEXs.
(A) PEX13 and PEX16 physically interact with DAU in Arabidopsis seedlings. The band at 50 kD indicates the heavy chain of anti-FLAG IgG. Wild-type seedlings were used as the negative control.
(B) to (E) N. benthamiana leaves coinfiltrated with Agrobacterium containing 35S-driven construct pairs as indicated were photographed with a charge-coupled device camera. CLuc-DAU interacts with PEX13-NLuc (B) and PEX16-NLuc (C). No interaction of DAU-NLuc with Cluc-PEX13 (D) is detected. DAU-NLuc interacts with Cluc-PEX16 (E). The pseudocolor bar shows the relative range of luminescence intensity in the image. Bars = 1 cm.
DAU Regulates Peroxisomal Protein Import in Pollen
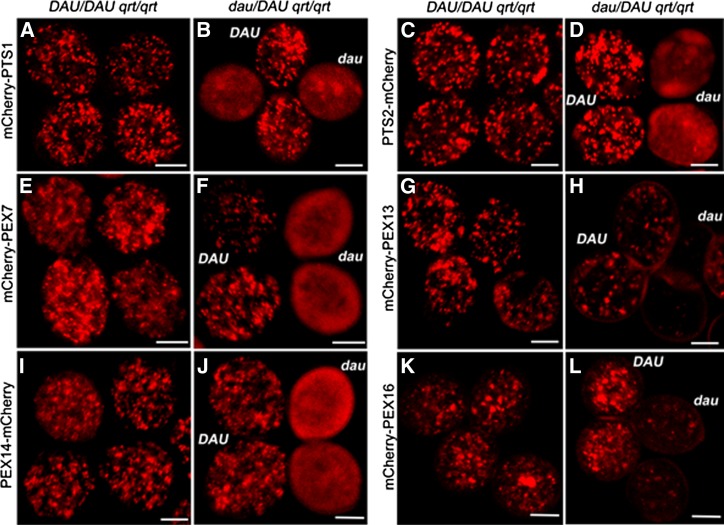
To further explore the peroxisomal protein import in dau mutant pollen, Arabidopsis peroxisomal markers fused with mCherry driven by the Lat52 promoter were transformed into wild-type and dau/DAU qrt/qrt mutant plants, respectively. As shown in Figure 6, mCherry-PTS1 exhibited a punctate peroxisome pattern in wild-type pollen (Figure 6A) but appeared as diffusely cytosolic localization with occasionally large aggregates in dau mutant pollen (Figure 6B). This is consistent with the result in sporophytic tissues of the apem9 mutant (Goto et al., 2011). In dau/DAU plants complemented with the genomic DNA fragment, mCherry-PTS1 showed a punctate pattern (Supplemental Figure 3F), as observed in wild-type pollen (Figure 6A). Moreover, PTS2-mCherry and mCherry-PEX7 were localized to peroxisomes in wild-type pollen (Figures 6C and 6E) but appeared cytosolic in dau pollen grains (Figures 6D and 6F). The amount and intensity of punctuate particles representing mCherry-PEX13 were significantly decreased in dau mutant pollen (Figure 6H), compared with the wild type (Figure 6G). The mCherry-PEX13–labeled peroxisomes in dau pollen were reduced to 7.3% of that of the DAU pollen. Unlike PEX13, the fluorescence representing PEX14-mCherry was diffusely distributed throughout the cytosol in dau pollen (Figure 6J). Overexpression of mCherry-PEX16 caused peroxisome aggregation in wild-type pollen (Figure 6K), but the intensity of fluorescence was largely decreased in dau pollen (Figure 6L). Together, these data suggest that the amount and function of peroxisomes are severely disrupted in dau pollen, which result in defective proxisomal protein import.
Figure 6.
PTS1- and PTS2-Dependent Protein Import into Peroxisomes Is Impaired in dau Mutant Pollen.
Constructs as indicated at the left were transformed into DAU/DAU qrt/qrt plants and dau/DAU qrt/qrt plants, respectively. Homozygous lines were obtained in the T2 generation. Naturally dehisced quartet pollen grains were viewed with confocal microscopy.
(A) and (B) mCherry-PTS1 displayed punctate peroxisomal localization in DAU pollen and diffuse cytosolic localization in dau pollen.
(C) and (D) PTS2-mCherry displayed dot-like pattern in DAU pollen and a diffuse pattern in dau pollen.
(E) and (F) Peroxisome-localized mCherry-PEX7 in DAU pollen and cytosol-localized mCherry-PEX7 with some large aggregates in dau pollen.
(G) and (H) Peroxisome-localized mCherry-PEX13 in DAU pollen and weak or lack of mCherry-PEX13 in dau pollen.
(I) and (J) Peroxisome-localized PEX14-mCherry in DAU pollen and weakly diffuse PEX14-mCherry in dau pollen.
(K) and (L) Aggregation of mCherry-PEX16 in DAU pollen and weak or lack of mCherry-PEX16 in dau pollen.
Bars = 10 µm.
The Male Sterility of dau Mutant Is Partially Caused by Reduced JA Synthesis
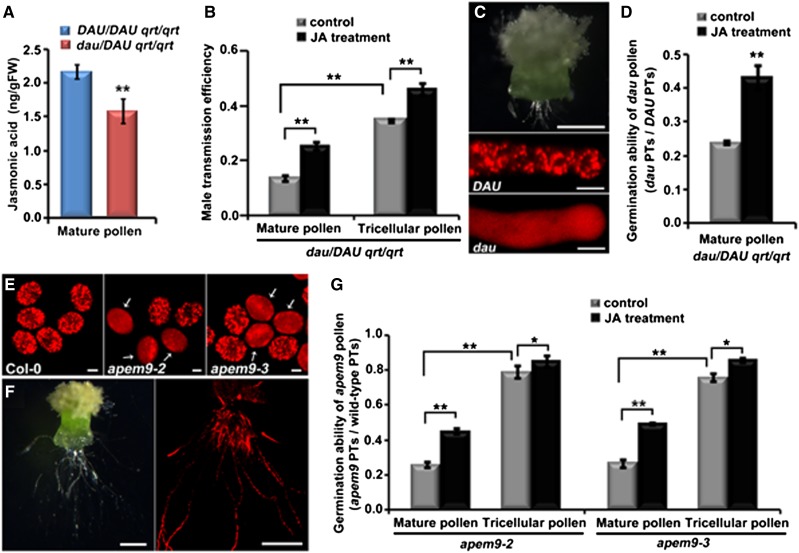
One notable function of plant peroxisomes is to generate JA. In the dad1 mutant, which is defective in JA biosynthesis, in vivo pollen germination is inhibited (Ishiguro et al., 2001). Thus, we speculate that the male sterility may be caused by JA deficiency in dau pollen. The JA levels of pollen from DAU/DAU qrt/qrt and dau/DAU qrt/qrt plants were quantified using ultra-high performance liquid chromatography–triple quadrupole mass spectrometry. Indeed, the JA level in mature pollen from dau/DAU qrt/qrt plants was decreased to 73.11% of that from DAU/DAU qrt/qrt plants (Figure 7A), indicating that the amount of JA is significantly reduced in dau pollen. We next explored whether exogenous JA can rescue the sterility of dau pollen. Methyl jasmonate was applied to the bud clusters of dau/DAU qrt/qrt plants as described (Ishiguro et al., 2001). Two days after treatment, the mature or tricellular pollen grains from dau/DAU qrt/qrt plants were pollinated on the wild-type stigmas and the male transmission efficiency (TE) was scored. After JA treatment, the TE of the mature dau/DAU qrt/qrt pollen was raised to an average of 25% (n > 300) compared with 13% without JA treatment (Figure 7B), and the TE of the tricellular dau/DAU qrt/qrt pollen was raised to an average of 46% (n > 300) compared with 35% without treatment (Figure 7B). These data indicate that the sterility of dau pollen can be partially rescued by exogenous JA.
Figure 7.
Level of JA Is Decreased in dau Mutant Pollen and Exogenous Application of JA Can Partially Rescue the Male Sterility of dau Mutants.
(A) JA levels in mature pollen of DAU/DAU qrt/qrt and dau/DAU qrt/qrt plants. FW, fresh weight. Error bars represent sd.
(B) Statistical Comparison of male TE of dau/DAU qrt/qrt mutant, using mature or tricellular pollen with or without JA treatment. Data presented are means ± sd of three independent experiments (n > 300).
(C) Semi–in vivo pollen tube growth assay showing dau/DAU pollen tubes emerging from a Landsberg erecta pistil (top panel). mCherry-PTS1 displayed a punctate pattern in the DAU wild-type pollen tube but a diffuse pattern in the dau mutant pollen tube (bottom panels).
(D) Statistical analysis of germination ratios between dau and DAU pollen tubes (PTs) in semi–in vivo pollen tube growth assay, which represented the germination ability of dau mature pollen in the absence or presence of exogenous JA. Data presented are means ± sd of three independent experiments (n > 300).
(E) mCherry-PTS1 exhibited a punctate peroxisomal localization in Col-0 pollen but diffusely cytosolic localization with occasional large aggregates (arrows) in apem9-2 and apem9-3 mutant pollen.
(F) Semi–in vivo pollen tube germination assay showing pollen tubes of the apem9-2 mutant emerging from a Col-0 pistil. The wild-type and apem9-2 mutant pollen tubes can be distinguished by distinct patterns of mCherry-PTS1.
(G) Quantitative assessment of germination ratios between apem9 and wild-type pollen tubes in a semi–in vivo pollen tube growth assay. Mature or tricellular pollen from apem9-2 and apem9-3 plants were previously treated with or without JA. Data presented are means ± sd of three independent experiments (n > 1000).
**Student’s t test, P < 0.01; *Student’s t test, 0.01 < P < 0.05. Bars = 200 µm in top panel of (C) and (F) and 10 µm in bottom panels of (C) and (E).
To assess whether the increased TE of dau pollen is due to the increased germination ability on stigmas, we used a semi–in vivo pollen tube growth system (Qin et al., 2009) to monitor pollen germination. Pollen grains from dau/DAU qrt/qrt plants carrying the ProLat52:mCherry-PTS1 transgene, which distinguishes the mutant and wild-type pollen, were used to pollinate wild-type stigmas. The numbers of the dau mutant and wild-type pollen tubes were scored (Figure 7C). The ratio was expected to be close to 1.00 if the dau mutation did not affect pollen germination. As shown in Figure 7D, the ratio was ∼0.24 (n = 612) when mature pollen from dau/DAU qrt/qrt plants were used as pollen donors, confirming that pollen germination on the stigma was impaired in the dau mutant (Figure 1J). The ratio was increased to 0.43 (n = 575) when using mature pollen from JA-treated dau/DAU qrt/qrt plants (Figure 7D), indicating that the germination of dau pollen was enhanced by exogenous JA. This suggests that exogenous application of JA promotes the germination of dau pollen on the stigma and thus increases the male TE.
Furthermore, we analyzed the pollen phenotype of two heterozygous T-DNA insertion lines, namely, apem9-2 and apem9-3 (Goto et al., 2011), which are null alleles of the DAU/APEM9 gene. In apem9-2 and apem9-3 pollen, mCherry-PTS1 exhibited a diffusely cytosolic distribution (Figure 7E). The mature pollen from wild-type, apem9-2, and apem9-3 plants showed no significant differences in pollen germination in vitro (Supplemental Table 2). However, a semi–in vivo pollen tube growth assay (Figure 7F) showed that, in apem9-2 and apem9-3, the mature pollen grains were defective in germination on the stigma and the tricellular pollen in undehisced anthers had better germination ability than the mature pollen (n > 1000) (Figure 7G). Besides, the germination ability of the mature pollen and tricellular pollen was greatly enhanced after JA treatment (n > 1000) (Figure 7G). These data demonstrate that apem9-2 and apem9-3 mutants display similar phenotypes to the dau mutant.
PTS1-Dependent Protein Import and Peroxisome Structure Are Differentially Disrupted in pex Mutant Pollen
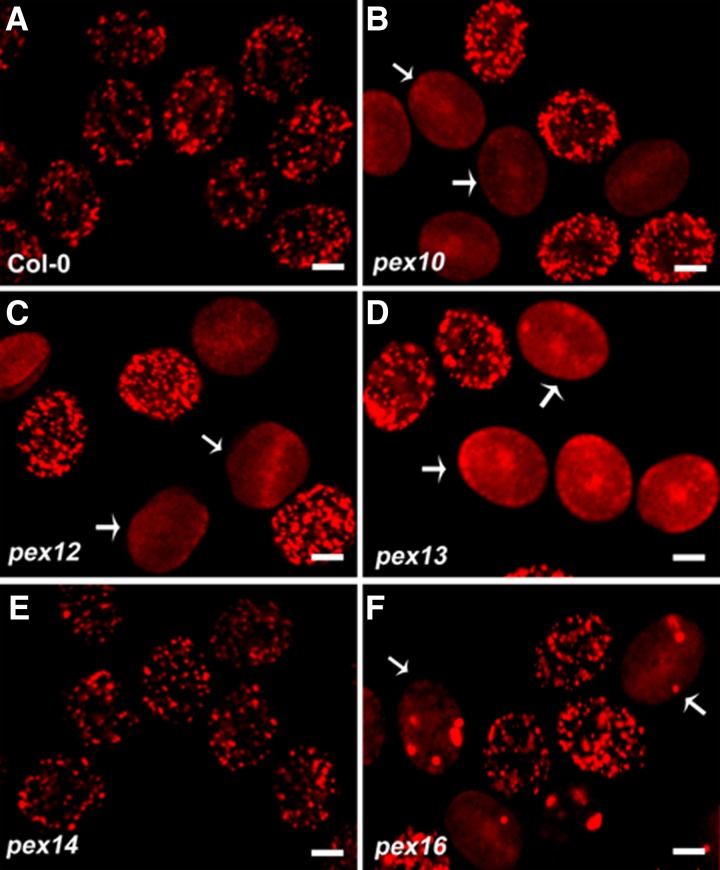
Previous studies showed that pex mutants are defective in PTS1-dependent protein import and peroxisome structure (Hayashi et al., 2000; Schumann et al., 2003; Fan et al., 2005; Mano et al., 2006; Nito et al., 2007; Boisson-Dernier et al., 2008; Monroe-Augustus et al., 2011). However, most of the observations were performed in leaf, root, or embryo cells, except for pex13 (Boisson-Dernier et al., 2008). Therefore, we first examined PTS1-dependent protein import in the pollen of pex10, pex12, pex13, pex14, and pex16 heterozygous plants. In wild-type pollen, peroxisomes appeared as dot-like structures (Figure 8A), while in pex10 and pex12 mutant pollen, mCherry-PTS1 appeared mainly cytosolic, with a few dot-like structures (Figures 8B and 8C), similar to that in PEX10 and PEX12 RNA interference (RNAi) lines (Nito et al., 2007). In pex13 mutant pollen, PTS1 import was severely disrupted (Figure 8D), as previously reported (Boisson-Dernier et al., 2008). In comparison, all of the fluorescent pollen grains from pex14 heterozygous plants uniformly displayed a punctate pattern (Figure 8E), although impaired PTS1 import was observed in the leaf and root cells of pex14 mutants (Hayashi et al., 2000; Monroe-Augustus et al., 2011). In pex16 mutant pollen, fluorescent spots representing mCherry-PTS1 were larger and fewer than those in the wild type, with weaker signal in the cytosol (Figure 8F), consistent with the observations in PEX16 RNAi lines (Nito et al., 2007). These data demonstrate that PTS1-dependent protein import is abolished in pex10, pex12, and pex13 pollen, unaffected in pex14 pollen, and impaired in pex16 pollen.
Figure 8.
PTS1-Dependent Protein Import in Pollen Grains from the Wild Type and pex Heterozygous Mutants.
(A) mCherry-PTS1 displayed punctate peroxisomal localization in wild-type pollen.
(B) to (D) mCherry-PTS1 displayed a diffuse pattern in pex10 (B), pex12 (C), and pex13 (D) pollen (white arrows).
(E) Peroxisomal localization of mCherry-PTS1 in pex14 pollen.
(F) Aggregated and reduced mCherry-PTS1 localization in pex16 pollen (white arrows).
Bars = 10 µm.
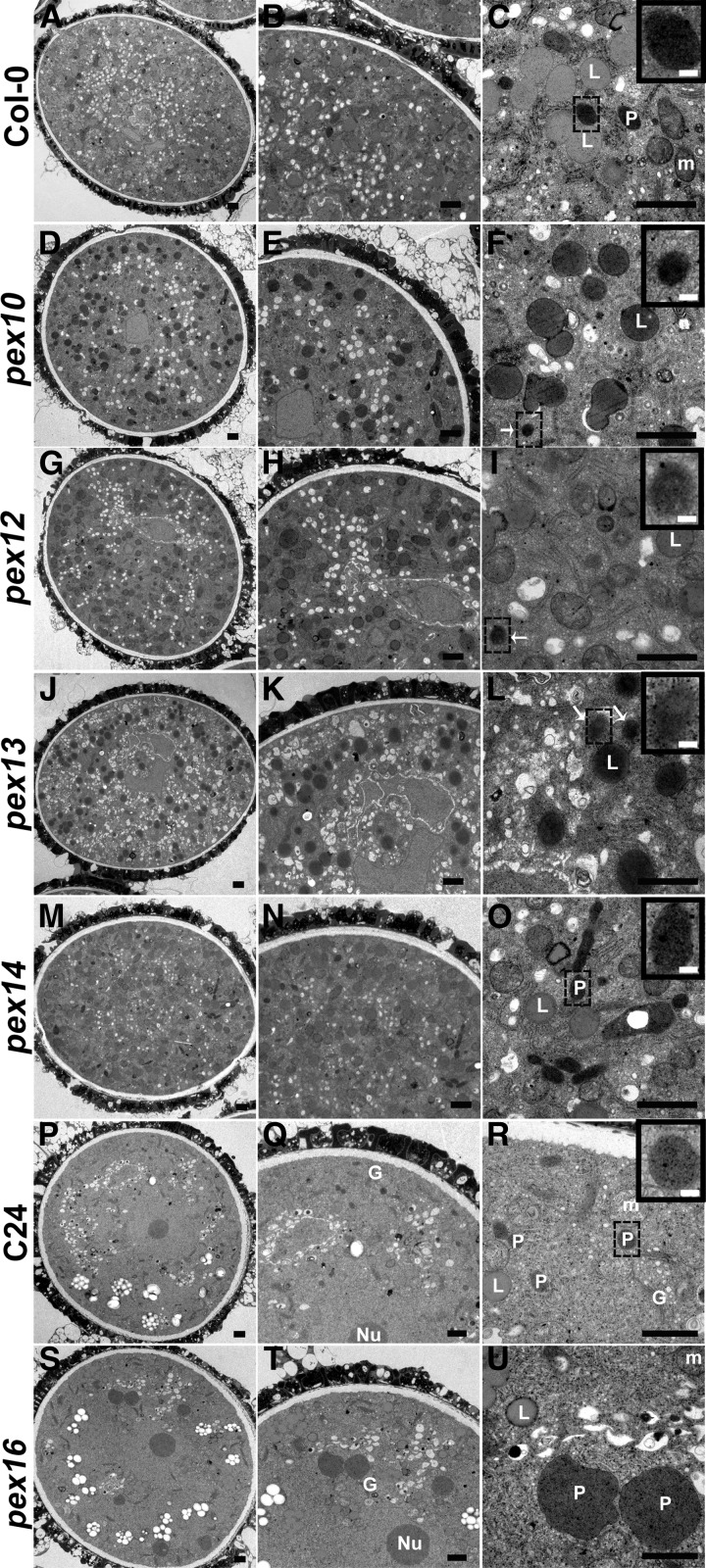
To investigate whether the mCherry-PTS1 patterns are associated with peroxisome biogenesis defects, the ultrastructures of the pex tricellular pollen were analyzed by TEM. The peroxisomes from wild-type pollen were darkly stained with a clear boundary after DAB staining (Figures 9A to 9C). In pex10, pex12, and pex13 mutant pollen, we could only scarcely observe the peroxisome-like structures lacking clear membrane (Figures 9F, 9I, and 9L). Besides, lipid bodies in pex10, pex12, and pex13 mutant pollen (Figure 9F, 9I, and 9L) were stained darker than the wild-type pollen (Figure 9C). This phenotype is in agreement with previous reports that peroxisome biogenesis and lipid bodies are impaired in pex10 and pex12 mutants (Schumann et al., 2003; Fan et al., 2005). In pex14 mutant pollen, heavily stained intact peroxisomes were present (Figure 9M to 9O), and there was no obviously morphological difference from those in wild-type pollen (Figures 9A to 9C). Since pex16 mutants are in the C24 background, we compared the ultrastructure of pollen from C24 and pex16 plants. Pollen peroxisomes from C24 plants were darkly stained with a smooth boundary (Figures 9P to 9R), while peroxisomes in pex16 mutant pollen were much larger and with an irregular boundary (Figures 9S to 9U), as reported in PEX16 RNAi lines (Nito et al., 2007). The TEM analysis indicated that the abolished PTS1-dependent protein import in pex10, pex12, and pex13 mutant pollen (Figures 8B to 8D) is most likely due to the defects in peroxisome assembly and integrity, and the impaired PTS1-dependent protein import in pex16 mutant pollen (Figure 8F) is likely caused by reduced and deformed peroxisomes.
Figure 9.
TEM of pex Tricellular Pollen Compared with Wild-Type Pollen.
(A) to (C) Observation of a wild-type pollen grain, showing mitochondria, lipid bodies, and heavily stained peroxisomes with a clear boundary (inset). The boxed region is enlarged as the inset.
(D) to (L) TEM micrographs of pex10 ([D] to [F]), pex12 ([G] to [I]), and pex13 ([J] to [L]) pollen. Note darkly stained peroxisome-like structures (white arrow and insets) and heavily stained lipid bodies. Insets depict enlarged views of boxed regions.
(M) to (O) Observation of a pex14 pollen grain, showing heavily stained peroxisomes with a clear boundary (inset). The inset is an enlarged view of the boxed region.
(P) to (R) Observation of a C24 wild-type pollen grain, showing mitochondria, lipid bodies, and heavily stained peroxisomes with a clear boundary (inset). The boxed region is enlarged as the inset.
(S) to (U) TEM micrographs of pex16 pollen, showing huge peroxisomes with an irregular boundary.
L, lipid body; m, mitochondrion; P, peroxisome, G, Golgi apparatus. Bars = 1 µm in (A) to (U) and 0.1 µm in the insets of (C), (F), (I), (L), (O), and (R).
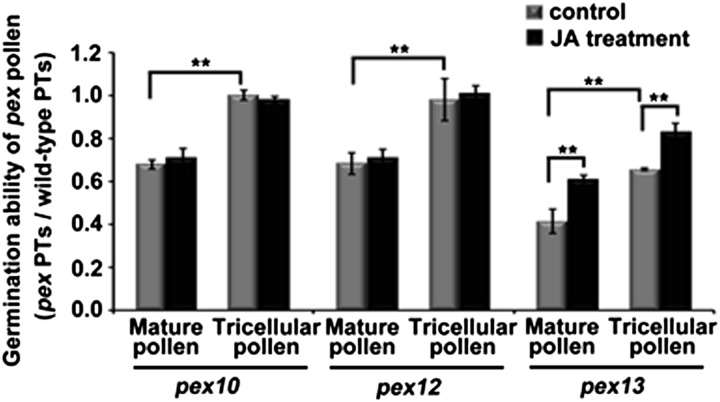
Exogenous Application of JA Can Partially Rescue the Male Sterility of pex13 but Not pex10 or pex12 Mutant
Since pex10, pex12, and pex13 mutant pollen display a similar peroxisome-defective phenotype as dau mutant pollen, a semi–in vivo pollen tube growth assay was performed to investigate the male TE. In pex10 and pex12 mutants, when mature pollen were used, the ratio of mutant to wild-type pollen tubes was around 0.68 (Figure 10, left and middle), indicating that pollen germination in pex10 and pex12 is not impaired as severely as in the dau mutant (Figure 7C); when using pex10 and pex12 tricellular pollen in undehisced anthers as pollen donors, the ratio was around 1.00, indicating that the tricellular pollen of pex10 and pex12 show no defect in pollen germination (Figure 10, left and middle). This indicates that undehisced mutant pollen perform better than mature pollen. In addition, exogenous JA had little effect on promoting pollen germination in pex10 and pex12 mutants (Figure 10, left and middle). However, when using pex13 mature pollen as donors, the ratio between mutant and wild-type pollen tube numbers was around 0.41 (Figure 10, right), indicating that in vivo pollen germination in pex13 mutants is dramatically impaired. Moreover, the pex13 tricellular pollen in undehisced anthers germinated better than pollen grains from dehisced anthers and the germination ability of pex13 tricellular and mature pollen were both promoted by exogenous JA (Figure 10). These results indicate that pex10 or pex12 mutation does not have remarkable effects on JA-mediated pollen germination, but pex13 plants show apparent defects in JA-mediated pollen maturation and in vivo germination.
Figure 10.
Statistical Comparison of Pollen Germination Ability in pex10, pex12, and pex13 Mutants.
Germination ratios between pex and wild-type pollen tubes were scored in a semi–in vivo pollen tube (PT) growth assay. Mature or tricellular pollen from pex10, pex12, and pex13 plants were previously treated with or without JA. Data are presented as mean values of sd from three independent experiments (n > 300). **Student’s t test, P < 0.01.
DISCUSSION
DAU Is a Key Regulator of Peroxisome Biogenesis and Matrix Protein Transport
Recently, Goto et al. (2011) reported that DAU/APEM9 was a peroxisomal membrane protein involved in PTS1 matrix protein import and associated with the PEX1-PEX6 complex. Our data suggested that DAU/APEM9 also plays important roles in peroxisome biogenesis and peroxisomal protein import, including membrane and matrix proteins. First, peroxisomes with clear membrane structures were not observed in dau pollen grains, and only a few peroxisome-like DAB-staining structures were present. Consistently, the import of PTS1- and PTS2-containing matrix proteins was impaired in the mutant. Second, the localization of peroxisomal membrane proteins was differentially affected in dau pollen. On one hand, PEX14-mCherry was unable to target to the peroxisomal membrane and appeared cytosolic in dau pollen. On the other hand, mCherry-PEX13 and mCherry-PEX16 appeared as punctuated structures, though with much reduced number in dau pollen. Furthermore, DAU interacts with PEX16, which has been implicated in early peroxisome biogenesis (Kim and Mullen, 2013). These findings suggest that DAU/APEM9 is most likely involved in early peroxisome biogenesis and required for matrix protein import.
DAU Has Two TMDs and Both Termini Face the Cytosol
DAU/APEM9 contains two hydrophobic regions that may serve as transmembrane domains. Goto et al. (2011) proposed that only the C-terminal hydrophobic region functions as a TMD. Our protease protection assays showed that DAU/APEM9 most likely contains two TMDs with both the N and C terminus extending to the cytosol. In addition, the C-terminal portion of DAU, containing the C-terminal TMD, was able to target specifically to peroxisomes, indicating the peroxisome membrane targeting signal indeed exists in or is adjacent to the C-terminal TMD. Furthermore, overexpression of DAU(267-333)-EGFP causes peroxisome elongation and tabulation. This is consistent with the previous report that peroxisome morphology and the localization of DAU/APEM9 were altered by a point mutation in the C-terminal TMD (Goto et al., 2011). The functional significance of the N-terminal TMD requires more investigation. Together, our data suggest that DAU/APEM9 most likely contains two TMDs and the C terminus is essential for its peroxisome targeting.
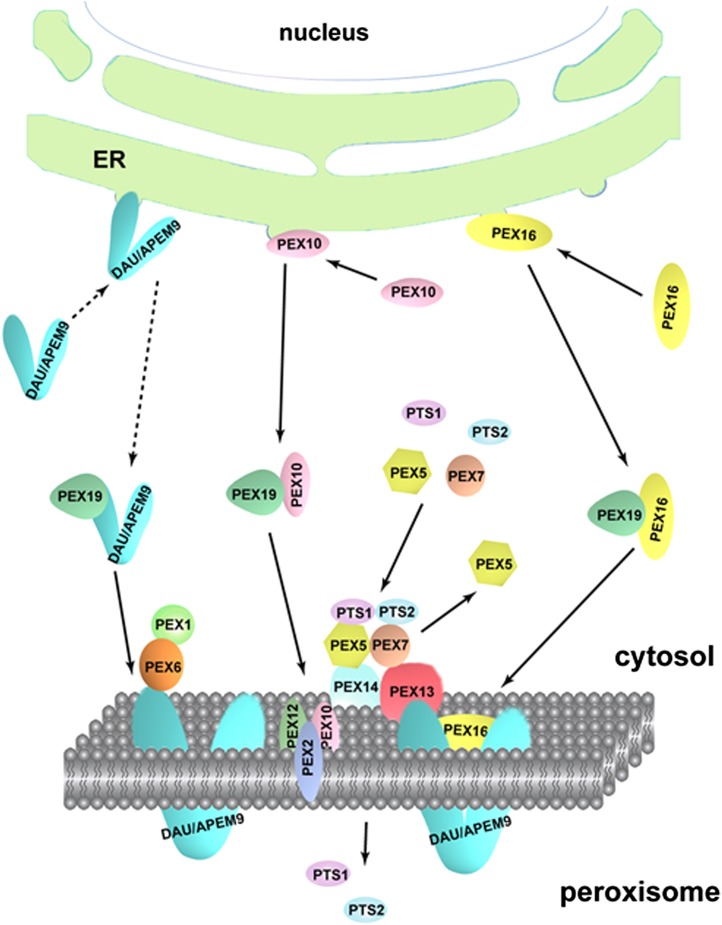
Model of DAU Function
Our data showed that DAU/APEM9 interacts with PEX13 and PEX16 and is required for peroxisome biogenesis and structure maintenance. The defect of matrix and membrane protein import in the mutant may be caused by the reduced number and disrupted membrane integrity of peroxisomes. Based on available data, a tentative model is proposed here to explain DAU/APEM9 function in peroxisome biogenesis and protein import (Figure 11). DAU/APEM9 first interacts with PEX19 in the cytosol as suggested previously (Goto et al., 2011) and then is transported to the peroxisomal membrane, probably via a similar mechanism as PEX16 or PEX10 (Hu et al., 2012). From our TEM results, it appeared that DAU possibly functions in the membrane assembly or membrane structure maintenance, synergistically with PEX13, because both dau and pex13 mutations impair peroxisome generation and disrupt the membrane integrity. Whether PEX10 and PEX12 also work together or independently with DAU and PEX13 needs further investigation. DAU/APEM9 interacts with PEX13 and PEX16 in the peroxisomal membrane. It is also essential for the targeting of PEX14 and PEX7 to the peroxisome and further for matrix protein import, but not essential for the targeting of PEX13 and PEX16. This further indicates that PEX16 and PEX13 are early PEXs assembled to peroxisomes. It has been shown that PEX14 and PEX13 accept matrix protein receptors PEX5 and PEX7 by direct interaction, respectively (Nito et al., 2002; Mano et al., 2006). Therefore, the matrix protein import defect may result from the defective targeting of PEX14 and reduced level of PEX13.
Figure 11.
Working Model of Peroxisomal Protein Import.
DAU/APEM9 is transported to the peroxisomal membrane by PEX19, probably via a similar mechanism as PEX16 and PEX10 (Hu et al., 2012). DAU/APEM9 regulates PTS1- and PTS2-containing peroxisomal matrix protein import via PEX14, which interacts directly with the PTS1 receptor PEX5 and another membrane protein PEX13, which binds to the PTS2 receptor PEX7. The docked receptor-cargo complex translocates the cargo PTS1- and PTS2-containing protein into the peroxisome matrix, and the receptors are recycled back to the cytosol in aid of the RING-finger proteins PEX2, PEX10, and PEX12 and the AAA-ATPase PEX1-PEX6. DAU/APEM9 tethers the PEX1-PEX6 complex to the peroxisomal membrane.
DAU/APEM9 Regulates Pollen Maturation and Germination via JA Biosynthesis
JA production is one of the important roles of plant peroxisomes. It is known that JA is required for anther dehiscence (Wilson et al., 2011) and pollen maturation (Ishiguro et al., 2001) and regulates a battery of genes, including 365 JA-regulated genes in pollen (Mandaokar et al., 2006). JA level is reduced in dau pollen and the in vivo pollen germination defect of dau mutants was amended by the exogenous application of JA, indicating that the male sterility of dau mutants partially resulted from JA deficiency. Consistently, PEX6 was shown to be involved in JA biosynthesis after wounding, possibly by affecting the import of the enzymes involved JA production into the peroxisome (Delker et al., 2007). Since peroxisome biogenesis and membrane integrity are disrupted in dau pollen, and the peroxisome targeting of PEX6 is abolished in the apem9 mutant (Goto et al., 2011), it is plausible to speculate that the import of OPR3 and other β-oxidation enzymes into the peroxisome might also be affected, resulting in JA deficiency. Taken together, we conclude that DAU/APEM9 regulates pollen maturation and in vivo germination via JA biosynthesis indirectly by affecting peroxisomal function. Because exogenous JA just partially rescues dau sterility, other abolished peroxisome function may also contribute to the dau pollen defect.
PEXs Play Different Roles in Pollen
PEXs are proteins required for peroxisome biogenesis and function. Although the basic design of peroxisome biogenesis and peroxisomal import machinery is conserved in eukaryotes, mutants of PEXs display different defects in peroxisome biogenesis, morphology, and function. Functional study of PEX genes in Arabidopsis somatic cells suggested that they can be divided into two distinct functional groups: Group 1, including PEX1, PEX2, PEX4, PEX5, PEX7, PEX10, PEX12, PEX13, and PEX14, whose mutation impairs peroxisome function due to misdistribution of peroxisomal matrix proteins in the cytosol; and Group 2, including PEX3, PEX11, PEX16, and PEX19, whose mutation causes reduced peroxisome function due to impaired peroxisome morphology (Nito et al., 2007). During pollen development, PEX also play differential roles. First, peroxisome biogenesis and matrix protein import defects in pex14 and pex16 pollen are quite different from those in dau, pex10, pex12, and pex13 pollen. Second, the loss of PEX16/SSE1 function causes deformed peroxisomes in the root cells of PEX16 knockdown plants (Nito et al., 2007) and the pex16 pollen (Figures 9P to 9U) and alters seed storage composition in shrunken sse1 seeds, which is lethal upon desiccation (Lin et al.,1999, 2004). Intriguingly, pex16 does not show defects in pollen germination, indicating the morphological changes of peroxisomes may be less detrimental to pollen. Third, the pex14/ped2 homozygous plants show reduced growth, while the pollen and homozygous embryo are viable although the matrix protein import is reduced, indicating that PEX14 plays an important but not essential role in peroxisomal function (Hayashi et al., 2000; Monroe-Augustus et al., 2011). Our results also show that the matrix protein import is not obviously affected in pex14 pollen in contrast with that of somatic cells, indicating that the function of PEX14 is not essential in pollen. Fourth, both pex10 and pex12 mutants show an embryo-lethal phenotype (Sparkes et al., 2003; Fan et al., 2005) and the integrity and numbers of peroxisomes are impaired in pex10 and pex12 pollen, suggesting that PEX10 and PEX12 are important during pollen and embryo development. In addition, gain-of-function mutation of pex2 interferes with peroxisome function in photomorphogenesis and development (Hu et al., 2002). PEX2, PEX10, and PEX12 all exhibit basal E3 ligase activity in yeast and plants and form a complex with enhanced activity in an E2-selective manner, suggesting that they may function synergistically (El Magraoui et al., 2012; Kaur et al., 2013). pex10 and pex12 show milder pollen defects than pex13 and dau, although all these mutants show peroxisome biogenesis and structural defect. This indicates that the peroxisome function in these mutants may be disrupted differently. The function of PEX10 and PEX12 might be, although unbelievable but not absolutely impossible, redundant to some degree in pollen. On the other hand, RING PEXs have been show to play different functions in peroxisome structure and matrix import by mutation in the Zn2+ binding motif (Prestele et al., 2010). Prestele et al. showed that PEX10-∆Zn causes deformed peroxisome shape, while such mutation in PEX2 does not cause peroxisome deformation, but matrix protein import defects. Interestingly, PEX12-∆Zn neither causes peroxisome defect nor impaired matrix import. Finally, the expression pattern and levels of PEXs are quite different in pollen (Supplemental Figure 5). Among the known PEXs, PEX13 and DAU are the most highly expressed, while PEX10, PEX12, PEX14, and PEX16 are weakly expressed in pollen. Additionally, Boisson-Dernier et al. (2008) reported that in the pex13/amc mutant, the male transmission is significantly reduced to 51% with a novel pollen tube reception phenotype only in selfed pistils, suggesting that the reduced male transmission is likely caused by defects in either pollen germination or pollen tube growth. We also observed reduced in vivo pollen germination in pex13/amc, while we did not observe abnormal pollen tube guidance or reception in the dau mutant.
In conclusion, we showed that DAU/APEM9 encodes a peroxisomal membrane protein with dual transmembrane domains, which is involved in peroxisome biogenesis and matrix protein import. DAU/APEM9 most likely functions via its interaction with PEX13 and PEX16. DAU/APEM9 is required for pollen maturation and in vivo germination via its role in peroxisomal function, which partially involves JA biosynthesis. DAU/APEM9 and peroxins most likely play distinct roles in pollen. Further study on functional specificity of these peroxins will provide insight into peroxisome biogenesis and their roles in plant development.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Landsberg erecta, Columbia-0 (Col-0), and C24 plants were grown in an air-conditioned room at 22°C under a 16-h-light/8-h-dark cycle. Tetrad pollen plants of dau/DAU qrt/qrt were obtained by crossing dau heterozygous plants to qrt1/qrt1 homozygous plants (Landsberg erecta background) (Preuss et al., 1994). The T-DNA insertion lines SALK_132193 (apem9-2), SALK_022380 (apem9-3), SALK_007838 (pex10), SALK_013612 (pex12), SALK_055083 (pex13), SALK_007441 (pex14), and CS6000 (pex16) were obtained from the ABRC.
Genetic Analysis
The screen of dau from Ds insertion lines was conducted as described previously by Sundaresan et al. (1995). Thermal asymmetric interlaced PCR was performed to isolate genomic sequences flanking the Ds according to previous reports (Liu et al., 1995). The insertion positions of T-DNA insertion lines were confirmed using the T-DNA left border primer LBa1 and gene-specific primers (Supplemental Table 3).
Light Microscopy
For light and fluorescent microscopy, specimens were observed using a Zeiss Axioskop II microscope, and images were acquired with a Cannon PowerShot G6. Staining assays with Alexander, DAPI, and aniline blue were performed as described previously (Johnson-Brousseau and McCormick, 2004).
For semithin sections, anthers were fixed with 4% glutaraldehyde in 25 mM sodium phosphate buffer, pH 6.8, and were kept in the fixative at 4°C from 4 h to overnight after infiltration. The samples were dehydrated with a conventional ethanol series with 30 min for each step and then infiltrated and embedded with Historesin according to the manufacturer’s instructions (Leica). Sections (5 to 6 μm) were made with a microtome (Leica). Before observation, 0.5 μg/mL of DAPI solution was added to the slides and stained for 30 min. The samples were then rinsed briefly and examined under a Zeiss Axioskop II microscope with epifluorescence optics.
Electron Microscopy
For scanning electron microscopy, pollen from dehiscing anthers was stuck onto double-sided tape. After critical point dry, the samples was coated with gold and observed with an S-3000N scanning electron microscope (Hitachi).
For TEM, anthers were fixed at 4°C for 8 to 12 h with 2.5% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.2. After three washes with cacodylate buffer, the anthers were postfixed in 1% buffered osmium tetroxide, washed three times in distilled water, and dehydrated in an ethanol series. Then, the buffer was exchanged with 100% propylene oxide, propylene oxide/Epon812 series, and 100% Epon812 for 2 d. Anthers were embedded in Epon812 and polymerized at 60°C. Ultrathin sections were stained with 1% uranyl acetate and lead citrate. Specimens were examined using a JEM-1400 electron microscope (JEOL).
For catalase detection, anthers were fixed at 4°C for 8 to 12 h with 2.5% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.2. After three washes with cacodylate buffer, anthers were incubated for 120 min at room temperature in the dark in a solution containing 0.2% of DAB (Sigma-Aldrich) and 0.02% H2O2 in 50 mM Tris-HCl, pH 3.9. After being rinsed with cacodylate buffer, the anthers were postfixed in 1% buffered osmium tetroxide, as described above.
Cloning of the DAU Genomic Fragment
A 2.8-kb DAU (At3g10572) genomic fragment (from 668 bp upstream of the start codon to 201 bp downstream of the DAU stop codon) was cloned into pCAMBIA1300 (Cambia) at the XbaI and KpnI sites, and the construct was verified by sequencing.
GUS Activity Assay
A 668-bp fragment upstream of the ATG start codon and a 201-bp fragment were inserted separately into pBI101 (Clontech) flanking the GUS reporter gene. The method for GUS staining was described previously (Ding et al., 2006).
Subcellular Localization
The 35S promoter and the NOS terminator sequences were inserted into pCAMBIA1300 to produce pCAM1300-35S-NOS. N- and C-terminal EGFP fragments were inserted into pCAM1300-35S-NOS to give rise to pCAM1300-35S-N-EGFP-NOS and pCAM1300-35S-C-EGFP-NOS. To produce 35S-EGFP-DAU and 35S-DAU-EGFP, the full-length coding sequence of DAU was cloned into pCAM1300-35S-N-EGFP-NOS and pCAM1300-35S-C-EGFP-NOS at PstI and XbaI sites. DAU(1-115) was cloned into pCAM1300-35S-N-EGFP-NOS at SalI and BamHI sites to produce 35S-EGFP-DAU(1-115). DAU(267-333) was cloned into pCAM1300-35S-C-EGFP-NOS at PstI and BamHI sites to produce 35S-DAU(267-333)-EGFP.
mCherry-PTS1 plasmid, initially named px-rk CD3-983 (Nelson et al., 2007), was obtained from the ABRC. The N-terminal mCherry fragment without a stop codon was amplified and inserted into pCAM1300-35S-NOS following digestion with SalI to produce pCAM1300-35S-N-mCherry-NOS. A C-terminal mCherry fragment without an initiation codon was amplified and inserted into pCAM1300-35S-NOS following digestion with SmaI to produce pCAM1300-35S-C-mCherry-NOS.
To produce 35S-mCherry-PEX7, 35S-mCherry-PEX13, and 35S-mCherry-PEX16, the full-length coding sequence of PEX13 and PEX16 were amplified with gene-specific primers (Supplemental Table 3) and inserted into pCAM1300-35S-N-mCherry-NOS. To produce 35S-PTS2-mCherry, 35S-PEX12- mCherry, and 35S-PEX14-mCherry, the PTS2 signal sequence and the full-length coding sequence of PEX12 and PEX14 were amplified with gene-specific primers (Supplemental Table 3) and inserted into pCAM1300-35S-C-mCherry-NOS following digestion with SalI and SmaI.
The above constructs were transformed into Agrobacterium tumefaciens strain GV3101. Bacterial suspensions were infiltrated into leaves of 7-week-old Nicotiana benthamiana plants using a needleless syringe. After infiltration, plants were grown in 16 h light/8 h darkness for 3 d at 22°C. The leaves were observed using a Zeiss LSM510 META laser scanning microscope.
The Lat52 promoter sequence was amplified and inserted into the pCAMBIA1300-NOS construct at HindIII and PstI sites to produce pCAM1300-Lat52pro-NOS. The mCherry-PTS1 fragment was amplified from px-rk CD3-983 using primers N-mCherry-F and PTS1-R and inserted into pCAM1300-Lat52pro-NOS to produce Lat52-mCherry-PTS1. mCherry-PEX7, mCherry-PEX13, mCherry-PEX16, PTS2-mCherry, and PEX14-mCherry were inserted into pCAM1300-Lat52pro-NOS to produce Lat52-mCherry-PEX7, Lat52-mCherry-PEX13, Lat52-mCherry-PEX16, Lat52-PTS2-mCherry, and Lat52-PEX14-mCherry. These constructs were transformed into Agrobacterium strain GV3101 and transformed into dau/DAU qrt/qrt and DAU/DAU qrt/qrt plants. Images of transgenic pollen were captured with a Zeiss LSM510 META laser scanning microscope.
Whole Mount Clearing of Embryos
The method for phenotypic analysis of mutant embryos was described previously (Ding et al., 2006).
Proteinase Protection Assay and Coimmunoprecipitation
The peroxisome isolation and proteinase protection assays were performed according to Lisenbee et al. (2003). To produce 35S-FLAG-PEX13 and 35S-FLAG-PEX16, the full-length PEX13 and PEX16 coding sequences were fused in-frame to a 3 X FLAG tag and then inserted into the pWM101 plasmid (Ding et al., 2006) between KpnI and XbaI. A coimmunoprecipitation assay was performed as reported (Zhang and Hu, 2010). The coding sequence of the first 268 amino acids of DAU was cloned into pET28a (Novagen). Purified 6xHis-DAU(1-168) recombinant protein was used to immunize mice to produce the DAU antibody.
Firefly Luciferase Complementation Imaging Assay
To generate DAU-NLuc, PEX13-NLuc, and PEX16-NLuc, the corresponding coding sequences were subcloned into pCAMBIA-NLuc (Chen et al., 2008) at the KpnI and SalI sites. To produce CLuc-DAU, CLuc-PEX13, and CLuc-PEX16, the corresponding coding sequences were subcloned into pCAMBIA-CLuc at the KpnI and SalI sites. The constructs were transformed into Agrobacterium strain GV3101. Bacterial suspensions were infiltrated into leaves of 7-week-old N. benthamiana plants using a needleless syringe. After infiltration, plants were grown with 16 h light/8 h darkness for 3 d at 22°C. Images were captured by a low-light cooled charge-coupled device imaging apparatus (NightOWL II LB983 with indiGO software).
In Situ Hybridization
In situ hybridization and signal detection were performed according to previous reports (Shi et al., 2005; Ding et al., 2006).
Semi–in Vivo Pollen Germination Assay
The method was modified from (Palanivelu and Preuss, 2006). After pollination, pistils were cut off and placed horizontally on solid pollen germination medium (Fan et al., 2001) at 22°C for 4 to 6 h. Pollen tubes emerging from the pistils were visualized using a Zeiss LSM510 META laser scanning microscope.
Measurement of JA
Fresh mature pollen from dau/DAU qrt/qrt and DAU/DAU qrt/qrt plants were collected separately using a vacuum cleaner (Johnson-Brousseau and McCormick, 2004). The extraction and quantification of JA was performed as described previously (Fu et al., 2012).
Application of Methyl Jasmonate
Methyl jasmonate application was conducted as described (Ishiguro et al., 2001).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL or Arabidopsis Genome Initiative database under the following accession numbers: DAU (At3g10572), PEX7 (At1g29260), PEX12 (At3g04460), PEX13 (At3g07560), PEX14 (At5g62810), and PEX16 (At2g45690).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Mutant Embryos Were Arrested before the Heart Stage.
Supplemental Figure 2. DAPI Staining of dau/DAU qrt/qrt Quartet Pollen.
Supplemental Figure 3. Peroxisomal Localization of DAU-EGFP.
Supplemental Figure 4. The Specificity Determination of DAU Antibody by Immunoblot.
Supplemental Figure 5. Expression Levels of PEX10, PEX12, and PEX13 in Pollen Development and Germination.
Supplemental Table 1. Complementation Analysis of ProDAU:DAU-EGFP Transgenic Plants.
Supplemental Table 2. In Vitro Germination Ratio of apem9-2 and apem9-2.
Supplemental Table 3. Sequences of Primers Used in This Work.
Supplementary Material
Acknowledgments
We thank De Ye at the China Agricultural University for initial help in the mutant screen. We thank the expertise of Jinfang Chu, Xiaohong Sun, and Cunyu Yan (National Centre for Plant Gene Research, Beijing, and Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China) in JA measurement. We also thank Kang Chong (Institute of Botany), Yongbiao Xue (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences), and Jianping Hu (Michigan State University) for invaluable suggestions and support. W.-C.Y. was supported by a grant (2007CB947600) from the Ministry of Science and Technology, China, and projects (30830063 and 30921003) from National Science Foundation of China.
AUTHOR CONTRIBUTIONS
X.-R.L., H.-J.L., L.Y., and W.-C.Y. designed the experiments and analyzed the data. L.Y. was involved in initial phenotypic characterization. X.-R.L. performed the peroxisome analysis. H.-J.L. carried out the coimmunoprecipitation experiment. M.L., J.L., and D.-Q.S. provided assistance during the experimentation. X.-R.L., H.-J.L., L.Y., and W.-C.Y. wrote the article.
Glossary
- JA
jasmonic acid
- ER
endoplasmic reticulum
- Kanr
kanamycin-resistant
- Kans
kanamycin-sensitive
- DAPI
4′,6-diamidino-2-phenylindole
- TEM
transmission electron microscopy
- DAB
3,3′-diaminobenzidine tetrahydrochloride
- TMD
transmembrane domains
- TE
transmission efficiency
- RNAi
RNA interference
- Col-0
Columbia-0
Footnotes
Online version contains Web-only data.
References
- Alexander M.P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44: 117–122. [DOI] [PubMed] [Google Scholar]
- Bechtold N., Pelletier G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82: 259–266. [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier A., Frietsch S., Kim T.H., Dizon M.B., Schroeder J.I. (2008). The peroxin loss-of-function mutation abstinence by mutual consent disrupts male-female gametophyte recognition. Curr. Biol. 18: 63–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J. (2009). Jasmonate passes muster: A receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60: 183–205. [DOI] [PubMed] [Google Scholar]
- Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., Tang X., Zhou J.M. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146: 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copenhaver G.P., Keith K.C., Preuss D. (2000). Tetrad analysis in higher plants. A budding technology. Plant Physiol. 124: 7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammai V., Subramani S. (2001). The human peroxisomal targeting signal receptor, Pex5p, is translocated into the peroxisomal matrix and recycled to the cytosol. Cell 105: 187–196. [DOI] [PubMed] [Google Scholar]
- Delker C., Zolman B.K., Miersch O., Wasternack C. (2007). Jasmonate biosynthesis in Arabidopsis thaliana requires peroxisomal beta-oxidation enzymes—Additional proof by properties of pex6 and aim1. Phytochemistry 68: 1642–1650. [DOI] [PubMed] [Google Scholar]
- Ding Y.H., Liu N.Y., Tang Z.S., Liu J., Yang W.C. (2006). Arabidopsis GLUTAMINE-RICH PROTEIN23 is essential for early embryogenesis and encodes a novel nuclear PPR motif protein that interacts with RNA polymerase II subunit III. Plant Cell 18: 815–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Magraoui F., Bäumer B.E., Platta H.W., Baumann J.S., Girzalsky W., Erdmann R. (2012). The RING-type ubiquitin ligases Pex2p, Pex10p and Pex12p form a heteromeric complex that displays enhanced activity in an ubiquitin conjugating enzyme-selective manner. FEBS J. 279: 2060–2070. [DOI] [PubMed] [Google Scholar]
- Fan J., Quan S., Orth T., Awai C., Chory J., Hu J. (2005). The Arabidopsis PEX12 gene is required for peroxisome biogenesis and is essential for development. Plant Physiol. 139: 231–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L.M., Wang Y.F., Wang H., Wu W.H. (2001). In vitro Arabidopsis pollen germination and characterization of the inward potassium currents in Arabidopsis pollen grain protoplasts. J. Exp. Bot. 52: 1603–1614. [PubMed] [Google Scholar]
- Fu J., Chu J., Sun X., Wang J., Yan C. (2012). Simple, rapid, and simultaneous assay of multiple carboxyl containing phytohormones in wounded tomatoes by UPLC-MS/MS using single SPE purification and isotope dilution. Anal. Sci. 28: 1081–1087. [DOI] [PubMed] [Google Scholar]
- Ghaedi K., Tamura S., Okumoto K., Matsuzono Y., Fujiki Y. (2000). The peroxin pex3p initiates membrane assembly in peroxisome biogenesis. Mol. Biol. Cell 11: 2085–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto S., Mano S., Nakamori C., Nishimura M. (2011). Arabidopsis ABERRANT PEROXISOME MORPHOLOGY9 is a peroxin that recruits the PEX1-PEX6 complex to peroxisomes. Plant Cell 23: 1573–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götte K., Girzalsky W., Linkert M., Baumgart E., Kammerer S., Kunau W.H., Erdmann R. (1998). Pex19p, a farnesylated protein essential for peroxisome biogenesis. Mol. Cell. Biol. 18: 616–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grou C.P., Carvalho A.F., Pinto M.P., Alencastre I.S., Rodrigues T.A., Freitas M.O., Francisco T., Sá-Miranda C., Azevedo J.E. (2009). The peroxisomal protein import machinery—A case report of transient ubiquitination with a new flavor. Cell. Mol. Life Sci. 66: 254–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M., Nito K., Toriyama-Kato K., Kondo M., Yamaya T., Nishimura M. (2000). AtPex14p maintains peroxisomal functions by determining protein targeting to three kinds of plant peroxisomes. EMBO J. 19: 5701–5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M., Yagi M., Nito K., Kamada T., Nishimura M. (2005). Differential contribution of two peroxisomal protein receptors to the maintenance of peroxisomal functions in Arabidopsis. J. Biol. Chem. 280: 14829–14835. [DOI] [PubMed] [Google Scholar]
- Hu J., Aguirre M., Peto C., Alonso J., Ecker J., Chory J. (2002). A role for peroxisomes in photomorphogenesis and development of Arabidopsis. Science 297: 405–409. [DOI] [PubMed] [Google Scholar]
- Hu J., Baker A., Bartel B., Linka N., Mullen R.T., Reumann S., Zolman B.K. (2012). Plant peroxisomes: Biogenesis and function. Plant Cell 24: 2279–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro S., Kawai-Oda A., Ueda J., Nishida I., Okada K. (2001). The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13: 2191–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson-Brousseau S.A., McCormick S. (2004). A compendium of methods useful for characterizing Arabidopsis pollen mutants and gametophytically-expressed genes. Plant J. 39: 761–775. [DOI] [PubMed] [Google Scholar]
- Kandasamy M.K., Nasrallah J.B., Nasrallah M.E. (1994). Pollen-pistil interactions and developmental regulation of pollen tube growth in Arabidopsis. Development 120: 3405–3418. [Google Scholar]
- Karnik S.K., Trelease R.N. (2005). Arabidopsis peroxin 16 coexists at steady state in peroxisomes and endoplasmic reticulum. Plant Physiol. 138: 1967–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim P.K., Mullen R.T. (2013). PEX16: A multifaceted regulator of peroxisome biogenesis. Front. Physiol. 4: 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim P.K., Mullen R.T., Schumann U., Lippincott-Schwartz J. (2006). The origin and maintenance of mammalian peroxisomes involves a de novo PEX16-dependent pathway from the ER. J. Cell Biol. 173: 521–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur N., Zhao Q., Xie Q., Hu J. (2013). Arabidopsis RING peroxins are E3 ubiquitin ligases that interact with two homologous ubiquitin receptor proteins(F). J. Integr. Plant Biol. 55: 108–120. [DOI] [PubMed] [Google Scholar]
- Lin Y., Cluette-Brown J.E., Goodman H.M. (2004). The peroxisome deficient Arabidopsis mutant sse1 exhibits impaired fatty acid synthesis. Plant Physiol. 135: 814–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Sun L., Nguyen L.V., Rachubinski R.A., Goodman H.M. (1999). The Pex16p homolog SSE1 and storage organelle formation in Arabidopsis seeds. Science 284: 328–330. [DOI] [PubMed] [Google Scholar]
- Lisenbee C.S., Heinze M., Trelease R.N. (2003). Peroxisomal ascorbate peroxidase resides within a subdomain of rough endoplasmic reticulum in wild-type Arabidopsis cells. Plant Physiol. 132: 870–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.G., Mitsukawa N., Oosumi T., Whittier R.F. (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8: 457–463. [DOI] [PubMed] [Google Scholar]
- Lorenzo C., Lucas M., Vivo A., De Felipe M. (1990). Effect of nitrate on peroxisome ultrastructure and catalase activity in nodules of Lupinus albus L. cv. Multolupa. J. Exp. Bot. 41: 1573–1578. [Google Scholar]
- Mandaokar A., Thines B., Shin B., Lange B.M., Choi G., Koo Y.J., Yoo Y.J., Choi Y.D., Choi G., Browse J. (2006). Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. Plant J. 46: 984–1008. [DOI] [PubMed] [Google Scholar]
- Mano S., Nakamori C., Nito K., Kondo M., Nishimura M. (2006). The Arabidopsis pex12 and pex13 mutants are defective in both PTS1- and PTS2-dependent protein transport to peroxisomes. Plant J. 47: 604–618. [DOI] [PubMed] [Google Scholar]
- Matsuzaki T., Fujiki Y. (2008). The peroxisomal membrane protein import receptor Pex3p is directly transported to peroxisomes by a novel Pex19p- and Pex16p-dependent pathway. J. Cell Biol. 183: 1275–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConn M., Browse J. (1996). The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 8: 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe-Augustus M., Ramón N.M., Ratzel S.E., Lingard M.J., Christensen S.E., Murali C., Bartel B. (2011). Matrix proteins are inefficiently imported into Arabidopsis peroxisomes lacking the receptor-docking peroxin PEX14. Plant Mol. Biol. 77: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen R.T., Trelease R.N. (2006). The ER-peroxisome connection in plants: Development of the “ER semi-autonomous peroxisome maturation and replication” model for plant peroxisome biogenesis. Biochim. Biophys. Acta 1763: 1655–1668. [DOI] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136. [DOI] [PubMed] [Google Scholar]
- Nito K., Hayashi M., Nishimura M. (2002). Direct interaction and determination of binding domains among peroxisomal import factors in Arabidopsis thaliana. Plant Cell Physiol. 43: 355–366. [DOI] [PubMed] [Google Scholar]
- Nito K., Kamigaki A., Kondo M., Hayashi M., Nishimura M. (2007). Functional classification of Arabidopsis peroxisome biogenesis factors proposed from analyses of knockdown mutants. Plant Cell Physiol. 48: 763–774. [DOI] [PubMed] [Google Scholar]
- Page D.R., Grossniklaus U. (2002). The art and design of genetic screens: Arabidopsis thaliana. Nat. Rev. Genet. 3: 124–136. [DOI] [PubMed] [Google Scholar]
- Palanivelu R., Preuss D. (2006). Distinct short-range ovule signals attract or repel Arabidopsis thaliana pollen tubes in vitro. BMC Plant Biol. 6: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.H., Halitschke R., Kim H.B., Baldwin I.T., Feldmann K.A., Feyereisen R. (2002). A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 31: 1–12. [DOI] [PubMed] [Google Scholar]
- Prestele J., Hierl G., Scherling C., Hetkamp S., Schwechheimer C., Isono E., Weckwerth W., Wanner G., Gietl C. (2010). Different functions of the C3HC4 zinc RING finger peroxins PEX10, PEX2, and PEX12 in peroxisome formation and matrix protein import. Proc. Natl. Acad. Sci. USA 107: 14915–14920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss D., Rhee S.Y., Davis R.W. (1994). Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science 264: 1458–1460. [DOI] [PubMed] [Google Scholar]
- Qin Y., Leydon A.R., Manziello A., Pandey R., Mount D., Denic S., Vasic B., Johnson M.A., Palanivelu R. (2009). Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, pointing to genes critical for growth in a pistil. PLoS Genet. 5: e1000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramón N.M., Bartel B. (2010). Interdependence of the peroxisome-targeting receptors in Arabidopsis thaliana: PEX7 facilitates PEX5 accumulation and import of PTS1 cargo into peroxisomes. Mol. Biol. Cell 21: 1263–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudall P.J., Bateman R.M. (2007). Developmental bases for key innovations in the seed-plant microgametophyte. Trends Plant Sci. 12: 317–326. [DOI] [PubMed] [Google Scholar]
- Sanders P.M., Lee P.Y., Biesgen C., Boone J.D., Beals T.P., Weiler E.W., Goldberg R.B. (2000). The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12: 1041–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann U., Wanner G., Veenhuis M., Schmid M., Gietl C. (2003). AthPEX10, a nuclear gene essential for peroxisome and storage organelle formation during Arabidopsis embryogenesis. Proc. Natl. Acad. Sci. USA 100: 9626–9631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi D.Q., Liu J., Xiang Y.H., Ye D., Sundaresan V., Yang W.C. (2005). SLOW WALKER1, essential for gametogenesis in Arabidopsis, encodes a WD40 protein involved in 18S ribosomal RNA biogenesis. Plant Cell 17: 2340–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh T., Hayashi M., Mano S., Arai Y., Goto S., Nishimura M. (2009). Molecular components required for the targeting of PEX7 to peroxisomes in Arabidopsis thaliana. Plant J. 60: 488–498. [DOI] [PubMed] [Google Scholar]
- Sparkes I.A., Brandizzi F., Slocombe S.P., El-Shami M., Hawes C., Baker A. (2003). An Arabidopsis pex10 null mutant is embryo lethal, implicating peroxisomes in an essential role during plant embryogenesis. Plant Physiol. 133: 1809–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A., Browse J. (2000). The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA 97: 10625–10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan V., Springer P., Volpe T., Haward S., Jones J.D., Dean C., Ma H., Martienssen R. (1995). Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9: 1797–1810. [DOI] [PubMed] [Google Scholar]
- Swanson R., Edlund A.F., Preuss D. (2004). Species specificity in pollen-pistil interactions. Annu. Rev. Genet. 38: 793–818. [DOI] [PubMed] [Google Scholar]
- Taylor L.P., Hepler P.K. (1997). Pollen germination and tube growth. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48: 461–491. [DOI] [PubMed] [Google Scholar]
- Turner J.G., Ellis C., Devoto A. (2002). The jasmonate signal pathway. Plant Cell 14 (suppl.): S153–S164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Malek B., van der Graaff E., Schneitz K., Keller B. (2002). The Arabidopsis male-sterile mutant dde2–2 is defective in the ALLENE OXIDE SYNTHASE gene encoding one of the key enzymes of the jasmonic acid biosynthesis pathway. Planta 216: 187–192. [DOI] [PubMed] [Google Scholar]
- Wilson Z.A., Song J., Taylor B., Yang C. (2011). The final split: The regulation of anther dehiscence. J. Exp. Bot. 62: 1633–1649. [DOI] [PubMed] [Google Scholar]
- Zhang X., Hu J. (2010). The Arabidopsis chloroplast division protein DYNAMIN-RELATED PROTEIN5B also mediates peroxisome division. Plant Cell 22: 431–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman B.K., Monroe-Augustus M., Silva I.D., Bartel B. (2005). Identification and functional characterization of Arabidopsis PEROXIN4 and the interacting protein PEROXIN22. Plant Cell 17: 3422–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.