This work shows that the ubiquitin receptor DA1 genetically and physically interacts with the ubiquitin-specific protease UBP15 to regulate seed size in Arabidopsis, suggesting that DA1 and UBP15 are promising targets for crop improvement.
Abstract
Although the control of organ size is a fundamental question in developmental biology, little is known about the genetic and molecular mechanisms that determine the final size of seeds in plants. We previously demonstrated that the ubiquitin receptor DA1 acts synergistically with the E3 ubiquitin ligases DA2 and ENHANCER1 OF DA1 (EOD1)/BIG BROTHER to restrict seed growth in Arabidopsis thaliana. Here, we describe UBIQUITIN-SPECIFIC PROTEASE15 (UBP15), encoded by SUPPRESSOR2 OF DA1 (SOD2), which acts maternally to regulate seed size by promoting cell proliferation in the integuments of ovules and developing seeds. The sod2/ubp15 mutants form small seeds, while overexpression of UBP15 increases seed size of wild-type plants. Genetic analyses indicate that UBP15 functions antagonistically in a common pathway with DA1 to influence seed size, but does so independently of DA2 and EOD1. Further results reveal that DA1 physically associates with UBP15 in vitro and in vivo and modulates the stability of UBP15. Therefore, our findings establish a genetic and molecular framework for the regulation of seed size by four ubiquitin-related proteins DA1, DA2, EOD1, and UBP15 and suggest that they are promising targets for increasing seed size in crops.
INTRODUCTION
Seed size in flowering plants is crucial for evolutionary fitness and is also an important agronomic trait (Alonso-Blanco et al., 1999; Moles et al., 2005; Fan et al., 2006; Song et al., 2007; Orsi and Tanksley, 2009; Gegas et al., 2010; Linkies et al., 2010). In flowering plants, a seed consists of three major components: the embryo, the endosperm, and the seed coat, each with different genetic compositions. Seed development involves a double-fertilization event. One sperm nucleus fertilizes the egg to produce the diploid embryo, while the other sperm nucleus fertilizes the central cell to form the triploid endosperm (Lopes and Larkins, 1993; Reiser and Fischer, 1993). The outermost layer of the seed is seed coat, which differentiates from maternal integuments. Therefore, the size of a seed is regulated by the coordinated growth of the embryo, endosperm, and maternal tissue.
Several factors that act maternally to regulate seed size have been described in Arabidopsis thaliana. For example, KLUH/CYTOCHROME P450 78A5 (CYP78A5) influences seed size by promoting cell proliferation in the maternal integuments (Adamski et al., 2009), while AUXIN RESPONSE FACTOR2 (ARF2)/MEGAINTEGUMENTA affects seed growth by restricting cell proliferation in the integuments (Schruff et al., 2006). By contrast, TRANSPARENT TESTA GLABRA2 (TTG2) promotes seed growth by increasing cell elongation in the maternal integuments (Garcia et al., 2005). APETALA2 (AP2) may repress seed growth by restricting cell elongation in the integuments (Jofuku et al., 2005; Ohto et al., 2005, 2009). Overexpression of EOD3/CYP78A6 promotes both cell proliferation and cell elongation in the maternal integuments, resulting in large seeds (Fang et al., 2012). Seed size is also influenced by zygotic tissues. Mutations in HAIKU (IKU) or MINISEED3 (MINI3) reduce seed size due to precocious cellularization of the endosperm (Garcia et al., 2003; Luo et al., 2005; Wang et al., 2010). SHORT HYPOCOTYL UNDER BLUE1 (SHB1) promotes seed growth by increasing endosperm proliferation (Zhou et al., 2009). SHB1 associates with the promoters of MINI3 and IKU2, and this association requires MINI3 (Kang et al., 2013). A recent study has shown that cytokinin acts downstream of the IKU pathway to regulate seed size (Li et al., 2013). In addition, the endosperm growth is influenced by epigenetic mechanisms associated with parent-of-origin effects (Scott et al., 1998; Xiao et al., 2006).
The ubiquitin pathway has been known to play an important part in seed size determination in plants. The ubiquitin receptor DA1 acts maternally to affect seed growth by restricting cell proliferation in the integuments (Li et al., 2008; Xia et al., 2013). In a screen for modifiers of da1-1, we identified enhancer of da1-1 (eod1), which has a mutation in the E3 ubiquitin ligase BIG BROTHER (BB) (Disch et al., 2006; Li et al., 2008). DA1 functions synergistically with EOD1 to restrict seed growth (Li et al., 2008). Similarly, we recently reported that DA1 acts synergistically with another E3 ubiquitin ligase DA2 to regulate seed growth by restricting cell division in the maternal integuments (Xia et al., 2013). Interestingly, DA2 shares high similarity with rice (Oryza sativa) GRAIN WIDTH AND WEIGHT2 (GW2), encoded by a quantitative trait locus for grain size, which influences grain growth by restricting cell proliferation (Song et al., 2007; Xia et al., 2013). Another quantitative trait locus, SEED WIDTH ON CHROMOSOME 5 (qSW5/GW5), affects grain width by limiting cell proliferation in rice (Shomura et al., 2008; Weng et al., 2008). The qSW5/GW5 gene encodes an unknown protein that interacts physically with polyubiquitin, suggesting that qSW5/GW5 might be related to the ubiquitin-proteasome protein degradation pathway (Weng et al., 2008). However, the target substrates of these ubiquitin-related regulators remain to be discovered.
To identify the downstream targets of DA1, we isolated suppressors of the large-seed phenotype of da1-1. Here, we describe sod2 mutants, which suppress the seed size phenotype of da1-1. SOD2 encodes the ubiquitin-specific protease UBP15, which has known roles in leaf development (Liu et al., 2008) but no previously identified function in seed size determination. UBP15 influences seed size by promoting cell proliferation in the maternal integuments. Genetic analyses show that SOD2/UBP15 functions in a common pathway with the ubiquitin receptor DA1 to regulate seed growth. We further demonstrate that DA1 physically interacts with UBP15 and modulates the stability of UBP15. Thus, our findings define the genetic and molecular mechanisms of four ubiquitin-related proteins, DA1, DA2, EOD1, and UBP15, in seed size determination.
RESULTS
The sod2 Mutations Suppress the Seed Size Phenotype of da1-1
To identify other components in the DA1 pathway, we initiated a genetic screen for modifiers of da1-1 in seed size. From this screen, we identified several suppressors of da1-1 (sod) from ethyl methanesulfonate–treated M2 populations of da1-1. We designated two of these suppressors sod2-1 and sod2-2, which are allelic to each other. The sod2 da1-1 seeds were smaller and lighter than da1-1 seeds (Figures 1A to 1C and 1J). In addition, sod2 da1-1 plants produced small flowers and leaves compared with da1-1 plants (Figures 1D to 1I, 1K and 1L). The size of cells in sod2 da1-1 petals and leaves was similar to that in wild-type petals and leaves (Figures 1K and 1L), indicating that mutations in SOD2 cause a decrease in cell number. Taken together, these results show that the sod2 mutations suppressed the seed and organ size phenotypes of da1-1.
Figure 1.
sod2 Suppresses the Seed Size Phenotype of da1-1.
(A) to (C) Seeds of Col-0 (A), da1-1 (B), and sod2-1 da1-1 (C).
(D) to (F) Ten-day-old seedlings of Col-0 (D), da1-1 (E), and sod2-1 da1-1 (F).
(G) to (I) Flowers of Col-0 (G), da1-1 (H), and sod2-1 da1-1 (I).
(J) Seed area (SA) and seed weight (SW) of Col-0, da1-1, sod2-1 da1-1, and sod2-2 da1-1 plants.
(K) Fifth leaf area (LA) and leaf palisade cell area (LCA) of Col-0, da1-1, sod2-1 da1-1, and sod2-2 da1-1 plants.
(L) Petal area (PA), petal length (PL), petal width (PW), and petal cell area (PCA) of Col-0, da1-1, sod2-1 da1-1, and sod2-2 da1-1 plants.
Values in (J) to (L) are given as mean ± se relative to the respective wild-type values, set at 100%. **P < 0.01 compared with the da1-1 (Student’s t test). Bars = 0.5 mm in (A) to (C) and 1 mm in (D) to (I).
SOD2 Encodes the Ubiquitin-Specific Protease UBP15
The sod2-1 mutation was identified by map-based cloning in an F2 population of a cross between sod2-1 da1-1 and da1-1Ler. The sod2-1 mutation was mapped into an interval between markers F19K19-122 and F11A6-129 on chromosome I (Supplemental Figure 1A). This mapping region includes UBIQUITIN-SPECIFIC PROTEASE15 (UBP15; At1g17110), which is known to influence leaf growth in Arabidopsis (Liu et al., 2008), yet no function for it in determination of seed size has been described. UBP15 contains a signature MYND-type zinc finger domain (Zf-MYND), which has been suggested to be a protein–protein interaction domain in mammalian cells (Lutterbach et al., 1998), and a ubiquitin-specific protease (UBP) domain that is required for deubiquitination activity (Figure 2B) (Liu et al., 2008). To define the molecular basis for the sod2 mutations, the At1g17110 genomic DNA sequences of two sod2 mutant alleles were amplified by PCR and sequenced. We identified a single mutation in each of the mutant alleles. In sod2-1, the 5′-intron-exon boundary of intron 4 is changed from G to A (Figure 2A; Supplemental Figure 1B), replacing a G that is highly conserved at plant gene splice sites. Similarly, sod2-2 has a G-to-A transition in the 5′-intron-exon boundary of the last intron (Figure 2A; Supplemental Figure 1B). Mutations in sod2 alleles altered the splicing of At1g17110 mRNA (Supplemental Figures 1C and 1D).
Figure 2.
Identification and Molecular Characterization of the SOD2/UBP15 Gene.
(A) The SOD2/UBP15 gene structure. The start codon (ATG) and the stop codon (TAG) are indicated. Closed boxes indicate the coding sequence, open boxes indicate the 5′ and 3′ untranslated regions, and lines between boxes indicate introns. The mutation sites of sod2-1 and sod2-2 and the T-DNA insertion site in ubp15-1 are shown.
(B) The SOD2 protein contains a Zf-MYND and a UBP domain.
(C) Plants (21-d-old) of da1-1, sod2-2 da1-1, COM#1, and COM#2 (from left to right). COM is sod2-2 da1-1 transformed with the genomic sequence of the UBP15 gene (gUBP15).
(D) Flowers of da1-1, sod2-2 da1-1, COM#1, and COM#2 (from left to right).
(E) Seeds of da1-1, sod2-2 da1-1, COM#1, and COM#2 (from left to right).
(F) Seed area (SA) and seed weight (SW) of da1-1, sod2-2 da1-1, COM#1, and COM#2.
(G) Petal area (PA), petal length (PL) and petal width (PW) of da1-1, sod2-2 da1-1, COM#1, and COM#2.
(H) to (O) UBP15 expression activity was monitored by pUBP15:GUS transgene expression. Six GUS-expressing lines were observed, and all showed a similar pattern, although they differed slightly in the intensity of the staining. Histochemical analysis of GUS activity in an 8-d-old seedling (H), the developing sepals (I), the developing petals (J), the developing stamens (K), the developing carpels (L), and the developing ovules ([M] to [O]).
Values in (F) and (G) are given as mean ± se relative to the respective da1-1 values, set at 100%. **P < 0.01 compared with the sod2-2 da1-1 (Student’s t test). Bars = 5 cm in (C), 1 mm in (D) and (H), 0.5 mm in (E), and 0.1 mm in (I) to (O).
To further investigate whether mutations in At1g17110/UBP15 could suppress the seed and organ size phenotypes of da1-1, we obtained the ubp15-1 mutant harboring a T-DNA insertion in the At1g17110 gene (Liu et al., 2008) and generated the ubp15-1 da1-1 double mutant. Like sod2 mutations, the ubp15-1 mutation suppressed the seed and organ size phenotypes of da1-1 (Supplemental Figure 2), suggesting that UBP15 is the SOD2 gene. We further transformed sod2-2 da1-1 plants with a genomic fragment (gUBP15) that includes 2074-bp promoter and the At1g17110 gene. Most transgenic plants exhibited the seed and organ size phenotypes of da1-1 (Figures 2C to 2G), indicating that At1g17110/UBP15 is the SOD2 gene.
To evaluate the expression of UBP15, we generated transgenic Arabidopsis plants containing a UBP15 promoter:β-glucuronidase fusion (pUBP15:GUS). The tissue-specific expression patterns of UBP15 were investigated using a histochemical assay for GUS activity. In seedlings, GUS activity was detected in cotyledons, leaves, and roots (Figure 2H). In flowers, GUS activity was observed in sepals, petals, stamens, and carpels (Figures 2I to 2L). GUS activity was stronger in younger floral organs than older floral organs (Figures 2J to 2L). Similarly, relatively higher GUS activity was observed in younger ovules than older ones (Figures 2M to 2O). Thus, these results indicate that UBP15 expression is temporally and spatially regulated.
UBP15 Acts Maternally to Regulate Seed Size
The ubp15-1 single mutant formed small leaves and flowers compared with the wild type (Supplemental Figure 3) (Liu et al., 2008). Considering that the sod2/ubp15 mutations suppressed the seed size phenotype of da1-1 (Figures 2E and 2F; Supplemental Figure 2), we asked whether the ubp15 single mutant influences seed size. We therefore measured the size and weight of wild-type and ubp15-1 seeds. As shown in Figures 3A and 3E, seeds from ubp15-1 plants were significantly smaller and lighter than wild-type seeds. The embryo constitutes the major volume of a mature seed in Arabidopsis. The ubp15-1 embryos were obviously smaller than wild-type embryos (Figure 3B). The changes in seed size were also reflected in the size of seedlings (Figure 3C). Cotyledons of ubp15-1 seedlings were clearly smaller than those of wild-type seedlings (Figures 3C and 3F). Thus, these results indicate that UBP15 is required for seed size control.
Figure 3.
UBP15 Acts Maternally to Regulate Seed Size.
(A) Seeds of Col-0 (left) and ubp15-1 (right).
(B) Mature embryos of Col-0 (left) and ubp15-1 (right).
(C) Seven-day-old seedlings of Col-0 (left) and ubp15-1 (right).
(D) Mature ovules of Col-0 (left) and ubp15-1 (right).
(E) Seed area (SA) and seed weight (SW) of Col-0 and ubp15-1.
(F) Cotyledon area (CoA) of Col-0 and ubp15-1.
(G) Seed area of Col-0 × Col-0 F1, ubp15-1 × ubp15-1 F1, Col-0 × ubp15-1 F1, ubp15-1 × Col-0 F1, Col-0 × ubp15-1 F2, and ubp15-1 × Col-0 F2.
(H) The outer integument length of Col-0 and ubp15-1 at 0, 6, and 8 DAP. Ovules at 0 DAP are mature ovules from wild-type and ubp15-1 plants 2 d after emasculation.
(I) The number of cells in the outer integuments of Col-0 and ubp15-1 at 0, 6, and 8 DAP.
(J) The length of cells in the outer integuments of Col-0 and ubp15-1 at 0, 6, and 8 DAP.
Values in (E) to (J) are given as mean ± se relative to the respective wild-type values, set at 100%. **P < 0.01 and *P < 0.05 compared with the wild type (Student’s t test). Bars = 0.5 mm in (A), 0.25 mm in (B), 1 mm in (C), and 0.1 mm in (D).
As the size of a seed is controlled by the zygotic and/or maternal tissues, we investigated whether UBP15 acts zygotically or maternally. To test this, we conducted reciprocal cross experiments between the wild type and ubp15-1. We pollinated ubp15-1 plants with wild-type pollen. The size of seeds from such crosses was comparable to that from self-pollinated ubp15-1 mutant (Figure 3G). The size of seeds from wild-type plants pollinated by ubp15-1 mutant pollen was similar to that from the self-pollinated wild-type plants (Figure 3G). These results indicate that UBP15 functions maternally to regulate seed size. We further investigated the size of ubp15-1/Columbia-0 (Col-0) F2 and Col-0/ubp15-1 F2 seeds. Seeds from ubp15-1/Col-0 F1 and Col-0/ubp15-1 F1 plants have wild-type, ubp15-1, and ubp15/+ embryos within ubp15/+ seed coats, respectively. The size of ubp15-1/Col-0 F2 and Col-0/ubp15-1 F2 seeds was similar to that of wild-type seeds (Figure 3G). Thus, these results show that the embryo and endosperm genotypes for UBP15 do not influence seed size, and UBP15 is required in sporophytic tissue of the mother plant to determine seed size.
UBP15 Regulates Cell Proliferation in the Maternal Integuments
The reciprocal cross experiments demonstrated that UBP15 acts maternally to influence seed growth (Figure 3G). The ovule integuments are maternal tissues and develop into the seed coat after fertilization, which could physically limit seed growth (Adamski et al., 2009; Fang et al., 2012). Consistent with this notion, several previous studies indicated that the integument size of ovules influences the final size of seeds in Arabidopsis (Schruff et al., 2006; Adamski et al., 2009; Fang et al., 2012). We therefore examined mature ovules from wild-type and ubp15-1 plants 2 d after emasculation. The ubp15-1 ovules were obviously smaller than wild-type ovules (Figure 3D). The outer integument length of ubp15-1 ovules was significantly decreased, compared with that of wild-type ovules (Figure 3H).
The size of the integument is determined by cell number and cell size. We therefore investigated the number of outer integument cells in wild-type and ubp15-1 ovules. As shown in Figure 3I, the number of outer integument cells in ubp15-1 ovules was significantly reduced compared with that in wild-type ovules, indicating that UBP15 is required for cell proliferation in the maternal integuments of ovules. After fertilization, cells in the integuments predominantly carry on expansion but still undergo division (Garcia et al., 2005). We further counted the number of outer integument cells in wild-type and ubp15-1 seeds at 6 and 8 d after pollination (DAP). In wild-type seeds, the outer integument cell number at 6 DAP was comparable with that at 8 DAP (Figure 3I), indicating that cells in the outer integuments of wild-type seeds completely cease division by 6 DAP. In ubp15 seeds, the outer integument cell number at 6 DAP was similar to that at 8 DAP, showing that cells in the outer integuments of ubp15-1 seeds also stop dividing by 6 DAP (Figure 3I). As shown in Figure 3I, the number of outer integument cells in ubp15 seeds was clearly decreased, compared with that in wild-type seeds. We further measured the length of outer integument cells in wild-type and ubp15-1 seeds at 6 and 8 DAP. Cells in ubp15-1 outer integuments were slightly longer than those in wild-type outer integuments (Figure 3J), suggesting a possible compensation mechanism between cell number and cell size in the maternal integuments. Thus, these results indicate that UBP15 is required for cell proliferation in the maternal integuments of ovules and developing seeds.
Plants Overexpressing UBP15 Show Similar Phenotypes to da1-1
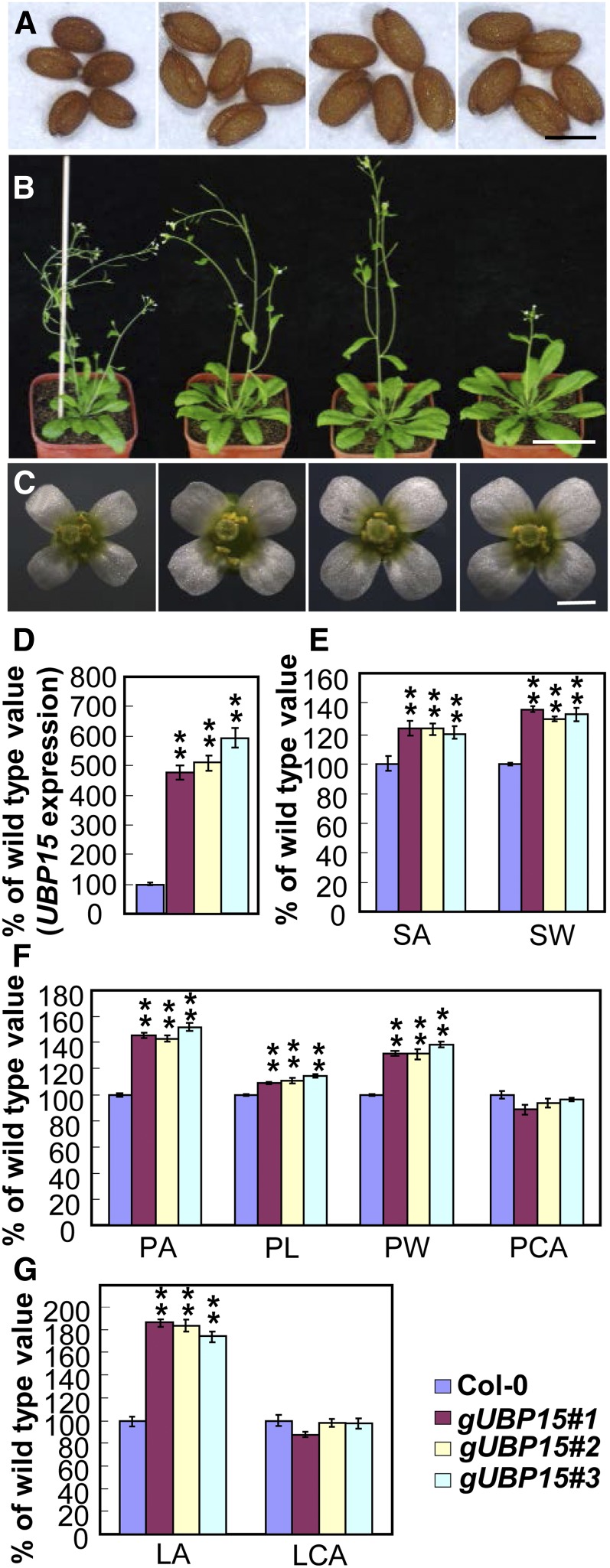
We transformed Col-0 plants with a genomic fragment (gUBP15). Interestingly, some transgenic plants had dramatic increases in UBP15 mRNA compared with wild-type plants (Figure 4D). It is possible that the integration regions of the transgenes in the genome affect the promoter activity of gUBP15. gUBP15 transgenic plants formed larger and heavier seeds than wild-type plants (Figures 4A and 4E). gUBP15 transgenic plants also produced larger flowers and leaves than wild-type plants (Figures 4B, 4C, 4F. and 4G). The increased size of petals and leaves in gUBP15 transgenic plants was not caused by larger cells (Figures 4F and 4G), indicating that it is the number of petal and leaf cells that is higher. We previously reported that the da1-1 mutant formed large seeds, flowers, and leaves by promoting cell proliferation (Li et al., 2008). Thus, the seed and organ size phenotypes of gUBP15 transgenic plants were similar to those observed in da1-1 plants (Li et al., 2008).
Figure 4.
Plants Overexpressing UBP15 Show Similar Phenotypes to da1-1.
(A) Seeds of Col-0, gUBP15#1, gUBP15#2, and gUBP15#3 plants (from left to right). gUBP15 is Col-0 transformed with the genomic sequence of the UBP15 gene.
(B) Plants (28 d old) of Col-0, gUBP15#1, gUBP15#2, and gUBP15#3 (from left to right).
(C) Flowers of Col-0, gUBP15#1, gUBP15#2, and gUBP15#3 (from left to right).
(D) Quantitative real-time RT-PCR analysis of the UBP15 gene expression in Col-0, gUBP15#1, gUBP15#2, and gUBP15#3 seedlings.
(E) Seed area (SA) and seed weight (SW) of Col-0, gUBP15#1, gUBP15#2, and gUBP15#3.
(F) Petal area (PA), petal length (PL), petal width (PW), and petal cell area (PCA) of Col-0, gUBP15#1, gUBP15#2, and gUBP15#3.
(G) The average area of the fifth leaves (LA) and palisade cells (LCA) of Col-0, gUBP15#1, gUBP15#2, and gUBP15#3.
Values in (D) to (G) are given as mean ± se relative to the respective wild-type values, set at 100%. **P < 0.01 compared with the wild type (Student’s t test). Bars = 0.5 mm in (A), 5 cm in (B), and 1 mm in (C).
UBP15 Acts in a Common Pathway with DA1 to Affect Seed Size, but Does so Independently of DA2 and EOD1
As sod2/ubp15 mutations suppressed the seed size phenotype of da1-1, we sought to determine genetic relationships between UBP15 and DA1 in seed size. We therefore measured the size of seeds from wild-type, ubp15-1, da1-1, and ubp15-1 da1-1 plants. As shown in Figures 5A and 5C, the average size and weight of ubp15-1 da1-1 seeds were indistinguishable from those of ubp15-1 single mutant seeds, indicating that ubp15-1 is epistatic to da1-1 with respect to seed size and weight. The changes in seed size were reflected in the size of the embryos (Figure 5B). The size of ubp15-1 da1-1 embryos was comparable with that of ubp15-1 embryos (Figure 5B). We further investigated mature ovules from wild-type, ubp15-1, da1-1, and ubp15-1 da1-1 plants. The outer integument length of ubp15-1 da1-1 ovules was similar to that of ubp15-1 ovules (Figure 5D). Similarly, the outer integument length of ubp15-1 da1-1 seeds was indistinguishable from that of ubp15-1 seeds at 6 and 8 DAP. In addition, the size of ubp15-1da1-1 flowers and petals was comparable with that of ubp15-1 flowers and petals (Supplemental Figure 4). Together, these genetic analyses show that ubp15-1 is epistatic to da1-1 with respect to seed and organ size, indicating DA1 and UBP15 function antagonistically in a common pathway to regulate seed and organ growth.
Figure 5.
ubp15-1 Is Epistatic to da1-1.
(A) Seeds of Col-0, ubp15-1, da1-1, and ubp15-1 da1-1.
(B) Mature embryos of Col-0, ubp15-1, da1-1, and ubp15-1 da1-1.
(C) Seed area (SA) and seed weight (SW) of Col-0, ubp15-1, da1-1, and ubp15-1 da1-1.
(D) The outer integument length of Col-0, ubp15-1, da1-1, and ubp15-1 da1-1 at 0, 6, and 8 DAP.
(E) The number of cells in the outer integuments of Col-0, ubp15-1, da1-1, and ubp15-1 da1-1 at 0, 6, and 8 DAP.
Values in (C) to (E) are given as mean ± se relative to the respective wild-type values, set at 100%. **P < 0.01 compared with the wild type (Student’s t test). Bars = 0.5 mm in (A) and 0.25 mm in (B).
To further investigate the cellular basis of epistatic interactions between DA1 and UBP15 in seed size, we counted the outer integument cell number of ovules and developing seeds in the wild type, ubp15-1, da1-1, and ubp15-1 da1-1. The number of outer integument cells in ubp15-1 da1-1 ovules was comparable to that in ubp15-1 ovules (Figure 5E). Similarly, the number of outer integument cells in ubp15-1 da1-1 developing seeds was indistinguishable from that in ubp15-1 developing seeds (Figure 5E). These results show that ubp15 is epistatic to da1-1 with respect to the number of outer integument cells. Thus, DA1 and UBP15 act antagonistically in a common pathway to regulate cell proliferation in the maternal integuments.
We previously reported that DA1 functions synergistically with EOD1 and DA2 in the control of seed and organ size in Arabidopsis (Li et al., 2008; Xia et al., 2013). We further asked whether UBP15 acts antagonistically with EOD1 or DA2 in a common pathway to regulate seed and organ growth. To test this, we generated ubp15-1 eod1-2 and ubp15-1 da2-1 double mutants and investigated their seed and organ size phenotypes. The genetic interaction between ubp15-1 and eod1-2 was essentially additive for both seed and petal size compared with their parental lines (Supplemental Figures 5A and 5B). Similarly, the seed and petal size phenotypes of ubp15-1 da2-1 were additive compared with those of ubp15-1 and da2-1 single mutants (Supplemental Figures 5C and 5D). Therefore, these genetic analyses suggest that UBP15 acts independently of EOD1 and DA2 to influence seed and organ growth.
DA1 Modulates UBP15 Stability
DA1 encodes a ubiquitin receptor that has been proposed to be involved in ubiquitin-mediated protein degradation (Li et al., 2008; Xia et al., 2013). The ubiquitin proteasome system is involved in the degradation of many proteins. To test whether UBP15 could be degraded in a proteasome-dependent manner, we treated the Arabidopsis gUBP15-GFP (green fluorescent protein) transgenic line with the proteasome inhibitor MG132. After MG132 treatment, the level of UBP15 protein was higher than that in untreated plants (Figure 6A), indicating that the ubiquitin proteasome influences the stability of UBP15. Considering that DA1 and UBP15 act antagonistically in a common genetic pathway to regulate seed and organ growth, we asked whether DA1 could affect the stability of UBP15. To test this, we crossed da1-1 with two independent gUBP15-GFP transgenic lines. After the selection of plants containing da1-1 and gUBP15-GFP transgene, we measured and compared the levels of UBP15 proteins in different genetic backgrounds. As shown in Figure 6B, the UBP15-GFP protein levels in the da1-1 mutant background were obviously higher than those in wild-type plants, while the transcript levels of UBP15 in gUBP15-GFP and gUBP15-GFP;da1-1 plants were similar (Figure 6E). Thus, these results indicate that DA1 influences the stability of UBP15 in Arabidopsis.
Figure 6.
DA1 Modulates UBP15 Stability.
(A) The proteasome inhibitor stabilizes UBP15. Ten-day-old gUBP15-GFP seedlings were treated with or without 20 μM MG132. Total protein extracts were subjected to immunoblot assays using anti-GFP and anti-RPN6 (as loading control) antibodies.
(B) The UBP15-GFP protein accumulates at higher levels in the da1-1 mutant. Total protein extracts were subjected to immunoblot assays using anti-GFP and anti-RPN6 (as loading control) antibodies.
(C) Seeds of gUBP15-GFP#7 and gUBP15-GFP#7;da1-1.
(D) Flowers of gUBP15-GFP#7 and gUBP15-GFP#7;da1-1.
(E) The relative expression of UBP15 in gUBP15-GFP#7 and gUBP15-GFP#7;da1-1. Data shown are mean ± sd of three replicates.
(F) Seed area (SA) and seed weight (SW) of gUBP15-GFP#7 and gUBP15-GFP#7;da1-1.
(G) Petal area (PA), petal length (PL), and petal width (PW) of gUBP15-GFP#7 and gUBP15-GFP#7;da1-1.
Values in (F) and (G) are given as mean ± se relative to the respective gUBP15-GFP#7 values, set at 100%. **P < 0.01 compared with gUBP15-GFP#7 (Student’s t test). Bars = 0.5 mm in (C) and 1 mm in (D).
As the accumulation of UBP15 was observed in the da1-1 mutant background, one would expect that the da1-1 allele should increase the seed and organ size phenotypes of gUBP15-GFP transgenic plants. We therefore measured the seed and organ size of gUBP15-GFP and gUBP15-GFP/da1-1 plants. As shown in Figures 6C, 6D, 6F, and 6G, the seed and organ size phenotypes of gUBP15-GFP transgenic plants were strongly enhanced by da1-1. Thus, our genetic analyses further support the role of DA1 in modulating UBP15 stability.
DA1 Interacts with UBP15 in Vitro and in Vivo
Considering that DA1 modulates UBP15 stability, we asked whether DA1 could physically interact with UBP15 and influence its degradation. To test this, we performed in vitro pull-down assays. DA1 was expressed as a glutathione S-transferase (GST) fusion protein, while UBP15 was expressed as a maltose binding protein (MBP) fusion. As shown in Figure 7A, GST-DA1 was pulled down with MBP-UBP15, but not with MBP alone, indicating that DA1 physically interacts with UBP15 in vitro. Considering that the da1-1 mutation (DA1R358K) affects UBP15 stability, we further asked whether the DA1R358K mutation influences the interaction between DA1 and UBP15. Using a GST-DA1R358K fusion protein in pull-down experiments with MBP-UBP15, we showed that the DA1R358K mutation does not affect the interaction between DA1 and UBP15 (Figure 7A).
Figure 7.
DA1 Physically Interacts with UBP15 in Vitro and in Vivo.
(A) DA1 directly interacts with UBP15 in vitro. GST-DA1 and GST-DA1R358K were pulled down (PD) by MBP-UBP15 immobilized on amylose resin and analyzed by immunoblotting (IB) using an anti-GST antibody.
(B) The C terminus of DA1 interacts with UBP15 in vitro. Both GST-DA1-LIM+C and GST-DA1-C were pulled down by MBP-UBP15 immobilized on amylose resin and analyzed by immunoblotting using an anti-GST antibody.
(C) The C-terminal region of UBP15 interacts with DA1 in vitro. GST-DA1 was pulled down by MBP-UBP15-C immobilized on amylose resin and analyzed by immunoblotting using an anti-GST antibody.
(D) UBP15 interacts with DA1 in vivo. N. benthamiana leaves were transformed by injection of Agrobacterium tumefaciens EHA105 cells harboring 35S:GFP-UBP15 and 35S:Myc-DA1 plasmids. Myc-DA1 was detected in the immunoprecipitated GFP-UBP15 complex, indicating that there is a physical association between DA1 and UBP15 in vivo.
(E) Schematic diagram of DA1 and its derivatives containing specific domains. The DA1 contains two UIM motifs in the N terminus, a single LIM domain, and the C-terminal region. aa, amino acid.
(F) Schematic diagram of UBP15, UBP15-N, and UBP15-C. The UBP15 contains one predicted Zf-MYND domain in the N terminus and the UBP domain in the C-terminal region.
(G) A genetic and molecular framework for DA1, DA2, EOD1/BB, and UBP15/SOD2-mediated regulation of seed and organ size. DA1 and DA2 act synergistically to restrict seed and organ size, suggesting that DA1 and DA2 may have a common downstream substrate. Similarly, DA1 and EOD1 may share a common target. However, DA2 acts independently of EOD1 to influence seed and organ size, suggesting that DA2 and EOD1 may target distinct cell proliferation stimulators (substrate 1 and substrate 2) for degradation, with common regulation via DA1. DA1 acts the upstream of UBP15 and modulates UBP15 stability. However, UBP15 acts independently of DA2 and EOD1, suggesting that UBP15 is not the target of DA2 or EOD1 for degradation.
DA1 contains two ubiquitin interaction motifs (UIMs) in the N terminus, a single LIM (Lin-11, Isl-1, and Mec3) domain, and the C-terminal region (Figure 7E) (Li et al., 2008). We further investigated which domain of DA1 is responsible for the interaction between DA1 and UBP15. The UIM domains (DA1-UIM), the LIM domain (DA1-LIM), the C-terminal region (DA1-C), and the LIM domain and C terminus (DA1-LIM+C) were expressed as GST fusion proteins in Escherichia coli (Figure 7E). UBP15 was expressed as an MBP fusion protein and used in pull-down experiments. As shown in Figure 7B, GST-DA1-LIM+C and GST-DA1-C were pulled down with MBP-UBP15, but not with MBP alone. However, GST-DA1-UIM and GST-DA1-LIM did not bind to MBP-UBP15. Thus, these results show that the C-terminal region of DA1 is required for the interaction with UBP15 in vitro.
UBP15 contains a signature Zf-MYND near the N terminus, which has been proposed to be a protein–protein interaction domain in mammalian cells (Lutterbach et al., 1998), and a UBP domain in the C terminus (Figure 7F). We further asked which region of UBP15 is responsible for the interaction between DA1 and UBP15. The N-terminal region containing Zf-MYND domain (UBP15-N) and the C-terminal region (UBP15-C) were expressed as MBP fusion proteins in E. coli. DA1 was expressed as a GST fusion protein and used in pull-down experiments. As shown in Figure 7C, GST-DA1 was pulled down with MBP-UBP15-C, but not with MBP-UBP15-N or MBP alone. Thus, these results show that the C-terminal region of UBP15 is required for the interaction between DA1 and UBP15 in vitro.
To investigate possible association between DA1 and UBP15 in planta, we used coimmunoprecipitation assays to detect their interactions in vivo. We transiently coexpressed 35S:GFP-UBP15 and 35S:Myc-DA1 in Nicotiana benthamiana leaves. 35S:GFP coexpressed with 35S:Myc-DA1 was used as a negative control. Although the majority of GFP-UBP15 fusion proteins were degraded, a very weak band corresponding to GFP-UBP15 could be detected (Figure 7D). The coimmunoprecipitation analysis indicated that Myc-DA1 was detected in the immunoprecipitated GFP-UBP15 complex but not in the negative control (GFP) (Figure 7D), revealing a physical association between DA1 and UBP15 in vivo. We further transiently coexpressed 35S:GFP-UBP15-C and 35S:Myc-DA1 in N. benthamiana leaves. The coimmunoprecipitation analysis showed that the C-terminal region of UBP15 is required for the interaction with DA1 in vivo (Supplemental Figure 6).
DISCUSSION
Seed size in flowering plants is an important trait for plant fitness and agricultural purposes (Alonso-Blanco et al., 1999; Orsi and Tanksley, 2009). Although several factors that function maternally to influence seed size were described in Arabidopsis, including TTG2, AP2, ARF2, CYP78A5/KLUH, CYP78A6/EOD3, DA1, and DA2, little is known about their genetic and molecular mechanisms in seed size determination. We previously demonstrated that the ubiquitin receptor DA1 acts synergistically with the E3 ubiquitin ligases DA2 and EOD1 to restrict seed growth in Arabidopsis (Li et al., 2008; Xia et al., 2013), but their target substrates remain to be discovered. Here, we show that DA1 physically associates with the ubiquitin-specific protease UBP15/SOD2 and modulates the stability of UBP15. Further results reveal that UBP15 and DA1 act antagonistically in a common genetic pathway to regulate seed size. Thus, our findings define the genetic and molecular mechanisms of four ubiquitin-related proteins: DA1, DA2, EOD1, and UBP15 in seed size control.
UBP15 Functions Maternally to Regulate Seed Size
The sod2/ubp15 mutants were identified as suppressors of the large seed and organ phenotypes of da1-1. The ubp15 single mutants produced small seeds and organs, whereas overexpression of UBP15 resulted in large seeds and organs (Figures 3 and 4; Supplemental Figure 3), indicating that UBP15 is a positive regulator of seed and organ size. Several studies suggested that there is a possible correlation between organ size and seed growth. For example, arf2, da1-1, and da2-1 mutants with large organs produced large seeds (Schruff et al., 2006; Li et al., 2008; Xia et al., 2013), while klu mutants with small organs formed small seeds (Adamski et al., 2009). However, organ and seed size is not always positively related. For example, several other mutants with altered organ size had normal-sized seeds (Horiguchi et al., 2005; White, 2006; Lee et al., 2009; Xu and Li, 2011). Thus, these studies suggest that organs and seeds may possess both common and distinct mechanisms to determine their respective size.
The size of a seed is determined by maternal sporophytic and/or zygotic tissues. Our reciprocal cross experiments indicated that UBP15 functions maternally to promote seed growth, and the embryo and endosperm genotypes for UBP15 do not determine seed size (Figure 3G). Several previous studies demonstrated that the maternal integuments are crucial for determining seed size in Arabidopsis. For example, the arf2, da1-1, da2-1, and eod3-1D mutants produced large seeds due to the enlargement of the maternal integuments (Schruff et al., 2006; Li et al., 2008; Fang et al., 2012; Xia et al., 2013), whereas klu mutants formed small seeds as a result of reduced integument size (Adamski et al., 2009). The ubp15-1 integuments were smaller than wild-type integuments (Figures 3D and 3H). The ubp15-1 mutation also suppressed the large integument size phenotype of da1-1 (Figure 5D). Thus, these studies suggest that the regulation of maternal integument size is one of the important mechanisms for control of seed size.
The size of the integument or seed coat is coordinately determined by cell proliferation and cell expansion. The number of cells in ubp15-1 integuments was less than that in wild-type integuments (Figure 3I), indicating that UBP15 is required for cell proliferation in the maternal integuments. By contrast, the size of cells in ubp15-1 integuments was slightly increased compared with that in wild-type integuments (Figure 3J), suggesting a possible compensation mechanism between cell proliferation and cell expansion in maternal integuments. The integuments of da1-1 had more cells than those of the wild type (Xia et al., 2013). Genetic analyses show that ubp15-1 is epistatic to da1-1 with respect to cell number in the integuments (Figure 5E), further supporting the role of UBP15 in cell proliferation. Consistent with this, expression of UBP15 in younger ovules was higher than that in older ones (Figures 2H to 2O). Thus, it is possible that the integument cell number may determine seed size by setting the growth potential of the seed coat.
A Genetic Framework for DA1, DA2, EOD1, and UBP15-Mediated Control of Seed Size
Several factors involved in ubiquitin-related activities have been reported to affect seed and organ growth in plants. We previously identified the ubiquitin receptor DA1 as a negative factor of seed and organ size (Li et al., 2008). A modifier screen identified an enhancer of da1-1 (EOD1) (Li et al., 2008), which encodes the E3 ubiquitin ligase BB (Disch et al., 2006). DA1 acts synergistically with EOD1 to restrict seed and organ growth, suggesting that DA1 and EOD1 might share a common downstream target. DA1 functions synergistically with another E3 ubiquitin ligase DA2 to influence seed and organ growth (Li et al., 2008; Xia et al., 2013). However, genetic analyses show that DA2 and EOD1 act in different pathways to regulate seed and organ size (Li et al., 2008; Xia et al., 2013), suggesting that DA2 and EOD1 may target distinct growth stimulators for degradation, with common regulation via DA1. In this study, we identified two allelic suppressors of da1-1 (sod2) mutated in the gene encoding UBP15, which possesses deubiquitination activity (Liu et al., 2008). With respect to seed and organ size, ubp15-1 is epistatic to da1-1 (Figure 5), and overexpression of UBP15 caused similar large seed and organ phenotypes to da1-1 (Figure 4). In addition, UBP15 and DA1 showed similar expression patterns. For example, UBP15 and DA1 were highly expressed during early stages of organ formation, but levels were largely reduced at the later stages of organ development (Figures 2H to 2O) (Li et al., 2008). Thus, these results support that DA1 and UBP15 act antagonistically in a common genetic pathway to regulate seed and organ growth. However, our genetic analyses show that UBP15 acts independently of EOD1 and DA2 to influence seed and organ size, suggesting that UBP15 is not the substrate of the E3 ubiquitin ligases DA2 or EOD1 for degradation. Thus, our findings define a framework for the control of seed and organ size by four ubiquitin-related proteins DA1, DA2, EOD1, and UBP15 in Arabidopsis (Figure 7G).
DA1 Modulates UBP15 Stability
Our genetic analyses showed that DA1 and UBP15 act antagonistically in a common pathway to control seed and organ growth (Figure 5). DA1 encodes a ubiquitin receptor with two UIMs that can bind ubiquitin in vitro (Li et al., 2008). Ubiquitin receptors have been proposed to interact with polyubiquitinated substrates via their UIM domains and facilitate substrate degradation by the proteasome (Verma et al., 2004). In this study, we showed that UBP15 stability is affected by the proteasome (Figure 6A). A recent proteomic study has shown that UBP15 is a ubiquitination target (Kim et al., 2013). Thus, it is possible that DA1 could be involved in mediating the degradation of UBP15 by the proteasome. Supporting this, the level of UBP15 protein was obviously increased in the da1-1 mutant compared with that in the wild type (Figure 6B). We further revealed that DA1 physically associates with UBP15 in vitro and in vivo (Figures 7A to 7D). The interaction between DA1 and UBP15 may help DA1 specifically recognize UBP15 for proteasomal degradation.
Currently, plant seeds and organs are not only used as foods, but also used as sustainable fuel and energy sources. Our findings establish a genetic and molecular framework for the regulation of seed and organ size by DA1, DA2, EOD1, and UBP15 in Arabidopsis. Interestingly, overexpression of the maize da1-1 mutant allele (Zea mays da1-1) has been known to increase seed mass of maize (Wang et al., 2012), and the homolog of DA2 in rice is GW2, a negative regulator of rice grain size (Song et al., 2007). Thus, our current understanding of DA1, DA2, EOD1, and UBP15 functions suggests that these four ubiquitin-related proteins and their orthologs in other plant species could be used to engineer large seed size and increased biomass.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Col-0 was used as the wild-type line. All mutants used in this study were in the Col-0 background. sod2-1 and sod2-2 were identified as suppressors of da1-1 from an ethyl methanesulfonate–treated M2 population of da1-1. The ubp15-1 (SALK_018601) was obtained from the Nottingham Arabidopsis Stock Centre. Arabidopsis plants were grown under long-day conditions (16 h light/8 h dark) at 22°C.
Map-Based Cloning, Constructs, and Plant Transformation
The sod2-1 mutation was mapped in the F2 population of a cross between sod2-1 da1-1 and da1-1Ler. Molecular markers were developed using public database (Supplemental Table 1).
The 6424-bp genomic sequence containing 2074-bp promoter, the UBP15 gene, and 276-bp 3′ untranslated region was amplified using the primers gUBP15 (99)-F and gUBP15 (99)-R. PCR products were then cloned to pDONR207 using BP enzyme (Invitrogen). The UBP15 genomic DNA was then subcloned to the pMDC99 binary vector by LR reaction to generate the gUBP15 construct. The plasmid gUBP15 was introduced into the sod2-2 da1-1 mutant plants using Agrobacterium tumefaciens GV3101, and transformants were selected on medium containing hygromycin (30 μg/mL).
The gUBP15-GFP construct was made using a PCR-based Gateway system. The 6145-bp genomic sequence containing 2074-bp promoter and the UBP15 gene without the stop code was amplified using the primers gUBP15 (99)-F and gUBP15 (107)-R. PCR products were cloned to pDONR207 using BP enzyme (Invitrogen). The UBP15 genomic DNA was then subcloned to the pMDC107 binary vector with the GFP gene to generate the gUBP15-GFP construct. The plasmid gUBP15-GFP was introduced into Col-0 plants using Agrobacterium GV3101, and transformants were selected on medium containing hygromycin (30 μg/mL).
The 2111-bp promoter sequence of UBP15 was amplified using the primers pUBP15-F and pUBP15-R (Supplemental Table 1). PCR products were cloned into pDONR207 using BP enzyme. The UBP15 promoter was then subcloned to the binary vector pGWB3 with the GUS reporter gene to generate the transformation plasmid pUBP15:GUS. The plasmid pUBP15:GUS was introduced into Col-0 plants using Agrobacterium GV3101, and transformants were selected on medium containing hygromycin (30 μg/mL).
Morphological and Cellular Analysis
To determine seed size, we photographed mature dry seeds under a Leica microscope (Leica S8APO) using a Leica charge-coupled device camera (DFC420). The projective area of seeds was measured using Image J software. Average seed weight was determined by weighing mature dry seeds in batches of 500 using an electronic analytical balance (Mettler Moledo AL104). The weights of five sample batches were measured for each seed lot.
Petals (stage 14), leaves, and cotyledons were scanned to produce digital images, respectively. The average area of petals, leaves, and cotyledons was calculated using Image J software. Leaf and petal cell sizes were measured from differential interference contrast images.
GUS Staining
Seedlings, sepals, petals, stamens, carpels, and ovules of pUBP15:GUS transgenic plants were stained in a GUS staining solution (1 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid, 100 mM Na3PO4 buffer, 3 mM each K3Fe(CN)6/K4Fe(CN)6, 10 mM EDTA, and 0.1% Nonidet P-40) and incubated at room temperature for 5 h. After GUS staining, chlorophyll was removed using 70% ethanol.
RNA Isolation, RT-PCR, and Quantitative Real-Time RT-PCR Analysis
Total RNA was extracted from Arabidopsis seedlings using an RNAprep pure plant kit (Tiangen). Total mRNA was reverse transcribed into cDNA using SuperScript III reverse transcriptase (Invitrogen). Quantitative real-time RT-PCR analysis was performed with a Lightcycler 480 engine (Roche) using the Lightcycler 480 SYBR Green Master (Roche). The primers used for RT-PCR and quantitative real-time RT-PCR are described in Supplemental Table 1.
In Vitro Protein–Protein Interaction
The coding sequences of UBP15 and its N terminus (UBP15-N) were subcloned into XbaI and SbfI sites of the pMAL-c2 vector to generate MBP-UBP15 and MBP-UBP15-N constructs. The coding sequence of the C terminus (UBP15-C) was subcloned into XbaI and HindIII sites of the pMAL-c2 vector to generate the MBP-UBP15-C construct. The specific primers for MBP-UBP15, MBP-UBP15-N, and MBP-UBP15-C constructs were MBP-UBP15-F, MBP-UBP15-R, MBP-UBP15N-R, MBP-UBP15C-F, and MBP-UBP15C-R, respectively (Supplemental Table 1). GST-DA1, GST-DA1R358K, GST-DA1-UIM, GST-DA1-LIM+C, GST-DA1-LIM, and GST-DA1-C constructs were made as previously described (Xia et al., 2013).
For in vitro pull-down assay, bacterial lysates containing ∼15 μg MBP-UBP15, MBP-UBP15-N, or MBP-UBP15-C fusion proteins were mixed with lysates containing ∼30 μg GST-DA1, GST-DA1R358K, GST-DA1-UIM, GST-DA1-LIM, GST-DA1-LIM+C, or GST-DA1-C fusion proteins. Amylose resins (25 μL; New England Biolabs) were added into each combined solution with rocking at 4°C for 30 min. Beads were washed five times with the buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, pH 8.0, 1% Triton X-100, 10% glycerol, 1 mM PMSF, and 1× Complete protease inhibitor cocktail [Roche]), and the isolated proteins were further separated on a 10% SDS-polyacrylamide gel and detected by immunoblot analysis with anti-GST (Abmart) and anti-MBP antibodies (New England Biolabs), respectively.
Coimmunoprecipitation
The coding sequence of UBP15 was amplified using the primers GFP-UBP15-F and GFP-UBP15-R. PCR products were subcloned into the pCR8/GW/TOPO TA vector (Invitrogen) using TOPO enzyme. UBP15 was then subcloned into the Gateway binary vector pMDC43 containing the 35S promoter and the GFP gene to generate the 35S:GFP-UBP15 construct. The 35S:Myc-DA1 construct was made as previously described (Xia et al., 2013).
Nicotiana benthamiana leaves were transformed by injection of Agrobacterium EHA105 cells harboring 35S:GFP-UBP15, 35S:Myc-DA1, and 35S:GFP plasmids. Total protein was extracted with extraction buffer (50 mM Tris/HCl, pH 7.5, 150 mM NaCl, 20% glycerol, 2% Triton X-100, 1 mM EDTA, 1× Complete protease inhibitor cocktail, and 20 μM MG132) and incubated with GFP-Trap_A (Chromotek) for 30 min at 4°C. Beads were washed three times with wash buffer (50 mM Tris/HCl, pH 7.5, 150 mM NaCl, 20% glycerol, 0.1% Triton X-100, 1 mM EDTA, pH 8.0, 1× Complete protease inhibitor cocktail, and 20 μM MG132). The immunoprecipitates were separated in 10% SDS-polyacrylamide gel and detected by immunoblot analysis with anti-Myc (Abmart) and anti-GFP (Beyotime) antibodies, respectively.
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for genes mentioned in this article are as follows: At1g19270 (DA1), At1g17110 (SOD2/UBP15), At3g63530 (EOD1/BB), and At1g78420 (DA2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Identification and Molecular Characterization of SOD2/UBP15.
Supplemental Figure 2. ubp15-1 Suppresses the Seed- and Organ-Size Phenotypes of da1-1.
Supplemental Figure 3. ubp15-1 Forms Small Organs.
Supplemental Figure 4. ubp15-1 Is Epistatic to da1-1.
Supplemental Figure 5. UBP15 Acts Independently of EOD1 and DA2 to Regulate Seed and Organ Size.
Supplemental Figure 6. The C-Terminal Region of UBP15 Interacts with DA1 in Vivo.
Supplemental Table 1. Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank the Nottingham Arabidopsis Stock Centre for the ubp15-1 mutant. This work was supported by the grants from National Basic Research Program of China (2009CB941503), the National Natural Science Foundation of China (91017014, 31221063, 30870215, and 31300242), and the Hundred Talent Program of the Chinese Academy of sciences.
AUTHOR CONTRIBUTIONS
L.D., N.L., and Y.H.L. designed the research. Y.H.L. conceived and supervised the project. L.D. performed most of the experiments. N.L. and L.D. conducted pull-down assays. L.C. performed coimmunoprecipitation experiments. Y.X. identified the ubp15-1 da1-1 double mutant. Y.L. and Y.Z. helped do cellular analysis. L.D., N.L., C.L., and Y.H.L. analyzed data. Y.H.L. wrote the article.
Glossary
- Zf-MYND
MYND-type zinc finger domain
- Col-0
Columbia-0
- DAP
days after pollination
- UIM
ubiquitin interaction motif
Footnotes
Online version contains Web-only data.
References
- Adamski N.M., Anastasiou E., Eriksson S., O’Neill C.M., Lenhard M. (2009). Local maternal control of seed size by KLUH/CYP78A5-dependent growth signaling. Proc. Natl. Acad. Sci. USA 106: 20115–20120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C., Blankestijn-de Vries H., Hanhart C.J., Koornneef M. (1999). Natural allelic variation at seed size loci in relation to other life history traits of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96: 4710–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disch S., Anastasiou E., Sharma V.K., Laux T., Fletcher J.C., Lenhard M. (2006). The E3 ubiquitin ligase BIG BROTHER controls arabidopsis organ size in a dosage-dependent manner. Curr. Biol. 16: 272–279. [DOI] [PubMed] [Google Scholar]
- Fan C., Xing Y., Mao H., Lu T., Han B., Xu C., Li X., Zhang Q. (2006). GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 112: 1164–1171. [DOI] [PubMed] [Google Scholar]
- Fang W., Wang Z., Cui R., Li J., Li Y. (2012). Maternal control of seed size by EOD3/CYP78A6 in Arabidopsis thaliana. Plant J. 70: 929–939. [DOI] [PubMed] [Google Scholar]
- Garcia D., Fitz Gerald J.N., Berger F. (2005). Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell 17: 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D., Saingery V., Chambrier P., Mayer U., Jürgens G., Berger F. (2003). Arabidopsis haiku mutants reveal new controls of seed size by endosperm. Plant Physiol. 131: 1661–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegas V.C., Nazari A., Griffiths S., Simmonds J., Fish L., Orford S., Sayers L., Doonan J.H., Snape J.W. (2010). A genetic framework for grain size and shape variation in wheat. Plant Cell 22: 1046–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G., Kim G.T., Tsukaya H. (2005). The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 43: 68–78. [DOI] [PubMed] [Google Scholar]
- Jofuku K.D., Omidyar P.K., Gee Z., Okamuro J.K. (2005). Control of seed mass and seed yield by the floral homeotic gene APETALA2. Proc. Natl. Acad. Sci. USA 102: 3117–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X., Li W., Zhou Y., Ni M. (2013). A WRKY transcription factor recruits the SYG1-like protein SHB1 to activate gene expression and seed cavity enlargement. PLoS Genet. 9: e1003347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.Y., Scalf M., Smith L.M., Vierstra R.D. (2013). Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis. Plant Cell 25: 1523–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.H., Ko J.H., Lee S., Lee Y., Pak J.H., Kim J.H. (2009). The Arabidopsis GRF-INTERACTING FACTOR gene family performs an overlapping function in determining organ size as well as multiple developmental properties. Plant Physiol. 151: 655–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Nie X., Tan J.L., Berger F. (2013). Integration of epigenetic and genetic controls of seed size by cytokinin in Arabidopsis. Proc. Natl. Acad. Sci. USA 110: 15479–15484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zheng L., Corke F., Smith C., Bevan M.W. (2008). Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev. 22: 1331–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkies A., Graeber K., Knight C., Leubner-Metzger G. (2010). The evolution of seeds. New Phytol. 186: 817–831. [DOI] [PubMed] [Google Scholar]
- Liu Y., Wang F., Zhang H., He H., Ma L., Deng X.W. (2008). Functional characterization of the Arabidopsis ubiquitin-specific protease gene family reveals specific role and redundancy of individual members in development. Plant J. 55: 844–856. [DOI] [PubMed] [Google Scholar]
- Lopes M.A., Larkins B.A. (1993). Endosperm origin, development, and function. Plant Cell 5: 1383–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Dennis E.S., Berger F., Peacock W.J., Chaudhury A. (2005). MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc. Natl. Acad. Sci. USA 102: 17531–17536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutterbach B., Sun D., Schuetz J., Hiebert S.W. (1998). The MYND motif is required for repression of basal transcription from the multidrug resistance 1 promoter by the t(8;21) fusion protein. Mol. Cell. Biol. 18: 3604–3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moles A.T., Ackerly D.D., Webb C.O., Tweddle J.C., Dickie J.B., Westoby M. (2005). A brief history of seed size. Science 307: 576–580. [DOI] [PubMed] [Google Scholar]
- Ohto M.A., Fischer R.L., Goldberg R.B., Nakamura K., Harada J.J. (2005). Control of seed mass by APETALA2. Proc. Natl. Acad. Sci. USA 102: 3123–3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto M.A., Floyd S.K., Fischer R.L., Goldberg R.B., Harada J.J. (2009). Effects of APETALA2 on embryo, endosperm, and seed coat development determine seed size in Arabidopsis. Sex. Plant Reprod. 22: 277–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi C.H., Tanksley S.D. (2009). Natural variation in an ABC transporter gene associated with seed size evolution in tomato species. PLoS Genet. 5: e1000347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser L., Fischer R.L. (1993). The ovule and the embryo sac. Plant Cell 5: 1291–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schruff M.C., Spielman M., Tiwari S., Adams S., Fenby N., Scott R.J. (2006). The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 133: 251–261. [DOI] [PubMed] [Google Scholar]
- Scott R.J., Spielman M., Bailey J., Dickinson H.G. (1998). Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 125: 3329–3341. [DOI] [PubMed] [Google Scholar]
- Shomura A., Izawa T., Ebana K., Ebitani T., Kanegae H., Konishi S., Yano M. (2008). Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 40: 1023–1028. [DOI] [PubMed] [Google Scholar]
- Song X.J., Huang W., Shi M., Zhu M.Z., Lin H.X. (2007). A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39: 623–630. [DOI] [PubMed] [Google Scholar]
- Verma R., Oania R., Graumann J., Deshaies R.J. (2004). Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell 118: 99–110. [DOI] [PubMed] [Google Scholar]
- Wang A., Garcia D., Zhang H., Feng K., Chaudhury A., Berger F., Peacock W.J., Dennis E.S., Luo M. (2010). The VQ motif protein IKU1 regulates endosperm growth and seed size in Arabidopsis. Plant J. 63: 670–679. [DOI] [PubMed] [Google Scholar]
- Wang X., Liu B., Huang C., Zhang X., Luo C., Cheng X., Yu R., Wu Z. (2012). Over expression of Zmda1-1 gene increases seed mass of corn. Afr. J. Biotechnol. 11: 13387–13395. [Google Scholar]
- Weng J., et al. (2008). Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 18: 1199–1209. [DOI] [PubMed] [Google Scholar]
- White D.W. (2006). PEAPOD regulates lamina size and curvature in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 13238–13243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia T., Li N., Dumenil J., Li J., Kamenski A., Bevan M.W., Gao F., Li Y. (2013). The ubiquitin receptor DA1 interacts with the E3 ubiquitin ligase DA2 to regulate seed and organ size in Arabidopsis. Plant Cell 25: 3347–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W., Brown R.C., Lemmon B.E., Harada J.J., Goldberg R.B., Fischer R.L. (2006). Regulation of seed size by hypomethylation of maternal and paternal genomes. Plant Physiol. 142: 1160–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R., Li Y. (2011). Control of final organ size by Mediator complex subunit 25 in Arabidopsis thaliana. Development 138: 4545–4554. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Zhang X., Kang X., Zhao X., Zhang X., Ni M. (2009). SHORT HYPOCOTYL UNDER BLUE1 associates with MINISEED3 and HAIKU2 promoters in vivo to regulate Arabidopsis seed development. Plant Cell 21: 106–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.